The IVc subgroup bHLH transcription factor bHLH104 interacts with ILR3 to regulate the Arabidopsis Fe deficiency responses via targeting the Ib subgroup bHLH genes and PYE expression.
Abstract
Iron (Fe) is an indispensable micronutrient for plant growth and development. The regulation of Fe homeostasis in plants is complex and involves a number of transcription factors. Here, we demonstrate that a basic helix-loop-helix (bHLH) transcription factor, bHLH104, belonging to the IVc subgroup of bHLH family, acts as a key component positively regulating Fe deficiency responses. Knockout of bHLH104 in Arabidopsis thaliana greatly reduced tolerance to Fe deficiency, whereas overexpression of bHLH104 had the opposite effect and led to accumulation of excess Fe in soil-grown conditions. The activation of Fe deficiency-inducible genes was substantially suppressed by loss of bHLH104. Further investigation showed that bHLH104 interacted with another IVc subgroup bHLH protein, IAA-LEUCINE RESISTANT3 (ILR3), which also plays an important role in Fe homeostasis. Moreover, bHLH104 and ILR3 could bind directly to the promoters of Ib subgroup bHLH genes and POPEYE (PYE) functioning in the regulation of Fe deficiency responses. Interestingly, genetic analysis showed that loss of bHLH104 could decrease the tolerance to Fe deficiency conferred by the lesion of BRUTUS, which encodes an E3 ligase and interacts with bHLH104. Collectively, our data support that bHLH104 and ILR3 play pivotal roles in the regulation of Fe deficiency responses via targeting Ib subgroup bHLH genes and PYE expression.
INTRODUCTION
Iron (Fe) is a necessary micronutrient for both plants and animals. It is required as an essential cofactor for a number of cellular enzymatic reactions, such as photosynthesis, mitochondrial respiration, hormone biosynthesis, and nitrogen fixation (Hänsch and Mendel, 2009). Although Fe is one of the most abundant elements on earth, it often exists as insoluble ferric hydroxides in neutral and basic soils, resulting in low Fe bioavailability. Therefore, it is critical to determine the mechanisms underlying the uptake and trafficking of Fe, which is vital to improve Fe bioavailability and content of crops.
To optimize the Fe acquisition from soil, plants have evolved two major strategies to take up Fe: the reduction-based strategy (Strategy I) in dicotyledonous plants and nongraminaceous monocots, and the chelation-based strategy (Strategy II) specific to graminaceous monocots (Walker and Connolly, 2008; Hindt and Guerinot, 2012). Strategy I involves the extrusion of protons to decrease the pH of rhizosphere and the reduction of Fe3+ by FERRIC REDUCTASE OXIDASE2 (FRO2) to more soluble Fe2+, which are subsequently imported across the root epidermal cell membrane via IRON REGULATED TRANSPORTER1 (IRT1) (Curie and Briat, 2003; Hell and Stephan, 2003); while the graminaceous monocots release phytosiderophores, such as mugineic acids, to chelate Fe from Fe-limited soil (Kobayashi et al., 2010).
Due to the important biological functions and special redox properties of Fe, plants have developed a series of sophisticated regulatory systems at transcriptional and posttranscriptional levels to maintain Fe homeostasis. A number of transcription factors involved in Fe homeostasis have been identified. In Arabidopsis thaliana, two regulatory networks, the FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) network and the POPEYE (PYE) network, were verified to modulate Fe deficiency responses (Ivanov et al., 2012). Both FIT and PYE are members of the basic helix-loop-helix (bHLH) transcription factor family, suggesting the importance of bHLH proteins in Fe homeostasis. FIT was identified via its ortholog FER in tomato (Solanum lycopersicum) (Ling et al., 2002; Colangelo and Guerinot, 2004; Yuan et al., 2005; Bauer et al., 2007). The FIT-null mutation, fit-1, is lethal at the seedling stage without extra Fe supply, and approximately half of the Fe deficiency-inducible genes are deregulated in fit-1 Fe-deficient roots (Colangelo and Guerinot, 2004). However, overexpression of FIT had no effect on the expression of its targets FRO2 and IRT1, since their induction depends upon the dimerization of FIT with other four Ib subgroup bHLH proteins, bHLH38, bHLH39, bHLH100, and bHLH101, whose expression is dramatically induced by low Fe stress and repressed under Fe overload (Yuan et al., 2008; Wang et al., 2013b). These four Ib bHLH genes are induced independently from FIT, and the regulatory mechanism that acts upon FIT and Ib subgroup bHLH genes remains unclear.
The second regulatory system for Fe deficiency responses is mediated by the PYE bHLH protein, which is specifically induced in root pericycle under Fe-deficient conditions. The pye-1 mutant is sensitive to Fe deficiency, and three Fe transport-related genes, NA SYNTHASE4 (NAS4), FRO3, and ZINC-INDUCED FACILITATOR1 (ZIF1), are highly upregulated under Fe-deficient conditions in pye-1 mutant and identified as direct targets of PYE (Long et al., 2010). Additionally, a yeast two-hybrid assay showed that PYE can interact with IAA-LEUCINE RESISTANT3 (ILR3), which is also a bHLH transcription factor and may mediate metal homeostasis by regulating transporter levels (Rampey et al., 2006). In rice (Oryza sativa), a set of bHLH transcription factors have also been demonstrated to regulate Fe homeostasis. A homolog of Arabidopsis bHLH38 and bHLH39 in rice, IRON-RELATED TRANSCRIPTION FACTOR2 (IRO2), is essential for positively regulating genes involved in mugineic acid biosynthesis under Fe deficiency, and overexpression of IRO2 improved both Fe uptake and translocation to seeds (Ogo et al., 2006, 2007, 2011). Moreover, a PYE homologous protein in rice, IRO3, is induced 20- to 70-fold upon Fe deficiency. IRO3-overexpressing lines are hypersensitive to low Fe, and the genes induced by Fe deficiency are inhibited in these plants, suggesting that IRO3 is a negative regulator of Fe deficiency responses in rice (Zheng et al., 2010). Recently, rice bHLH133 was characterized to play a role in the regulation of Fe transportation from roots to shoots, further demonstrating the roles for bHLH proteins in Fe homeostasis (Wang et al., 2013a).
Despite the fact that plants are frequently challenged by Fe deficiency, Fe can be toxic by generating hydroxyl radicals via the Fenton reaction when it is accumulated at high levels (Thomine and Vert, 2013). Two pea (Pisum sativum) mutants, bronze and degenerative leaves, were the first mutants identified to be able to accumulate excess Fe in plants, causing leaf bronzing or degeneration. However, the causative genes have not been identified (Grusak et al., 1990; Kneen et al., 1990). In Arabidopsis, knockdown of the phloem-specific Fe transporter OLIGOPEPTIDE TRANSPORTER3, which loads Fe into the phloem to regulate shoot-to-root signaling, leads to Fe overload in most organs of the plants, except in embryos (Stacey et al., 2008; Mendoza-Cózatl et al., 2014; Zhai et al., 2014). Recently, the lesion mutant of SHK1 BINDING PROTEIN1, which mediates symmetric histone dimethylation, was found to accumulate high levels of Fe in shoots and exhibit tolerance to Fe deficiency. This reveals that epigenetic control is involved in Fe homeostasis (Fan et al., 2014). By coexpression analysis of PYE, Long et al. (2010) found the E3 ligase BRUTUS (BTS) as a putative negative regulator of Fe absorption. The knockdown mutant of BTS shows increased tolerance compared with the wild type on Fe-deficient media (Long et al., 2010). BTS possesses three putative hemerythrin domains, which were reported to contain μ-oxo diiron centers that could reversibly bind O2, functioning as metal storage or O2 transport proteins (Wirstam et al., 2003). In mammals, the E3 ubiquitin ligase F-BOX AND LEUCINE-RICH REPEAT PROTEIN5 (FBXL5) was characterized to harbor a hemerythrin domain and promote Fe-dependent ubiquitination and degradation of IRON REGULATORY PROTEIN2 (Salahudeen et al., 2009; Vashisht et al., 2009). Most recently, knockdown of rice HEMERYTHRIN MOTIF-CONTAINING RING- AND ZINC-FINGER PROTEIN1 (HRZ1) or HRZ2, the orthologs of BTS, caused increased accumulation of Fe and enhanced tolerance to Fe deficiency (Kobayashi et al., 2013). These results further suggest a role for BTS in negatively regulating Fe acquisition in Arabidopsis.
In this study, we characterized an Arabidopsis transcription factor belonging to the IVc subgroup of bHLH family, bHLH104, which plays an important role in Fe homeostasis. Mutation of bHLH104 led to significantly reduced tolerance to Fe deficiency, while overexpression of bHLH104 strongly promoted Fe accumulation. Besides bHLH104, another IVc subgroup bHLH transcription factor, ILR3, was also found to be required for Fe deficiency responses and could form heterodimers with bHLH104. Chromatin immunoprecipitation (ChIP) assays demonstrated that bHLH104 and ILR3 regulated the expression of four Ib subgroup bHLH genes and PYE by binding to specific elements in their promoters, indicating a cascade of bHLH transcription factors during the control of Fe homeostasis. Further genetic analysis suggested that bHLH104 might function in the same pathway of BTS in Fe signaling.
RESULTS
The bHLH104 Knockout Mutant Plants Have Altered Fe Homeostasis
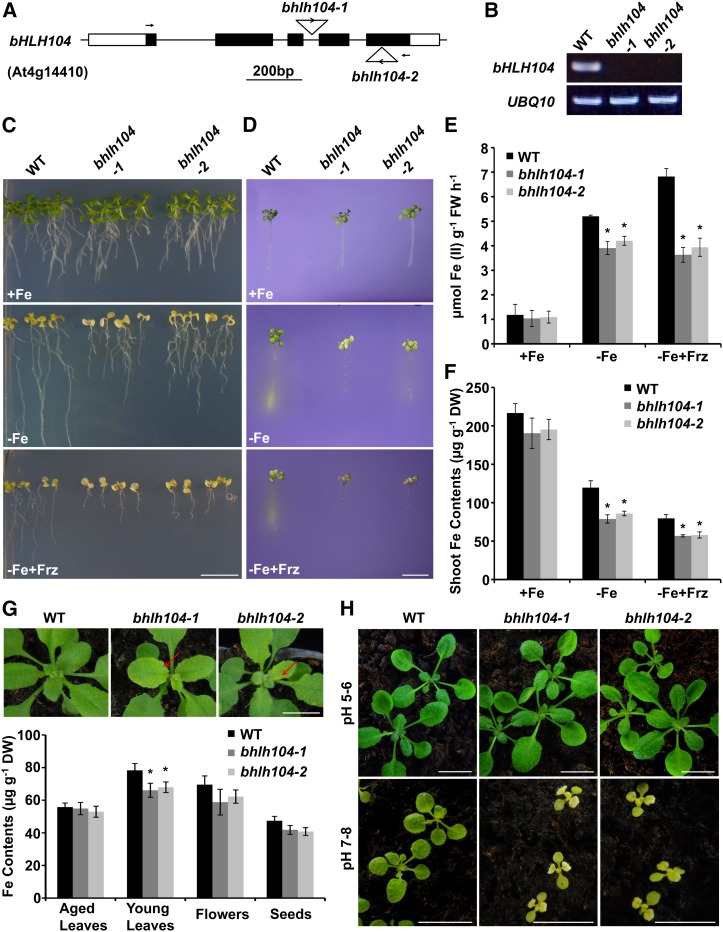
Previous studies have shown that the regulation of genes that control Fe uptake and distribution by transcription factors such as bHLH family proteins is crucial for Fe homeostasis (Walker and Connolly, 2008; Hindt and Guerinot, 2012). Members in the IVc subgroup of bHLH transcription factor family were reported to interact with Fe deficiency response regulator PYE (Long et al., 2010), suggesting a putative role of this bHLH subgroup (bHLH34, bHLH104, ILR3, and bHLH115) in Fe homeostasis regulation. Thus, we screened several T-DNA insertion lines of these genes from the ABRC; however, only homozygous mutants of bHLH104, one harboring a T-DNA in the third intron (bhlh104-1), the other in the last exon (bhlh104-2), were identified as null mutants, exhibiting loss of the full-length bHLH104 transcript (Figures 1A and 1B). Interestingly, when these bhlh104 mutants were grown on the Fe-limited media (−Fe, without Fe; −Fe+Frz, with 50 μM ferrozine to chelate the trace amounts of Fe from agar), they showed an extreme inhibition of root growth and smaller, chlorotic cotyledons compared with larger and light-green cotyledons of the wild type. By contrast, when Fe was present at normal concentrations (+Fe, with 100 μM Fe), both bhlh104-1 and bhlh104-2 had no discernable differences from the wild type (Figure 1C). It is noteworthy that during the first few days, the growth of wild-type and bhlh104 plants showed no obvious difference upon Fe deficiency. However, from the fifth day, the root growth of bhlh104 mutants was significantly repressed and almost stopped at 1 week on Fe-deficient media with ferrozine (Supplemental Figure 1A). This might be due to the fact that the initial nutrition for the growth of bhlh104 mutants came from the embryos, which did not rely on the exogenous Fe supply. In parallel, plants were first cultivated on Fe-sufficient media for 4 d and then exposed to Fe deficiency. Consistent with the results above, the leaves of bhlh104 mutants became more chlorotic than the wild type 10 d after transfer (Supplemental Figure 1B). Besides of Fe deficiency, we also tested the response of bhlh104 mutants to Zn, Cu, and Mn deficiency. The mutants displayed no observable phenotype under these stresses, indicating that the susceptibility of bhlh104 mutants to Fe deficiency was specific (Supplemental Figure 1C).
