Highlights
-
•
Fungi possess a variable often expanded cytochrome b5 reductase (CBR) gene family.
-
•
In the phytopathogen Zymoseptoria tritici ZtCBR1 is essential for full virulence.
-
•
Sphingolipid, sterol and fatty acid biosynthesis were altered in ΔZtCBR1.
-
•
First report of a CBR impacting directly upon fungal sterol biosynthesis in situ.
Abbreviations: CBR, cytochrome b5 reductase; b5, cytochrome b5; CPR, cytochrome P450 reductase; CYP, cytochrome P450; STB, Septoria tritici blotch; DPI, days post inoculation; WT, wild-type; HPLC, high pressure liquid chromatography; GC–MS, gas chromatography–mass spectrometry; FAME, fatty acid methyl ester; FPKM, fragments per kilobase per million mapped fragments; RNA-seq, RNA sequencing; LCB, sphingolipid long chain base
Keywords: Septoria tritici, Mycosphaerella graminicola, Dimorphic fungi, Fatty acids, Cytochrome P450, CYP51
Abstract
Septoria tritici blotch (STB) caused by the Ascomycete fungus Zymoseptoria tritici is one of the most economically damaging diseases of wheat worldwide. Z. tritici is currently a major target for agricultural fungicides, especially in temperate regions where it is most prevalent. Many fungicides target electron transfer enzymes because these are often important for cell function. Therefore characterisation of genes encoding such enzymes may be important for the development of novel disease intervention strategies. Microsomal cytochrome b5 reductases (CBRs) are an important family of electron transfer proteins which in eukaryotes are involved in the biosynthesis of fatty acids and complex lipids including sphingolipids and sterols. Unlike the model yeast Saccharomyces cerevisiae which possesses only one microsomal CBR, the fully sequenced genome of Z. tritici bears three possible microsomal CBRs. RNA sequencing analysis revealed that ZtCBR1 is the most highly expressed of these genes under all in vitro and in planta conditions tested, therefore ΔZtCBR1 mutant strains were generated through targeted gene disruption. These strains exhibited delayed disease symptoms on wheat leaves and severely limited asexual sporulation. ΔZtCBR1 strains also exhibited aberrant spore morphology and hyphal growth in vitro. These defects coincided with alterations in fatty acid, sphingolipid and sterol biosynthesis observed through GC–MS and HPLC analyses. Data is presented which suggests that Z. tritici may use ZtCBR1 as an additional electron donor for key steps in ergosterol biosynthesis, one of which is targeted by azole fungicides. Our study reports the first functional characterisation of CBR gene family members in a plant pathogenic filamentous fungus. This also represents the first direct observation of CBR functional ablation impacting upon fungal sterol biosynthesis.
1. Introduction
Septoria tritici blotch (STB) caused by the wheat leaf-specific Ascomycete fungus Zymoseptoria tritici is one of the most economically damaging diseases of wheat worldwide. The most significant ‘within field’ damage caused by Z. tritici is mediated through asexual spores. These spores are rain splash propagated throughout the wheat canopy where they attach to leaf surfaces and then germinate into infectious hyphae which penetrate the plant through stomata. This takes place within 24 h of spores landing on the leaf’s surface and is followed by a symptomless phase of slow intercellular colonisation within the leaf lasting approximately 10 days (Orton et al., 2011). Following this initial symptomless period the fungus elicits a rapid onset of host cell necrosis, which bears hallmarks of plant programmed cell death. At this point Z. tritici switches to a necrotrophic mode of feeding and begins accumulating biomass rapidly, generating asexual fruiting bodies (termed pycnidia) within the developing necrotic lesions (Keon et al., 2007). Following infection of a susceptible wheat cultivar by a wild-type (WT) strain the complete disease cycle takes approximately 21 days, culminating in the development of mature pycnidia on infected leaves. The masses of asexual spores produced by the pycnidia (pycnidiospores) are multicellular units most frequently composed of four to six cells (Eyal et al., 1987).
There is currently a major deficit in commercially relevant STB-resistant germplasm. As a result Z. tritici is a major target for fungicides, especially in temperate regions where it is most prevalent: approximately 70% of fungicides (equating to a cost of >€400 m) sold in the EU are used to prevent STB (O’Driscoll et al., 2014). Due in part to the intense selective pressure caused by the widespread use of just a handful of antifungal chemistries, fungicide resistance in Z. tritici is a major problem (Cools and Fraaije, 2008; Siah et al., 2014; Taher et al., 2014). Many fungicides target enzymes involved in electron transfer systems as such systems are often essential for cell function. Therefore, analysis of genes encoding hitherto uncharacterised electron transfer enzymes in Z. tritici may be relevant for future development of novel antifungal chemistries.
Aside from those present on the chloroplastic and mitochondrial membranes, the two major eukaryotic electron transfer systems are the cytochrome P450 reductase (CPR)-dependent and microsomal cytochrome b5 reductase (CBR)-dependent pathways. The former provides the electrons necessary for the function of cytochrome P450 (CYP) enzymes, which are involved in various biological processes such as detoxification of xenobiotic compounds and biosynthesis and metabolism of lipids and secondary metabolites (George et al., 1998; Mutch et al., 2007; Lepesheva and Waterman, 2007; Richter et al., 2008). The latter usually involves transfer of electrons through CBR then cytochrome b5 (b5) to terminal electron acceptor desaturase or hydroxylase enzymes. The major functions of the desaturases and hydroxylases in this system are to catalyse double bond formation between carbon atoms and addition of hydroxyl groups during the biosynthesis of unsaturated fatty acids (UFAs) and the more complex lipids the sphingolipids and sterols (Huang et al., 1999; Knutzon et al., 1998; Michaelson et al., 2013; Grinstead and Gaylor, 1982; Moreno-Perez et al., 2011). In addition to its major role in electron transfer to desaturases and hydroxylases, cytochrome b5 is also known to be involved in electron transfer to some CYP enzymes (Henderson et al., 2013; Gan et al., 2009), though the functional relationship between CYPs and the CBR-b5 pathway remains largely elusive. In addition to the microsomal CBR-b5 system there is also a mitochondrial CBR-b5 system (Hahne et al., 1994). Though the function of this system also remains largely elusive, it has been linked with metabolism of xenobiotics and lipid biosynthesis (Nikiforova et al., 2014; Neve et al., 2012; Glory and Thiruvenangdam, 2011).
To date studies on microsomal CBR enzymes have been limited to the model yeast Saccharomyces cerevisiae, the industrial arachidonic acid-producing fungus Mortierella alpina (Class: Zygomycota), the model white-rot fungus Phanerochaete chrysosporium (Order: Basidiomycota), and the filamentous fungus Mucor racemosus (Class: Zygomycota). In S. cerevisiae it was demonstrated that the sole microsomal CBR present in the fully sequenced genome is able to provide the reducing power necessary for the function of a CYP enzyme (albeit in a reconstituted system), CYP51, needed for biosynthesis of the main fungal sterol, ergosterol, rendering CPR dispensable (Lamb et al., 1999; Sutter and Loper, 1989). Similarly, studies in P. chrysosporium showed that the CBR-b5 system is able to efficiently provide electrons to the enzyme CYP63A2, a multifunctional CYP (Syed et al., 2011). These findings offered some insight into the potential overlap in function between the CPR and CBR systems in fungi. In M. alpina two CBRs were identified and cloned in the late 1990s. The first of these was heterologously expressed in Aspergillus oryzae leading to an increase in ferricyanide reduction activity (Sakuradani et al., 1999), though no analyses of the in situ function of this enzyme were carried out. The second of these was used in conjunction with the first in a phylogenetic analysis, which demonstrated evolutionary divergence of mammalian CBR enzymes from those of fungi and plants (Certik et al., 1999). In M. racemosus, CBR was cloned and expressed in E. coli (Mirzaei et al., 2010), though again no analyses of the biological function of the enzyme in the host organism were conducted.
Whilst much research has been carried out on mammalian (Celik et al., 2013; Elahian et al., 2014) and plant CBRs (Wayne et al., 2013; Kumar et al., 2006; Shockey et al., 2005; Bagnaresi et al., 2000), there have been no further investigations into the roles of these enzymes in fungi. In order to address this, we have analysed members of the CBR gene family in Z. tritici which is a plant pathogen in the Dothideomycete class of fungi. In the last decade the molecular interaction between this fungus and its host has come under greater scrutiny (Gohari et al., 2014; Lee et al., 2014; Suffert et al., 2013; do Amaral et al., 2012; Motteram et al., 2009; Marshall et al., 2011). However, there have been little in the way of investigations into the metabolic processes important for growth and plant pathogenesis. Based upon analysis of the fully sequenced genome of the Z. tritici reference isolate IPO323 (Goodwin et al., 2011), and recent RNA sequencing (RNA-seq) data (Rudd et al., 2015) we determined that only one of the three putative CBRs in Z. tritici, ZtCBR1, was highly expressed both in vitro and throughout plant infection. By generating targeted ZtCBR1 disruption strains it was shown that this gene is essential for full virulence in wheat. In vitro observation of ΔZtCBR1 strains revealed various morphological and biochemical defects including reduced spore size, reduced filamentous growth and almost a complete lack of asexual sporulation at the end of the infection cycle in planta. Perturbations in sphingolipid, sterol and fatty acid biosynthesis pathways were identified using GC–MS and HPLC analyses in the ΔZtCBR1 strain. This study represents the first functional analysis of members of the CBR gene family in a plant pathogenic Ascomycete fungus, and highlights several CBR1-regulated functions that underpin virulence in Z. tritici.
2. Materials and methods
2.1. Identification of sequence homologues of yeast cytochrome b5 reductase (ScCBR1) in Z. tritici and other filamentous fungi
In order to identify putative microsomal CBR genes in several unrelated Ascomycete and Basidiomycete fungi including Z. tritici, BLASTp analyses were carried out via the NCBI BLASTp suite (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) using the S. cerevisiae CBR sequence, ScCBR1 (GenBank accession: CAA82214.1), as a query (fungal genomes queried are detailed in Supplementary Table 2).
BLAST hits above 30% amino acid (aa) identity with an e value lower than e−10 were retrieved and subjected to PFam domain prediction (Finn et al., 2014) to determine whether they contained flavin adenine dinucleotide (FAD)-binding (PFam identifier: PF00667) and nicotinamide adenine dinucleotide (NAD)-binding domains (PFam identifier: PF08030), which are both universally present in CBRs. Sequences that contained additional molybdopterin-binding domains (PFam identifier: PF00174) were excluded from further analyses as this is a feature common to nitrate reductases, which are structurally closely related to CBRs but involved in different processes (Truong et al., 1991).
The putative microsomal CBR sequences were then subjected to a TargetP analysis (Emanuelsson et al., 2000) (accessed via: http://www.cbs.dtu.dk/services/TargetP/) to determine whether the predicted proteins were potentially localised to mitochondria or the endoplasmic reticulum. The S. cerevisiae mitochondrial CBR sequence, ScMCR1 (GenBank accession: NP_012221.2), was then used as a query in further BLASTp analyses to determine whether CBRs identified with a TargetP prediction of localisation to mitochondria were indeed more closely related to this sequence than SsCBR1. This step was needed to differentiate between sequence homologues as mitochondrial CBRs are highly similar to microsomal CBR sequences owing to the presence of the highly conserved FAD- and NAD-binding domains necessary for their functions.
2.2. In vitro culture conditions and fungal strains
Fungal spores were routinely cultured on yeast peptone dextrose (YPD) agar for six days at 18 °C. For RNA-seq analysis spores of the WT reference isolate IPO323 were grown in Czapek Dox broth (CDB) minimal medium and potato dextrose broth (PDB) nutrient-rich medium. Fungal cultures were propagated in shaking flasks at 220 rpm and 18 °C for 3 days for PDB or 5 days for CDB and then harvested via vacuum filtration, as detailed in Rudd et al. (2015). These incubation periods were determined to be within the logarithmic growth phase for Z. tritici. For analysis of fatty acid methyl ester (FAME), sphingolipid long chain base (LCB) and sterol content the ΔKu70 strain (treated as WT) Bowler et al., 2010 and the ΔZtCBR1-1 strain generated in this study were propagated under the same conditions in yeast peptone dextrose (YPD) liquid medium. Spores were harvested via vacuum filtration after 4 days of growth and snap-frozen in liquid nitrogen. Prior to analysis of sterol content, spores were freeze-dried. The same culture conditions were used to grow spores for microscopy. To induce filamentous growth in vitro, spores were spot-inoculated onto 1% agar from a spore suspension of 1 × 106 spores ml−1 according to (Motteram et al., 2011).
