Abstract
We have developed a novel approach for DNA detection as well as genetic screening of mutations by uniquely combining DNA-responsive and optically diffracting materials. This approach entails the polymerization of a photonic crystal within a hydrogel network that alters the diffraction of light in response to a target DNA strand. The utility of this approach, which permits label-free sensing, was demonstrated via the detection of a target sequence from the DNA binding domain of the major tumor suppressor protein p53. Using a complementary capture probe strand, we were able to detect down to picomole concentrations of the target p53 sequence. Moreover, we demonstrated that this approach could readily detect a single base pair mutation in the target strand, which corresponds to the hotspot cancer mutation R175H in p53. The sensitivity of detection was increased by lowering the rate of annealing of the target strand and adjusting the solution ionic strength during optical characterization. Changes in ionic strength during characterization impact the melting temperature of the bound target DNA and the Donnan potential between the hydrogel and solution, which influence detection. We further showed that this approach is sensitive to epigenetic changes via the detection of a fully methylated form of the target p53 sequence. Ultimately, this approach represents a new paradigm for DNA detection and specifically genetic screening of p53 as well as other disease markers and nucleotide modifications that alter the properties of DNA (e.g., epigenetic alterations and adducts with chemical carcinogens).
INTRODUCTION
Due to the importance of DNA detection in a myriad of fields, including genetic screening, forensics, pathogen identification, and biotechnology (i.e., genome engineering), the development of new technologies for DNA sensing is critical. Of particular interest is the development of new approaches that accelerate DNA detection with high fidelity and reduce the cost of traditional DNA sequencing. In addition to traditional DNA sequencing (e.g., capillary electrophoresis), which, although precise, requires specialized instrumentation, other methods of detecting DNA include electrochemical impedance spectroscopy,1–3 surface enhanced Raman spectroscopy,4 nanoparticle aggregation assays,5–7 analysis by quartz crystal microbalance,8–9 surface plasmon resonance,10–12 and scanning tunneling spectroscopy.13–15 Additionally, fluorescent16–17 and chemiluminescent18–19 based techniques for DNA sensing have been reported. However, many of these techniques, as with traditional sequencing, require highly specialized instrumentation as well as exogenous labels or reagents and, moreover, are unable to detect down to single nucleotide changes.20–21
A novel approach for the detection of DNA, which may overcome many of the limitations of current sensing methods, entails combining DNA-responsive and optically diffracting materials. In one such approach, a photonic crystal may be polymerized within a hydrogel matrix that can swell or contract in response to the presence of an analyte. The hydrogel matrix can be rationally tuned to change volume in response to a specific analyte by tethering a receptor for the analyte within the hydrogel along with the photonic crystal.22–25 Depending on the properties of the analyte (i.e., charge or hydrophobic character), receptor binding may trigger a volume change of the hydrogel by creating a Donnan potential or altering the interaction of the hydrogel with water. Changes in hydrogel volume are accompanied by an alteration in the lattice spacing of the photonic crystal that may be readily measured by reflectance spectroscopy or, if large enough, visually through changes in coloration of the hydrogel. We and others have previously exploited this approach to develop sensors for a broad spectrum of small molecule analytes,26–28 metals,29–31 changes in solution conditions,32–33 and, more recently, protein kinase activity.34–35 Notably, in all cases, this approach enabled the detection of environmental cues in the absence of exogenous reagents, using changes in optical properties of the hydrogel as the primary readout.
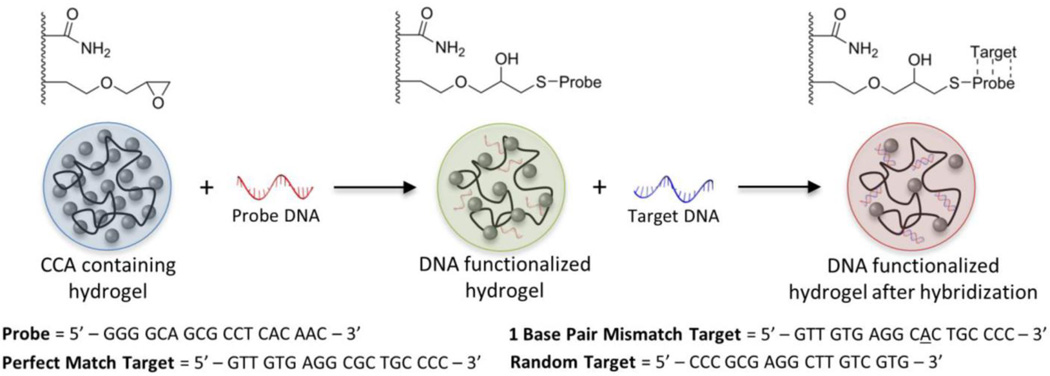
In this work, we explored the utility of this approach as a sensing platform for label-free DNA detection via encapsulation of a crystalline colloidal array (CCA) within a DNA-responsive hydrogel. Specifically, we fabricated DNA-responsive hydrogel films that alter the diffraction of light upon hybridization of a specific “target” DNA strand to a capture “probe” sequence (Figure 1). We reasoned that hybridization of the target strand would cause a change in the volume of the hydrogel network by increasing the concentration of immobilized negative charges within the hydrogel. The addition of negative charges upon hybridization is due to the backbone of the target DNA being comprised of negatively-charged phosphate groups. Once these charges become immobilized within the hydrogel network, the Donnan potential between the hydrogel and surrounding solution is modified, resulting in swelling of the hydrogel and ultimately a change in the diffraction spectrum of the encapsulated CCA.
Figure 1.
Schematic of hydrogel functionalization with “probe” DNA and subsequent hybridization of “target” DNA strands. Color changes in the optically diffracting hydrogel are representative of those observed upon functionalization and hybridization due to changes in the lattice spacing of the encapsulated CCA. The sequences of the probe and target DNA strands that were used are shown below the schematic.
The utility of this approach for DNA sensing was demonstrated via the detection of the gene for the major tumor suppressor protein p53. A key transcription factor involved in cell regulation, p53, which is inactivated in virtually all human cancers, is of specific interest as a marker for early cancer detection.36–41 For sensing, a short 18-mer sequence that is complementary to the DNA-binding domain of the p53 gene was used as the capture probe and was conjugated to the hydrogel network (Figure 1). The perfect matching 18-mer sequence from the wild-type p53 gene was used as the target strand. Furthermore, we were interested in determining if the sensing approach was sensitive to mutations in p53, which is the most frequently mutated gene in cancer. To determine the sensitivity of the optical response of the detection approach to DNA mutations, a single base in the target strand was changed (G→A). This genetic alteration corresponds to mutation of arginine at position 175 to a histidine (i.e., R175H), which is one of the more frequent oncogenic mutations in p53. Finally, we tested if the detection scheme was also sensitive to DNA methylation by using a methylated form of the wild-type target strand (Table 1). We show that changes in methylation can also be readily detected, which may have additional implications in screening for epigenetic disease markers.
