A genetic and molecular mechanism involving the ubiquitin receptors DA1, DAR1, and DAR2 and the transcription factors TCP14 and TCP15 links cell and organ growth with the regulation of endoreduplication.
Abstract
Organ growth involves the coordination of cell proliferation and cell growth with differentiation. Endoreduplication is correlated with the onset of cell differentiation and with cell and organ size, but little is known about the molecular mechanisms linking cell and organ growth with endoreduplication. We have previously demonstrated that the ubiquitin receptor DA1 influences organ growth by restricting cell proliferation. Here, we show that DA1 and its close family members DAR1 and DAR2 are redundantly required for endoreduplication during leaf development. DA1, DAR1, and DAR2 physically interact with the transcription factors TCP14 and TCP15, which repress endoreduplication by directly regulating the expression of cell-cycle genes. We also show that DA1, DAR1, and DAR2 modulate the stability of TCP14 and TCP15 proteins in Arabidopsis thaliana. Genetic analyses demonstrate that DA1, DAR1, and DAR2 function in a common pathway with TCP14/15 to regulate endoreduplication. Thus, our findings define an important genetic and molecular mechanism involving the ubiquitin receptors DA1, DAR1, and DAR2 and the transcription factors TCP14 and TCP15 that links endoreduplication with cell and organ growth.
INTRODUCTION
Organ growth is determined by the spatial and temporal coordination of cell division, cell growth, and cell differentiation, but relatively little is known about how cell growth and size are regulated during organogenesis. During organ growth, the initiation of cell differentiation is often accompanied by a switch from the mitotic cell cycle to the endoreduplication cycle or endocycle, during which nuclear DNA content is increased by additional rounds of full-genome duplication without intervening cell division, giving rise to cells with higher ploidy (Sugimoto-Shirasu and Roberts, 2003; Breuer et al., 2010, 2014; Edgar et al., 2014). Altered levels of endoreduplication lead to changes in cell number and cell size, thereby influencing organ growth in both plants and animals (Dewitte and Murray, 2003; Dewitte et al., 2003; Breuer et al., 2014; Edgar et al., 2014). For example, during leaf development, cells in leaf primordia actively proliferate, and the growth of leaves is often associated with a switch for most cells to begin endoreduplication with a basipetal polarity. In animals, endoreduplication has crucial roles in establishing organ and body size and in maintaining tissue or organ growth in response to stresses (Flemming et al., 2000; Lee et al., 2009).
Some of the regulatory mechanisms influencing the endocycle are shared between animals and plants, and a crucial step to promote the endocycle is downregulation of the activity of the cyclin-dependent kinase (CDK) complexes associated with mitotic cyclins (Wuarin et al., 2002; Costanzo et al., 2004; Inzé and De Veylder, 2006; Breuer et al., 2014; Edgar et al., 2014). For example, CDKB1;1 interacts with CYCLIN A (CYCA2s) to repress the endoreduplication in Arabidopsis thaliana (Boudolf et al., 2009). Loss-of-function of CYCA2;3 increases the endoreduplication levels in mature organs, whereas coexpression of CYCA2;3 with CDKB1;1 results in ectopic cell divisions and inhibits endoreduplication (Imai et al., 2006; Boudolf et al., 2009). The CYCD3 genes influence cell number in developing leaves by regulating the duration of the mitotic phase and the timing of the transition to endoreduplication (Dewitte et al., 2007). Loss of CYCD3 function results in larger cells with increased ploidy levels (Dewitte et al., 2007), whereas overexpression of CYCD3;1 causes increased numbers of smaller cells with reduced endoreduplication (Dewitte et al., 2003). A key phosphorylation target of d-cyclin kinases appears to be the RETINOBLASTOMA-RELATED (RBR) protein. Inactivation of RBR results in extra endoreduplication in Arabidopsis rosette leaves (Desvoyes et al., 2006). RBR binds to the heterodimeric transcription factors E2F/DP and masks their activation domains. E2F/DP regulate the expression of genes involved in DNA biosynthesis, DNA replication licensing, and DNA replication and repair (Egelkrout et al., 2001; De Veylder et al., 2002; del Mar Castellano et al., 2004). In addition, the posttranslational modification of cyclins influences endoreplication. For example, the anaphase-promoting complex/cyclosome (APC/C) is a multisubunit E3 ubiquitin ligase complex that has a central role in the degradation of CYC proteins. CELL CYCLE SWITCH52 (CCS52) and CELL CYCLE DIVISION20, which activate APC/C, have conserved functions in plants, yeast, and animals (Li et al., 2007; Kevei et al., 2011; Breuer et al., 2012; Guttery et al., 2012). Arabidopsis CCS52A proteins regulate endocycle onset and progression (Lammens et al., 2008; Larson-Rabin et al., 2009; Breuer et al., 2012). UV-B INSENSITIVE4, a plant-specific inhibitor of the APC/C activity, interacts with CCS52A1 to suppress the endocycle by inhibiting CYCA2;3 destruction (Hase et al., 2006; Heyman et al., 2011). Although a number of core cell cycle regulators involved in endoreduplication have been identified, relatively little is known about how their activities contribute to cell and organ growth.
The TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) family of plant-specific transcription factors regulate plant growth and development by influencing cell proliferation and cell differentiation (Cubas et al., 1999; Nath et al., 2003; Palatnik et al., 2003; Li et al., 2005). Class I TCP genes are thought to promote cell proliferation and growth, while class II TCP genes are proposed to repress cell proliferation and growth (Martín-Trillo and Cubas, 2010). The class I gene TCP15 has been reported to regulate endoreduplication by directly binding to the promoters of two cell-cycle genes, RBR and CYCA2;3, and increasing their expression (Li et al., 2012). Arabidopsis plants expressing TCP15SRDX, in which TCP15 is fused with a repressive SRDX domain, form larger cells with increased DNA ploidy, while plants overexpressing TCP15 have smaller cells with decreased DNA ploidy (Li et al., 2012). Another study shows that expression of TCP14SRDX (TCP14 is closely related to TCP15) or TCP15SRDX induces endoreduplication and cell proliferation in leaves but represses cell proliferation in internodes, indicating that the influence of TCP factors on cell proliferation is context-dependent (Kieffer et al., 2011). Thus, TCP14/15 may provide a link between the regulation of endoreduplication and cell and organ growth.
We have previously revealed that the ubiquitin receptor DA1 regulates organ growth by limiting cell proliferation (Li et al., 2008). DA1 contains two ubiquitin-interacting motifs (UIM) typical of ubiquitin receptors (Hicke et al., 2005) and a single LIM domain defined by its conservation with the canonical Lin-11, Isl-1, and Mec-3 domains (Freyd et al., 1990; Hiyama et al., 1999; Li et al., 2008). In Arabidopsis, seven other predicted proteins share extensive sequence similarity in addition to their shared UIM and LIM domains with DA1 and have been named DA1-related (DAR) proteins (Li et al., 2008). Here, we show that DA1 and DA1-related proteins (DAR1 and DAR2) act redundantly to regulate endoreduplication during leaf development. Our findings define a genetic and molecular mechanism linking the activities of DA1, DAR1, and DAR2 with the levels of TCP14/15 proteins and the control of endoreduplication. This mechanism may play a role in coordinating the extent of endoreduplication during cell and organ growth.
RESULTS
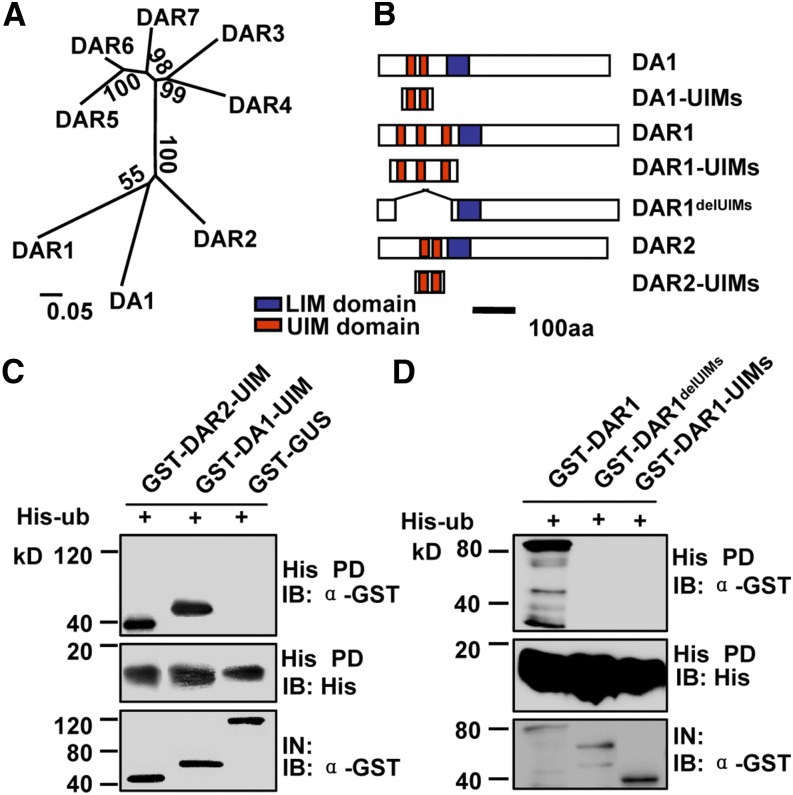
UIMs of DAR1 and DAR2 Are Required for Ubiquitin Binding
DA1, DAR1, and DAR2 are the most closely related family members in Arabidopsis and contain multiple UIMs in their respective N-terminal regions (Figures 1A and 1B). DA1 UIMs bind ubiquitin in vitro (Figure 1C) (Li et al., 2008). Therefore, we tested the ubiquitin binding activity of DAR1 and DAR2. DA1-UIMs, DAR1-UIMs, and DAR2-UIMs were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli. Ubiquitin was expressed in E. coli as a His fusion protein. As shown in Figures 1B to 1D, GST-DA1-UIMs and GST-DAR2-UIMs bound to His-ubiquitin, but GST-DAR1-UIMs and a negative control GST-GUS (for β‑glucuronidase) protein did not. We then tested whether the full-length DAR1 could bind to ubiquitin using DAR1 and DAR1delUIMs (without UIMs) expressed as GST fusion proteins. As shown in Figure 1D, GST-DAR1 bound to His-ubiquitin but DAR1delUIMs did not, indicating that the UIMs of DAR1 are essential but not sufficient for binding ubiquitin. Thus, these results suggest that DAR1 and DAR2 may also act as ubiquitin receptors.
Figure 1.
