Abstract
Chronic treatment with the β-blocker carvedilol has been shown to reduce established maladaptive left ventricle (LV) hypertrophy and to improve LV function in experimental heart failure. However, the detailed mechanisms by which carvedilol improves LV failure are incompletely understood. We previously showed that carvedilol is a β-arrestin-biased β1-adrenergic receptor ligand, which activates cellular pathways in the heart independent of G protein-mediated second messenger signaling. More recently, we have demonstrated by microRNA (miR) microarray analysis that carvedilol upregulates a subset of mature and pre-mature miRs, but not their primary miR transcripts in mouse hearts. Here, we next sought to identify the effects of carvedilol on LV gene expression on a genome-wide basis. Adult mice were treated with carvedilol or vehicle for 1 wk. RNA was isolated from LV tissue and hybridized for microarray analysis. Gene expression profiling analysis revealed a small group of genes differentially expressed after carvedilol treatment. Further analysis categorized these genes into pathways involved in tight junction, malaria, viral myocarditis, glycosaminoglycan biosynthesis, and arrhythmogenic right ventricular cardiomyopathy. Genes encoding proteins in the tight junction, malaria, and viral myocarditis pathways were upregulated in the LV by carvedilol, while genes encoding proteins in the glycosaminoglycan biosynthesis and arrhythmogenic right ventricular cardiomyopathy pathways were downregulated by carvedilol. These gene expression changes may reflect the molecular mechanisms that underlie the functional benefits of carvedilol therapy.
Keywords: beta-blocker, biased G protein-coupled receptor signaling, gene regulation, transcriptome, left ventricle
heart failure (HF) is a major public health problem affecting more than 5 million Americans. The total healthcare costs of HF are over $30 billion annually and are projected to increase as the population ages (6). Left ventricular failure (LVF) is the most common cause of HF, but effective therapeutic options for LVF are still lacking. Previous studies reported that treatment with the nonselective beta-adrenergic receptor antagonist (β-blocker) Carvedilol (Carv) reverses established LVF in different experimental HF models and that this improvement in LV function was associated with reduced hypertrophy, fibrosis, and apoptosis (5, 37, 38). These changes were partially explained by positive effects of Carv on β-arrestin-mediated cardioprotective signaling pathways, which we have identified (10, 11). We previously showed that Carv is a β-arrestin-biased β1-adrenergic receptor (β1AR) ligand, which activates cellular pathways in the heart independent of G protein-mediated second messenger accumulation, a concept known as biased signaling (10, 22). Carv has also been shown to have pleiotropic cardioprotective effects beyond heart-rate control. Indeed, a spectrum of anti-inflammatory, antioxidant, and antiapoptotic actions of Carv was reported in vitro and in vivo (19, 36). More recently, we have demonstrated by microRNA (miR) microarray analysis that Carv upregulates a subset of mature and pre-mature miRs, but not their primary miR transcripts in mouse hearts (11). This suggests that Carv could regulate specific genes and signaling pathways linked to physiological outcomes, such as protection against apoptosis, which may underlie the mechanisms of its beneficial effects against LVF.
Here, we performed global gene expression profiling in the LV of mice treated with or without Carv. Out of 25,376 mouse genes examined, we found that Carv treatment upregulated 49 genes involved primarily in the tight junction, malaria, and viral myocarditis pathways, while it downregulated 34 genes involved in the glycosaminoglycan biosynthesis and arrhythmogenic right ventricular cardiomyopathy (ARVC) pathways. Our microarray and real-time PCR data provide evidence that the biased β-blocker Carv regulates the expression of unique subsets of LV genes, which may be mechanistically linked to its cardioprotective effects.
MATERIALS AND METHODS
Animal Study
We used 8 to 12 wk old C57BL/6 mice for this study. Research with animals carried out for this study was handled according to approved protocols and animal welfare regulations of Georgia Regents University's Institutional Animal Care and Use Committee. All animal procedures were performed to conform with National Institutes of Health guidelines (Guide for the Care and Use of Laboratory Animals). Carv was dissolved in 10% DMSO, and mini-osmotic pumps (Alzet model 2001, DURECT) were then filled to deliver at the rate of 19 mg/kg/day (which is the maximum dose we can administer due to its solubility) for Carv over a period of 7 days (when the relative expression levels of several chosen cardiac-enriched miRs and transcripts reached the maximum in our time-course experiments from 1 to 7 days of Carv treatment) as previously described (11). In control mice, vehicle (10% DMSO) was used. All used mice injected with Carv via mini-osmotic pumps displayed no significant changes in cardiac function compared with DMSO controls as described previously (11). Seven days after drug administration, all mice were euthanized within 2 h of each other by thoracotomy with 1–4% inhalant isoflurane, and LV tissues were excised and flash-frozen in liquid N2 for mRNA analysis as described previously (9–11).
RNA Samples for Microarray Analysis
Total RNA from six independent mouse LVs (3 vehicle controls and 3 Carv-treated samples) was prepared as described previously (9, 11). RNA quantity and quality were measured by NanoDrop ND-1000. RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Intact total RNA run on a denaturing gel had sharp 28S and 18S rRNA bands. The 28S rRNA band was approximately twice as intense as the 18S rRNA band, further confirming that the RNA was intact. MRNA levels were validated by custom-designed Taqman PCR array (Applied Biosystems) as described below.
DNA Microarray
The experiments were conducted using the Arraystar Mouse Microarray v2.0 (8 × 60K, Arraystar) designed for the global profiling of mouse protein-coding transcripts as previously described (2, 18, 29, 39). Using the second-generation microarray we detected 25,376 coding transcripts. Each transcript was represented by a specific exon or splice junction probes, which can identify individual transcripts accurately. We also printed 15 positive probes for housekeeping genes and 20 negative probes onto the array for hybridization quality control.
RNA Labeling and Array Hybridization
Sample labeling and array hybridization for six independent LV samples were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) with minor modifications. To reduce possible bias due to different dyes, we used a single color system as described previously (23). The Agilent Quick Amp Labeling Kit was used for sample labeling. Hybridization was performed in Agilent's SureHyb Hybridization Chambers. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured using NanoDrop ND-1000. We fragmented 1 μg of each labeled cRNA by adding 5 μl of 10× blocking agent and 1 μl of 25× fragmentation buffer, and then the mixture was heated at 60°C for 30 min. Finally, 25 μl of 2× GE hybridization buffer was added to dilute the labeled cRNA. We dispensed 50 μl of hybridization solution into the gasket slide and assembled it to the gene expression microarray slide. The slides were incubated for 17 h at 65°C in an Agilent hybridization oven. The hybridized arrays were washed, fixed, and scanned with using the Agilent DNA Microarray Scanner (part number G2505C).
