The small GTPase ROP10 regulates the reestablishment of cell polarity, leading to root hair deformation in legumes during root nodule symbiosis.
Abstract
Rhizobia preferentially enter legume root hairs via infection threads, after which root hairs undergo tip swelling, branching, and curling. However, the mechanisms underlying such root hair deformation are poorly understood. Here, we showed that a type II small GTPase, ROP10, of Medicago truncatula is localized at the plasma membrane (PM) of root hair tips to regulate root hair tip growth. Overexpression of ROP10 and a constitutively active mutant (ROP10CA) generated depolarized growth of root hairs, whereas a dominant negative mutant (ROP10DN) inhibited root hair elongation. Inoculated with Sinorhizobium meliloti, the depolarized swollen and ballooning root hairs exhibited extensive root hair deformation and aberrant infection symptoms. Upon treatment with rhizobia-secreted nodulation factors (NFs), ROP10 was transiently upregulated in root hairs, and ROP10 fused to green fluorescent protein was ectopically localized at the PM of NF-induced outgrowths and curls around rhizobia. ROP10 interacted with the kinase domain of the NF receptor NFP in a GTP-dependent manner. Moreover, NF-induced expression of the early nodulin gene ENOD11 was enhanced by the overexpression of ROP10 and ROP10CA. These data suggest that NFs spatiotemporally regulate ROP10 localization and activity at the PM of root hair tips and that interactions between ROP10 and NF receptors are required for root hair deformation and continuous curling during rhizobial infection.
INTRODUCTION
ROP GTPases (Rho-related GTPases from plants) constitute a plant-specific clade of the conserved Rho family of Ras-related small G proteins in plants (Yang and Fu, 2007). These small GTPases act as molecular switches by cycling between a GDP-bound inactive form and a GTP-bound active form. The membrane-associated activated GTP-bound forms interact with cellular effectors, which in turn trigger multiple extracellular signal-induced downstream responses such as leaf cell morphogenesis, polarized cell growth, pathogen defense reactions, abiotic stress-related responses, and plant hormone responses (Nibau et al., 2006; Craddock et al., 2012).
Regulatory functions have been attributed to ROP GTPases during polarized growth of plant cells, particularly with respect to tip growth of pollen tubes and root hairs (Nibau et al., 2006; Kost, 2008). A subset of type I ROP GTPases induces the depolarized growth of pollen tubes and root hairs when they are activated in these cells (Nibau et al., 2006; Yang and Fu, 2007). In Arabidopsis thaliana, overexpression of ROP1 or ROP5 as well as constitutively active mutant proteins can induce depolarized pollen tube growth, whereas overexpression of dominant negative mutants inhibits pollen tube growth (Kost et al., 1999; Li et al., 1999). Analogously, overexpression of ROP2, ROP4, or a ROP6 constitutively active mutant causes either isotropic growth or increased length of root hairs, whereas overexpression of a dominant negative mutant of ROP2 results in tip growth inhibition (Molendijk et al., 2001; Jones et al., 2002). These ROP GTPases of Arabidopsis are localized at the plasma membrane (PM) of growing tips of pollen tubes and root hairs. Altered PM localization of ROP GTPases in cells overexpressing these proteins correlates with uncontrolled growth, indicating that polarized tip growth depends on ROP GTPases (Kost et al., 1999; Li et al., 1999; Molendijk et al., 2001; Jones et al., 2002). Various regulators that modulate the shuttling between inactive and active forms of ROP GTPases have been identified, including guanine nucleotide exchange factors (RopGEFs) that stimulate GDP-to-GTP exchange to activate ROP GTPases, ROP GTPase-activating proteins (RopGAPs) that accelerate GTP hydrolysis, and guanine nucleotide dissociation inhibitors (RhoGDIs) that inhibit GDP release, thus shifting ROP GTPases toward the inactive state. Some of these regulators have been shown to modulate ROP GTPase activity in pollen tubes and root hairs (Carol et al., 2005; Gu et al., 2006; Klahre et al., 2006; Klahre and Kost, 2006; Zhang and McCormick, 2007; Hwang et al., 2008, 2010; Riely et al., 2011).