UIMs of DA1, DAR1, and DAR2 Are Required for Ubiquitin Binding.
(A) Phylogenetic tree of the DA1 family members in Arabidopsis. The phylogenetic tree was constructed using the neighbor-joining method of the MEGA 4.1 program. Values at nodes represent percentages of 1000 bootstrap replicates. Alignments used to generate the phylogeny are presented in Supplemental Data Set 1. The scale bar at bottom represents genetic distance.
(B) Schematic diagram of DA1, DAR1, and DAR2 and their derivatives with different domains. aa, amino acids.
(C) UIMs of DA1 and DAR2 interact with ubiquitin in vitro. GST-DA1-UIMs and GST-DAR2-UIMs were pulled down (PD) by His-ubiquitin immobilized on Ni-NTA agarose and analyzed by immunoblotting (IB) using an anti-GST antibody.
(D) UIMs of DAR1 are required for ubiquitin binding. GST-DAR1, GST-DAR1-UIMs, and DAR1delUIMs without UIM domains were pulled down by His-ubiquitin immobilized on Ni-NTA agarose and analyzed by immunoblotting using an anti-GST antibody.
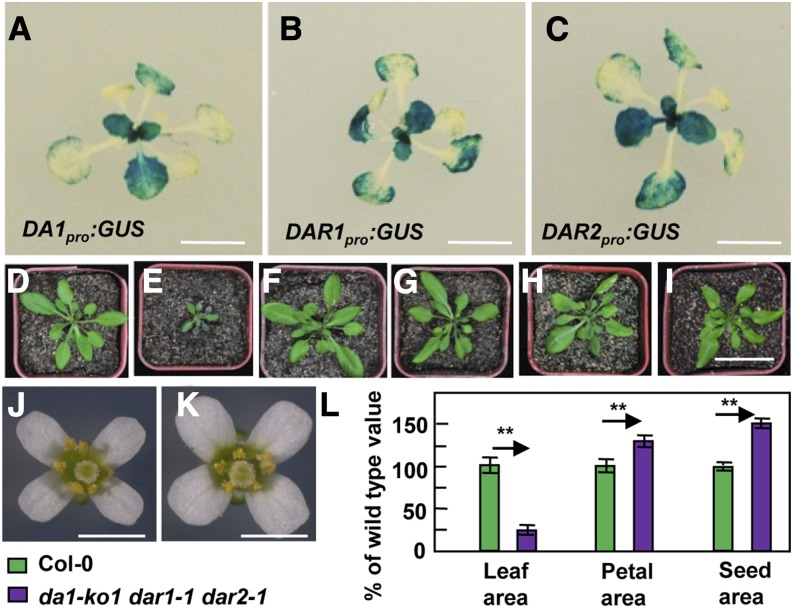
DA1, DAR1, and DAR2 Are Redundantly Required for Plant Growth and Development in an Organ-Dependent Manner
As DA1, DAR1, and DAR2 are closely related family members in Arabidopsis, we tested whether they have redundant functions in organ growth. We first investigated the tissue-specific expression patterns of DA1, DAR1, and DAR2 using a histochemical assay for GUS activity of transgenic plants containing DA1pro:GUS, DAR1pro:GUS, and DAR2pro:GUS, respectively. As shown in Figures 2A to 2C, DA1, DAR1, and DAR2 had similar expression patterns during leaf development, suggesting that they might possess overlapping functions in leaf growth.
Figure 2.
DA1, DAR1, and DAR2 Redundantly Regulate Plant Growth and Development in a Context-Dependent Manner.
(A) to (C) The expression activity of DA1, DAR1, and DAR2 was monitored by DA1pro:GUS, DAR1pro:GUS, and DAR2pro:GUS transgene expression. Histochemical analysis of GUS activity in 16-d-old plants is shown.
(D) to (I) Thirty-day-old plants of Col-0 (D), da1-ko1 dar1-1 dar2-1 (E), 35S:DA1;da1-ko1 dar1-1 dar2-1 (F), 35S:DAR1;da1-ko1 dar1-1 dar2-1 (G), 35S:DAR2;da1-ko1 dar1-1 dar2-1 (H), and gDAR2;da1-ko1 dar1-1 dar2-1 (I). 35S:DA1;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with the DA1 coding sequence driven by the 35S promoter. 35S:DAR1;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with the DAR1 coding sequence driven by the 35S promoter. 35S:DAR2;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with the DAR2 coding sequence driven by the 35S promoter. gDAR2;da1-ko1 dar1-1 dar2-1 is da1-ko1 dar1-1 dar2-1 transformed with a DAR2 genomic copy. Expression of DA1, DAR1, DAR2, or gDAR2 rescued the small-plant phenotype of da1-ko1 dar1-1 dar2-1.
(J) and (K) Flowers of Col-0 and da1-ko1 dar1-1 dar2-1 plants.
(L) Leaf area, petal area, and seed area of Col-0 and da1-ko1 dar1-1 dar2-1 plants. Values are given as means ± se relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
Bars in (A) to (C) = 2 cm; bar in (D) to (I) = 5 cm; bars in (J) and (K) = 2 mm.
The da1-1 mutant produces large seeds and organs due to increased cell proliferation (Li et al., 2008), and the mutant protein encoded by the da1-1 allele was proposed to have negative activity toward DA1 and DAR1 (Li et al., 2008). Although da1-ko1 and dar1-1 single mutants did not show any obvious growth phenotypes, the da1-ko1 dar1-1 double mutant produced larger seeds and flowers than did the wild type (Supplemental Figure 1A) (Li et al., 2008; Xia et al., 2013). Interestingly, the da1-ko1 dar1-1 double mutant formed slightly small leaves compared with the wild type (Supplemental Figure 1C), although da1-ko1 dar1-1 plants looked normal. By contrast, the double mutants da1-ko1 dar2-1 and dar1-1 dar2-1 did not exhibit altered flower and leaf size (Supplemental Figure 1). We then generated a da1-ko1 dar1-1 dar2-1 triple mutant. Unexpectedly, the da1-ko1 dar1-1 dar2-1 triple mutant exhibited very small plants and leaves compared with wild-type plants (Figures 2D, 2E, and 2L). The da1-ko1 dar1-1 dar2-1 mutant still formed larger flowers and seeds than the wild type (Figures 2J to 2L). Therefore, the simultaneous disruption of DA1, DAR1, and DAR2 affects plant growth and development in an organ-dependent manner.
We then introduced 35S:DA1, 35S:DAR1, and 35S:DAR2 constructs into da1-ko1 dar1-1 dar2-1 plants and isolated 21, 30, and 28 transgenic plants, respectively. Most transgenic lines exhibited a complementation of the small plant phenotype of da1-ko1 dar1-1 dar2-1 (Figures 2D to 2I; Supplemental Table 1). Complementation of da1-ko1 dar1-1 dar2-1 with 35S:DA1, 35S:DAR1, and 35S:DAR2 also restored the petal and seed size phenotypes of dar1-1 dar2-1, da1-ko1 dar2-1, and da1-ko1 dar1-1, respectively (Supplemental Figure 2). However, overexpression of DA1, DAR1, or DAR2 in wild-type Columbia-0 (Col-0) plants did not affect leaf and petal size (Supplemental Figure 3). Transformation of da1-ko1 dar1-1 dar2-1 with a DAR2 genomic fragment (gDAR2) also restored the phenotypes of da1-ko1 dar1-1 (Figure 2I; Supplemental Figure 2C). These results demonstrate that the simultaneous disruption of DA1, DAR1, and DAR2 results in small plant and leaf phenotypes and also indicate that DA1, DAR1, and DAR2 are redundantly required for plant growth and development in a context-dependent manner.
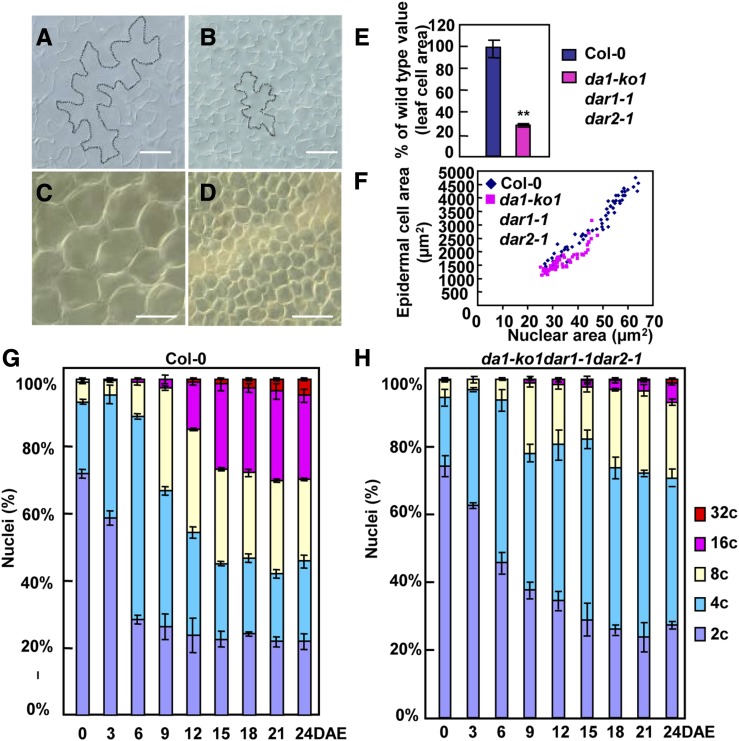
DA1, DAR1, and DAR2 Are Redundantly Required for Normal Endoreduplication Levels during Leaf Development
During organogenesis, the final size of an organ is determined by cell number and cell size. To assess their contributions to leaf size in da1-ko1 dar1-1 dar2-1, we first measured epidermal and palisade cell sizes in wild-type and da1-ko1 dar1-1 dar2-1 fifth leaves. Epidermal and palisade cells in da1-ko1 dar1-1 dar2-1 fifth leaves were dramatically smaller than those in wild-type fifth leaves (Figures 3A to 3F); the average area of palisade cells in da1-ko1 dar1-1 dar2-1 fifth leaves was ∼29.2% of that in wild-type fifth leaves (Figure 3E). By contrast, the average area of cells in da1-ko1 dar1-1 dar2-1 petals was comparable with that in the wild type or single mutants (Supplemental Figure 1B), indicating that the triple mutant da1-ko1 dar1-1 dar2-1 had an organ-dependent reduction in cell size.
Figure 3.
DA1, DAR1, and DAR2 Act Redundantly to Regulate Endoreduplication during Leaf Development.