2.3. Growth and inoculation of plants
Seventeen day old seedlings of the STB-susceptible wheat cultivar Riband were used for all plant infection assays and RNA-seq analysis. Seeds were pre-germinated on wet sand at 10% relative humidity for 3 days prior to potting and subsequently kept with a 16 h daylight cycle. Adaxial surfaces of second leaves were inoculated according to (Keon et al., 2007) with spore suspensions at a density of 2 × 106 spores ml−1 in 0.1% Silwet in sterile water. For infection assays, mock leaves were inoculated with 0.1% Silwet only. Plant inoculation for RNA-seq analysis conducted in the previous study (Rudd et al., 2015) followed the same procedure, without the pre-germination step, using a spore density of 106 spores ml−1 in 0.1% Tween20 in sterile water.
For analysis of asexual fungal sporulation, 8–12 replicate leaves from independently inoculated wheat seedlings randomly distributed in a walk-in temperature, humidity and light-controlled artificial environment were collected at either 21 or 34 days post inoculation (DPI). For the previously published RNA-seq data, each of two biological replicate plant samples were made up of 5 leaves collected from independent plants randomly distributed in a single walk-in temperature and humidity-controlled glasshouse. Samples were collected at one, four, nine, 14 and 21 DPI. Leaves collected for RNA-seq were immediately frozen in liquid nitrogen, freeze-dried, then ground to fine powder in liquid nitrogen before RNA extraction.
2.4. RNA extraction and RNA sequencing
All aspects relating to RNA-seq analysis of fungal gene expression during growth in Czapek-Dox and Potato Dextrose broths and at five time points of plant infection are described in detail in a previous study (Rudd et al., 2015). To summarise, the following principle procedures were followed: Total RNA was isolated from freeze-dried tissues using the Trizol procedure (Chomczynski and Sacchi, 1987) incorporating a final LiCl2 precipitation. All samples (single-end) were mapped with TopHat (v2.0.6) against the Z. tritici genome (-G Mycosphaerella_graminicola.MG2.16.gtf) (Trapnell et al., 2012). Cufflinks (v2.1.1) was used to calculate FPKM values for reference annotations (-G Mycosphaerella_graminicola.MG2.16.gtf) but excluding genes annotated with rRNA (-M rRNA_genes.gtf). Differential expression analysis was done with cuffdiff (cuffdiff -u -M rRNA_genes.gtf -b Mycosphaerella_graminicola.MG2.16.dna.toplevel.fa).
2.5. Agrobacterium tumefaciens – mediated targeted disruption of fungal genes
In order to target ZtCBR1 for gene function ablation the plasmid pNOV2114 was used (Motteram et al., 2009). Two sequences flanking ZtCBR1 were obtained from the JGI genome for Z. tritici (http://genome.jgi.doe.gov/Mycgr3/Mycgr3.home.html) and PCR amplified. Amplified sequences were then purified and inserted into pNOV2114 either side of a hygromycin resistance cassette (hph) (also inserted as purified PCR product) under a trpC promoter. The same procedure was used to generate transformation vectors for ZtCBR2 and ZtCYP-24, though the plasmid used was pCHYG (Motteram et al., 2009), which already contained hph under the trpC promoter. All primers and added restriction sites used for these procedures are detailed in Supplementary Table S1 and Supplementary Fig. S1, A shows diagrams indicating positions and sizes of flanking sequences relative to genes of interest.
Agrobacterium tumefaciens – mediated transformation of Z. tritici spores was carried out according to (Zwiers and De Waard, 2001) with slight modifications. Instead of using the antibiotic Cefotaxim, Timentin was used in transformant selection plates as it was found to be more efficient for removal of residual Agrobacterium tumefaciens after transformation. All transformations were carried out in a ΔKu70 background as disruption of this gene has been shown to prevent ectopic insertion whilst maintaining WT growth and virulence (Bowler et al., 2010); the ΔKu70 strain used in transformation was treated as WT in all subsequent experiments.
Transformant colonies were sub-cultured twice on hygromycin-selective agar (50 ug ml−1). To confirm integration of hph+trpC at the desired locus and in the correct orientation, a primer from within hph and within 200 bp of a flanking region used to guide insertion were used to amplify a diagnostic region of approximately 1.5 kb. A single band in gDNA of mutant strains and lack thereof in gDNA of the WT was considered representative of successful targeted disruption. All primers used for these procedures are detailed in Supplementary Table 1. At least three disruption strains were generated for each gene of interest (Supplementary Fig. S1, B). Two ΔZtCBR1 strains were carried forward for further analysis and arbitrarily named ΔZtCBR1-1 and ΔZtCBR1-2. Only single ΔZtCBR2 and ΔZtCYP-24 strains were fully analysed in planta as preliminary testing of several strains showed no apparent reductions in virulence.
2.6. Quantification of asexual sporulation in infected leaf tissue
To assess the degree of asexual sporulation exhibited by Z. tritici strains at the stated time points post inoculation, leaf samples were kept for a further 48 h at 100% relative humidity in darkness at 18 °C. Leaves were then submerged in 1 ml of distilled water and left overnight. Submerged leaves were vortexed for ∼10 s and spores released into the water were counted using a haemocytometer. A total of 8–12 leaves each taken from individual wheat seedlings were analysed this way for each fungal strain tested. Analysis of variance (ANOVA) was used on raw data for ΔZtCYP-24 and ΔZtCBR2 and loge2-transformed data for the ΔZtCBR1 strains alongside the WT to assess differences in the amount of asexual sporulation. When a significant difference (p < 0.05, F-test) was found, Fisher’s least significant difference (LSD) test was used to determine significant (p < 0.05) pairwise differences between strains.
Micrographs of mutant and WT spores were generated via bright field imaging using a Zeiss LSM 780 microscope (Carl Zeiss AG, Oberkochen, Germany). In order to visualise cell boundaries within spores, spore-suspensions (106 spores ml−1 in sterile distilled water) were stained for five minutes at room temperature with the fluorescent cell wall stain calcofluor white at a 1% concentration and imaged using the same microscope with a 405 nm laser. Distance between septa and number of individual cells in each spore were then assessed using ImageJ software (Schneider et al., 2012). Spores from three replicate cultures were included in these analyses and for each replicate culture a minimum of 11 spores were assessed. Differences in mean cell length and number of spores between strains were assessed using ANOVA on loge2-transformed data treating each individual spore as a technical replicate and each culture as a biological replicate. This was followed by Fisher’s LSD test to determine significant pairwise differences between strains. To determine differences in mean proportion of spores with only one cell present between strains, two sample binomial tests were used to compare the WT with each individual strain, ΔZtCBR1-1 and ΔZtCBR1-2. Statistical analyses were carried out using the GenStat (2014, 17th edition, © VSN international Ltd, Hemel Hempstead, UK) statistics package.
2.7. GC–MS analysis of sterol content of ΔZtCBR1-1
Samples from four independent liquid cultures were analysed. Each sample of 20 mg fresh weight (freeze-dried) was used for replicate analyses and results from WT and the ΔZtCBR1-1 mutant strain were compared using Student’s t-tests. Non-saponifiable lipids were extracted as reported previously (Kelly et al., 1995). Samples were dried in a vacuum centrifuge (Heto) and derivatized by addition of 100 μl of 90% bis(trimethylsilyl)-trifluoroacetamide (BSTFA) – 10% trimethylsilyl (TMS) (Sigma–Aldrich) and 200 μl anhydrous pyridine (Sigma–Aldrich) and heating for 2 h at 80 °C. Gas chromatography–mass spectrometry was performed using a VG12-250 mass spectrometer (VG Biotech) with splitless injection. Individual sterols were identified by reference to relative retention times, mass ions, and fragmentation patterns. Data were analysed using MSD Enhanced ChemStation (Agilent Technologies). Statistical analyses were carried out using the GenStat (2014, 17th edition, 297 © VSN international Ltd, Hemel Hempstead, UK) statistics package.
2.8. GC–MS analysis of fatty acid methyl ester content of ΔZtCBR1-1
Samples from five independent liquid cultures were analysed. Each sample of 20 mg fresh weight was used for replicate analyses, and results from WT and the ΔZtCBR1-1 mutant strain were compared using Student’s t-tests. Lipids were extracted and methylated as described (Garces and Mancha, 1993) with minor modifications. Methyl heptadecanoate (C17:0) was added to samples as an internal standard. Following methylation the heptane fraction was concentrated and re-suspended in 300 μl solvent prior to injection of 1 μl onto the GC column. Methyl ester derivatives of total fatty acids extracted were analysed by GC (Agilent 7890A) using an Agilent DB-225 column (30 m × 0.32 mm × 0.3 μm). Inlet and detector temperature was set to 250 °C and 1 μl of each sample was analysed using splitless injection and a constant flow rate of 2 ml min−1. The oven temperature cycle was set as follows: a start temperature of 50 °C was held for 1 min to allow vaporised samples and the solvent (hexane) to condensate at the front of the column. Oven temperature was then increased rapidly to 190 °C at a rate of 40 °C min−1 followed by a slower increase to 220 °C at a rate of 1.5 °C min−1. The final temperature of 220 °C was held for 1 min giving a total run time of 25 min 50 s per sample. Fatty acid methyl esters (FAMEs) were detected using a Flame Ionisation Detector (FID). Chromatograms were analysed using the offline session of the Agilent ChemStation software (Agilent Technologies). The retention time and identity of each fatty acid methyl ester (FAME) peak was calibrated using the FAME Mix Rapeseed oil standard (Supelco). Statistical analyses were carried out using the GenStat (2014, 17th edition, © VSN international Ltd, Hemel Hempstead, UK) statistics package.
2.9. HPLC analysis of sphingolipid long chain 320 base content of ΔZtCBR1-1
Samples from four independent liquid cultures were analysed. Each sample of 20 mg fresh weight was used for replicate analyses and results from WT and the ΔZtCBR1-1 mutant strain were compared using a Student’s t-test. A 2 μg aliquot of d20:0 LCB was used as the internal standard. Long chain bases (LCBs) were liberated from material by an alkaline hydrolysis extraction method based on (Sperling et al., 1998). Briefly, this was performed using 10% BaOH and dioxane 1:1 v/v in capped tubes overnight at 110 °C. These were then cooled and extracted with chloroform/dioxane/water (8/3/8, v/v/v). The LCB fraction was converted to dinitrophenyl derivatives with 0.2 ml 0.5% (v/v) methanolic 1-fluoro-2,4-dinitrobenzene and 0.8 ml 2 M boric acid/KOH at 60 °C for 30 min. LCBs were then extracted by phase partitioning with CHCl3/methanol/H2O, 2:1:1 (v/v/v). The organic phase was removed and washed with an equal volume of 0.1 M KOH and 0.5 M KCl. The organic phase was then blown down and resuspended in 200 μl MeOH for analysis. Analysis by reverse-phase HPLC was performed using a C18 RP 250 × 4 mm column with a flow rate of 1 ml min−1 and a concave gradient from 80% to 100% methanol/acetonitrile/2-propanol, 10:3:1 (v/v/v), against water in 45 min. The elution was monitored with ESI-MS/MS MRM on a 4000 QTRAP and at a wavelength of 350 nm on an Agilent 1200 HPLC. Statistical analyses were carried out using the GenStat (2014, 17th edition, © VSN international Ltd, Hemel Hempstead, UK) statistics package.
3. Results
3.1. The Z. tritici genome encodes three putative microsomal CBRs, alike many other genomes of filamentous fungi
In order to investigate the presence of putative microsomal CBR sequences in 21 fungal genomes derived from pathogens and non-pathogens, in both the Basidiomycete and Ascomycete phyla, BLASTp analyses were conducted using the S. cerevisiae CBR sequence ScCBR1 as a query. Sequences retrieved were subjected to a TargetP analysis to determine whether the predicted proteins were likely to be localised to mitochondria or microsomes. This analysis identified three putative microsomal CBR proteins in Z. tritici. The putative microsomal CBR sequences were named ZtCBR1-3 in order of similarity to SsCBR1 (GenBank accessions: XP_003854385.1, XP_003852872.1 and XP_003847738.1; and Ensembl identifiers: Mycgr3T69942, Mycgr3T57682 and Mycgr3T51378, respectively for ZtCBR1, ZtCBR2 and ZtCBR3). These three sequences contained both NAD- and FAD-binding domains canonical for CBRs (Fig. 1); the ZtCBR2 sequence also contained a cytochrome b5 fusion domain (PFam identifier: PF00173) at the N-terminus (Table 1 and Fig. 1).
Fig. 1.
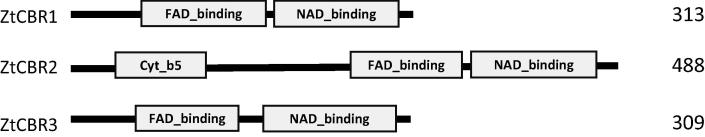
Structural characteristics of the three putative Z. tritici microsomal CBR proteins. PFam domains of the three Z. tritici putative microsomal CBR sequences retrieved. Each predicted protein sequence contains FAD-binding (PFam identifier: PF00667) and NAD-binding (PFam identifier: PF08030) domains canonical for CBR sequences. ZtCBR2 also contains a b5-fusion domain (PFam identifier: PF00173) at the N-terminus. Amino acid sequence length is given to the right.