Table 1.
Names and sequences of DNA oligos
| Name | Sequence |
|---|---|
| Probe | 5’ – GGG GCA GCG CCT CAC AAC – 3’ |
| Perfect Match (PM) Target | 5’ – GTT GTG AGG CGC TGC CCC – 3’ |
| 1 Base Pair Mismatch (1bpMM) Target | 5’ – GTT GTG AGG CAC TGC CCC – 3’ |
| Random Target | 5’ – CCC GCG AGG CTT GTC GTG – 3’ |
| Methylated Perfect Match (mPM)Target | 5’ – GTT GTG AGG mCGmC TGmC mCmCmC – 3’ |
EXPERIMENTAL METHODS
Materials
Acrylamide (AA), N,N’-methylenebis(acrylamide) (BA), and allyl glycidyl ether (AGE) monomers were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. DNA oligos (Table 1) were purchased and used as received from Integrated DNA Technologies (Coralville, IA). The crosslinker SPDP-PEG4-NHS was purchased and used without further purification from Conju-Probe (San Diego, CA).
Synthesis of Polystyrene Nanospheres
Highly-charged polystyrene (PS) nanospheres were synthesized via emulsion polymerization as previously described.42–43 The PS nanospheres used in all experiments had a concentration of 12 wt% in water and were 87 nm in diameter with a polydispersity of 4.9%, as determined by dynamic light scattering (Titan DynaPro with Dyna V6.3.4 software package, Wyatt Technology, Inc.; Santa Barbara, CA). Particles were stored at room temperature over a BioRad (Hercules, CA) AG501-X8 mixed bed resin.
Hydrogel Polymerization and Crystalline Colloidal Array Formation
Hydrogels were photopolymerized by solubilizing 35 mg (0.98 M) AA in 480 µL of the CCA. To this mixture, a solution of 1 mg (0.015 M) BA and 24 mg (0.42 M) AGE in 20 µL DMSO was added. The photoinitiator Irgacure 2959 (BASF; Florham Park, NJ) (10 wt% in DMSO) was added at a final concentration of 0.05 wt% to the CCA-monomer solution. The solution was then pipetted into a mold formed by two glass slides separated by a 273 ± 2 µm Parafilm spacer. Samples were flood exposed with 365 nm light at 15 mW/cm2 from a UV mercury lamp for 45 min. Films were subsequently equilibrated and stored in ultrapure water for a minimum of 24 h prior to DNA functionalization.
Hydrogel Functionalization with DNA Probe
DNA probe was solubilized in 100 mM NaPO4 buffer, pH 8, at a concentration of 5 mM. A fifteen-fold molar excess of the SPDP-PEG4-NHS linker relative to the DNA probe was solubilized in 10 µL DMSO and added to the DNA solution. The solution was incubated at 4°C and reacted for 2 h. The solution was then desalted to remove excess linker after which 100 mM tris(2-carboxyethyl)phosphine (TCEP) was added and reacted at room temperature for 1 h to reduce the linker disulfide bond. To determine the amount of linker containing DNA, the absorbance of the cleaved pyridine-2-thione was measured at 343 nm. The concentration of the reduced linker was calculated using the pyridine-2-thione extinction coefficient of 8080 M−1 cm−1.44 After determining the linker concentration, a final desalt was performed to remove the cleaved pyridine-2-thione group.
Prior to functionalization, hydrogels were equilibrated in 100 mM NaPO4 buffer, pH 8. To each sample, 25 µL of 75 µM linker-modified DNA probe in 100 mM NaPO4 buffer was added and reacted at room temperature for 36 h. Post-functionalization, the samples were thoroughly rinsed and stored in 100 mM NaCl, pH 6.
DNA Hybridization
Samples to be annealed were equilibrated in 100 mM NaCl solution, pH 6, prior to the addition of perfect match (PM), single base pair mismatch (1bpMM), random DNA, or methylated perfect match (mPM) target DNA. Target DNA was added to the hydrogels at concentrations ranging from 0.5 to 1000 µM and annealed by heating the hydrogel samples to 85 °C, holding that temperature for 30 min, and ramping from 85 to 55 °C at rates ranging from 0.05 to 1 °C/min. Once the temperature reached 55 °C, the system temperature was no longer controlled and samples were permitted to naturally cool to room temperature.
Measurement of DNA Melting Point
The melting point for both the PM and 1bpMM samples was determined by first annealing samples with 50 µM PM target or 500 µM 1bpMM target in 100 mM NaCl followed by equilibration in 2.5 mM NaCl solution. Melting points were subsequently measured by submerging the hydrogels a in 2.5 mM NaCl solution and increasing the solution temperature from room temperature to 65 °C at a rate of 0.25 °C/min.
Optical Diffraction Measurements
The optical response of the equilibrated hydrogel-encapsulated CCA biosensors was measured with an Ocean Optics (Dunedin, FL) USB-4000 fiber-optic spectrophotometer operated in reflectance mode set to an angle of incidence of 15° from the sample surface normal. This optical setup allowed for characterization of the peak diffraction wavelength in situ and in real-time, enabling measurements as a function of temperature (e.g., to generate melting curves). In all experiments, the hydrogels were initially rinsed with 100 mM NaCl solution, pH 6.0, to remove non-hybridized DNA. For experiments utilizing a single ionic strength condition, samples were introduced to the desired ionic strength by a stepwise decrease in NaCl concentration. For experiments investigating sensor response to solution ionic strength, samples were first equilibrated and characterized in 10 mM NaCl, pH 6, followed by a stepwise reduction in ionic strength to 0.01 mM NaCl with measurements taken after equilibration at each condition.