(A) and (B) Epidermal cells in Col-0 (A) and da1-ko1 dar1-1 dar2-1 (B) fifth leaves.
(C) and (D) Palisade cells in Col-0 (C) and da1-ko1 dar1-1 dar2-1 (D) fifth leaves.
(E) Average area of palisade cells in Col-0 and da1-ko1 dar1-1 dar2-1 fifth leaves. Values are given as means ± se relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
(F) Nuclear size distribution of epidermal cells in Col-0 and da1-ko1 dar1-1 dar2-1 fifth leaves.
(G) and (H) Nuclear DNA ploidy distribution of Co-0 (G) and da1-ko1 dar1-1 dar2-1 (H) fifth leaves measured over a period of 24 DAE. At 0 DAE, the fifth leaves (∼0.5 mm2) were visible. The values represent averages of three independent biological replicates.
Bars in (A) and (B) = 50 μm; bars in (C) and (D) = 100 μm.
A reduction in ploidy levels is often correlated with a decrease in cell size (Sugimoto-Shirasu and Roberts, 2003). To explore whether the reduced cell size in da1-ko1 dar1-1 dar2-1 leaves was associated with a decrease in ploidy levels, we performed a time-course analysis of ploidy levels in wild-type and da1-ko1 dar1-1 dar2-1 fifth leaves (Figures 3G and 3H; Supplemental Figure 4A). At 0, 3, and 6 d after emergence (DAE), most cells in wild-type and da1-ko1 dar1-1 dar2-1 fifth leaves exhibited a 2C or 4C DNA content, suggesting a high mitotic activity at this stage of development. The da1-ko1 dar1-1 dar2-1 mutant shifted the balance of 2C and 4C toward 2C compared with the wild type at 3 and 6 DAE. At 6 DAE, 8C and 16C cells were detected in wild-type leaves, indicating the onset of endoreduplication. By contrast, in da1-ko1 dar1-1 dar2-1 leaves, the switch from the mitotic cycle to the endocycle was delayed, showing reduced levels of 8C nuclei and an absence of 16C nuclei (Figures 3G and 3H). At 9 DAE and subsequent time points during leaf growth, the 8C to 32C fractions in da1-ko1 dar1-1 dar2-1 were much lower than those in the wild type, while the fractions of 2C and 4C DNA were much higher in da1-ko1 dar1-1 dar2-1 than those in the wild type. At 15 DAE and subsequent time points, the fraction of 2C DNA in da1-ko1 dar1-1 dar2-1 was similar to that in the wild type, whereas the fraction of 4C DNA was obviously higher in da1-ko1 dar1-1 dar2-1 than that in the wild type, suggesting an arrest of G2/M phase. At 24 DAE, ∼54% of the cells in wild-type leaves had 8C to 32C nuclei, while only 29% of cells in da1-ko1 dar1-1 dar2-1 leaves had 8C to 32C nuclei. Ploidy level distribution measured by flow cytometry can be expressed as an endoreduplication index (Noir et al., 2013), which represents the average number of endocycles undergone by a given nucleus. During leaf growth, the average number of endocycles in da1-ko1 dar1-1 dar2-1 was reduced compared with that in the wild type (Supplemental Figure 4B). As reduced ploidy is often associated with decreased nuclear size, we measured nuclear size in epidermal cells from fifth leaves. As shown in Figure 3F, the area of nuclei in epidermal cells of da1-ko1 dar1-1 dar2-1 leaves was smaller than that in the wild type. In addition, we measured ploidy levels in wild-type and da1-ko1 dar1-1 dar2-1 petals. As shown in Supplemental Figure 5, the ploidy levels in da1-ko1 dar1-1 dar2-1 petals were similar to those in wild-type petals. Taken together, these results indicated that DA1, DAR1, and DAR2 are redundantly required for endoreduplication during leaf growth.
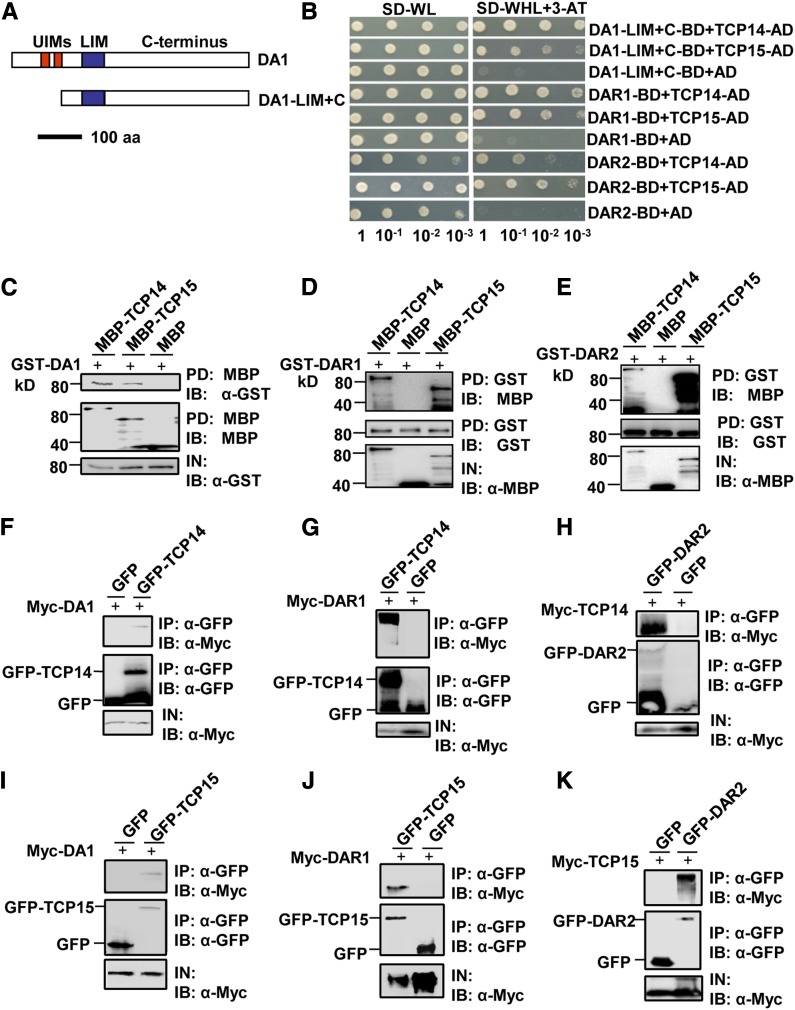
DA1, DAR1, and DAR2 Physically Interact with TCP14 and TCP15 in Vitro and in Vivo, Respectively
To understand how DA1, DAR1, and DAR2 act redundantly to influence endoreduplication during leaf growth, we performed a yeast two-hybrid screen to identify putative DA1 binding proteins. DA1 contains two UIMs, a single LIM domain, and the conserved C-terminal region (Figure 4A). Because DA1 autoactivated the reporter gene when fused to the GAL4 DNA binding domain (BD), a truncated version of DA1, DA1-LIM+C, containing the LIM domain and the C-terminal region, was fused to the GAL4 DNA binding domain and used as a bait. Among several interacting proteins, four different clones corresponding to TCP15 were identified in this screen. TCP15 has recently been shown to regulate endoreduplication by promoting the expression of several cell-cycle genes (Kieffer et al., 2011; Li et al., 2012), suggesting that TCP15 may be involved in the control of endoreduplication by DA1, DAR1, and DAR2. As both TCP15 and its closest homolog TCP14 act redundantly to influence endoreduplication in Arabidopsis (Kieffer et al., 2011; Li et al., 2012), we then tested whether DA1 could interact with TCP14. As shown in Figure 4B, DA1-LIM+C also interacted with TCP14 in a yeast two-hybrid assay. Because DA1, DAR1, and DAR2 are redundantly required for endoreduplication, we then asked whether DAR1 and DAR2 also interact with TCP14 and TCP15. As expected, DAR1 and DAR2 interacted with both TCP14 and TCP15 (Figure 4B).
Figure 4.
DA1, DAR1, and DAR2 Interact with TCP14 and TCP15 in Vitro and in Vivo.
(A) Schematic diagram of DA1 and DA1-LIM+C. The predicted DA1 protein contains two UIM motifs, a single LIM domain, and the C-terminal region. aa, amino acids.
(B) DA1-LIM+C, DAR1, and DAR2 interact with TCP14 and TCP15 in yeast.
(C) DA1 directly interacts with TCP14 and TCP15 in vitro. GST-DA1 was pulled down (PD) by MBP-TCP14 and MBP-TCP15 immobilized on amylose resin and analyzed by immunoblotting (IB) using an anti-GST antibody.
(D) DAR1 directly interacts with TCP14 and TCP15 in vitro. MBP-TCP14 and MBP-TCP15 were pulled down by GST-DAR1 immobilized on GST beads and analyzed by immunoblotting using an anti-MBP antibody.
(E) DAR2 directly interacts with TCP14 and TCP15 in vitro. MBP-TCP14 and MBP-TCP15 were pulled down by GST-DAR2 immobilized on GST beads and analyzed by immunoblotting using an anti-MBP antibody.
(F) and (I) DA1 interacts with TCP14 and TCP15 in vivo. N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring 35S:Myc-DA1 and 35S:GFP-TCP14 or 35S:GFP-TCP15 plasmids. Total proteins were immunoprecipitated (IP) with GFP-Trap-A, and the immunoblot was probed with anti-GFP and anti-Myc antibodies, respectively. Myc-DA1 was detected in the immunoprecipitated GFP-TCP14 or GFP-TCP15 complex.
(G) and (J) DAR1 interacts with TCP14 and TCP15 in vivo. N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring 35S:Myc-DAR1 and 35S:GFP-TCP14 or 35S:GFP-TCP15 plasmids. Total proteins were immunoprecipitated with GFP-Trap-A, and the immunoblots were probed with anti-GFP and anti-Myc antibodies. Myc-DAR1 was detected in the immunoprecipitated GFP-TCP14 or GFP-TCP15 complex.
(H) and (K) DAR2 interacts with TCP14 and TCP15 in vivo. N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring 35S:GFP-DAR2 and 35S:Myc-TCP14 or 35S:My-TCP15 plasmids. Total proteins were immunoprecipitated with GFP-Trap-A, and the immunoblots were probed with anti-GFP and anti-Myc antibodies. Myc-TCP14 and Myc-TCP15 were detected in the immunoprecipitated GFP-DAR2 complex.
