Abstract
In our previous work, indolizinoquinolinedione derivative 1 was identified as a Top1 catalytic inhibitor. Herein, a series of 6-substituted indolizinoquinolinedione derivatives were synthesized through modification of the parent compound 1. Top1 cleavage and relaxation assays indicate that none of these novel compounds act as classical Top1 poison, and that the compounds with alkylamino terminus at C-6 side chain, including 8, 11–16, 18–21, 25, 26 and 28–30, are the most potent Top1 catalytic inhibitors. Top1-mediated unwinding assay demonstrated that 14, 22 and 26 were Top1 catalytic inhibitors without Top1-mediated unwinding effect. Moreover, MTT results showed that compounds 26, 28–30 exhibit significant cytotoxicity against human leukemia HL-60 cells, and that compound 26 exerts potent cytotoxicity against A549 lung cancer cells at nanomolar range.
Keywords: Indolizinoquinolinedione, DNA topoisomerase, Inhibitor, Anticancer
1. Introduction
DNA topoisomerase I (Top1) is an essential nuclear enzyme regulating the topology of DNA in many cellular metabolic processes [1,2]. To perform its functions, Top1 cleaves one strand of DNA to form a transient Top1 covalent complex (Top1cc) that allows the relaxation of DNA supercoils by controlled rotation. Afterward, religation of the DNA single-strand break releases Top1 and the relaxed DNA [3]. Inhibition of Top1, and especially drug trapping of Top1cc results in DNA damage, which triggers cell death. Hence, Top1 is a validated target for anticancer agents [4-6].
Top1 inhibitors are classified as Top1 “poisons” and “catalytic inhibitors”. Top1 poison traps Top1cc generally by inhibiting the religation of the DNA single-strand break generated by Top1. In contrast with Top1 poisons, a Top1 catalytic inhibitor inhibits Top1-mediated DNA cleavage. Among the classical Top1 poisons, three camptothecins, topotecan, irinotecan and belotecan (Fig. 1) are widely used clinically for cancer treatment. In spite of their effectiveness in solid tumors (ovary, colon, lung), camptothecins have known limitations, including chemically instability, doselimiting side effects and drug efflux-mediated resistance [6,7].
Fig. 1.
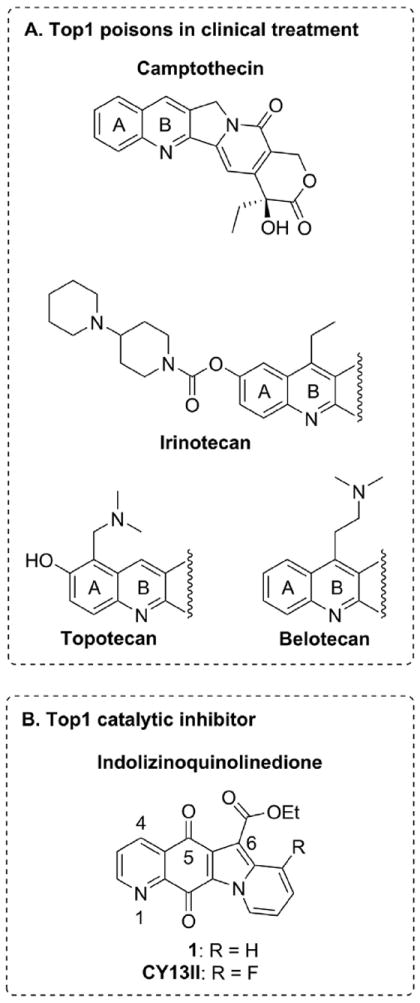
Structures of Top1 poisons and our catalytic inhibitors. (A) The typical Top1 poisons, camptothecin and its clinical derivatives. (B) Our previously developed catalytic inhibitors.
In a recent study, we reported a novel kind of Top1 catalytic inhibitors, the indolizinoquinolinedione derivatives, which inhibit Top1-mediated DNA cleavage reactions and kill cancer cells by arresting their cell cycle progression in G2/M phase [8,9]. Structure activity relationships (SAR) evaluation indicated that the nitrogen atomin the A ring played an important role for cytotoxicity. N,N-syn isomer exhibited higher cytotoxicity and Top1 inhibitory activity than N,N-trans isomer or the corresponding derivatives with two nitrogen atoms in the A ring, such as indolizinophthalazinediones and indolizinoquinoxalinediones [10,11]. This led us to propose that the aminoalkyl side chain enhanced the cytotoxicity and Top1 inhibitory activity of the parent molecule possibly because the cationic side chain under physiological condition could improve cellular uptake and interactions with the Top1-DNA complex [12-15]. In the present study, to obtain compounds with enhanced cytotoxicity and Top1 inhibitory activity, we synthesized a series of 6-substituted indolizinoquinolinedione derivatives and evaluated for their biochemical and biological activities.
2. Results and discussion
2.1. Chemistry
Similar to our reported compound CY13II [8], compound 1 (Fig. 1) is a Top1 catalytic inhibitor, which shows equipotent Top1 inhibitory activity to CPT, and produces no detectable Top1-mediated DNA cleavage and unwinding effects (Supplementary Material). As shown in Scheme 1, compound 1 was modified as a parent molecule to give a series of indolizinoquinolinediones 3–21 with amide side chain at C-6 position. Compound 2 was obtained by hydrolysis of compound 1, which was prepared according to our previously published method [9]. The target amides 3–21 were obtained through acid chloride formation and subsequent amination of compound 2 because it was difficult to synthesize them according to Defant’s one-pot method [15]. According to this synthetic procedure, ester 22 was obtained using 2-(pyridin-2-yl) ethanol as material.
Scheme 1.
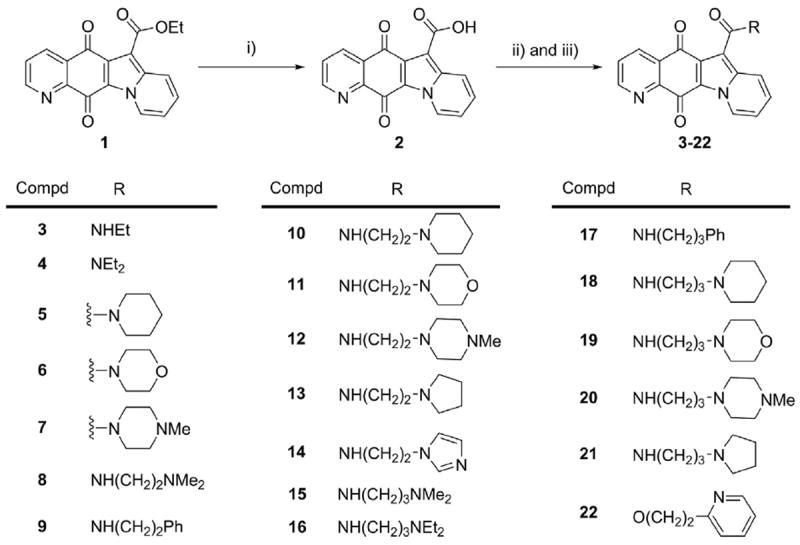
Synthesis of compounds 3–22.
Reagents and conditions: i) 15% K2CO3, i-propanol, refluxing, 24 h. ii) SOCl2, Et3N, CHCl3, refluxing, 5 h. iii) amine or alcohol materials, DMAP, CHCl3, refluxing, 5 h.
To assess the effect of alkylamino terminus of 6-alkyloxy carbonyl substituent, esters 25–30 were synthesized as shown in Scheme 2. Following chlorination, acid 2 was esterified with bromoethanol or bromopropanol to give intermediate 23 or 24, respectively. And then, bromide intermediate reacted with excess amines to give the targets 25–30. The structures of all synthesized compounds were elucidated by using NMR and HRMS spectra.
Scheme 2.
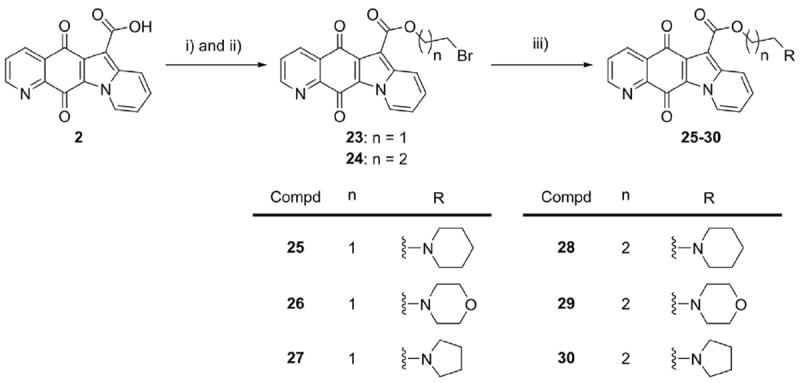
Synthesis of compounds 25–30.