Figure 1.
The bHLH104 Knockout Mutants Have Decreased Tolerance to Fe-Deficient Conditions.
(A) The position and orientation of the T-DNA insertion site for bhlh104-1 (Salk_099496C) and bhlh104-2 (Salk_043862C) mutants.
(B) Testing for the full-length bHLH104 transcript in the wild type and the bhlh104-1 and bhlh104-2 mutants grown for 4 weeks in normal soil by RT-PCR. Primers used for amplification of bHLH104 are indicated in (A). Amplification of UBIQUITIN10 (UBQ10) is shown as a control.
(C) Phenotypes of the wild type and the bhlh104 mutants grown for 2 weeks on Fe-sufficient (+Fe, 100 μM Fe) or Fe-deficient (−Fe, without Fe; −Fe+Frz, with 50 μM ferrozine) media. Bar = 1 cm.
(D) Rhizosphere acidification by 7-d-old wild type and bhlh104 mutants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe and −Fe+Frz) media. Four plants were transferred to agar plates containing bromocresol purple for 24 h. Acidification is indicated by a yellow color around the roots. Bar = 1 cm.
(E) Ferric-chelate reductase activity of the wild type and bhlh104 mutants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe and −Fe+Frz) media for 7 d. The ferrozine assay was performed on 10 pooled plant roots. Values are means ± sd of three independent experiments. Significant differences from the wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test. FW, fresh weight.
(F) Fe contents in shoots of plants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe and −Fe+Frz) media for 8 d. The data represent means ± sd of three independent experiments. Significant differences from the wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test. DW, dry weight.
(G) Representative images of the wild type and bhlh104 mutants grown in normal soil for 25 d. Arrows indicate slight chlorosis in young leaves of bhlh104 plants compared with the wild type. Below the leaf graphs are the Fe contents in rosette leaves, flowers, and seeds of the wild type and bhlh104 mutants. For 4-week-old plants, aged leaves refer to the third true leaf to the sixth leaf, while young leaves refer to the seventh leaf to the tenth leaf. Rosette leaves and flowers were harvested at 1 week after bolting. The results are means ± sd of three independent experiments. Significant differences from the wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test. Bar = 1 cm.
(H) Phenotypes of 3-week-old wild type and bhlh104 mutants germinated on either normal soil (pH 5 to 6) or alkaline soil (pH 7 to 8). Bars = 1 cm.
We then investigated the H+-ATPase activity and ferric-chelate reductase activity in bhlh104 plants, which are typical indicators of Fe deprivation (Yi and Guerinot, 1996; Fox and Guerinot, 1998). Seven-day-old seedlings grown under Fe-sufficient or Fe-deficient conditions were tested by their rhizosphere acidification. As expected, the wild-type plants showed visible coloration around roots caused by activation of H+-ATPase under Fe deficiency. By contrast, the bhlh104 roots displayed an obvious decrease in acidification (Figure 1D). In addition, the ferric-chelate reductase activity was increased in both wild-type and bhlh104 roots in response to Fe deficiency; however, the activity in the bhlh104 mutants was decreased compared with the wild type (Figure 1E). These results indicated a defect in the Strategy I response due to the loss of bHLH104. To assess the Fe contents in wild-type and bhlh104 plants, inductively coupled plasma-atomic emission spectrometry was used to quantify the Fe contents in shoots. In accordance with the inferior growth of bhlh104 plants under Fe-deficient conditions, their shoots showed ∼70% Fe relative to the wild type (Figure 1F). Furthermore, we also did Perls Fe staining in roots of wild-type and bhlh104 plants. Similarly strong Fe staining was seen between them under Fe-sufficient conditions in which Fe was abundant in their roots; however, no signal was observed in their Fe-deficient roots. We noted that the root cells of bhlh104 Fe-deficient plants showed no obvious alterations compared with the wild type, indicating that the reduced growth of bhlh104 roots was not caused by inhibition of cell elongation (Supplemental Figure 1D).
We next examined the phenotypes of soil-grown bhlh104 mutants. A slight chlorosis was observed in the young leaves of bhlh104 plants at the time of bolting when grown in normal soil (Figure 1G). Fe content measurements revealed that the young leaves of bhlh104 mutants showed 15.5% lower Fe levels than those in the wild type, whereas the adult leaves had similar Fe levels to the wild type (Figure 1G). We then germinated the plants in alkaline soil (pH 7 to 8), which makes Fe prone to form ferric hydroxides that cannot be easily used by plants (Guerinot and Yi, 1994). After growth for 2 weeks, the bhlh104 mutants showed overall growth arrest, with only two pieces of bleached true leaves (Figure 1H). Taken together, these results suggest that bHLH104 is crucial for the efficient uptake of Fe under Fe deficiency.
bHLH104-Overexpressing Plants Exhibit Increased Tolerance to Fe Deficiency
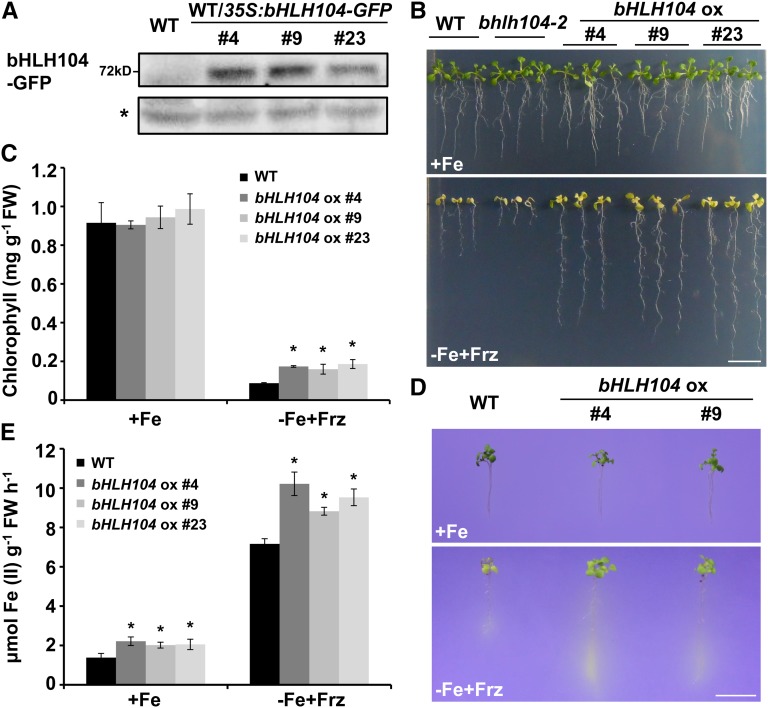
The physiological role of bHLH104 in Fe homeostasis was further investigated by analyzing the phenotypes of bHLH104-overexpressing (bHLH104 ox) plants, in which a 35S promoter drove the expression of a bHLH104-GFP fusion gene. This tagged version could complement bhlh104-2 mutants, indicating that the GFP tag did not interfere with the function of bHLH104 (Supplemental Figure 2). Three independent bHLH104 ox lines were chosen for subsequent analyses by detecting fusion protein expression (Figure 2A). In contrast to the bhlh104 mutants, the bHLH104 ox plants showed significant tolerance of Fe-deficient (−Fe+Frz) media, with almost 2-fold root length and larger, greener leaves than the wild type (Figure 2B; Supplemental Figure 3A). The total chlorophyll content of the bHLH104 ox plants was higher than that of the wild type under Fe-deficient conditions (Figure 2C). To determine whether bHLH104 ox plants altered their H+-ATPase and ferric-chelate reductase activities in response to Fe deficiency, we also detected rhizosphere acidification and conducted ferrozine assays. When cultivated under Fe-deficient conditions, the bHLH104 ox plants exhibited obvious coloration around the roots more than the wild type, indicating increased H+-ATPase activity with overexpression of bHLH104 during Fe deficiency responses. By contrast under normal conditions, no distinct coloration was observed in the wild-type and bHLH104 ox plants, which suggested that the H+-ATPase activity of bHLH104 ox plants was not constitutively activated, irrespective of Fe supply (Figure 2D). Furthermore, the ferric-chelate reductase activity of bHLH104 ox roots was also higher under Fe deficiency and displayed a slight increase under Fe-sufficient conditions compared with the wild type (Figure 2E). Taken together, these results further indicated a positive role of bHLH104 in Fe deficiency responses.
Figure 2.
Plants Overexpressing bHLH104 Exhibit Enhanced Tolerance to Fe Deficiency.
(A) The level of the bHLH104-GFP fusion protein in shoots of 7-d-old bHLH104 ox plants grown on half-strength MS media, as determined by immunoblot analysis using an anti-GFP antibody. The asterisk labels the nonspecific signal used as loading control.
(B) Phenotypes of wild-type and bHLH104 ox plants grown for 2 weeks on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media. Bar = 1 cm.
(C) Total chlorophyll contents in wild-type and bHLH104 ox plants grown for 2 weeks on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media. The data represent means ± sd of three independent experiments. Significant differences from the wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
(D) Rhizosphere acidification of 7-d-old wild-type and bHLH104 ox plants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media. Bar = 1 cm.
(E) Ferric-chelate reductase activity of wild-type and bHLH104 ox plants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media for 7 d. The data represent means ± sd of three independent experiments. Significant differences from the wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
Overexpression of bHLH104 Leads to Fe Overload under Normal Soil Cultivation
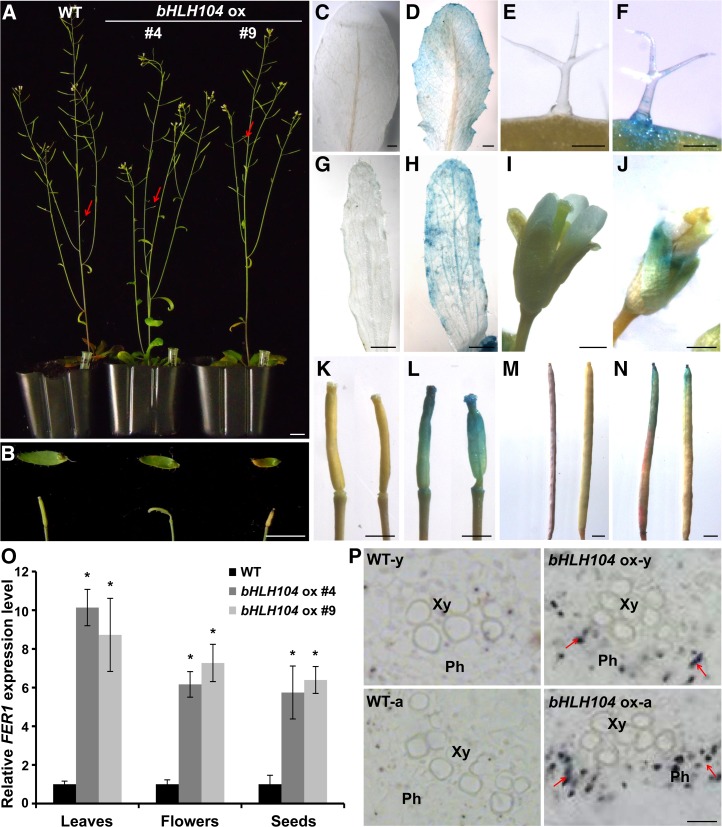
Due to the strikingly enhanced tolerance of bHLH104 ox plants under conditions of Fe deficiency, we wondered whether growth features were altered in bHLH104 ox plants under soil cultivation. Within the 4-week vegetative growth period of bHLH104 ox plants in normal soil, no macroscopic alterations in phenotype were observed. However, around 2 weeks after bolting, a portion of siliques in bHLH104 ox plants, especially those located at the bottom of the stem, appeared to be aborted. Furthermore, necrotic lesions were seen at the ends of the cauline leaves of bHLH104 ox plants (Figures 3A and 3B). These necrotic lesions were verified by Trypan blue staining to detect cell death (Supplemental Methods). Strong staining was clearly observed where the hydathodes were localized and also at the end of the vasculature (Supplemental Figures 3B to 3G). These toxic symptoms prompted us to detect the location of Fe in bHLH104 ox plants compared with the wild type using Perls staining. As shown in Figure 3, the rosette leaves of the wild type did not show detectable amounts of Fe3+ staining, whereas the bHLH104 ox leaves exhibited high levels of Fe3+ along the margins, especially in the hydathodes (Figures 3C and 3D). Moreover, high levels of stainable Fe were also observed in the trichomes of bHLH104 ox leaves (Figures 3E and 3F). Similar to the rosette leaves, the cauline leaves of bHLH104 ox plants exhibited strong Fe3+ staining (Figures 3G and 3H) and the reproductive tissues of bHLH104 ox plants also possessed more Fe3+ than their counterparts in the wild type. As with leaves, the wild type had no detectable Fe3+ in the flowers; by contrast, large amounts of Fe3+ were observed in the sepals of bHLH104 ox flowers (Figures 3I and 3J). To investigate whether the Fe overaccumulation resulted in abortion of siliques, we performed Fe3+ staining in mature siliques and aborted siliques of bHLH104 ox plants, respectively. Interestingly, the mature siliques had detectable Fe3+ only in their distal ends, while significant Fe staining was observed throughout the aborted siliques (Figures 3K to 3N), which hinted that the abortion likely resulted from Fe toxicity. This Fe overaccumulation was also reflected by significantly higher transcript levels of FERRETIN1 (FER1), encoding one of the ferritin isoforms, in bHLH104 ox leaves, flowers, and seeds compared with the wild type (Figure 3O), supporting the view that ferritin functions in protecting cells against oxidative damage resulting from Fe overload (Ravet et al., 2009).