The establishment of nodule symbiosis between legumes and nitrogen-fixing rhizobia depends on the ability of the bacteria to infect host roots via root hairs. The root hair infection process is intimately associated with the reorientation of polarized tip growth of root hairs in the susceptible root zone close to the root tip. The root hairs of the infection zone exhibit altered growth behavior, called root hair deformation or root hair curling (Heidstra et al., 1994). Root hair deformation is the result of isotropic growth, by which the tip of the hair swells and then grows in an altered direction (Esseling et al., 2003). To entrap the invading bacteria, the growth direction of the root hairs must be constantly redirected toward the bacteria, which suggests that bacteria secrete positional signals, namely lipochitooligosaccharidic nodulation factors (NFs). The perception of NFs by the cognate NF receptors in the host plant initiates symbiotic signaling. NF signaling precedes the invasion of rhizobia into root hair cells and is required for both the formation of infection threads containing entrapped bacteria and the initiation of a nodule primordium. Early responses in root hairs triggered by NFs include increased intracellular Ca2+ levels, a Ca2+ spiking response, and rapid changes of the cytoskeleton (Cárdenas et al., 1998; de Ruijter et al., 1998; Timmers et al., 1999; Amor et al., 2003; Geurts et al., 2005). These cellular responses are associated with the deformation and curling of root hairs as well as the specific activation of early nodulin genes (Esseling et al., 2003; Jones et al., 2007). NF-induced symbiotic signaling in the model legume Medicago truncatula depends on NF receptors, namely NFP and LYK3. These receptors are LysM domain receptor-like kinases with an extracellular domain and an intracellular kinase domain located in epidermal cells, which perceive sulfated NFs secreted by the symbiont Sinorhizobium meliloti (Limpens et al., 2003; Smit et al., 2007). NFP lacks protein kinase activity, which is required for all early NF-induced responses (Madsen et al., 2003; Radutoiu et al., 2003), whereas LYK3 has an active kinase activity allowing downstream signaling via phosphorylation, but its mutant is still sensitive to NFs and shows Ca2+ spiking and root hair deformation responses. Therefore, LYK3 is thought to mediate bacterial entry into root hairs (Catoira et al., 2001; Limpens et al., 2003; Smit et al., 2007). Downstream components of the NF signal transduction pathway in M. truncatula include DMI2, a leucine-rich repeat receptor kinase (Esseling et al., 2004), DMI1, a potassium ion channel (Ané et al., 2004; Riely et al., 2007), NENA, a nuclear pore component (Groth et al., 2010), CCaMK/DMI3, a Ca2+ and calmodulin-dependent protein kinase (Lévy et al., 2004; Mitra et al., 2004), the CCaMK-regulated transcriptional regulator CYCLOPS/IPD3 (Messinese et al., 2007; Yano et al., 2008; Singh et al., 2014), and various transcriptional regulators such as NSP1, NSP2, ERN1, and NIN (Kaló et al., 2005; Smit et al., 2005; Marsh et al., 2007, Middleton et al., 2007). As a consequence of NF signaling, the expression of many symbiotic genes such as the early nodulin gene ENOD11 is induced (Andriankaja et al., 2007; Hirsch et al., 2009).
Recent studies indicated that ROP GTPases of legumes participate in nodule symbiosis by regulating rhizobial infection and nodulation (Ke et al., 2012; Kiirika et al., 2012). Whether ROP GTPases participate in the regulation of root hair deformation induced by NFs or rhizobial infection was not clearly established, however. Given the central roles for ROP GTPases in polarized cell growth, we hypothesized that ROP GTPases may regulate the root hair deformation required for initiation of the rhizobial infection process. In a previous study, we identified seven ROP genes in M. truncatula and found that ROP10 is transiently induced by rhizobial infection (Liu et al., 2010), implying that ROP10 likely plays a unique function during early preinfection stages. Here, we report that a type II small GTPase, ROP10, of M. truncatula is a major regulator of the polarized growth of root hairs and is involved in a legume-specific root hair deformation process. We found that overexpression of ROP10 and a constitutively active mutant form (ROP10CA) causes depolarized root hair growth, whereas overexpression of a dominant negative mutant form (ROP10DN) inhibits root hair elongation. Expression of fluorescent fusion proteins indicated that ROP10 is localized at the PM in the apex of root hair tips, where it assists in defining tip growth. In response to applied NFs, ROP10 is transiently upregulated in root hairs, and ROP10 proteins are ectopically localized at the PM of NF-induced outgrowths and curls induced by rhizobial inoculation. We also demonstrated that ROP10 interacts with the NF receptors. Using M. truncatula plants carrying the early nodulin gene ENOD11 promoter reporter gene construct, we showed that overexpression of ROP10 or ROP10CA enhances NF signaling. Thus, our study provides important information about root hair development as well as root hair deformation responses induced by NFs in the early stage of symbiotic interaction.