Table 1.
Number and distribution of Saccharomyces cerevisiae CBR homologues in filamentous fungal genome sequences Below left: four plant endophytic fungi that had no sequences homologous to S. cerevisiae CBRs in their genomes. Fungal lifestyles were derived from Urban et al., 2015 (footnote a).
| Sequence | Species | ScCBR1 aa identity (%) | ScCBR1 e value | TargetP mitochondrial prediction? | ScMCR1 aa identity (%) | ScMCR1 e value | b5 fusion domain? | Fungal lifestyle |
|---|---|---|---|---|---|---|---|---|
| XP_003854385.1 (ZtCBR1) | Zymoseptoria tritici | 49 | 1.00E−82 | N | – | – | N | Hemibiotrophic plant pathogen |
| XP_003852872.1 (ZtCBR2) | Zymoseptoria tritici | 39 | 1.00E−63 | N | – | – | Y | Hemibiotrophic plant pathogen |
| XP_003852910.1 | Zymoseptoria tritici | 39 | 3.00E−57 | Y | 47 | 2.00E−78 | N | Hemibiotrophic plant pathogen |
| XP_003847738.1 (ZtCBR3) | Zymoseptoria tritici | 33 | 1.00E−30 | N | – | – | N | Hemibiotrophic plant pathogen |
| XP_756793.1 | Ustilago maydis | 46 | 6.00E−82 | N | – | – | N | Biotrophic plant pathogen |
| XP_759922.1 | Ustilago maydis | 40 | 8.00E−41 | Y | 36 | 3.00E−62 | N | Biotrophic plant pathogen |
| XP_001799967.1 | Stagonospora nodorum | 51 | 5.00E−95 | N | – | – | N | Necrotrophic plant pathogen |
| XP_001806619.1 | Stagonospora nodorum | 46 | 2.00E−70 | N | – | – | Y | Necrotrophic plant pathogen |
| XP_001801691.1 | Stagonospora nodorum | 39 | 3.00E−51 | Y | 46 | 5.00E−76 | N | Necrotrophic plant pathogen |
| XP_003322373.1 | Puccinia graminis | 48 | 5.00E−84 | N | – | – | N | Biotrophic plant pathogen |
| XP_003319934.2 | Puccinia graminis | 38 | 1.00E−47 | Y | 40 | 6.00E−62 | N | Biotrophic plant pathogen |
| CCA68189.1 | Piriformospora indica | 49 | 6.00E−93 | N | – | – | N | Endophyte |
| CCA67532.1 | Piriformospora indica | 32 | 1.00E−46 | Y | 46 | 2.00E−69 | N | Endophyte |
| XP_009850995.1 | Neurospora tetrasperma | 50 | 1.00E−80 | N | – | – | N | Saprophyte |
| XP_009854993.1 | Neurospora tetrasperma | 46 | 2.00E−72 | N | – | – | Y | Saprophyte |
| EGZ77533.1 | Neurospora tetrasperma | 42 | 1.00E−53 | Y | 49 | 3.00E−84 | N | Saprophyte |
| XP_009856344.1 | Neurospora tetrasperma | 41 | 2.00E−53 | N | – | – | N | Saprophyte |
| XP_009849163.1 | Neurospora tetrasperma | 32 | 2.00E−32 | N | – | – | N | Saprophyte |
| XP_956601.1 | Neurospora crassa | 50 | 2.00E−80 | N | – | – | N | Saprophyte |
| XP_965191.1 | Neurospora crassa | 46 | 4.00E−72 | N | – | – | Y | Saprophyte |
| XP_964971.1 | Neurospora crassa | 42 | 1.00E−53 | Y | 43 | 1.00E−83 | N | Saprophyte |
| XP_961775.1 | Neurospora crassa | 33 | 6.00E−31 | N | – | – | N | Saprophyte |
| XP_007925194.1 | Mycosphaerella fijiensis | 44 | 9.00E−80 | N | – | – | N | Hemibiotrophic plant pathogen |
| XP_007926128.1 | Mycosphaerella fijiensis | 40 | 3.00E−55 | Y | 45 | 2.00E−73 | N | Hemibiotrophic plant pathogen |
| XP_007922376.1 | Mycosphaerella fijiensis | 30 | 2.00E−29 | Y | 32 | 2.00E−50 | N | Hemibiotrophic plant pathogen |
| XP_385028.1 | Fusarium graminearum | 46 | 1.00E−71 | N | – | – | Y | Nectrotrophic plant pathogen |
| ESU10740.1 | Fusarium graminearum | 46 | 4.00E−59 | N | – | – | Y | Nectrotrophic plant pathogen |
| XP_383723.1 | Fusarium graminearum | 43 | 9.00E−69 | N | – | – | Y | Nectrotrophic plant pathogen |
| XP_382313.1 | Fusarium graminearum | 42 | 4.00E−65 | N | – | – | Y | Nectrotrophic plant pathogen |
| XP_381102.1 | Fusarium graminearum | 39 | 2.00E−56 | Y | 46 | 2.00E−75 | N | Nectrotrophic plant pathogen |
| XP_387123.1 | Fusarium graminearum | 32 | 1.00E−34 | N | – | – | N | Nectrotrophic plant pathogen |
| XP_385079.1 | Fusarium graminearum | 30 | 4.00E−27 | N | – | – | N | Nectrotrophic plant pathogen |
| EME45766.1 | Dothistroma septosporum | 44 | 5.00E−84 | N | – | – | N | Hemibiotrophic plant pathogen |
| EME46111.1 | Dothistroma septosporum | 38 | 1.00E−48 | Y | 42 | 3.00E−69 | N | Hemibiotrophic plant pathogen |
| EME38673.1 | Dothistroma septosporum | 33 | 5.00E−37 | Y | 31 | 4.00E−46 | N | Hemibiotrophic plant pathogen |
| EFQ24978.1 | Colletotrichum graminicola | 50 | 1.00E−81 | N | – | – | N | Hemibiotrophic plant pathogen |
| EFQ25098.1 | Colletotrichum graminicola | 46 | 8.00E−72 | N | – | – | Y | Hemibiotrophic plant pathogen |
| EFQ27839.1 | Colletotrichum graminicola | 41 | 2.00E−57 | Y | 46 | 2.00E−76 | N | Hemibiotrophic plant pathogen |
| EFQ36452.1 | Colletotrichum graminicola | 33 | 1.00E−33 | Y | 34 | 3.00E−53 | N | Hemibiotrophic plant pathogen |
| XP_007700426.1 | Cochliobolus sativus | 54 | 5.00E−89 | N | – | – | N | Nectrotrophic plant pathogen |
| XP_007700318.1 | Cochliobolus sativus | 44 | 2.00E−69 | N | – | – | Y | Nectrotrophic plant pathogen |
| XP_007704621.1 | Cochliobolus sativus | 40 | 8.00E−53 | Y | 46 | 2.00E−76 | N | Nectrotrophic plant pathogen |
| EMD92868.1 | Cochliobolus heterostrophus | 53 | 8.00E−89 | N | – | – | N | Nectrotrophic plant pathogen |
| EMD88525.1 | Cochliobolus heterostrophus | 44 | 2.00E−69 | N | – | – | Y | Nectrotrophic plant pathogen |
| EMD86858.1 | Cochliobolus heterostrophus | 40 | 7.00E−53 | Y | 46 | 2.00E−76 | N | Nectrotrophic plant pathogen |
| EPQ65715.1 | Blumeria graminis | 45 | 3.00E−66 | N | – | – | N | Biotrophic plant pathogen |
| CCU81251.1 | Blumeria graminis | 44 | 2.00E−64 | N | – | – | N | Biotrophic plant pathogen |
| EPQ62420.1 | Blumeria graminis | 43 | 2.00E−58 | N | – | – | N | Biotrophic plant pathogen |
| CCU78450.1 | Blumeria graminis | 42 | 3.00E−58 | N | – | – | N | Biotrophic plant pathogen |
| XP_001208762.1 | Aspergillus terreus | 51 | 7.00E−88 | N | – | – | N | Facultative parasite/saprophyte |
| XP_001218611.1 | Aspergillus terreus | 45 | 1.00E−74 | N | – | – | Y | Facultative parasite/saprophyte |
| XP_001215899.1 | Aspergillus terreus | 44 | 1.00E−69 | N | – | – | Y | Facultative parasite/saprophyte |
| Q0CRD8.2 | Aspergillus terreus | 41 | 3.00E−49 | Y | 49 | 2.00E−86 | N | Facultative parasite/saprophyte |
| XP_001212924.1 | Aspergillus terreus | 39 | 5.00E−47 | Y | 48 | 7.00E−83 | N | Facultative parasite/saprophyte |
| XP_001214268.1 | Aspergillus terreus | 31 | 8.00E−31 | Y | 39 | 1.00E−67 | N | Facultative parasite/saprophyte |
| XP_663970.1 | Aspergillus nidulans | 51 | 6.00E−88 | N | – | – | N | Facultative parasite/saprophyte |
| Q5AZB4.2 | Aspergillus nidulans | 51 | 7.00E−88 | N | – | – | N | Facultative parasite/saprophyte |
| XP_661466.1 | Aspergillus nidulans | 47 | 3.00E−77 | N | – | – | Y | Facultative parasite/saprophyte |
| CBF75218.1 | Aspergillus nidulans | 47 | 5.00E−77 | N | – | – | Y | Facultative parasite/saprophyte |
| XP_658036.1 | Aspergillus nidulans | 43 | 4.00E−57 | Y | 48 | 1.00E−86 | N | Facultative parasite/saprophyte |
| XP_682189.1 | Aspergillus nidulans | 38 | 5.00E−59 | N | – | – | Y | Facultative parasite/saprophyte |
| CBF82301.1 | Aspergillus nidulans | 37 | 6.00E−38 | N | – | – | Y | Facultative parasite/saprophyte |
| XP_663990.1 | Aspergillus nidulans | 32 | 9.00E−31 | N | – | – | N | Facultative parasite/saprophyte |
| XP_755738.2 | Aspergillus fumigatus | 49 | 2.00E−86 | N | – | – | N | Facultative parasite/saprophyte |
| XP_753636.1 | Aspergillus fumigatus | 45 | 4.00E−75 | N | – | – | Y | Facultative parasite/saprophyte |
| XP_748717.1 | Aspergillus fumigatus | 44 | 1.00E−70 | N | – | – | Y | Facultative parasite/saprophyte |
| XP_750202.1 | Aspergillus fumigatus | 43 | 4.00E−50 | Y | 47 | 5.00E−79 | N | Facultative parasite/saprophyte |
| Endophytes with no sequences retrieved by SsCBR1 | ||||||||
| Epichloe festucae | Microsomal | |||||||
| Ascocoryne sarcoides | Mitochondrial | |||||||
| Penicillium aurantiogriseum | ||||||||
| Harpophora oryzae | ||||||||
Fungal lifestyle source, Urban et al., 2015 Nucleic Acids Research. Names given to the Z. tritici CBR genes referred to throughout the rest of this study are given underneath GenBank accessions.
This higher number of microsomal CBR sequences relative to S. cerevisiae was also seen amongst distantly related fungi. For example the non-pathogenic fungus Aspergillus nidulans contained seven putative microsomal CBRs, the highest number observed for the fungal species analysed in this study. The lowest number of putative microsomal CBRs found was in Mycosphaerella musiva, a plant pathogenic Dothideomycete fungus related to Z. tritici which contained only a single CBR sequence lacking a TargetP-predicted mitochondrial localisation. All sequences identified that were predicted to be localised to mitochondria through TargetP analyses showed higher similarity to SsMCR1 than SsCBR1, which was initially used to retrieve them. In the species A. terreus, Colletotrichum graminicola, Dothistroma septosporum and Mycosphaerella fijiensis, two to three CBR sequences with TargetP-predicted mitochondrial localisation and higher similarity to SsMCR1 were retrieved using SsCBR1 as a query sequence. In addition to NAD and FAD-binding domains, PFam analyses also identified b5 fusion domains in a number of putative microsomal CBR sequences other than ZtCBR2; these were all present at the N-terminus.
No obvious link between number of CBR sequences and fungal lifestyle was apparent except for the lack of CBR sequences observed in four of the five endophytic fungal species analysed (Table 1). The data suggest that many filamentous fungi (perhaps excluding certain endophytic species) have a greater diversity of CBRs relative to the Ascomycete yeast S. cerevisiae.
