Abstract
Background
Immunohistochemical analysis of cellular interactions in the bone marrow in situ is demanding, due to its heterogeneous cellular composition, the poor delineation and overlap of functional compartments and highly complex immunophenotypes of several cell populations (e.g. regulatory T-cells) that require immunohistochemical marker sets for unambiguous characterization. To overcome these difficulties, we herein present an approach to describe objects (e.g. cells, bone trabeculae) by a scalar field that can be propagated through registered images of serial histological sections.
Methods
The transformation of objects within images (e.g. cells) to a scalar field was performed by convolution of the object’s centroids with differently formed radial basis function (e.g. for direct or indirect spatial interaction). On the basis of such a scalar field, a summation field described distributed objects within an image.
Results
After image registration i) colocalization analysis could be performed on basis scalar field, which is propagated through registered images, and - due to the shape of the field – were barely prone to matching errors and morphological changes by different cutting levels; ii) furthermore, depending on the field shape the colocalization measurements could also quantify spatial interaction (e.g. direct or paracrine cellular contact); ii) the field-overlap, which represents the spatial distance, of different objects (e.g. two cells) could be calculated by the histogram intersection.
Conclusions
The description of objects (e.g. cells, cell clusters, bone trabeculae etc.) as a field offers several possibilities: First, co-localization of different markers (e.g. by immunohistochemical staining) in serial sections can be performed in an automatic, objective and quantifiable way. In contrast to multicolour staining (e.g. 10-colour immunofluorescence) the financial and technical requirements are fairly minor. Second, the approach allows searching for different types of spatial interactions (e.g. direct and indirect cellular interaction) between objects by taking field shape into account (e.g. thin vs. broad). Third, by describing spatially distributed groups of objects as summation field, it gives cluster definition that relies rather on the bare object distance than on the modelled spatial cellular interaction.
Electronic supplementary material
The online version of this article (doi:10.1186/s13000-015-0383-0) contains supplementary material, which is available to authorized users.
Background
Histological interpretation of lympho-hematopoietic tissues (e.g. bone marrow, lymph nodes, thymus) is a demanding task in many haematological diseases due to a highly complex composition of these tissue comprising lymphoid, myeloid, dendritic and eventually epithelial cells and bony structures in addition to notoriously “fuzzy borders” even of well defined functional structures (e.g. lymphoid follicles, niches [1–6]). Against this background, several intricate issues need to be addressed [7, 8]: a) The quantitative evaluation of distinct cell populations per area, e.g. CD4+, CD25+, Foxp3+ regulatory T-cells (Tregs) [9] that may need a set of immunohistochemical markers for identification. b) The spatial distribution of different cell populations in relation to each other and c) to functional regions (e.g. paratrabecular or perivascular niches) [7, 8].
In the routine diagnostic setting, the issues are currently addressed by rough visual estimation of cellular contents and locations. Reliable counting of cellular infiltrates and manual delineation of regions in combination with sophisticated multiplex immunohistochemical staining or confocal microscopy are usually reserved to scientific questions.
By exploring the bone marrow histology in chronic myeloid leukaemia (CML) with regard to the immunological milieu in the context of an on-going study [10], we have been facing all of the above-mentioned issues. Niches that are supposed to harbour leukaemia stem cells [5] and the tumour microenvironment that comprises mesenchymal stromal cells and various immune cells are of particular interest [11] in relation to the hypothetical impact of immunity on the eradication of CML and the modulation of the immunological milieu by antibodies [12] and other drugs (e.g. kinase inhibitors) [13, 14].
To more objectively describe the complex cellular composition of the bone marrow in CML and the interaction of cell populations that need definition by a plethora of immunohistochemical markers, we herein propose a method to annotate cells or rather cell cluster by scalar fields and to propagate these fields through several registered images. By doing so, quantification (a), localization in regard to niches (b) and spatial interaction analysis (c) could be addressed achieved.
Methods
Patient collective
For this technically oriented, proof-of-principle study we used two instructive cases of formalin-fixed bone marrow (trephine) biopsies from the archive of the Institute of Pathology, University Medical Centre Mannheim: One depicts the classical paratrabecular infiltrates of a follicular lymphoma; the other one shows scattered lymphoid cells among the dense granulocytic infiltrates in a case of CML.
Immunohistochemistry staining
For immunohistochemistry (IHC) staining, the following antibodies were applied on formalin-fixed, paraffin-embedded tissue using a routine immunoperoxidase technique [15, 16] and experimental sequential IHC [17]: anti-CD3 (Dako M7254), anti-CD4 (Dako M7319), anti-CD8 (Dako M7103), anti-CD20 (Dako M0755), anti-Bcl2 (DakoM0887), anti-MPO (Dako C7246), anti-CD61 (Dako C7280). Two chromogens were used, DAB (DAB chromogen) as fix and NovaRed (VECTOR NovaRed) as removable chromogen for sequential IHC [17].
Comment on image ethics
The histological images shown and used within this work underwent image registration and cropping as part of the method described herein. However, image compression was not applied and image augmenting or manipulating techniques were not performed (e.g. contrast enhancement, gamma setting) according to Digital Image Ethics [18–20].
Image acquisition and pre-processing (Additional file 1: Figure S1)
Differently stained histological slides of bone marrow biopsies were fully digitalized using an Aperio ScanScope (Aperio/Leica biosystems) and saved in the proprietary svs-format. These files (circa 1GB per file) were decomposed into two regions (each composed of 11,000x10,000 pixel, i.e. about 450 MB per file) and saved in the tagged image file format (tiff) to be manageable with a standard desktop computer. A pathologist approximately chose the regions of interest in serial sections (= rough manual matching).
Then, all files per case were loaded into Fiji [21] and registered with several, established plugin-functions (“Register Virtual Stack Slices” [22, 23], „Template Matching” [24], „StackReg” [25, 26]). Also in Fiji, a colour deconvolution plugin (“Colour Deconvolution” [27–29]) was used to separate the brown (immunoperoxidase staining) and blue areas (DAB background staining) per image. Subsequently, these registered image sets were transferred to MATLAB (version R2014a, Mathworks, Natick, MA, USA).
Definition and segmentation of spaces and niches
In bone marrow histology, there are several spaces that are defined via their distance to certain easily recognizable structures e.g. paratrabecular or perivascular spaces. The cellular composition of these spaces and the infiltration by certain neoplastic populations in the course of haematological diseases (e.g. follicular lymphoma, CML) are of high diagnostic relevance [7]. Some of these anatomic regions overlap with functional microenvironments for stem cells called niches [3, 5, 30].
In the current study, we focused on the paratrabecular region for the sake of simplicity. This was achieved by manually segmenting in Fiji the bone trabeculae (in the images within this work visualized as green area) and their subsequent description as scalar field by the convolution approach described below.
Image processing in MATLAB to detect cells (Additional file 1: Figure S1)
Registered images were loaded into the MATLAB environment and underwent further processing by custom-arranged standard image processing procedures. (link to a corresponding Git-repository: https://bitbucket.org/CATWeis/on-the-representation-of-cells-in-bone-marrow-pathology-by-a): image segmentation (application of thresholding algorithms), object detection (application of cross-correlation algorithms) and separation (application of watershed algorithms). At the end a detected nucleus/particle is represented by a centroid. Centroids that are within a certain range (application of distance measure algorithm) to immunoperoxidase-stained areas are referred as “stained” whereas the remaining are referred as “non-stained”.
