Abstract
The large (L) proteins of non-segmented, negative-strand RNA viruses, a group that includes Ebola and rabies viruses, catalyze RNA-dependent RNA polymerization with viral ribonucleoprotein as template, a noncanonical sequence of capping and methylation reactions, and polyadenylation of viral messages. We have determined by electron cryomicroscopy the structure of the vesicular stomatitis virus (VSV) L protein. The density map, at a resolution of 3.8 Å, has led to an atomic model for nearly all of the 2109-residue polypeptide chain, which comprises three enzymatic domains [RNA-dependent RNA polymerase (RdRp), polyribonucleotidyl transferase (PRNTase), and methyl transferase] and two structural domains. The RdRp resembles the corresponding enzymatic regions of dsRNA virus polymerases and influenza virus polymerase. A loop from the PRNTase (capping) domain projects into the catalytic site of the RdRp, where it appears to have the role of a priming loop and to couple product elongation to large-scale conformational changes in L.
Keywords: RNA-dependent RNA polymerase, RNA capping, cryoEM single-particle analysis
Introduction
The non-segmented negative-strand (NNS) RNA viruses include some of the most lethal human and animal pathogens, including Ebola virus and rabies virus. Their multifunctional, large (L) polymerase proteins, carried within the virions (Baltimore et al., 1970), have biochemical properties that distinguish them from most other RNA polymerases of viruses or of their hosts. In addition to their RdRp activity (Emerson and Wagner, 1973), NNS RNA virus L proteins catalyze an unusual sequence of mRNA capping reactions (Hercyk et al., 1988), and the RdRp itself polyadenylates the viral message (Hunt et al., 1984). A nucleocapsid (N) protein sheath coats the genomic RNA, and the viral polymerase uses this N-RNA complex as template, rather than uncoated RNA.
Our understanding of RNA synthesis in NNS RNA viruses comes principally from studies of vesicular stomatitis virus (VSV) -- an enveloped, bullet-shaped, rhabdovirus, closely related to rabies virus. VSV causes an acute disease of livestock. The VSV L protein does not bind the N-RNA template directly but requires a cofactor, the viral phosphoprotein (P), as a bridge (Green and Luo, 2009). Delivery of the N-RNA-L-P complex into the cytoplasm when the virus enters a cell initiates infection. Transcription starts at the 3′ end of the genome and produces a triphosphate 47-nucleotide leader RNA, followed by sequential transcription of five capped and polyadenylated mRNAs (Abraham and Banerjee, 1976; Ball and White, 1976; Whelan and Wertz, 2002). At each gene junction, L terminates synthesis of the upstream gene, adding a polyadenylate (polyA) tail by iterative transcription of a U7 tract (Barr and Wertz, 2001; Barr et al., 1997; Stillman and Whitt, 1997). It then transcribes the downstream gene. Roughly 30% attenuation occurs at each successive gene, with dissociation of the template (Iverson and Rose, 1981). Replication also starts at the 3′ end of the genome, but encapsidation by newly synthesized N accompanies synthesis of the nascent RNA strand, and in this mode, L ignores all the cis-acting signals that dictate sequential transcription of mRNAs (La Ferla and Peluso, 1989; Patton et al., 1984; Peluso and Moyer, 1983).
The various enzymatic activities of L are tightly linked. A GDP: polyribonucleotidyltransferase (PRNTase) adds the cap structure (Ogino and Banerjee, 2007) when the nascent RNA chain length has reached 31 nucleotides, as shown by artificially stalling transcription at various chain lengths (Tekes et al., 2011). The unconventional mechanism of cap addition proceeds through a covalent adduct between a histidine residue on L (H1227) and the monophosphate nascent RNA, which is transferred onto a GTP-derived GDP acceptor (Li et al., 2008; Ogino and Banerjee, 2007). Failure to cap results in premature termination of transcription, linking cap addition to RdRp processivity (Li et al., 2008; Stillman and Whitt, 1999; Wang et al., 2007). Subsequent methylation is first at the 2′O position on the ribose of the first nucleotide and then on the N7 of the capping guanylate -- opposite to the typical order of modification (Rahmeh et al., 2009). Failure to methylate, either by exogenously manipulating the concentration of S-adenosyl homocysteine (SAH) to compete with the S-adenosyl methionine methyl donor or by mutating critical catalytic residues in the methylase domain, can result in hyper-polyadenylation of mRNA transcripts, linking the methylation activity to the RdRp (Galloway and Wertz, 2008; Li et al., 2009; Rose et al., 1977). Both cap addition and subsequent methylation require specific sequence elements within the first 10-nt of the mRNA (Wang et al., 2007). The uncapped leader lacks those signals (Li et al., 2008; Ogino and Banerjee, 2007).
During RNA synthesis, a few subunits of N dissociate from the template RNA for access to the catalytic site of RdRp and then reassociate as the process continues (Albertini et al., 2006; Green et al., 2006). P may help coordinate these events (Green and Luo, 2009). There are 9 nucleotides associated with each N subunit in the RNP, but we do not yet know the number of transiently dissociated N subunits (and hence the length of the RNA segment inserted into the polymerase). Full polymerase processivity and correct recognition of the cis-acting signals in the viral genome both require N (Morin et al., 2012).
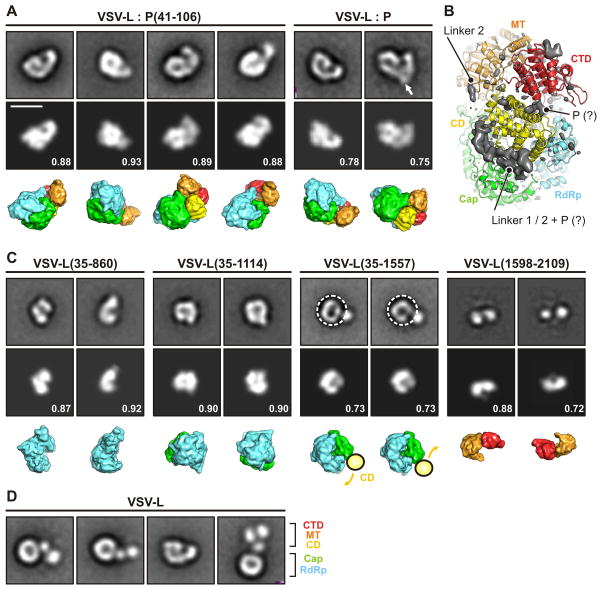
Images from negative-stain electron microscopy (EM) of purified VSV L show a ring-like “core” decorated by a set of 3 variably oriented globular appendages (Rahmeh et al., 2010). Truncations of L map the RdRp to the ring-like domain. Formation of a complex with P causes the globular appendages to reorganize into a compact tail, wrapped onto one side of the ring (Rahmeh et al., 2010). Complex formation with P enhances RdRp initiation and processivity, but P itself has no enzymatic activity. A fragment of P comprising residues 35–106 is sufficient to induce the conformational rearrangement (Rahmeh et al., 2012). Segments in the center and at the C-terminal end of the P polypeptide chain mediate dimerization and N-RNA binding, respectively (Ding et al., 2006; Green and Luo, 2009). Images from negative-stain EM of purified L-P complexes are a mixture of single and dimeric L species, in which the two L molecules have variable relative orientation (Rahmeh et al., 2010).