3.2. RNA sequencing analysis demonstrates high constitutive expression of ZtCBR1 and lower expression with transient up-regulation of ZtCBR2 in planta
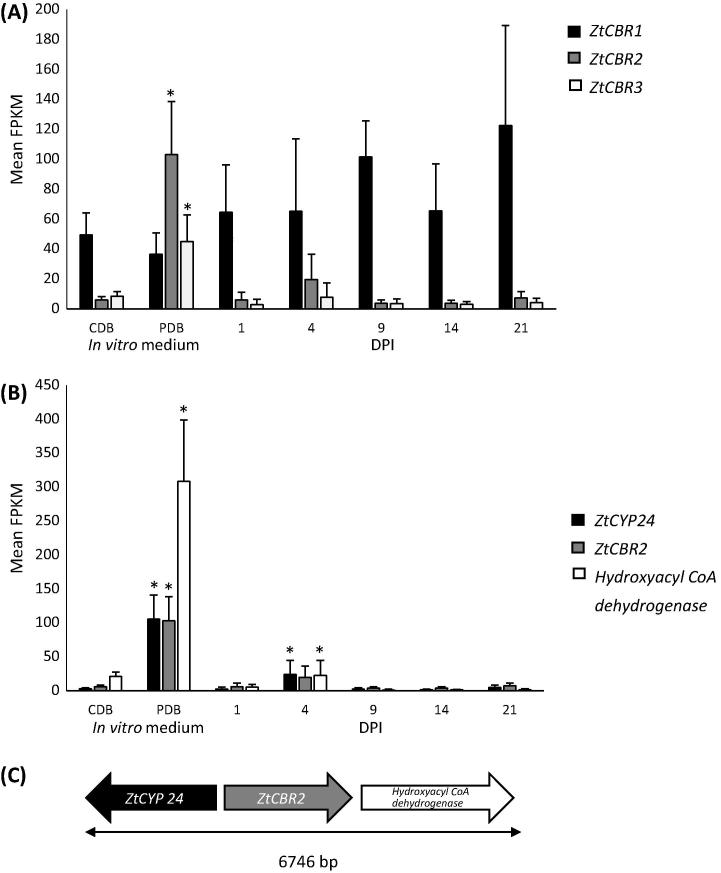
In order to identify genes that might be important for infection, an RNA-seq analysis was conducted in a previous study (for details see Rudd et al., 2015) on samples taken from two in vitro growth conditions and five in planta infection time points including one, four, nine, 14 and 21 days post inoculation (DPI). These time points are representative of major transitions in fungal growth; day one and day four representing the symptomless phase, day nine representing the transition to necrosis, day 14 necrotrophic growth and day 21 asexual sporulation. Analysis of mean FPKM values showed that ZtCBR1 was highly expressed in both in vitro conditions and at all tested infection time points (Fig. 2A). ZtCBR2, though generally less expressed overall, was up-regulated in the nutrient-rich medium PDB (p < 0.05) relative to during growth in CDB, and again (although data were not significant) on day four of infection relative to during growth in CDB. In addition, two genes neighbouring ZtCBR2 in the Z. tritici genome, annotated as a predicted CYP, ZtCYP-24 (GenBank accession: XP_003853538.1), and a predicted hydroxyacyl coA dehydrogenase (GenBank accession: XP_003852872.1), showed similar expression profiles to ZtCBR2. Both genes exhibited a significant up-regulation in PDB (p < 0.05) and up-regulation on day four of infection (p < 0.05) either relative to during growth in CDB for ZtCYP-24 or to all other infection time points for the hydroxyacyl CoA dehydrogenase (Fig. 2B). ZtCBR3 displayed a similar (albeit lower level) expression pattern to ZtCBR2 and was significantly up-regulated during growth in PDB relative to during growth in CDB (p < 0.05) (Fig. 2A).
Fig. 2.
Expression profiles of CBR and related genes in Z. tritici. (A) Mean FPKM values showing expression of Z. tritici CBRs in Czapek Dox broth (CDB), potato dextrose broth (PDB) and at one, four, nine, 14 and 21 days post inoculation (DPI) of wheat leaves; significant differences in expression of ZtCBR2 and ZtCBR3 (* p < 0.05) were found during growth in PDB relative to during growth in CDB. (B) Mean FPKM values showing expression profile of ZtCBR2 and the two neighbouring genes, ZtCYP-24 and a putative hydroxyacyl CoA dehydrogenase across the same set of conditions. (C) A diagram showing gene organisation across the region with the total size of the putative three gene cluster in base pairs indicated below. The two genes neighbouring ZtCBR2 were significantly up-regulated both in PDB and on day four of infection (* p < 0.05) relative to during growth in CDB; ZtCBR2 exhibited a similar expression profile though apparent up-regulation was only significant in PDB (p < 0.05).
3.3. Fungal gene deletion and wheat leaf infection assays demonstrate an important role for ΔZtCBR1 in disease progression and asexual sporulation in Z. tritici
Based on the previous gene expression analysis, and in order to assess the roles for different CBRs and CYPs in fungal growth and virulence, various gene disruption strains were generated in the ΔKu70 strain of Z. tritici IPO323 and PCR-verified (Supplementary Fig. 1). These were then tested for the ability to cause disease on the STB-susceptible wheat cultivar Riband (Keon et al., 2007).
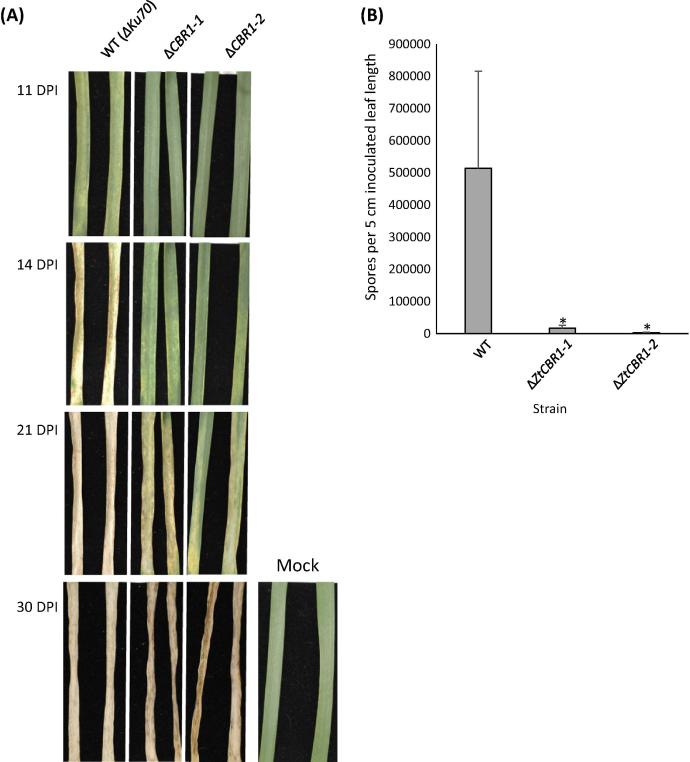
Two independent CBR1 mutants, ΔZtCBR1-1 and ΔZtCBR1-2, both caused delayed symptom manifestation in planta. At 11 DPI when symptoms first appeared in the WT, neither ΔZtCBR1-1 nor ΔZtCBR1-2 had caused any symptoms. At 14 DPI when the WT had caused substantial host necrosis, only limited chlorosis was apparent in leaves infected with ΔZtCBR1-1 and ΔZtCBR1-2. After 21 days, leaf necrosis and pycnidiation were apparent in the WT but only patchy chlorosis was apparent in ΔZtCBR1-1 and ΔZtCBR1-2. After 30 days, necrosis was observed in leaves inoculated with ΔZtCBR1-1 and ΔZtCBR1-2, though pycnidia were not visible (Fig. 3A). In contrast to wild-type infections, no pycnidia were visualised on ΔZtCBR1-infected leaves even when assays were allowed to proceed for a further 14 days (at 44 DPI – data not shown). Quantitative analysis of asexual sporulation performed at 34 DPI, demonstrated that both ΔZtCBR1-1 and ΔZtCBR1-2 produced significantly reduced asexual spore-numbers (p < 0.05) relative to the WT (Fig. 3B).
Fig. 3.
ΔZtCBR1 mutants show delayed disease symptom induction and strongly reduced asexual sporulation on wheat leaves. (A) Leaves infected with WT, ΔCBR1-1 and ΔZtCBR1-2 after 11, 14, 21 and 30 DPI; mock-inoculated control leaves at 30 DPI are shown to the right. A total of eight leaves per strain/mock were inoculated and two representative leaves are shown. (B) Mean number of spores (recovered by washing) per 5 cm length of inoculated leaf for WT, ΔZtCBR1-1 and ΔZtCBR1-2 after 34 DPI showing a significant reduction in asexual sporulation of the mutant strains relative to the WT (* p < 0.05).
In contrast all other mutant strains except for ΔZtCBR1-1 and ΔZtCBR1-2 caused WT symptoms in planta with no changes in asexual spore counts (Supplementary Fig. S2). After 14 days cell death was apparent in leaves inoculated with WT, ΔZtCYP-24 and ΔZtCBR2 strains. After 21 days symptoms had progressed to widespread necrosis at the site of inoculation, and necrotic lesions contained numerous pycnidia. For all strains a total of 12 leaves were evaluated, each showing symptoms consistent with the next. Two representative leaves are shown for each assay in Supplementary Fig. S2, A. Asexual sporulation did not appear to be affected by functional ablation of ZtCYP-24 and ZtCBR2, as evidenced by retrieval of a WT amount of asexual spores from infected leaves after 21 days (Supplementary Fig. 2B).
3.4. ΔZtCBR1 spores have altered morphology and transition more slowly to filamentous growth
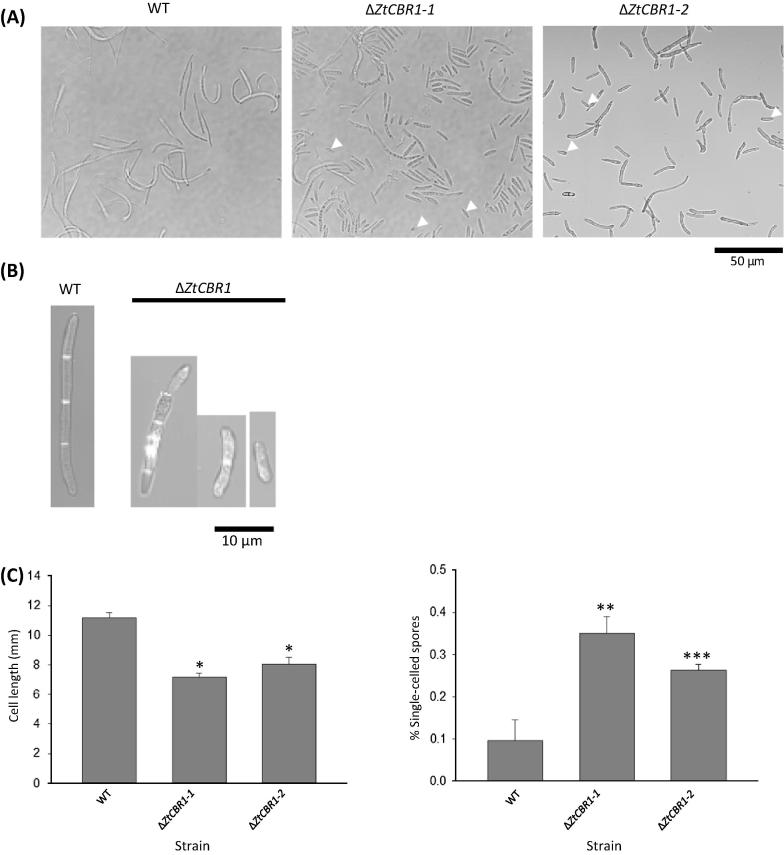
In order to capture morphological defects observed from bright field and laser scanning microscopic imaging of ΔZtCBR1 strains (Fig. 4A and B), both individual cell length (defined as distance between septa in the multicellular spores) and the number of cells per spore were assessed. These microscopic analyses revealed that both ΔZtCBR1 strains exhibited a significant overall decrease in individual cell length (mean length, WT = 11.14 μm, ΔZtCBR1-1 = 7.17 μm, ΔZtCBR1-2 = 7.97 μm, p < 0.05) and increase in the proportion of single-celled spores (binomial test ΔZtCBR1-1 vs WT, p = 0.003, ΔZtCBR1-2 vs WT p < 0.001) (Fig. 4C and D).
Fig. 4.
ΔZtCBR1 mutants show abnormal spore morphologies. (A) Bright field imaging of WT and ΔZtCBR1 spores. Arrowheads highlight examples of single-celled spores more frequently observed in ΔZtCBR1. (B) Representative spores of WT and ΔZtCBR1 strains stained with calcofluor white. (C) Mean cell length for WT and two ΔZtCBR1 strains. Bars represent standard error (* p < 0.05). (D) Mean percentage of single-celled spores for WT and two ΔZtCBR1 strains. Bars represent standard error (** p < 0.01, *** p < 0.001).