Introduction of a field (Additional file 1: Figure S1)
The basic assumption of this work is that every object (cells, bone trabeculae etc.) can be represented by a scalar field with its maximum at the object’s centroid. Mathematically, this approach is related to multidimensional kernel estimation [31, 32].
The scalar field of an object is represented by a radial basis function (RBF), in this work, by an inverse multiquadric RBF
| 1 |
with x and y as variables and with xP and yP as coordinates of the centroid. The invers multiquadric RBF was chosen among the many other RBF due to its feasible shape for this project and due to its many changeable parameters. However, many other RBF would be likewise applicable. The parameter α is chosen to have an arbitrary maximum of 100 [arbitrary unit] at the coordinates of the centroid. Hereby, the shape of the RBF, which is defined by the parameters and , could be adapted to different cell interaction models [33]: Hypothesis 1) Direct spatial cell-cell- and cell-niche-interaction (henceforth called ‘direct interaction’) is represented by a RBF (henceforth denoted by ’RBFdirect’) with high values in the area of the nucleus (>90), mediate values in the area of the cell (>80) and with a step, asymptotic decrease towards 0 (black solid line Fig. 1). Hypothesis 2) Indirect spatial interactions via proposed secretory factors (henceforth called ‘indirect interaction’) are represented by a RBF (henceforth denoted by ‘RBFindirect’) with, again, high values at the centroid position (>90) but a broad shape and medium values within the proposed 250 μm range for paracrine interaction [33, 34] (solid red line Fig. 1).
Fig. 1.
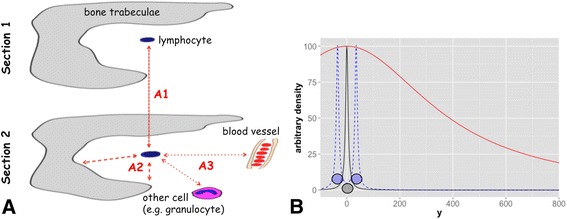
Illustration of the task and the solution via a RBF. a: Sketch of two serial sections where a single lymphocyte in the centre could be appreciated in both sections. (A1) One task is to colocalize this cell/its immunohistochemical staining in both sections. (A2) Another task is to measure its distance to the bone trabeculae. (A3) Furthermore, is should be allocated to niches via measuring its distance to certain objects (bone trabeculae, other cells, vessels). b: By usage of different parameters (β and γ in equation 1) a RBF for direct and indirect spatial interaction could be modelled according to the particular image resolution/pixel-distance relation. The parameters are here chosen to accomplish a shape for RBFdirect, which represents potential direct object-object interaction, and for RBFindirect, which represents paracrine interaction. Black solid line: One point at position xp = 0 (representing a nucleus) is convoluted with an inverse, multiquadric basis function for direct interaction (RBFdirect with β = 3 and γ = 0.01). For illustration purposes the RBF is only shown with one variable. As a consequence, every point in this figure (xЄ [−100,800]) has an arbitrary scalar field value [arbitrary density]. Blue dashed line: Overlay of two fields for direct interaction, where one represents a centroid at xp = −50 and one at xp = 50.Red solid line: The single point at position xp = 0 is convoluted with an inverse, multiquadric basis function for indirect interaction (RBFindirect with β = 3 and γ = 0.00002)
On the basis of this function, for every image a kernel (application of frequency space convolution algorithm) is calculated that needs to have the size of the image and that must not have the value 0 at any point. The latter requirement is important to allow distance calculations as described below.
Normalization of field values
The cell centroids and the bone areas were convoluted with these RBF, which are additive to highlight regions with higher concentrations, respectively, clusters (blue dashed line Fig. 1 for two adjacent nuclei). Since every point has a field with values >0 in every pixel of the image, the summation fields after convolution easily yields high scalar values >10,000 depending on the overall object number. To normalize the scalar fields, the linear mapping
| 2 |
with C(x y) as the local scalar field (equation 1), Cmin as the absolute minimum and with Cmax as the absolute maximum of the scalar field is applied.
Calculation of the gradient and the divergence
One further hypothesis of this work is that the gradient and the divergence of the applied scalar field are meaningful and, respectively, are useful to detect and describe object clusters. This hypothesis is in line with approaches in biophysics, e.g. fluid dynamics [34, 35] and earth science, e.g. digital elevation models [36]. The gradient can be calculated by functions implemented in MATLAB. For example, the normalized gradient is calculated by dividing it by the mean gradient per field according to
| 3 |
with C(x y) as the local scalar field (equation 1), G as the gradient of C(x y) and with as the mean gradient of C(x y).The divergence can also easily be calculated by in MATLAB implemented functions and is likewise normalized.
Statistical evaluation (Additional file 1: Figure S1)
Representation of the field as histograms and statistical evaluation of the distributions were performed in MATLAB. Another assumption is that several readings routinely used for co-localization analysis in fluorescence microscopy could be used for similar analysis with the fields that are produced herein: Calculation of correlation coefficients (Pearson’s correlation coefficient (PCC), overlap coefficient according to Manders (MOC), overlap coefficients M1and M2 [37, 38]) and graphical presentation were performed with R [39], in particular with the ggplot2 package [40].
Results
Proof of principle for the description of colocalization and possible spatial interaction via a scalar field
To directly test whether the chosen approach is able to describe proximity of cells or “degree of co-localization” with regard to the above-defined direct interaction, a set of artificial binary test images was analysed. These images were matrices of 500×500pixel that contain one point and/or a set of points; the positions of which were changed in a stepwise manner. The images were convoluted with the above-described RBFdirect (hypothesis 1 in the method section that a sharp function could map direct cellular contact) and further processed. The matrices containing the scalar values could be linearized and plotted against each other as routinely performed in co-localization analysis: For a single point versus another single point (Fig. 2a-c) and for a single point versus a cluster (Fig. 2d-f) the width of the point cloud depends on the distance. Furthermore, the shape of the point cloud also depends on the number of points per matrix. Of note, the absolute number of pairs of variants in this case depends on the image size (e.g. for a 10×10 pixel image 100 pairs of variants and not on the number of points.
Fig. 2.