Data Analysis
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. We also included 15 positive probes for housekeeping genes and 20 negative probes onto the array for hybridization quality control. In addition to these normalization methods, we performed more robust “quantile normalization” across all six samples, rather than normalizing using one/some control probes. That is, the entire intensity distributions are normalized across the arrays, not just some reference points. Quantile normalization and subsequent data processing were performed using GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, mRNAs, that in at least one out of six samples had flags in present or marginal (“all targets value”), were chosen for further data analysis. Differentially expressed mRNAs with statistical significance were identified through Volcano Plot filtering between two groups of samples. Using Agilent GeneSpring Software GX v11.5.1 and the latest KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg) database, we performed pathway analysis for genes that are differentially expressed (DE) in both microarray and real-time PCR verification analyses, to gain insights on the biological pathways significantly enriched in DE mRNAs. Fisher's exact test was used to calculate the P values of the pathway ID and converted to enrichment score by negative log10 transformation. Multiple test correction by Benjamini-Hochberg procedure was applied to calculate false discovery rate (FDR). Gene Ontology (GO) analysis was performed to associate DE mRNAs with GO categories. The GO categories are derived from Gene Ontology (http://www.geneontology.org), which comprises three structured networks of defined terms that describe gene product attributes. We used the most updated version of GO classifications with 20,000 GO terms. The TopGO package is used for GO enrichment calculation. The P value denotes the significance of GO term enrichment in the DE mRNA list. Finally, hierarchical clustering was performed to distinguish mRNA expression patterns among samples and genes using the Euclidean distance algorithm and the R heat-map package.
RNA Isolation and Taqman PCR Array Analysis
Total RNA from 12 independent mouse LVs (6 vehicle controls and 6 Carv-treated samples) was prepared using Trizol (Life Technologies) and treated with RNase-free DNase I to remove genomic DNA as described (11). cDNA for detection of mRNAs was synthesized using Invitrogen (Carlsbad, CA) SuperScript II reverse transcriptase and oligo(dT) primers. Expression of mRNAs was detected using Taqman Gene expression assays. Custom-designed Taqman PCR array analyses were performed in a 96-well format as described previously (31) with gene probes including two housekeeping genes shown in Supplementary Table S9.1 Real-time PCR reactions were amplified and analyzed using an ABI Sequence Detection System as described previously (12). PCR reaction conditions were as follows: step 1: 50°C for 2 min, step 2: 95°C for 10 min, step 3: 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Expression relative to endogenous controls was calculated using 2−ΔΔCt, and levels were normalized to vehicle controls. We performed independent experiments in triplicate using different batches of RNAs each time.
Statistical Analysis
Data are expressed as means ± SE from at least three independent experiments with different mice per group. Statistical significance was determined by using both Student unpaired t-tests without correction (P values) and t-test with correction for multiple tests on all genes by Benjamini-Hochberg procedure (FDRs). A P value < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Carv Induces the Expression of a Relatively Small Number of Genes in Mouse LVs
To test whether the β-arrestin-biased β-blocker Carv (10, 34) can regulate gene expression, we performed gene microarray profiling in mouse LVs following Carv treatment. We used 8 to 12 wk old wild-type mice and infused them with DMSO (vehicle control) or Carv (19 mg/kg/day). Among 25,376 mouse genes that we profiled, 49 genes were upregulated (Fig. 1, Fig. 2, and Table 1; >2-fold and P < 0.05), and 34 genes were downregulated (Fig. 1, Fig. 2, and Table 2; >2-fold and P < 0.05) by treatment with Carv. In addition, there was a subset of LV genes differently expressed between the DMSO-control and Carv-treated mice with a magnitude of 1.50–1.99-fold (Supplementary Tables S1, S2). Using custom-designed Taqman PCR arrays (Supplementary Table S9), we validated key gene expression changes with real-time PCR analysis on independent LV samples (Tables 1 and 2, Supplementary Tables S1 and S2).
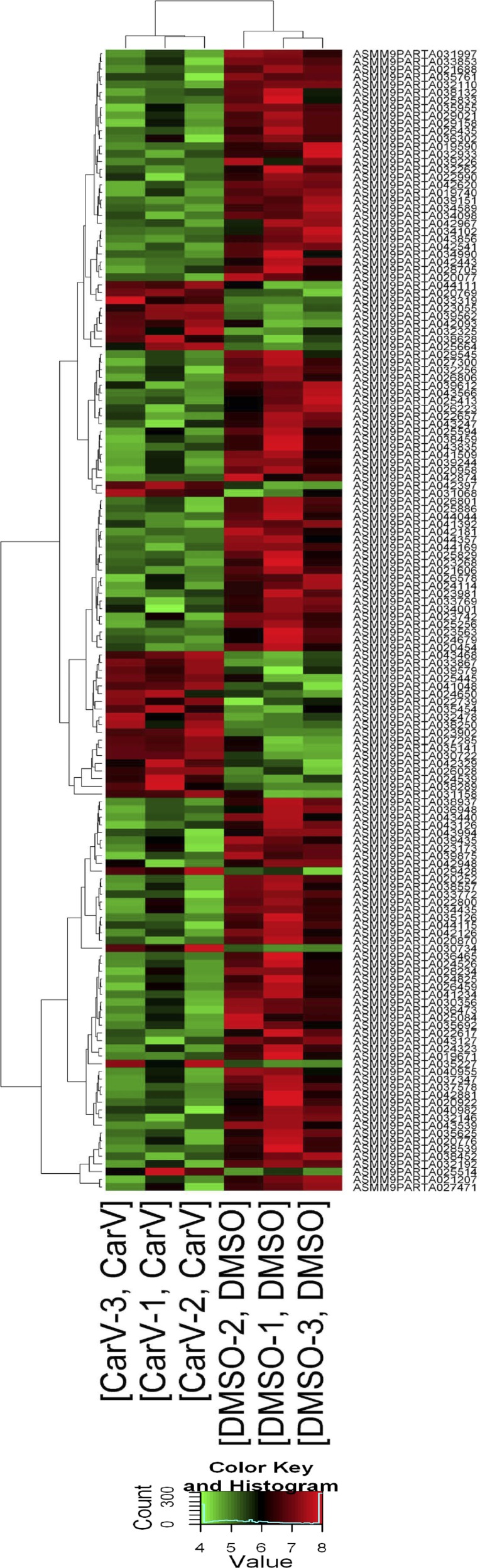
Fig. 1.
Gene array analysis in mouse hearts. Wild-type (WT) mice were infused with 10% DMSO (vehicle control) or carvedilol (Carv, 19 mg/kg/day) for 7 days via micro-osmotic pumps. Microarray experiments were performed in mouse left ventricles. The heat map represents mRNA expression values in 2 conditions (DMSO vs Carv). The expression level was visualized via colors. The dendrogram shows the relationships among the expression levels of samples. Hierarchical clustering was performed on both the samples and the genes based on “differentially expressed mRNAs.” Red indicates high relative expression, and green indicates low relative expression.