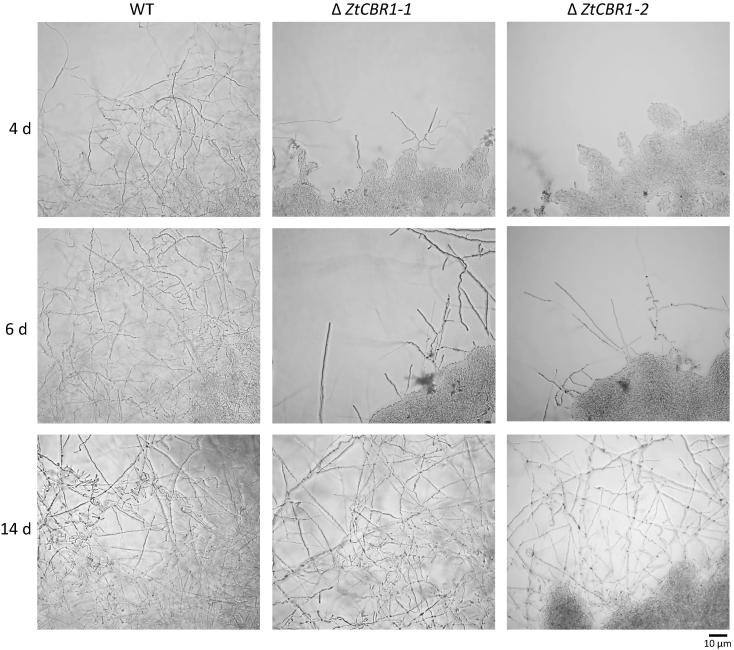
In order to infect wheat leaves, Z. tritici spores require differentiation into hyphae. It is thought that hyphal growth is induced when Z. tritici is exposed to low nutrient environments such as on the leaf surface, or in sterilised water culture and water agar in vitro. In order to assess defects in hyphal growth in both ΔZtCBR1 strains, spores were spot inoculated onto 1% water agar plates and grown for two weeks. After four days WT spores had formed an extensive hyphal network, whereas ΔZtCBR1 spores had produced no hyphae. However, after six days limited hyphal growth was observed for both ΔZtCBR1 strains, and after two weeks the mycelium exhibited almost WT filamentous growth (Fig. 5).
Fig. 5.
ΔZtCBR1 mutants show reduced frequency and rate of hyphal growth. Micrograph showing the appearance of radial hyphal growth produced from the edge of a 5 μl spore droplet for WT and the two ΔZtCBR1 strains after four, six and fourteen days (d) of growth on 1% water agar.
3.5. ΔZtCBR1-1 exhibits an altered fatty acid methyl ester profile
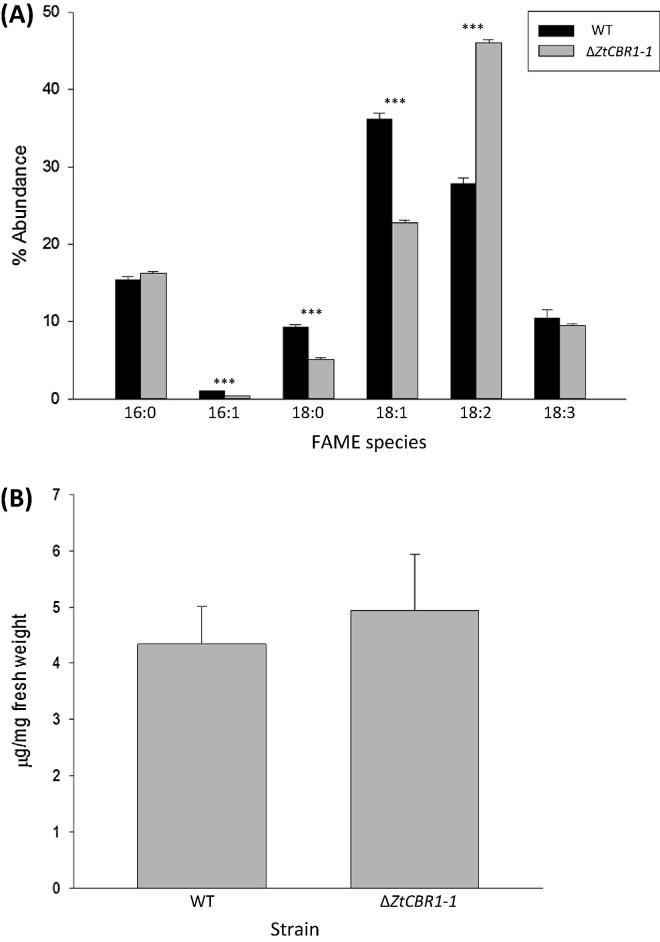
In order to assess whether ZtCBR1 ablation had impacted on fatty acid biosynthesis by fungal cells, the fatty acid methyl ester (FAME) profile of the WT strain and ΔZtCBR1-1 were analysed using GC–MS. ΔZtCBR1-1 displayed a significant decrease in relative abundance of the fatty acid species 16:1, 18:0 and 18:1 (p < 0.001). This strain also displayed a significant increase in relative abundance of the polyunsaturated species 18:2 (p < 0.001). The relative abundances of the species 16:0 and 18:3 were not significantly changed relative to WT relative abundances (p > 0.05) (Fig. 6A). The total amount of FAMEs in ΔZtCBR1-1 was not significantly altered relative to WT levels (p > 0.05) (Fig. 6B).
Fig. 6.
ΔZtCBR1 mutants have altered fatty acid methyl ester (FAME) profiles. (A) Mean relative abundance of the fatty acid methyl ester (FAME) species 16:0, 16:1, 18:0, 18:1, 18:2 and 18:3 in the WT and a ΔZtCBR1 strain. Bars represent standard error (** p < 0.001). (B) Total FAME content of WT and the same ΔZtCBR1 strain expressed in *** μg mg−1 of fresh weight. Bars represent standard error.
3.6. ΔZtCBR1-1 displays an altered sphingolipid long chain base (LCB) profile
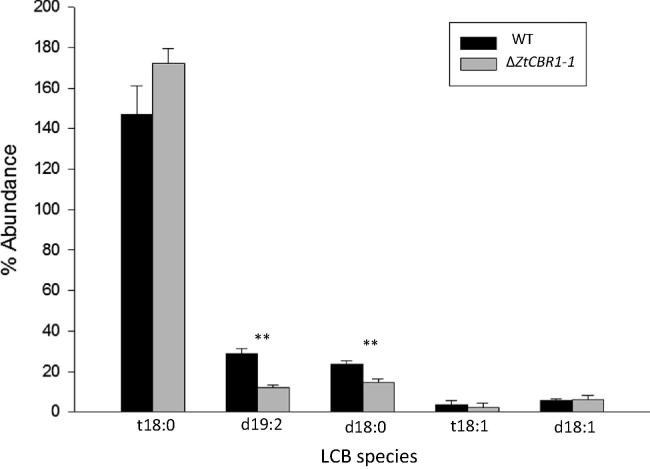
In order to assess the effect of ZtCBR1 ablation on sphingolipid biosynthesis, sphingolipid LCBs were analysed in the WT strain and ΔZtCBR1-1 using HPLC. The ΔZtCBR1-1 mutant exhibited a significant decrease in relative abundance of the LCB species dihydroxy 19:2 (d19:2) and dihydroxy 18:0 (d18:0) compared to the WT (p < 0.01). This strain also displayed an increase in the relative abundance of trihydroxy 18:0 (t18:0) that was approaching significance at the 5% level (p = 0.054) (Fig. 7).
Fig. 7.
ΔZtCBR1 mutants have altered sphingolipid profiles. (A) Mean relative abundance of the sphingolipid long chain base (LCB) species trihydroxy 18:0 (t18:0), dihydroxy 19:2 (d19:2), dihydroxy 18:0 (d18:0), trihydroxy 18:1 (t18:1) and dihydroxy 18:1 (d18:1) in a WT and ΔZtCBR1 mutant strain. Bars represent standard error (** p < 0.01).
3.7. ΔZtCBR1-1 displays an altered sterol profile
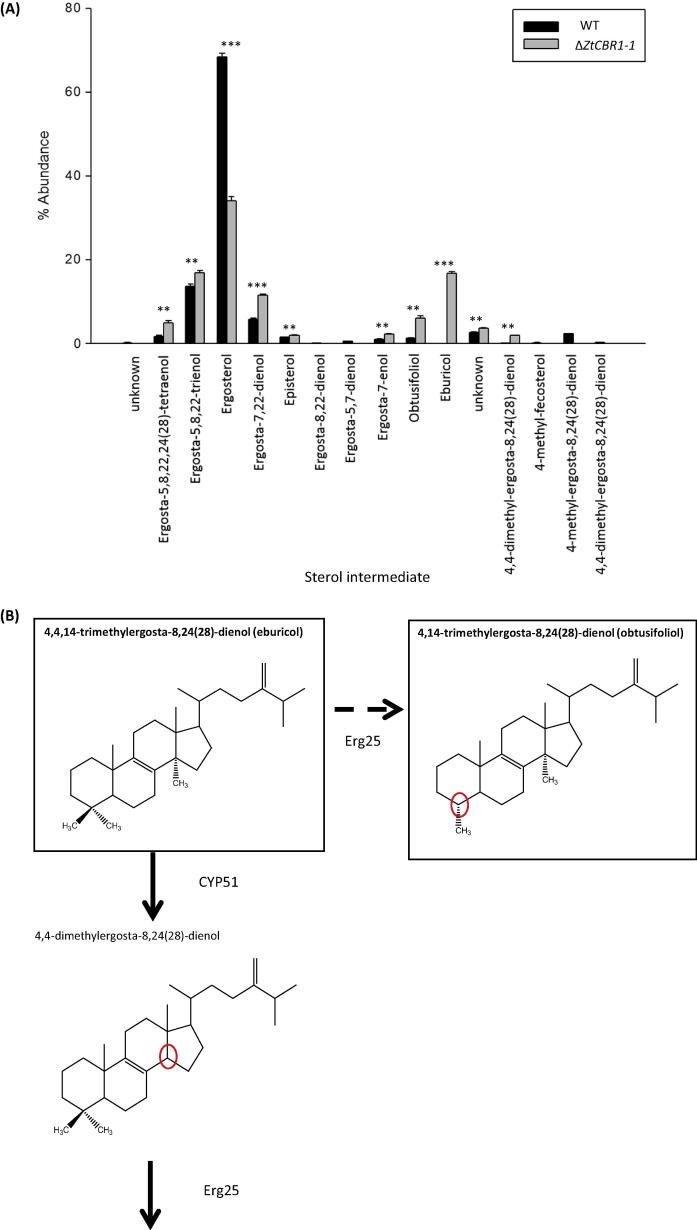
In order to assess the effect of ZtCBR1 ablation on sterol biosynthesis, the sterol profile of the WT and ΔZtCBR1-1 strains were analysed using GC–MS. ΔZtCBR1-1 displayed a significant reduction in the relative abundance of the final product of the sterol pathway, ergosterol (ergosta-5,7,22-trienol), relative to the WT (p < 0.001). Several intermediate compounds in the sterol biosynthesis pathway including ergosta-5,8,22,24(28)-tetraenol, ergosta-5,8,22-trienol, ergosta-7,22-dienol and obtusifoliol (14α-dimethyl-5α-ergosta-8,24(28)-dienol) were significantly increased in relative abundance in the ΔZtCBR1-1 strain compared to the WT (p < 0.01, p < 0.01, p < 0.001 and p < 0.01 respectively). The substrate of the enzyme CYP51 (eburicol (4,4,14-trimethylergosta-8,24(28)-dienol)), which is a target of azole antifungals, accumulated in ΔZtCBR1-1 but was not detected in the WT (p < 0.001) (Fig. 8).
Fig. 8.
ΔZtCBR1 mutants have altered sterol profiles. (A) Mean relative abundance of all ergosterol and all intermediates in the ergosterol biosynthetic pathway identified for WT and a ΔZtCBR1 strain. Bars represent standard error, all deviations from WT levels in the ΔZtCBR1 strain were significant (** p < 0.01, *** p < 0.001). (B) Diagram depicting the reactions catalysed by the enzymes CYP51 and Erg25 during sterol biosynthesis. Solid arrow represents usual direction of biosynthetic pathway. Perforated arrow represents an alternative route for the CYP51 substrate, eburicol, which may be followed more frequently if CYP51 activity is compromised. Solid boxes surround compounds that accumulated in ΔZtCBR1 relative to the WT. Red circles mark the sites of enzymatic alterations at each sterol biosynthesis step. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Unlike S. cerevisiae many filamentous fungal genomes have more than one microsomal CBR sequence
In several eukaryotes including plants and animals, the microsomal CBR-b5 electron transfer system has been shown to be important for the function of desaturase and hydroxylase enzymes involved in the biosynthesis of unsaturated fatty acids (UFAs), sterols and sphingolipids (Sperling et al., 1998; Uttaro, 2006; Poklepovich et al., 2012). Additionally, it is thought to be involved in certain CYP-catalysed reactions (Lamb et al., 1999, 2001). The genome of the model yeast S. cerevisiae contains only a single copy microsomal CBR sequence (Csukai et al., 1994; Truan et al., 1994). In the previously analysed filamentous fungus M. alpina, which is used in the industrial production of arachidonic acid, two microsomal CBRs have been identified in biochemical studies and structurally characterised (Sakuradani et al., 1999; Certik et al., 1999), though their functional importance to the organism is not known and the biochemical role of the secondary CBR is also unclear.