Reagents and conditions: i) SOCl2, Et3N, CHCl3, refluxing, 5 h. ii) 2-bromoethanol or 3-bromopropanol, DMAP, CHCl3, refluxing, 5 h. iii) amines, EtOH, refluxing, 18-24 h.
2.2. Top1 inhibitory activities
The synthesized compounds were tested for induction of Top1 cleavage complexes in Top1 cleavage assay [16]. None of the compounds showed significant activity, which means the synthesized compounds are not Top1 poison. A representative gel of Top1 cleavage assay result is shown in SFig. 2 (Supplementary Material).
The Top1 inhibitory activities of the synthesized compounds were studied using Top1 relaxation assay. As a well-known Top1 inhibitor, camptothecin (CPT) was used as a positive control. The IC50 of CPT (the concentration inhibiting 50% of Top1 catalytic activity) was 25 ± 5.8 μM. The inhibitory activities of the synthesized compounds were semiquantitatively expressed relative to CPT at 25 μM as follows: +, less than 40% of the activity of CPT; ++, between 41% and 80% of the activity of CPT; +++, between 81% and 120% of the activity of CPT; ++++, more than 121% of the activity of CPT. The results, summarized in Table 1, show that the compounds with a terminal alkylamino group at C-6 side chain, including 8, 11–16, 18–21, 25, 26 and 28–30, exhibit enhanced Top1 inhibitory activity. All of them had a Top1 inhibition of ++++ compared to parent compound 1 with Top1 inhibition of +++. As shown in Fig. 2, these compounds exhibited significant Top1 inhibition in dose-dependent manner. On the contrary, the compounds with 6- alkylamino carbonyl group without an alkylamino terminal had decreased Top1 inhibition. For example, compounds 4–6, 9 had Top1 inhibition of +, and compounds 7, 17 had Top1 inhibition of ++. Compound 22, with 2′-pyridinyl ethoxy carbonyl substituent at C-6 position, showed equipotent Top1 inhibitory activity with parent compound 1.
Table 1.
Top1 inhibitory activities and unwinding effect and cytotoxicities of the synthesized compounds.
| Cpd. | Relaxation assaya | Unwinding effect | Cytotoxicity [GI50 (μM)]b
|
|||
|---|---|---|---|---|---|---|
| HL-60 | CA46 | HeLa | A549 | |||
| 1 | +++ | No | 0.30 ± 0.029 | 3.7 ± 1.4 | 22 ± 6.3 | 1.4 ± 0.30 |
| 4 | + | −c | 14 ± 1.1 | 15 ± 2.0 | 46 ± 16 | 64 ± 45 |
| 5 | + | − | 52 ± 7.6 | 13 ± 3.1 | 19 ± 4.6 | 76 ± 36 |
| 6 | + | − | 22 ± 2.2 | 13 ± 1.0 | >100 | 77 ± 6.2 |
| 7 | ++ | − | 5.7 ± 1.9 | 2.6 ± 0.88 | 62 ± 4.3 | 30 ± 1.8 |
| 8 | ++++ | Yes | 1.2 ± 0.18 | 5.9 ± 2.7 | 0.92 ± 0.034 | 0.61 ± 0.20 |
| 9 | + | − | 14 ± 4.1 | 7.9 ± 4.5 | 17 ± 2.2 | 19 ± 13 |
| 11 | ++++ | Yes | 1.2 ± 0.038 | 3.4 ± 0.0089 | 1.6 ± 0.25 | 0.89 ± 0.18 |
| 12 | ++++ | Yes | 2.8 ± 0.60 | 1.1 ± 0.17 | 1.0 ± 0.23 | 0.44 ± 0.34 |
| 13 | ++++ | Yes | 1.2 ± 0.065 | 15 ± 1.2 | 0.78 ± 0.20 | 0.49 ± 0.41 |
| 14 | ++++ | No | 2.3 ± 1.7 | 2.9 ± 1.5 | 5.8 ± 0.58 | 2.3 ± 0.33 |
| 15 | ++++ | Yes | 2.3 ± 1.8 | 1.2 ± 0.086 | 0.96 ± 0.12 | 0.37 ± 0.10 |
| 16 | ++++ | Yes | 1.4 ± 0.043 | 1.2 ± 0.063 | 1.3 ± 0.15 | 1.0 ± 0.055 |
| 17 | ++ | − | 75 ± 22 | 14 ± 0.66 | 29 ± 11 | 15 ± 2.2 |
| 18 | ++++ | Yes | 1.3 ± 0.089 | 1.2 ± 0.086 | 1.2 ± 0.18 | 3.3 ± 5.5 |
| 19 | ++++ | Yes | 1.3 ± 0.040 | 1.1 ± 0.00029 | 5.9 ± 1.2 | 4.2 ± 0.52 |
| 20 | ++++ | Yes | 2.2 ± 1.4 | 3.8 ± 1.1 | 8.9 ± 1.2 | 5.5 ± 2.1 |
| 21 | ++++ | Yes | 1.9 ± 1.1 | 2.7 ± 0.22 | 1.8 ± 0.11 | 1.2 ± 0.066 |
| 22 | +++ | No | 0.11 ± 0.0064 | 2.9 ± 0.19 | 1.1 ± 0.17 | 7.8 ± 0.89 |
| 25 | ++++ | Yes | 0.32 ± 0.070 | 3.0 ± 0.56 | 2.2 ± 0.55 | 0.52 ± 0.54 |
| 26 | ++++ | No | 0.034 ± 0.0012 | 5.8 ± 2.7 | 10 ± 0.18 | <0.01 |
| 28 | ++++ | Yes | 0.025 ± 0.0047 | 0.90 ± 0.036 | 0.48 ± 0.19 | 1.0 ± 0.025 |
| 29 | ++++ | Yes | 0.064 ± 0.0008 | 1.4 ± 0.059 | 2.2 ± 0.43 | 1.9 ± 2.3 |
| 30 | ++++ | Yes | 0.026 ± 0.0021 | 1.1 ± 0.042 | 0.94 ± 0.13 | 0.14 ± 0.15 |
| CPT | +++ | − | 0.015 ± 0.0017 | 0.52 ± 0.064 | 14 ± 4.0 | 1.6 ± 0.23 |
Top1 inhibitory activity was semiquantitatively expressed relative to CPT at 25 μM of tested concentration as follows: +, less than 40% of the activity; ++, between 41% and 80% of the activity; +++, between 81% and 120% of the activity; ++++, more than 121% of the activity. Every experiment was repeated at least two times independently.
GI50 values were defined as the concentrations of compounds that resulted in 50% cell growth inhibition, shown as means ± SD. Every experiment was repeated at least three times.
“−” means “not determined”.
Fig. 2.
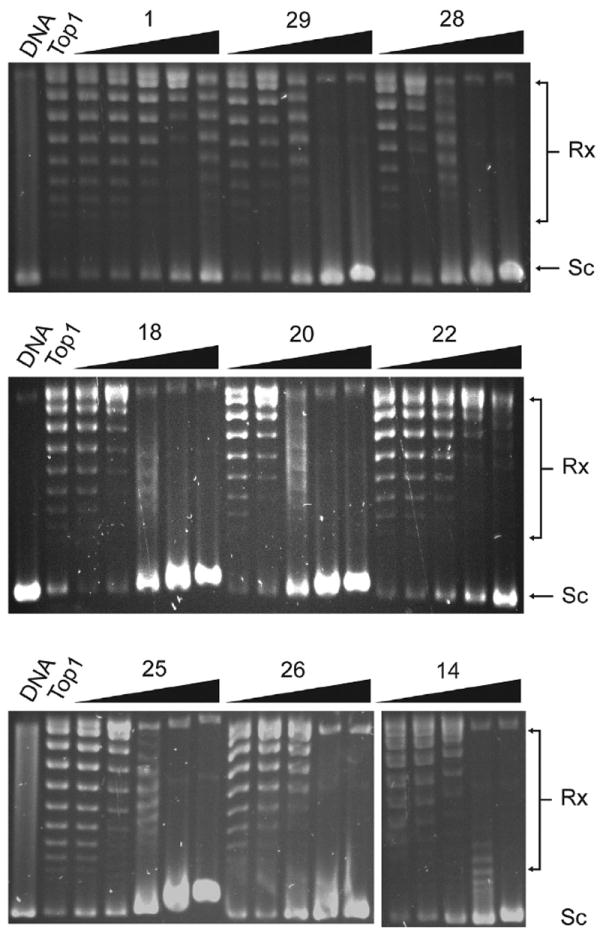
Representative Top1 relaxation assay. Lane 1: pBR322 DNA alone; Lane 2: pBR322 DNA and enzyme without compound; Lanes 3–17: pBR322 DNA and Top1 and tested compounds at 0.2, 1, 5, 25, 125 μM, respectively. Rx, relaxed DNA; Sc, supercoiled DNA.