We have determined the structure of a complex of L with P(35–106) by electron cryomicroscopy (cryoEM). Into a density map at 3.8 Å resolution, we have built a nearly complete model of the 2109 residue polypeptide chain. The result shows that even for a fully asymmetric structure with molecular mass less than 250 kDa, the features in a density map from single-particle cryoEM can be well enough defined for a complete de novo chain trace. We distinguish five domains: three -- an RNA-dependent RNA polymerase (RdRp), an mRNA capping domain, and a methyltransferase domain -- with assigned enzymatic activity and two -- a connector between the capping and methyltransferase domains and a C-terminal domain -- that appear to have largely organizational roles. We suggest that the conformation of the protein in the complex we have examined corresponds to an initiation state, ready to accept the 3′ end of a template. Elongation beyond one or two nucleotides will require a large-scale domain reorganization.
Results
CryoEM structure determination
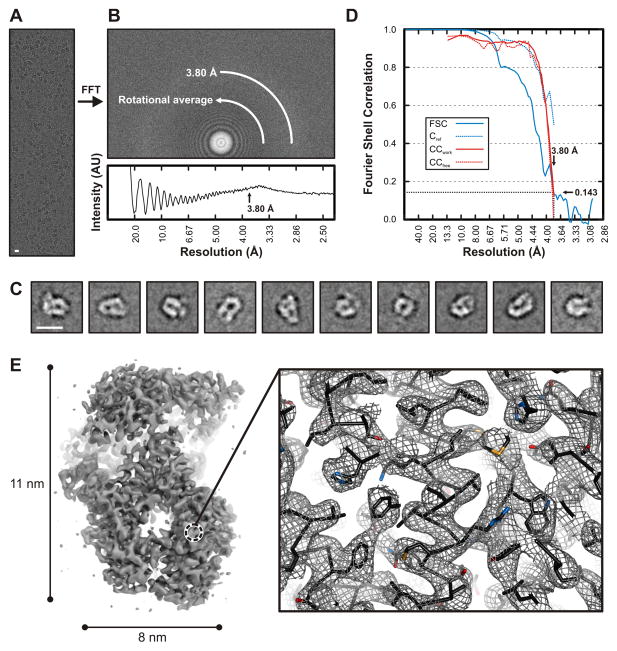
We recorded images from VSV L bound with P(35–106), from grids prepared for cryoEM as described in Experimental Procedures. All data were taken on an FEI F20 microscope with a Gatan K2 Summit detector. Following two-dimensional classification with IMAGIC (van Heel et al., 1996) and TIGRIS (http://tigris.sourceforge.net) and calculation of an initial three-dimensional reference density with EMAN2 (Tang et al., 2007), we used FREALIGN (Lyumkis et al., 2013) for refinement and three-dimensional classification (see Experimental Procedures). Fig. 1 shows images, summarizes stages of the analysis, and illustrates the final 3.8-Å resolution density map.
Figure 1. Electron cryomicroscopic reconstruction of VSV-L at 3.8 Å resolution.
(A) Raw image of VSV-L particles in vitreous ice recorded at 1.8 μm defocus (scale bar = 10 nm).
(B) Power spectrum of the image shown in (A), with plot of the rotationally averaged intensity versus resolution. Arrow indicates the spatial frequency corresponding to 3.8 Å resolution.
(C) Representative class averages (scale bar = 10 nm).
(D) Fourier shell correlation analysis: FSC, correlation between the half-set 3-D reconstructions (solid blue line); Cref, estimated correlation between the final map and a perfect reference map containing no errors, calculated from FSC (dotted blue line) (Rosenthal and Henderson, 2003); CCwork (solid red line) and CCfree (dotted red line), correlation between the final map and refined model for working and test set of structure factors, respectively.
(E) Right, overview of VSV-L reconstruction. In the view shown, the particle (240 kDa) is molecular weight of the VSV-L protein is approximately 110 Å long and 80 Å wide. Right, close-up view of a representative region in the polymerase domain (RdRp). The volume shown in close-up is from the protein interior, not on the RdRp surface. Density is shown as gray mesh; polypeptide-chain backbone of the refined model, as black ribbon; side-chain atoms, as sticks (carbons, black; nitrogen, blue; oxygen, red; sulfur, orange).
Note the continuous backbone density, α-helical grooves and resolution of bulky side chains -- features that allowed building and stereochemical refinement of the atomic model.
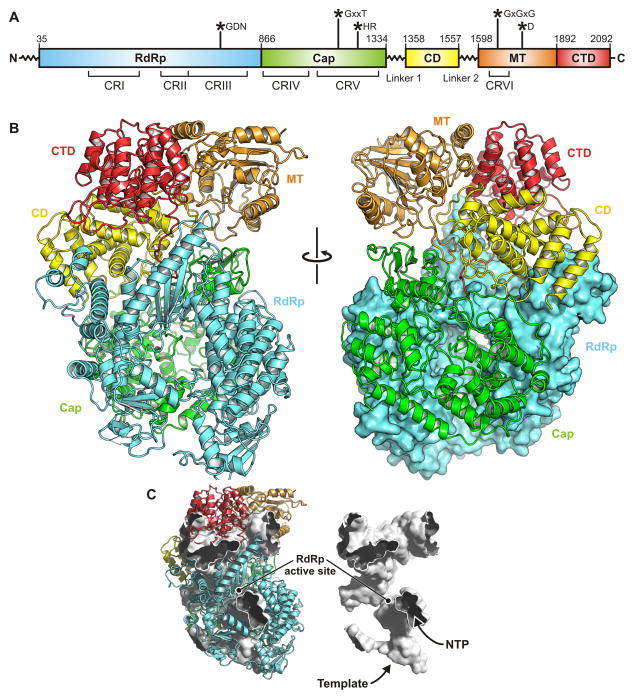
Domain organization of VSV-L
We traced the polypeptide chain from residue 35 to the carboxy terminus, residue 2109, leaving out poorly ordered linker segments 1335–1357 and 1558–1597 and a short, poorly ordered loop 1840–1849. Some segments of approximately 10–15 residues between 1100 and 1334 and between 1358 and 1557 are in poor density, and the chain trace in those regions is approximate. Because the beginning and end of the segment and “puddles” of density leave little uncertainty about the overall course of the segment in question, we have kept those residues in the model. We list the specific segments in question in the Experimental Procedures section. We paid considerable attention to correct stereochemistry during model building, but to adjust both the fit and the stereochemistry beyond the capacity of visual inspection, we carried out one round of refinement, by calculating structure factors from the map and using both amplitudes and phases in the target function, with secondary-structure hydrogen bonds imposed as restraints. After some minor adjustments and one more cycle, Rfree and Rwork were 29.6 and 26.2, respectively (Table S1 and Fig. S1).