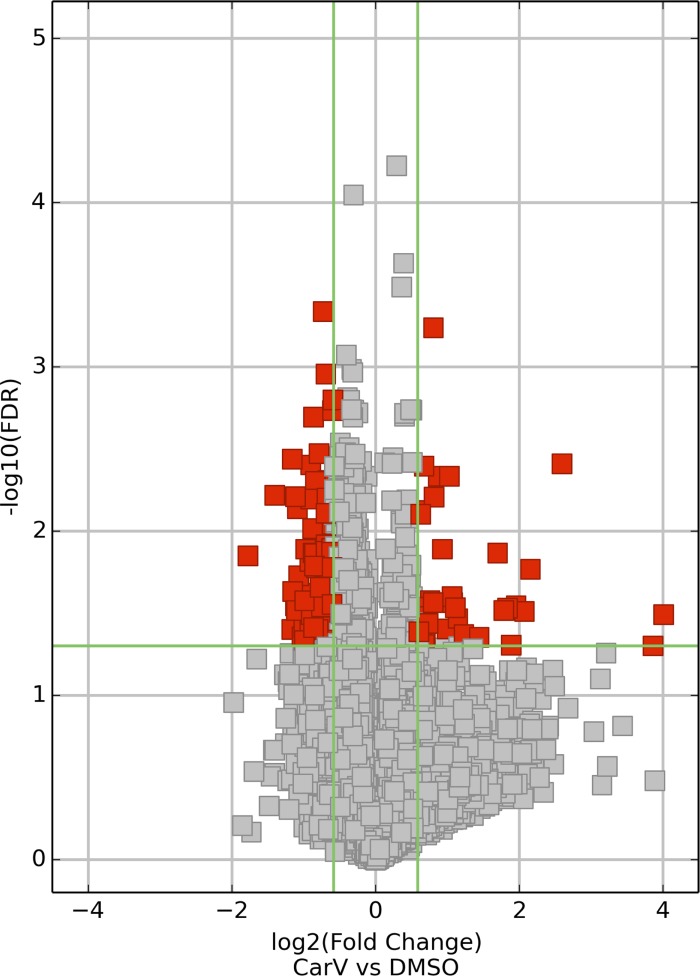
Fig. 2.
Volcano plot analysis of differentially expressed (DE) mRNAs. Volcano plots are constructed from fold-change values and false discovery rate (FDR) values, allowing one to visualize the relationship between fold-change and statistical significance (which takes both magnitude of change and variability into consideration). The vertical lines correspond to 1.5-fold up and down, and the horizontal line represents an FDR value of 0.05, so the red points in the plot represent the differentially expressed (DE) mRNAs with statistical significance.
Table 1.
Genes differentially upregulated in carvedilol-treated mouse hearts
| (Carv/DMSO) Fold Change |
|||
|---|---|---|---|
| Seq. Name | Gene Symbol | Microarray | Real-time PCR |
| NM_133186 | Steap3 | 16.08 | 0.88 |
| NM_011315 | Saa3 | 14.51 | 9.6† |
| NM_023220 | Sppl2a | 7.29 | 0.97 |
| NM_016768 | Pbx3 | 6.69 | 0.86 |
| NM_025706 | Tbc1d15 | 6.47 | 0.96 |
| NM_016875 | Ybx2 | 6.45 | 0.88 |
| NM_011091 | Pira4 | 6.03 | 2.11* |
| NM_011088 | Pira11 | 6.01 | 1.79* |
| NM_011331 | Ccl12 | 4.45 | 2.61* |
| NM_001159567 | Meis2 | 4.2 | 0.96 |
| NM_001177417 | Gm6792 | 4.12 | UD |
| NM_001164329 | Gm6904 | 3.88 | 0.69 |
| NM_008624 | Mras | 3.87 | 0.9 |
| NM_175273 | Fam219b | 3.7 | 0.88 |
| NM_001115085 | Tssc4 | 3.67 | 0.86 |
| NM_001199940 | Serpina3i | 3.57 | 4.96† |
| NM_011094 | Pira7 | 3.47 | 1.64* |
| NM_026405 | Rab32 | 3.45 | 1.2 |
| NM_027118 | Cdk13 | 3.4 | 0.96 |
| NM_030729 | Nckipsd | 3.35 | 0.92 |
| NM_207245 | Zfp870 | 3.35 | 0.95 |
| NM_008933 | Prm2 | 3.34 | UD |
| NM_021895 | Actn4 | 3.31 | 0.77 |
| NM_011116 | Pld3 | 3.24 | 0.93 |
| NM_001167913 | Smok2b | 3.24 | UD |
| NM_001178012 | Sfxn3 | 3.14 | 0.71 |
| NM_026136 | Gm3404 | 2.92 | UD |
| NM_011250 | Rbl2 | 2.91 | 0.81 |
| NM_001142337 | Lmo2 | 2.9 | 0.87 |
| NM_207659 | Hook3 | 2.74 | 0.88 |
| NM_001085419 | Gm13102 | 2.71 | 2.03* |
| NM_207671 | Zfp318 | 2.62 | 0.88 |
| NM_009879 | Ift81 | 2.51 | 0.86 |
| NM_177191 | Sycp2 | 2.38 | 0.82 |
| NM_153778 | Atoh8 | 2.36 | 0.74 |
| NM_026573 | Upf3b | 2.34 | 1.17 |
| NM_027980 | 2310003H01Rik | 2.32 | 0.84 |
| NM_146331 | Olfr954 | 2.27 | UD |
| NM_001033245 | Hk3 | 2.25 | 1 |
| NM_134065 | Epdr1 | 2.21 | 0.91 |
| NM_028009 | Rpusd1 | 2.21 | 0.9 |
| NM_080728 | Myh7 | 2.16 | 2.09† |
| NM_021398 | Slc43a3 | 2.1 | 0.82 |
| NM_026644 | Agpat4 | 2.1 | 0.73 |
| NM_026909 | Thap7 | 2.09 | 1.23 |
| NM_080448 | Srgap3 | 2.04 | 1.07 |
| NM_023292 | Pus3 | 2.04 | 0.85 |
| NM_008277 | Hpd | 2.01 | 2.70* |
| NM_011144 | Ppara | 2.01 | 0.81 |
The expression of genes identified by microarray analysis (Fig. 1) was verified by real-time RT-PCR analysis. Fold change values represent mean of expression levels from 3 (microarray) or 6 (real-time PCR) independent mice per group. The genes, which are significantly upregulated by carvedilol (Carv) compared with vehicle control (DMSO) in both microarray and real-time PCR analyses, include * or †.
UD, undetectable.
P < 0.05 vs. DMSO;
P < 0.01 vs. DMSO.
Table 2.