RESULTS AND DISCUSSION
Effect of Target DNA Concentration on Sensor Response
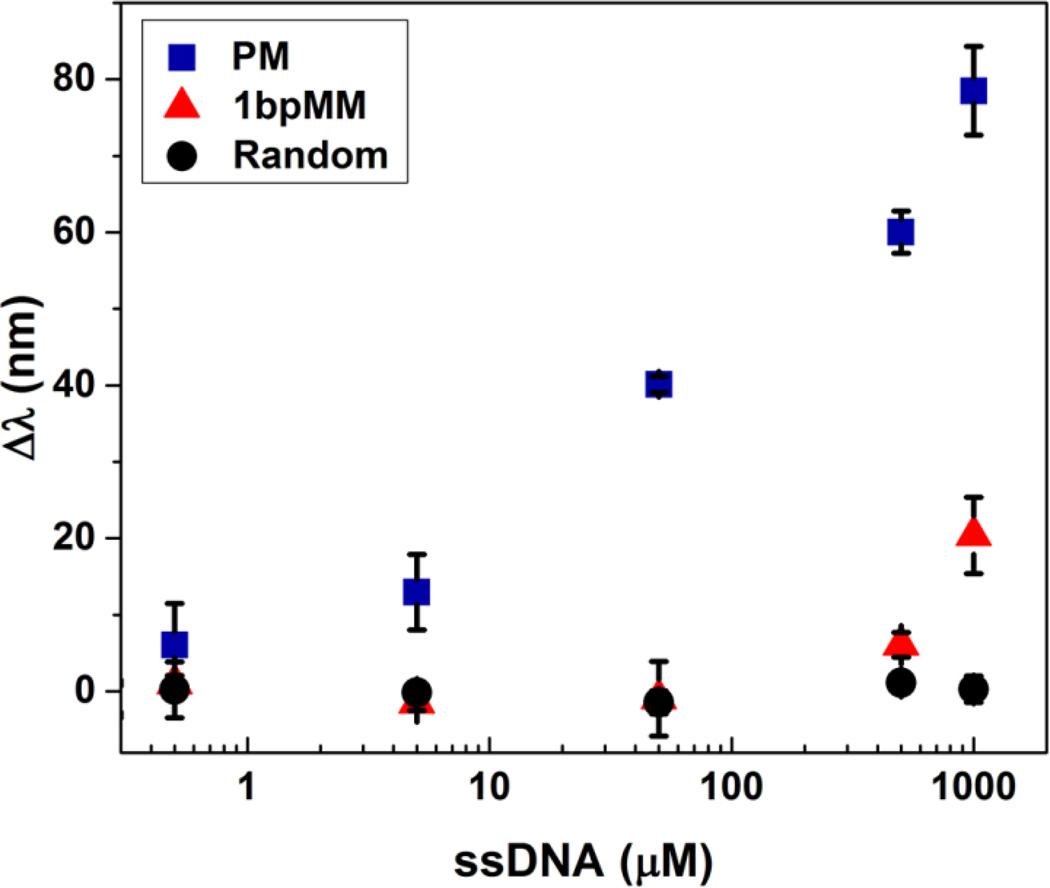
Upon hydrogel functionalization with the p53 probe strand, the sensitivity and selectivity of the sensing platform was investigated using a fully complementary (i.e., perfect match) and random sequence. Figure 2 shows the sensor response to concentrations of the perfect match (PM) and random sequence ranging from 0.5 to 1000 µM, which corresponds to 25 pmole to 50 nmole of target DNA. Addition of the PM causes a dose-dependent redshift in the diffraction peak with increasing target concentrations, resulting in a nearly 8-fold change in signal over the concentration range. Conversely, the random sequence, which has the same GC content as the PM, does not elicit a response at even the highest concentrations used. These results highlight the sensitivity and selectivity of the approach, which presumably is due to differences in the hybridization of the PM relative to the random sequence. Hybridization of the PM would result in the immobilization of negative charges from the target DNA strand within the hydrogel, triggering a change in the Donnan potential of the system. This change would, as a result, cause the hydrogel network to expand and, in turn, the lattice spacing of the embedded CCA to increase, leading to the observed redshift in peak diffraction. Accordingly, given that the random sequence would not be expected to hybridize with the capture probe, the lack of response that was generated by the random sequence is not surprising. In these measurements, the amount of target DNA was less than that of the probe strand within the hydrogel such that the immobilization of additional probe would not enhance the response.
Figure 2.
Change in wavelength of peak diffraction relative to that of the probe functionalized sensor as a function of the concentration of target ssDNA. Samples were annealed at a rate of 0.2 °C/min with PM, 1bpMM, or random target DNA concentrations ranging from 0.5 to 1000 µM. Diffraction measurements were taken in a 0.25 mM NaCl solution at pH 6 and at room temperature. Error bars represent ±1σ from the mean of 3 samples.
Due to the sensitivity of the approach to the hybridization between the target and probe strands, we also hypothesized that mutations in the target that weakened hybridization would be detectable. Of particular interest was if single base pair mutations could be detected using this approach. The detection of single base pair missense mutations is of practical importance for the identification and screening of genetic makers that are associated with various diseases. In the case of p53, screening for specific known hotspot mutations allows for the detection of genetic hallmarks of cancer, which permits rational treatment using cancer-specific therapies. To understand if our approach is sensitive to single base pair mutations, the optical response of functionalized hydrogels to the target sequence with a base pair mismatch (1bpMM) was measured. Notably, the mutation that was introduced results in the hotspot mutation R175H, which perturbs the structure of the DNA binding domain of p53, resulting in a loss of p53 function.45–46 Although a redshift in the diffraction peak relative to the probe functionalized sensor (i.e., the signal Δλ) was not observed at low concentrations of 1bpMM, a significant change in the diffraction peak was apparent at higher concentrations, indicating that, despite weaker affinity for the probe, a target with a single base pair substitution can be detected. Presumably, because the binding affinity would increase, the response to a base pair change with longer target and probe sequences would theoretically increase relative to the control. Moreover, the apparent selectivity of the approach for the PM relative to 1bpMM suggests that, in principle, a target strand may be differentiated from similar sequences in complex DNA mixtures. Such mixtures may include a multitude of strands with differing sequences and lengths, which may arise from the digestion of cellular DNA.
Effect of Annealing Rate and Temperature on Sensor Response
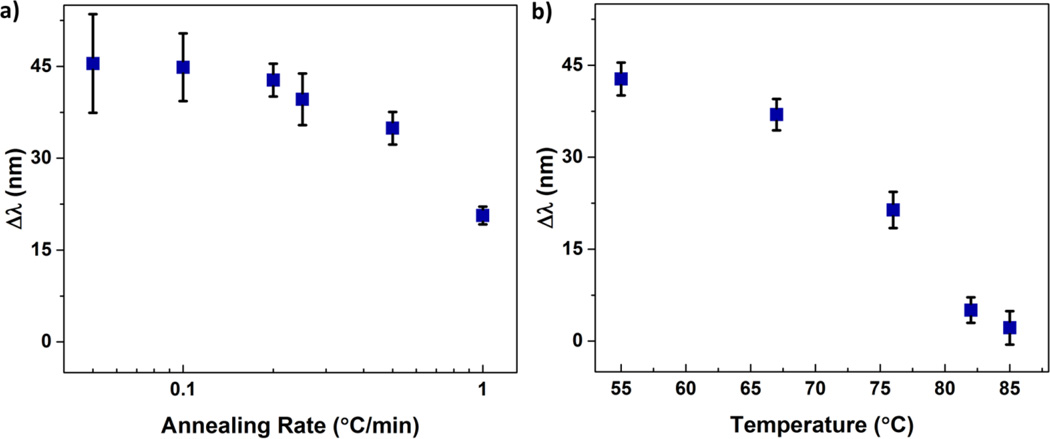
Annealing conditions can greatly affect the ability of DNA to form the most thermodynamically favorable duplexes. Accordingly, the impact of annealing conditions on the sensitivity of the detection of the target sequence was investigated. The conditions for annealing were varied by heating hydrogels that contained the probe in the presence of the target DNA to approximately 10 °C above the theoretical Tm of the bound PM (~76 °C in 100 mM NaCl). After heating, the solution containing the free PM target and hydrogel was cooled to well below the Tm at different cooling rates. The final temperature to which the solution was cooled was 55 °C at which the PM sequence should be mostly bound within the hydrogel. As shown in Figure 3a, the magnitude of the redshift in diffraction is greatest at low cooling rates and drops off as the rate of cooling increases beyond 0.2 °C/min. The decline in sensitivity at high cooling rates may be attributed to imperfect hybridization of the PM to the probe sequence, which would result in increased dissociation of the PM strand.