We have assigned residues to domains and linkers as follows: RdRp, 35–865; capping domain, 866–1334; linker 1, 1335–1357; connector domain, 1358–1557; linker 2, 1558–1597; methyl transferase, 1598–1892; C-terminal domain, 1893–2109 (Figs. 2 and S2; Data S1). The boundaries between the capping domain and the connector domain and between the latter and the methyl transferase are evident from the disordered linkers that intervene. The boundary between the RdRp and the capping domain corresponds to a previously identified tryptic cleavage site. Although the interface between the two domains is relatively extensive and well packed, we have shown that fragment 1–860 can be expressed independently (Rahmeh et al., 2010). The boundary between the connector domain and the methyltransferase domain also corresponds to two independently stable fragments: 1–1593 and 1594–2109. Between the methyltransferase domain and the C-terminal domain is an extended but relatively polar interface. Negative-stain electron microscopy of a fragment that includes residues 1594–2109 shows that stain can penetrate between the two domains and that their connection might be flexible (Rahmeh et al., 2010).
Figure 2. Structure of VSV-L.
(A) Domain organization of VSV-L. The polymerase domain (RdRp) is in cyan; capping domain (Cap), green; connector domain (CD), yellow; methyl transferase (MT), orange; C-terminal domain (CTD), red. Amino-acid residue numbers indicate functional domain boundaries. Flexible linkers 1 and 2 connect Cap to CD and CD to MT domain, respectively. Conserved regions within L proteins of non-segmented negative-strand (NNS) RNA viruses are labeled CR I – VI. Asterisks indicate the position of active site residues.
(B) Ribbon diagram of VSV-L polypeptide chain; domains colored as in (A).
(C) Substrate channels and internal cavities of VSV-L, depicted as white surface enclosed by the structure in ribbon representation. In this orientation, the entrance to the template channel leading to the active site faces down; the channel runs between the RdRp and capping domains. Nucleotides can access the RdRp active site through the channel in the foreground.
See also Figure S2.
There are well defined secondary-structure features in the density for each of the domains, leading us to believe that poor density for certain segments reflects local disorder, rather than failure of 3-D classification algorithms to detect overall variation in position or orientation of any particular domain with respect to the others.
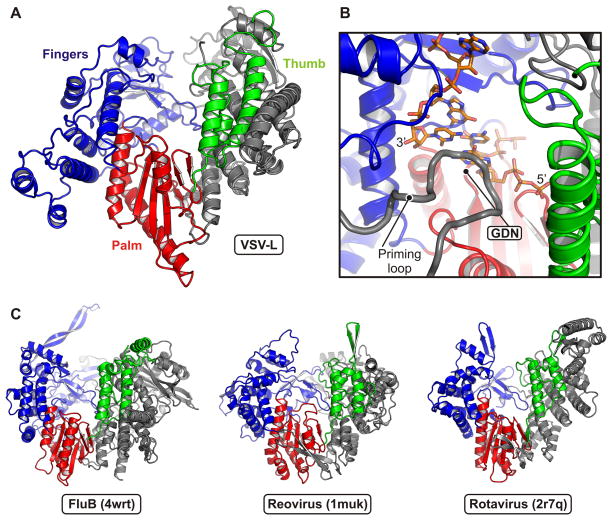
RdRp
The RdRp has at its core a right-hand, “fingers-palm-thumb” structure (residues 360–865), common to a very large group of RNA and DNA polymerases (Fig. 3). The catalytic site on the palm is in a deep channel between the fingers and thumb subdomains (extended from the palm as if in a loose hand grip). Appended to the core on the N-terminal side is a globular region (residues 1–359) that closes the channel on one end and reinforces the relatively slender thumb subdomain.
Figure 3. Polymerase (RdRp) domain.
(A) Structure of the RdRp domain. Residues 35–865 are shown in ribbon representation in conventional orientation (viewed from inside the surrounding “cage” and upside down with respect to the view in Fig. 2). The palm subdomain is in red; the fingers, blue; the thumb, green. The N-terminal region is gray.
(B) Close-up view of the active site. Palm, fingers and thumb are colored as in (A). The GDN active site motif is at a β-hairpin in the palm domain. A model for the positions of the template RNA strand and two nucleotides is derived from the reovirus λ3 initiation complex (PDB 1n1h) after superposition on VSV-L RdRp. The priming loop (residues 1157–1173) intruding from the capping domain (gray) positions the initial nucleotide of the transcript.
(C) Similarity of the VSV-L RdRp domain to those of other viral polymerases. Structures of influenza virus B (PDB 4wrt; PA residues 248–716 and PB1 residues 1–616), reovirus λ3 (PDB 1muk; residues 2–890) and rotavirus VP1 (PDB 2r7q; residues 2–778) are shown with the same orientation and coloring scheme as the VSV-L RdRp in (A).
From the appearance of VSV-L in negative-stain electron microscopy, and in particular from the size and staining of a “doughnut-like” part, we suggested that the RdRp might be similar in cage-like structure to the dsRNA virus polymerases (Rahmeh et al., 2010). Those enzymes have their catalytic sites at the center of an enclosed cavity, connected to the exterior by four channels, for template entrance, template exit, transcript exit, and NTP access (Lu et al., 2008; Tao et al., 2002). Comparison of the chain trace with their structures shows that this suggestion was correct, with one modification. The dsRNA virus RdRps have a C-terminal “bracelet” domain that encircles the exit path for the template and includes a site for binding the methyl G cap on the non-template, plus-sense strand (Lu et al., 2008; Tao et al., 2002). In VSV L, the capping domain, which has no structural similarity to the bracelet domain of the dsRNA virus RdRps, occupies the corresponding space. That is, residue 865, which we have taken as the end of the RdRp, is at the C-terminus of the thumb.
We compared the positions of secondary structural elements in VSV L, reovirus λ3 (Tao et al., 2002), rotavirus VP1 (Lu et al., 2008), and the heterotrimeric influenza virus polymerase (Pflug et al., 2014; Reich et al., 2014). The secondary structural elements with correspondences in the three other polymerases extend from about residue 107 in VSV-L to the end of the RdRp domain (Fig. 3). The analogous parts of reovirus λ3 encompass residues 150 to 860 (approximately); those of rotavirus VP1, residues 135 to 750; those of human influenza virus B polymerase, residues 415 to the C-terminus (714) of the PA subunit and residues 8–586 of the PB1 subunit. The homology thus extends from the middle of PA into PB1. The region in common between VSV-L and influenza virus PB1 corresponds to the fingers-palm-thumb core structure, and the region shared with PA is a large part of the RdRp N-terminal domain.