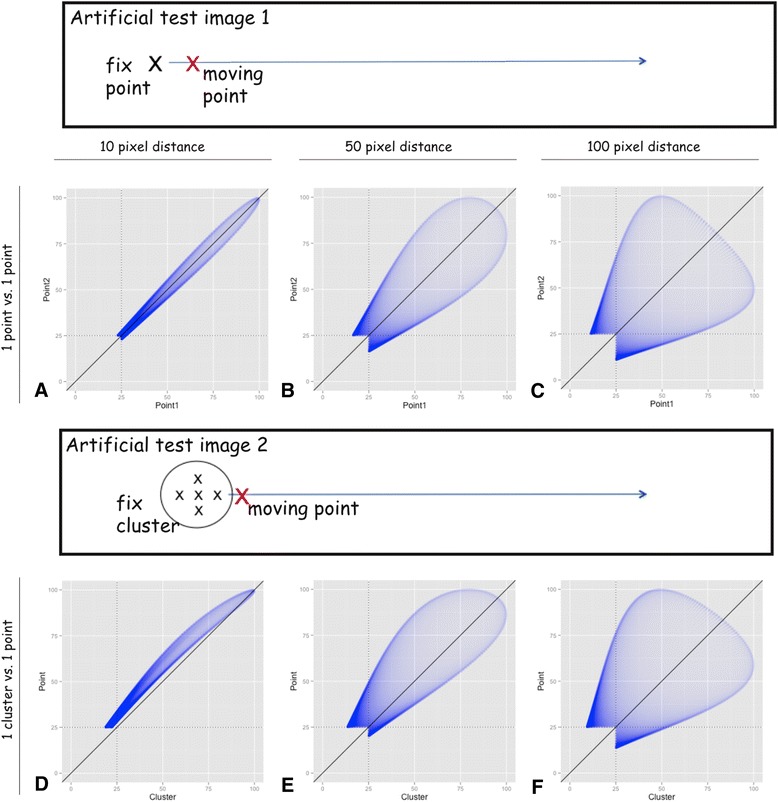
Scatter plots of several artificial test images. Binary test images containing one point and/or a set of points were generated and convoluted with the RBFdirect. The values of a pair of normalized matrices (compare equation 2) are plotted against each other, resulting in a scatter plot. The sketches above the scatter plots represent the artificial test images (black box) with one fix (black X) and one moving point (red X) and, respectively, one fix cluster of five points (black Xs) and one moving point (red X). For Matlab, these images were realized as matrices of 500x500pixel is one, respectively, five fix points and one moving point (in the supplement). Density values below a threshold of 25 are excluded. a-c: In the upper line, two matrices each containing one single point are plotted against each other. The position or respectively the distance of these two are changed as described (e.g. point at M(1,1) against N(1,11) etc.). d-f: In analogy, one point in the lower line is plotted against one cluster consisting of 5 points
To characterize these resulting point clouds, several well-established correlation coefficients (e.g. Pearson’s correlation coefficient (PCC) and overlap coefficient according to Manders (MOC) [37, 38]) could be applied after thresholding to avoid false high co-localization due to near zero values (e.g. threshold of 25 in Fig. 2). The PCC and the MOC are inversely correlated with the spatial distance of the points/clusters, since the values of both coefficients decrease with increasing distance: E.g. one point versus one point showed at the distance of 10 pixel PCC = 0.96 and M = 1.00 and accordingly at 100 pixel distance PCC = 0.31 and M = 0.83.
For comparison, without the convolution, the standard colocalization method, at a distance of 0 pixel PCC = 1 and M = 1 and respectively at a distance of 1 pixel PCC = 0 and M = 0.
Furthermore, the established overlap coefficients M1 and M2 [37, 38] could be obtained after setting a threshold (e.g. 50 to get the overlap of 50-perzentile). These values also correlated with the distance and showed a relation with both the number of points and the distance of points within a cluster: E.g. one cluster and one point at a distance of 50 pixel show Manders coefficients of MCluster = 0.83 and MPoint = 0.59, while the values were Mcluster = 0.03 and MPoint = 0.02 at a distance of 200 pixel.
Comparison to colocalization via colour channel analysis
The golden standard of colocalization analysis in fluorescence microscopy is to analyse and plot against each other the different colour channels (e.g. red and green) [38]. By comparing and analysing the “brown” channel obtained by colour deconvolution [28], we tried to adapt this approach to IHC images and subsequently compared the results of both methods. First we compared the results for analysis of one CD61-positive megakaryocyte translated one cell width to left (Fig. 3a): Whereas parts of the point cloud are arranged around the bisecting line (Fig. 3a upper scatter plot), the density function-based analysis shows a clearly forked point cloud (lower scatter plot). Second we analysed the same region now stained for MPO by sequential IHC [17] for colocalization of megakaryocytes and granulocytes (Fig. 3b): The point cloud for the colour channel analysis is mostly distributed around the bisecting line (Fig. 3b upper scatter plot). This does not fit to the images where the brown colour is placed in different areas (one brown megakaryocyte in A and many brown granulocytes in B). In comparison, the lower scatter plot shows that there is mostly no correlation between the CD61- and the MPO-positive cells. However, there is one branch of the point cloud that is due to the direct spatial contact of cell populations (Fig. 3b white arrows).
Fig. 3.
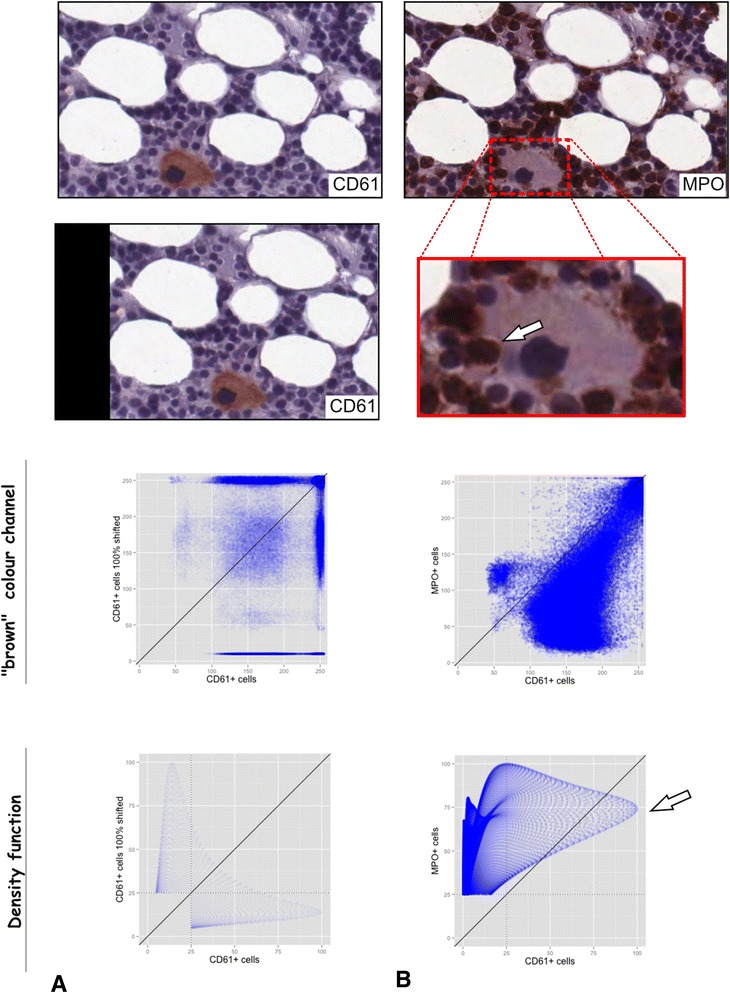
Comparison of the colocalization via RBF to the standard colocalization analysis in fluorescence microscopy. The golden standard of colocalization analysis in fluorescence microscopy is to analyse the different colour channels (e.g. red and green). In this figure we simply matched this method to IHC by comparing and analysing the “brown” channel obtained by colour deconvolution. One small region of a slide sequentially stained for CD61 (A) and MPO (A) was analysed. Subsequently, colocalization on basis of the herein described RBF was compared to analysis of colour chancels. a One small region with one CD61-positive megakaryocyte (upper image) was compared to the same region shifted one cell width to left (middle image). The upper scatter plot shows the brown colour channels of the images plotted against each other, whereas the lower one shows the scatter plot for the RBF. b The same small region now stained for MPO was used to check for colocalization/direct spatial interaction of the CD61-positive megakaryocyte and the MPO-positive granulocytes (upper image). The arrow in the zoomed part of the image highlight direct spatial interaction of the megakaryocyte and the granulocytes. The upper scatter plot again shows the brown colour channels plotted against each other and the lower one shows the RBFs plotted against each other
Proof of principle for valid distance measurements through histogram intersection
The Euclidian distance is usually applied to measure the distance between two objects in an histological sections. Therefore, Euclidian distance is regarded as the gold standard. However, with vast numbers of cells this approach is not feasible for manual counting. Herein, we apply the histogram intersection of scalar fields, which represent objects, as surrogate for distances between different objects. The histogram intersection is widely applied in the comparison of multidimensional distributions [31, 41].