Figure 3.
Overexpression of bHLH104 Results in Fe Overaccumulation in Soil Growth Conditions.
(A) Phenotypes of wild-type and bHLH104 ox plants at the reproductive stage. Bar = 1 cm.
(B) Magnified image of cauline leaves and siliques (marked by arrowheads in [A]) in wild-type and bHLH104 ox plants. Bar = 500 μm.
(C) to (N) Perls Fe stain signals in rosette leaves ([C] and [D]), trichomes ([E] and [F]), cauline leaves ([G] and [H]), flowers ([I] and [J]), young siliques ([K] and [L]), and mature siliques ([M] and [N]) of wild-type ([C], [E], [G], [I], [K], and [M]) and bHLH104 ox ([D], [F], [H], [J], [L], and [N]) plants. Bars = 1 mm in (C), (D), (G), (H), and (K) to (N), 100 μm in (E) and (F), and 500 μm in (I) and (J).
(O) The relative expression of FER1 in wild-type and bHLH104 ox leaves, flowers, and seeds. Plants were grown for 6 weeks in soil. The gene expression level in the wild type was set to 1. The data represent means ± sd of three independent experiments. Significant differences from the corresponding wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
(P) Perls/DAB stain for Fe3+ signals in 4-μm cross sections of wild-type and bHLH104 ox young (Y) and aged (A) leaves, showing Fe deposits in phloem of the vessels in bHLH104 ox plants (indicated by arrows). Ph, phloem; Xy, xylem. Bar = 10 μm.
To study the Fe distribution in leaves in more detail, Perls staining and diaminobenzidine (DAB) intensification assays were conducted on cross sections of leaves. It was remarkable that there were Fe deposits in the phloem of both young and aged leaves of bHLH104 ox plants, whereas no signal was observed in the wild type (Figure 3P). This indicates that the excessively absorbed Fe may be retained in the phloem and could not be employed by mesophyll cells; thus, the phloem appears to be an important tissue for Fe transport. Taken together, overexpression of bHLH104 results in overaccumulation of Fe in various organs of plants and the excess Fe in the vasculature is mainly enriched in the phloem.
Expression of Genes Responding to Fe Deficiency Is Regulated by bHLH104
Changes in Fe availability could release signals to regulate the expression of genes involved in Fe acquisition and transport (Thimm et al., 2001). To examine the effect of bHLH104 deletion or overexpression on the regulation of genes related to Fe deficiency, several representative genes were selected for analysis under Fe-sufficient (+Fe) and Fe-deficient (−Fe+Frz) conditions using quantitative real-time PCR (qPCR). As shown in Table 1, the expression of bHLH34 and ILR3 did not show significant differences among these plants under both conditions, while bHLH115 showed a 2-fold induction upon Fe deficiency; however, this expression was not affected by loss or overexpression of bHLH104. These indicated that the level of bHLH104 did not influence the expression of other three IVc subgroup bHLH genes. The expression of Fe uptake machinery genes, FIT, FRO2, and IRT1, whose transcript abundance could be induced by low Fe (Robinson et al., 1999; Vert et al., 2002; Colangelo and Guerinot, 2004), was limited in bhlh104 mutants. Moreover, the upregulated expression of four subgroup Ib bHLH genes under Fe-deficient conditions (Wang et al., 2007; Sivitz et al., 2012) was dramatically suppressed in bhlh104 plants. In addition, under Fe-sufficient conditions, the expression levels of these four genes in bhlh104 mutants were also lower than the wild type, with ∼2-fold reduction. Induction of PYE and BTS by Fe deficiency, which is considered to represent another pathway that regulates Fe utilization (Long et al., 2010), was also lower in the bhlh104 mutants compared with the wild type. In addition, the expression levels of Fe transport-related genes, such as NAS4, FERRIC REDUCTASE DEFECTIVE3 (FRD3), and ZIF1 (Green and Rogers, 2004; Haydon and Cobbett, 2007; Klatte et al., 2009), were all deregulated by the loss of bHLH104.
Table 1. Differential Expression of Fe Deficiency-Responsive Genes in Roots of the Wild Type, bhlh104 Mutants, and bHLH104 ox Plants.
| +Fe Roots |
−Fe+Frz Roots |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genes | bhlh104-1 | bhlh104-2 | bHLH104 ox#4 | bHLH104 ox#9 | Wild Type | bhlh104-1 | bhlh104-2 | bHLH104 ox#4 | bHLH104 ox#9 |
| Subgroup IVc bHLH genes | |||||||||
| bHLH104 | 0.04 ± 0.01* | 0.05 ± 0.01* | 6.56 ± 0.76* | 6.82 ± 0.55* | 1.12 ± 0.03 | 0.03 ± 0.01* | 0.04 ± 0.01* | 4.52 ± 0.58* | 4.18 ± 0.84* |
| bHLH34 | 0.93 ± 0.11 | 0.99 ± 0.12 | 1.08 ± 0.11 | 0.96 ± 0.06 | 1.06 ± 0.08 | 0.92 ± 0.08 | 0.97 ± 0.15 | 0.89 ± 0.11 | 1.04 ± 0.06 |
| ILR3 | 0.84 ± 0.08 | 1.06 ± 0.22 | 0.92 ± 0.07 | 1.05 ± 0.07 | 0.97 ± 0.04 | 0.88 ± 0.06 | 0.80 ± 0.15 | 1.06 ± 0.12 | 1.03 ± 0.15 |
| bHLH115 | 0.97 ± 0.06 | 1.05 ± 0.12 | 1.24 ± 0.25 | 0.98 ± 0.08 | 1.85 ± 0.23 | 1.94 ± 0.26 | 1.89 ± 0.14 | 1.82 ± 0.20 | 1.65 ± 0.21 |
| Subgroup Ib bHLH genes | |||||||||
| bHLH38 | 0.43 ± 0.07* | 0.55 ± 0.03* | 18.70 ± 2.75* | 14.73 ± 3.92* | 107.39 ± 9.63 | 33.59 ± 5.58* | 20.71 ± 3.33* | 128.91 ± 11.88 | 123.08 ± 7.84 |
| bHLH39 | 0.39 ± 0.06* | 0.42 ± 0.04* | 12.25 ± 4.42* | 14.62 ± 3.15* | 46.85 ± 6.65 | 14.17 ± 3.06* | 10.02 ± 2.11* | 63.56 ± 6.86 | 65.80 ± 8.01 |
| bHLH100 | 0.36 ± 0.05* | 0.41 ± 0.06* | 45.64 ± 7.60* | 37.79 ± 5.82* | 237.84 ± 25.49 | 58.08 ± 4.06* | 34.89 ± 6.12* | 376.12 ± 26.61 | 317.4 ± 19.52 |
| bHLH101 | 0.37 ± 0.05* | 0.45 ± 0.06* | 161.24 ± 12.45* | 186.33 ± 18.20* | 52.90 ± 5.74 | 7.19 ± 1.64* | 7.42 ± 0.44* | 188.07 ± 10.97* | 186.76 ± 15.54* |
| Fe uptake | |||||||||
| FIT | 0.99 ± 0.06 | 1.15 ± 0.22 | 3.92 ± 0.62* | 2.24 ± 0.45* | 2.65 ± 0.68 | 2.17 ± 0.30 | 1.97 ± 0.49 | 8.29 ± 1.02* | 8.20 ± 2.62* |
| FRO2 | 0.32 ± 0.04* | 0.48 ± 0.07* | 56.11 ± 7.70* | 35.95 ± 9.52* | 86.53 ± 7.56 | 31.17 ± 4.92* | 25.15 ± 7.23* | 183.26 ± 11.31* | 172.45 ± 15.11* |
| IRT1 | 0.64 ± 0.09 | 0.61 ± 0.04 | 33.48 ± 6.46* | 19.29 ± 5.57* | 43.41 ± 7.99 | 16.97 ± 3.14* | 13.27 ± 2.11* | 72.26 ± 5.32* | 62.54 ± 8.56 |
| IRT2 | 1.11 ± 0.17 | 0.63 ± 0.06 | 1.58 ± 0.30 | 1.67 ± 0.38 | 1.41 ± 0.12 | 0.92 ± 0.08 | 0.88 ± 0.06 | 2.83 ± 0.58* | 3.07 ± 0.82* |
| Fe translocation | |||||||||
| YSL1 | 1.28 ± 0.13 | 0.78 ± 0.10 | 27.20 ± 3.39* | 22.43 ± 3.03* | 0.22 ± 0.02 | 0.28 ± 0.08 | 0.42 ± 0.07 | 2.83 ± 0.54* | 2.39 ± 0.32* |
| NAS4 | 0.94 ± 0.03 | 0.69 ± 0.10 | 45.77 ± 7.00* | 90.25 ± 9.49* | 84.66 ± 10.25 | 16.35 ± 3.57* | 12.14 ± 2.5* | 134.73 ± 11.01* | 161.15 ± 15.94* |
| ZIF1 | 0.89 ± 0.03 | 1.21 ± 0.11 | 6.32 ± 0.52* | 6.39 ± 0.89* | 10.78 ± 1.50 | 3.09 ± 0.64* | 2.18 ± 0.36* | 18.09 ± 2.64* | 16.51 ± 2.06* |
| FRD3 | 0.76 ± 0.07 | 0.87 ± 0.03 | 4.59 ± 0.34* | 5.29 ± 0.60* | 0.82 ± 0.10 | 0.17 ± 0.06* | 0.22 ± 0.12* | 1.41 ± 0.08* | 2.36 ± 0.21* |
| Fe homeostasis regulation | |||||||||
| PYE | 0.80 ± 0.06 | 0.78 ± 0.08 | 6.17 ± 1.14* | 7.42 ± 0.96* | 3.80 ± 0.83 | 1.76 ± 0.16* | 1.36 ± 0.23* | 9.09 ± 1.59* | 9.16 ± 0.98* |
| BTS | 1.06 ± 0.06 | 1.10 ± 0.06 | 3.06 ± 0.23* | 3.29 ± 0.50* | 7.26 ± 1.02 | 2.51 ± 0.37* | 1.72 ± 0.22* | 8.40 ± 2.54 | 9.07 ± 1.45 |
| MYB10 | 0.87 ± 0.08 | 0.96 ± 0.10 | 13.60 ± 3.54* | 12.09 ± 2.69* | 40.88 ± 8.16 | 6.43 ± 1.54* | 10.32 ± 1.48* | 69.55 ± 4.94* | 70.12 ± 8.32* |
| MYB72 | 0.46 ± 0.07* | 0.74 ± 0.08 | 276.29 ± 28.85* | 145.2 ± 19.31* | 560.15 ± 37.18 | 44.03 ± 5.35* | 55.63 ± 7.39* | 1710.28 ± 90.73* | 1525.01 ± 96.46* |
The relative gene expression levels measured by qPCR in roots of the wild type, bhlh104 mutants, and bHLH104 ox plants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media for 8 d. The expression of TUB2 was used to normalize mRNA levels, and the gene expression level in the wild type under Fe-sufficient conditions was set to 1. The data represent means ± sd of three independent experiments. Significant differences from the corresponding wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
By contrast, the expression of these Fe uptake- and transport-related genes in bHLH104 ox plants was upregulated compared with the wild type under both Fe-sufficient and Fe-deficient conditions (Table 1). Based on these expression analyses for Fe-related genes, it appears that bHLH104 functions relatively upstream in the transcriptional regulatory network of the Fe deficiency response.