Capping domain
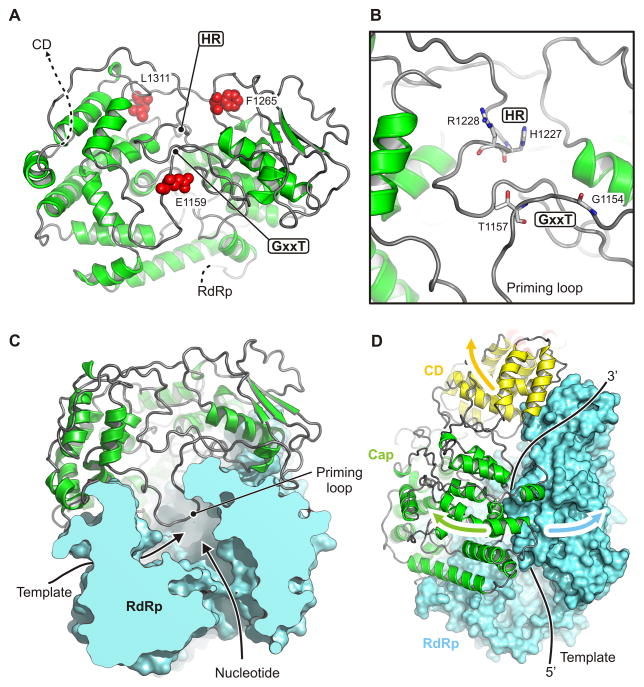
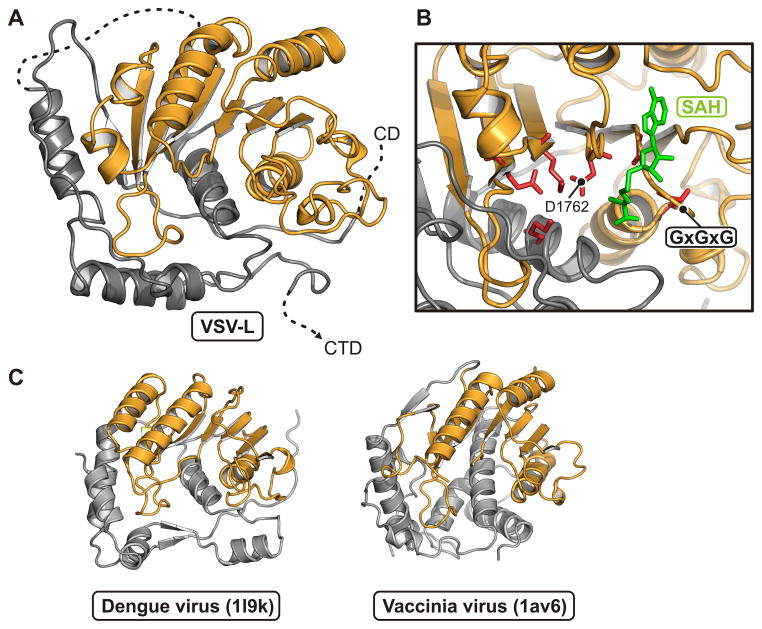
Unlike the corresponding host-cell process, the capping reaction of NNS RNA viruses proceeds from a covalent linkage between the 5′ end of the RNA and a histidine residue, with attack on that linkage by a guanosine nucleotide. The enzyme is thus a polyribonucleotidyl transferase (PRNTase) rather than a guanylyl transferase. Two conserved motifs -- GxxT and HR, separated by about 70 residues -- mark the catalytic site (Fig. 4A,B). The former participates in guanosine nucleotide binding; the latter is the site of covalent RNA attachment. The domain has no structural homologues that we could detect with standard search methods. The largely α-helical, N-terminal half (residues 866–1100), which abuts the polymerase domain, is well ordered. The C-terminal half (1100–1334) has several poorly defined segments, including the loop that bears the HR sequence. Despite uncertain definition of side chains, the separation of the two conserved sites is about 10 Å, appropriate if a GTP bound at the former is to attack the histidine-liganded 5′ phosphate of the nascent RNA at the latter (Fig. 4B). Positions corresponding to sites in human respiratory syncytial virus (hRSV) of resistance mutations to a small-molecule capping inhibitor (Liuzzi et al., 2005) impinge on the active site from three sides (Fig. 4A); their locations, and the relatively poor definition of the active site in the map, suggest that activation of the domain, perhaps by binding the 5′ end of the nascent message, induces a conformational rearrangement, similar to the domain closures seen in many enzymes when they bind their substrate. Two candidate Zn sites, one with clear density where two Cys (residues 1120 and 1123) and two His (1294 and 1296) ligand the likely Zn ion, and one with three Cys (residues 1081,1299, and 1302) and a Glu (1108), contribute structural integrity to the capping domain. The sites are close to each other and well outside the catalytic center. In both cases, the liganding residues are present as a conserved set in most NNS virus L proteins and absent as a set in the others.
Figure 4. Capping domain.
(A) Structure of the capping domain. Residues 866–1334 in ribbon representation. Motifs GxxT and HR are sites of guanosine nucleotide binding and of covalent RNA attachment, respectively. Residues corresponding to positions of inhibitor-resistance mutations in human RSV polymerase (Liuzzi et al., 2005) are shown as red spheres.
(B) Close-up of the active site.
(C) Configuration of the priming loop in VSV-L. Only the RdRp and capping domains are shown. The priming loop (residues 1157–1173) protrudes from the capping domain into the active site of the RdRp domain.
(D) Proposed domain shifts to allow transcript elongation and eventual template release.
A loop between residues 1157 (the threonine of the GxxT motif) and 1173 projects back into the cavity of the polymerase domain (Figs. 3B, 4C and S3). The poorly ordered tip of this loop occupies the same position as the priming loop in the reovirus polymerase (Tao et al., 2002). The loop in VSV-L polymerase domain that corresponds to the λ3 priming loop is shorter than its reovirus homologue, and the capping-domain loop projects over it. Neither polymerase requires a polynucleotide primer to initiate, and the priming loop in the reovirus polymerase supports the initiating nucleoside triphosphate. As elongation proceeds, the tip of the loop recedes to make room for the dsRNA replication product or for the short double-stranded region just upstream of the newly added nucleotide during transcription (Tao et al., 2002). This loop, which contacts the minor groove of the nascent product, may also enhance fidelity, by retarding elongation of mismatches detected by poor minor-groove geometry and allowing more time for ATP-based pyrophosphorolysis of the mismatch. The position of the likely priming loop of VSV-L on the capping domain, adjacent to the GxxT residues, suggests coupling of capping to initiation of polymerization.