To confirm the physical interactions of DA1, DAR1, and DAR2 with TCP14 and TCP15, we expressed DA1, DAR1, and DAR2 as GST fusion proteins in E. coli, while TCP14 and TCP15 were expressed as maltose binding protein (MBP) fusion proteins in E. coli. As shown in Figures 4C to 4E, GST-DA1, GST-DAR1, and GST-DAR2 all bound to both MBP-TCP14 and MBP-TCP15, while they did not bind to MBP alone. These results indicated that DA1, DAR1, and DAR2 all physically interact with both TCP14 and TCP15 in vitro, confirming the interactions observed in yeast cells.
Finally, to demonstrate whether DA1, DAR1, and DAR2 also physically associate with TCP14/15 in planta, we used coimmunoprecipitation analyses to detect their interactions in vivo. We transiently coexpressed 35S:Myc-DA1 with 35S:GFP-TCP14 or 35S:GFP-TCP15 in Nicotiana benthamiana leaves. Transiently expressed 35S:GFP and 35S:Myc-DA1 were used as negative controls. Total proteins were isolated and incubated with green fluorescent protein (GFP)-Trap-A agarose beads to immunoprecipitate GFP-TCP14, GFP-TCP15, and GFP. Immunoprecipitated proteins were detected with anti-GFP and anti-Myc antibodies, respectively. As shown in Figures 4F and 4I, Myc-DA1 was detected in the immunoprecipitated GFP-TCP14 and GFP-TCP15 complexes but not in the negative control (GFP), indicating that DA1 physically associates with TCP14 and TCP15 in planta. We then transiently coexpressed 35S:Myc-DAR1 with 35S:GFP-TCP14 or 35S:GFP-TCP15 in N. benthamiana leaves. Myc-DAR1 was detected in the immunoprecipitated GFP-TCP14 and GFP-TCP15 complexes (Figures 4G and 4J). 35S:GFP-DAR2 was also transiently coexpressed with 35S:Myc-TCP14 or 35S:Myc-TCP15 in N. benthamiana leaves. Coimmunoprecipitation analyses showed that Myc-TCP14 and Myc-TCP15 were detected in the immunoprecipitated GFP-DAR2 complex (Figures 4H and 4K). Together, these results showed that DA1, DAR1, and DAR2 all physically associated with both TCP14 and TCP15 in planta.
DA1, DAR1, and DAR2 Modulate the Stability of TCP14/15
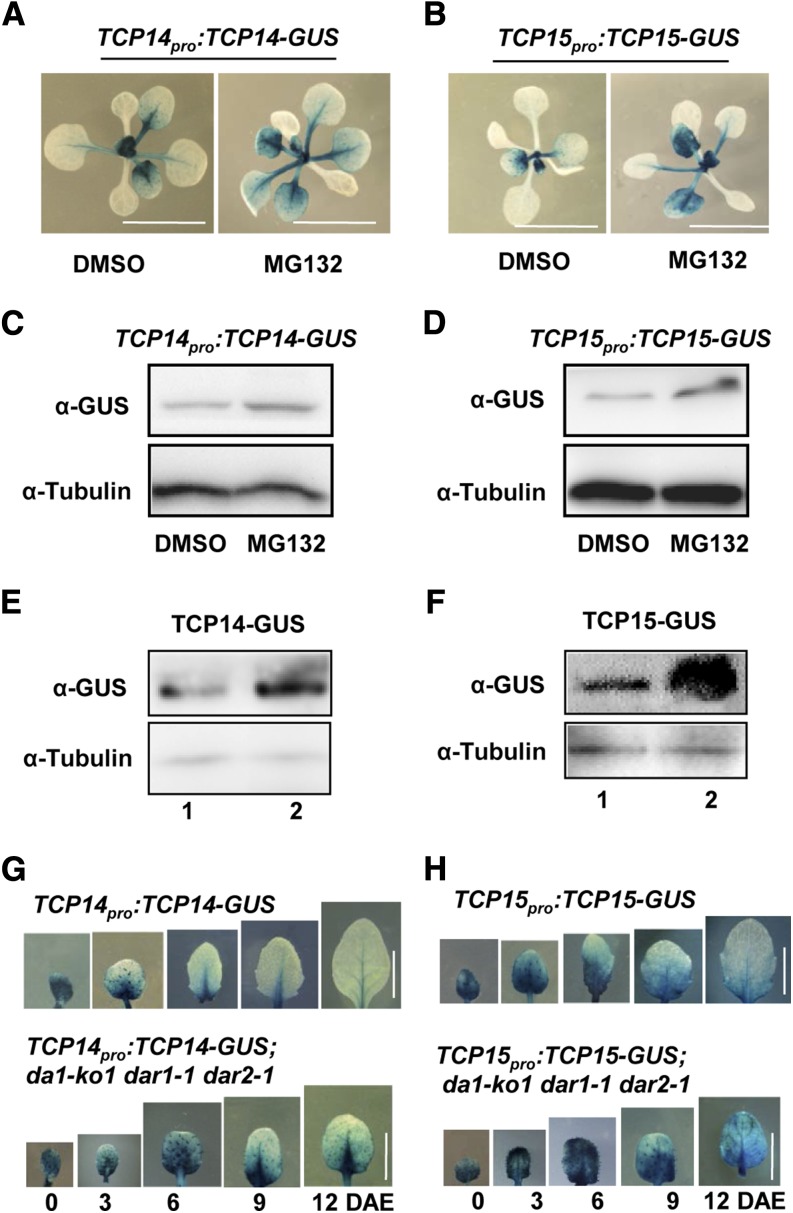
DA1 encodes a ubiquitin receptor that has been proposed to be involved in ubiquitin-mediated protein degradation (Li et al., 2008; Xia et al., 2013; Du et al., 2014). To test whether TCP14/15 protein levels might be affected by proteasome-dependent mechanisms, we treated Arabidopsis TCP14pro:TCP14-GUS and TCP15pro:TCP15-GUS transgenic lines (Kieffer et al., 2011; Li et al., 2012) with the proteasome inhibitor MG132. After MG132 treatment, GUS enzyme activity of the TCP14pro:TCP14-GUS and TCP15pro:TCP15-GUS transgenic plants was increased compared with that in untreated plants (Figures 5A and 5B). Furthermore, the levels of TCP14-GUS and TCP15-GUS fusion proteins detected in immunoblots in MG132-treated plants were higher than those in untreated plants (Figures 5C and 5D). These results indicated that the ubiquitin proteasome influences the stability of TCP14/15.
Figure 5.
DA1, DAR1, and DAR2 Modulate the Stability of TCP14/15.
(A) to (D) A proteasome inhibitor stabilizes TCP14 and TCP15. Histochemical analysis of GUS activity of 10-d-old TCP14pro:TCP14-GUS (A) and TCP15pro:TCP15-GUS (B) seedlings treated with or without 20 μM MG132 is shown. Total protein extracts from TCP14pro:TCP14-GUS (C) and TCP15pro:TCP15-GUS (D) were subjected to immunoblot assays using anti-GUS and anti-tubulin (as loading control) antibodies.
(E) The TCP14-GUS protein accumulates at higher levels in TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1 plants (2) than in TCP14pro:TCP14-GUS plants (1). Total protein extracts were subjected to immunoblot assays using anti-GUS and anti-tubulin (as loading control) antibodies.
(F) The TCP15-GUS protein accumulates at higher levels in TCP15pro:TCP15-GUS;da1-ko1 dar1-1 dar2-1 plants (2) than in TCP15pro:TCP15-GUS plants (1). Total protein extracts were subjected to immunoblot assays using anti-GUS and anti-tubulin (as loading control) antibodies.
(G) Histochemical analysis of GUS activity in TCP14pro:TCP14-GUS and TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1 transgenic plants measured over a period of 12 DAE.
(H) Histochemical analysis of GUS activity in TCP15pro:TCP15-GUS and TCP15pro:TCP15-GUS;da1-ko1 dar1-1 dar2-1 transgenic plants measured over a period of 12 DAE.
Bars in (A) and (B) = 1 cm; bars in (G) and (H) = 5 mm.
Considering that DA1, DAR1, and DAR2 physically associate with TCP14/15 (Figure 4), we then asked whether DA1, DAR1, and DAR2 could affect the stability of TCP14/15 proteins in Arabidopsis. To test this, we crossed da1-ko1 dar1-1 dar2-1 with TCP14pro:TCP14-GUS and TCP15pro:TCP15-GUS transgenic lines, respectively. After selection of plants containing TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1 and TCP15pro:TCP15-GUS;da1-ko1 dar1-1 dar2-1, we measured and compared the levels of TCP14-GUS or TCP15-GUS proteins in different genetic backgrounds. As shown in Figures 5E and 5F, TCP14-GUS and TCP15-GUS protein levels were clearly higher in the da1-ko1 dar1-1 dar2-1 mutant background than those in wild-type plants. By contrast, da1-ko1 dar1-1 dar2-1 did not strongly affect transcript levels of TCP14/15 (Supplemental Figure 6). These results indicated that DA1, DAR1, and DAR2 redundantly influence the stability of TCP14/15 in Arabidopsis.
We further performed a time-course analysis of the GUS enzyme activity in the fifth leaves of TCP14pro:TCP14-GUS, TCP15pro:TCP15-GUS, TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1, and TCP15pro:TCP15-GUS;da1-ko1 dar1-1 dar2-1 transgenic plants. At 0 DAE, the GUS activity was very high in both TCP14pro:TCP14-GUS and TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1 fifth leaves (Figure 5G). At 6 DAE and subsequent time points, the GUS activity in TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1 fifth leaves was obviously higher than that in TCP14pro:TCP14-GUS fifth leaves (Figure 5G). Similarly, the GUS activity in TCP15pro:TCP15-GUS;da1-ko1 dar1-1 dar2-1 fifth leaves was clearly stronger than that in TCP15pro:TCP15-GUS fifth leaves at 6 DAE and subsequent time points (Figure 5H). Thus, these results further indicate that DA1, DAR1, and DAR2 modulate the levels of TCP14/15 proteins.