Figure 3.
Optical response Δλ observed for DNA hybridization as a function of a) annealing rate and b) annealing temperature. All samples were annealed in the presence of 50 µM PM target DNA at rates ranging from 0.05 to 1 °C/min. In a) the annealing rate was maintained from 85 to 55 °C, followed by uncontrolled but consistent cooling from 55 °C to room temperature. Samples presented in b) were annealed at a rate of 0.2 °C/min and, at the specified temperatures, were removed and immediately cooled in ice water followed by rinsing in 100 mM NaCl to quench hybridization. Diffraction measurements were taken in a 0.5 mM NaCl solution at pH 6 and at room temperature. Error bars represent ±1σ from the mean of 3–6 samples.
The impact of annealing conditions on the sensitivity of target detection was also investigated by using a fixed cooling rate, but varying the annealing time. In this case, samples were heated to 85 °C and subsequently cooled at a rate of 0.2 °C/min for different times, which resulted in different final annealing temperatures, ranging from 85–55 °C. At the final annealing temperature, the hydrogels were quickly cooled in ice water followed by rinsing in 100 mM NaCl solution at room temperature to quench any further hybridization. Figure 3b shows the optical response to the PM sequence as a function of the final annealing temperature. As anticipated, at high final annealing temperatures, where the amount of bound PM is expected to be low, the response of the sensing approach to the PM is low. A significant increase in the detection of the PM sequence is observed at longer times and thus lower final annealing temperatures. Annealing of the hydrogels below 55 °C resulted in no further change in the sensor response, indicating further lowering the annealing temperature has negligible impact on hybridization and sensitivity of the approach.
Characterization of the Critical Melting Temperature of Target DNA
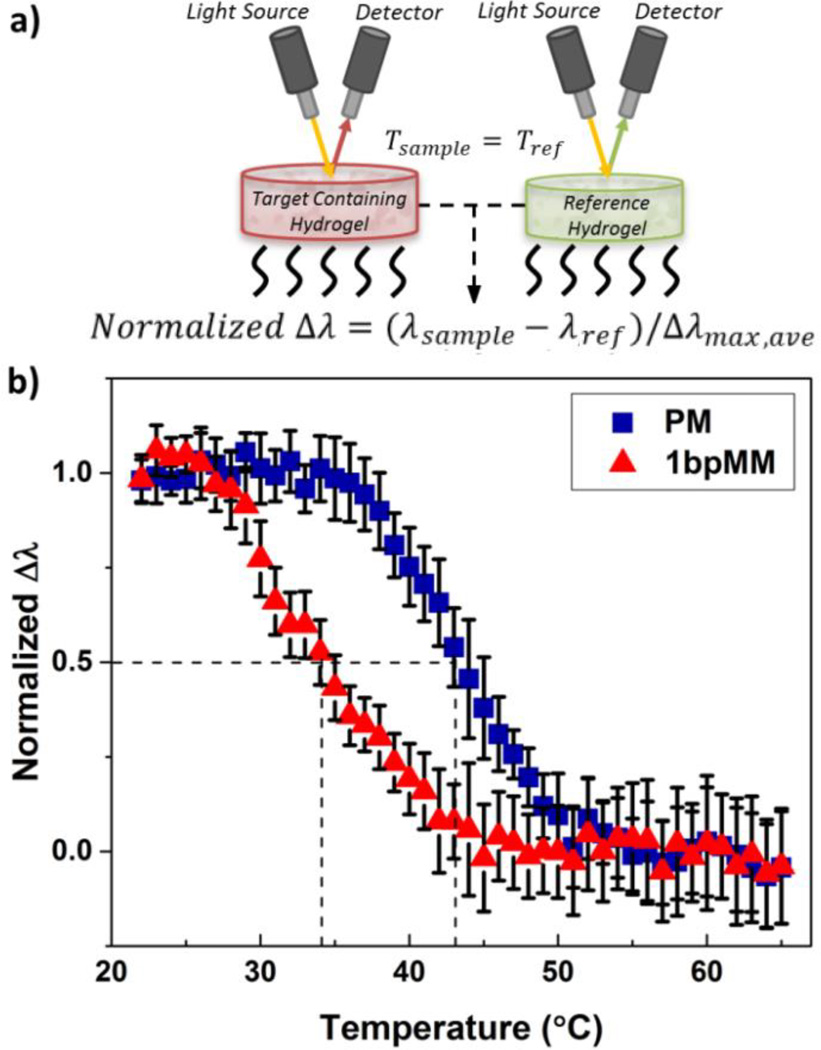
A fundamentally interesting question related to DNA detection using our sensing approach is if the Tm of the bound target DNA is the same in the hydrogel as in solution. Dramatic changes in Tm of the bound target DNA, relative to the annealing or characterization temperatures, may significantly lower the magnitude of the observed response to the target and thus the detection limit. To determine the Tm of the target sequences used in this work, the PM (at 50 µM) and 1bpMM (at 500 µM) sequences were annealed at a cooling rate of 0.2 °C/min to the probe immobilized in hydrogels. For reference measurements, hydrogels were subject to annealing under identical conditions without any target DNA. Of note, a larger concentration of the 1bpMM sequence relative to the PM sequence was used to obtain optical responses of similar magnitude for the two DNA targets. After annealing, the hydrogels were heated from room temperature to 65 °C at a rate of 0.25 °C/min and the diffraction response of each sample was measured over the entire temperature range. The diffraction response for the PM and 1bpMM sequences is reported as a normalized response, which was determined as the difference of the diffraction wavelength (i.e., Δλ) of the sample with target DNA from that of the reference (with no target DNA) divided by the average maximum shift in peak diffraction wavelength (i.e., Δλmax,ave) from the PM or 1bpMM. By reporting the normalized difference in diffraction wavelength, volume changes in the hydrogel related to temperature increases and DNA denaturation could be decoupled. In this case, changes in the equilibrium volume of the hydrogel at elevated temperatures may be attributed to changes in the solvent density and Flory-Huggins interaction parameter that alters the free energy of mixing of the system.47
From the melting curve of the normalized diffraction response for the PM and 1bpMM sequences (Figure 4b), the Tm of bound PM and 1bpMM could be approximated. The approximated Tm of the bound PM and 1bpMM were 43.5 and 34.3 °C, respectively, as determined from linear interpolation of the melting curves. For comparison, the theoretical Tm of bound PM and 1bpMM in solution are 43 and 35 °C, respectively,48 which are in good agreement with the experimentally determined values when bound in the hydrogel. This close agreement implies that the hybridization of the target DNA in the hydrogel is nearly identical to that in solution and, moreover, that the theoretical Tm of the target DNA-probe duplex in solution can be used to rationally design the probe strand to enhance sensitivity as well as to optimize the annealing and characterization temperatures.