Connector domain
The connector domain is a bundle of 8 helices (Fig. 2B); it appears to have largely an organizational role in positioning or spacing the catalytic domains. Disordered linkers, 23 and 40 residues long, respectively, lead into and out of the connector domain. The endpoints of these linkers in well-defined density show that they must occupy an extended groove between the capping and connector domains; the groove also extends into the interface between the capping and methyltransferase domains (Fig. 5). Strong, low resolution density features fill this groove, but they are not sharp enough to suggest particular linker conformations (Fig. 5B). The location of P indicated by negative-stain electron microscopy (Figs. 5A and S4 and Table S2) leads us to suggest that the groove also holds some or all of the P fragment present in the L-P complex we have imaged. Because P(35–106) locks the smaller domains of L into a fixed configuration, it is plausible that it might do so by stabilizing folded structures for the two linker segments -- gluing them down, so to speak, alongside the connector domain (Fig. 5B).
Figure 5. Domain reorganization.
(A) Projection angle matching between class averages of negatively stained complexes of VLS-L and P protein (top row) (Rahmeh et al., 2012; Rahmeh et al., 2010) and projections calculated from the model (middle row). The bottom row shows the model in the same orientation with the individual domains colored as in Fig. 2. Numbers are correlation coefficients between model and negative-stain class averages. VLS-L:P(35–106) corresponds to the structure determined here. In the panel for VLS-L:P, an arrow indicates additional density observed in some class averages that we attribute to the bound P dimer. Scale bar = 10 nm.
(B) Difference density map (mapobserved - mapmodel) calculated to 5 Å resolution and shown together with the model. The map shows density present in the image reconstruction that could not be fit with a molecular model. Strong density -- presumably from linkers 1 and 2, which enter and leave this density at defined points, and with potential contribution from P (tentative assignment indicated by “?”) -- lines the groove between capping and connector domains. We have not attempted to interpret the small, low-resolution feature at the upper right.
(C) Projection angle matching of VSV-L fragments. For VSV-L(35–1557), the negative-stain class averages suggest a conformationally variable connection between the connector domain (CD) and the polymerase (RdRP) and capping domain (Cap). We therefore selected only the “doughnut” part of the image and aligned residues 35–1334.
(D) Full-length VSV-L without P. CD, MT and CTD extend in variable orientation from the RdRp-Cap doughnut.
Methyl transferase domain
The methyltransferase domain has the structure characteristic of many other domains that catalyze transfer of a methyl group from S-adenosyl methionine (Figs. 6 and S5). It methylates both the ribose O2′ and the guanosine N7 (Rahmeh et al., 2009). Most of the domain superposes extremely well on the flavivirus methyltransferases, also dual specificity enzymes (Egloff et al., 2002; Ray et al., 2006; Zhou et al., 2007). Evidence for functional feedback from the VSV-L methyltransferase to the RdRp comes from the observation that addition of S-adenosyl homocysteine, which inhibits methylation, leads to hyperpolyadenylation; mutations that prevent methyl transfer have a similar effect (Galloway and Wertz, 2008; Li et al., 2009; Rose et al., 1977). The methyltransferase domain contacts both the connector and the capping domains, but it has no direct contact with the RdRp. Moreover, there is no obvious “tunnel” that would allow the 5′ end of the transcript to move from the catalytic site of the capping domain to the catalytic site of the methyltransferase domain. We conclude, as we discuss in greater detail below, that the L protein probably undergoes a substantial conformation change following initiation of polymerization and that the inter-domain communication we see in this structure is relevant to formation of the first one or two phosphodiester bonds, but not to subsequent elongation and 5′-end modification.
Figure 6. Methyl transferase domain.
(A) Structure of the methyl transferase: residues 1598–1892 in ribbon representation. The consensus fold of the S-adenosyl methionine-dependent methyl transferase subdomain is in orange. The N-terminal and C-terminal regions, in gray.
(B) Close-up of the active site. The SAM/SAH binding-site motif, GxGxG, is between β1 and αA. An SAH molecule (green) is derived from a superposition of its complex with dengue virus NS5 MT (PDB 1l9k). Residues that participate in the methyl transferase activity are in red.
(C) Comparison of VSV-L MT domain with other viral AdoMet-dependent methyl transferases. Structures of dengue virus NS5 MT (PDB 1l9k; residues 7–267) and vaccinia virus VP39 MT (PDB 1av6; residues 3–297) are shown in the same orientation and color scheme as in (A).
See also Figure S5.
C-terminal domain
Like the connector domain, the C-terminal domain, which terminates in a ca. 25-residue long C-terminal “arm”, appears to have an essentially organizational role (Fig. 2B). It is largely an α-helical bundle, but a projecting, almost beak-like, β-hairpin supported by a second interhelical loop, imparts a noticeable asymmetry. The C-terminal arm, a feature that appears from sequence alignments to be conserved among NNS viral polymerases, but variable in length, extends back against the RdRp, augmenting the β-hairpin that bears the catalytic Asp-Asn sequence at its tip, and terminates at the three-way junction of the capping, connector and methyltransferase domains, where it has one or more contacts with each. The arm thus contributes to closing the multi-domain structure we see in the L-P complex, further stabilized by the phosphoprotein, P.
Template channel
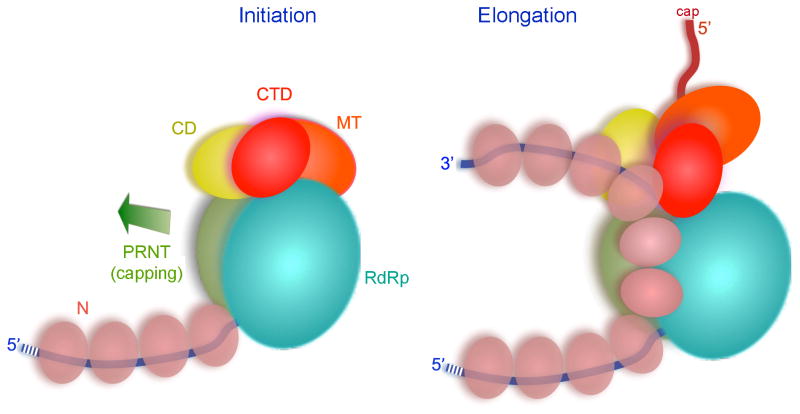
Superpositions of related positions in reovirus λ3 (Tao et al., 2002) and rotavirus VP1 (Lu et al., 2008) have allowed us to model a bound template and a template-primer-NTP complex, because λ3 was catalytically active in the crystals studied used to determine the structure and VP1 in its crystals incorporated template in a sequence-specific register (Tao et al., 2002). The template entrance channel is at the interface between the RdRp and the capping domain (Fig. 2C). Polar and especially basic residues project into the groove from both sides. As in all polymerases of this family, the template runs across a “fingers loop” (residues 523–545 in VSV-L) and twists sharply to present the templating base to the catalytic center. A hydrophobic residue in the loop (Phe 541 in VSV-L) bears on the templating base to enforce correct base pairing with the incoming nucleoside triphosphate. For initiation at the 3′ end of the viral RNA (either for replication or for transcription of leader RNA), the priming nucleoside triphosphate will rest against the loop from the capping domain described above (Fig. 3B). Any further elongation, after forming the initial phosphodiester bond, will require this loop to move, and substantial elongation will almost certainly require displacement of the entire capping domain (Fig. 4D). Indeed, we suggest that to accommodate transcriptional elongation, the entire array of smaller domains may reorganize.