bHLH104 Interacts with Another IVc Subgroup bHLH Transcription Factor ILR3
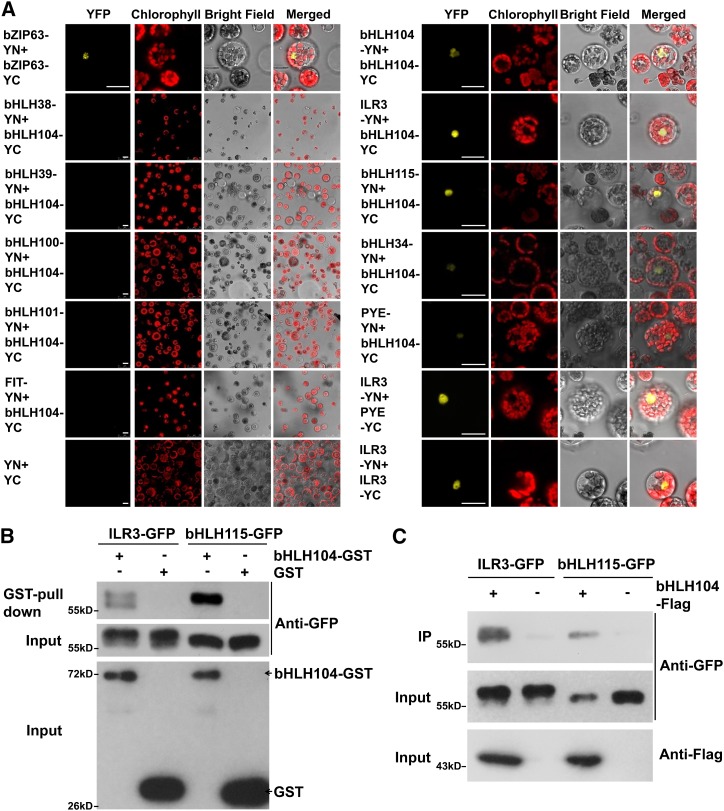
The IVc subgroup of bHLH transcription factor comprises four members, bHLH34, bHLH104, ILR3, and bHLH115. Given the potential for bHLH transcription factors to regulate the expression of downstream genes by forming diverse homodimers or heterodimers (Toledo-Ortiz et al., 2003), we then tested whether bHLH104 interacted with members of IVc subgroup bHLH transcription factors and other known bHLH proteins involved in the Fe homeostasis regulation using bimolecular fluorescence complementation (BiFC) assays. As shown in Figure 4A, bHLH104 showed no signal with Ib subgroup bHLH transcription factors or FIT. However, bHLH104 could interact with itself and bind strongly to ILR3, which was suggested to be involved in the regulation of metal homeostasis in a previous study (Rampey et al., 2006). Moreover, bHLH104 also showed an intense interaction with bHLH115, which shares 63% similarity with ILR3. We also observed a slight signal between bHLH104 and bHLH34, as well as with PYE. These results imply that bHLH104 may function with other members of IVc subgroup bHLH transcription factors, but not with Ib subgroup bHLH proteins. To further confirm the interaction of bHLH104 with ILR3 and bHLH115, a glutathione S-transferase (GST) pull-down assay was performed using a recombinant N-terminal GST-tagged bHLH104 protein. As shown in Figure 4B, the GFP-tagged ILR3 and bHLH115 protein could be pulled down by GST-bHLH104, but not by GST alone, indicating that bHLH104 interacts with ILR3 and bHLH115 in vitro. Consistent with the results of the pull-down assay, an in vivo coimmunoprecipitation (co-IP) assay also showed that bHLH104-Flag fusion proteins could be immunoprecipitated with ILR3-GFP or bHLH115-GFP when they were transiently coexpressed in Arabidopsis protoplasts (Figure 4C). These results indicate that bHLH104 may regulate Fe homeostasis by forming heterodimers with ILR3 and bHLH115.
Figure 4.
bHLH104 Interacts with Itself, ILR3, and bHLH115, but Not Ib Subgroup bHLH Proteins or FIT.
(A) BiFC analysis in Arabidopsis protoplasts transformed with different combinations of the constructs and visualized using confocal microscopy. Bars = 20 μm.
(B) Pull-down assay of GST-fused bHLH104 expressed in Escherichia coli with ILR3-GFP or bHLH115-GFP expressed in Arabidopsis protoplasts. Total proteins were pulled down by glutathione Sepharose 4B and detected using an anti-GFP antibody.
(C) Co-IP assay of bHLH104-Flag and ILR3-GFP or bHLH115-GFP coexpressed in Arabidopsis protoplasts. Total proteins were immunoprecipitated by anti-FLAG M2 affinity gel and detected using an anti-GFP antibody.
ILR3 Also Functions in Response to Fe Deficiency in Arabidopsis
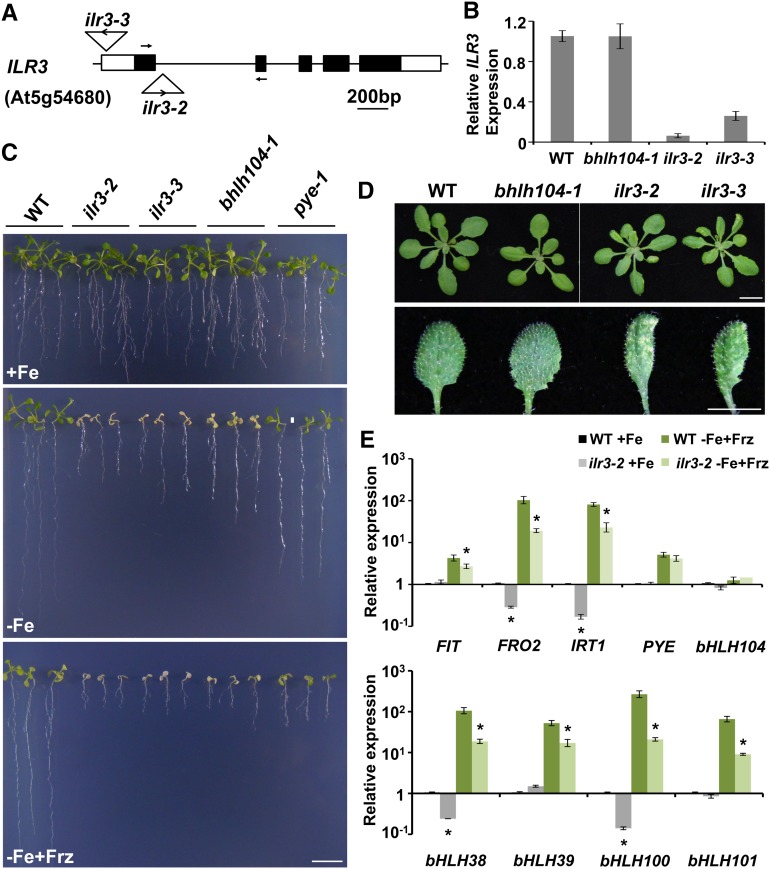
The interaction between bHLH104 and ILR3 raised the question of whether ILR3 was also required for the Fe deficiency responses. To this end, we examined the phenotype of an ILR3 knockout mutant, ilr3-2, on Fe-deficient media. Furthermore, we identified a partial loss-of-function allele and named it ilr3-3 (Figures 5A and 5B). Interestingly, both the ilr3-2 and ilr3-3 mutants displayed hypersensitivity on Fe-deficient (−Fe and −Fe+Frz) media, exhibiting chlorotic leaves and short roots, which seemed even more severe than the bhlh104-1 plants (Figure 5C). These two ilr3 mutants revealed interveinal chlorosis of young leaves under soil-growth conditions (Figure 5D), similar to the phenotype caused by Fe deficiency. We then analyzed the expression of several Fe deficiency-responsive genes in ilr3-2 roots. Similar to the bhlh104 mutants, the expression of Fe uptake genes, FIT, FRO2, and IRT1, as well as four Ib subgroup bHLH genes, all showed significant decrease in ilr3-2 roots upon Fe deficiency, compared with the wild type (Figure 5E). The expression of bHLH104 in ilr3-2 showed no significant difference compared with that in the wild type under both conditions, indicating that the mutation of ILR3 did not affect the expression of bHLH104. Taken together, these results suggest a critical role for ILR3 in the Fe deficiency responses.
Figure 5.
The ILR3 Knockout Mutants Show Sensitivity to Fe Deficiency and Decreased Expression of Fe Uptake-Related Genes.
(A) The position and orientation of the T-DNA insertion site for the ilr3-2 (Salk_004997C) and ilr3-3 (Salk_043690C) mutants.
(B) Determination of the relative ILR3 expression level in wild-type, bhlh104-1, ilr3-2, and ilr3-3 plants grown for 4 weeks in normal soil, assessed by qPCR. Primers used for amplification of ILR3 are indicated in (A). Values are means ± sd of three independent experiments.
(C) Phenotype of wild-type, ilr3-2, ilr3-3, bhlh104-1, and pye-1 mutants grown for 2 weeks on Fe-sufficient (+Fe) or Fe-deficient (−Fe and −Fe+Frz) media. Bar = 1 cm.
(D) Phenotypes of wild-type, bhlh104, and ilr3 mutants grown in normal soil for 4 weeks. The magnified picture of leaves shows interveinal leaf chlorosis of ilr3 mutants. Bars = 1 cm.
(E) The relative gene expression measured by qPCR in roots of wild-type and ilr3-2 plants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media for 8 d. The expression of TUB2 was used to normalize mRNA levels, and the gene expression level in the wild type under Fe-sufficient conditions was set to 1. The data represent means ± sd of three independent experiments. Significant differences from the corresponding wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
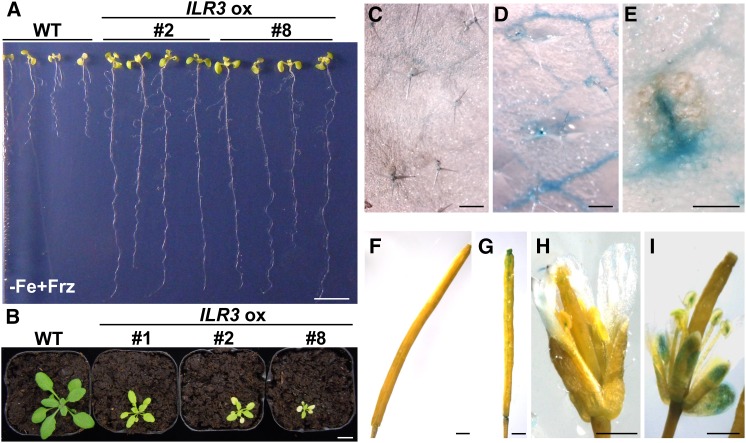
Having ascertained that ILR3 is involved in Fe homeostasis regulation, we then asked whether ILR3-overexpressing plants also had enhanced resistance to Fe deficiency. As expected, plants overexpressing 35S:ILR3-GFP (ILR3 ox), in which ILR3 was fused to a GFP tag, displayed longer roots and more leaves than the wild type on Fe-deficient (−Fe+Frz) media (Figure 6A; Supplemental Figures 4A to 4C). Interestingly, ILR3 ox plants showed small rosette and pale leaves compared with the wild type when grown in soil, with several leaves being chlorotic (Figure 6B). To further determine whether the ILR3 ox plants showed a similar Fe distribution pattern to bHLH104 ox plants, we localized Fe3+ in various organs of ILR3 ox plants. As shown in the bHLH104 ox plants, the rosette leaves of ILR3 ox plants also possessed large quantities of Fe3+ in the vasculature (Figures 6C and 6D). Moreover, several necrotic spots were seen scattered in the leaves where Fe was overaccumulated (Figure 6E; Supplemental Figures 4D to 4G). Furthermore, the sepals of flowers and the aborted siliques which were also observed in ILR3 ox plants accumulated extremely high levels of Fe (Figures 6F to 6I). Thus, consistent with bHLH104 ox plants, overexpression of ILR3 also gives rise to Fe overload in Arabidopsis.
Figure 6.
Plants Overexpressing ILR3 Exhibit Enhanced Fe Deficiency Tolerance.
(A) Phenotypes of wild-type and ILR3 ox plants grown for 2 weeks on Fe-deficient (−Fe+Frz) media. Bar = 1 cm.
(B) Phenotypes of wild-type and ILR3 ox plants grown in normal soil for 4 weeks. Bar = 1 cm.
(C) to (I) Perls Fe stain signals in rosette leaves ([C] to [E]), siliques ([F] and [G]), and flowers ([H] and [I]) of wild-type ([C], [F], and [H]) and ILR3 ox ([D], [E], [G], and [I]) plants.