Again, a set of artificial binary test images with variable object distances was analysed: The intersection of the histograms decreased with the distance of two points (for Fig. 2a-c: 0.98, 0.89 and 0.79) and of one point and one cluster respectively (for Fig. 2d-e: 1.00, 0.95 and 0.83). However, due to the shape of the applied RBF (bell-shape with asymptotically approach to zero; please compare equation 1 and Fig. 1), the distance measurement via histogram intersection is only meaningful within a certain range. Beyond a certain distance (for RBFdirect approximately 200 pixel/100 μm), the intersection function shows an asymptotic behaviour. Therefore, the distance and the histogram intersection cease correlating beyond a certain value (in this example beyond 0.4).
Proof of principle for description and detection of object clusters via calculation of the gradient and the divergence
On the basis of the applied scalar fields, several typical values for scalar and vector fields like the gradient (i) and the divergence (ii) could be calculated by standard mathematical operations [42].
Ad (i) The normalized gradient (equation 3) can be used to compare the structure of object clusters in different images regardless of the number of objects.
Ad (ii) In this approach, the object centroids and the clusters are represented by sinks: In an artificial test image containing a cluster of five points with variable distance, it could be observed, that the number of sinks depends on the distance between points: For a distance of 25 pixel or less, the sinks were fused to one single “compound sink” (Additional file 2: Figure S2A), whereas a distance of 125 pixel between each of the 5 points (Additional file 2: Figure S2D) resulted in one sink per point for the present example. These numerical values are examples that are intended to show that the divergence depends on the distance and the shape of the function; and that it can be used to describe clusters of objects.
Application on histological images
To test the presented approach under less artificial conditions, is was applied to the spatial characterization of lymphoid infiltrates in immunohistochemically stained bone marrow sections using follicular lymphoma as a model for spatially distinct neoplastic lymphoid infiltrates, and chronic myeloid leukaemia as a model of indistinct reactive lymphoid infiltrates among neoplastic cells.
Example 1: Spatial interaction of B-cell markers in a focal, dense infiltration
Tumour infiltrates of follicular lymphoma in the bone marrow typically form either dense intertrabecular nodules, or dense, linear, paratrabecular lymphoid aggregates along the bone. In sections stained for the markers CD20 and Bcl2, tumorous B cells typical show co-expression [7, 8].
Analysis of the co-localization with RBFdirect (Fig. 4f for CD20 and G for Bcl2). Theoretically, the two fields could be combined by addition to one field describing the density of the infiltration by CD20+ Bcl2+ B-cells (data not shown); as delineated in the method section (equation 4).
Fig. 4.
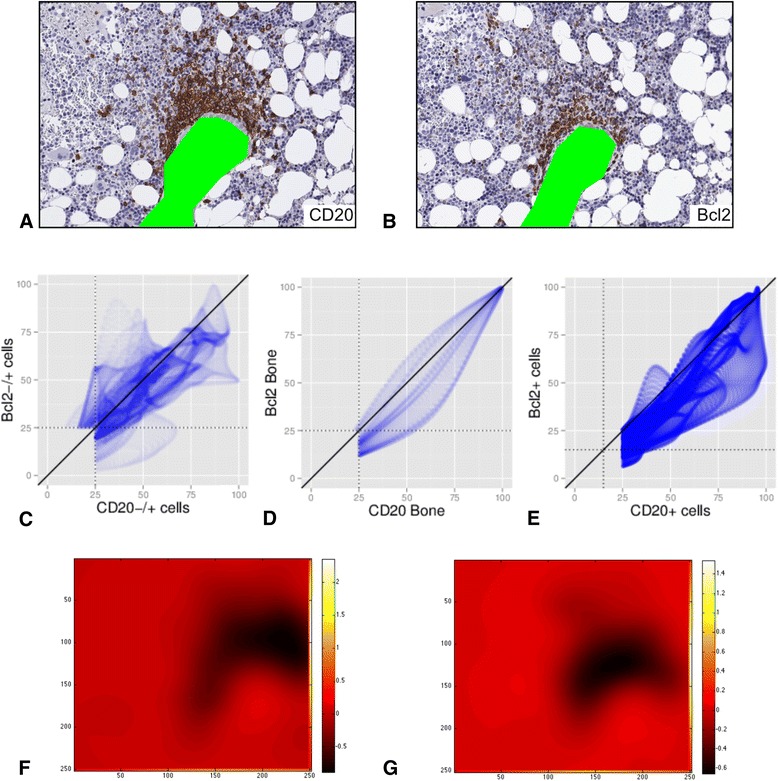
Direct spatial interaction in a case with one focal, dense infiltration. Serial sections of a trephine biopsy from a patient with a follicular lymphoma were stained for CD20 and Bcl2. Slides were fully digitalized, registered and analysed as described in Methods.A-B: Staining for CD20 and Bcl2. After segmentation of nuclei, there were 352 CD20+ and 859 CD20−/+ nuclei in a, and 233 Bcl2+ and 795 Bcl2−/+ nuclei in b. c Scatter plot for all nuclei (CD20−/+ and Bcl2−/+) with PCC = 0.74, MOC = 0.97 and MCD20−/+ = 0.90 and MBcl2−/+ = 0.95. d Scatter plot for the bone areas with PCC = 0.94, MOC = 0.98 and MBoneCD20 = 0.76 and MBoneBcl2 = 0.99. e Scatter plot for all positive nuclei (CD20+ and Bcl2+) with PCC = 0.90, MOC = 0.97 and MBcl2+ = 0.99 and MCD20+ = 0.71, respectively. f-g Heat map of the diversity for CD20 (F) and Bcl2 (G)
Example 2: Allocation and spatial interaction of B- and T-cells in a focal, dense infiltration
Lymphocytosis with lymphoid follicles are a common finding in bone marrow sections [7]. Follicles are usually composed of CD3+ T cells and CD20+ B cells* and are therefore an example for a focal, dense infiltration where immunohistochemical T- and B-cell markers co-localize within the infiltration but not within single cells.
Analysis of co-localization in serial sections (Fig. 4) stained repeatedly (serially) for CD3 and CD20 (sections CD3 I, CD3 II, CD20 I and CD20 II) showed, that there was a spatial overlap/a co-localization of these markers (Manders coefficients each >0.57). This is against the expectation, since CD3 and CD20 usually are not expressed by one cell. However, a closer look at the images reveals (small insets in Fig. 5), that brown stained areas do co-localize in the images (white arrows in the small insets). Furthermore, from the point of spatial interaction, there is of course an interaction of neighbouring T- and B-cells. Analysing a sketch of a lymphoid follicle composed of B- and T-cells drawn on basis of Fig. 4 carve this point out: In this case there is no overlap of cells but the cells are close neighbours. Therefore MT-cell = 0.34 and MT-cell = 0.67.
Fig. 5.