Domain reorganization
The configurations of VSV-L we have characterized in published work by negative-stain electron microscopy illustrate the potential for large-scale domain reorganization (Rahmeh et al., 2010). Images of L alone show a core “doughnut”, which admits stain at its center, three globular appendages, in apparently variable positions and orientations with respect to each other and to the core. Addition of P, or of the peptide, residues 35–106, that we have used to stabilize the complex studied here, locks the appendages in place (Rahmeh et al., 2012; Rahmeh et al., 2010). Many of the projections of this locked structure resemble a figure “6”. Class averages from these images agree extremely well with projections of the structure we describe (Fig. 5A), as do class averages from images of four different L fragments (Fig. 5C). One of these (1–860) corresponds precisely to the RdRp. Another, 1–1121, includes the RdRp and the largely helical, N-terminal half of the capping domain. The tryptic cleavage that initially generated that fragment is in a surface loop. The last fragment previously imaged by negative staining comprises the methyltransferase and C-terminal domains (Fig. 5C).
These comparisons show that we need to modify our initial assignment of the three globular appendages to the capping domain, the methyltrasferase domain, and one unassigned domain (Rahmeh et al., 2010). The capping domain is part of the doughnut, and the appendages correspond to the connector, methyltransferase and C-terminal domains, respectively (Fig. 5D). The linkers between the capping domain and the connector and between the connector and the methyltransferase clearly allow the latter two to move away from the rest of the molecule; good definition in negative stain for the third globular appendage suggests that in the unlocked structure, the C-terminal arm also pulls away from the RdRp. Many of its interactions, as it inserts back against the rest of the molecule in the structure we have determined, are indeed with the connector and methyltransferase domains.
Images of negatively stained complexes of L with inact, dimeric P often show two, linked, figure-“6” L molecules, but occasionally the P dimer does not recruit a second L and appears as a surface feature on the hook of the “6” (Fig. 5A). Comparison of the L structure with these projections is consistent with our proposal that the interaction with P(35–106) that stabilizes the “6” conformation is with the linker segments at either end of the connector domain (Fig. 5B).
Discussion
CryoEM
High-resolution cryoEM structure determination has until recently relied on either high symmetry or large size -- for example, icosahedral viruses, which have both, or ribosomes, which are large enough to produce reasonable contrast for getting started with iterative determination of particle orientations and centers (Grigorieff and Harrison, 2011). Developments in cryoEM during the past five years have now allowed us to determine the molecular structure of an asymmetric protein of total mass less than 250 kDa. Dose fractionation (“movies”), enabled by use of a direct electron detector, and refinement and maximum-likelihood classification procedures (Lyumkis et al., 2013), implemented in FREALIGN, were crucial for achieving a resolution adequate to build an atomic model.
Sequential transcription
A de novo initiation event with ATP as initiating nucleotide appears to start synthesis of each mRNA transcript. We interpret our structured as that of an early initiation state, representing an L-P complex ready for loading onto the end of the template to synthesize leader RNA. During the transition to elongation the priming loop – contributed by the capping domain – must shift out of the way to accommodate the product (Fig. 7). Inspection further suggests that after addition of only a few more nuceotides, the capping domain as a whole must withdraw from tight contact with the RdRp to allow further elongation, as there do not appear to be clear exit channels for transcript and template. Upon termination of a transcript, the polymerase reinitiates on the next gene, but the efficiency of producing the succeeding transcript is only about 70% (Iverson and Rose, 1981). The template entrance channel in VSV-L is at the interface of the capping domain and the RdRp, and dissociation of the template will be straightforward (when initiation or early elongation aborts) if that interface opens as suggested. Otherwise the entire template would have to thread through the active site and emerge through another channel. Transcription of the downstream gene probably requires reestablishing the inter-domain contact seen in our structure, so that the priming loop can reinsert into the active site of the RdRp domain for subsequent de novo initiation.
Figure 7. Model for transcription of an RNP complex by VSV-L.
Left: initiation complex, with domains organized as in the structure described here. Viral genomic RNA in blue, N-protein in beige, domains of VSV-L in the colors used in Fig. 2. Arrow shows direction of capping domain displacement required for the transition to an elongation complex. Right: elongation requires both displacement of the capping domain (with likely accompanying reorganization of the CD, MT, and CTD) and displacement of 2–3 N subunits from the template residues looped into the polymerase. The N subunits are shown linked as a continuous chain, as suggested by their structure (Green et al., 2006). The emerging transcript is in red.
See Experimental Procedures.
Coupling of capping, polymerase, and methyltransferase activities
Displacement of the capping domain from the RdRp as elongation proceeds might have two consequences. First, the active site of the PRNTase might reorganize (e.g., by “domain closure”) into a better ordered configuration than the one we see in the present structure. Second, because the capping domain faces both the connector and methyltransferase domains, its displacement might also induce rearrangement of the rest of the capping machinery. A large-scale reorganization of this kind could account for some of the observed functional crosstalk between the capping and polymerase activities.
A cap is added only when the length of a transcript has reached 31 nucleotides (Tekes et al., 2011). In vitro, very short (up to 5-nucleotide) transcripts can be capped in trans by L, but this process is inefficient and fails completely with longer transcripts (Li et al., 2008; Ogino and Banerjee, 2007). Conversely, mutations in L that disrupt cap addition cause premature termination (Li et al., 2009; Li et al., 2008). Mutations in the specific, cis-acting signals at the 5′ end of the nascent strand, which are absent from the leader RNA, also block cap addition and result in premature termination of that transcript (Li et al., 2008; Ogino and Banerjee, 2007; Stillman and Whitt, 1997; Wang et al., 2007). The precision of the 31-nucleotide requirement suggests that the reorganized structure that allows elongation is a well-defined state, rather than a loosely ordered one.
Reorganization of the capping machinery can also account for why mRNA cap methylation requires no additional chain length (Tekes et al., 2011). In the configuration represented by the structure we have determined, the catalytic sites for capping and methylation are distant from each other. If the smaller domains move away from the polymerase core, the capped, nascent RNA could probably release from the capping enzyme and gain immediate access to the methylase domain. Methylation in trans can occur under some circumstances, but previous work has shown that transcripts stalled at a chain length of 31-nt are fully methylated -- presumably in cis -- by the stalled L (Tekes et al., 2011).