RESULTS
ROP10 Regulates Polarized Tip Growth of Root Hairs
To better understand the function of ROP10, the ROP10 gene of M. truncatula was PCR-cloned from genomic DNA (∼4 kb). By aligning the genomic sequence to its cDNA, we found that the ROP10 gene contains eight exons and seven introns (Supplemental Figure 1A). This gene structure is consistent with the prediction by bioinformatic analysis (Liu et al., 2010). ROP10 belongs to the type II group of ROP GTPases, which has a long, extra exon compared with other members in the ROP GTPase family of M. truncatula (Liu et al., 2010). ROP10 is predicted to encode a protein with 210 amino acid residues and seven conserved domains (Supplemental Figure 1B). A comparison of the membrane localization domain of ROP10 with counterparts of known type II ROP GTPases in plants revealed that ROP10 has a typical C-terminal domain of the type II ROP GTPase group for membrane partitioning. This domain encompasses a polybasic region and a plant-specific motif called a GC-CG box with two cysteine (C) residues (Supplemental Figure 1C).
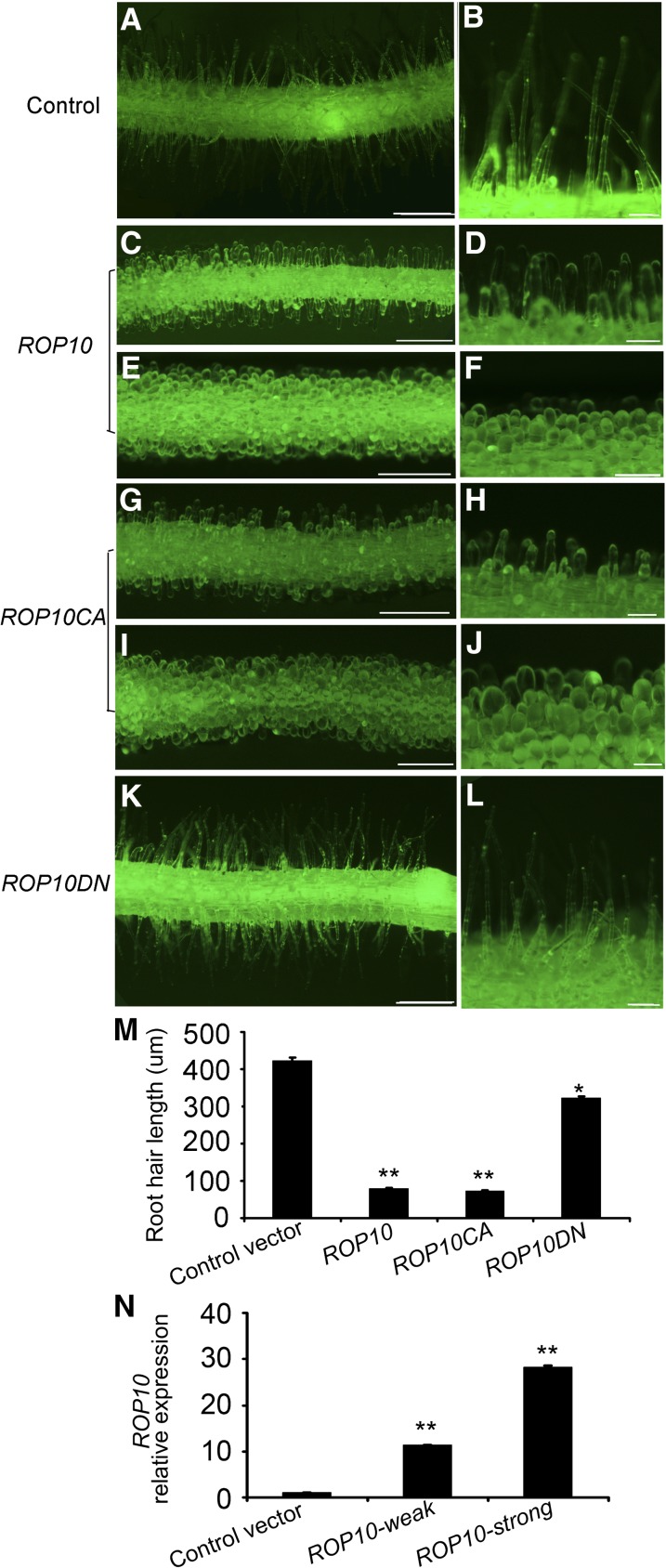
Some type I ROP GTPases are well-characterized regulators of the polarized growth of root hairs (Molendijk et al., 2001; Jones et al., 2002), but the involvement of type II ROP GTPases in the tip growth of root hairs remains to be clarified. To explore the role of ROP10 in root hair development, two ROP10 mutants that affect their abilities to shuttle between the GTP-bound active and the GDP-bound inactive forms, named ROP10CA and ROP10DN, were included in our analysis (Supplemental Figure 1B). These gain-of-function mutants were created by the replacement of specific amino acid residues (Li et al., 1999; Molendijk et al., 2001; Jones et al., 2002). The ROP10CA (G17V) mutant presumably locks ROP10 in the active GTP-bound state, and the ROP10DN (D123A) mutant locks ROP10 in an inactive state (Supplemental Figure 1B). Using an Agrobacterium rhizogenes-mediated transformation procedure, we generated transgenic