2.3. Top1-mediated unwinding effect
Because DNA intercalating agents appear to inhibit Top1 by unwinding closed circular DNA [17,18]. We investigated whether our new series of synthesized compounds produce DNA unwinding in the presence of excess Top1 [18]. Top1-mediated unwinding assays were performed for the compounds with more or equipotent Top1 inhibitory activities to parent compound 1, and ethidium bromide (EB) was used as a positive control. Representative gels with supercoiled pBR322 DNA as substrate are shown in Fig. 3A. As expected, EB exhibited clear unwinding effect. By contrast, compounds 14, 22 and 26 had no Top1-mediated unwinding effect. To confirm these results, Top1-mediated unwinding assay with relaxed pBR322 DNA as substrate was performed [18]. As shown in Fig. 3B, compounds 14, 22 and 26 indeed had no unwinding effect, indicating that they are Top1 catalytic inhibitors similar to the parent compound 1 (SFig. 1, Supplementary Material). As shown in Table 1, significant Top1-mediated unwinding effect was found for compounds 8, 12, 13, 15, 16, 18–21, 25, 28–30, and weak unwinding effect for compound 11, implying that their Top1 inhibitions could be partially attributed to the unwinding capabilities.
Fig. 3.
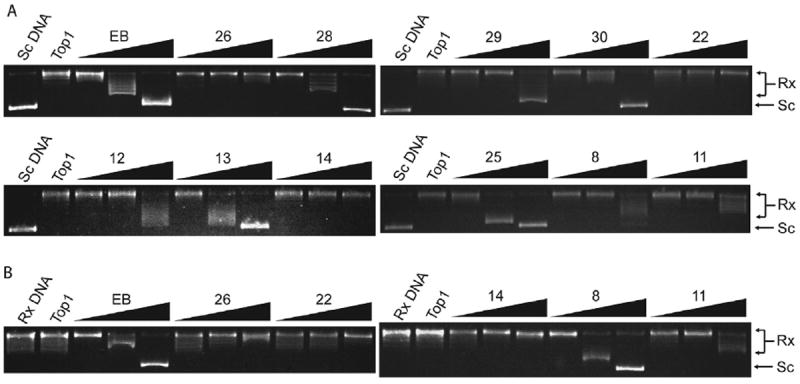
Representative Top1-mediated unwinding assay using supercoiled DNA (A) or relaxed DNA (B) as substrate, respectively. Lane 1: pBR322 DNA alone; Lane 2: pBR322 DNA and enzyme without compound; Lanes 3–11: pBR322 DNA and Top1 and tested compounds at 1, 5, 25 μM, respectively. EB was used as positive control at 0.6, 1.2, 2.4 μM, respectively. Rx, relaxed DNA; Sc, supercoiled DNA.
2.4. Cytotoxicities
The synthesized 6-substituted indolizinoquinolinediones, except compounds 3 and 10 due to their low solubility in DMSO and compound 27 due to its chemical instability, were assessed for cytotoxicity using the MTT method in four human cancer cell lines, human promyelocytic leukemia cells (HL-60), Burkitt’s lymphoma cells (CA46), cervical cancer HeLa cells and human lung adenocarcinoma epithelial cells (A549). The compounds were tested in a five-dose assay ranging from 10−8 to 10−4 M concentration. The GI50 values, defined as the concentrations of the tested compounds that resulted in 50% cell growth inhibition, are summarized in Table 1.
As shown in Table 1, the compounds with 6-alkylamino alkyloxy carbonyl group, including 26 and 28–30, had increased cytotoxicity. Compound 26 was most potent against A549 with GI50 less than 0.01 μM and Top1 inhibition of ++++. Compound 28 with Top1 inhibition of ++++ showed the highest potency against HL- 60, CA46 and Hela cells with GI50 of 0.025, 0.90 and 0.48 μM, respectively. Although the compounds with 6-alkylamino alkylamino carbonyl group, such as 8, 11–16 and 18–21, were more potent Top1 inhibition than parent compound 1, their cytotoxicities were not as good as expected against HL-60, CA46 and A549 cells, which might possibly due to their poor solubility and cellular permeability. The compounds with lower Top1 inhibition, such as 4–7, 9 and 17 generally exhibited lower cytotoxicity against HL-60, CA46 and A549 cell lines compared to compound 1. Compound 22 with equipotent Top1 inhibition exhibited equipotent cytotoxicity to compound 1 against HL-60 and CA46 cell lines.
3. Conclusion
Indolizinoquinolinedione derivative 1 served as a base to develop novel Top1 catalytic inhibitors. In this work, compound 1 was modified to give a series of 6-substituted indolizinoquinolinedione derivatives. Top1 cleavage and relaxation assays indicate that none of synthesized compounds acts as Top1 poison, and that the compounds with alkylamino terminus at C-6 side chain exhibit increased Top1 catalytic inhibition compared to derivative 1. Top1-mediated unwinding assay confirmed that three compounds 14, 22 and 26 are Top1 catalytic inhibitors without Top1-mediated unwinding effect. Compound 26 is the most cytotoxicity against lung carcinoma A549 cells with Top1 inhibition of ++++. Compound 28, with associated unwinding effect, was the most potent against human leukemia HL-60, Burkitt’s lymphoma CA46 and cervical cancer HeLa cells with potent Top1 inhibition (++++). This study demonstrates that the optimization of C-6 side chain of indolizinoquinolinedione could provide novel Top1 catalytic inhibitors with increased cytotoxicity and Top1 catalytic inhibitory activity.
4. Methods and materials
4.1. General experiments
The major chemical reagents for synthesis were purchased from Alfa Aesar, Sigma Aldrich Co or Aladdin Reagent Database Inc (Shanghai). Ethyl 5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline- 6-carboxylate (1) was prepared by our lab according to our reported method [9]. The common solvents were obtained from local commercial suppliers and used without further purification. Plasmid pBR322 DNA and purified calf thymus DNA topoisomerase I was purchased from TakaRa Biotechnology (Dalian) Co., Ltd., unless otherwise mentioned. One unit of Top1 was defined as the amount that relaxes 0.5 μg of pBR322 DNA at 37 °C for 30 min. Chemical reaction courses were monitored by silica gel GF254 thin layer chromatography. Melting points were determined in open capillary tubes on a MPA100 Optimelt Automated Melting Point System without being corrected. Nuclear magnetic resonance spectra were recorded on a Bruker AVANCE III 400 MHz spectrometer using tetramethylsilane as an internal reference. Mass spectra were analyzed on an Agilent 6120 (Quadrupole LC-MS) mass spectrometer. The high-resolution mass spectra were analyzed on an SHIMADZU LCMS-IT-TOF mass spectrometer. All compounds tested for biological activities were analyzed by HPLC and their purities were more than 95%.
4.2. Synthesis of compound 2
Compound 1 (1.28 g, 4 mmol) was hydrolyzed with K2CO3 (15%) in isopropanol solution (200 mL) to give aubergine solid 2 (1.05 g), yield 90%, mp = 269.5–271.2 °C. 1H NMR (CDCl3) δ 13.58 (s, br, 1H), 9.96 (d, J = 6.9 Hz, 1H), 9.13 (d, J = 4.7 Hz, 1H), 9.00 (d, J = 9.1 Hz, 1H), 8.65 (d, J = 7.9 Hz, 1H), 7.73 (dd, J = 7.9, 4.8 Hz, 1H), 7.68–7.58 (m, 1H), 7.39 (t, J = 6.9 Hz, 1H). HRMS (ESI) m/z: 291.0412 [M – H]−, calcd for C16H7N2O4 291.0411.
4.3. General procedure for the synthesis of compounds 3–22
At room temperature, to a red solution of compound 2 (150 mg, 0.5 mmol) and triethylamine (0.14 mL, 1.0 mmol) in chloroform (40 mL), thionyl chloride (2.5 mL) was added dropwise. The mixture was stirred and heated under reflux for 5 h. The mixture gradually became a red solution. The reaction solution was then cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was obtained under reduced pressure for a period to get rid of most of the residual SOCl2 to give an orange solid residue. 4-(Dimethylamino)pyridine (70 mg, 0.6 mmol) and different amine or alcohol derivative (1.80 mmol) in chloroform (30 mL) were added dropwise to the resultant residue. The reaction mixture instantaneously became a red solution. The reaction mixture was heated under reflux for 5 h, and cooled to room temperature. The solvent was evaporated under reduced pressure. The crude product was purified by silica gel column chromatography to give the target compound.