DA1, DAR1, and DAR2 Act in a Common Pathway with TCP14/15 to Regulate Endoreduplication during Leaf Development
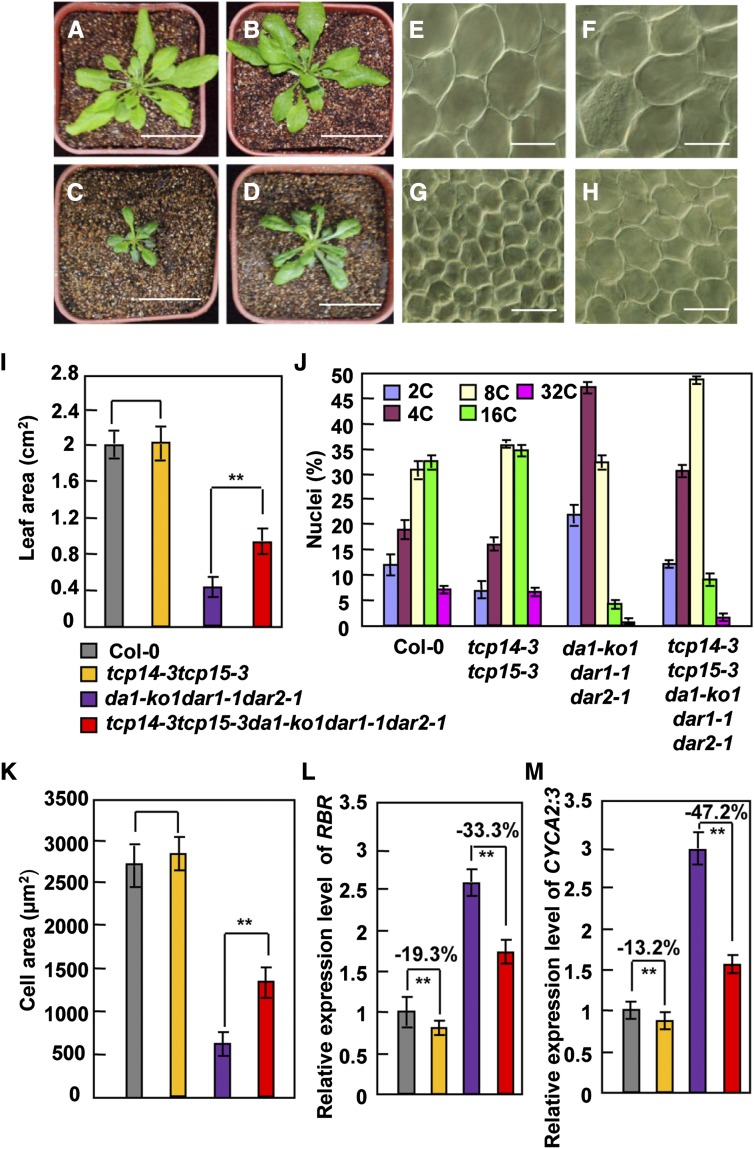
As DA1, DAR1, and DAR2 interact with TCP14/15 and modulate the stability of TCP14/15 proteins, we sought to establish genetic relationships between the DA1 family members (DA1, DAR1, and DAR2) and TCP14/15 in the regulation of endoreduplication, cell size, and organ growth. Therefore, we generated the pentuple tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 mutant. Rosette leaves of tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 plants were clearly larger than those of the da1-ko1 dar1-1 dar2-1 plants. By contrast, the size of tcp14-3 tcp15-3 rosette leaves was not significantly different from that of wild-type rosette leaves (Figures 6A to 6D and 6I) (Kieffer et al., 2011), indicating that tcp14-3 tcp15-3 partially suppressed the small-leaf phenotype of da1-ko1 dar1-1 dar2-1. Considering that da1-ko1 dar1-1 dar2-1 plants produced smaller leaves with small cells, we then tested whether tcp14-3 tcp15-3 could rescue the small cell size phenotype of da1-ko1 dar1-1 dar2-1 leaves. Interestingly, cells in tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 fifth leaves were significantly larger than those in da1-ko1 dar1-1 dar2-1 fifth leaves, although the size of cells in tcp14-3 tcp15-3 fifth leaves was comparable with that observed in wild-type fifth leaves (Figures 6E to 6H and 6K).
Figure 6.
DA1, DAR1, and DAR2 Act in a Common Pathway with TCP14/15 to Regulate Endoreduplication.
(A) to (D) Forty-day-old plants of Col-0 (A), tcp14-3 tcp15-3 (B), da1-ko1 dar1-1 dar2-1 (C), and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 (D).
(E) to (H) Palisade cells in Col-0 (E), tcp14-3 tcp15-3 (F), da1-ko1 dar1-1 dar2-1 (G), and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 (H) fifth leaves.
(I) Average area of Col-0, tcp14-3 tcp15-3, da1-ko1 dar1-1 dar2-1, and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 fifth leaves.
(J) Distribution of nuclear ploidy in Col-0, tcp14-3 tcp15-3, da1-ko1 dar1-1 dar2-1, and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 fifth leaves.
(K) Average area of palisade cells in Col-0, tcp14-3 tcp15-3, da1-ko1 dar1-1 dar2-1, and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 fifth leaves.
(L) Expression levels of RBR in Col-0, tcp14-3 tcp15-3, da1-ko1 dar1-1 dar2-1, and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 leaves.
(M) Expression levels of CYCA2;3 in Col-0, tcp14-3 tcp15-3, da1-ko1 dar1-1 dar2-1, and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 leaves.
Values in (I) to (M) are given as means ± se. **P < 0.01 compared with the wild type (Student’s t test). Bars in (A) to (D) = 5 cm; bars in (E) to (H) = 50 μm.
As da1-ko1 dar1-1 dar2-1 leaves had smaller cells with low ploidy levels, we performed flow cytometric examination of nuclei isolated from wild-type, tcp14-3 tcp15-3, da1-ko1 dar1-1 dar2-1, and tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 fifth leaves. The distributions of ploidy classes in tcp14-3 tcp15-3 were similar to those seen in the wild type (Figure 6J; Supplemental Figure 7), consistent with previous studies (Kieffer et al., 2011; Li et al., 2012). However, the proportions of 8C to 32C cells in tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 leaves were higher than those in da1-ko1 dar1-1 dar2-1 leaves (Figure 6J). By contrast, the proportions of 2C and 4C cells in tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 leaves were lower than those in da1-ko1 dar1-1 dar2-1 leaves (Figure 6J). These results revealed that tcp14-3 tcp15-3 partially suppressed the reduced ploidy levels of da1-ko1 dar1-1 dar2-1 leaves, indicating that DA1, DAR1, and DAR2 function in a common pathway with TCP14/15 to regulate endoreduplication during leaf growth in Arabidopsis.
A previous study demonstrated that TCP15 directly binds to the promoter regions of RBR and CYCA2;3 (Li et al., 2012), which are negative regulators of endoreduplication (Desvoyes et al., 2006; Imai et al., 2006). Considering that DA1, DAR1, and DAR2 interact genetically and physically with TCP14/15 and modulate their protein stabilities (Figures 4 to 6), we anticipated that expression of RBR and CYCA2;3 would be increased in da1-ko1 dar1-1 dar2-1. The expression of RBR and CYCA2;3 genes was measured by real-time quantitative RT-PCR, using RNA samples isolated from wild-type and da1-ko1 dar1-1 dar2-1 leaves. As shown in Figures 6L and 6M, transcript levels of RBR and CYCA2;3 in da1-ko1 dar1-1 dar2-1 leaves were dramatically elevated compared with those measured in wild-type leaves. By contrast, expression of RBR and CYCA2;3 genes was only slightly downregulated in tcp14-3 tcp15-3 leaves compared with that measured in wild-type leaves. Expression levels of RBR and CYCA2;3 in tcp14-3 tcp15-3 da1-ko1 dar1-1 dar2-1 leaves were much lower than those in da1-ko1 dar1-1 dar2-1 leaves, indicating that tcp14-3 tcp15-3 partially suppressed the effect of da1-ko1 dar1-1 dar2-1 on the expression of RBR and CYCA2;3. Thus, our genetic analyses show that TCP14/15 act downstream of DA1, DAR1, and DAR2 to influence endoreduplication by regulating the expression of at least two key cell-cycle regulators.
DISCUSSION
Organ growth is determined by the coordinated progression of cellular proliferation, growth, and differentiation. How the duration of cell proliferation, the extent of cell growth, and the onset of differentiation are coordinately regulated to determine the characteristic final size and shape of organs is a major question in biology. In animals and plants, endoreduplication has often been associated with cell and organ growth, and it is also correlated with the onset of cell differentiation (Dewitte and Murray, 2003; Dewitte et al., 2003; Breuer et al., 2014; Edgar et al., 2014). Although a number of core cell-cycle regulators involved in endoreduplication have been identified, how their activities are coordinated during cell and organ growth are not well understood. In this study, we show that the organ growth regulator DA1 and its close family members DAR1 and DAR2 are redundantly required for the progressive increase in cells with higher ploidy levels during leaf development. DA1, DAR1, and DAR2 physically associate with the transcription factors TCP14/15, which regulate endoreduplication by directly influencing the expression of several cell-cycle genes (Li et al., 2012). Our data reveal that DA1, DAR1, and DAR2 modulate the levels of TCP14/15 proteins in Arabidopsis. Genetic analyses further demonstrate that DA1, DAR1, and DAR2 function in a common pathway with TCP14/15 to regulate endoreduplication by influencing the expression of RBR and CYCA2;3. Thus, our findings establish genetic and molecular mechanisms linking cell and organ growth with the regulation of endoreduplication by the ubiquitin receptors DA1, DAR1, and DAR2 and the transcription factors TCP14/15.
DA1, DAR1, and DAR2 Are Redundantly Required for Endoreduplication during Leaf Development
We previously identified the ubiquitin receptor DA1 as a negative regulator of organ size (Li et al., 2008). DA1, DAR1, and DAR2 are close family members (Figure 1A), suggesting that they might act redundantly to influence organ growth. The da1-ko1 dar1-1 mutant produced larger seeds and flowers than the wild type or either of the single mutants (Li et al., 2008; Xia et al., 2013). Interestingly, the da1-ko1 dar1-1 mutant showed slightly small fifth leaves (Supplemental Figure 1C), although da1-ko1 dar1-1 plants look normal. By contrast, dar2-1, da1-ko1 dar2-1, and dar1-1 dar2-1 mutants did not show any obvious phenotypes in aerial organs (Supplemental Figure 1). Surprisingly, we observed that the da1-ko1 dar1-1 dar2-1 triple mutant produced larger seeds and flowers but had much smaller plants and leaves than the wild type (Figures 2D and 2E). These data indicated that the simultaneous disruption of DA1, DAR1, and DAR2 has organ-dependent effects on growth and also suggested that they are redundantly required for plant growth. Similarly, the simultaneous disruption of AHK2, AHK3, and AHK4 has context-dependent effects on growth, and the ahk2 ahk3 ahk4 triple mutant had small leaves and large seeds in Arabidopsis (Riefler et al., 2006). DA1, DAR1, and DAR2 had similar expression patterns during leaf growth (Figures 2A to 2C), consistent with their redundant functions in leaf development.