hairy roots of M. truncatula that express ROP10, ROP10CA, and ROP10DN under the control of a Lotus japonicus ubiquitin gene (Ljubq1) promoter. Green fluorescent protein (GFP) coexpression was used to visualize transformed roots. Control transgenic roots transformed with the empty vector produced normal root hairs with tubular extensions growing at the tips, straight and away from the primary root axis (Figures 1A and 1B). However, transgenic roots overexpressing ROP10 generated depolarized root hairs. The phenotypes ranged from short root hairs (Figures 1C and 1D) to swollen root hairs (Figures 1E and 1F). The depolarized degree of the root hairs positively correlated with the transcript levels of ROP10 in these plants (Figure 1N). Compared with the ROP10-overexpressing roots, the ROP10CA-overexpressing roots produced shorter, swollen root hairs (Figures 1G and 1H) and more strongly ballooning root hairs at the time of or shortly after the emergence of root hairs (Figures 1I and 1J). Thus, ROP10CA overexpression resulted in a similar but more pronounced phenotype than overexpression of ROP10. The ROP10DN-overexpressing roots formed morphologically normal root hairs similar to the control root hairs; however, root hair elongation was arrested (Figures 1K and 1L). In comparison with control root hairs (422.8 ± 8.9 μm), the average length of root hairs in the root elongation zone was significantly decreased for transgenic roots overexpressing ROP10 (80.0 ± 1.9 μm) or ROP10CA (72.4 ± 1.9 μm) and was slightly decreased for roots overexpressing ROP10DN (322.2 ± 4.3 μm) (Figure 1M). These results indicate that ROP10 is involved in the maintenance of cell polarity of root hair tip growth.
Figure 1.
Root Hair Phenotypes of Transformed M. truncatula Roots Overexpressing ROP10 and ROP10CA or ROP10DN.
(A) and (B) Roots harboring the empty vector showed normal tip growth of root hairs.
(C) to (F) Roots overexpressing ROP10 produced root hairs with weakly ([C] and [D]) to strongly ([E] and [F]) depolarized growth.