4.3.1. N-ethyl-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (3)
Using ethanamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 100:1 to give aubergine solid 3 (134 mg), yield 84%, mp = 302.7–304.4 °C. 1H NMR (CDCl3) δ 10.22 (s, br, 1H), 10.02 (d, J = 6.8 Hz, 1H), 9.18 (d, J = 9.2 Hz, 1H), 9.07 (s, br, 1H), 8.60 (d, J = 7.6 Hz, 1H), 7.67 (s, br, 1H), 7.54–7.47 (m, 1H), 7.30–7.27 (m, 1H), 3.63–3.54 (m, 2H), 1.39 (t, J = 7.3 Hz, 3H). 13C NMR (CDCl3) δ 183.4, 172.4, 162.6, 154.9, 149.9, 141.2,135.6,130.2,128.5,128.1,126.8,124.6,123.7,122.2,119.0,110.7, 34.4, 14.7. HRMS (ESI) m/z: 342.0847 [M + Na]+; calcd for C18H13N3O3Na 342.0849.
4.3.2. N,N-diethyl-5,12-dioxo-5,12-dihydroindolizino[2,3-g] quinoline-6-carboxamide (4)
Using diethylamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 100:1 to give aubergine solid 4 (80 mg), yield 46%, mp = 271.9–272.2 °C. 1H NMR (CDCl3) δ 9.80 (d, J = 7.0 Hz, 1H), 9.04 (d, J = 3.5 Hz, 1H), 8.51 (dd, J = 7.8, 1.3 Hz, 1H), 7.71 (d, J = 9.0 Hz, 1H), 7.63 (dd, J = 7.8, 4.7 Hz, 1H), 7.40–7.33 (m, 1H), 7.20 (t, J = 6.9 Hz, 1H), 3.95–3.85 (m, 1H), 3.64–3.55 (m, 1H), 3.43–3.33 (m, 1H), 3.32–3.23 (m, 1H),1.44 (t, J = 7.1 Hz, 3H),1.03 (t, J = 7.1 Hz, 3H). 13C NMR (CDCl3) δ 180.4, 171.9, 163.7, 154.2, 150.8, 136.8, 134.9, 130.3, 128.5, 126.9, 126.5, 125.1, 119.6, 117.9, 112.2, 43.2, 39.5, 14.5, 12.8. HRMS (ESI) m/z: 370.1156 [M + Na]+; calcd for C20H17N3O3Na 370.1162.
4.3.3. 6-(Piperidine-1-carbonyl)indolizino[2,3-g]quinoline-5,12-dione (5)
Using piperidine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 100:1 to give aubergine solid 5 (113 mg), yield 63%, mp = 289.4–290.6 °C. 1H NMR (CDCl3) δ 9.80 (d, J = 6.9 Hz, 1H), 9.03 (d, J = 4.2 Hz, 1H), 8.50 (d, J = 7.8 Hz, 1H), 7.76 (d, J = 9.0 Hz, 1H), 7.62 (dd, J = 8.0, 4.4 Hz, 1H), 7.36 (t, J = 7.8 Hz, 1H), 7.19 (t, J = 7.2 Hz, 1H), 4.15–4.08 (m, 1H), 3.76–3.69 (m, 1H), 3.40–3.34 (m, 2H), 1.93–1.83 (m, 1H), 1.75–1.67 (m, 3H), 1.41–1.33 (m, 2H). 13C NMR (CDCl3) δ 180.4, 172.0, 162.7, 154.2,150.7,137.1,134.9,130.3,128.4,127.0,126.6,125.3,121.2,119.8, 117.9, 111.6, 48.0, 43.1, 26.6, 25.6, 24.6. HRMS (ESI) m/z: 382.1170 [M + Na]+; calcd for C21H17N3O3Na 382.1162.
4.3.4. 6-(Morpholine-4-carbonyl)indolizino[2,3-g]quinoline-5,12-dione (6)
Using morpholine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give aubergine solid 6 (108 mg), yield 60%, mp = 308.9–309.5 °C. 1H NMR (CDCl3) δ 9.81 (dd, J = 7.0, 1.0 Hz, 1H), 9.04 (dd, J = 4.7, 1.7 Hz, 1H), 8.51 (dd, J = 7.9, 1.7 Hz, 1H), 7.88–7.75 (m, 1H), 7.64 (dd, J = 7.8, 4.7 Hz, 1H), 7.40 (ddd, J = 9.0, 6.9, 1.1 Hz, 1H), 7.22 (ddd, J = 8.2, 6.0, 2.5 Hz, 1H), 4.08–3.91 (m, 3H), 3.83–3.74 (m, 2H), 3.54–3.48 (m, 2H), 3.42–3.32 (m, 1H). 13C NMR (CDCl3) δ 180.4, 172.1, 163.0, 154.4, 150.6, 137.4, 134.9, 130.2, 128.5, 127.3, 126.7, 125.4, 121.2, 119.8, 118.1, 110.2, 66.8, 47.3, 42.6. HRMS (ESI) m/z: 384.0960 [M + Na]+; calcd for C20H15N3O4Na 384.0955.
4.3.5. 6-(4-Methylpiperazine-1-carbonyl)indolizino[2,3-g] quinoline-5,12-dione (7)
Using N-methylpiperazine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give aubergine solid 7 (78 mg), yield 42%, mp = 274.9–275.5 °C. 1H NMR (CDCl3) δ 9.81 (d, J = 7.0 Hz, 1H), 9.04 (dd, J = 8.4, 1.6 Hz, 1H), 8.51 (d, J = 7.8 Hz, 1H), 7.77 (d, J = 9.1 Hz, 1H), 7.63 (dd, J = 7.8, 4.7 Hz, 1H), 7.42–7.33 (m, 1H), 7.21 (t, J = 6.9 Hz, 1H), 4.13–4.06 (m, 1H), 3.95–3.88 (m, 1H), 3.52–3.46 (m, 1H), 3.44–3.37 (m, 1H), 2.77–2.72 (m, 1H), 2.52–2.44 (m, 2H), 2.34 (s, 3H), 2.21–2.14 (m, 1H). 13C NMR (CDCl3) δ 180.3, 172.0, 162.8, 154.3, 150.6, 137.2, 135.0, 130.2, 128.5, 127.2, 126.6, 125.4, 121.2, 119.8, 118.0, 110.7, 55.2, 54.6, 46.8, 46.0, 42.0. HRMS (ESI) m/z: 397.1294 [M + Na]+; calcd for C21H18N4O3Na 397.1271.
4.3.6. N-(2-(Dimethylamino)ethyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (8)
Using N,N-dimethylethanediamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 40:1 to give aubergine solid 8 (160 mg), yield 88%. 1H NMR (CDCl3) δ 10.29 (s, br, 1H), 10.01 (d, J = 6.9 Hz, 1H), 9.14 (d, J = 9.2 Hz, 1H), 9.06 (d, J = 3.1 Hz, 1H), 8.60 (d, J = 6.7 Hz, 1H), 7.67 (dd, J = 7.8, 4.7 Hz, 1H), 7.55–7.45 (m, 1H), 7.32–7.26 (m, 1H), 3.70 (q, J = 5.7 Hz, 2H), 2.73 (t, J = 6.4 Hz, 2H), 2.42 (s, 6H). The 1H NMR spectrum is similar to the reference [15]. HRMS (ESI) m/z: 363.1471 [M + H]+; calcd for C20H19N4O3 363.1452.
4.3.7. 5,12-Dioxo-N-phenethyl-5,12-dihydroindolizino[2,3-g] quinoline-6-carboxamide (9)
Using phenylethanamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 200:1 to give aubergine solid 9 (140 mg), yield 71%, mp = 263.0–264.5 °C. 1H NMR (CDCl3) δ 10.29 (s, br, 1H), 9.99 (d, J = 8.0 Hz, 1H), 9.15 (d, J = 8.0 Hz, 1H), 9.05 (d, J = 4.0 Hz, 1H), 8.53 (d, J = 8.0 Hz, 1H), 7.66 (dd, J = 8.0, 4.5 Hz, 1H), 7.55–7.46 (m, 1H), 7.38–7.19 (m, 6H), 3.83–3.77 (m, 2H), 3.06 (t, J = 8.0 Hz, 2H). 13C NMR (CDCl3) δ 183.2, 172.3, 162.8, 154.9, 149.7, 141.2, 139.4, 135.5, 130.1, 128.9, 128.6, 128.5, 128.0, 126.9, 126.3, 124.6, 123.6, 122.2, 119.1, 110.4, 41.1, 35.8. HRMS (ESI) m/z: 396.1341 [M + H]+; calcd for C24H18N3O3 396.1343.