Bars = 200 μm in (C) to (E) and 1 mm in (F) to (I).
bHLH104 and ILR3 Bind to the E-Box Motifs in the Promoters of Ib Subgroup bHLH Genes and PYE
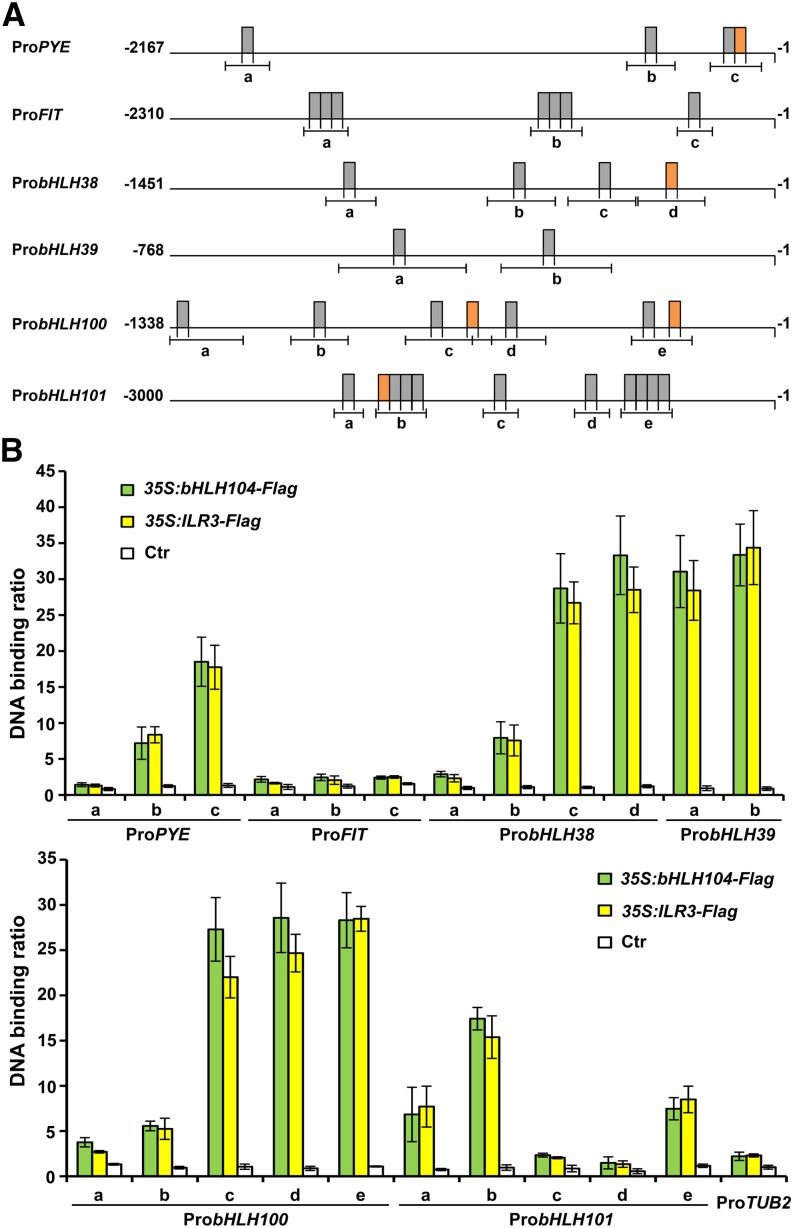
The bHLH transcription factors have been reported to be associated with the E-box (5′-CANNTG-3′) cis-element in the promoters of their target genes (Fisher and Goding, 1992). Considering that FIT and four Ib subgroup bHLH genes were downregulated in bhlh104 and ilr3 mutants, we speculated whether these known bHLH genes were directly regulated by bHLH104 or ILR3. Bioinformatics analysis showed several E-box motifs were in the putative promoter regions of FIT, Ib subgroup bHLH genes, and PYE (Figure 7A). To determine the binding capacity of bHLH104 and ILR3 to their promoters, ChIP experiments were conducted. 35S:bHLH104-Flag and 35S:ILR3-Flag were transformed into Arabidopsis protoplasts, and ChIP DNA was immunoprecipitated using anti-FLAG affinity gel. A fragment of the TUB2 promoter containing an E-box motif was used as a negative control. qPCR analyses showed that bHLH104 and ILR3 bound strongly to the promoters of Ib subgroup bHLH genes and PYE, but not to FIT (Figure 7B). Interestingly, the canonical E-box motifs (CACGTG, also referred to as G-box) in bHLH38, bHLH100, bHLH101, and PYE promoters were more enriched than their noncanonical E-box (CANNTG) motifs. However, bHLH104 and ILR3 could also recognize the chromatin region of the bHLH39 promoter, which only harbors noncanonical E-box sequences (Figure 7B). These results indicate that bHLH104 and ILR3 interact with the specific E-box regulatory elements of Ib subgroup bHLH genes and PYE to affect their transcription during the response to Fe deficiency.
Figure 7.
ChIP-qPCR Analyses of the Binding of bHLH104 and ILR3 to the Promoters of PYE, FIT, and Ib Subgroup bHLH Genes.
(A) Promoter structure diagrams for PYE, FIT, and Ib subgroup bHLH genes. Gray boxes show noncanonical E-boxes with sequence 5′-CANNTG-3′, while orange boxes show canonical E-boxes (5′-CACGTG-3′). Lines under the boxes indicate sequences detected by ChIP-qPCR assays.
(B) ChIP-qPCR analyses of the DNA binding ratio of bHLH104 and ILR3 to the promoters of PYE, FIT, and Ib subgroup bHLH genes. Chromatin from Arabidopsis protoplasts expressing 35S:bHLH104-Flag or 35S:ILR3-Flag was extracted by anti-FLAG M2 affinity gel. The protoplasts without transfection were used as negative control. qPCR was used to quantify enrichment of the PYE, FIT, and Ib subgroup bHLH gene promoters and a fragment of the TUB2 promoter containing an E-box motif was used as a negative control. The binding of TUB2 promoter fragment in protoplasts without transfection was set to 1 and used to normalize the DNA binding ratio of bHLH genes. The data represent means ± sd of three independent experiments.
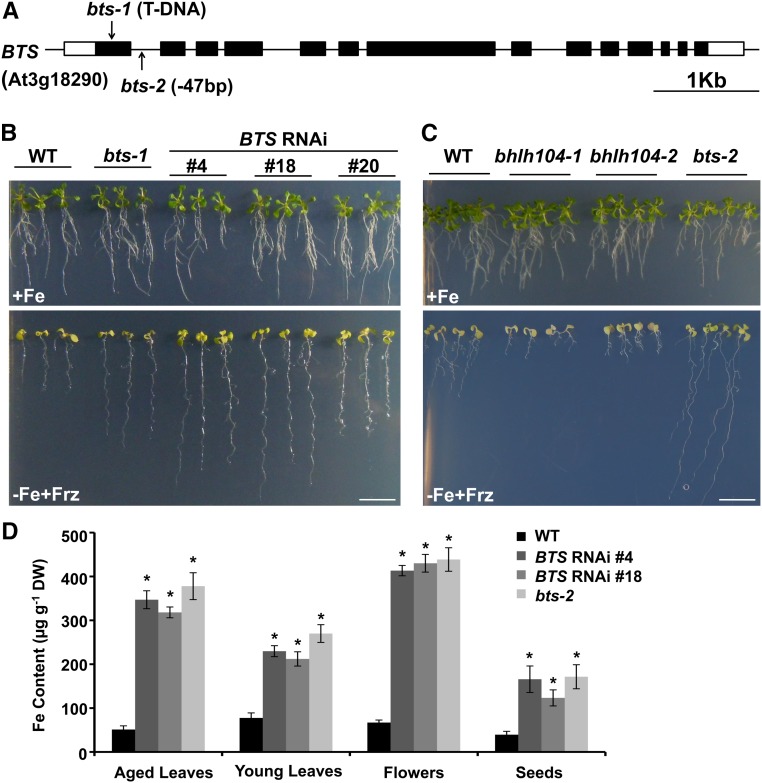
Loss of bHLH104 Partially Rescues the Phenotype Conferred by BTS Lesion
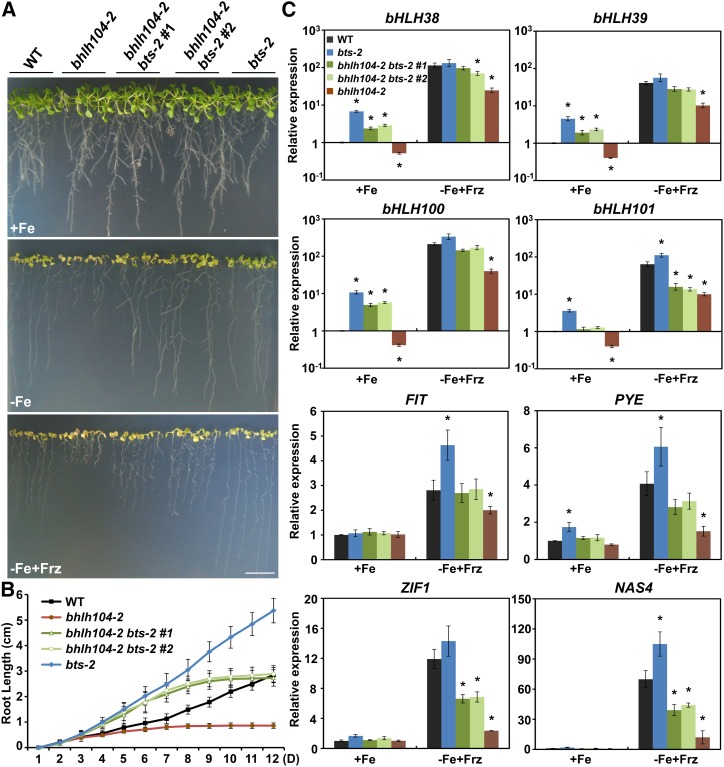
It has been reported that bHLH104 and ILR3 could interact with BTS in a yeast two-hybrid assay; moreover, the phenotype of increased tolerance on Fe-deficient media was also seen in a partial loss-of-function allele bts-1, which has 70% BTS expression (Long et al., 2010). To further confirm the function of BTS in Fe homeostasis, we performed RNA interference of BTS and obtained transgenic lines with only 20% BTS expression (Supplemental Figure 5A). Furthermore, we also identified an Fe deficiency-tolerant mutant (Salk_004748C). Positional cloning of this mutant identified the BTS locus, whose first intron has a 47-bp fragment deletion, thus destroying its open reading frame (Figure 8A). This mutant also harbors a T-DNA in At1g67520 that encodes a putative lectin protein kinase. Using a segregating population, the T-DNA insertion in At1g67520 was separated and a homozygous mutation for BTS was obtained, namely, bts-2. We then investigated the responses of BTS RNA interference (RNAi) and bts-2 plants to Fe deficiency. In accordance with the phenotype of bts-1 mutant, the BTS RNAi and bts-2 plants showed remarkably better growth on Fe-deficient (−Fe+Frz) media (Figures 8B and 8C; Supplemental Figure 5B). Moreover, the BTS RNAi and bts-2 plants had significantly higher Fe contents in their rosette leaves, flowers, and seeds compared with the wild type (Figure 8D). Thus, our results reinforce the view that BTS functions as a negative regulator of the Fe deprivation response and Fe uptake.
Figure 8.
The BTS Lesion Plants Display Increased Tolerance to Fe Deficiency.
(A) The position of the T-DNA insertion site for bts-1 (Salk_016526) mutants and the sequence loss site of bts-2 mutants.
(B) Phenotypes of wild-type and BTS RNAi plants grown for 2 weeks on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media. Bar = 1 cm.
(C) Phenotypes of wild-type and bts-2 plants grown for 2 weeks on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media. Bar = 1 cm.
(D) Fe contents in various tissues of the wild-type, BTS RNAi, and bts-2 plants. Results are means ± sd of three independent experiments. Significant differences from the wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
We also tested the phenotype of BTS RNAi plants under normal soil cultivation. During vegetative development, the BTS RNAi plants were unaffected, whereas at the late reproductive stage, aborted siliques and necrotic lesions in cauline leaves were observed in BTS RNAi plants (Supplemental Figures 5C and 5D). Further detailed analysis of Fe distribution verified that the leaves and flowers of BTS RNAi plants overaccumulated Fe. Interestingly, the embryo of BTS RNAi plants showed obvious Fe staining in the funiculus, indicating excess Fe was imported into the seeds (Supplemental Figures 6A to 6Q). Furthermore, a closer examination of Fe distribution also revealed excess Fe deposition in the phloem of BTS RNAi plants (Supplemental Figure 6R).
The findings regarding Fe overaccumulation in BTS RNAi and bts-2 plants were similar to those of bHLH104 and ILR3 ox plants, suggesting that BTS may antagonize with them to modulate Fe homeostasis. Thus, we performed a crossing assay and obtained the double mutants of bhlh104-2 and bts-2. When cultivated under Fe-deficient conditions for 2 weeks, the bhlh104-2 bts-2 lines showed a similar phenotype to the wild type, with shorter roots and more chlorotic leaves compared with bts-2 (Figure 9A). Interestingly, in the initial stage, the root growth of bhlh104-2 bts-2 plants was comparable to bts-2 plants, which was faster than the wild type. However, the root growth of the double mutants was significantly repressed at the beginning of the eighth day, which was also observed in bhlh104-2 mutants (Figure 9B). This indicates that loss of bHLH104 could decrease the tolerance of bts-2 plants to Fe deficiency and the faster growth of bhlh104-2 bts-2 plants within 1 week might be due to the overaccumulated Fe in their embryos.
Figure 9.
Loss of bHLH104 Decreases the Tolerance of bts-2 Plants to Fe Deficiency.
(A) Phenotypes of wild-type and bhlh104-2 bts-2 plants grown for 2 weeks on Fe-sufficient (+Fe) and Fe-deficient (−Fe and −Fe+Frz) media. Bar = 1 cm.
(B) Quantification of root length of plants on Fe-deficient media (−Fe+Frz) according to the time course. Values are means ± sd of 10 plants for each line.
(C) The relative expression levels of selected Fe deficiency-responsive genes, as measured by qPCR, in roots of the wild type, bts-2 mutants, and bhlh104-2 bts-2 double mutants grown on Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) media for 8 d. The expression of TUB2 was used to normalize mRNA levels, and the gene expression levels in the wild type under Fe-sufficient conditions were set to 1. The data represent means ± sd of three independent experiments. Significant differences from the corresponding wild type are indicated by an asterisk (P < 0.05), as determined by Student’s t test.