(G) to (J) Roots overexpressing ROP10CA produced short swollen root hairs ([G] and [H]) as well as completely ballooning root hairs ([I] and [J]).
(K) and (L) Roots overexpressing ROP10DN produced short root hairs with almost normal tip growth.
About 120 composite plants were obtained for each construct in each experiment, and over 40% of composite plants generated transformed roots. The hairy root transformation experiment for each construct was repeated at least five times. Bars in (A), (C), (E), (G), (I), and (K) = 500 µm; bars in in (B), (D), (F), (H), (J), and (L) = 100 µm.
(M) Quantitative analysis of the average length of root hairs in transformed roots overexpressing the indicated proteins. Ten root hair cells were measured per transformed root, and 15 transformed roots were scored. Error bars indicate se. Statistical significance (*P < 0.05, **P < 0.01) was evaluated by Student’s t test.
(N) Quantitative RT-PCR of ROP10 mRNA levels in transformed roots overexpressing ROP10 that showed weakly depolarized root hairs (ROP10-weak) and strongly depolarized root hairs (ROP10-strong). Statistical significance (**P < 0.01) was evaluated by Student’s t test. Error bars indicate se. Data presented are representative of three independent experiments.
To further ascertain the role of ROP10, we obtained a homozygous Tnt1 line of the ROP10 gene (NF2968, designated rop10; Supplemental Figure 2) from a Tnt1 retrotransposon-tagged mutant population of M. truncatula (Tadege et al., 2008). The Tnt1 insertion was inserted in the second intron at position 459 bp downstream from the translation initiation codon of ROP10 in the rop10 mutant, which was confirmed by PCR (Supplemental Figures 2A and 2B). In the mutant, the expression of ROP10 was significantly impaired (Supplemental Figure 2C). However, under our growth conditions, there was no observable developmental defect in root hair tip growth compared with the wild type (Supplemental Figures 2D and 2E), suggesting that ROP genes possibly have functional redundancy or overlap in the regulation of root hair tip growth in M. truncatula.
The actin cytoskeleton is essential for tip growth because it promotes short-distance vesicle transport to the PM at the apex (Kost, 2008). Therefore, we speculated that the actin organization of root hairs is affected by overexpression of the different ROP10 forms. AlexaFluor 488-conjugated phalloidin was used to stain F-actin in root hairs of M. truncatula roots overexpressing ROP10, ROP10CA, and ROP10DN under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The control root hairs transformed with the empty vector predominantly showed actin filaments that were oriented parallel to the longitudinal axis of the root hairs (Supplemental Figure 3A). The transformed root hairs overexpressing ROP10 showed disordered actin filament arrangements, and the extent of cytoskeletal disruption in root hairs correlated directly with the depolarized growth phenotypes. Weakly tip-swollen root hairs exhibited longitudinal actin filaments in the shanks and a transverse actin filament arrangement in swollen tips (Supplemental Figure 3B). Short, strong, swollen root hairs displayed small and disorganized actin fragments (Supplemental Figure 3C), whereas ballooning root hairs showed mostly transverse or web-like actin filament arrangements (Supplemental Figure 3D). Transformed ballooning root hairs overexpressing ROP10CA also produced web-like actin structures (Supplemental Figure 3E). However, root hairs overexpressing ROP10DN exhibited no apparent alteration of the cytoskeleton arrangement (Supplemental Figure 3F). We further detected the expression of ACTINB in transformed roots and found that the transcript levels of ACTINB in transformed roots overexpressing ROP10 and ROP10CA are similar to those in control transformed roots (Supplemental Figure 3G), suggesting that overexpression of ROP10 and ROP10CA did not affect the expression levels of ACTINB in root hairs. These results demonstrate that ROP10 influences root hair tip growth by regulating the organization of the actin cytoskeleton.