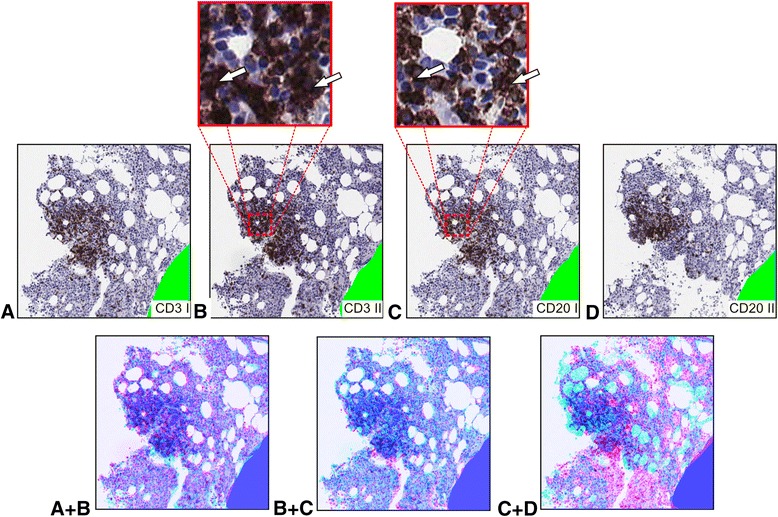
Direct spatial interaction in a lymphoid follicle. Serial sections of a trephine biopsy from a patient with bone marrow lymphocytosis were stained for CD3, CD3, CD20 and CD20 (a-d). Subsequently, slides were fully digitalized, registered and analysed as described. By segmentation, there are 320 CD3+ and 1,156 CD3−/+ nuclei in a, 392 CD3+ and 1,139 CD3−/+ nuclei in b, 316 CD20+ and 1,176 CD20−/+ nuclei in c and respectively 202 CD20+ and 1,182 CD20−/+ nuclei in d. Overlap readings are calculated for direct interaction (RBFdirect). A + B: Overlap of both images with A (first section stained for CD3) drawn in red and respectively B (second section stained for CD3) drawn in blue. For stained cells, PCC = 0.95, MOC = 0.99 and M1st section CD3+ = 0.98 and M2nd section CD3 = 0.81. B + C: Overlap of both images with B (second section stained for CD3) drawn in red and respectively C (first section stained for CD20) drawn in blue. For stained cells, PCC = 0.97, MOC = 0.99 and M2ndsection CD3+ = 0.83 and M1stsection CD20 = 0.97. C + D: Overlap of both images with C (first section stained for CD20) drawn in red and respectively D (second section stained for CD20) drawn in blue. For stained cells, PCC = 0.67, MOC = 0.92 and M1st section CD20+ = 0.57 and M2nd section CD20 = 0.97
Example 3: Co-localization of T-cell markers in a lose infiltration
In CML, neoplastic myeloid infiltrates in the bone marrow are commonly intermixed with a minority of dispersed lymphocytes of unknown significance [7, 8]. Antibodies to three T cells markers (CD3, CD4 and CD8 [7]) and one B cell marker (CD20) [7] were applied to serial CML bone marrow sections to test for the expected strong overlap between T cell markers (e.g. CD3 and CD8) but for a minor overlap between T and B cell markers (e.g. CD3 and CD20). The resulting overlap coefficients are shown in Table 1 for direct and indirect interaction (furthermore the images and three resulting scatter plots are shown in Additional file 3: Figure S3).
Table 1.
Comparison of direct and indirect interaction for three registered sections in a case with loose lymphoid infiltrates
| Direct interaction/RBFdirect | Indirect interaction/RBFindirect | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3 | CD3 Bone | CD4 | CD4 Bone | CD8 | CD8 Bone | CD20 | CD20 Bone | CD3 | CD3 Bone | CD4 | CD4 Bone | CD8 | CD8 Bone | CD20 | CD20 Bone | |
| CD3 | MCD3 = 0.00 | MCD3 = 0.00b | MCD3 = 0.02 | MCD3 = 0.00b | MCD3 = 0.02 | MCD3 = 0.10a | MCD3 = 0.00 | MCD3 = 0.60 | MCD3 = 0.54 | MCD3 = 0.36 | MCD3 = 0.54 | MCD3 = 0.36 | MCD3 = 0.69a | MCD3 = 0.60 | ||
| CD3 Bone | MCD3Bone = 0.00 | MCD3Bone = 0.03 | MCD3Bone = 0.43a | MCD3Bone = 0.00 | MCD3Bone = 0.43b | MCD3Bone=0.00 | MCD3Bone=0.88 | MCD3Bone = 0.89 | MCD3Bone = 0.71 | MCD3Bone = 0.50 | MCD3Bone = 0.71 | MCD3Bone = 0.50 | MCD3Bone=0.59 | MCD3Bone=0.98 | ||
| CD4 | MCD4 = 0.00b | MCD4 = 0.61 | MCD4 = 0.00 | MCD4 = 0.00 | MCD4 = 0.00 | MCD4 = 0.00 | MCD4 = 0.39 | MCD4 = 0.91 | MCD4 = 0.80 | MCD4 = 0.24 | MCD4 = 0.72 | MCD4 = 0.21 | MCD4 = 0.69 | MCD4 = 0.24 | ||
| CD4 Bone | MCD4Bone = 0.00 | MCD4Bone = 0.79a | MCD4Bone = 0.00 | MCD4Bone = 0.00 | MCD4Bone=0.84 | MCD4Bone=0.01 | MCD4Bone=0.31 | MCD4Bone = 0.88 | MCD4Bone = 0.83 | MCD4Bone = 0.36 | MCD4Bone = 0.66 | MCD4Bone=0.92 | MCD4Bone=0.54 | MCD4Bone=0.82 | ||
| CD8 | MCD8 = 0.00b | MCD8 = 0.00 | MCD8 =0.00 | MCD8 = 0.00 | MCD8 = 0.00 | MCD8 = 0.00 | MCD8 = 0.00 | MCD8 = 0.91 | MCD8 = 0.80 | MCD8 = 0.50 | MCD8 = 0.31 | MCD8 = 0.27 | MCD8 = 0.80 | MCD8 = 0.51 | ||
| CD8 Bone | MCD8Bone = 0.00 | MCD8Bone = 0.71b | MCD8Bone = 0.00 | MCD8Bone = 0.96 | MCD8Bone = 0.00 | MCD8Bone=0.01 | MCD8Bone=0.25 | MCD8Bone = 0.89 | MCD8Bone = 0.83 | MCD8Bone = 0.21 | MCD8Bone = 1.00 | MCD8Bone = 0.64 | MCD8Bone=0.51 | MCD8Bone=0.83 | ||
| CD20 | MCD20=0.15 a | MCD20=0.00 | MCD20 = 0.00 | MCD20 = 0.06 | MCD20 = 0.00 | MCD20 = 0.07 | MCD20 = 0.00 | MCD20=0.99 a | MCD20=0.57 | MCD20 = 0.59 | MCD20 = 0.31 | MCD20 = 0.99 | MCD20 = 0.27 | MCD20 = 0.58 | ||
| CD20 Bone | MCD20Bone=0.00 | MCD20Bone=0.91 | MCD20Bone=0.03 | MCD20Bone=0.46 | MCD20Bone=0.00 | MCD20Bone=0.44 | MCD20Bone=0.00 | MCD20Bone=0.90 | MCD20Bone=0.99 | MCD20Bone=0.36 | MCD20Bone=0.50 | MCD20Bone=0.68 | MCD20Bone=0.46 | MCD20Bone=0.60 | ||
Three sequential sections stained for CD3, CD20, CD4 and CD8 were registered and subsequently the overlap coefficients were calculated for direct and indirect interaction (application of RBFdirect with its sharp shape and respectively RBFindirect with its broad silhouette)
aAs expected, small overlap/direct interaction between CD3+ and CD20+ cells and noticeable indirect, paracrine interaction
bUnexpected low overlap between the T cell markers CD3, CD4 and CD8 assumedly due to morphological changes in the course of serial sectioning (see arrows in Additional file 3: Figure S3). Modification of the RBFdirect would overcome this problem, however, at the cost of reduced specificity
The images are fairly registered with regard to the bone trabeculae (Manders coefficients for bone 0.5-0.92 and for all cells >0.95). The reduced overlap is due to morphological changes that inevitably occur when sectioning through different levels of a core biopsy. As expected and shown in Table 1, there is no direct interaction between CD3+ and CD20+ (MCD3 = 0.10 versus MCD20 = 0.15). Due to morphological changes (the changes are highlighted by arrows in Additional file 3: Figure S3), there is also no spatial interaction between CD3, CD4 and CD8 (Manders coefficients each 0.00). For the indirect interaction, the Manders coefficients are each higher but still less than for CD3 and CD20. This finding will be picked up in the Discussion.