Inhibition of mRNA cap methylation by high concentrations of S-adenosyl homocysteine can result in hyper-polyadenylation, demonstrating a linkage between the methylase and RdRp domains (Galloway and Wertz, 2008; Li et al., 2009; Rose et al., 1977). The RdRp domain of L carries out polyadenylation by iterative transcription of a gene-end U tract element. Some but not all mutations that inhibit methylation result in the hyper-polyadenylation phenotype (Galloway and Wertz, 2008), indicating that the crosstalk mechanism is not a readout of cap modification but probably a consequence of interactions between domains and between the protein and the nascent transcript.
The cap methylase of VSV participates in both ribose 2′O and guanine-N7 methylation reactions. The preferred substrate for all other ribose 2′O methyltransferases is 7mGpppN, and like other proteins that recognize the mRNA cap structure – such as eIF-4E – 2′O methylases -- those enzymes position the ribose in the active site by π-π stacking interactions with the 7mG RNA. The order of cap methylation in VSV is reversed. Methylation of 2′O precedes and facilitates subsequent methylation of guanine-N7. The absence of aromatic residues that could participate in such interactions with a 7mGpppN RNA in the VSV methyl transferase is consistent with this altered reaction sequence.
The N protein
The template for polymerase is not naked RNA, but a complex in which the template RNA is encased within the nucleocapsid protein sheath (Fig. 7). In that complex the RNA bases are not accessible to the RdRp of L, and the N protein must transiently dissociate from the RNA for the RdRp to proceed (Green et al., 2006). The structure of L allows us to estimate that 20–25 nucleotides of the template strand are threaded through the polymerase domain. Accordingly, because each molecule of N covers nine nucleotides of RNA, two or three molecules of N must be displaced from the template strand at any one time. Adjacent N subunits in the RNP interact stably, embracing each other through N- and C-terminal extensions (Green et al., 2006). Thus, looping out of template RNA need not entail dissociation of N from the RNP coil. Indeed, if we consider the linked chain of N subunits as the analog of a complementary RNA strand, then the displaced N is the counterpart of a looped-out plus-sense strand during transcription by the related polymerases of dsRNA viruses. This N-protein bridge could account for the precision of the 31-nucleotide length of nascent transcript required for cap addition, perhaps by creating a defined spacing between the RdRp and the popped-out capping domain. The N protein influences L activity, as recognition of the cis-acting signals in the genome requires it and as its presence influences incorporation by L of substituted nucleotide analogs (Morin and Whelan, 2014). It may also be necessary for capping.
The P protein
P is an adaptor that engages both the N-RNA template complex and the L protein. A small, globular domain at the C-terminal end of P (residues 195–265) interacts with the N-RNA complex (Green and Luo, 2009). This domain could in principle move from one subunit to the next as polymerization proceeds. The structure we describe here contains only part of the N-terminal region of P. Although it is poorly ordered in our density map, we suggest that P(35–106) may occupy some of the strong, low resolution density features between the capping and connector domains, locking in the linker segments at both ends of the latter. Depending on the chain polarity and on the flexibility of intervening segments, the C-terminal domain of P could lie near the opening through which RNA enters the active site of the RdRp and thus, through its interaction with N, be part of the process that feeds template rapidly through the polymerase channel.
Homologies and comparisons
Homologous L proteins include those of rabies, Ebola, measles and respiratory syncytial viruses. Alignment of their sequences (Data S1) shows the same overall arrangement of the various domains, identifies the active site residues of the protein for RdRp, PRNTase and methyl transferase activities, and suggests domain boundaries for expressing various fragments of the proteins from VSV and related viruses. All NNS RNA viruses have a polymerase complex that comprises the enzymatic subunit, L, and an equivalent of the VSV phosphoprotein, P. In some cases, additional viral proteins (VP24 in the case of the filoviruses, M2-1 in the case of respiratory syncytial virus) are necessary for full polymerase processivity. The three-dimensional interconnections among domains in the VSV polymerase suggest that binding of these accessory proteins to any of the smaller domains might influence large-scale rearrangements and affect enzyme processivity.
Parts of L are structurally similar to the heterotrimeric polymerase of influenza virus. The major differences are the capping machineries, reflecting the distinction between influenza-virus cap-snatching and NNS RNA-virus cap synthesis (Reich et al., 2014). A structural feature of the flu polymerase that is also absent from the VSV polymerase is the long PB1 arm, which is probably required for a step of mRNA transcription (Pflug et al., 2014; Reich et al., 2014).
The VSV-L structure allows preliminary interpretation of mechanisms for inhibitors of its homologues. For measles virus, resistance to a non-nucleoside analog inhibitor that blocks gene expression maps to the polymerase domain (Krumm et al., 2014). The positions of those mutations, mapped onto VSV-L, flank the GDNQ motif at the RdRp catalytic site, suggesting an allosteric mechanism, like that of the non-nucleoside analogue reverse transcriptase inhibitors for HIV. A compound active against RSV targets the capping domain. The locations of positions in VSV-L that correspond to sites of resistance mutations in RSV-L are consistent with our proposal that a domain closure accompanies activation of the capping activity after polymerization has commenced (Liuzzi et al., 2005).
Experimental Procedures
Protein expression and purification
We expressed N-terminally 6xHis-tagged L protein from the Indiana strain of VSV in Sf21 insect cells and purified the protein as described previously (Li et al., 2008; Rahmeh et al., 2010). We expressed VSV-P(35–106) in E. coli, purified the fragment, and combined it with VSV-L, as described in Supplemental Experimental Procedures.
Electron microscopy
Images of VSV-L:P(35–106) were recorded with a Tecnai F20 electron microscope (FEI) operated at 200 kV, using UCSF Image4 (courtesy Yuemin Li, UCSF) to collect movies on a K2 Summit direct detector (Gatan), operated in superresolution mode with dose fractionation. For each 6 s exposure, we collected 30 frames of 200 ms each, with a total electron dose of 31 e/Å2. Full details are in Supplemental Experimental Procedures.
Image processing
From 1272 movies we picked 356,611 particles, carried out two-dimensional classification with multivariate statistical analysis (MSA) in IMAGIC (van Heel et al., 1996) and K-means classification in TIGRIS (http://tigris.sourceforge.net), determined defocus with CTFFIND3 (Mindell and Grigorieff, 2003), calculated class averages with full contrast transfer function (CTF) correction, and selected good class sums as references for particle alignment and subsequent re-classification, iterating twice. An initial model, calculated with EMAN2 (e2initialmodel.py (Tang et al., 2007)) used 292 class averages. Refinement and three-dimensional classification (3 classes) in FREALIGN (Lyumkis et al., 2013) of the 292 CTF-corrected class averages yielded one class with about 10 Å resolution, which we used as an initial reference for refinement and classification of the full particle stack.