Thus, we chose the 8-d-old seedlings grown under Fe-sufficient (+Fe) or Fe-deficient (−Fe+Frz) conditions to test whether the reduced tolerance of bhlh104-2 bts-2 double mutants relative to bts-2 plants resulted from impaired transcription of Fe deficiency-responsive genes. Intriguingly, the bts-2 mutants showed extremely enhanced expression of Ib subgroup bHLH genes under Fe-sufficient conditions, and this upregulation was also seen in bhlh104-2 bts-2 double mutants; however, the degree of elevation was lower than in the bts-2 mutants (Figure 9C). Under Fe-deficient conditions, levels of these Ib subgroup bHLH genes were slightly higher in bts-2 mutants compared with the wild type, while in the bhlh104-2 bts-2 double mutants, the expression of bHLH39 and bHLH101 was not induced to the same levels as in the wild type; however, they remained higher than in the bhlh104-2 mutants (Figure 9C). In addition, induction of PYE, ZIF1, and NAS4 by Fe deficiency was also lower in the bhlh104-2 bts-2 double mutants compared with the wild type (Figure 9C). Therefore, loss of bHLH104 could partially block the activation of Fe deficiency-related genes conferred by BTS lesion. Taken together, these genetic studies suggest that bHLH104 may function downstream of BTS to regulate the Fe deficiency responses.
DISCUSSION
It is becoming increasingly clear that a complicated regulatory network is involved in modulating Fe homeostasis. In Arabidopsis, a model requiring two pathways mediated by bHLH family proteins, FIT and PYE, is responsible for the Fe deficiency responses. In this study, we identified two members belonging to the IVc subgroup of bHLH transcription factors, bHLH104 and ILR3, which function as essential regulators in Fe homeostasis.
bHLH104 and ILR3 Are Required to Maintain Fe Homeostasis in Arabidopsis
In Arabidopsis, the large bHLH family contains 162 members, which have been reportedly known to regulate myriad of developmental and physiological responses, one of which is the regulation of Fe deprivation signaling (Bailey et al., 2003; Heim et al., 2003; Li et al., 2006; Pires and Dolan, 2010). In this study, we found that both knockout mutants of bHLH104 and ILR3 exhibited severely attenuated responses to Fe deficiency, displaying shorter roots and chlorotic cotyledons (Figures 1C and 5C). By contrast, overexpression of bHLH104 or ILR3 in Arabidopsis conferred remarkable tolerance to Fe deficiency (Figures 2 and 6A). Analysis of the expression profiles of known regulatory genes in Fe uptake such as FIT, FRO2, IRT1, and Ib subgroup bHLH genes revealed the downregulation of these genes in bhlh104 and ilr3 mutants (Table 1, Figure 5E). These results indicate that bHLH104 and ILR3 function positively in the Fe deficiency responses. It is worth mentioning that knockout or overexpression of bHLH104 and ILR3 had severe effects on root development in Fe-limited conditions (Figures 1C, 2B, 5C, and 6A), which might have a further effect on Fe absorption. Therefore, the Fe deficiency tolerance and higher Fe contents of bHLH104 and ILR3 overexpression plants may be partially ascribed to their large root mass.
Overexpression of bHLH104 led to excessive Fe accumulation in plants, especially in the phloem (Figure 3P). A similar Fe overload in the phloem was also seen in nas4x-2, a nicotianamine loss mutant that cannot remobilize Fe from the phloem to the sink organs, thus resulting in Fe being retained in the phloem and the young leaves being Fe deficient (Klatte et al., 2009; Schuler et al., 2012). However, new leaves of bHLH104 ox plants did not display an Fe-deficient phenotype, and the expression of NAS4, which is involved in nicotianamine biosynthesis, was upregulated in bHLH104 ox plants (Table 1). This upregulation of NAS4 could further enhance the transport of Fe from the phloem to mesophyll cells. However, the excessively absorbed Fe is too much to be completely employed, leading to the observed Fe overaccumulation in the phloem of bHLH104 ox plants.
The transcription factors bHLH104 and ILR3 belong to the IVc subgroup of bHLH family, which has been found widely conserved in flowering plants, such as rice, tomato, soybean (Glycine max), and grape (Vitis vinifera; Pires and Dolan, 2010). Analysis of miRNA macroarrays predicted that Phaseolus vulgaris miR1509 might target bHLH104 during the Mn stress response (Valdés-López et al., 2010; Gupta et al., 2014). Moreover, the Chlamydomonas reinhardtii bHLH protein Cre24.g770450, which is similar to the Arabidopsis subgroup IVc transcription factors, is induced in Fe-poor conditions (Urzica et al., 2012). Recently, a bHLH transcription factor in chrysanthemum (Chrysanthemum × morifolium), Cm-bHLH1, which is highly similar to ILR3, was shown to regulate Fe intake under Fe deficiency (Zhao et al., 2014). These findings hint that the IVc subgroup bHLH transcription factors may have conserved functions in plant metal homeostasis.
Both bHLH104 and ILR3 Regulate the Promoters of Ib Subgroup bHLH Genes
The bHLH domain comprises two regions. The C-terminal HLH region is responsible for forming homo- or heterodimers, while the N-terminal basic region determines the DNA-protein interaction by binding to canonical E-box (G-box) or noncanonical E-box motifs (Toledo-Ortiz et al., 2003; Li et al., 2006). For example, CRYPTOCHROME-INTERACTING bHLH1 (CIB1) can form heterodimers with CIB2, CIB4, and CIB5 and bind to the noncanonical E-box sequence of the FT promoter in vivo (Liu et al., 2013). The Arabidopsis tapetum development regulator DYSFUNCTIONAL TAPETUM1 forms homodimers and also interacts with three other bHLH proteins, bHLH10, bHLH89, and bHLH91 (Feng et al., 2012). These results indicate that dimerization of bHLH proteins is important for specifically binding the promoters of target genes.
Here, we demonstrated that bHLH104 could interact with ILR3 and both of them bound to the canonical E-box of bHLH38, bHLH100, bHLH101, and PYE with much higher affinity than with noncanonical E-box DNA sequences. In addition, they also associated with the chromatin region of the bHLH39 promoter, which only contains noncanonical E-box sequences (Figure 7). In previous studies, these Ib subgroup bHLH genes were demonstrated to be widely expressed in various tissues, especially in the epidermis of roots and veins of leaves (Wang et al., 2007). Similarly, bHLH104 is ubiquitously expressed in both roots and leaves. In particular, it is highly expressed in the root stele and leaf vasculature (Supplemental Figure 7). This tissue expression pattern was also observed for ILR3 using β-glucuronidase (GUS) activity analysis of its promoter (Rampey et al., 2006). Furthermore, the induction of four Ib subgroup bHLH genes was extremely repressed upon Fe deficiency in the absence of bHLH104 or ILR3 (Table 1, Figure 5E). Therefore, we conclude that bHLH104 and ILR3 are the direct upstream activators of Ib subgroup bHLH genes. Although the upregulation of FIT under Fe deficiency was also inhibited in both bhlh104 and ilr3 mutants (Table 1, Figure 5E), there was no detectable binding to the FIT promoter by bHLH104 or ILR3. This suggested the existence of other intermediate transcription factors that act upon FIT but are downstream regulators of bHLH104 and ILR3. bHLH104 could also interact with bHLH115 and a slight interaction was seen between bHLH104 and bHLH34 (Figure 4), suggesting putative roles of bHLH115 and bHLH34 in Fe homeostasis regulation. Moreover, this also implies that bHLH104 may dimerize with more than one partner to specifically regulate target genes.
Although bHLH104 and ILR3 could form heterodimers, and both of them targeted Ib subgroup bHLH gene promoters, the BiFC assay also suggested that bHLH104 and ILR3 could interact with themselves (Figure 4A). Furthermore, the overexpression of bHLH104 and ILR3 exhibited more tolerance to Fe deficiency stress and resulted in Fe overload (Figures 2 and 6). This is quite different from FIT. Despite the fact that FIT could form homodimers, overexpression of FIT alone did not lead to any obvious phenotype or detectable change of Fe contents in shoots, since FIT must interact with a Ib subgroup bHLH protein to drive the expression of FRO2 and IRT1 (Yuan et al., 2008). These findings indicated that bHLH104 and ILR3 could both promote Fe accumulation without necessarily relying on each other. Comparison of the physiological response to Fe deficiency revealed that ilr3 mutants appeared more sensitive than bhlh104, since the bhlh104 mutants could develop true leaves, while the ilr3 mutants were stunted with only two pieces of chlorotic cotyledons (Figure 5C). Moreover, the overexpression of ILR3 resulted in chlorotic leaves that were not seen in bHLH104 ox plants (Figure 6B), suggesting that they might act independently on different downstream targets. Thus, the response to Fe deficiency requires the activities of both bHLH104 and ILR3.
BTS and bHLH104 Probably Function in the Same Pathway in Fe Homeostasis
To date, several candidates for a long-distance Fe signal have been characterized in Arabidopsis; however, the mechanisms of sensing cellular Fe status remain unclear (Walker and Connolly, 2008; Hindt and Guerinot, 2012). In Arabidopsis, BTS is a homolog of HRZs and FBXL5, the known Fe sensors in rice and mammals, which negatively regulate Fe acquisition (Salahudeen et al., 2009; Vashisht et al., 2009). In a previous study, a partial loss-of-function allele of BTS, bts-1, was reported to confer tolerance to Fe deficiency (Long et al., 2010); however, the Fe content and gene expression regulation still remain unclear. Here, we showed that plants with a lesion in BTS were capable to accumulate excess Fe and the expression of Fe deficiency-inducible genes was substantially enhanced (Figures 8D and 9C). These characteristics are very similar to those of the rice HRZs knockdown plants (Kobayashi et al., 2013), confirming their conserved functions of negatively regulating Fe uptake in plants. Furthermore, HRZ1/2 and BTS can all bind Fe and Zn and have E3 ligase activity (Kobayashi et al., 2013). Thus, BTS may function as a Fe sensor in Arabidopsis by regulating the stability of downstream factors.
Loss of bHLH104 could decrease the tolerance to Fe deficiency conferred by BTS lesion, which suggested an opposite function between BTS and bHLH104 in Fe homeostasis. Both of them are located in the nucleus and have been found to interact with each other in a yeast two-hybrid assay (Long et al., 2010; Kobayashi et al., 2013). Moreover, genes responding to Fe deficiency that were downregulated in bhlh104 mutants were upregulated in BTS RNAi lines (Table 1, Figure 9C). These traits indicate that bHLH104 and BTS might work in a common pathway to regulate Fe homeostasis. In Arabidopsis, several E3 ligases have been reported to regulate the abundance of transcription factors. For example, the RING-type E3 ligase, KEEP ON GOING, is a negative regulator of abscisic acid and ubiquitinates the transcription factor ABSCISIC ACID INSENSITIVE5 following abscisic acid treatment (Liu and Stone, 2010). Hence, BTS might control Fe contents in plants by regulating the abundance of bHLH104 and ILR3. Recently, a study by Selote et al. (2015) demonstrated that BTS could target ILR3 and bHLH115 but not bHLH104 for the 26S proteasome-mediated degradation, using a cell-free degradation assay. However, the mutation of bHLH104 could partially complement the phenotype of BTS lesion (Figure 9), which suggested that the loss of bHLH104 might disrupt the formation of heterodimers with ILR3 and bHLH115 that function to activate the Fe deficiency responses in bts-2 plants. In future studies, a triple or tetra mutant should be constructed to further clarify the regulatory mechanism between BTS and these IVc subgroup bHLH proteins.
This regulatory network involving BTS-bHLH104/ILR3-Ib subgroup bHLH transcription factors/PYE in Arabidopsis raised the question of whether this regulatory mechanism also exists in Strategy II plants, since the Fe deficiency response has been demonstrated to be partially conserved between graminaceous and nongraminaceous plants (Kobayashi and Nishizawa, 2012). To date, the downstream regulator of rice HRZs has not been identified. However, knockdown of HRZs resulted in the upregulation of rice IRO2 and IRO3, which are the orthologs of Arabidopsis bHLH38/39 and PYE, respectively (Ogo et al., 2006; Zheng et al., 2010; Kobayashi et al., 2013). bHLH38/39 and PYE are both downstream regulators of bHLH104 and ILR3; therefore, we wondered whether there also exist transcription factors acting upstream of IRO2 and IRO3 in rice, which play a similar role to bHLH104 and ILR3. Interestingly, the core sequence for rice IRO2 promoter binding was determined to be CACGTGG in a biochemical analysis (Ogo et al., 2006), implying that the transcriptional activation of IRO2 may also be via bHLH-associated E-box elements in its promoter. Thus, further study on the regulatory network of Strategy II plants would be of interest.
In summary, our results provide evidence that the IVc subgroup bHLH transcription factors bHLH104 and ILR3 act as positive regulators in the Fe deficiency responses. bHLH104 and ILR3 could form heterodimers, and both of them bind to the promoters of subgroup Ib bHLH genes and PYE, functioning as upstream regulators of these two transcriptional regulation pathways. We also demonstrate that bHLH104 acts downstream of BTS, which negatively regulate Fe uptake in Arabidopsis (Figure 10). This regulatory network provides insight into the mechanism of plants’ responses to Fe deficiency and also offers a strategy for the design and breeding of Fe-deficient crops by genetic manipulation of regulatory genes. However, it is important to ensure reasonable Fe absorption, since excess Fe can cause a decrease in fertility. Future investigations of other two transcription factors belonging to the IVc subgroup will be meaningful, and clarification of the relationship between BTS and the IVc subgroup bHLH transcription factors would be of great interest.