4.3.8. 5,12-Dioxo-N-(2-(piperidin-1-yl)ethyl)-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (10)
Using 2-(piperidin-1-yl)ethanamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 25:1 to give aubergine solid 10 (170 mg), yield 85%, mp = 244.5–247.2 °C. 1H NMR (CDCl3) δ 10.30 (s, br, 1H), 10.02 (d, J = 7.0 Hz, 1H), 9.15 (d, J = 9.2 Hz, 1H), 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.59 (dd, J = 7.9, 1.7 Hz, 1H), 7.67 (dd, J = 7.9, 4.7 Hz, 1H), 7.54–7.45 (m, 1H), 7.31–7.27 (m, 1H), 3.71 (q, J = 5.9 Hz, 2H), 2.78–2.69 (m, 2H), 2.63–2.57 (m, 4H), 1.77–1.62 (m, 6H). 13C NMR (CDCl3) δ 183.0, 172.4, 162.7, 154.8, 149.8, 141.2, 135.5, 130.2, 128.5, 128.0, 126.8, 124.7, 123.6, 122.2, 118.9, 110.4, 57.7, 54.6, 37.0, 25.9, 24.3. HRMS (ESI) m/z: 403.1787 [M + H]+; calcd for C23H23N4O3 403.1765.
4.3.9. N-(2-Morpholinoethyl)-5,12-dioxo-5,12-dihydroindolizino [2,3-g]quinoline-6-carboxamide (11)
Using 2-morpholinoethanamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 20:1 to give aubergine solid 11 (162 mg), yield 80%, mp = 217.9–218.8 °C. 1H NMR (CDCl3) δ 10.35 (s, br, 1H), 10.02 (d, J = 7.2 Hz, 1H), 9.14 (d, J = 9.0 Hz, 1H), 9.07 (d, J = 4.6 Hz, 1H), 8.58 (d, J = 7.9 Hz, 1H), 7.68 (dd, J = 7.6, 4.8 Hz, 1H), 7.54–7.47 (m, 1H), 7.32–7.26 (m, 1H), 3.87–3.78 (m, 4H), 3.71 (q, J = 6.0 Hz, 2H), 2.75 (t, J = 6.3 Hz, 2H), 2.65 (s, br, 4H). 13C NMR (CDCl3) δ 183.0, 172.4, 162.8, 154.8, 149.7, 141.2, 135.5, 130.2, 128.5, 128.0, 126.8, 124.7, 123.6, 122.2, 119.0, 110.3, 67.0, 57.3, 53.5, 36.5. HRMS (ESI) m/z: 427.1365 [M + Na]+; calcd for C22H20N4O4Na 427.1377.
4.3.10. N-(2-(4-Methylpiperazin-1-yl)ethyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (12)
Using 2-(4-methylpiperazin-1-yl)ethanamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 30:1 to give aubergine solid 12 (159 mg), yield 76%, mp = 254.5–256.3 °C. 1H NMR (CDCl3) δ 10.30 (s, br, 1H), 10.03 (d, J = 6.7 Hz, 1H), 9.15 (d, J = 9.5 Hz, 1H), 9.07 (s, 1H), 8.58 (d, J = 7.2 Hz, 1H), 7.71–7.66 (m, 1H), 7.54–7.49 (m, 1H), 7.31–7.27 (m, 1H), 3.74–3.66 (m, 2H), 2.85–2.76 (m, 10H), 2.49 (s, 3H). 13C NMR (CDCl3) δ 183.2, 172.4, 162.8, 154.9, 149.8, 141.22, 135.5, 130.2, 128.6, 128.1, 126.9, 124.7, 123.6, 119.0, 110.4, 106.6, 56.7, 54.7, 52.0, 45.3, 39.6. HRMS (ESI) m/z: 418.1893 [M + H]+; calcd for C23H24N5O3 418.1874.
4.3.11. N-(2-(Pyrrolidin-1-yl)ethyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (13)
Using 2-(pyrrolidin-1-yl)ethanamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give aubergine solid 13 (132 mg), yield 68%, mp = 209.2–210.1 °C. 1H NMR (CDCl3) δ 10.33 (s, br, 1H), 10.00 (d, J = 6.9 Hz, 1H), 9.12 (d, J = 8.9 Hz, 1H), 9.06 (d, J = 4.7 Hz, 1H), 8.58 (d, J = 7.9 Hz, 1H), 7.67 (dd, J = 7.9, 4.7 Hz, 1H), 7.55–7.46 (m, 1H), 7.33–7.25 (m, 1H), 3.77 (q, J = 6.4 Hz, 2H), 2.96 (t, J = 6.6 Hz, 2H), 2.81 (s, br, 4H), 1.90 (s, br, 4H). 13C NMR (CDCl3) δ 182.1, 171.3, 161.9,153.8,148.7,140.1,134.6,129.2,127.6,127.0,125.9,123.7,122.5, 121.2, 118.0, 109.2, 54.0, 53.2, 37.4, 22.6. HRMS (ESI) m/z: 389.1600 [M + H]+; calcd for C22H21N4O3 389.1608.
4.3.12. N-(2-(1H-Imidazol-1-yl)ethyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (14)
Using 2-(1H-imidazol-1-yl)ethanamine hydrochloride as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 200:1 to give aubergine solid 14 (81 mg), yield 42%, mp = 249.5–250.7 °C. 1H NMR (DMSO-d6) δ 9.89 (t, J = 6.0 Hz, 1H), 9.81 (d, J = 8.0 Hz, 1H), 9.04 (dd, J = 4.0, 1.6 Hz, 1H), 8.72 (d, J = 8.0 Hz, 1H), 8.48 (dd, J = 8.0, 1.6 Hz, 1H), 8.11 (s, 1H), 7.84 (dd, J = 8.0, 4.7 Hz, 1H), 7.66–7.58 (m, 1H), 7.50–7.41 (m, 2H), 7.12 (s, 1H), 4.31 (t, J = 6.0 Hz, 2H), 3.79 (q, J = 6.0 Hz, 2H). 13C NMR (CD3OD) δ 183.2, 172.6, 163.9, 155.6, 149.7, 141.6, 136.6, 130.7, 130.5, 128.9, 128.7, 126.6, 125.1, 123.3, 122.6, 121.8, 120.6, 109.8, 47.9, 41.0. HRMS (ESI) m/z: 386.1244 [M + H]+; calcd for C21H16N5O3 386.1248.
4.3.13. N-(3-(Dimethylamino)propyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (15)
Using N,N-dimethylpropane-1,3-diamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 40:1 to give red solid 15 (147 mg), yield 78%, mp = 166.7–168.0 °C. 1H NMR (CDCl3) δ 10.31 (s, br, 1H), 10.02 (d, J = 4.0 Hz, 1H), 9.11 (d, J = 8.0 Hz, 1H), 9.07 (d, J = 4.0 Hz, 1H), 8.59 (dd, J = 8.0, 1.4 Hz, 1H), 7.68 (dd, J = 8.0, 4.6 Hz, 1H), 7.55–7.48 (m, 1H), 7.30 (d, J = 8.0 Hz, 1H), 3.68–3.58 (m, 2H), 2.84–2.74 (m, 2H), 2.53 (s, 6H), 2.12–2.05 (m, 2H). 13C NMR (CDCl3) δ 183.4, 172.4, 162.8, 154.9, 149.8, 141.2, 135.6, 130.2, 128.5, 128.1, 126.8, 124.6, 123.6, 122.2, 119.0, 110.5, 57.1, 45.2, 37.5, 27.3. HRMS (ESI) m/z: 377.1608 [M + H]+, calcd for C21H21N4O3 377.1608.
4.3.14. N-(3-(Diethylamino)propyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (16)
Using N,N-diethylpropane-1,3-diamine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 25:1 to give aubergine solid 16 (166 mg), yield 82%, mp = 184.5–187.1 °C. 1H NMR (CDCl3) δ 10.27 (s, br, 1H), 10.00 (d, J = 8.0 Hz, 1H), 9.13 (d, J = 8.0 Hz, 1H), 9.06 (dd, J = 4.0, 1.7 Hz, 1H), 8.57 (dd, J = 8.0, 1.7 Hz, 1H), 7.67 (dd, J = 8.0, 4.7 Hz, 1H), 7.50 (ddd, J = 9.1, 6.8, 1.1 Hz, 1H), 7.31–7.26 (m, 1H), 3.59 (q, J = 6.2 Hz, 2H), 2.81–2.66 (m, 6H),1.99 (quint, J = 7.2 Hz, 2H),1.14 (t, J = 7.2 Hz, 6H). 13C NMR (CDCl3) δ 183.3, 172.3, 162.7, 154.9, 149.8, 141.2, 135.6, 130.2, 128.6, 128.0, 126.9, 124.6, 123.6, 122.2, 119.1, 110.6, 50.4, 46.9, 38.0, 26.8, 11.6. HRMS (ESI) m/z: 405.2198 [M + H]+; calcd for C23H25N4O3 405.2198.