We used FREALIGN to refine and 3D classify (3 classes) the full-dose particle stack, initially using 3x binned images (high resolution limit 10 Å) for 11 cycles of refinement and classification, followed by 160 cycles with unbinned images. At this point, the resolution was about 7 Å. We extracted the best set of 155,443 particles and carried out 100 more cycles of refinement and classification (3 classes), extending the resolution to 6 Å. We then extracted 74,940 particles with the best scores from the two best classes. A final 7 cycles of refinement of angles and shifts (using a 6 Å reference model) alternated between the full-dose (31 e/Å2) stack and a low-dose (12 e/Å2) stack. The final map (3.8 Å resolution at FSC=0.143 criterion) was calculated from the low-dose images.
Model building
We traced the polypeptide chain of the RdRp using the programs O (Jones et al., 1991) and Coot (Emsley et al., 2010), with the reovirus and rotavirus polymerases (λ3 and VP1, respectively) as connectivity guides, and traced the methyltransferase domain, with the consensus fold of S-adenosylmethionine (SAM)-dependent transferases as guide. For the capping domain, connector domain, and C-terminal domain, with no known homologs, we built initial models to fit the density, confirming and adjusting connectivity by reference to the amino-acid sequence. Side-chain density was strong enough in secondary structure elements of each domain to establish sequence register. We checked and corrected the entire structure with O. For the following segments of the capping and connector domains, the density did not allow confident assignment of backbone stereochemistry, and Cα positions will have larger errors than in the rest of the model: 1159–1171; 1210–1226; 1308–1334; 1512–1518; 1534–1541 (see Fig. S6 and Supplementral Experimental Procedures for further details).
Structure refinement
We refined the structure to improve the fit and to optimize stereochemistry, by calculating structure factors from the final map and using them as input to standard crystallographic refinement procedures in PHENIX (Adams et al., 2010). We flagged 4% of the structure factors for cross validation and estimated figures of merit from the phase angle difference between the two half-set reconstructions used to estimate FSC. We carried out several rounds of individually restrained positional and B-factor refinement, including one round of torsion-angle simulated annealing and real-space refinement. We applied secondary structure restraints throughout the refinement and Ramachandran restraints in the final round. We analyzed the final model with MolProbity (Chen et al., 2010). Refinement and model statistics are in Table S1, and a complete account of the refinement protocol is in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Robin Ross for protein expression and purification and Thomas Walz for advice and encouragement. The research was supported by NIH grants GM62580 (to SCH), AI059371 (to SPJW), and AI057159 (New England Regional Center of Excellent for Biodefense and Emerging Infectious Diseases). NG and SCH are Investigators in the Howard Hughes Medical Institute.
Footnotes
Accession Numbers
The EMDB accession code for the cryoEM map reported in this paper is EMD-6337, and the Protein Data Bank (PDB) accession code for the refined atomic coordinates is 5a22.
Author contributions
B.L. and Z.L. designed experiments and acquired and analyzed data; S.J. developed strategies for computational data analysis and analyzed data; A.R. and B.M. helped design experiments and interpret data; N.G. contributed software developments and image analysis; S.C.H. and S.P.J.W. conceived and co-directed the project and contributed to the experimental design; B.L., Z.L., S.J., N.G., S.C.H. and S.P.J.W. all participated in data interpretation and in writing and revising the MS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham G, Banerjee AK. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Ball LA, White CN. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Huang AS, Stampfer M. Ribonucleic acid synthesis of vesicular stomatitis virus, II. An RNA polymerase in the virion. Proc Natl Acad Sci USA. 1970;66:572–576. doi: 10.1073/pnas.66.2.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Wertz GW. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J Virol. 2001;75:6901–6913. doi: 10.1128/JVI.75.15.6901-6913.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Whelan SP, Wertz GW. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Green TJ, Lu S, Luo M. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol. 2006;80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU, Wagner RR. L protein requirement for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1973;12:1325–1335. doi: 10.1128/jvi.12.6.1325-1335.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta cryst D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway SE, Wertz GW. S-adenosyl homocysteine-induced hyperpolyadenylation of vesicular stomatitis virus mRNA requires the methyltransferase activity of L protein. J Virol. 2008;82:12280–12290. doi: 10.1128/JVI.01225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci USA. 2009;106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Current opinion in structural biology. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk N, Horikami SM, Moyer SA. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Smith EF, Buckley DW. Aberrant polyadenylation by a vesicular stomatitis virus mutant is due to an altered L protein. J Virol. 1984;52:515–521. doi: 10.1128/jvi.52.2.515-521.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson LE, Rose JK. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta cryst A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Krumm SA, Yan D, Hovingh ES, Evers TJ, Enkirch T, Reddy GP, Sun A, Saindane MT, Arrendale RF, Painter G, et al. An orally available, small-molecule polymerase inhibitor shows efficacy against a lethal morbillivirus infection in a large animal model. Science Transl Med. 2014;6:232ra252. doi: 10.1126/scitranslmed.3008517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ferla FM, Peluso RW. The 1:1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J Virol. 1989;63:3852–3857. doi: 10.1128/jvi.63.9.3852-3857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Rahmeh A, Brusic V, Whelan SP. Opposing effects of inhibiting cap addition and cap methylation on polyadenylation during vesicular stomatitis virus mRNA synthesis. J Virol. 2009;83:1930–1940. doi: 10.1128/JVI.02162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Rahmeh A, Morelli M, Whelan SP. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, et al. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. The EMBO journal. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin B, Whelan SP. Sensitivity of the polymerase of vesicular stomatitis virus to 2′ substitutions in the template and nucleotide triphosphate during initiation and elongation. J Biol Chem. 2014;289:9961–9969. doi: 10.1074/jbc.M113.542761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Patton JT, Davis NL, Wertz GW. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol. 1984;49:303–309. doi: 10.1128/jvi.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso RW, Moyer SA. Initiation and replication of vesicular stomatitis virus genome RNA in a cell-free system. Proc Natl Acad Sci USA. 1983;80:3198–3202. doi: 10.1073/pnas.80.11.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Morin B, Schenk AD, Liang B, Heinrich BS, Brusic V, Walz T, Whelan SP. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc Natl Acad Sci USA. 2012;109:14628–14633. doi: 10.1073/pnas.1209147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- Rose JK, Lodish HF, Brock ML. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in vitro in the presence of S-adenosylhomocysteine. J Virol. 1977;21:683–693. doi: 10.1128/jvi.21.2.683-693.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Stillman EA, Whitt MA. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman EA, Whitt MA. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73:7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage--structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- Tekes G, Rahmeh AA, Whelan SP. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS pathogens. 2011;7:e1002073. doi: 10.1371/journal.ppat.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Wang JT, McElvain LE, Whelan SP. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J Virol. 2007;81:11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Wertz GW. Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc Natl Acad Sci USA. 2002;99:9178–9183. doi: 10.1073/pnas.152155599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.