The small-leaf phenotype of da1-ko1 dar1-1 dar2-1 was mainly caused by a reduction in cell size (Figures 3A to 3E). Endoreduplication is an important determinant of cell and organ size, shown by the correlation of cell size and ploidy levels in Arabidopsis and other plant species (Dewitte and Murray, 2003; Dewitte et al., 2003; Sugimoto-Shirasu and Roberts, 2003; Breuer et al., 2014; Gegas et al., 2014). Consistent with these observations, the overall ploidy levels in da1-ko1 dar1-1 dar2-1 leaves were dramatically reduced compared with those in wild-type leaves (Figures 3G and 3H; Supplemental Figure 4). A time-course analysis of ploidy levels in developing leaves indicated that da1-ko1 dar1-1 dar2-1 delayed the switch from the mitotic cycle to the endocycle at early stages of leaf development (Figures 3G and 3H). At the late stages of leaf development, we also observed that the fraction of 2C DNA in da1-ko1 dar1-1 dar2-1 was similar to that in the wild type, while the number of 4C cells in da1-ko1 dar1-1 dar2-1 was higher than that in the wild type (Figures 3G and 3H), suggesting an arrest of G2/M phase. At 6 DAE and subsequent time points, da1-ko1 dar1-1 dar2-1 had reduced proportions of cells with higher ploidy (Figures 3G and 3H; Supplemental Figure 4). The endoreduplication index in da1-ko1 dar1-1 dar2-1 leaves was decreased in comparison with that in wild-type leaves (Supplemental Figure 4B). Considering that da1-ko1, dar1-1, dar2-1, da1-ko1 dar2-1, and dar1-1 dar2-1 mutants did not influence endoreduplication in leaves and da1-ko1 dar1-1 showed slightly reduced ploidy levels in leaves (Supplemental Figure 8A), the disruption of DA1, DAR1, and DAR2 may affect endoreduplication in a dosage-dependent manner.
We have previously shown that da1-1 formed large leaves with more cells (Li et al., 2008). Kinematic analysis of leaf growth showed that da1-1 formed larger leaves with more normal-sized cells due to the increased cell proliferation, possibly by influencing the proportions of mitotic and endocycling cells during leaf development. However, leaves of the triple da1-ko1 dar1-1 dar2-1 mutant have reduced cell numbers and smaller cells (Figures 3A to 3E; Supplemental Figure 9). These seemingly opposite effects might be related to the much reduced activity in the functionally redundant triple mutant; this leads to very small cells with reduced competence to divide, resulting in reduced cell numbers. Small decreases in activity seen in the da1-1 allele do not strongly reduce cell size, and these cells may have normal competence to divide. Similar possible dosage-dependent effects were seen for strong and mild CCS52A1-overexpressing (CCS52A1-OE) lines (Larson-Rabin et al., 2009). Cells in highly expressing CCS52A1-OE lines had both reduced proliferation and endoreduplication, while cells with less high levels of CCS52A1 expression showed decreased proliferation but allowed extra endoreduplication. Similarly, strong KRP2-overexpressing (KPR2-OE) lines exhibited decreases in both cell number and cell size, whereas weak KPR2-OE lines had decreased cell number and increased cell size (Verkest et al., 2005). It is possible, therefore, that DA1, DAR1, and DAR2 may act redundantly to regulate endoreduplication levels during leaf development, thereby influencing cell number and cell size in a dosage-dependent manner.
DA1, DAR1, and DAR2 Regulate Endoreduplication by Modulating the Stability of TCP14/15
In a yeast two-hybrid screen, we showed that DA1 interacts with TCP15, which regulates endoreduplication in Arabidopsis (Li et al., 2012). We further demonstrated that DA1, DAR1, and DAR2 physically interact with TCP15 and its closest homolog TCP14 in vitro and in vivo, respectively (Figure 4). TCP14 and TCP15 act redundantly to control endoreduplication (Kieffer et al., 2011), suggesting a functional significance of the interactions of DA1, DAR1, and DAR2 with both TCP14 and TCP15 proteins. DA1 encodes a ubiquitin receptor with two UIMs that bind ubiquitin in vitro (Li et al., 2008). The UIM motifs of DAR1 and DAR2 were also required for ubiquitin binding (Figures 1C and 1D), suggesting that DAR1 and DAR2 are also ubiquitin receptors. Ubiquitin receptors have many functions, including facilitating substrate degradation by the proteasome (Verma et al., 2004). We showed that the stability of TCP14/15 proteins is affected by the proteasome (Figures 5A to 5D). In the da1-ko1 dar1-1 dar2-1 mutant, TCP14 and TCP15 protein levels were higher than those in the wild type (Figures 5E and 5F), showing that DA1, DAR1, and DAR2 are involved in regulating TCP14/15 protein stability (Figure 7). The da1-ko1 dar1-1 dar2-1 triple mutant had a much stronger effect on the stability of TCP14/15 than the da1-ko1 dar1-1 double mutant (Figures 5E and 5F; Supplemental Figures 8B and 8C), suggesting that DA1, DAR1, and DAR2 regulate TCP14/15 protein stability in a dosage-dependent manner. Relatively higher expression of the DA1 family members (DA1, DAR1, and DAR2) and TCP14/15 was observed in younger leaves than in older ones (Figures 2A to 2C) (Li et al., 2008, 2012; Kieffer et al., 2011), suggesting that they have similar expression domains during leaf development and also supporting their overlapping functions. The interactions of DA1, DAR1, and DAR2 with TCP14/15 may facilitate recognition by the proteasomal machinery.
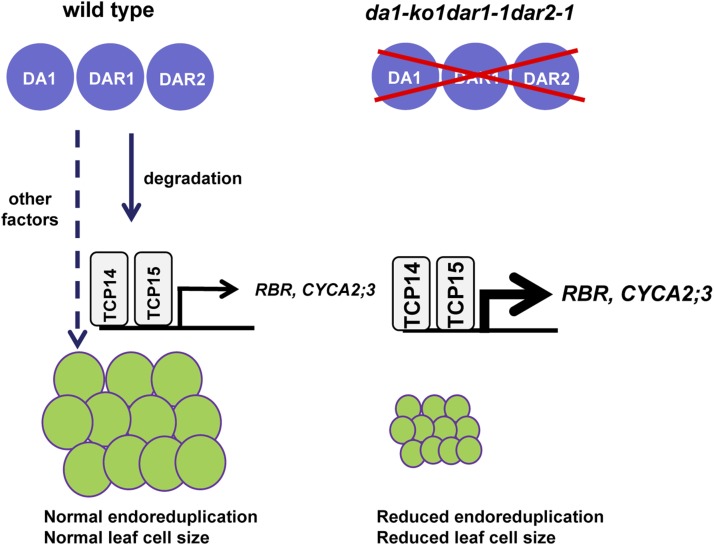
Figure 7.
A Model for DA1, DAR1, and DAR2 Function in Endoreduplication.
In the wild type, DA1, DAR1, and DAR2 act redundantly to regulate endoreduplication by modulating the stability of the transcription factors TCP14/15. TCP14/15 repress endoreduplication by directly regulating the expression of several cell-cycle genes (e.g., RBR and CYCA3;2). Other factors might also mediate the effects of DA1, DAR1, and DAR2 on endoreduplication. In da1-ko1 dar1-1 dar2-1, the transcription factors TCP14/15 are accumulated, resulting in increased expression of RBR and CYCA2;3 and strong repression of endoreduplication.
TCP14 and TCP15 were recently shown to regulate endoreduplication in Arabidopsis (Kieffer et al., 2011; Li et al., 2012). Plants expressing TCP15SRDX showed increased cell size and endoreduplication in leaves, whereas plants overexpressing TCP15 exhibited reduced cell size and endoreduplication in leaves (Li et al., 2012). Although tcp14 tcp15 double mutants did not have any obvious effects on leaf development, expression of TCP14pro:TCP14SRDX in tcp14 tcp15 double mutants (Kieffer et al., 2011) did show increased leaf shape and curling phenotypes compared with expression in wild-type leaves. Consistent with these observations, we found that tcp14-3 tcp15-3 double mutants did not influence leaf size, leaf cell size, and ploidy levels of leaf cells (Figures 6A to 6K). However, tcp14-3 tcp15-3 partially suppressed the decreased leaf size, cell size, and ploidy level phenotypes seen in da1-ko1 dar1-1 dar2-1 triple mutants, indicating that DA1, DAR1, and DAR2 act in a common pathway with TCP14/15 to regulate endoreduplication and cell growth during leaf development.
TCP14/15 have been reported to promote cell proliferation in internodes and to repress cell proliferation in leaves and floral tissues (Kieffer et al., 2011). Overexpression of TCP15 (pTA:TCP15-EYFP with dexamethasone induction) suppressed endoreduplication, while overexpression of TCP15 reduced cell proliferation in roots (Li et al., 2012). TCP14/15 also have been shown to promote cell proliferation in the shoot apex (Davière et al., 2014). These results suggest that the influence of TCP factors on cell proliferation is organ-dependent. Most 35S:TCP15 plants showed a growth arrest after germination (Li et al., 2012), suggesting that the overaccumulation of TCP15 causes defects in plant growth and cell proliferation. It is possible that the accumulation of TCP14/15 in da1-ko1 dar1-1 dar2-1 leaves results in the small-leaf phenotype. By contrast, the weak dominant-negative da1-1 mutant formed large flowers and leaves by increasing cell proliferation (Li et al., 2008). The da1-1 mutant might not affect endoreduplication and the levels of TCP14/15 proteins in petals and leaves. Thus, tcp14-3 tcp15-3 could not suppress the large petal and leaf size phenotypes of the da1-1 mutant (Supplemental Figure 10). Consistent with this, we previously demonstrated that SOD2/UBP15 acts downstream of DA1 to influence seed and flower size by regulating cell proliferation (Du et al., 2014).