4.3.15. 5,12-Dioxo-N-(3-phenylpropyl)-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (17)
Using 3-phenylpropan-1-amine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give aubergine solid 17 (178 mg), yield 87%, mp = 233.2–234.0 °C. 1H NMR (CDCl3) δ 10.30 (s, 1H), 10.01 (d, J = 4.0 Hz, 1H), 9.16 (d, J = 8.0 Hz, 1H), 9.06 (dd, J = 4.0, 1.6 Hz, 1H), 8.59 (dd, J = 8.0, 1.6 Hz, 1H), 7.67 (dd, J = 8.0, 4.7 Hz, 1H), 7.53–7.47 (m, 1H), 7.34–7.27 (m, 5H), 7.22–7.17 (m, 1H), 3.58 (q, J = 6.4 Hz, 2H), 2.84 (t, J = 7.8 Hz, 2H), 2.09 (quint, J = 7.6 Hz, 2H). 13C NMR (CDCl3) δ 183.3, 172.3, 162.7, 154.9, 149.8, 141.7, 141.2, 135.6, 130.2, 128.5, 128.4, 128.0, 126.8, 125.9, 124.5, 123.7, 122.2, 119.1, 110.6, 39.1, 33.5, 31.2. HRMS (ESI) m/z: 410.1495 [M + H]+, calcd for C25H20N3O3 410.1499.
4.3.16. 5,12-Dioxo-N-(3-(piperidin-1-yl)propyl)-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (18)
Using 3-(piperidin-1-yl)propan-1-amine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 25:1 to give aubergine solid 18 (170 mg), yield 82%, mp = 246.6–252.3 °C. 1H NMR (CD3OD) δ 9.56 (d, J = 8.0 Hz, 1H), 8.86 (dd, J = 4.0, 1.4 Hz, 1H), 8.56 (d, J = 8.0 Hz, 1H), 8.33 (dd, J = 8.0, 1.4 Hz, 1H), 7.71 (dd, J = 8.0, 4.7 Hz, 1H), 7.45–7.40 (m, 1H), 7.23 (t, J = 8.0 Hz, 1H), 3.39–3.35 (m, 2H), 2.75–2.61 (m, 6H), 1.98–1.88 (m, 2H), 1.72–1.69 (m, 4H), 1.59–1.55 (m, 2H). 13C NMR (CD3OD) δ 184.5, 174.0, 163.7, 155.5, 149.8, 141.6, 136.7, 131.6, 130.3, 128.8, 128.7, 125.2, 123.5, 122.7, 120.5, 110.2, 57.6, 55.4, 38.6, 26.8, 26.3, 24.9. HRMS (ESI) m/z: 417.1924 [M + H]+, calcd for C24H25N4O3 417.1921.
4.3.17. N-(3-Morpholinopropyl)-5,12-dioxo-5,12-dihydroindolizino [2,3-g]quinoline-6-carboxamide (19)
Using 3-morpholinopropan-1-amine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 20:1 to give aubergine solid 19 (175 mg), yield 84%, mp = 200.9–203.9 °C. 1H NMR (CDCl3) δ 10.25 (s, br, 1H), 9.98 (d, J = 4.0 Hz, 1H), 9.13 (d, J = 8.0 Hz, 1H), 9.06 (dd, J = 6.0, 1.6 Hz, 1H), 8.55 (dd, J = 8.0, 1.6 Hz, 1H), 7.67 (dd, J = 8.0, 4.7 Hz, 1H), 7.52–7.47 (m, 1H), 7.31–7.25 (m, 1H), 3.78–3.73 (m, 4H), 3.59 (q, J = 6.4 Hz, 2H), 2.60–2.53 (m, 6H), 1.95 (quint, J = 7.2 Hz, 2H). 13C NMR (CDCl3) δ 183.3, 172.2, 162.7, 154.9, 149.7, 141.1, 135.5, 130.1, 128.6, 128.0, 126.8, 124.5, 123.6, 122.2, 119.1, 110.5, 67.0, 56.4, 53.7, 37.7, 26.4. HRMS (ESI) m/z: 419.1716 [M + H]+, calcd for C23H23N4O4 419.1714.
4.3.18. N-(3-(4-Methylpiperazin-1-yl)propyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (20)
Using 3-(4-methylpiperazin-1-yl)propan-1-amine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 30:1 to give aubergine solid 20 (140 mg), yield 65%, mp = 165.6–171 °C. 1H NMR (CDCl3) δ 10.26 (s, br, 1H), 10.01 (d, J = 8.0 Hz, 1H), 9.14 (d, J = 12.0 Hz, 1H), 9.06 (dd, J = 6.0, 1.7 Hz, 1H), 8.57 (dd, J = 8.0, 1.7 Hz, 1H), 7.68 (dd, J = 8.0, 4.7 Hz, 1H), 7.50 (ddd, J = 9.2, 6.9, 1.1 Hz, 1H), 7.30–7.27 (m, 1H), 3.59 (q, J = 6.4 Hz, 2H), 2.68–2.51 (m, 10H), 2.31 (s, 3H), 1.95 (quint, J = 7.2 Hz, 2H). 13C NMR (CD3OD) δ 183.6, 172.7, 163.8, 155.5, 150.0, 141.7, 136.8, 131.1, 130.3, 128.8, 125.4, 123.6, 122.9, 120.5, 110.3, 56.9, 55.7, 53.7, 45.9, 38.7, 27.2. HRMS (ESI) m/z: 432.2034 [M + H]+, calcd for C24H26N5O3 432.2030.
4.3.19. 5,12-Dioxo-N-(3-(pyrrolidin-1-yl)propyl)-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (21)
Using 3-(pyrrolidin-1-yl)propan-1-amine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 25:1 to give red solid 21 (157 mg), yield 78%, mp = 189.8–191.4 °C. 1H NMR (CDCl3) δ 10.26 (s, br, 1H), 9.99 (d, J = 8.0 Hz, 1H), 9.12 (d, J = 8.0 Hz, 1H), 9.05 (dd, J = 4.0, 1.7 Hz, 1H), 8.57 (dd, J = 8.0, 1.6 Hz, 1H), 7.67 (dd, J = 8.0, 4.7 Hz, 1H), 7.53–7.46 (m, 2H), 7.31–7.27 (m, 1H), 3.61 (q, J = 6.4 Hz, 2H), 2.77–2.69 (m, 6H), 2.04 (quint, J = 7.2 Hz, 2H), 1.85 (s, br, 4H). 13C NMR (CD3OD) δ 182.1, 171.6, 163.1, 154.3, 148.7, 140.4, 135.2, 129.6, 129.2, 128.4, 127.6, 124.2, 122.0, 121.5, 119.3, 108.8, 53.9, 52.7, 35.9, 26.1, 22.8. HRMS (ESI) m/z: 403.1769 [M + H]+, calcd for C23H23N4O3 403.1765.
4.3.20. 2-(Pyridin-2-yl)ethyl5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxylate (22)
Using 2-(pyridin-2-yl)ethanol as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give orange solid 22 (115 mg), yield 58%, mp = 167.6–173 °C. 1H NMR (CDCl3) δ 9.93 (d, J = 8.0 Hz, 1H), 9.02 (dd, J = 4.0, 1.6 Hz, 1H), 8.61 (d, J = 4.0 Hz, 1H), 8.54 (dd, J = 6.0, 1.6 Hz, 1H), 8.12 (d, J = 8.0 Hz, 1H), 7.67–7.61 (m, 2H), 7.42–7.38 (m, 1H), 7.33 (d, J = 8.0 Hz, 1H), 7.24–7.17 (m, 2H), 4.88 (t, J = 6.0 Hz, 2H), 3.39 (t, J = 6.0 Hz, 2H). 13C NMR (CDCl3) δ 179.0, 173.1, 162.6, 158.3, 154.1, 149.5, 149.4, 139.9, 136.5, 135.4, 130.9, 128.8, 128.6, 128.5, 128.3, 127.0, 123.7, 123.1, 121.7, 121.2, 118.0, 64.3, 37.3. HRMS (ESI) m/z: 398.1133 [M + H]+, calcd for C23H16N3O4 398.1135.
4.4. Synthesis of compound 23 and 24
At room temperature, to a red solution of compound 2 (0.14 g, 0.48 mmol) and triethylamine (0.14 mL, 1.00 mmol) in chloroform (40 mL), thionyl chloride (5 mL) was added dropwise. The mixture was stirred and heated under reflux for 5 h. The reaction solution was then cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was obtained under reduced pressure for a period to get rid of most of the residual SOCl2 to give an orange solid residue. A solution of 4-(dimethylamino)pyridine (0.07 g, 0.6 mmol) and 2-bromoethanol or 3-bromopropanaol (1.8 mmol) in CHCl3 (30 mL) was added dropwise to the resultant residue. The reaction mixture instantaneously became a red solution. The reaction mixture was heated under reflux for 5 h, and cooled to room temperature. The solvent was evaporated under reduced pressure. The resulting residue was purified by silica gel column chromatography to give the target compound.