Genes differentially downregulated in carvedilol-treated mouse hearts
| (Carv/DMSO) Fold Change |
|||
|---|---|---|---|
| Seq. Name | Gene Symbol | Microarray | Real-time PCR |
| NM_001159627 | Heph | 0.24 | 0.90 |
| NM_178671 | Ubxn10 | 0.29 | UD |
| NM_001136068 | Klrc1 | 0.31 | 1.30 |
| NM_001025435 | BC053393 | 0.32 | UD |
| NM_178655 | Ank2 | 0.37 | 0.87 |
| NM_001044697 | Zfp2 | 0.38 | 0.92 |
| NM_175408 | Tmem139 | 0.38 | 1.12 |
| NM_027728 | Enkur | 0.38 | 0.19‡ |
| NM_023063 | Lima1 | 0.41 | 0.65* |
| NM_178444 | Egfl7 | 0.43 | 0.64* |
| NM_001033382 | Cacna2d4 | 0.43 | UD |
| NM_026661 | Aar2 | 0.43 | 0.94 |
| NM_175280 | 4930529M08Rik | 0.43 | 0.28‡ |
| NM_001001650 | Prss48 | 0.44 | UD |
| NM_029654 | Atg2b | 0.44 | 0.94 |
| NM_009541 | Zbtb17 | 0.44 | 0.84 |
| NM_025496 | Cdrt4 | 0.44 | 0.37* |
| NM_029614 | Prss23 | 0.44 | 0.31* |
| NM_001081196 | Hnrnpul2 | 0.45 | 0.96 |
| NM_021325 | Cd200r1 | 0.45 | 1.26 |
| NM_024474 | Col26a1 | 0.45 | 0.16‡ |
| NM_001167879 | Gareml | 0.45 | 0.31† |
| NM_011097 | Pitx1 | 0.45 | UD |
| NM_001114179 | Slc25a53 | 0.46 | 1.41 |
| NM_177657 | D630003M21Rik | 0.46 | 0.51† |
| NM_177854 | Sertm1 | 0.47 | 0.63* |
| NM_001163569 | Kif9 | 0.47 | 0.50* |
| NM_028326 | Zfp618 | 0.47 | 1.05 |
| NM_001024846 | Zfp62 | 0.48 | 0.91 |
| NM_173375 | Fam180a | 0.48 | 0.14‡ |
| NM_019577 | Ccl24 | 0.49 | 0.52* |
| NM_027025 | Adora3 | 0.49 | 0.56* |
| NM_177919 | Tceal5 | 0.49 | 0.37† |
| NM_148944 | Chrnb4 | 0.49 | UD |
The expression of genes identified by microarray analysis (Fig. 1) was verified by real-time RT-PCR analysis. Fold change values represent mean of expression levels from 3 (microarray) or 6 (real-time PCR) independent mice per group. The genes, which are significantly downregulated by Carv compared with vehicle control (DMSO) in both microarray and real-time PCR analyses, include *, †, or ‡.
P < 0.05 vs. DMSO;
P < 0.01 vs. DMSO;
P < 0.001 vs. DMSO.
Carv-responsive Genes are Ubiquitous in Cells and have Diverse Functions
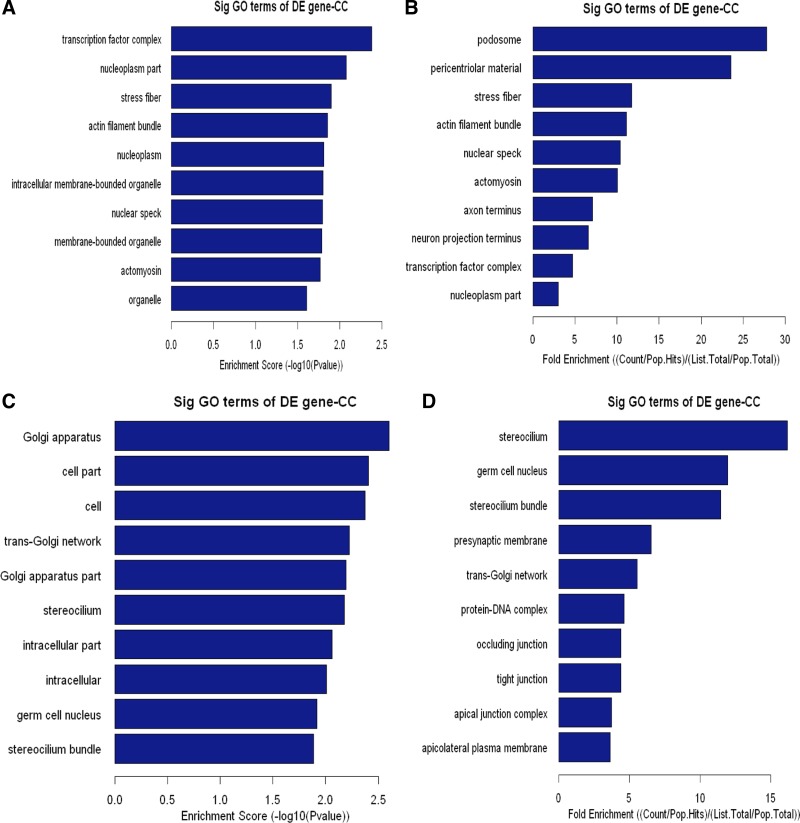
Our data analysis demonstrates that Carv-responsive genes are widely distributed in the cell. The breakdown of subcellular localizations and cellular component analysis of upregulated genes are shown in Fig. 3, A and B, and Supplementary Table S3. The breakdown of subcellular localizations and cellular component analysis of downregulated genes are shown in Fig. 3, C and D, and Supplementary Table S4.
Fig. 3.
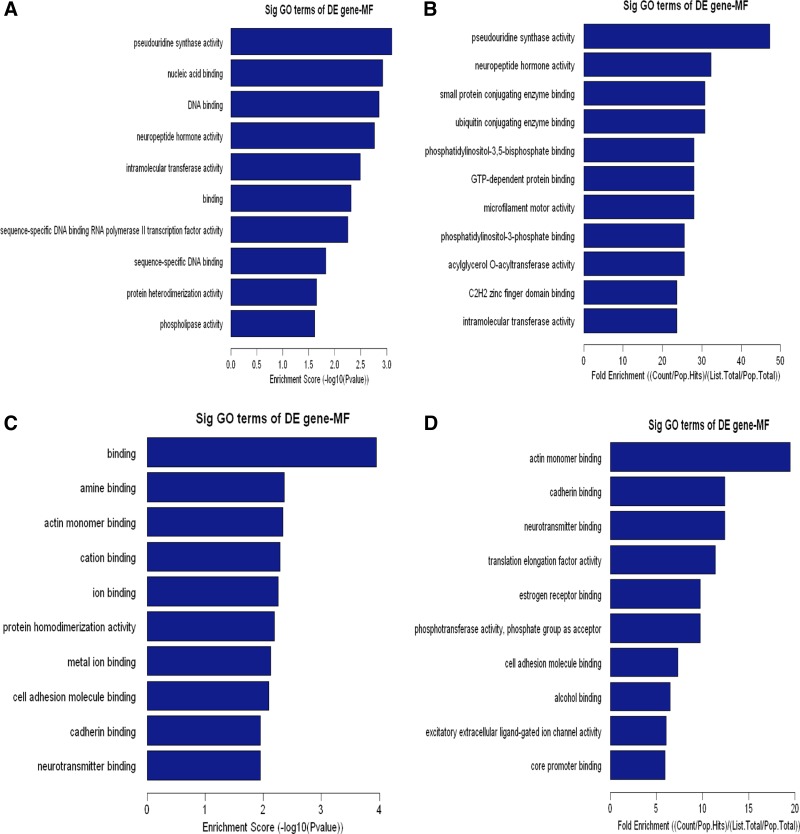
Cellular component (CC) analysis of DE genes. The upregulated (A, B) or downregulated (C, D) genes were classified by CC by the Gene Ontology (GO) classification system. Fisher's exact test is used to find if there is more overlap between the DE list and the GO annotation list than would be expected by chance. The P value denotes the significance of GO terms enriched in the DE genes. The top 10 terms with the lowest P value are shown as bar plots. A or C: Enrichment Score, the GO ID's enrichment score value, and it equals [−log10(P value)]. B or D: Fold Enrichment, the GO ID's fold enrichment value, and it equals (Count/Pop.Hits)/(List.Total/Pop.Total). Pop.Hits, the number of background population genes associated with the listed GO ID. List.Total, the total number of DE genes. Pop.Total, The total number of background population genes.