In the current study putative microsomal CBR sequences encoded in a range of fully sequenced fungal genomes were identified from a number of distantly related fungi including plant pathogens (biotrophs, hemibiotrophs and necrotrophs) and saprophytes. The mean number of putative microsomal CBR sequences identified was three, as was observed for Z. tritici, though some species contained considerably more. For instance A. nidulans contained seven and Fusarium graminearum contained six copies. However others, including M. fijiensis and D. septosporum, were more similar to S. cerevisiae having only a single predicted microsomal CBR sequence (Table 1). This is perhaps a little surprising given that these two fungi are also Dothideomycetes and are members of the genus Mycosphaerellaceae alongside Z. tritici. Intriguingly out of the five endophytic species analysed only the genome of one contained sequences similar to the S. cerevisiae CBRs. Though it is not possible to identify the precise reason for a larger number of microsomal CBR enzymes in certain fungal species without further functional studies, we could speculate that this larger number might be associated with a larger diversity of the terminal CBR electron acceptors. In particular the CYPs are known to be highly diversified amongst fungi, where they are thought to be important for metabolism of the diverse array of xenobiotics to which the organism may be exposed (Chen et al., 2014). It is interesting to note that the largest number of microsomal CBRs was found in the species A. nidulans, which is capable of colonising numerous environmental niches and therefore may require the ability to metabolise a more diverse array of xenobiotic compounds.
Our genome-wide analysis overall suggested no clear links between the numbers of predicted CBR genes with either particular pathogenic or saprophytic lifestyles. The exception to this is the five endophytic species analysed which frequently returned no significant sequence homologues (highest e value cut off used = e−10). Contrarily for one of these species, the rice endophyte Harpophora oryzae, the predominant difference between its genome and those of non-endophytic species that has been observed is relative expansion of numerous gene families, in particular those associated with transposable elements and carbohydrate metabolism. In fact, in this species only 10 gene families were found to be contracted relative to non-endophytes (though details of these families are not presented in the cited study) (Xu et al., 2014). Though only a speculation, it is possible that the selective pressures of an endophytic lifestyle may lead to a loss of genes associated with lipid metabolism like the CBRs. However, such an observation has yet to be formally tested.
4.2. ZtCBR1 is constitutively expressed and required for full virulence on wheat leaves and asexual sporulation whereas ZtCBR2 is not
In addition to the presence of canonical CBR domains in the retrieved sequences, PFam analysis also identified numerous putative microsomal CBRs with N-terminal b5 fusions. Though such fusions have been reported before (Yantsevich et al., 2008; Davis et al., 2002), little is known about their functions. ZtCBR2 in Z. tritici was found to contain a b5 fusion domain. Intriguingly this gene shared a similar expression pattern to two neighbouring genes, including a putative CYP, annotated in the Z. tritici genome sequence as CYP-24, and a putative hydroxyacyl coA dehydrogenase (Fig. 2B). Given the presence of a b5 fusion domain and the observation that the CBR-b5 system may transfer electrons to CYPs, it is possible that this represents a discrete, co-regulated electron transfer chain.
Genes in this putative cluster were significantly up-regulated relative to their expression during growth in CDB in PDB and relative to either during growth in CDB or growth at all other infection time points on the fourth day of infection (p < 0.05) (Fig. 2B and C). Up-regulation of the genes in this cluster in the nutrient-rich medium PDB relative to CDB would suggest that this micro-region might be involved in metabolism of complex nutrient sources. However, low levels of expression of this cluster at later time points during plant infection when complex nutrient sources are released from necrotic host tissue would suggest that this is not the case.
An alternative hypothesis would be that this cluster is responsible for degradation of a compound common to numerous plant species, as PDB is derived from plant material. Degradation or metabolism of host-derived molecules by CYP enzymes has been shown to be important in various plant pathogenic fungi (Coleman et al., 2011; Pedrini et al., 2013; Miao et al., 1991). However, targeted deletion of the two genes ZtCBR2 and ZtCYP-24 did not lead to any reduction in virulence or asexual sporulation in planta (Supplementary Fig. S2) or any clearly evident phenotypic change in vitro. If ZtCBR2 is the primary electron donor for the enzymes in this cluster, it may be that the cluster is functionally redundant for plant infection. Furthermore, even if ZtCYP-24 is able to receive electrons from an alternate redox partner, this observation would indicate that alone it is not essential. Further characterisation of this cluster, including single and double deletions of both the CYP and the putative hydroxyacyl coA dehydrogenase would be needed to characterise its potential role in plant infection.
Despite the apparent high number and potential diversification of CBR sequences in filamentous fungi, in Z. tritici only one CBR, ZtCBR1, was highly expressed under both in vitro conditions and at all infection time points tested (Fig. 2A). This may indicate that it is the major Z. tritici CBR involved in processes described in other eukaryotes for members of this gene family. Further evidence from this comes from the observed biochemical and growth defects in ΔZtCBR1 strains and the delayed virulence and absence of asexual sporulation in infected wheat leaves (Figs. 4–8).
4.3. ZtCBR1 is involved in fatty acid, sphingolipid and sterol metabolism in Z. tritici
In accordance with the various roles of the CBR-b5 electron transfer system in other eukaryotes, unsaturated fatty acid (UFA), sphingolipid and sterol profiles of ΔZtCBR1 strains were analysed and aberrations in all three of these pathways were observed (Figs. 6–8). Biosynthesis of UFAs proceeds via the insertion of double bonds between carbons of fatty acyl chains by desaturases, which are reliant on the CBR-b5 electron transfer system for reducing power. The first double bond is normally formed between the 9th and 10th carbons (the Δ9 position) of palmitic (16:0) or stearic (18:0) acid to make palmitoleic (16:1) or oleic (18:1) acid respectively. In all eukaryotes, this is carried out by a Δ9 desaturase (which is fused to b5 in fungi) that receives electrons from the CBR system (Uttaro, 2006; Tamura et al., 1976).This is the only desaturation event that takes place in S. cerevisiae as it only contains a single fatty acid desaturase, OLE1 (Stukey et al., 1990).
Many plants and fungi can carry out further desaturations, including M. alpina which contains Δ5, Δ6 and Δ12 desaturases, as well as a multifunctional desaturase and two additional Δ9 desaturases with differing substrate specificity (Knutzon et al., 1998; Sakuradani et al., 1999; Sakuradani and Shimizu, 2003; Kikukawa et al., 2013; MacKenzie et al., 2002; Wongwathanarat et al., 1999). In the current study, fatty acid methyl ester (FAME) derivatives of fatty acid species with either 16 or 18 carbons and up to 3 double bonds were investigated in a ΔZtCBR1 mutant strain, ΔZtCBR1-1. It was found that the relative abundances of 18:1 and 16:1 were significantly depleted in this strain, suggesting that ZtCBR1 is an important redox partner for the Z. tritici Δ9 desaturase. However, the Δ9 desaturase substrates 18:0 and 16:0 did not significantly accumulate in ΔZtCBR1-1 relative to the WT. Conversely 18:0 decreased and 18:2, the product of Δ12 desaturation of 18:1, increased in relative abundance in this strain (Fig. 6A). It may be that the increase in 18:2 observed was a compensation for the perturbations in sterol and sphingolipid biosynthesis that were also observed in this strain. However, at this point and without protein enzyme activity studies, it is not possible to conclusively determine the precise fatty acid desaturase enzymes to which ZtCBR1 transfers electrons.
Sphingolipid biosynthesis also involves the activity of desaturase enzymes. Sphingolipids are composed of two distinct portions, a fatty acid and a long chain base (LCB), which are amide-linked via a variety of possible head groups. The LCB is an aliphatic amino alcohol which may vary in the number of double bonds or hydroxyl groups that it possesses. In addition to the desaturases involved in LCB biosynthesis, the hydroxylases that are involved also rely on the CBR-b5 system for electrons. In this study sphingolipid LCBs were analysed. Two distinct LCB desaturases are known to exist in fungi, the Δ4 and the Δ8 desaturase (Oura and Kajiwara, 2008; Beckmann et al., 2003). It is possible that ZtCBR1 is involved in electron transfer to these enzymes in Z. tritici as it was found that dihydroxy 19:2 (d19:2) was depleted in ΔZtCBR1-1 relative to the WT. Though there was no increase in the precursor of this compound, d18:1, a close to statistically significant increase in the relative abundance of trihydroxy 18:0 (t18:0) was observed (p = 0.054) (Fig. 7). This may be a compensation for the lack of d19:2 as an increase in hydroxylated LCBs may offset some of the effects of depletion of saturated LCBs. A reduction in the amount of dihydroxy 18:0 (d18:0) provides further evidence for this as it is the precursor of t18:0, which may be produced via additional hydroxylation. However, again it is not possible to precisely determine which sphingolipid desaturase or hydroxylase enzymes ZtCBR1 provides electrons to without further functional studies.
Sterol biosynthesis involves various desaturases and hydroxylases and the two cytochrome P450s (CYPs), CYP51 and CYP61 (Lepesheva and Waterman, 2007; Alcazar-Fuoli et al., 2006; Bjorkhem and Leitersdorf, 2000; Kelly et al., 1997). S. cerevisiae only has three CYPs, including the sterol biosynthetic CYPs and CYP56, a dityrosine hydroxylase involved in sporulation (Briza et al., 1994). Intriguingly CPR, the cytochrome P450 reductase, usually thought to be the primary redox partner for CYPs, has been shown to be dispensable in S. cervisiae (Sutter and Loper, 1989). This may be due to use of the CBR-b5 system as an alternative redox partner for CYPs as it has been demonstrated that it can fully support CYP51 activity in a reconstituted cell-free system (Lamb et al., 1999). Disruption of ZtCBR1 had a major effect on sterol biosynthesis. In ΔZtCBR1-1 the final product ergosterol was significantly depleted in relative abundance compared to the WT. The two pathway intermediates that increased in relative abundance most prominently in this strain were eburicol, the substrate of CYP51, and obtusifoliol, the product of 4 α demethylation of eburicol by Erg25, which usually acts downstream of CYP51 (Bard et al., 1996) (Fig. 8). This may indicate that ZtCBR1 is not only an alternative redox partner for CYP51 in Z. tritici but that it is necessary for its function.
ΔZtCBR1 was also found to accumulate ergosta-5,8,22 trienol which has been shown to accumulate in some CYP61-inhibited fungal strains (Loto et al., 2012). This may indicate that ZtCBR1 is also important for the function of this enzyme in Z. tritici. Finally, another intermediate, ergosta-7,22-dienol, was also shown to accumulate in ΔZtCBR1. This is likely a result of decreased activity of the sterol Δ5 desaturase enzyme, ERG3, which requires CBR for electron transfer (Poklepovich et al., 2012; Kawata et al., 1985; Arthington et al., 1991) (Fig. 8).
Little is understood about the direct involvement of CBR in electron transfer to the fungal sterol biosynthesis CYPs CYP51 and CYP61, though it has been shown that disruption of the intermediate electron transfer enzyme b5 leads to an accumulation of a similar array of sterol intermediates in the Ascomycete yeast Candida albicans (Rogers et al., 2004). Further indication of the importance of this system for sterol biosynthesis comes from the observation that in Schizosaccharomyces pombe, Sre1, which is known to regulate sterol biosynthetic enzymes, also regulates CBR and b5 (Todd et al., 2006), and in humans CYP51 catalysis has been shown to be enhanced by the presence of b5 (Lamb et al., 2001). However, b5 has been shown to enhance CYP catalysed reactions independently of CBR via allosteric interaction (Porter, 2002), leading to the question of potential functional redundancy between the two electron donors CPR and CBR for CYP catalysis during sterol biosynthesis. In the current study, CBR enzyme functional ablation was shown to strongly affect both CYP-catalysed and the Δ5 desaturase-catalysed steps of the sterol biosynthetic pathway, which ultimately led to a reduction in relative abundance of the final product ergosterol. To our knowledge this represents the first direct observation of the effects of CBR functional ablation on sterol biosynthesis in a natural live cell system.
4.4. Growth and virulence defects in ΔZtCBR1 strains may be attributable to one or more of the various lipid metabolism abnormalities
Though it may not be possible to pinpoint the precise biochemical basis of the morphological, growth and virulence defects observed in ΔZtCBR1, we can speculate that alterations in sphingolipid and sterol content were a major contributory factor. Sterols and sphingolipids are known to group together in biological membranes to form specific regions that have been termed lipid rafts (for a review see Simons and Sampaio, 2011). Lipid raft regions are hypothesised to be important sites for the attachment of specific membrane proteins involved in various cellular processes. For example, in S. cerevisiae several different sphingolipid and sterol biosynthesis mutants have shown deficiencies in Golgi trafficking (Proszynski et al., 2005).