4.4.1. 2-Bromoethyl 5,12-dioxo-5,12-dihydroindolizino[2,3-g] quinoline-6-carboxylate (23)
Using 2-bromoethanol as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give orange solid 23 (142 mg), yield 74%. 1H NMR (CDCl3) δ 9.97 (d, J = 7.0 Hz, 1H), 9.03 (dd, J = 4.7, 1.7 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 8.48–8.44 (m, 1H), 7.66 (dd, J = 7.9, 4.7 Hz, 1H), 7.56–7.52 (m, 1H), 7.31–7.24 (m, 1H), 4.78 (t, J = 6.1 Hz, 2H), 3.78 (t, J = 6.1 Hz, 2H). ESI-MS m/z: 399.0 (100%), 401.0 (100%) [M + H]+.
4.4.2. 3-Bromopropyl 5,12-dioxo-5,12-dihydroindolizino[2,3-g] quinoline-6-carboxylate (24)
Using 2-bromoethanol as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 100:1 to give orange solid 24 (151 mg), yield 76%. 1H NMR (CDCl3) δ 9.98 (d, J = 7.0 Hz, 1H), 9.05 (dd, J = 4.7, 1.7 Hz, 1H), 8.57 (dd, J = 7.9, 1.7 Hz, 1H), 8.42 (d, J = 9.1 Hz, 1H), 7.67 (dd, J = 7.9, 4.7 Hz, 1H), 7.54 (ddd, J = 9.1, 6.9, 1.1 Hz, 1H), 7.32–7.23 (m, 1H), 4.61 (t, J = 5.8 Hz, 2H), 3.76 (t, J = 6.8 Hz, 2H), 2.44 (quint, J = 6.2 Hz, 2H). ESI-MS m/z: 413.1 (100%), 415.1 (100%) [M + H]+.
4.5. General procedure for the synthesis of compounds 25–30
At room temperature, to a stirred suspension of compound 23 or 24 (0.4 mmol) in ethanol (20 mL), reagent amine (8 mmol) was added. The reaction mixture was heated under reflux for 18–24 h. After being cooled to room temperature, the solvent was evaporated in vacuo. The resulting residue was purified by silica gel column chromatography to give the target compound.
4.5.1. 2-(Piperidin-1-yl)ethyl5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxylate (25)
Using compound 23 and piperidine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 60:1 to give orange solid 25 (69 mg), yield 43%, mp = 332.6–337 °C. 1H NMR (CD3OD) δ 9.73 (d, J = 8.0 Hz, 1H), 8.98 (d, J = 4.0 Hz, 1H), 8.21–8.16 (m, 2H), 7.68 (dd, J = 7.7, 4.0 Hz, 1H), 7.62–7.56 (m, 1H), 7.42 (t, J = 8.0 Hz, 1H), 3.85–3.80 (m, 2H), 3.18–3.15 (m, 2H) 2.12–2.08 (m, 4H), 1.75–1.71 (m, 6H). 13C NMR (D2O) δ 177.8, 171.1, 161.2, 154.0, 146.8, 139.4, 134.9, 131.4, 128.5, 128.5, 127.7, 125.5, 121.1, 119.9, 119.8, 102.9, 58.1, 55.3, 53.9, 22.8, 21.1. HRMS (ESI) m/z: 404.1607 [M + H]+, calcd for C23H22N3O4 404.1605.
4.5.2. N-(3-(4-methylpiperazin-1-yl)propyl)-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxamide (26)
Using compound 23 and morpholine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give orange solid 26 (108 mg), yield 67%, mp = 165.6–171 °C. 1H NMR (CDCl3) δ 9.96 (d, J = 8.0 Hz, 1H), 9.03 (dd, J = 6.0, 1.7 Hz, 1H), 8.55 (dd, J = 8.0, 1.7 Hz, 1H), 8.51 (d, J = 8.0 Hz, 1H), 7.66 (dd, J = 8.0, 4.7 Hz, 1H), 7.51 (ddd, J = 9.0, 6.9, 1.0 Hz, 1H), 7.28–7.24 (m, 1H), 4.59 (t, J = 6.0 Hz, 2H), 3.78–3.73 (m, 4H), 2.88 (t, J = 6.0 Hz, 2H), 2.63 (s, br, 4H). 13C NMR (CDCl3) δ 179.1, 173.1, 162.7, 154.1, 149.5, 140.0, 135.3, 130.8, 128.6, 128.1, 127.0, 123.1, 121.3, 118.0, 106.1, 67.0, 61.7, 56.9, 53.7. HRMS (ESI) m/z: 406.1400 [M + H]+, calcd for C22H20N3O5 406.1397.
4.5.3. 2-(Pyrrolidin-1-yl)ethyl 5,12-dioxo-5,12-dihydroindolizino [2,3-g]quinoline-6-carboxylate (27)
Using compound 23 and pyrrolidine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give orange solid 27 (75 mg), yield 48%, mp = 190–192 °C. 1H NMR (CDCl3) δ 9.93 (d, J = 8.0 Hz, 1H), 9.05 (d, J = 4.0 Hz, 1H), 8.49 (d, J = 8.0 Hz, 1H), 8.43 (d, J = 8.8 Hz, 1H), 7.66 (dd, J = 8.0, 4.7 Hz, 1H), 7.58–7.52 (m,1H), 7.31–7.27 (m, 1H), 4.84 (t, J = 6.0 Hz, 2H), 3.37 (t, J = 7.1 Hz, 2H), 3.27 (s, br, 4H), 2.07 (s, br, 4H). HRMS (ESI) m/z: 390.1451 [M + H]+, calcd for C22H20N3O4 390.1448.
4.5.4. 3-(Piperidin-1-yl)propyl-5,12-dioxo-5,12-dihydroindolizino [2,3-g]quinoline-6-carboxylate (28)
Using compound 24 and piperidine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 20:1 to give orange solid 28 (153 mg), yield 92%, mp = 247.6–247.9 °C. 1H NMR (CD3OD) δ 9.86 (d, J = 6.5 Hz, 1H), 8.96 (d, J = 3.0 Hz, 1H), 8.52 (d, J = 7.7 Hz, 1H), 8.35 (d, J = 9.6 Hz, 1H), 7.80 (dd, J = 7.8, 4.7 Hz, 1H), 7.66–7.60 (m, 1H), 7.39 (t, J = 6.9 Hz, 1H), 4.49 (t, J = 6.0 Hz, 2H), 3.46–3.43 (m, 2H), 3.19–3.10 (m, 4H), 2.31–2.27 (m, 2H), 1.91–1.87 (m, 6H). 13C NMR (D2O) δ 177.0, 170.7, 161.7, 153.6, 146.5, 139.2, 134.8, 130.8, 128.7, 128.1, 127.5, 125.5, 121.1, 119.8, 119.3, 102.6, 62.1, 53.7, 53.3, 22.8, 21.5, 21.2. HRMS (ESI) m/z: 418.1761 [M+ H]+, calcd for C24H24N3O4 418.1761.
4.5.5. 3-Morpholinopropyl-5,12-dioxo-5,12-dihydroindolizino[2,3-g]quinoline-6-carboxylate (29)
Using compound 24 and morpholine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 50:1 to give orange solid 29 (141 mg), yield 84%, mp = 177.4–183.0 °C. 1H NMR (CDCl3) δ 9.76 (d, J = 7.0 Hz, 1H), 9.00 (dd, J = 4.6, 1.5 Hz, 1H), 8.43 (dd, J = 7.9, 1.5 Hz, 1H), 8.23 (d, J = 9.1 Hz, 1H), 7.82 (dd, J = 7.9, 4.7 Hz, 1H), 7.69–7.62 (m, 1H), 7.45 (t, J = 6.8 Hz, 1H), 4.37 (t, J = 6.5 Hz, 2H), 3.59 (t, J = 4.4 Hz, 4H), 2.55–2.49 (m, 6H), 1.95 (quint, J = 6.8 Hz, 2H). 13C NMR (CDCl3) δ 179.1, 173.1, 163.2, 154.1, 149.5, 140.1, 135.4, 130.9, 128.7, 128.6, 128.0, 127.0, 123.0, 121.1, 118.1, 106.0, 70.0, 63.5, 55.5, 53.7, 25.9. HRMS (ESI) m/z: 420.1559 [M + H]+, calcd for C23H22N3O5 420.1554.