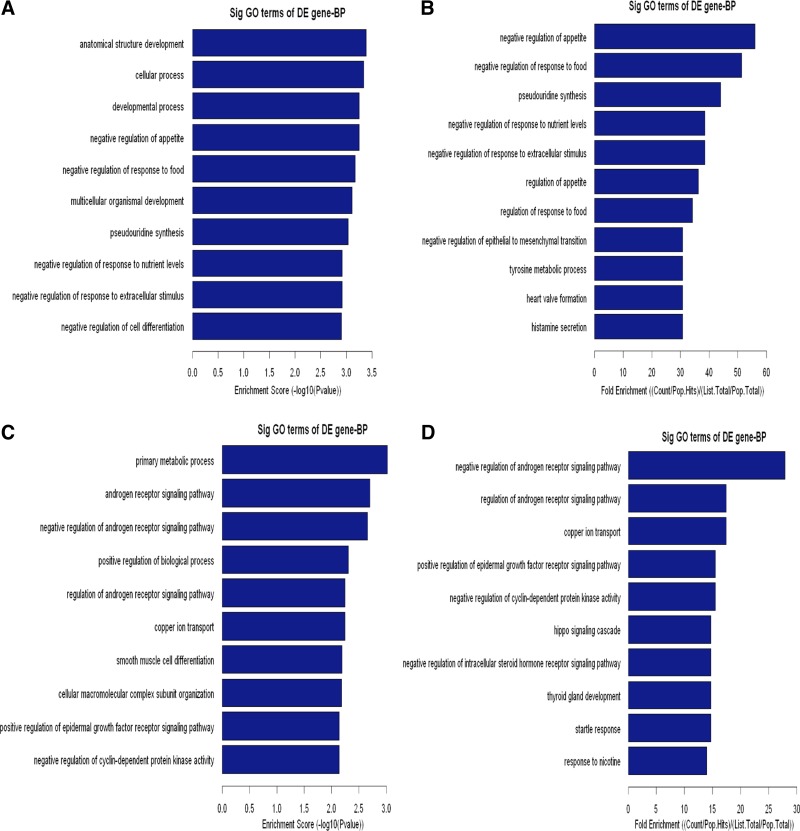
To elaborate the functional scope of Carv, we next grouped the identified Carv-responsive genes according to their GO biological and molecular functions. Upregulated genes were overrepresented in cellular and metabolic processes (Fig. 4, A and B, and Supplementary Table S5), as well as protein and nucleic acid binding (Fig. 5, A and B, and Supplementary Table S7). Downregulated genes were highly involved in metabolic processes and biological regulation (Fig. 4, C and D, and Supplementary Table S6) as well as binding and catalytic activity (Fig. 5, C and D, and Supplementary Table S8). In agreement with our findings, metabolic proteins were shown to account for the majority of DE proteins in a previous proteomic profiling study of cellular responses to Carv in vascular smooth muscle cells (33), supporting the hypothesis that Carv regulates metabolic processes.
Fig. 4.
Biological process (BP) analysis of DE genes. The upregulated (A, B) or downregulated (C, D) genes were classified by BP by the GO classification system. Fisher's exact test is used to find if there is more overlap between the DE list and the GO annotation list than would be expected by chance. The P value denotes the significance of GO terms enriched in the DE genes. The top 10 terms with the lowest P value are shown as bar plots.
Fig. 5.
Molecular function (MF) analysis of DE genes. The upregulated (A, B) or downregulated (C, D) genes were classified by MF by the GO classification system. Fisher's exact test is used to find if there is more overlap between the DE list and the GO annotation list than would be expected by chance. The P value denotes the significance of GO terms enriched in the DE genes. The top 10 terms with the lowest P value are shown as bar plots.
Other functional categories of the Carv transcriptome include multicellular organismal processes, nitrogen or nucleobase-containing compound metabolic processes, developmental processes (Fig. 4, A and B, and Supplementary Table S5 as well as Fig. 4, C and D, and Supplementary Table S6), metal ion binding, cation binding, transcription factor activity (Fig. 5, A and B, and Supplementary Table S7, as well as Fig. 5, C and D, and Supplementary Table S8), positive regulation of biological processes, cellular component organization or biogenesis (Fig. 4, C and D, and Supplementary Table S6), hydrolase activity and protein dimerization activity (Fig. 5, C and D, and Supplementary Table S8). Interestingly, Carv [but not another beta-blocker metoprolol, which is also being used in clinic (24)] significantly enhanced Serca2 gene transcription under oxidative stress and two Sp1 sites in the Serca2 gene promoter region mediated the response to Carv (13). This suggests that Carv specifically regulates gene transcription, which is also supported by our data on transcriptional regulation (Fig. 5, A and B, and Supplementary Table S7). Having a wide subcellular and functional distribution of genes in its transcriptome, Carv is involved in a wide range of cellular processes in diverse cellular compartments.