Figure 4.
a) Schematic of the optical setup for characterizing melting curves and b) normalized melting curves for samples annealed with PM or 1bpMM target DNA. The normalized optical response was calculated as the difference in peak diffraction between the target containing samples and reference samples, normalized to the average maximum shift for the PM and 1bpMM samples. The Tm for the hybridized PM (blue squares) and 1bpMM (red triangles) was found to be 43.5 and 34.3 °C, respectively. Sample annealing was performed prior to melting with 50 µM PM or 500 µM 1bpMM target DNA at a rate of 0.2 °C/min. Melting was performed by ramping samples from room temperature to 65 °C at a rate of 0.25 °C/min. Diffraction measurements were taken in a 2.5 mM NaCl solution at pH 6. Error bars represent ±1σ from the mean of 3 samples.
Characterization of Sensor Response to Solution Ionic Strength
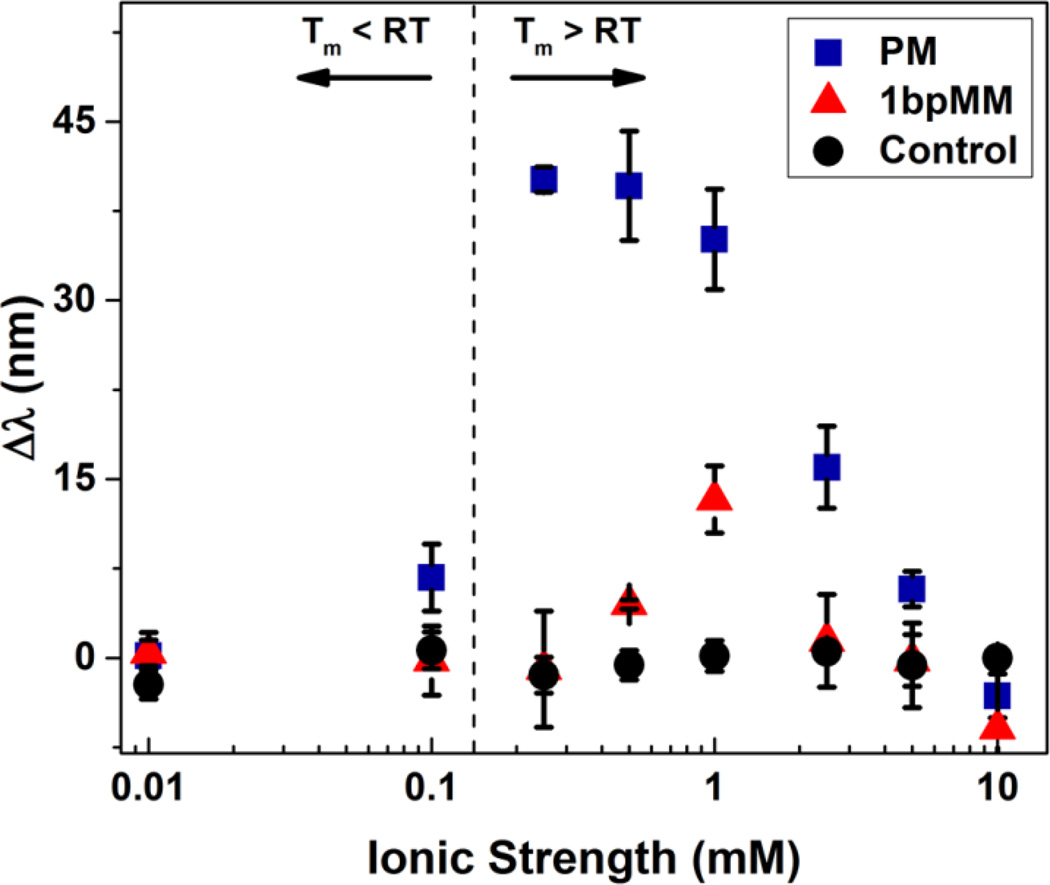
Due to the effect of ionic strength on DNA melting and the Donnan potential between the hydrogel and the surrounding environment, the detection of target DNA is strongly dependent on ionic strength during the diffraction measurements. To understand the magnitude of this effect, the sensor response as a function of ionic strength of the optical characterization solution was investigated for PM, 1bpMM, and control samples (Figure 5). For both PM and 1bpMM samples, starting at high ionic strengths, the response of the sensor increased significantly as the ionic strength of the characterization solution was lowered. However, as the ionic strength was further lowered, the sensor response decreased, resulting in two discernable regimes that describe the effect of ionic strength on the sensor response.
Figure 5.
Optical response Δλ as a function of ionic strength for samples annealed with 50 µM PM (blue squares) or 1bpMM (red triangles) target DNA annealed at a ramp rate of 0.2 °C/min. Control samples (black circles) included probe functionalized hydrogels annealed in the absence of target DNA and hydrogels that were not functionalized with probe but annealed in the presence of target DNA. The vertical dashed line indicates the ionic strength conditions at which the melting temperature of the PM target DNA is equal to the optical characterization temperature. Diffraction measurements were taken after sample equilibration in 0.01 to 10 mM NaCl solutions at pH 6 and at room temperature. Error bars represent ±1σ from the mean of 3–6 samples.
The distinction between these regimes is most notable for the PM sequence for which a maximum response was observed at an ionic strength of 0.25 mM. A decrease in the sensor response below this ionic strength is presumably due to the reduction in Tm for the hybridized PM to less than the characterization temperature, which results in weaker binding of the target strand. Notably, the predicted salt adjusted Tm for the hybridized PM is equivalent to room temperature at 0.15 mM salt (dashed line), which corresponds to the characterization temperature for these samples. As such, at ionic strengths less than 0.15 mM, one would expect that the amount of bound PM within the hydrogel at room temperature is very low. Despite the formation of the duplex being favored, the decrease in sensor response above the optimum ionic strength can be explained by a reduction in Donnan potential upon hybridization. This reduction is due to a smaller gradient in the concentration of mobile ions from the interior to the exterior of the hydrogel with increasing ionic strength.35 For the 1bpMM sample, the optimum ionic strength for detection was significantly higher than for the PM as expected given that the Tm for bound 1bpMM is presumably lower (than for bound PM) at all ionic strengths.