PM-Localized Activated ROP10 Determines the Extent of Depolarized Root Hair Growth
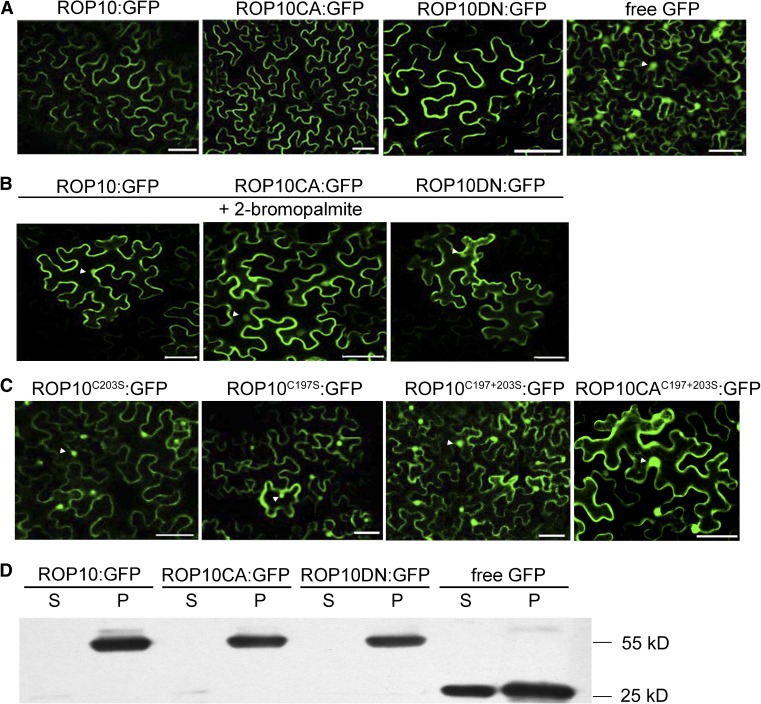
Membrane targeting is usually a prerequisite for the activation of ROP GTPases or their interaction with effectors (Yalovsky et al., 2008). Therefore, we investigated the subcellular localization of ROP10 in plant cells. We expressed ROP10 and mutant proteins fused with GFP under the control of the CaMV 35S promoter (ROP10:GFP, ROP10CA:GFP, and ROP10DN:GFP). Agrobacterium tumefaciens carrying these constructs was used for the infiltration of Nicotiana benthamiana leaves, and transgenic epidermal cells were analyzed by confocal laser scanning microscopy. GFP-tagged ROP10 and the mutant proteins were only detected in the PM of epidermal cells, whereas GFP alone as a control was distributed throughout the PM, cytoplasm, and the nuclei (Figure 2A). To further define the PM localization of different ROP10 forms, we performed membrane fractionation of N. benthamiana leaf cells for transformed ROP10 and mutant proteins followed by immunoblotting with anti-GFP antibodies. In accordance with the fluorescence data (Figure 2A), protein immunoblots showed that GFP-tagged ROP10 and the mutant proteins were localized exclusively in the PM, whereas GFP only was detected in both membrane and soluble fractions (Figure 2D). Immunoblot analysis further confirmed that both active and inactive forms of ROP10 are localized at the PM.
Figure 2.
Subcellular Localization of ROP10:GFP and Mutant Forms in N. benthamiana Leaf Epidermal Cells.
(A) ROP10:GFP, ROP10CA:GFP, and ROP10DN:GFP were localized exclusively at the PM, whereas free GFP was distributed in the PM, cytoplasm, and nuclei. Bars = 50 μm.
(B) Inhibition of S-acylation of ROP10:GFP, ROP10CA:GFP, and ROP10DN:GFP by a treatment with 2-bromopalmitate resulted in release of the expressed proteins from the PM. Fluorescence is observed in the cytoplasm and nuclei (arrowheads). Bars = 50 μm.
(C) ROP10 proteins with mutations in the GC-CG box (ROP10C203S:GFP, ROP10C197S:GFP, and ROP10C197+203S:GFP) and ROP10CA mutant with mutations (ROP10CAC197+203S:GFP) showed reduced association with the PM, and fluorescence is observed in the cytoplasm and nuclei (arrowheads). Bars = 50 μm.
(D) Immunoblotting using GFP monoclonal antibody. Soluble (S) and insoluble membrane (P) fractions were separated from N. benthamiana leaf cells overexpressing different forms of ROP10 proteins. ROP10, ROP10CA, and ROP10DN were detected in the membrane fraction, and free GFP was detected in both the soluble and membrane fractions.