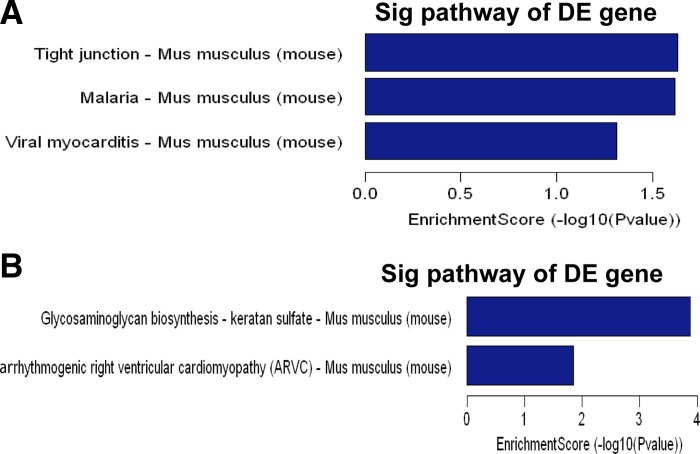
Carv Regulates the Expression of Genes Involved in Tight Junction, Malaria, Viral Myocarditis, Glycosaminoglycan Biosynthesis, and ARVC
Our signaling pathway analysis of DE genes by Carv further demonstrated that upregulated genes are involved in tight junction, malaria, and viral myocarditis pathways (Fig. 6A), while downregulated genes are involved in glycosaminoglycan biosynthesis and ARVC (Fig. 6B). The potential functional specialization of the Carv transcriptome in terms of cellular roles of the differentially regulated genes is further discussed below.
Fig. 6.
Signaling pathway analysis of DE genes. Pathway analysis is a functional analysis mapping genes to KEGG pathways. The P value denotes the significance of the pathway correlated to the conditions [upregulated (A) or downregulated (B) in Carv compared with DMSO control]. Enrichment score is the value of the Pathway ID, and it equals “−log10(P value)”. The bar plot shows the top 5 enrichment score value of the significant enrichment pathway. DE genes identified in both microarray and real-time PCR verification analyses (Tables 1 and 2, Supplementary Tables S1 and S2) are only included for pathway analysis.
Tight junction, malaria, and viral myocarditis.
We classified the upregulated genes by the KEGG signaling pathway classification system. The top three canonical signaling pathways are tight junction, malaria, and viral myocarditis, which are illustrated in Fig. 6A. In both microarray and real-time PCR verification data, the expression of genes involved in tight junction (e.g., myh7), malaria (e.g., ccl12 and itgb2), and viral myocarditis (e.g., itgb2 and myh7) was increased after Carv treatment (Table 1 and Supplementary Table S1).
Glycosaminoglycan biosynthesis and ARVC.
We classified downregulated genes using the KEGG signaling pathway classification system. The top two canonical pathways are glycosaminoglycan biosynthesis and ARVC, which are illustrated in Fig. 6B. In both microarray and real-time PCR verification data, genes involved in glycosaminoglycan biosynthesis-keratan sulfate (e.g., chst1 and chst2) were decreased in expression after Carv treatment (Table 2 and Supplementary Table S2). We also showed that genes involved in ARVC (e.g., cacnb1 and ctnna2) were decreased in expression after Carv treatment (Table 2 and Supplementary Table S2). Carv was shown to attenuate the downregulation of fatty acid and adult enriched-glycolytic enzyme encoding genes with the induction of pressure overload-induced hypertrophy in rats (27), suggesting that Carv therapy regulates metabolic enzyme-encoding genes during cardiac hypertrophy, which is in agreement with our results (Fig. 6B). Taken together, our transcriptome data indicate that Carv regulates the expression of a subset of genes/signaling pathways that regulate cardiac structure, function, and metabolism.
Interpretation of the Current Findings Compared with Previous Publications
Carv is one of three β-blockers approved for HF in the US and has many documented actions including antagonism of β1AR, β2AR, and α1AR as well as antioxidant effects (20, 26). In a recent meta-analysis, Carv led to less sudden cardiac death and all-cause mortality in patients with acute myocardial infarction (MI) and those with HF compared with other β-blockers (3). We previously showed that Carv stimulates β-arrestin-mediated β1AR cardioprotective signaling without activating G proteins, providing an additional potential mechanism for its clinical efficacy (10). Identifying additional beneficial downstream signaling pathways regulated by Carv should lead to a better understanding of how β-arrestin biased ligands exert their cardioprotective effects.
By microarray-based transcriptome and real-time PCR verification approaches, we have identified 10 mouse LV genes (Table 1, Supplementary Table S1) that were upregulated and 17 genes (Table 2, Supplementary Table S2) that were downregulated in response to Carv stimulation. These genes were ubiquitously distributed in the cell and were enriched in previously known and unknown pathways for Carv, including tight junction, malaria, viral myocarditis, glycosaminoglycan biosynthesis, and ARVC. This suggests that Carv's cardioprotective effects may in part be due to modulation of gene expression linked to the identified signaling pathways.
Previous studies as below have suggested that Carv has a superior cardioprotective effect in experimental myocarditis compared with other β-blockers by reducing LV inflammation and oxidative stress. Carv was shown to inhibit the activation of p38MAPK pathway through β1AR and β2AR (32). Carv led to an increase in the production of anti-inflammatory cytokines IL-12 and IFN-gamma (21) and a decrease of inflammatory cytokines IL-6 and TNF-α by inducing the cardiac cAMP response element binding protein expression and phosphorylation (17), which resulted in a stronger effect in reducing myocardial virus replication compared with the selective β1-blocker metoprolol and the nonselective β-blocker propranolol. Carv, but not metoprolol, also improved survival, reduced lipid peroxidation, and increased antioxidant enzyme activities with amelioration of acute viral myocarditis (16). Our gene profiling and real-time PCR analyses showed that the expression of genes encoding proteins in the viral myocarditis pathway (e.g., itgb2 and myh7) was increased in the Carv-treated LV tissues when compared with the vehicle controls (Fig. 6A, Table 1, and Supplementary Table S1). Altogether, our data along with previous studies suggest that Carv regulates the viral myocarditis pathway to reduce LV inflammation and oxidative stress.
Similarly, it has been shown that Carv not only is useful for controlling arrhythmia but also improves LV function in some patients with ARVC (7). Carv treatment significantly improved d-galactose-induced senescence in mice by attenuating the elevated level of acetylcholine esterase (14), suggesting that these two pathways are regulated by Carv in part due to its antioxidant effect and modulation of mitochondrial permeability transition. In agreement with these previous studies, we showed that the expression of genes encoding proteins in ARVC pathway (e.g., cacnb1 and ctnna2) was decreased in the Carv-treated LV tissues when compared with the vehicle controls (Fig. 6B, Table 2, and Supplementary Table S2).
On the other hand, little has been documented about Carv-mediated effects on the genes encoding proteins of tight junction (e.g., myh7), malaria (e.g., ccl12 and itgb2), and glycosaminoglycan biosynthesis pathways (e.g., chst1 and chst2), which we identified in this study. Low-molecular-weight glycosaminoglycans are considered cardioprotective in isoproterenol-induced MI in rats by maintaining lactate dehydrogenase-isoenzyme (28), and it has been acknowledged that heparin, a glycosaminoglycan, protects the ischemic myocardium (8). Bongo et al. (1) recently reviewed the neuroimmune axon guidance cue netrin-1 and its cardioprotective role in ischemia/reperfusion injury. A recent study also showed that the netrin-1 receptor mediates netrin-1/NO-dependent cardioprotection (15). Our findings and the three previous studies imply that glycosaminoglycan biosynthesis and axon guidance pathways may play important parts in heart remodeling. Additional studies will be needed to further clarify the relationship between Carv's cardioprotective actions and the pathways identified in our study.