These results ultimately show the importance of considering ionic strength and characterization temperature, which are intimately related, when expanding this sensing approach to other sequences and mutations. For example, for probe and target strands with a lower GC content, that when hybridized have a lower Tm, a lower characterization temperature or a lower ionic strength could be used. Similarly, if longer probe and target strands are used, the characterization temperature or ionic strength (or both) could be increased to enable detection. However, the use of shorter probe and target strands enables a wider range of characterization conditions due to a greater difference in the Tm between a strand that is a perfect match and one that has a base pair mutation. The flexibility to rationally alter the characterization conditions for the detection of virtually any sequence or length target strand represents a major strength of the approach. In the case of longer target strands that form secondary structures (e.g., hairpin loops), the annealing conditions could be altered to ensure melting of the target and hybridization with the probe. Additionally, the location of a base pair mutation has little effect on the Tm of hybridization unless the mutation is present at one of the end positions. The location of the mutation at an end position would, in theory, result in a decrease in the difference in the response between the perfect match and mutated strand. As such, this suggests that the probe strand should be designed such that the anticipated mutation is internal within the probe sequence.
Detection of Methylated DNA
Having demonstrated the utility of our sensing approach to detect single base pair mutations, an interesting question to ask is if this approach is also sensitive to epigenetic DNA modifications. Such modifications include DNA methylation and hydroxymethylation, which play a crucial role in gene regulation and thus the development and progression of a variety of diseases.49–50 Currently, the primary methods for detecting such modifications include mass spectrometry51–52 and methylation-specific PCR,53–54 although newer methods, including photopolymerization-based amplification,55 have recently been reported. For conventional DNA sensing methods, these modifications are difficult to detect due to the often negligible impact these modifications generally have on DNA melting and thus hybridization.56–57
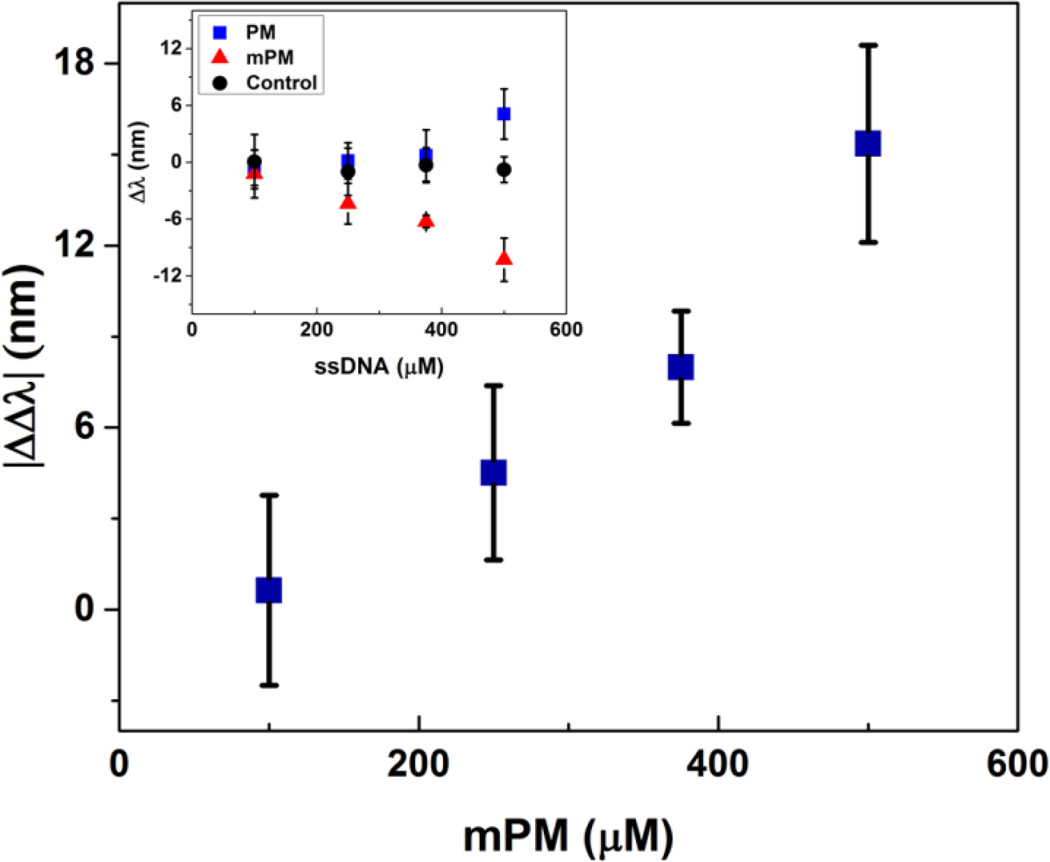
To explore the question of sensitivity to epigenetic changes, we characterized the optical response using our approach to the fully methylated form of the PM sequence (mPM). The PM sequence, when fully methylated, contains six methylated cytosine bases (Table 1). We hypothesized that the methylated moieties in the mPM sequence would reduce the relative hydrophilicity of the hydrogel (i.e., increase the strength of the Flory-Huggins interaction parameter χ) and the extent of mixing in water, thereby causing the hydrogel to contract, rather than swell. As expected, a dose-dependent response to the addition of the mPM target sequence annealed at a rate of 0.2 °C/min was observed when measured at 10 mM ionic strength and room temperature. This response, which is shown in Figure 6, is reported as the difference between the optical peak shift due to hybridization of the mPM target strand and hybridization of the PM target strand (|ΔΔλ|). The raw response generated by the addition of the mPM sequence is shown in the inset. Of note, optical characterization was performed in a high ionic strength solution to minimize the electrostatic contributions to the observed response upon hybridization of the mPM and PM sequences. By minimizing the contribution of electrostatics, the response that is observed is primarily due to volume changes arising from modulation of the Flory-Huggins interaction parameter and the addition of the methyl groups to the target DNA. Moreover, as anticipated, the response generated by the addition of the mPM sequence resulted in a blueshift in the diffraction peak of the encapsulated CCA, which is consistent with the hydrogel shrinking.
Figure 6.
Optical response as a function of the concentration of methylated DNA target. The reported response (|ΔΔλ|) is the difference between the optical shift in the wavelength of peak diffraction upon hybridization of the methylated target and the detected shift upon hybridization of the PM target. The inset shows the raw response generated by hybridization of the mPM target. Samples were annealed from 85 to 55 °C utilizing a ramp rate of 0.2 °C/min and target DNA concentrations ranging from 100 to 500 µM. Diffraction measurements were taken in a 10 mM NaCl solution at pH 6 and at room temperature. Error bars represent ±1σ from the mean of 3–6 samples.