Another important process in S. cerevisiae that involves formation of lipid rafts is mating. This involves polarisation of the plasma membrane to form a ‘schmoo tip’, in which lipid raft domains have been observed (Bagnat and Simons, 2002). Echoing this process, the fungus C. albicans has been shown to utilise lipid raft domains to form hyphae. Evidence for this comes from the observation of these regions at hyphal tips and from the formation of aberrant hyphae in spores exposed to sterol or sphingolipid biosynthesis-disrupting compounds (Martin and Konopka, 2004).
The morphological defects observed in ΔZtCBR1 strains could be representative of an underlying defect in lipid raft formation. Filamentous growth in this strain, though WT in appearance, was substantially slowed (Fig. 5). Due to their reduced LCB and ergosterol content ΔZtCBR1 spores may have been less frequently able to produce lipid raft domains, leading to less frequent hyphal extension resulting in slower overall growth. This is consistent with observations of reductions in particular sphingolipid LCB species and ergosterol rather than total ablation.
Other defects brought on by aberrations in lipid raft formation may be more specific to a pathogenic lifestyle. For instance, in several mammalian-pathogenic fungi, lipid raft-embedded proteins have been shown to be essential for adherence to host cells (Humen et al., 2011; Mittal et al., 2008). In the plant pathogen F. graminearum, the importance of lipid rafts in infection was demonstrated through disruption of the ceramide synthase gene (Bar1), essential for sphingolipid biosynthesis. The F. graminearum ΔBar1 mutant strains were unable to produce perithecia though hyphal differentiation and leaf penetration were still observed. Intriguingly in the ΔBar1 mutant strains generated in this study, sporulation resulted in the formation of shorter, less uniform spores with fewer cells than the WT (Rittenour et al., 2011). A similar observation was made for the ΔZtCBR1 mutants presented in the current study, which also showed these morphological defects (Fig. 4).
The highly reduced amount of sporulation in ΔZtCBR1 is particularly interesting given that this strain was eventually able to produce an extensive hyphal network in vitro (Fig. 5) and induce full necrosis of leaf tissue (Fig. 3A). This suggests that the normal virulence mechanisms that may elicit host cell death are still functionally intact in the ΔZtCBR1 strains but occur later, possibly as a consequence of the reduced hyphal growth rate. There are various possible explanations for subsequent loss of asexual sporulation in diseased leaves, including the influence of lipid signalling on developmental processes such as growth and proliferation. For instance, it has been demonstrated that in S. cerevisiae sphingolipid LCBs interact with the protein kinase Pkh1, which controls numerous processes including cell wall integrity and growth (Liu et al., 2005). Furthermore, lipid rafts are also known to mediate localisation of H-Ras, a key element of the mitogen activated protein kinase (MAPK) signalling pathway, to the correct sites in the cellular membrane systems (Anderson, 2006). This is intriguing because in Z. tritici disruption of the MAPK-encoding gene MgFus3 led to a lack of pycnidiation in vitro (Cousin et al., 2006). In light of data presented in the current study, it is possible that this MAPK is influenced by cellular lipid content. Future analysis should involve sequential disruption of Z. tritici genes involved in biosynthesis of specific lipids and lipid-derived signalling molecules.
4.5. Conclusion
By characterising members of the CBR family in Z. tritici this study has demonstrated for the first time the importance of these genes in regulating infection-related processes in a plant pathogenic fungus. To our knowledge, this is the first time that these genes have been functionally characterised in fungi other than S. cerevisiae and M. alpina. Defects in pathways thought to require enzymes that receive electrons from CBR-b5 observed in ΔZtCBR1 ultimately led to an almost complete lack of asexual sporulation in planta. Thus, processes dependent upon particular CBR-b5 electron transfers in Z. tritici may represent important new targets for future disease intervention.
Acknowledgments
This research was carried out as part of a Biotechnology and Biological Sciences Research Council (BBSRC) (UK) Collaborative Award in Science and Engineering (CASE) studentship, in collaboration with Syngenta, Jealott’s Hill, Bracknell, UK. Kim Hammond-Kosack and Jason Rudd are supported by the BBSRC through the Institute Strategic Program Grant 20:20 Wheat® (BB/J/00426X/1). All experiments involving Z. tritici WT and transgenic isolates were conducted in biological containment facilities under FERA licence number 101948/11982851/2. This work was supported in part by the European Regional Development Fund/Welsh Government funded BEACON research program (Swansea University). Many thanks to Na Li (Syngenta) for her contribution to the generation of Z. tritici mutant strains.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fgb.2015.05.008.
Contributor Information
Mark C. Derbyshire, Email: markcharder@gmail.com.
Jason Rudd, Email: jason.rudd@rothamsted.ac.uk.
Appendix A. Supplementary material
References
- Alcazar-Fuoli L. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: Role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 2006;50(2):453–460. doi: 10.1128/AAC.50.2.453-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.H. Role of lipids in the MAPK signaling pathway. Prog. Lipid Res. 2006;45(2):102–119. doi: 10.1016/j.plipres.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Arthington B.A. Cloning, disruption and sequence of the gene encoding the yeast C-5-sterol desaturase. Gene. 1991;102(1):39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- Bagnaresi P. Tonoplast subcellular localization of maize cytochrome b(5) reductases. Plant J. 2000;24(5):645–654. doi: 10.1046/j.1365-313x.2000.00914.x. [DOI] [PubMed] [Google Scholar]
- Bagnat M., Simons K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 2002;99(22):14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard M. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc. Natl. Acad. Sci. USA. 1996;93(1):186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C. Stereochemistry of a bifunctional dihydroceramide Delta(4)-desaturase/hydroxylase from Candida albicans; a key enzyme of sphingolipid metabolism. Org. Biomol. Chem. 2003;1(14):2448–2454. doi: 10.1039/b303939k. [DOI] [PubMed] [Google Scholar]
- Bjorkhem I., Leitersdorf E. Sterol 27-hydroxylase deficiency: a rare cause of xanthomas in normocholesterolemic humans. Trends Endocrinol. Metab. 2000;11(5):180–183. doi: 10.1016/s1043-2760(00)00255-1. [DOI] [PubMed] [Google Scholar]
- Bowler J. New capabilities for Mycosphaerella graminicola research. Mol. Plant Pathol. 2010;11(5):691–704. doi: 10.1111/j.1364-3703.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza P., Eckerstorfer M., Breitenbach M. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a 2-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc. Natl. Acad. Sci. USA. 1994;91(10):4524–4528. doi: 10.1073/pnas.91.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik H., Kosar M., Arinc E. In vitro effects of myricetin, morin, apigenin, (+)-taxifolin, (+)-catechin, (−)-epicatechin, naringenin and naringin on cytochrome b5 reduction by purified NADH-cytochrome b5 reductase. Toxicology. 2013;308:34–40. doi: 10.1016/j.tox.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Certik M. Characterization of the second form of NADH-cytochrome b(5) reductase gene from arachidonic acid-producing fungus Mortierella alpina 1S-4. J. Biosci. Bioeng. 1999;88(6):667–671. doi: 10.1016/s1389-1723(00)87098-x. [DOI] [PubMed] [Google Scholar]
- Chen W. Fungal cytochrome P450 monooxygenases: their distribution, structure, functions, family expansion, and evolutionary origin. Geno. Biol. Evol. 2014;6(7):1620–1634. doi: 10.1093/gbe/evu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coleman J.J. Characterization of the gene encoding pisatin demethylase (FoPDA1) in fusarium oxysporum. Mol. Plant Microbe Interact. 2011;24(12):1482–1491. doi: 10.1094/MPMI-05-11-0119. [DOI] [PubMed] [Google Scholar]
- Cools H.J., Fraaije B.A. Are azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola. Pest Manag. Sci. 2008;64(7):681–684. doi: 10.1002/ps.1568. [DOI] [PubMed] [Google Scholar]
- Cousin A. The MAP kinase-encoding gene MgFus3 of the non-appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 2006;7(4):269–278. doi: 10.1111/j.1364-3703.2006.00337.x. [DOI] [PubMed] [Google Scholar]
- Csukai M., Murray M., Orr E. Isolation and complete sequence of a CBR, a gene encoding a putative cytochrome b reductase in Saccharomyces cerevisiae. Eur. J. Biochem. 1994;219(1–2):441–448. doi: 10.1111/j.1432-1033.1994.tb19957.x. [DOI] [PubMed] [Google Scholar]
- Davis C.A. Heterologous expression of an endogenous rat cytochrome b(5)/cytochrome b(5) reductase fusion protein: Identification of histidines 62 and 85 as the heme axial ligands. Arch. Biochem. Biophys. 2002;400(1):63–75. doi: 10.1006/abbi.2002.2783. [DOI] [PubMed] [Google Scholar]
- do Amaral A.M. Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola. PLoS One. 2012;7(12):e49904. doi: 10.1371/journal.pone.0049904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahian F. Human cytochrome b5 reductase: structure, function, and potential applications. Crit. Rev. Biotechnol. 2014;34(2):134–143. doi: 10.3109/07388551.2012.732031. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Eyal Z., Scharen A.L., Prescott J.M., van Ginkel M. The International Maize and Wheat Improvement Center; Mexico: 1987. The Septoria Diseases of Wheat: Concepts and Methods of Disease Management. [Google Scholar]
- Finn R.D. Pfam: the protein families database. Nucleic Acids Res. 2014;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L. Role of NADPH-cytochrome P450 reductase and cytochrome-b(5)/NADH-b(5) reductase in variability of CYP3A activity in human liver microsomes. Drug Metab. Dispos. 2009;37(1):90–96. doi: 10.1124/dmd.108.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces R., Mancha M. One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal. Biochem. 1993;211(1):139–143. doi: 10.1006/abio.1993.1244. [DOI] [PubMed] [Google Scholar]
- George H.L., Hirschi K.D., VanEtten H.D. Biochemical properties of the products of cytochrome P450 genes (PDA) encoding pisatin demethylase activity in Nectria haematococca. Arch. Microbiol. 1998;170(3):147–154. doi: 10.1007/s002030050627. [DOI] [PubMed] [Google Scholar]
- Glory M.D.D., Thiruvenangdam Chrysin attenuates the instability of xenobiotic metabolizing and mitochondrial enzymes during Diethyl nitrosamine induced liver carcinoma. J. Pharm. Res. 2011;4(6):1839–1842. [Google Scholar]
- Gohari A.M. Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 2014;15(4):394–405. doi: 10.1111/mpp.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S.B. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7(6):e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstead G.F., Gaylor J.L. Total enzymatic-synthesis of cholesterol from 4,4,14-alpha-trimethyl-5-alpha-cholesta-8,24-dien-3-beta-ol – solubilization, resolution, and reconstitution of delta-7-sterol 5-desaturase. J. Biol. Chem. 1982;257(23):3937–3944. [PubMed] [Google Scholar]
- Hahne K. Incomplete arrest in the outer-membrane sorts NADH-cytochrome-b(5) reductase to 2 different submitochondrial compartments. Cell. 1994;79(5):829–839. doi: 10.1016/0092-8674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Henderson C.J., McLaughlin L.A., Wolf C.R. Evidence that cytochrome b(5) and cytochrome b(5) reductase can act as sole electron donors to the hepatic cytochrome p450 systems. Mol. Pharmacol. 2013;83(6):1209–1217. doi: 10.1124/mol.112.084616. [DOI] [PubMed] [Google Scholar]
- Huang Y.S. Cloning of Delta 12-and Delta 6-desaturases from Mortierella alpina and recombinant production of gamma-linolenic acid in Saccharomyces cerevisiae. Lipids. 1999;34(7):649–659. doi: 10.1007/s11745-999-0410-8. [DOI] [PubMed] [Google Scholar]
- Humen M.A., Perez P.F., Lievin-Le V. Moal, lipid raft-dependent adhesion of Giardia intestinalis trophozoites to a cultured human enterocyte-like Caco-2/TC7 cell monolayer leads to cytoskeleton-dependent functional injuries. Cell. Microbiol. 2011;13(11):1683–1702. doi: 10.1111/j.1462-5822.2011.01647.x. [DOI] [PubMed] [Google Scholar]
- Kawata S., Trzaskos J.M., Gaylor J.L. Microsomal enzymes of cholesterol biosynthesis from lanosterol – purification and characterization of delta-7-sterol 5-desaturase of rat liver microsomes. J. Biol. Chem. 1985;260(11):6609–6617. [PubMed] [Google Scholar]