Example 4: Localization of cellular infiltrates in relation to bone trabeculae
While the above paragraphs describe the direct spatial interaction/co-localization of certain cell populations or rather stained and non-stained cells, we next addressed the allocation of cells to a pre-defined niche. The periosteal niche is defined as the space in close proximity to bone trabeculae [30, 43, 44]. Therefore, depending on its distance to the closest bone, a given cell or population could be allocated (or not) to this niche.
One way to implicitly address distance is to run a co-localization/interaction analysis on basis of RBFindirect. By doing so, for the dense lymphoid infiltration in example 1 (follicular lymphoma) there is more indirect spatial interaction (kCD20+ = 0.91) than for the “loose” infiltrate in example 2 (kCD20+ = 0.73) between CD20+ cells and the bone trabeculae.
Distance could also be measured by another approach, namely by calculating the histogram intersection (as depicted above) for bone trabeculae and a cell population. Drawn on basis of the bone marrow region Fig. 3, the sketches in Fig. 6 show different types of bone marrow infiltration by a lymphoma. For example, 33 cells located only on the blue dashed have a mean histogram intersection of 0.81 ± 0.07 for direct interaction (and of 0.91 ± 0.06 for indirect interaction). Assuming that these values as threshold for allocation into the periostal region, the sketched infiltrations within Fig. 4 could be analysed accordingly: The mean histogram intersection for the dense infiltration in A is 0.81 ± 0.05 (respectively 0.88 ± 0.03 for indirect interaction), while it is 0.51 ± 0.05 (respectively 0.77 ± 0.02) for the non-paratrabecular infiltration in B, and 0.66 ± 0.013 (respectively 0.82 ± 0.05) for the diffuse, mixed infiltration in C. Hence, the infiltration in Fig. 4a could be allocated by the histogram intersection to the periostal region. Applying the intersection values of cells on the blue line as threshold (0.81 ± 0.07), object per object revealed a significantly different mean intersection of 0.82 ± 0.07 for the dense infiltration in example 1 compared to 0.77 ± 0.15 for the “loose” infiltrate in example 2 (p = 0.017).
Fig. 6.
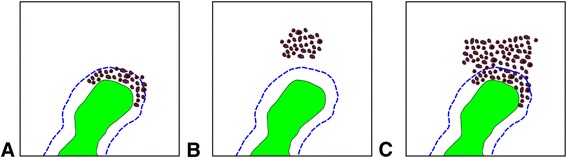
Sketches of different bone marrow infiltration by a lymphoma. Sketch based on the region shown in Fig. 3 to visualize different patterns of bone marrow infiltration. The bone trabeculae are drawn in green. The blue dashed line delineates a region with a maximal sagittal distance of approximately 20 μm around the bone that is regarded as the location of the periosteal niche (compare [50]). Histogram intersection is calculated for direct and indirect interaction (RBFdirect and RBFindirect). a Nodular, peritrabecular infiltration (intersection 0.81 ± 0.05 for direct and 0.88 ± 0.03 for indirect interaction). b Nodular, non-peritrabecular infiltration (intersection 0.51 ± 0.05 for direct and 0.77 ± 0.02 for indirect interaction). c Diffuse bone marrow infiltration (intersection 0.66 ± 0.013 for direct and 0.82 ± 0.05 for indirect interaction)
Discussion
The herein presented approach (compare Additional file 1: Figure S1) enables to run morphological analysis of bone marrow infiltrates, which is a demanding task due to – amongst other – highly complex immunophenotypes (e.g. Treg are defined by CD4 CD25 and FoxP3). Immunohistochemical staining of serial sections or sequential IHC can be performed to detect such cells that are defined by a marker set. (1) Herein we present an approach to propagate the cell position through serial slides by a scalar field function (Fig. 1a task A1). By doing so colocalization methods established for immunofluorescence could by applied. (2) Besides this, the scalar field function introduces a new perspective to objectively and automatically interpret complex objects (e.g. cellular infiltrates) and different modes of their spatial relationships (e.g. direct vs. paracrine) in histological sections (Fig. 1 task A2). Furthermore it allows for (3) a qualitative assessment of e.g. direct cellular contact or paracrine contact (Fig. 1 task A3) and by doing so also enables to (4) detect cell clusters.
1) Is co-localization analysis on basis of scalar fields in serial sections possible?
The co-localization/spatial interactions (direct or indirect) could be measured by using several well-established correlation coefficients (e.g. Manders’ overlap coefficient [37]) and by overlap coefficients M1 and M2 [38]. However, since these methods are initially defined for scatter plots of intensity values of different channels (e.g. green and red) and not for scalar field values, the interpretation of the resulting graphs and values need to be adapted: There is no relation between the number of particles and the points in the scatter plot similar to a scatterplot of two colour channels of one immunofluorescence image. The shape and position of the point cloud encodes the co-localization and also the clustering (see Fig. 2). In this context, especially the overlap coefficients M1 and M2 seem to fit best for the spatial interaction/co-localization analysis; by setting a threshold, one obtains the overlap of the corresponding percentiles (e.g. 50 %-percentile in Fig. 3 and Additional file 3: Figure S3).
At first glance, one may assume that it is a drawback of the approach described here that absolute distance values of single cells and clusters (e.g. in [m]) are lost. However, it is possible to state the overlap in form of overlap coefficients (e.g. Manders overlap coefficients M1 and M2) or as histogram intersection. These measurements may be more significant in regard to biology as pure metric measurements, since they already incorporate interaction models depending on distances for direct and indirect cellular interaction [33].
2) Is there are difference between co-localization and spatial interaction?
Throughout this work, the terms co-localization and spatial interaction are used more or less synonymously: According to their shape the scalar fields could describe different interactions models with (hypothesis 1) RBFdirect for spatial object-object interaction (maximum value at the centroid, medium values at the cell edge and then a step slope; compare black solid line in Fig. 1b) and (hypothesis 2) RBFindirect for indirect interaction (medium values within the proposed 250 μm range for paracrine interaction [33, 34] and a slight slope; compare red solid line in Fig. 1b); these fields could overlap and, therefore, could describe co-localization on basis of overlap. Thus, an huge overlap of RBFdirect points to spatial co-localization of two objects whereas an intermediate (e.g. histogram intersection of >0.95 for direct spatial contact and of 0.81 for 20 μm distance) overlap rather points to a close neighbourhood of them. Whether two objects are co-localized or occur as neighbours mainly depends on the chosen shape of the radial basis function; a broad radial basis function is less prone to registration errors whereas a narrow function is more specific for real overlap of objects. This trade off comes to effect in example 3 and in the linked sketch in Additional file 4: Figure S4: On the one hand, the narrow RBF results in very specific co-localization of markers; on the other hand this function is very prone to morphological changes throughout serial sections. This limitation can almost certainly be overcome by applying repetitive cycles of staining/de-staining using a spectrum of different antibodies on one given section [17, 45–47].