To become functional and attached to the PM, type I ROP GTPases are prenylated on the C residue in the C-terminal CaaL box, whereas type II ROP GTPases undergo posttranslational S-acylation on conserved C residues in the GC-CG box (Lavy et al., 2002; Lavy and Yalovsky, 2006; Sorek et al., 2011). To determine whether ROP10 is also an S-acylated protein, GFP-tagged ROP10 and mutant proteins were expressed in N. benthamiana leaves that were treated with an S-acylation inhibitor, 2-bromopalmitate, which was used to detect protein S-acylation in vivo (Lavy et al., 2002). In the presence of 2-bromopalmitate, the mislocalization of ROP10:GFP, ROP10CA:GFP, and ROP10DN:GFP from the PM to cytoplasm and nuclei was observed (Figure 2B), whereas a control treatment with DMSO showed no visible effects (Supplemental Figure 4), suggesting that S-acylation is required for the association of ROP10 and mutant proteins with the PM. To substantiate that ROP10 is S-acylated, we mutated the conserved S-acylated C residues at positions 197 and 203 in the GC-CG box of ROP10 and ROP10CA (Supplemental Figure 1C) and expressed them as fusion proteins with GFP under the control of the CaMV 35S promoter (ROP10C197+203S:GFP and ROP10CAC197+203S:GFP, respectively). Analysis of the protein distribution in N. benthamiana epidermal cells with confocal laser scanning microscopy showed that both ROP10C197+203S:GFP and ROP10CAC197+203S:GFP were primarily localized in the nuclei, and concentrated regions of fluorescence were observed in the cytoplasm, which is a typical cytoplasmic distribution (Figure 2C). The mutant proteins with a single mutation in the GC-CG box of ROP10 (ROP10C197S:GFP or ROP10C203S:GFP) also showed cytoplasmic and nuclear localization (Figure 2C), suggesting that both the 197th and 203rd C residues in the GC-GC box function synergistically to assist the association of ROP10 with the PM. Protein immunoblot analyses also corroborated that these mutant proteins were localized in both membrane and soluble fractions (Supplemental Figure 5), complementing the fluorescence data (Figure 2C). The distribution of these mutant proteins suggested that mutations of these C residues of ROP10 compromised the association of the protein with the PM, and this distribution correlated with the S-acylation status of the mutants. These findings indicate that both C residues in the GC-CG box of ROP10 are required for targeting this protein to the PM.
To determine whether altered PM localization of ROP10 affects the polarized growth of root hairs, we generated transgenic hairy roots of M. truncatula that expressed the constructs ROP10C203S, ROP10C197S, and ROP10C197+203S under the control of the CaMV 35S promoter. β‑glucuronidase (GUS) staining was used for the identification of transformed roots. Compared with control plants transformed with the empty vector, transgenic roots overexpressing proteins with mutations in the GC-CG box produced swollen root hairs of significantly reduced length (Supplemental Figures 6A and 6B). However, the degree of root hair swelling was less than that of root hairs of transgenic roots overexpressing ROP10 or ROP10CA (Figure 1; Supplemental Figure 6C). The average length of root hairs expressing the proteins with mutations in the GC-CG box was slightly longer than that of transgenic roots overexpressing ROP10 or expressing ROP10CA but shorter than that of transgenic roots expressing ROP10DN (144.5 ± 2.6 μm for ROP10C203S, 178.2 ± 3.7 μm for ROP10C197S, and 139.7 ± 3.4 μm for ROP10C197+203S; compare Supplemental Figure 6C with Figure 1M). Hence, the root hair length reflected the degree of depolarized growth of root hairs in transformed roots expressing different ROP10 mutant proteins. These data show that the alteration of ROP10 in PM localization caused by mutations of C residues in the GC-CG box compromises the protein’s capability to induce depolarized growth and that the degree of depolarized growth correlates with ROP10 distribution at the PM.