- Kelly S.L. Mode of action and resistance to azole antifungals associated with the formation of 14-alpha-methylergosta-8,24(28)-dien-3-beta,6-alpha-diol. Biochem. Biophys. Res. Commun. 1995;207(3):910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- Kelly S.L. Characterization of Saccharomyces cerevisiae CYP61, sterol Delta(22)-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 1997;272(15):9986–9988. doi: 10.1074/jbc.272.15.9986. [DOI] [PubMed] [Google Scholar]
- Keon J. Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant Microbe Interact. 2007;20(2):178–193. doi: 10.1094/MPMI-20-2-0178. [DOI] [PubMed] [Google Scholar]
- Kikukawa H. Characterization of a trifunctional fatty acid desaturase from oleaginous filamentous fungus Mortierella alpina 1S-4 using a yeast expression system. J. Biosci. Bioeng. 2013;116(6):672–676. doi: 10.1016/j.jbiosc.2013.05.023. [DOI] [PubMed] [Google Scholar]
- Knutzon D.S. Identification of Delta 5-desaturase from Mortierella alpina by heterologous expression in bakers’ yeast and canola. J. Biol. Chem. 1998;273(45):29360–29366. doi: 10.1074/jbc.273.45.29360. [DOI] [PubMed] [Google Scholar]
- Kumar R. A mutation in arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation. Plant J. 2006;48(6):920–932. doi: 10.1111/j.1365-313X.2006.02925.x. [DOI] [PubMed] [Google Scholar]
- Lamb D.C. Biodiversity of the P450 catalytic cycle: yeast cytochrome b(5)/NADH cytochrome b(5) reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 1999;462(3):283–288. doi: 10.1016/s0014-5793(99)01548-3. [DOI] [PubMed] [Google Scholar]
- Lamb D.C. Human sterol 14 alpha-demethylase activity is enhanced by the membrane-bound state of cytochrome b(5) Arch. Biochem. Biophys. 2001;395(1):78–84. doi: 10.1006/abbi.2001.2566. [DOI] [PubMed] [Google Scholar]
- Lee W.S. Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 2014;27(3):236–243. doi: 10.1094/MPMI-07-13-0201-R. [DOI] [PubMed] [Google Scholar]
- Lepesheva G.I., Waterman M.R. Sterol 14 alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta-Gen. Sub. 2007;1770(3):467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 2005;280(24):22679–22687. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- Loto, I., et al., 2012. Enhancement of Carotenoid Production by Disrupting the C22-Sterol Desaturase Gene (CYP61) in Xanthophyllomyces dendrorhous. BMC Microbiology. p. 12. [DOI] [PMC free article] [PubMed]
- MacKenzie D.A. A third fatty acid Delta 9-desaturase from Mortierella alpina with a different substrate specificity to ole1p and ole2p. Microbiol. Sgm. 2002;148:1725–1735. doi: 10.1099/00221287-148-6-1725. [DOI] [PubMed] [Google Scholar]
- Marshall R. Analysis of two in planta expressed LysM Effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 2011;156(2):756–769. doi: 10.1104/pp.111.176347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.W., Konopka J.B. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell. 2004;3(3):675–684. doi: 10.1128/EC.3.3.675-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao V.P.W., Matthews D.E., Vanetten H.D. Identification and chromosomal locations of a family of cytochrome P450 genes for pisatin detoxification in the fungus Nectria haematococca. Mol. Gen. Genet. 1991;226(1–2):214–223. doi: 10.1007/BF00273606. [DOI] [PubMed] [Google Scholar]
- Michaelson L.V. Identification of a cytochrome b5-fusion desaturase responsible for the synthesis of triunsaturated sphingolipid long chain bases in the marine diatom Thalassiosira pseudonana. Phytochemistry. 2013;90:50–55. doi: 10.1016/j.phytochem.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Mirzaei S.A., Yazdi M.T., Sepehrizadeh Z. Secretory expression and purification of a soluble NADH cytochrome b5 reductase enzyme from Mucor racemosus in Pichia pastoris based on codon usage adaptation. Biotechnol. Lett. 2010;32(11):1705–1711. doi: 10.1007/s10529-010-0348-z. [DOI] [PubMed] [Google Scholar]
- Mittal K., Welter B.H., Temesvari L.A. Entamoeba histolytica: lipid rafts are involved in adhesion of trophozoites to host extracellular matrix components. Exp. Parasitol. 2008;120(2):127–134. doi: 10.1016/j.exppara.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Perez A.J. Sphingolipid base modifying enzymes in sunflower (Helianthus annuus): Cloning and characterization of a C4-hydroxylase gene and a new paralogous Delta 8-desaturase gene. J. Plant Physiol. 2011;168(8):831–839. doi: 10.1016/j.jplph.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Motteram J. Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 2009;22(7):790–799. doi: 10.1094/MPMI-22-7-0790. [DOI] [PubMed] [Google Scholar]
- Motteram J. Aberrant protein N-glycosylation impacts upon infection-related growth transitions of the haploid plant-pathogenic fungus Mycosphaerella graminicola. Mol. Microbiol. 2011;81(2):415–433. doi: 10.1111/j.1365-2958.2011.07701.x. [DOI] [PubMed] [Google Scholar]
- Mutch D.M. The disruption of hepatic cytochrome P450 reductase afters mouse lipid metabolism. J. Proteome Res. 2007;6(10):3976–3984. doi: 10.1021/pr0700448. [DOI] [PubMed] [Google Scholar]
- Neve E.P.A. Amidoxime reductase system containing cytochrome b(5) type B (CYB5B) and MOSC2 Is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 2012;287(9):6307–6317. doi: 10.1074/jbc.M111.328237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova A.B., Saris N.-E.L., Kruglov A.G. External mitochondrial NADH-dependent reductase of redox cyclers: VDAC1 or Cyb5R3? Free Radical Biol. Med. 2014;74:74–84. doi: 10.1016/j.freeradbiomed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- O’Driscoll A. The wheat-Septoria conflict: a new front opening up? Trends Plant Sci. 2014;19(9):602–610. doi: 10.1016/j.tplants.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Orton E.S., Deller S., Brown J.K.M. Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 2011;12(5):413–424. doi: 10.1111/j.1364-3703.2010.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura T., Kajiwara S. Disruption of the sphingolipid Delta(8)-desaturase gene causes a delay in morphological changes in Candida albicans. Microbiol. -Sgm. 2008;154:3795–3803. doi: 10.1099/mic.0.2008/018788-0. [DOI] [PubMed] [Google Scholar]
- Pedrini N. Targeting of insect epicuticular lipids by the entomopathogenic fungus Beauveria bassiana: hydrocarbon oxidation within the context of a host-pathogen interaction. Front. Microbiol. 2013:4. doi: 10.3389/fmicb.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poklepovich T.J. The cytochrome b(5) dependent C-5(6) sterol desaturase DES5A from the endoplasmic reticulum of Tetrahymena thermophila complements ergosterol biosynthesis mutants in Saccharomyces cerevisiae. Steroids. 2012;77(13):1313–1320. doi: 10.1016/j.steroids.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter T.D. The roles of cytochrome b(5) in cytochrome P450 reactions. J. Biochem. Mol. Toxicol. 2002;16(6):311–316. doi: 10.1002/jbt.10052. [DOI] [PubMed] [Google Scholar]
- Proszynski T.J. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. USA. 2005;102(50):17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M.E.A. Sequential asymmetric polyketide heterocyclization catalyzed by a single cytochrome P450 monooxygenase (AurH) Angew. Chem. Int. Ed. 2008;47(46):8872–8875. doi: 10.1002/anie.200803714. [DOI] [PubMed] [Google Scholar]
- Rittenour W.R. Control of glucosylceramide production and morphogenesis by the bar1 ceramide synthase in Fusarium graminearum. PLoS One. 2011;6(4):e19385. doi: 10.1371/journal.pone.0019385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K.M. Disruption of the Candida albicans CYB5 gene results in increased azole sensitivity. Antimicrob. Agents Chemother. 2004;48(9):3425–3435. doi: 10.1128/AAC.48.9.3425-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd J.J. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 2015;167(3):1158–1185. doi: 10.1104/pp.114.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuradani E., Shimizu S. Gene cloning and functional analysis of a second Delta 6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Biosci. Biotechnol. Biochem. 2003;67(4):704–711. doi: 10.1271/bbb.67.704. [DOI] [PubMed] [Google Scholar]
- Sakuradani E., Kobayashi M., Shimizu S. Identification of an NADH-cytochrome b(5) reductase gene from an arachidonic acid-producing fungus, Mortierella alpina 1S-4, by sequencing of the encoding cDNA and heterologous expression in a fungus, Aspergillus oryzae. Appl. Environ. Microbiol. 1999;65(9):3873–3879. doi: 10.1128/aem.65.9.3873-3879.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuradani E. Identification of Delta 12-fatty acid desaturase from arachidonic acid-producing Mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur. J. Biochem. 1999;261(3):812–820. doi: 10.1046/j.1432-1327.1999.00333.x. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J.M. Cloning, functional analysis, and subcellular localization of two isoforms of NADH: cytochrome b(5) reductase from developing seeds of tung (Vernicia fordii) Plant Sci. 2005;169(2):375–385. [Google Scholar]
- Siah A. QoI Resistance and Mitochondrial Genetic Structure of Zymoseptoria tritici in Morocco. Plant Dis. 2014;98(8):1138–1144. doi: 10.1094/PDIS-10-13-1057-RE. [DOI] [PubMed] [Google Scholar]
- Simons K., Sampaio J.L. Membrane organization and lipid rafts. Cold Spring Harbor Perspect. Biol. 2011;3(10):17. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling P., Zahringer U., Heinz E. A sphingolipid desaturase from higher plants – identification of a new cytochrome b(5) fusion protein. J. Biol. Chem. 1998;273(44):28590–28596. doi: 10.1074/jbc.273.44.28590. [DOI] [PubMed] [Google Scholar]
- Stukey J.E., McDonough V.M., Martin C.E. The OLE1 gene of Saccharomyces cerevisiae encodes the delta-9 fatty acid desaturase and can be functionally replaced by the rat steroyl coA desaturase gene. J. Biol. Chem. 1990;265(33):20144–20149. [PubMed] [Google Scholar]
- Suffert F., Sache I., Lannou C. Assessment of quantitative traits of aggressiveness in Mycosphaerella graminicola on adult wheat plants. Plant. Pathol. 2013;62(6):1330–1341. [Google Scholar]
- Sutter T.R., Loper J.C. Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome-P450 reductase causes increased sensitivity to ketoconazole. Biochem. Biophys. Res. Commun. 1989;160(3):1257–1266. doi: 10.1016/s0006-291x(89)80139-1. [DOI] [PubMed] [Google Scholar]
- Syed K. Cytochrome b(5) reductase-cytochrome b(5) as an active P450 redox enzyme system in Phanerochaete chrysosporium: a typical properties and in vivo evidence of electron transfer capability to CYP63A2. Arch. Biochem. Biophys. 2011;509(1):26–32. doi: 10.1016/j.abb.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher K. Sensitivity of Zymoseptoria tritici isolates from tunisia to pyraclostrobin, fluxapyroxad, epoxiconazole, metconazole, prochloraz and tebuconazole. J. Phytopathol. 2014;162(7–8):442–448. [Google Scholar]
- Tamura Y. Fatty acid desaturase system of yeast microsomes – involvement of cytochrome b5-containing electron transport chain. Arch. Biochem. Biophys. 1976;175(1):284–294. doi: 10.1016/0003-9861(76)90510-5. [DOI] [PubMed] [Google Scholar]
- Todd B.L. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 2006;26(7):2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truan G. Cloning and characterization of a yeast cytochrome b(5)-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P450 reductase-deficient strain. Gene. 1994;142(1):123–127. doi: 10.1016/0378-1119(94)90366-2. [DOI] [PubMed] [Google Scholar]
- Truong H.N., Meyer C., Danielvedele F. Characteristics of Nicotiana tobacum reductase protein produced in Saccharomyces cerevisiae. Biochem. J. 1991;278:393–397. doi: 10.1042/bj2780393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttaro A.D. Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life. 2006;58(10):563–571. doi: 10.1080/15216540600920899. [DOI] [PubMed] [Google Scholar]
- Wayne L.L. Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in arabidopsis. Plant Cell. 2013;25(8):3052–3066. doi: 10.1105/tpc.113.113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongwathanarat P. Two fatty acid Delta 9-desaturase genes, ole1 and ole2, from Mortierella alpina complement the yeast ole1 mutation. Microbiol. -Sgm. 1999;145:2939–2946. doi: 10.1099/00221287-145-10-2939. [DOI] [PubMed] [Google Scholar]
- Xu X.-H. The rice endophyte Harpophora oryzae genome reveals evolution from a pathogen to a mutualistic endophyte. Sci. Rep. 2014:4. doi: 10.1038/srep05783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yantsevich A.V., Gilep A.A., Usanov S.A. Mechanism of electron transfer in fusion protein cytochrome b(5)-NADH-cytochrome b(5) reductase. Biochem. Moscow. 2008;73(10):1096–1107. doi: 10.1134/s0006297908100052. [DOI] [PubMed] [Google Scholar]
- Zwiers L.H., De Waard M.A. Efficient agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Curr. Genet. 2001;39(5–6):388–393. doi: 10.1007/s002940100216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.