TCP15 binds directly to the promoter regions of RBR and CYCA2;3 genes and promotes their expression (Li et al., 2012). RBR and its targets, E2F transcription factors, are known to take part in the regulation of endoreduplication (Desvoyes et al., 2006; Jordan et al., 2007; Magyar et al., 2012). CYCA2;3 also regulates endoreduplication and acts as a key regulator of ploidy levels in Arabidopsis (Imai et al., 2006; Vanneste et al., 2011). By directly modulating the expression of RBR and CYCA2;3, TCP15 may orchestrate cell-cycle genes to regulate endoreduplication. Our results showed that the expression of RBR and CYCA2;3 in da1-ko1 dar1-1 dar2-1 was greatly increased compared with that in the wild type (Figures 6L and 6M). Furthermore, tcp14-3 tcp15-3 suppressed the higher expression levels of RBR and CYCA2;3 in da1-ko1 dar1-1 dar2-1 (Figures 6L and 6M). These results support a model in which TCP14/15 act downstream of DA1, DAR1, and DAR2 and partially mediate the effect of DA1, DAR1, and DAR2 on the expression of RBR and CYCA2;3 (Figure 7). Because tcp14-3 tcp15-3 could not completely suppress the phenotypes of da1-ko1 dar1-1 dar2-1 (Figure 6), it is likely that other factors may also act downstream of DA1, DAR1, and DAR2 to regulate endoreduplication (Figure 7). Therefore, identifying other targets of DA1, DAR1, and DAR2 and understanding their functions will reveal new mechanisms linking endoreduplication and cell and organ growth.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana mutants used in this study were in the Col-0 background. da1-ko1 (SALK_126092), dar1-1 (SALK_067100), dar2-1 (SALK_016122), tcp14-3 (SM_3_19812), and tcp15-3 (SALK_011491) were obtained from the Nottingham Arabidopsis Stock Centre. The T-DNA insertions were identified by PCR and sequencing using the primers described in Supplemental Table 2. TCP14pro:TCP14-GUS (Kieffer et al., 2011) and TCP15pro:TCP15-GUS (Li et al., 2012) transgenic lines were used in this study. Arabidopsis plants were grown in soil at 22°C under long-day conditions (a 16-h-light/8-h-dark cycle).
Morphological and Cellular Analyses
Fully expanded petals (stage 14), leaves, and dry seeds were scanned to produce digital images. The organ and seed sizes were then measured using ImageJ software. Before analyzing cell number and cell size, organs were mounted in a clearing solution (chloral hydrate:water:glycerol, 8:3:1). Cleared samples were imaged using differential interference contrast optics on a Leica microscope (DM2500) and photographed with a SPOT FLEX Cooled CCD Digital Image System.
Ploidy levels were measured using a flow cytometer (BD FACS) according to methods described previously (Verkest et al., 2005; Noir et al., 2013). Briefly, leaves were chopped with a razor blade in buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM MOPS, and 1% Triton X-100), filtered through a cell strainer with a 30-mm mesh, and stained with 20 μg/mL 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) buffer. At least 15,000 nuclei isolated from ∼10 to 20 leaves were used for each ploidy measurement. Flow cytometry experiments were repeated at least three times for each genotype using independent biological replicates.
RNA Isolation and Quantitative Real-Time RT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings and leaves using the RNeasy Plant Mini kit (Tiangen). RT-PCR was performed using SuperScript III reverse transcriptase (Invitrogen). ACTIN2 mRNA was used as an internal control. Quantitative real-time RT-PCR analysis was performed with a LightCycler 480 machine (Roche) using the LightCycler 480 SYBR Green I Master (Roche). At least three biological replicates of each sample were performed in all experiments. The specific primers used for RT-PCR and real-time RT-PCR are described in Supplemental Table 2.
Ubiquitin Binding Assays
The coding sequences of DAR1, DA1-UIMs, DAR1-UIMs, and DAR2-UIMs were cloned into BamHI and XhoI sites of the vector pGEX-4T-1 (Amersham-Pharmacia) to generate GST-DAR1, GST-DA1-UIMs, GST-DAR1-UIMs, and GST-DAR2-UIMs constructs, respectively. The specific primers for GST-DAR1, GST-DA1-UIMs, GST-DAR1-UIMs, and GST-DAR2-UIMs constructs were GST-DAR1-F and GST-DAR1-R, GST-DA1-UIMs-F and GST-DA1-UIMs-R, GST-DAR1-UIMs-F and GST-DAR1-UIMs-R, and GST-DAR2-UIMs-F and GST-DAR2-UIMs-R, respectively (Supplemental Table 2). DAR1delUIMs with the deletion in UIMs was generated by following the instruction manual of PfuUltraII Fusion HS DNA Polymerase (Stratagene). DAR1delUIMs was subcloned into the BamHI and XhoI sites of the vector pGEX-4T-1 (Amersham-Pharmacia) to generate GST-DAR1delUIMs.
To test protein-protein interaction, bacterial lysates containing ∼15 μg of GST-DAR1, GST-DA1-UIMs, GST-DAR1-UIMs, GST-DAR2-UIMs, or GST-DAR1delUIMs fusion proteins were mixed with lysates containing ∼10 μg of His-ubiquitin fusion proteins. Ni-NTA agarose (20 μL; Qiagen) was added into each combined solution with continued rocking at 4°C for 2 h. Ni-NTA agarose was washed five times with buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, pH 8.0, 1% Triton X-100, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1× Complete Protease Inhibitor cocktail [Roche]), and the isolated proteins were separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GST (Abmart) and anti-His (Abmart) antibodies.
Yeast Two-Hybrid Assays
The coding sequences of DA1-LIM+C, DAR1, and DAR2 were cloned into NotI and SalI sites of the bait vector pDBleu (Invitrogen) to generate DA1-LIM+C-BD, DAR1-BD, and DAR2-BD constructs, respectively. The specific primers for DA1-LIM+C-BD, DAR1-BD, and DAR2-BD constructs were DA1-LIM+C-SalI-LP and DA1-LIM+C-NotI-RP, DAR1-SalI-LP and DAR1-NotI-RP, and DAR2-SalI-LP and DAR2-NotI-RP, respectively (Supplemental Table 2).
The coding sequences of TCP14 and TCP15 were cloned into EcoRI and NotI sites of the prey vector pEXP-AD502 (Invitrogen) to generate TCP14-AD and TCP15-AD constructs. The specific primers for TCP14-AD and TCP15-AD constructs were TCP14-EcoRI-LP and TCP14-NotI-RP and TCP15-EcoRI-LP and TCP15-NotI-RP, respectively (Supplemental Table 2). The bait and prey constructs were cotransformed into the yeast strain PJ69-4A to test their interactions.
In Vitro Protein-Protein Interaction
The coding sequence of DA1 was cloned into the BamHI and NotI sites of the pGEX-4T-1 vector to generate the GST-DA1 construct. The coding sequences of DAR1 and DAR2 were cloned into the BamHI and XhoI sites of the pGEX-4T-1 vector to generate GST-DAR1 and GST-DAR2 constructs, respectively. The coding sequences of TCP14 and TCP15 were subcloned into the pMAL-c2 vector to generate MBP-TCP14 and MBP-TCP15 constructs, respectively. The specific primers for GST-DA1, GST-DAR1, GST-DAR2, MBP-TCP14, and MBP-TCP15 constructs are described in Supplemental Table 2.
To test protein-protein interaction, bacterial lysates containing ∼15 μg of GST-DA1 fusion proteins were mixed with lysates containing ∼30 μg of MBP-TCP14 or MBP-TCP15 fusion proteins. Amylose resin (20 μL; New England Biolabs) was added into each combined solution with continued rocking at 4°C for 1 h. Bacterial lysate containing ∼30 μg of MBP-TCP14 or MBP-TCP15 was mixed with lysate containing ∼15 μg of GST-DAR1 or GST-DAR2 fusion proteins, respectively. Glutathione Sepharose 4B (20 μL; GE Healthcare) was added into each combined solution with continued rocking at 4°C for 1 h. Beads were washed five times with the buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, pH 8.0, 1% Triton X-100, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1× Complete Protease Inhibitor cocktail [Roche]), and the isolated proteins were separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GST (Abmart) and anti-MBP (Abmart) antibodies.
Coimmunoprecipitation
The coding sequence of DA1 was cloned into the KpnI and BamHI sites of the pCAMBIA1300-221-Myc vector to generate the transformation plasmid 35S:Myc-DA1. The coding sequence of DAR1 was cloned into the BamHI and SpeI sites of the pCAMBIA1300-221-Myc vector to generate the transformation plasmid 35S:Myc-DAR1. The coding sequence of TCP14 was cloned into the SacI and BamHI sites of the pCAMBIA1300-221-Myc vector to generate the transformation plasmid 35S:Myc-TCP14. The coding sequence of TCP15 was cloned into the KpnI and SacI sites of the pCAMBIA1300-221-Myc vector to generate the transformation plasmid 35S:Myc-TCP15. The specific primers used for 35S:Myc-DA1, 35S:Myc-DAR1, 35S:Myc-TCP14, and 35S:Myc-TCP15 constructs are described in Supplemental Table 2.
The 35S:GFP-DAR2, 35S:GFP-TCP14, and 35S:GFP-TCP15 constructs were made using a PCR-based Gateway system. The specific primers used for 35S:GFP-DAR2, 35S:GFP-TCP14, and 35S:GFP-TCP15 constructs were DAR2-GFP-FP and DAR2-GFP-RP, TCP14-GFP-LP and TCP14-GFP-RP, and TCP15-GFP-LP and TCP15-GFP-RP, respectively (Supplemental Table 2). PCR products were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. DAR2, TCP14, and TCP15 were then subcloned into Gateway binary vector pMDC43 containing the 35S promoter and GFP to generate 35S:DAR2-GFP, 35S:GFP-TCP14, and 35S:GFP-TCP15 constructs.
Nicotiana benthamiana leaves were transformed by injection of Agrobacterium tumefaciens GV3101 cells harboring different combinations of 35S:Myc-DA1, 35S:Myc-DAR1, 35S:GFP-DAR2, 35S:GFP-TCP14, 35S:GFP-TCP15, 35S:Myc-TCP14, 35S:Myc-TCP15, and 35S:GFP plasmids. Total protein was isolated with extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20% glycerol, 2% Triton X-100, 1 mM EDTA, 1× Complete Protease Inhibitor cocktail [Roche], and 20 μM MG132) and incubated with GFP-Trap_A (Chromotek) for 30 min at 4°C. Beads were washed three times with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20% glycerol, 0.1% Triton X-100, 1 mM EDTA, pH 8.0, 1× Complete Protease Inhibitor cocktail [Roche], and 20 μM MG132). The immunoprecipitates were separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-Myc (Abmart) and anti-GFP (Beyotime) antibodies.