The relative impact of even a single methylation site on the target DNA may be predicted based on a theoretical model for hydrogel swelling,47 in which the osmotic pressure term associated with hydrogel mixing in an aqueous solution scales as ΠM ∝ χ. To the simplest approximation χwater−hydrogel ∝ (δwater − δhydrogel)2 where δ are semiempirical solubility parameters related to the cohesive energy density as based on the approaches of Hildebrand or Hansen.58–59 Often, solubility parameters for polymers may be well estimated by using a group contribution approach, in which δ = (Σ F)/V where F are tabulated values of the molar attraction constants for common functional groups60 and V is the molar volume of the repeat unit. Therefore, the dependence of the interaction parameter on the number of methylation sites n on the DNA target may be estimated by , and assuming a linear dependence of the optical response on χ (a reasonable approximation for small changes in χ, see Ref. 35), the blueshift in optical response based on a single methyl group may be estimated to be 2 ~ 3 nm. Based on the error of the optical response in Figure 6, the methylation of as few as two sites may be reasonably detected. One way to potentially enhance the response to a single methylation site may be to encapsulate the CCA in a hydrogel with a lower inherent χ.
Similarly, it may be predicted that other chemical modifications to DNA, including hydroxymethylation or more significantly functionalities that are highly hydrophobic, will also be readily detectable using this approach. For example, a single phenyl modification is predicted to have approximately three times the effect as a methyl modification. Chemical carcinogens that form DNA adducts may likewise be detected, including the classic example of benzo[a]pyrene which through a series of chemical reactions may be covalently linked to guanines in DNA.61–63 In fact, the effect of benzo[a]pyrene on the p53 oncogene has been shown to lead to transversion mutations, such as the single base pair mutation considered here.64 The DNA biosensing scheme presented here may then also provide opportunities to screen for chemical carcinogens and DNA adducts that lead to mutations from which cancer originates.
CONCLUSIONS
In summary, we have developed and demonstrated the utility of optically diffracting hydrogels for the label-free detection of DNA, as well as missense mutations and methylated sites ubiquitous to genes associated with a variety of diseases. Specifically, we showed that a short target DNA sequence from p53 could be readily distinguished from an analogous sequence that has a single base pair mutation that corresponds with the cancer hotspot mutation R175H in a highly selective and dose-dependent manner. Furthermore, methylation of the native target sequence could be detected, indicating the feasibility of using this approach to screen for epigenetic modifications. Differences in the detection of the native versus mutant and methylated sequences can be attributed to alterations in hybridization and polymer-solvent interactions, respectively, and are sensitive to changes in ionic strength and hybridization conditions. This approach ultimately represents a new paradigm for screening oncogenic hotspot mutations in p53 and other cancer-associated proteins. More broadly, this approach may be extended to screen for genetic markers for other diseases as well as nucleotide modifications, stemming from epigenetic changes as well as chemical modification. The high selectivity in differentiating between a target strand and similar sequences would, in principle, permit the detection of the target strand from complex DNA mixtures.
Highlight.
A novel approach for the optical detection of DNA was developed via polymerization of a photonic crystal within DNA-responsive hydrogel films, allowing for screening of genetic and epigenetic modifications.
ACKNOWLEDGMENTS
Support was provided by the Univ. of Colorado Liquid Crystal Material Research Center (NSF DMR0820579) and NIH Pharmaceutical Biotechnology Training Program (NIH 5T32GM8732).
REFERENCES
- 1.Benvidi A, Firouzabadi AD, Moshtaghiun SM, Mazloum-Ardakani M, Tezerjani MD. Anal Biochem. 2015 doi: 10.1016/j.ab.2015.05.009. (In Press) [DOI] [PubMed] [Google Scholar]
- 2.Ianeselli L, Grenci G, Callegari C, Tormen M, Casalis L. Biosens Bioelectron. 2014;55:1–6. doi: 10.1016/j.bios.2013.11.067. [DOI] [PubMed] [Google Scholar]
- 3.Zhu BC, Booth MA, Shepherd P, Sheppard A, Travas-Sejdic J. Biosens Bioelectron. 2015;64:74–80. doi: 10.1016/j.bios.2014.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Barhoumi A, Halas NJ. J Am Chem Soc. 2010;132(37):12792–12793. doi: 10.1021/ja105678z. [DOI] [PubMed] [Google Scholar]
- 5.Chak CP, Lai JMY, Sham KWY, Cheng CHK, Leung KCF. Rsc Adv. 2011;1(7):1342–1348. [Google Scholar]
- 6.Bu T, Zako T, Fujita M, Maeda M. Chem Commun. 2013;49(68):7531–7533. doi: 10.1039/c3cc44182b. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Liang PP, Yu HX, Xu XW, Liu Y, Lou XH, Xiao Y. Anal Chem. 2014;86(7):3461–3467. doi: 10.1021/ac4040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeri AS, Gao LZ, Gao D. J Phys Chem B. 2010;114(2):1064–1068. doi: 10.1021/jp909501h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Xiao SY, Yao DB, Lam MHW, Liang HJ. Chem Commun. 2015;51(22):4670–4673. doi: 10.1039/c4cc10114f. [DOI] [PubMed] [Google Scholar]
- 10.Cheng XR, Hau BY, Endo T, Kerman K. Biosens Bioelectron. 2014;53:513–518. doi: 10.1016/j.bios.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Ding XJ, Yan YR, Li SQ, Zhang Y, Cheng W, Cheng Q, Ding SJ. Anal Chim Acta. 2015;874:59–65. doi: 10.1016/j.aca.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Zagorodko O, Spadavecchia J, Serrano AY, Larroulet I, Pesquera A, Zurutuza A, Boukherroub R, Szunerits S. Anal Chem. 2014;86(22):11211–11216. doi: 10.1021/ac502705n. [DOI] [PubMed] [Google Scholar]
- 13.Shapir E, Cohen H, Calzolari A, Cavazzoni C, Ryndyk DA, Cuniberti G, Kotlyar A, Di Felice R, Porath D. Nat Mater. 2008;7(1):68–74. doi: 10.1038/nmat2060. [DOI] [PubMed] [Google Scholar]