3) What are the advantages of the presented field approach?
After segmentation of e.g. cell nuclei in an image by standard image processing techniques, there are usually some distributed or clustered objects in the resulting image. To describe this distribution, clustering analysis could be applied [48, 49]. Therefore, a distance (usually spatial distance) measurement needs to be formulated and subsequently objects within a certain distance are (or not) assigned to a cluster. Although this approach appears straightforward and comprehensive, it depends on each single cell and is therefore prone to image artefacts and registration errors.
To avoid this disadvantage of spatial distance measurement, we herein describe each object (e.g. cell nucleus) by a scalar field on the basis of an inverse multiquadric radial basis function. These fields can overlap and interact in such a way, that the scalar values are summed. Consequently, not single objects but populations represented by a summation field are subject to the subsequent analysis, making this approach more robust against e.g. registration errors. The resulting field can be propagated through several matched sections of one tissue and can therefore be used to analyse co-localization and spatial interaction, notwithstanding the fact that a given single cell may not be represented at the same location in all slides due to image registration errors or due to section thickness. Furthermore, not only the position of a cell could be propagated and described: In the future we plan to describe more abstract properties of objects, such as relative object sizes, as field, that can be propagated and analysed subsequently, respectively. Furthermore, the description as scalar field will allow to use the whole body of field theory in future analyses.
4) Is it possible to describe clustering via scalar fields?
A great advantage of scalar fields is that they code the number of objects and their clustering. By calculating on this basis the gradient field and especially its divergence, cell clusters can be described as sources/sinks (compare Additional file 2: Figure S2). This approach does not need an explicit definition of a cluster e.g. by measuring absolute distances, but does implicitly define clusters by the shape of the RBF. Again, in contrast to direct measurements, this may incorporate already quasi-functional aspects by applying RBFs with different silhouettes according to different modes of interaction.
Conclusions
The description of objects in a histological section as scalar fields (e.g. cells of a certain type) opens several new perspectives: i) Co-localization of different marker (e.g. immunohistochemical staining) in serial sections becomes feasible. ii) Different types of spatial interaction (e.g. direct cellular and paracrine communication) could be modelled and subsequently analysed. iii) Description of aggregated objects via summation fields leads to an implicitly cluster definition.
Acknowledgements
The authors express their great gratitude to Prof. Dr. med. Christian Schultz, head of Neuroanatomy of the Medical Faculty Mannheim of the University of Heidelberg, for his promotion and experience concerning slide digitalization. Furthermore, the authors are greatly indebt to Ms. Katrin Wolk for performing myriads of reliable immunohistochemical staining on difficult decalcified sections.
The authors (CW, EP, AM) also would state their great gratitude for funding by the Jose-Carreras-Leukämiestiftung (grant: R10/17).
Abbreviations
- CD
Cluster of differentiation
- CML
Chronic myeloid leukemia
- Foxp3
Forkhead box P3
- MOC
Mander’s overlap coefficient
- MPO
Myeloperoxidase
- PCC
Pearson’s correlation coefficient
- RBF
Radial basis function
- Treg
Regulatory T-cell
Additional files
Flow chart for the presented approach. The main steps are “preprocessing” in Fiji, “cell segmentation” in Matlab, “introduction of the RBF” in Matlab and “statistical evaluation” in Matlab and R. The preprocessing and the cell segmentation are performed by custom arranged standard methods like colour deconvolution, thresholding, cross correlation etc. (PNG 220 kb)
Heat map of diversity for a cluster of five points. A cluster of five points is convoluted with the RBFdirect to obtain a scalar field. For this field the gradient field and - after normalization (compare equation 3) hereof - the diversity were calculated. In this approach, a sink represents a centroid. The distance of the points composing the cluster is changed from 25 pixel in A, to 38 pixel in B, to 50 pixel in C and to 125 pixel in D. (PNG 177 kb)
Direct spatial interaction in a case with a loose infiltration. Sections from a patient with CML were stained for CD3, CD20, CD4 and CD28. Slides were fully digitalized and registered like in the method section described. After segmentation there were 417 CD3+ and 6,187 CD3−/+ nuclei; 17 CD20+ and 5,770 CD20−/+ nuclei; 57 CD4+ and 6,394 CD4−/+ nuclei; and respectively 126 CD8+ and 5,696 CD8−/+ nuclei. A: Registered sections IHC-stained for CD3, CD20, CD4 and CD8. The registration between CD3 and CD20 and respectively between CD4 and CD8 is visually pretty good. However, due to different cuttings levels during processing, there is a continuous change of morphology. The white and the yellow arrow visualize these changes for one trabeculae and, respectively, for a focal lymphoid infiltrate. B: Scatter plot for all nuclei (CD3−/+, CD20−/+, CD4−/+ and CD8−/+). For CD3 vs. CD20 MCD3−/+ = 0.90 and MCD20−/+ = 0.90; for CD3 vs. CD4 MCD3−/+ = 0.90 and MCD4−/+ = 0.79; and respectively for CD4 vs. CD8 MCD4−/+ = 0.95 and MCD8−/+ = 0.80. C: Scatter plot for positive nuclei (CD3+, CD20+, CD4+ and CD8+). The respective overlap coefficients are shown in Table 1. (PNG 3007 kb)
Sketch of a lymphoid follicle. Sketch based on the region shown in Fig. 4 to visualize a non-malignant lymphoid follicle, which is composed of a mixture of T and B cells. A: T cells. B: B cells. (PNG 41 kb)
Footnotes
Alexander Marx and Frank Gerrit Zoellner are shared senior authorship.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CW developed the approach, wrote the algorithms and drafted the manuscript. BG carried out the staining, acquired the images and contributed to the programming. CS implemented sequential staining, acquired images and participated drafting the manuscript. CD also implemented sequential staining, participated drafting the manuscript and contributed to statistical analysis. EP participated in the design and the realization of the study. AM participated in the development of the approach, the study design, in its coordination and drafted the manuscript. FZ contributed to development of the approach, to the study design and drafted the manuscript. All authors read and approved the final manuscript.
References
- 1.Sison EA, Brown P. The bone marrow microenvironment and leukemia: biology and therapeutic targeting. Expert review of hematology. 2011;4:271–283. doi: 10.1586/ehm.11.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsson SK, Johnston HM, Coverdale JA. Spatial localization of transplanted hemopoietic stem cells: inferences for the localization of stem cell niches. Blood. 2001;97:2293–2299. doi: 10.1182/blood.V97.8.2293. [DOI] [PubMed] [Google Scholar]
- 3.Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8:290–301. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]
- 5.Doan PL, Chute JP. The vascular niche: home for normal and malignant hematopoietic stem cells. Leukemia. 2012;26:54–62. doi: 10.1038/leu.2011.236. [DOI] [PubMed] [Google Scholar]
- 6.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 7.Bain BJ, Clark DM, Wilkins BS. Bone Marrow Pathology. Chichester, West Sussex, UK: Wiley-Blackwell; 2010.
- 8.Swerdlow SH, Cancer IAfRo, Organization WH: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer. Chichester, West Sussex, UK: Wiley-Blackwell; 2008.
- 9.Dasgupta A, Saxena R. Regulatory T cells: a review. Natl Med J India. 2012;25:341–351. [PubMed] [Google Scholar]
- 10.Pfister E, Belhazarem D, Becker A, Weis C-A, Pfister F, Küffer S, et al.: Lacking Impact of Polymorphisms of Immunoregulatory Candidate Gens and of the Anti-apoptotic cFLIP Gene on the Response to Interferon-alpha in CML. Chichester, West Sussex, UK: Wiley-Blackwell. In review.