In conclusion, our data identify gene signatures regulated by Carv in mouse LVs and provide mechanistic clues that explain its powerful effects on LV function. It is becoming increasingly evident that the biological functions of Carv are much broader than we currently understand. Further studies are required to characterize the regulation between Carv and individual target genes and define the cellular consequences of their regulation.
Limitations
In vivo HF models [e.g., MI, ischemia-reperfusion (I/R), or transverse aortic constriction] may be required to understand the cardioprotective effects of Carv and to test the relevance of the proposed gene regulatory mechanism by which Carv offers added benefit in disease models. Moreover, extensive further studies such as time-course, dose response, and prevention or treatment protocols for Carv may be warranted to identify detailed gene regulatory mechanisms by Carv, because 1) recent studies demonstrated that expression levels of genes during I/R injury and HF progression are dynamically regulated (25, 35) and 2) an experimental right ventricular failure (RVF) study in rat showed that gene expression profile of Carv-treated RVs for 4 wk resembled the RVF prediction set, although Carv improved RV function (4). However, using healthy mice treated with Carv (19 mg/kg per day) for 1 wk, our current findings identify the novel gene regulatory mechanism by Carv, which may be linked, in part, to its mechanism for cell survival. Notably, using this study design, we were also able to discover a novel miR regulatory mechanism by Carv (11). In a later recent study using MI models, we indeed demonstrated that one of the Carv-responsive miRs, miR-150, acts a cardioprotective miR (30).
GRANTS
This work was supported by American Heart Association Grant-in-Aid 12GRNT12100048, Scientist Development Grant 14SDG18970040, and National Institutes of Health R01 HL-124251 to I.-m. Kim.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.-p.T. and I.-m.K. conception and design of research; J.-p.T., K.-m.P., Z.B., and I.-m.K. performed experiments; J.-p.T., K.-m.P., Z.B., F.R.J., A.S.B., K.A., and I.-m.K. analyzed data; J.-p.T., K.-m.P., Z.B., F.R.J., A.S.B., K.A., H.S., J.J., N.L.W., Y.T., and I.-m.K. interpreted results of experiments; J.-p.T., K.-m.P., Z.B., and I.-m.K. prepared figures; J.-p.T., H.S., J.J., N.L.W., Y.T., and I.-m.K. drafted manuscript; J.-p.T., F.R.J., A.S.B., K.A., H.S., J.J., N.L.W., Y.T., and I.-m.K. edited and revised manuscript; I.-m.K. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Sangmi Kim for critically reviewing the manuscript.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Bongo JB, Peng DQ. The neuroimmune guidance cue netrin-1: a new therapeutic target in cardiovascular disease. J Cardiol 63: 95–98, 2014. [DOI] [PubMed] [Google Scholar]
- 2.Cossette SM, Gastonguay AJ, Bao X, Lerch-Gaggl A, Zhong L, Harmann LM, Koceja C, Miao RQ, Vakeel P, Chun C, Li K, Foeckler J, Bordas M, Weiler H, Strande J, Palecek SP, Ramchandran R. Sucrose non-fermenting related kinase enzyme is essential for cardiac metabolism. Biol Open 4: 48–61, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinicolantonio JJ, Lavie CJ, Fares H, Menezes AR, O'Keefe JH. Meta-analysis of carvedilol versus beta 1 selective beta-blockers (atenolol, bisoprolol, metoprolol, and nebivolol). Am J Cardiol 111: 765–769, 2013. [DOI] [PubMed] [Google Scholar]
- 4.Drake JI, Gomez-Arroyo J, Dumur CI, Kraskauskas D, Natarajan R, Bogaard HJ, Fawcett P, Voelkel NF. Chronic carvedilol treatment partially reverses the right ventricular failure transcriptional profile in experimental pulmonary hypertension. Physiol Genomics 45: 449–461, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feuerstein GZ, Hamburger SA, Smith EF 3rd, Bril A, Ruffolo RR Jr. Myocardial protection with carvedilol. J Cardiovasc Pharmacol 19, Suppl 1: S138–S141, 1992. [DOI] [PubMed] [Google Scholar]
- 6.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation 127: e6–e245, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiroi Y, Fujiu K, Komatsu S, Sonoda M, Sakomura Y, Imai Y, Oishi Y, Nakamura F, Ajiki K, Hayami N, Murakawa Y, Ohno M, Hirata Y, Ohtomo K, Nagai R. Carvedilol therapy improved left ventricular function in a patient with arrhythmogenic right ventricular cardiomyopathy. Jpn Heart J 45: 169–177, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Kilgore KS, Tanhehco EJ, Naylor KB, Lucchesi BR. Ex vivo reversal of heparin-mediated cardioprotection by heparinase after ischemia and reperfusion. J Pharmacol Exp Ther 290: 1041–1047, 1999. [PubMed] [Google Scholar]
- 9.Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem 280: 22278–22286, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Kim IM, Tilley DG, Chen J, Salazar NC, Whalen EJ, Violin JD, Rockman HA. Beta-blockers alprenolol and carvedilol stimulate beta-arrestin-mediated EGFR transactivation. Proc Natl Acad Sci USA 105: 14555–14560, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim IM, Wang Y, Park KM, Tang Y, Teoh JP, Vinson J, Traynham CJ, Pironti G, Mao L, Su H, Johnson JA, Koch WJ, Rockman HA. β-arrestin1-biased β1-adrenergic receptor signaling regulates microRNA processing. Circ Res 114: 833–844, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ Res 106: 1233–1243, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koitabashi N, Arai M, Tomaru K, Takizawa T, Watanabe A, Niwano K, Yokoyama T, Wuytack F, Periasamy M, Nagai R, Kurabayashi M. Carvedilol effectively blocks oxidative stress-mediated downregulation of sarcoplasmic reticulum Ca2+-ATPase 2 gene transcription through modification of Sp1 binding. Biochem Biophys Res Commun 328: 116–124, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A, Dogra S, Prakash A. Effect of carvedilol on behavioral, mitochondrial dysfunction, and oxidative damage against d-galactose induced senescence in mice. Naunyn Schmiedebergs Arch Pharmacol 380: 431–441, 2009. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Wang P, Ye K, Cai H. Central role of SIAH inhibition in DCC-dependent cardioprotection provoked by netrin-1/NO. Proc Natl Acad Sci USA 112: 899–904, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li YC, Ge LS, Yang PL, Tang JF, Lin JF, Chen P, Guan XQ. Carvedilol treatment ameliorates acute coxsackievirus B3-induced myocarditis associated with oxidative stress reduction. Eur J Pharmacol 640: 112–116, 2010. [DOI] [PubMed] [Google Scholar]