Figure 10.
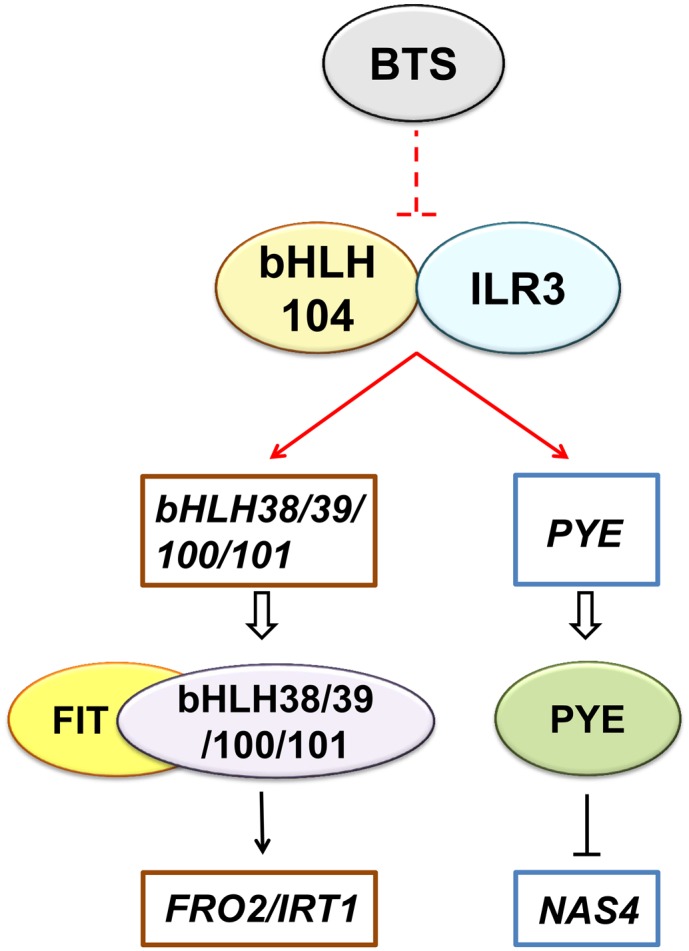
Model of bHLH104- and ILR3-Mediated Fe Deficiency Responses.
The model depicts the molecular function of bHLH104 and ILR3 in Arabidopsis Fe homeostasis relative to other known bHLH regulators. bHLH104 interacts with ILR3 and both of them regulate the expression of Ib subgroup bHLH genes and PYE by binding to their promoters. bHLH104 and ILR3 might be the downstream targets of an E3 ubiquitin ligase, BTS, which negatively regulates Fe absorption and was previously found to interact with bHLH104 and ILR3. The data generated in this study are indicated by red lines, while those from previous studies are indicated by black lines. Ovals represent central regulatory proteins and squares represent the downstream target genes.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Columbia-0 was used as the wild type in this study. The T-DNA insertion lines for bhlh104-1 (Salk_099496C), bhlh104-2 (Salk_043862C), ilr3-2 (Salk_004997C), and ilr3-3 (Salk_043690C) were confirmed using PCR with a T-DNA primer (LbaI) and gene-specific primers (Supplemental Table 1). The bhlh104-2 bts-2 double mutant plants were generated by crossing bhlh104-2 as the female parent, to bts-2 mutants; and two double mutant lines were studied in this work, named bhlh104-2 bts-2 #1 and bhlh104-2 bts-2 #2. For normal soil cultivation, plants were grown under light at 120 μM photons m−2 s−1, a 16-h/8-h light/dark cycle, 23°C /19°C day/night, and relative air humidity of 50 to 70%. High-pH soil (pH 7 to 8) was generated by adding CaO to soil, as described previously (Kim et al., 2006).
For phenotypic analyses of the Fe deprivation response, seeds were surface sterilized using 70% ethanol for 1 min followed by 20% bleach for 20 min and sown on Fe-sufficient media [+Fe, half Murashige and Skoog (MS) media containing 100 μM Fe(II)-EDTA with 1% sucrose and 1% agar] or Fe-deficient media [−Fe, without Fe(II)-EDTA; −Fe+Frz, with 50 μM ferrous chelate ferrozine]. After vernalization in darkness at 4°C for 3 d, the plates were placed in light with an irradiance of 120 μM photons m−2 s−1 for the indicated days at 23°C/19°C day/night.
Plasmid Construction and Plant Transformation
To construct the plants overexpressing bHLH104 or ILR3, the coding sequences of bHLH104 and ILR3 and a C-terminal GFP fusion sequence were amplified with gene-specific primers (Supplemental Table 1). The open reading frame was cloned into pRi35S and then cleaved by StuI/KpnI and inserted into pCAMBIA1301 to generate 35S:bHLH104-GFP and 35S:ILR3-GFP constructs. To produce BTS-silenced plants, 1000-bp sense and antisense sequences of BTS were cloned into pRi35S and then inserted into pCAMBIA1301. The different constructs were introduced into Agrobacterium tumefaciens strain EHA105 by heat shock. The transformants were then transformed into Columbia-0 plants through the floral dipping method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS agar plates with hygromycin, and homozygous T3 lines were used for subsequent analyses.
Rhizosphere Acidification, Ferric-Chelate Reductase Assays, and Tissue Elemental Analyses
The rhizosphere acidification assays were conducted as previously described (Yi and Guerinot, 1996). Seeds were germinated on Fe-sufficient or Fe-deficient media for 7 d, and four plants of every line were transferred to 1% agar plates containing 0.006% bromocresol purple and 0.2 mM CaSO4 (pH adjusted to 6.5 with NaOH) for 24 h.
Root ferric-chelate reductase activity was measured spectrophotometrically as described previously (Lucena et al., 2006). Ten intact plants of every line were pretreated for 30 min in plastic vessels with 4 mL half-strength MS solution without micronutrients (pH 5.5) and then soaked into 4 mL Fe(III) reduction assay solution [half-strength MS without micronutrients, 100 μM Fe(III)-EDTA, and 300 μM ferrozine, pH adjusted to 5.0 with KOH] for 30 min in darkness. An identical assay solution containing no plants was used as a blank. The purple-colored Fe(II)-ferrozine complex was quantified at 562 nm using a molar extinction coefficient of 28.6 mM−1 cm−1.
For tissue elemental analyses, seeds were placed on the Fe-sufficient or Fe-deficient media in a single row at a density of ∼15 seeds/cm for 8 d. Shoots of seedlings of four plates were collected to generate at least 200 mg of fresh weight. Tissue samples were dried at 80°C for 24 h to yield ∼20 to 40 mg tissue for elemental analyses. After cooling, all samples were digested with 10 mL concentrated nitric acid/perchloric acid (3:1) and diluted to 10 mL in 18 Ω water. The Fe contents were analyzed by inductively coupled plasma-atomic emission spectrometry, as described previously (Herbik et al., 2002).
Perls Staining for Fe, Tissue Sectioning, and DAB/H2O2 Intensification
For Fe3+ localization, plant tissues were vacuum infiltrated with Perls stain solution (equal volumes of 4% [v/v] HCl and 4% [w/v] K-ferrocyanide) for 30 min. Then the plant samples were incubated for another 1 h in the stain solution and rinsed three times with distilled water. For leaf staining, plant tissues were first incubated with fixative solution (methanol/chloroform/acetic acid, 6:3:1) for 1 h at room temperature and then added to the Perls stain solution. Localization of Fe3+ was observed using a stereomicroscope (SteREO Lumar.V12).
For tissue sectioning, leaves from adult plants were obtained at bolting and were fixed overnight at 4°C in FAA fixing buffer (containing 2% formaldehyde, 50% ethanol, and 5% acetic acid). Fixed tissues were dehydrated in 10, 30, 50, 60, 70, 80, 90, and 100% ethanol for 1 h at each concentration and then embedded in paraffin (Leica). Transverse sections (4 μm) were obtained using a Leica RM 2145 microtome.
The DAB intensification reaction was performed as previously described (Meguro et al., 2007; Roschzttardtz et al., 2009). The paraffin sections were dewaxed and rehydrated, placed into Perls stain solution for 30 min, and subsequently washed with distilled water. The sections were then incubated in a methanol solution containing 0.01 M NaN3 and 0.3% (v/v) H2O2 for 1 h and washed with 0.1 M phosphate buffer (pH 7.4). Then the sections were incubated in an intensification solution (0.1 M phosphate buffer solution containing 0.025% [w/v] DAB, 0.005% [v/v] H2O2, and 0.005% [w/v] CoCl2, pH 7.4) for 20 min and washed with distilled water to stop the reaction.
Total Chlorophyll Content Analyses
Total chlorophyll was extracted from leaves with 80% acetone in 2.5 mM HEPES-KOH (pH 7.5), and the chlorophyll content was determined as previously described (Wellburn, 1994).
RT-PCR and qPCR
To test for full-length bHLH104 transcript in the wild type and the bhlh104 mutants, leaves of plants grown for 4 weeks in normal soil were collected. Total RNA was extracted using a Plant RNA Kit (Omega) and 5 μg total RNA was used for cDNA synthesis using the PrimeScript RT reagent kit (Takara). The amplification of full-length bHLH104 transcript was performed using gene-specific primers (Supplemental Table 1), and UBQ10 was used as a control gene. PCR amplifications were done for 28 and 25 cycles for bHLH104 and UBQ10, respectively. For qPCR analyses, roots of seedlings grown on Fe-sufficient or Fe-deficient media were collected respectively to generate ∼50 mg fresh weight for RNA extraction. qPCR was performed using SYBR Premix Ex Taq (Takara), and amplification was monitored in real-time on the LightCycler 480 (Roche). TUBULIN2 (TUB2) was amplified as an internal control, and gene copy number was normalized to that of TUB2. TUB2 was relatively stable during different Fe supply when compared with the relative gene expression of ACTIN2 and UBQ4; thus, TUB2 was used as an internal control in this study. The primers used for qPCR analyses are listed in Supplemental Table 1.
Protein Extraction and Immunoblotting
To analyze the protein expression in transgenic plants, total proteins were extracted with protein extraction buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, and protease inhibitor cocktail [Roche]). The extracts were subsequently centrifuged at 18,000g for 10 min at 4°C to collect the supernatants for immunoblot analyses. Total proteins (200 μg) were separated by SDS-PAGE. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Millipore) and probed using antibodies specifically directed against GFP (Abmart).
Arabidopsis Protoplast Preparation and Transfection
The protoplast isolation from Arabidopsis leaves was performed as previously described (Zhai et al., 2009). Briefly, 14-d-old Arabidopsis leaves were cut into strips in TVL solution (0.3 M sorbitol and 50 mM CaCl2) and then incubated in enzyme solution (0.5 M sucrose, 10 mM MES-KOH at pH 5.7, 20 mM CaCl2, 40 mM KCl, 1% Cellulase R-10, and 1% Macerozyme R-10) with gentle shaking for 16 h in the dark. The protoplasts were collected by centrifugation at 100g for 7 min and washed twice with W5 solution (0.1% glucose, 0.08% KCl, 0.9% NaCl, 1.84% CaCl2, and 2 mM MES at pH 5.7). For BiFC analyses, 10 μg plasmid DNA per 100 μL (∼5 × 104 cells) Arabidopsis protoplasts was used for transfection. As a positive control, both bZIP63 fused with YN and bZIP63 fused with YC were cotransfected into protoplasts. After incubation for 12 h, the protoplasts were harvested and fluorescence emission of YFP in protoplasts was observed under a confocal microscope (TCS-SP5; Leica) as previously described (Yuan et al., 2008).
GST Pull-Down and Co-IP Assays
For pull-down assays, 4 mL Arabidopsis protoplasts (∼2 ×106 cells) was transfected with 400 μg 35S:ILR3-GFP or 35S:bHLH115-GFP plasmids and cultured in the dark for 16 h. The protoplasts were then harvested by centrifugation at 100g for 7 min. Total proteins were extracted with 1 mL protein extraction buffer. A portion of the protein extracts (500 μL) was mixed with 50 μg purified recombinant GST-bHLH104 and the remaining extract was mixed with 50 μg GST as a negative control. Then, 30 μL glutathione Sepharose 4B (GE Healthcare) was added to the samples. After incubation for 6 h with constant rotation at 4°C, the Sepharose was washed five times with ice-cold PBS buffer (pH 7.4), and bound proteins were eluted with 30 μL elution buffer (30 mM glutathione in PBS buffer). For immunoblotting analyses, 20 μL protein extract before incubation was used as input and 30 μL eluate was loaded as the immunoprecipitate.
For co-IP assays, 4 mL Arabidopsis protoplasts was cotransfected with 400 μg 35S:bHLH104-Flag and 400 μg 35S:ILR3-GFP or 400 μg 35S:ILR3-GFP alone (same for 35S:bHLH115-GFP). Total proteins were extracted with 500 μL protein extraction buffer and 20 μL protein extract was used as input. The total cell extracts were further incubated with 30 μL anti-FLAG M2 affinity gel (Sigma-Aldrich) for 6 h at 4°C with rotation. After washing five times with ice-cold TBS buffer (pH 7.4), the bound proteins were eluted by boiling the gel using 30 μL SDS-PAGE sample buffer without β-mercaptoethanol and loaded onto SDS-PAGE for immunoblotting.