4.5.6. 3-(Pyrrolidin-1-yl)propyl-5,12-dioxo-5,12-dihydroindolizino [2,3-g]quinoline-6-carboxylate (30)
Using compound 24 and pyrrolidine as material, the product was purified by silica gel column chromatography with eluent CH2Cl2:CH3OH = 20:1 to give orange solid 30 (153 mg), yield 95%, mp = 326.7–332.7 °C. 1H NMR (CD3OD) δ 9.67 (d, J = 8.0 Hz, 1H), 8.90 (d, J = 4.0 Hz, 1H), 8.31 (d, J = 6.4 Hz, 1H), 8.17 (d, J = 8.8 Hz, 1H), 7.72 (dd, J = 8.0, 4.7 Hz, 1H), 7.55–7.52 (m, 1H), 7.31 (t, J = 6.4 Hz, 1H), 4.46 (t, J = 6.0 Hz, 2H), 3.69–3.58 (m, 6H), 2.36–2.31 (m, 2H), 2.21–2.18 (m, 4H). 13C NMR (CD3OD) δ 179.0, 172.4, 162.9, 153.5, 148.7, 140.4, 135.2, 130.6, 129.5, 128.2, 127.5, 127.1, 122.7, 120.3, 118.5, 104.4, 61.4, 54.1, 52.4, 25.2, 22.8. HRMS (ESI) m/z: 404.1604 [M + H]+, calcd for C23H22N3O4 404.1605.
4.6. Top1 relaxation assay
The Top1 relaxation assay was carried out as described with slight modifications [19]. Briefly, reaction (20 μL) mixture containing 0.5 μg of plasmid pBR322 DNA in relaxation buffer (10 mM Tris, pH 7.5, 0.1 mM EDTA, 5 mM MgCl2, 50 mM KCl, 1 mM DTT, 15 μg/mL acetylated BSA) was incubated with 1 unit of calf thymus Top1 in the absence or in the presence of compound, previously dissolved in DMSO solution, for 30 min at 37 °C. Top1 was preincubated with compound for 15 min prior to the addition of plasmid pBR322 DNA, and the reaction was started by the addition of Top1 enzyme. The reaction was terminated by the addition of 4 μL of loading buffer (30% sucrose, 0.5% bromophenol blue, and 0.5% xylene cyanole FF in 10mMTris–HCl, pH 7.9). Then the sample was analyzed using a 0.8% agarose gel in 40 mM Tris-acetate (pH 8.0) and 1 mM EDTA (TAE buffer) at 5 V/cm. Gel was stained with ethidium bromide (EB) and visualized with a UV transilluminator. Image was acquired and quantified through AlphaEaseFC software.
4.7. Top1 cleavage assay
DNA cleavage assays were performed in briefly as follows [16]. A 3′-[32P]-labeled 117-bp DNA oligonucleotide was prepared as previously described. Approximately 2 nM radiolabeled DNA substrate was incubated with recombinant Top1 in 20 μL of reaction buffer (10 mM Tris–HCl pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 μg/mL BSA) at 25 °C for 20 min in the presence of various concentrations of test compounds. The reactions were terminated by adding SDS (0.5% final concentration) followed by the addition of two volumes of loading dye (80% formamide, 10 mM sodium hydroxide, 1 mM sodium EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue). Aliquots of each reaction mixture were subjected to 20% denaturing PAGE. Gels were dried and visualized by using a phosphoimager and ImageQuant software (Molecular Dynamics). For simplicity, cleavage sites were numbered as previously described in the 161-bp fragment.
4.8. Top1-mediated unwinding assay
The unwinding assay was performed in 20 μL reaction volume containing 0.05 μg of supercoiled or relaxed pBR322 DNA and excess Top1 (4 units) in relaxing buffer [18]. The DNAwas incubated with compound at room temperature for 10 min prior to the addition of Top1. After being incubated for 30 min at 37 °C, reaction was terminated by the addition of 4 μL loading buffer. The results were analyzed using 1% agarose gel in TAE buffer at 5 V/cm. The gel was stained with EB and visualized with a UV transilluminator.
4.9. Cell culture and MTT assay
Four different human cancer cell lines, GLC-82, NCI-H460, MCF-7 and MCF-7/ARD, were obtained from Laboratory Animal Center, Sun Yat-sen University, and cultured on RPMI-1640 medium at 37 °C in a humidified atmosphere with 5% CO2. All cells to be tested in the following assays had a passage number of 3–6.
For the drug treatment experiments, the cancer cells were treated with the compounds (predissolved in DMSO) at a five-dose assay ranging from 10−8 to 10−4 M concentration for a period of 2 days. At the end of the drug treatment period, MTT solution (50 μL, 1 mg/mL) in PBS (PBS without MTT as the blank) was fed to each well of the culture plate (containing 100 μL medium). After 4 h incubation, the formazan crystal formed in the well was dissolved with 100 μL of DMSO for optical density reading at 492 nm [20]. GI50 values were calculated by nonlinear regression analysis (GraphPad Prism).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81373257), Guangdong Natural Science Fund (No. S2013010015609) and Intramural Research Program of the National Institutes of Health National Cancer Institute (Z01 BC006161), Center of Cancer Research (No. Z01 BC006161). We gratefully thank Prof. Ding Li, School of Pharmaceutical Sciences, Sun Yat-sen University for the manuscript preparation.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmech.2015.07.007.
References
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 3.Stewart L, Redinbo MR, Qui X, Hol WGJ, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu N, Wu XW, Agama K, Pommier Y, Du J, Li D, Gu LQ, Huang ZS, An LK. A novel DNA topoisomerase I inhibitor with different mechanism from camptothecin induces G2/M phase cell cycle arrest to K562 cells. Biochemistry. 2010;49:10131–10136. doi: 10.1021/bi1009419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, An LK, Wu N, Wang XD, Bu XZ, Huang ZS, Gu LQ. Synthesis, cytotoxic activities and structure-activity relationships of topoisomerase I inhibitors: indolizinoquinoline-5,12-dione derivatives. Bioorg Med Chem. 2008;16:4617–4625. doi: 10.1016/j.bmc.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Shen DQ, Wu ZP, Wu XW, An ZY, Bu XZ, Gu LQ, Huang ZS, An LK. Synthesis and antiproliferative activity of indolizinophthalazine-5,12-dione derivatives, DNA topoisomerase IB inhibitors. Eur J Med Chem. 2010;45:3938–3942. doi: 10.1016/j.ejmech.2010.05.048. [DOI] [PubMed] [Google Scholar]
- 11.Shen DQ, Wu N, Li YP, Wu ZP, Zhang HB, Huang ZS, Gu LQ, An LK. Design, synthesis, and cytotoxicity of indolizinoquinoxaline-5,12-dione derivatives, novel DNA topoisomerase IB inhibitors. Aust J Chem. 2010;63:1116–1121. [Google Scholar]
- 12.Song Y, Shao Z, Dexheimer TS, Scher ES, Pommier Y, Cushman M. Structure- based design, synthesis, and biological studies of new anticancer norindenoisoquinoline topoisomerase I inhibitors. J Med Chem. 2010;53:1979–1989. doi: 10.1021/jm901649x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagarajan M, Morrell A, Ioanoviciu A, Antony S, Kohlhagen G, Agama K, Hollingshead M, Pommier Y, Cushman M. Synthesis and evaluation of indenoisoquinoline topoisomerase I inhibitors substituted with nitrogen heterocycles. J Med Chem. 2006;49:6283–6289. doi: 10.1021/jm060564z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Delcros JG, Cannon L, Konate F, Carias H, Biggerstaff J, Gardner RA, IV OP. Defining the molecular requirements for the selective delivery of polyamine conjugates into cells containing active polyamine transporters. J Med Chem. 2003;46:5129–5138. doi: 10.1021/jm030223a. [DOI] [PubMed] [Google Scholar]
- 15.Defant A, Guella G, Mancini I. Synthesis and in-vitro cytotoxicity evaluation of novel naphtindolizinedione derivatives, part II: improved activity for azaanalogues. Arch Pharm Chem Life Sci. 2009;342:80–86. doi: 10.1002/ardp.200800177. [DOI] [PubMed] [Google Scholar]
- 16.Dexheimer TS, Pommier Y. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat Protoc. 2008;3:1736–1750. doi: 10.1038/nprot.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman HM, Young PR. The interaction of intercalating drugs with nucleic acids. Ann Rev Biophys Bioeng. 1981;10:87–114. doi: 10.1146/annurev.bb.10.060181.000511. [DOI] [PubMed] [Google Scholar]
- 18.Pommier Y, Covey JM, Kerngan D, Markovits J, Pham R. DNA unwinding and inhibition of mouse leukemia L1210 DNA topoisomerase I by intercalators. Nucleic Acids Res. 1987;15:6713–6731. doi: 10.1093/nar/15.16.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu LF, Miller KG. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci U S A. 1981;78:3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetrie assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