- 14.Ryndyk DA, Shapir E, Porath D, Calzolari A, Di Felice R, Cuniberti G. Acs Nano. 2009;3(7):1651–1656. doi: 10.1021/nn800238g. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka H, Kawai T. Nat Nanotechnol. 2009;4(8):518–522. doi: 10.1038/nnano.2009.155. [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Yao J, Yao T, Shi S. Analyst. 2013;138(2):421–424. doi: 10.1039/c2an36142f. [DOI] [PubMed] [Google Scholar]
- 17.Qiu S, Li X, Xiong W, Xie L, Guo L, Lin Z, Qiu B, Chen G. Biosens Bioelectron. 2013;41:403–408. doi: 10.1016/j.bios.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Ma C, Zeng X, Li C, Deng Y, He N. J Biomed Nanotechnol. 2012;8(5):786–790. doi: 10.1166/jbn.2012.1446. [DOI] [PubMed] [Google Scholar]
- 19.Freeman R, Liu X, Willner I. J Am Chem Soc. 2011;133(30):11597–11604. doi: 10.1021/ja202639m. [DOI] [PubMed] [Google Scholar]
- 20.Sassolas A, Leca-Bouvier BD, Blum LJ. Chem Rev. 2008;108(1):109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 21.Tosar JP, Branas G, Laiz J. Biosens Bioelectron. 2010;26(4):1205–1217. doi: 10.1016/j.bios.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 22.Cai Z, Smith NL, Zhang JT, Asher SA. Anal Chem. 2015;87(10):5013–5025. doi: 10.1021/ac504679n. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JT, Wang L, Luo J, Tikhonov A, Kornienko N, Asher SA. J Am Chem Soc. 2011;133(24):9152–9155. doi: 10.1021/ja201015c. [DOI] [PubMed] [Google Scholar]
- 24.Kamenjicki M, Kasavamoorthy R, Asher S. Ionics. 2005;10:233–236. [Google Scholar]
- 25.Nair RV, Vijaya R. Prog Quant Electron. 2010;34(3):89–134. [Google Scholar]
- 26.Sharma AC, Jana T, Kesavamoorthy R, Shi L, Virji MA, Finegold DN, Asher SA. J Am Chem Soc. 2004;126(9):2971–2977. doi: 10.1021/ja038187s. [DOI] [PubMed] [Google Scholar]
- 27.Alexeev VL, Sharma AC, Goponenko AV, Das S, Lednev IK, Wilcox CS, Finegold DN, Asher SA. Anal Chem. 2003;75(10):2316–2323. doi: 10.1021/ac030021m. [DOI] [PubMed] [Google Scholar]
- 28.Tian ET, Wang JX, Zheng YM, Song YL, Jiang L, Zhu DB. J Mater Chem. 2008;18(10):1116–1122. [Google Scholar]
- 29.Yan FY, Asher S. Anal Bioanal Chem. 2007;387(6):2121–2130. doi: 10.1007/s00216-006-1088-8. [DOI] [PubMed] [Google Scholar]
- 30.Asher SA, Sharma AC, Goponenko AV, Ward MM. Anal Chem. 2003;75(7):1676–1683. doi: 10.1021/ac026328n. [DOI] [PubMed] [Google Scholar]
- 31.Arunbabu D, Sannigrahi A, Jana T. Soft Matter. 2011;7(6):2592–2599. [Google Scholar]
- 32.Xu M, Goponenko AV, Asher SA. J Am Chem Soc. 2008;130(10):3113–3119. doi: 10.1021/ja077979+. [DOI] [PubMed] [Google Scholar]
- 33.Lee K, Asher SA. J Am Chem Soc. 2000;122(39):9534–9537. [Google Scholar]
- 34.MacConaghy KI, Geary CI, Kaar JL, Stoykovich MP. J Am Chem Soc. 2014;136(19):6896–6899. doi: 10.1021/ja5031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacConaghy KI, Chadly DM, Stoykovich MP, Kaar JL. Anal Chem. 2015;87(6):3467–3475. doi: 10.1021/acs.analchem.5b00442. [DOI] [PubMed] [Google Scholar]
- 36.Joerger AC, Fersht AR. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 37.Joerger AC, Fersht AR. Oncogene. 2007;26(15):2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 38.Freed-Pastor WA, Prives C. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane DP, Benchimol S. Genes Dev. 1990;4(1):1–8. doi: 10.1101/gad.4.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Steele RJ, Thompson AM, Hall PA, Lane DP. Br J Surg. 1998;85(11):1460–1467. doi: 10.1046/j.1365-2168.1998.00910.x. [DOI] [PubMed] [Google Scholar]
- 41.Soussi T, Wiman KG. Cancer Cell. 2007;12(4):303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Arunbabu D, Sannigrahi A, Jana T. J Appl Polym Sci. 2008;108(4):2718–2725. [Google Scholar]
- 43.Reese CE, Guerrero CD, Weissman JM, Lee K, Asher SA. J Colloid Interf Sci. 2000;232(1):76–80. doi: 10.1006/jcis.2000.7190. [DOI] [PubMed] [Google Scholar]
- 44.Stuchbury T, Shipton M, Norris R, Malthouse JPG, Brocklehurst K, Herbert JAL, Suschitzky H. Biochem J. 1975;151(2):417–432. doi: 10.1042/bj1510417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu DP, Song H, Xu Y. Oncogene. 2010;29(7):949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soussi T, Lozano G. Biochem Biophys Res Commun. 2005;331(3):834–842. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- 47.Flory PJ. Principles of polymer chemistry. Ithaca: Cornell University Press; 1953. p. 672. [Google Scholar]
- 48.Howley PM, Israel MA, Law MF, Martin MA. Journal of Biological Chemistry. 1979;254(11):4876–4883. [PubMed] [Google Scholar]
- 49.Robertson KD. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 50.Shukla A, Sehgal M, Singh TR. Gene. 2015;564(2):109–118. doi: 10.1016/j.gene.2015.03.075. [DOI] [PubMed] [Google Scholar]
- 51.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Nucleic Acids Res. 2007;35(18):e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evron E, Dooley WC, Umbricht CB, Rosenthal D, Sacchi N, Gabrielson E, Soito AB, Hung DT, Ljung B, Davidson NE, Sukumar S. Lancet. 2001;357(9265):1335–1336. doi: 10.1016/s0140-6736(00)04501-3. [DOI] [PubMed] [Google Scholar]
- 55.Heimer BW, Shatova TA, Lee JK, Kaastrup K, Sikes HD. Analyst. 2014;139(15):3695–3701. doi: 10.1039/c4an00667d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Severin PM, Zou X, Gaub HE, Schulten K. Nucleic Acids Res. 2011;39(20):8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahl C, Guldberg P. Biogerontology. 2003;4(4):233–250. doi: 10.1023/a:1025103319328. [DOI] [PubMed] [Google Scholar]
- 58.Hildebrand JH, Scott RL. Regular solutions. Englewood Cliffs, N.J.: Prentice-Hall; 1962. p. 180. [Google Scholar]
- 59.Hansen C. Hansen Solubility Parameters - A User's Handbook. CRC Press; 1999. [Google Scholar]
- 60.Cowie JMG, Arrighi V. Polymers: Chemistry and Physics of Modern materials. 3 ed. CRC Press; 2008. [Google Scholar]
- 61.Jeffrey AM, Jennette KW, Blobstein SH, Weinstein IB, Beland FA, Harvey RG, Kasal H, Miura I, Nakanishi K. J Am Chem Soc. 1976;98(18):5714–5715. doi: 10.1021/ja00434a060. [DOI] [PubMed] [Google Scholar]
- 62.Osborne MR, Harvey RG, Brookes P. Chem Biol Interact. 1978;20(1):123–130. doi: 10.1016/0009-2797(78)90087-x. [DOI] [PubMed] [Google Scholar]
- 63.Kriek E, Den Engelse L, Scherer E, Westra JG. Biochim Biophys Acta. 1984;738(4):181–201. doi: 10.1016/0304-419x(83)90003-3. [DOI] [PubMed] [Google Scholar]
- 64.Denissenko MF, Pao A, Tang M, Pfeifer GP. Science. 1996;274(5286):430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]