Constructs and Plant Transformation
The 35S:DA1, 35S:DAR1, and 35S:DAR2 constructs were made using a PCR-based Gateway system. The specific primers used for 35S:DA1, 35S:DAR1, and 35S:DAR2 constructs are DA1CDS-F and DA1CDS-R, DAR1CDS-F and DAR1CDS-R, and DAR2CDS-F and DAR2CDS-R, respectively (Supplemental Table 2). PCR products were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. The DA1, DAR1, and DAR2 genes were subcloned into the Gateway binary vector pMDC32 containing the 35S promoter. The 6985-bp genomic sequence containing a 3311-bp promoter and the DAR2 gene was amplified using primers gDAR2-F and gDAR2-R. PCR products were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. The DAR2 genomic DNA was subcloned into the Gateway binary vector pMDC99 to generate the gDAR2 construct. The plasmids 35S:DA1, 35S:DAR1, 35S:DAR2, and gDAR2 were introduced into wild-type and da1-ko1 dar1-1 dar2-1 plants using Agrobacterium GV3101, and transformants were selected on hygromycin (30 μg/mL)-containing medium.
The DA1pro:GUS construct was made as described previously (Li et al., 2008). The DAR1pro:GUS and DAR1pro:GUS constructs were made using a PCR-based Gateway system. The specific primers used for DAR1pro:GUS and DAR1pro:GUS constructs are DARpro-F and DARpro-R and DAR2pro-F and DAR2pro-R, respectively (Supplemental Table 2). PCR products were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. The promoter sequences of DAR1 and DAR2 were then subcloned into the Gateway binary vector pMDC164 containing the GUS reporter gene. The plasmids DAR1pro:GUS and DAR1pro:GUS were introduced into Col-0 plants using Agrobacterium GV3101, and transformants were selected on hygromycin (30 μg/mL)-containing medium.
GUS Staining
Samples were stained in 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 50 mM NaPO4, 0.4 mM K3Fe(CN)6, 0.4 mM K4Fe(CN)6, and 0.1% (v/v) Triton X-100 and incubated at 37°C for 0.5 to 10 h in the dark. After GUS staining, chlorophyll was removed by 70% ethanol.
Proteasome Inhibitor Treatment and Immunoblot Assays
TCP14pro:TCP14-GUS and TCP15pro:TCP15-GUS seedlings were grown at 22°C on half-strength Murashige and Skoog (MS) medium for 8 d and then transferred to liquid half-strength MS medium with or without 20 μM MG132 for 48 h. Total protein extracts were separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GUS and anti-tubulin (as loading control) antibodies.
TCP14pro:TCP14-GUS, TCP15pro:TCP15-GUS, TCP14pro:TCP14-GUS;da1-ko1 dar1-1 dar2-1, and TCP15pro:TCP15-GUS;da1-ko1 dar1-1 dar2-1 seedlings were grown at 22°C on half-strength MS medium. Total protein extracts were separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GUS and anti-tubulin (as loading control) antibodies.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At1g19270 (DA1), At4g36860 (DAR1), At2g39830 (DAR2), At3g47620 (TCP14), and At1g69690 (TCP15).
Supplemental Data
Supplemental Figure 1. DA1, DAR1, and DAR2 Act Redundantly to Affect Organ Growth.
Supplemental Figure 2. Complementation Test of da1-ko1 dar1-1 dar2-1.
Supplemental Figure 3. Overexpression of DA1, DAR1 or DAR2 Does Not Affect Organ Size.
Supplemental Figure 4. DA1, DAR1, and DAR2 Act Redundantly to Influence Endoreduplication during Leaf Development.
Supplemental Figure 5. da1-ko1 dar1-1 dar2-1 Is Not Altered in Endoreduplication in Petal Cells.
Supplemental Figure 6. Expression of TCP14 and TCP15 in Col-0 and da1-ko1 dar1-1 dar2-1 Leaves.
Supplemental Figure 7. tcp14-3 tcp15-3 Partially Suppresses the Reduced Ploidy Level Phenotype of da1-ko1 dar1-1 dar2-1.
Supplemental Figure 8. DA1, DAR1, and DAR2 Redundantly Affect Endoreduplication.
Supplemental Figure 9. The Simultaneous Disruption of DA1, DAR1, and DAR2 Affects Cell Proliferation.
Supplemental Figure 10. tcp14-3 tcp15-3 Does Not Suppress the Petal and Leaf Size Phenotypes of da1-1.
Supplemental Table 1. Complementation Test of da1-ko1 dar1-1 dar2-1.
Supplemental Table 2. List of Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 1A.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers and the editor for their critical comments on this article, John Doonan and Robert Sablowski for helpful discussions, and Brendan Davies and Aiwu Dong for kindly providing the TCP14pro:TCP14-GUS and TCP15pro:TCP15-GUS seeds used in this study. We thank the Nottingham Arabidopsis Stock Centre for tcp14-3 and tcp15-3 seeds. This work was supported by the National Natural Science Foundation of China (Grants 91417304, 31425004, 91017014, and 31221063), the National Basic Research Program of China (Grant 2009CB941503), the Ministry of Agriculture of China (Grant 2013ZX08009-003-003), and the Biotechnology and Biological Science Research Council (GRO strategic program Grant BB/J0045881 to M.B. and a CASE studentship to J.D.).
AUTHOR CONTRIBUTIONS
Y.P., L.C., M.W.B., and Y.H.L. designed the research. Y.H.L. conceived and supervised the project. Y.P. and L.C. performed most of the experiments. L.C. and J.D. performed the yeast two-hybrid assay. Y.P. and L.C. conducted pull-down and coimmunoprecipitation assays. Y.P., L.C., Y.L., and Y.W. performed genetic and cellular analyses. Y.P., L.C., M.W.B., Z.Z., and Y.H.L. analyzed data. Y.P., L.C., M.W.B., and Y.H.L. wrote the article.
Glossary
- APC/C
anaphase-promoting complex/cyclosome
- Col-0
Columbia-0
- DAE
days after emergence
- MS
Murashige and Skoog
References
- Boudolf V., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C., Braidwood L., Sugimoto K. (2014). Endocycling in the path of plant development. Curr. Opin. Plant Biol. 17: 78–85. [DOI] [PubMed] [Google Scholar]
- Breuer C., Ishida T., Sugimoto K. (2010). Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 13: 654–660. [DOI] [PubMed] [Google Scholar]
- Breuer C., Morohashi K., Kawamura A., Takahashi N., Ishida T., Umeda M., Grotewold E., Sugimoto K. (2012). Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 31: 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa J.L., Tang X., Millman J.S., Schub O., Breitkreuz K., Dewar D., Rupes I., Andrews B., Tyers M. (2004). CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913. [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18: 215–222. [DOI] [PubMed] [Google Scholar]
- Davière J.M., Wild M., Regnault T., Baumberger N., Eisler H., Genschik P., Achard P. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24: 1923–1928. [DOI] [PubMed] [Google Scholar]
- del Mar Castellano M., Boniotti M.B., Caro E., Schnittger A., Gutierrez C. (2004). DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B., Ramirez-Parra E., Xie Q., Chua N.H., Gutierrez C. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140: 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T., de Almeida Engler J., Ormenese S., Maes S., Naudts M., Van Der Schueren E., Jacqmard A., Engler G., Inzé D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21: 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Murray J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54: 235–264. [DOI] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J.M., Jacqmard A., Kilby N.J., Murray J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., Murray J.A. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Li N., Chen L., Xu Y., Li Y., Zhang Y., Li C., Li Y. (2014). The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26: 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Zielke N., Gutierrez C. (2014). Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Egelkrout E.M., Robertson D., Hanley-Bowdoin L. (2001). Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13: 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming A.J., Shen Z.Z., Cunha A., Emmons S.W., Leroi A.M. (2000). Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc. Natl. Acad. Sci. USA 97: 5285–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G., Kim S.K., Horvitz H.R. (1990). Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344: 876–879. [DOI] [PubMed] [Google Scholar]
- Gegas V.C., Wargent J.J., Pesquet E., Granqvist E., Paul N.D., Doonan J.H. (2014). Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J. Exp. Bot. 65: 2757–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttery D.S., et al. (2012). A putative homologue of CDC20/CDH1 in the malaria parasite is essential for male gamete development. PLoS Pathog. 8: e1002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase Y., Trung K.H., Matsunaga T., Tanaka A. (2006). A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J. 46: 317–326. [DOI] [PubMed] [Google Scholar]
- Heyman J., Van den Daele H., De Wit K., Boudolf V., Berckmans B., Verkest A., Alvim Kamei C.L., De Jaeger G., Koncz C., De Veylder L. (2011). Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23: 4394–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Schubert H.L., Hill C.P. (2005). Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6: 610–621. [DOI] [PubMed] [Google Scholar]
- Hiyama H., Yokoi M., Masutani C., Sugasawa K., Maekawa T., Tanaka K., Hoeijmakers J.H., Hanaoka F. (1999). Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 274: 28019–28025. [DOI] [PubMed] [Google Scholar]
- Imai K.K., Ohashi Y., Tsuge T., Yoshizumi T., Matsui M., Oka A., Aoyama T. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Jordan C.V., Shen W., Hanley-Bowdoin L.K., Robertson D.N. (2007). Geminivirus-induced gene silencing of the tobacco retinoblastoma-related gene results in cell death and altered development. Plant Mol. Biol. 65: 163–175. [DOI] [PubMed] [Google Scholar]
- Kevei Z., Baloban M., Da Ines O., Tiricz H., Kroll A., Regulski K., Mergaert P., Kondorosi E. (2011). Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 6: e20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T., Boudolf V., Kheibarshekan L., Zalmas L.P., Gaamouche T., Maes S., Vanstraelen M., Kondorosi E., La Thangue N.B., Govaerts W., Inzé D., De Veylder L. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 105: 14721–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Rabin Z., Li Z., Masson P.H., Day C.D. (2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.O., Davidson J.M., Duronio R.J. (2009). Endoreplication: Polyploidy with purpose. Genes Dev. 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., York J.P., Zhang P. (2007). Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell. Biol. 27: 3481–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.Y., Li B., Dong A.W. (2012). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5: 270–280. [DOI] [PubMed] [Google Scholar]
- Magyar Z., Horváth B., Khan S., Mohammed B., Henriques R., De Veylder L., Bakó L., Scheres B., Bögre L. (2012). Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 31: 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Noir S., Bömer M., Takahashi N., Ishida T., Tsui T.L., Balbi V., Shanahan H., Sugimoto K., Devoto A. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161: 1930–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553. [DOI] [PubMed] [Google Scholar]
- Vanneste S., et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30: 3430–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A., Manes C.L., Vercruysse S., Maes S., Van Der Schueren E., Beeckman T., Genschik P., Kuiper M., Inzé D., De Veylder L. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Oania R., Graumann J., Deshaies R.J. (2004). Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118: 99–110. [DOI] [PubMed] [Google Scholar]
- Wuarin J., Buck V., Nurse P., Millar J.B. (2002). Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111: 419–431. [DOI] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., Gao F., Li Y. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.