- 17.Li-Sha G, Yi-He C, Na-Dan Z, Teng Z, Yue-Chun L. Effects of carvedilol treatment on cardiac cAMP response element binding protein expression and phosphorylation in acute coxsackievirus B3-induced myocarditis. BMC Cardiovasc Disord 13: 100, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao JM, Zeng SX, Zhou X, Lu H. Global effect of inauhzin on human p53-responsive transcriptome. PLoS One 7: e52172, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma XL, Yue TL, Lopez BL, Barone FC, Christopher TA, Ruffolo RR Jr, Feuerstein GZ. Carvedilol, a new beta adrenoreceptor blocker and free radical scavenger, attenuates myocardial ischemia-reperfusion injury in hypercholesterolemic rabbits. J Pharmacol Exp Ther 277: 128–136, 1996. [PubMed] [Google Scholar]
- 20.Mochizuki M, Yano M, Oda T, Tateishi H, Kobayashi S, Yamamoto T, Ikeda Y, Ohkusa T, Ikemoto N, Matsuzaki M. Scavenging free radicals by low-dose carvedilol prevents redox-dependent Ca2+ leak via stabilization of ryanodine receptor in heart failure. J Am Coll Cardiol 49: 1722–1732, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Nishio R, Shioi T, Sasayama S, Matsumori A. Carvedilol increases the production of interleukin-12 and interferon-gamma and improves the survival of mice infected with the encephalomyocarditis virus. J Am Coll Cardiol 41: 340–345, 2003. [DOI] [PubMed] [Google Scholar]
- 22.Noma T, Lemaire A, Naga Prasad SV, Barki-Harrington L, Tilley DG, Chen J, Le Corvoisier P, Violin JD, Wei H, Lefkowitz RJ, Rockman HA. Beta-arrestin-mediated beta1-adrenergic receptor transactivation of the EGFR confers cardioprotection. J Clin Invest 117: 2445–2458, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR, Walker SJ, Zhang L, Hurban P, de Longueville F, Fuscoe JC, Tong W, Shi L, Wolfinger RD. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol 24: 1140–1150, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Poole-Wilson PA, Swedberg K, Cleland JG, Di Lenarda A, Hanrath P, Komajda M, Lubsen J, Lutiger B, Metra M, Remme WJ, Torp-Pedersen C, Scherhag A, Skene A. Comparison of carvedilol and metoprolol on clinical outcomes in patients with chronic heart failure in the Carvedilol Or Metoprolol European Trial (COMET): randomised controlled trial. Lancet 362: 7–13, 2003. [DOI] [PubMed] [Google Scholar]
- 25.Port JD, Walker LA, Polk J, Nunley K, Buttrick PM, Sucharov CC. Temporal expression of miRNAs and mRNAs in a mouse model of myocardial infarction. Physiol Genomics 43: 1087–1095, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruffolo RR Jr, Feuerstein GZ. Pharmacology of carvedilol: rationale for use in hypertension, coronary artery disease, and congestive heart failure. Cardiovasc Drugs Ther 11, Suppl 1: 247–256, 1997. [DOI] [PubMed] [Google Scholar]
- 27.Sack MN, Harrington LS, Jonassen AK, Mjos OD, Yellon DM. Coordinate regulation of metabolic enzyme encoding genes during cardiac development and following carvedilol therapy in spontaneously hypertensive rats. Cardiovasc Drugs Ther 14: 31–39, 2000. [DOI] [PubMed] [Google Scholar]
- 28.Saravanan R, Shanmugam A, Rajkumar D. Preventive effect of glycosaminoglycans from Amussium pleuronectus (Linne) on biomolecules, lactate dehydrogenase-isoenzyme and electrocardiographic patterns in isoproterenol-induced myocardial infarction in Wistar rats. Indian J Pharmacol 44: 602–606, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stueckle TA, Lu Y, Davis ME, Wang L, Jiang BH, Holaskova I, Schafer R, Barnett JB, Rojanasakul Y. Chronic occupational exposure to arsenic induces carcinogenic gene signaling networks and neoplastic transformation in human lung epithelial cells. Toxicol Appl Pharmacol 261: 204–216, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y, Wang Y, Park KM, Hu Q, Teoh JP, Broskova Z, Ranganathan P, Jayakumar C, Li J, Su H, Ramesh G, Kim IM. MicroRNA-150 protects the mouse heart from ischaemic injury by regulating cell death. Cardiovasc Res 106: 387–397, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teoh JP, Park KM, Wang Y, Hu Q, Kim S, Wu G, Huang S, Maihle N, Kim IM. Endothelin-1/endothelin A receptor-mediated biased signaling is a new player in modulating human ovarian cancer cell tumorigenesis. Cell Signal 26: 2885–2895, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Chen Y, Jiang J, Zhou A, Pan L, Chen Q, Qian Y, Chu M, Chen C. Carvedilol has stronger anti-inflammation and anti-virus effects than metoprolol in murine model with coxsackievirus B3-induced viral myocarditis. Gene 547: 195–201, 2014. [DOI] [PubMed] [Google Scholar]
- 33.Wang M, Wang X, Ching CB, Chen WN. Proteomic profiling of cellular responses to Carvedilol enantiomers in vascular smooth muscle cells by iTRAQ-coupled 2-D LC-MS/MS. J Proteom 73: 1601–1611, 2010. [DOI] [PubMed] [Google Scholar]
- 34.Wisler JW, DeWire SM, Whalen EJ, Violin JD, Drake MT, Ahn S, Shenoy SK, Lefkowitz RJ. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proc Natl Acad Sci USA 104: 16657–16662, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129: 1009–1021, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue TL, Cheng HY, Lysko PG, McKenna PJ, Feuerstein R, Gu JL, Lysko KA, Davis LL, Feuerstein G. Carvedilol, a new vasodilator and beta adrenoceptor antagonist, is an antioxidant and free radical scavenger. J Pharmacol Exp Ther 263: 92–98, 1992. [PubMed] [Google Scholar]
- 37.Yue TL, Ma XL, Gu JL, Ruffolo RR Jr, Feuerstein GZ. Carvedilol inhibits activation of stress-activated protein kinase and reduces reperfusion injury in perfused rabbit heart. Eur J Pharmacol 345: 61–65, 1998. [DOI] [PubMed] [Google Scholar]
- 38.Yue TL, Ma XL, Wang X, Romanic AM, Liu GL, Louden C, Gu JL, Kumar S, Poste G, Ruffolo RR Jr, Feuerstein GZ. Possible involvement of stress-activated protein kinase signaling pathway and Fas receptor expression in prevention of ischemia/reperfusion-induced cardiomyocyte apoptosis by carvedilol. Circ Res 82: 166–174, 1998. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, Han C, Lu D, Wu T. miR-17-92 cluster promotes cholangiocarcinoma growth: evidence for PTEN as downstream target and IL-6/Stat3 as upstream activator. Am J Pathol 184: 2828–2839, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.