- 11.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 12.Houot R, Kohrt H, Goldstein MJ, Levy R. Immunomodulating antibodies and drugs for the treatment of hematological malignancies. Cancer Metastasis Rev. 2011;30:97–109. doi: 10.1007/s10555-011-9274-3. [DOI] [PubMed] [Google Scholar]
- 13.Fava C, Cortes JE, Kantarjian H, Jabbour E. Standard management of patients with chronic myeloid leukemia. Clin Lymphoma Myeloma. 2009;9(Suppl 4):S382–390. doi: 10.3816/CLM.2009.s.038. [DOI] [PubMed] [Google Scholar]
- 14.Jabbour E, Fava C, Kantarjian H. Advances in the biology and therapy of patients with chronic myeloid leukaemia. Best Pract Res Clin Haematol. 2009;22:395–407. doi: 10.1016/j.beha.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Immunohistochemical Staining Methods. 6th edn: Dako Denmark A/S, An Agilent Technologies Company. Chichester, West Sussex, UK: Wiley-Blackwell; 2013.
- 16.Welsch U, Mulisch M. Romeis Mikroskopische Technik. Spektrum Akademischer Verlag; 2010
- 17.van den Brand M, Hoevenaars BM, Sigmans JHM, Meijer JWR, van Cleef PHJ, Groenen PJTA, et al. Sequential immunohistochemistry: a promising new tool for the pathology laboratory. Histopathol. 2014;65:651–657. doi: 10.1111/his.12446. [DOI] [PubMed] [Google Scholar]
- 18.Couzin J. Scientific publishing. Don't pretty up that picture just yet. Science (New York, NY) 2006;314:1866–186. doi: 10.1126/science.314.5807.1866. [DOI] [PubMed] [Google Scholar]
- 19.Cromey DW. Avoiding twisted pixels: ethical guidelines for the appropriate use and manipulation of scientific digital images. Sci Eng Ethics. 2010;16:639–667. doi: 10.1007/s11948-010-9201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson H. Image manipulation: CSI: cell biology. Nature. 2005;434:952–953. doi: 10.1038/434952a. [DOI] [PubMed] [Google Scholar]
- 21.Okumura M. The World Health Organization Histologic Classification System Reflects the Oncologic Behavior of Thymoma. Cancer. 2002;94:581–583. doi: 10.1002/cncr.10226. [DOI] [PubMed] [Google Scholar]
- 22.BUnwarpJ [http://fiji.sc/BUnwarpJ]
- 23.Arganda-carreras I, Sorzano COS, Marabini R: Consistent and Elastic Registration of Histological Sections using Vector-Spline Regularization. Chichester, West Sussex, UK: Wiley-Blackwell.
- 24.Template Matching and Slice Alignment [https://sites.google.com/site/qingzongtseng/template-matching-ij-plugin]
- 25.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Transactions on image processing: a publication of the IEEE Signal Processing Society. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 26.Thevenaz L. Advanced Fiber Optics. Lausanne, Switzerland: EFPL Press; 2011.
- 27.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- 28.Landini G. Colour Deconvolution. In Book Colour Deconvolution (Editor ed.^eds.). City. Chichester, West Sussex, UK: Wiley-Blackwell; 2007.
- 29.Fiji Colour Deconvolution plugin [http://fiji.sc/Colour_Deconvolution]
- 30.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willard KE, Connelly DP. Nonparametric probability density estimation: improvements to the histogram for laboratory data. Comput Biomed Res. 1992;25:17–28. doi: 10.1016/0010-4809(92)90032-6. [DOI] [PubMed] [Google Scholar]
- 32.Rossiter JE. Calculating centile curves using kernel density estimation methods with application to infant kidney lengths. Stat Med. 1991;10:1693–1701. doi: 10.1002/sim.4780101107. [DOI] [PubMed] [Google Scholar]
- 33.Francis K, Palsson BO. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12258–12262. doi: 10.1073/pnas.94.23.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adam G, Läuger P, Stark G: Physikalische Chemie und Biophysik. Lausanne, Switzerland: Springer; 2009.
- 35.Huttner K. Fluid- und Thermodynamik Eine Einführung. 2. edition edn; 2002.
- 36.Trauth M, Gebbers R, Sillmann E, Marwan N. MATLAB® Recipes for Earth Sciences. Heidelberg: Springer Berlin; 2010. [Google Scholar]
- 37.Manders EM. Measurement of co-localization of objects in dualcolor confocal images. J Microsc. 1993;1993(169):375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 38.Zinchuk V, Grossenbacher-Zinchuk O. Recent advances in quantitative colocalization analysis: focus on neuroscience. Prog Histochem Cytochem. 2009;44:125–172. doi: 10.1016/j.proghi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Wang LS, Huang MH, Lin TS, Huang BS, Chien KY. Malignant thymoma. Cancer. 1992;70:443–450. doi: 10.1002/1097-0142(19920715)70:2<443::AID-CNCR2820700212>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 40.Regnard JF, Magdeleinat P, Dromer C, Dulmet E, de Montpreville V, Levi JF, et al. Prognostic factors and long-term results after thymoma resection: a series of 307 patients. J Thorac Cardiovasc Surg. 1996;112:376–384. doi: 10.1016/S0022-5223(96)70265-9. [DOI] [PubMed] [Google Scholar]
- 41.Rubner Y, Tomasi C, Guibas LJ. The Earth Mover's Distance as a Metric for Image Retrieval.
- 42.Papula L. Mathematik für Ingenieure und Naturwissenschaftler 2. Vieweg; 2007
- 43.Liu Y, Sattarzadeh A, Diepstra A, Visser L, van den Berg A. The microenvironment in classical Hodgkin lymphoma: an actively shaped and essential tumor component. SEMIN CANCER BIOL. 2014;24:15–22. doi: 10.1016/j.semcancer.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Rosenquist R, Davi F, Ghia P. The microenvironment in lymphomas--dissecting the complex crosstalk between tumor cells and 'by-stander' cells. Sem Cancer Biol. 2014;24:1–2. doi: 10.1016/j.semcancer.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Frisch J, Houchins JP, Grahek M, Schoephoerster J, Hagen J, Sweet J, Mendoza L, Schwartz D, Kalyuzhny AE. Novel multicolor immunofluorescence technique using primary antibodies raised in the same host species. Methods Mol Biol. 2011;717:233–244. doi: 10.1007/978-1-61779-024-9_13. [DOI] [PubMed] [Google Scholar]
- 46.Osman TA, Øijordsbakken G, Costea DE, Johannessen AC. Successful triple immunoenzymatic method employing primary antibodies from same species and same immunoglobulin subclass. Eur J Histochem. 2013;57 doi: 10.4081/ejh.2013.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 48.Baraldi A, Blonda P. A survey of fuzzy clustering algorithms for pattern recognition. II. IEEE transactions on systems, man, and cybernetics Part B, Cybernetics: a publication of the IEEE Systems, Man, and Cybernetics Society. 1999;29:786–801. doi: 10.1109/3477.809033. [DOI] [PubMed] [Google Scholar]
- 49.Uchida S. Image processing and recognition for biological images. Dev Growth Differ. 2013;55:523–549. doi: 10.1111/dgd.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohler A, Schmithorst V, Filippi MD, Ryan MA, Daria D, Gunzer M, et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood. 2009;114:290–298. doi: 10.1182/blood-2008-12-195644. [DOI] [PMC free article] [PubMed] [Google Scholar]