ChIP
ChIP was performed following a previously described protocol (Saleh et al., 2008), using 10 mL Arabidopsis protoplasts transfected with 1 mg 35S:bHLH104-Flag or 35S:ILR3-Flag plasmids. Ten milliliters of protoplasts without transfection was used as a negative control. After transfection for 24 h, the protoplasts were fixed with 1% formaldehyde for 15 min and neutralized with 0.125 M glycine for 5 min. After washing twice with W5 solution, the protoplasts were harvested by centrifugation at 100g for 3 min and suspended in SDS lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM PMSF, 1 mM EDTA, 1% SDS, 1% Triton X-100, and 0.1% sodium deoxycholate) for sonication. Then, 50 μL anti-FLAG M2 affinity gel (Sigma-Aldrich) was used for each immunoprecipitation and the enriched DNA fragments were analyzed by qPCR using the primers listed in Supplemental Table 2. The amount of ChIP-DNA coprecipitated by anti-FLAG M2 affinity gel was first normalized by comparing to the total input DNA used for each immunoprecipitation CT = CT(ChIP) − CT(input). Then, these normalized ChIP signals were compared between the detecting targets and the E-box motif of TUB2, which was used as a negative control. The binding of TUB2 promoter in the protoplast without transfection was set to 1, and the DNA binding ratio was given as the fold increase in signal relative to TUB2’s binding. A 3-fold DNA binding ratio relative to TUB2’s binding was used as the threshold in this study.
Histochemical GUS Staining Assays
To investigate the bHLH104 expression pattern, the 1474-bp sequence upstream of the ATG codon in the bHLH104 gene was amplified with specific primers (Supplemental Table 1). The promoter was cloned into pCAMBIA1381 to drive the expression of the GUS reporter gene. For the GUS staining, tissues were soaked overnight in GUS-staining buffer as previous described (Jefferson et al., 1987) and then washed with 70% ethanol. Images were captured under a stereomicroscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: bHLH104 (At4g14410), ILR3 (At5g54680), bHLH34 (At3g23210), bHLH115 (At1g51070), bHLH38 (At3g56970), bHLH39 (At3g56980), bHLH100 (At2g41240), bHLH101 (At5g04150), PYE (At3g47640), BTS (At3g18290), FIT (At2g28160), FRO2 (At1g01580), IRT1 (At4g19690), IRT2 (At4g19680), FER1 (At5g01600), YSL1 (At4g24120), NAS4 (At1g56430), ZIF1 (At5g13740), FRD3 (At3g08040), MYB10 (At3g12820), MYB72 (At1g56160), UBQ10 (At4g05320), and TUB2 (At5g62690).
Supplemental Data
Supplemental Figure 1. Phenotypic analyses of the wild type, bhlh104-1, and bhlh104-2 in response to Fe deficiency.
Supplemental Figure 2. Phenotypic analyses of wild-type and bhlh104-2 complemented plants in response to Fe deficiency.
Supplemental Figure 3. Quantification of root length of bHLH104 ox plants and Trypan blue staining of necrosis in bHLH104 ox leaves.
Supplemental Figure 4. Identification of ILR3 ox plants and Trypan blue staining of necrosis in ILR3 ox leaves.
Supplemental Figure 5. Plants silenced with BTS exhibit reduced fertility.
Supplemental Figure 6. Perls staining for Fe3+ of BTS RNAi plants.
Supplemental Figure 7. Expression pattern analysis of bHLH104.
Supplemental Table 1. Primers used to identify T-DNA insertion mutants, gene cloning, and qPCR.
Supplemental Table 2. Primers used for ChIP-qPCR.
Supplemental Methods. Trypan blue staining.
Supplementary Material
Acknowledgments
We thank Philip N. Benfey (University of Illinois at Chicago) for kindly sending us the bts-1 mutant seeds. We appreciate the gift of ilr3-2 seeds from Bonnie Bartel (Rice University at Houston). We thank Terri A. Long (North Carolina State University) for kindly providing experimental guidance. We also thank the ABRC for the seed stocks. This investigation was supported by the National Natural Science Foundation of China (No. 31370297 and No. 31425003), the Natural Science Foundation of Guangdong Province, PR China (No. S2013010012682), and the Fundamental Research Funds for the Central Universities.
AUTHOR CONTRIBUTIONS
H.-B.W. and J.Z. designed the study. J.Z. and M.L. performed research. J.Z., B.L., D.F., and J.W. analyzed data. J.Z. and H.-B.W. wrote the article. B.L., H.J., P.W., J.L., and F.X. revised the article.
Glossary
- bHLH
basic helix-loop-helix
- ChIP
chromatin immunoprecipitation
- DAB
diaminobenzidine
- qPCR
quantitative real-time PCR
- BiFC
bimolecular fluorescence complementation
- co-IP
coimmunoprecipitation
- RNAi
RNA interference
- MS
Murashige and Skoog
References
- Bailey P.C., Martin C., Toledo-Ortiz G., Quail P.H., Huq E., Heim M.A., Jakoby M., Werber M., Weisshaar B. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P., Ling H.Q., Guerinot M.L. (2007). FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol. Biochem. 45: 260–261. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Briat J.F. (2003). Iron transport and signaling in plants. Annu. Rev. Plant Biol. 54: 183–206. [DOI] [PubMed] [Google Scholar]
- Fan H., Zhang Z., Wang N., Cui Y., Sun H., Liu Y., Wu H., Zheng S., Bao S., Ling H.Q. (2014). SKB1/PRMT5-mediated histone H4R3 dimethylation of Ib subgroup bHLH genes negatively regulates iron homeostasis in Arabidopsis thaliana. Plant J. 77: 209–221. [DOI] [PubMed] [Google Scholar]
- Feng B., Lu D., Ma X., Peng Y., Sun Y., Ning G., Ma H. (2012). Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 72: 612–624. [DOI] [PubMed] [Google Scholar]
- Fisher F., Goding C.R. (1992). Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 11: 4103–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T.C., Guerinot M.L. (1998). Molecular biology of cation transport in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 669–696. [DOI] [PubMed] [Google Scholar]
- Green L.S., Rogers E.E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136: 2523–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak M.A., Welch R.M., Kochian L.V. (1990). Physiological characterization of a single-gene mutant of Pisum sativum exhibiting excess iron accumulation: I. Root iron reduction and iron uptake. Plant Physiol. 93: 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot M.L., Yi Y. (1994). Iron: nutritious, noxious, and not readily available. Plant Physiol. 104: 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta O.P., Sharma P., Gupta R.K., Sharma I. (2014). MicroRNA mediated regulation of metal toxicity in plants: present status and future perspectives. Plant Mol. Biol. 84: 1–18. [DOI] [PubMed] [Google Scholar]
- Hänsch R., Mendel R.R. (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12: 259–266. [DOI] [PubMed] [Google Scholar]
- Haydon M.J., Cobbett C.S. (2007). A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143: 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Hell R., Stephan U.W. (2003). Iron uptake, trafficking and homeostasis in plants. Planta 216: 541–551. [DOI] [PubMed] [Google Scholar]
- Herbik A., Bölling C., Buckhout T.J. (2002). The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol. 130: 2039–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt M.N., Guerinot M.L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R., Brumbarova T., Bauer P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5: 27–42. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.A., Punshon T., Lanzirotti A., Li L., Alonso J.M., Ecker J.R., Kaplan J., Guerinot M.L. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298. [DOI] [PubMed] [Google Scholar]
- Klatte M., Schuler M., Wirtz M., Fink-Straube C., Hell R., Bauer P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150: 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen B.E., Larue T.A., Welch R.M., Weeden N.F. (1990). Pleiotropic effects of brz: A mutation in Pisum sativum (L.) cv; Sparkle’ conditioning decreased nodulation and increased iron uptake and leaf necrosis. Plant Physiol. 93: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N.K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63: 131–152. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Nakanishi H., Nishizawa N.K. (2010). Recent insights into iron homeostasis and their application in graminaceous crops. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86: 900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nagasaka S., Senoura T., Itai R.N., Nakanishi H., Nishizawa N.K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4: 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H.Q., Bauer P., Bereczky Z., Keller B., Ganal M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. USA 99: 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li X., Li K., Liu H., Lin C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.A., Tsukagoshi H., Busch W., Lahner B., Salt D.E., Benfey P.N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C., Waters B.M., Romera F.J., García M.J., Morales M., Alcántara E., Pérez-Vicente R. (2006). Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J. Exp. Bot. 57: 4145–4154. [DOI] [PubMed] [Google Scholar]
- Meguro R., Asano Y., Odagiri S., Li C., Iwatsuki H., Shoumura K. (2007). Nonheme-iron histochemistry for light and electron microscopy: a historical, theoretical and technical review. Arch. Histol. Cytol. 70: 1–19. [DOI] [PubMed] [Google Scholar]
- Mendoza-Cózatl D.G., Xie Q., Akmakjian G.Z., Jobe T.O., Patel A., Stacey M.G., Song L., Demoin D.W., Jurisson S.S., Stacey G., Schroeder J.I. (2014). OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol. Plant 7: 1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y., Itai R.N., Kobayashi T., Aung M.S., Nakanishi H., Nishizawa N.K. (2011). OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75: 593–605. [DOI] [PubMed] [Google Scholar]
- Ogo Y., Itai R.N., Nakanishi H., Kobayashi T., Takahashi M., Mori S., Nishizawa N.K. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51: 366–377. [DOI] [PubMed] [Google Scholar]
- Ogo Y., Itai R.N., Nakanishi H., Inoue H., Kobayashi T., Suzuki M., Takahashi M., Mori S., Nishizawa N.K. (2006). Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 57: 2867–2878. [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R.A., Woodward A.W., Hobbs B.N., Tierney M.P., Lahner B., Salt D.E., Bartel B. (2006). An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K., Touraine B., Boucherez J., Briat J.F., Gaymard F., Cellier F. (2009). Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57: 400–412. [DOI] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697. [DOI] [PubMed] [Google Scholar]
- Roschzttardtz H., Conéjéro G., Curie C., Mari S. (2009). Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol. 151: 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen A.A., Thompson J.W., Ruiz J.C., Ma H.W., Kinch L.N., Li Q., Grishin N.V., Bruick R.K. (2009). An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326: 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schuler M., Rellán-Álvarez R., Fink-Straube C., Abadía J., Bauer P. (2012). Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24: 2380–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D., Samira R., Matthiadis A., Gillikin J.W., Long T.A. (2015). Iron-binding E3 ligase mediates iron response in plants by targeting bHLH transcription factors. Plant Physiol. 167: 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A.B., Hermand V., Curie C., Vert G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey M.G., Patel A., McClain W.E., Mathieu M., Remley M., Rogers E.E., Gassmann W., Blevins D.G., Stacey G. (2008). The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Essigmann B., Kloska S., Altmann T., Buckhout T.J. (2001). Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol. 127: 1030–1043. [PMC free article] [PubMed] [Google Scholar]
- Thomine S., Vert G. (2013). Iron transport in plants: better be safe than sorry. Curr. Opin. Plant Biol. 16: 322–327. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzica E.I., Casero D., Yamasaki H., Hsieh S.I., Adler L.N., Karpowicz S.J., Blaby-Haas C.E., Clarke S.G., Loo J.A., Pellegrini M., Merchant S.S. (2012). Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24: 3921–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O., Yang S.S., Aparicio-Fabre R., Graham P.H., Reyes J.L., Vance C.P., Hernández G. (2010). MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 187: 805–818. [DOI] [PubMed] [Google Scholar]
- Vashisht A.A., et al. (2009). Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326: 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E.L., Connolly E.L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11: 530–535. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Klatte M., Jakoby M., Bäumlein H., Weisshaar B., Bauer P. (2007). Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908. [DOI] [PubMed] [Google Scholar]
- Wang L., Ying Y., Narsai R., Ye L., Zheng L., Tian J., Whelan J., Shou H. (2013a). Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ. 36: 224–236. [DOI] [PubMed] [Google Scholar]
- Wang N., Cui Y., Liu Y., Fan H., Du J., Huang Z., Yuan Y., Wu H., Ling H.Q. (2013b). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6: 503–513. [DOI] [PubMed] [Google Scholar]
- Wellburn A.R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144: 307–313. [Google Scholar]
- Wirstam M., Lippard S.J., Friesner R.A. (2003). Reversible dioxygen binding to hemerythrin. J. Am. Chem. Soc. 125: 3980–3987. [DOI] [PubMed] [Google Scholar]
- Yi Y., Guerinot M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10: 835–844. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., Wang D., Ling H.Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18: 385–397. [DOI] [PubMed] [Google Scholar]
- Yuan Y.X., Zhang J., Wang D.W., Ling H.Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15: 613–621. [DOI] [PubMed] [Google Scholar]
- Zhai, Z., Jung, H.I., and Vatamaniuk, O.K. (2009). Isolation of protoplasts from tissues of 14-day-old seedlings of Arabidopsis thaliana J. Vis. Exp. pii: 1149. [DOI] [PMC free article] [PubMed]
- Zhai Z., et al. (2014). OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Song A., Li P., Chen S., Jiang J., Chen F. (2014). A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci. Rep. 4: 6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Ying Y., Wang L., Wang F., Whelan J., Shou H. (2010). Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.