To determine the effect of activated GTP-bound ROP10 at the PM on root hair tip growth, we compared the extent of the depolarized growth of M. truncatula root hairs expressing ROP10CAC197+203S and ROP10CA under the control of the Ljubq1 promoter. Transgenic roots overexpressing ROP10CAC197+203S showed weakly swollen and strongly ballooning root hairs (Supplemental Figures 6D and 6E), similar to the root hair phenotypes in transformed roots overexpressing ROP10CA (Figures 1H and 1J). However, the rate of strongly ballooning root hairs generated in transformed roots overexpressing ROP10CAC197+203S was significantly lower than that of roots overexpressing ROP10CA (Supplemental Figure 6F), suggesting that the impairment of GTP-bound ROP10 in the PM reduced the effect of depolarized growth caused by the activated ROP10CA. Therefore, these results suggest that the amount of PM-localized activated ROP10 specifies the extent of depolarized growth of root hairs.
ROP10 Is Localized at the PM of the Root Hair Tips to Regulate Polar Growth
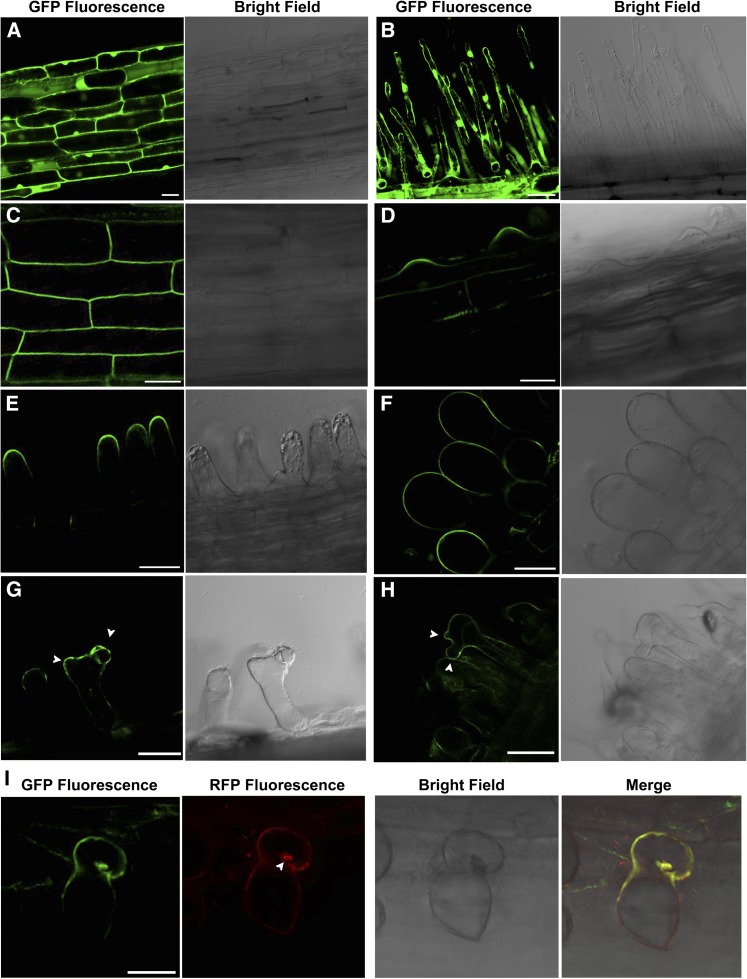
Type I ROP GTPases regulating polar tip growth of pollen tubes and root hairs have been shown to be localized in the tips of these cells (Li et al., 1999; Molendijk et al., 2001). To determine the localization of ROP10 in root hairs, we generated hairy roots expressing ROP10:GFP driven by the CaMV 35S promoter. In control roots expressing GFP alone, fluorescence was distributed throughout the PM, cytoplasm, and nuclei of root epidermal cells and root hairs (Figures 3A and 3B). In transformed roots expressing ROP10:GFP, fluorescence (in cells close to the root tip) was observed at the PM of root epidermal cells and at the apex of the root hair bulge (Figures 3C and 3D). In the young and elongating root hairs expressing ROP10:GFP that were comparable to the control root hairs, GFP fluorescence was observed at the PM in the apex of root hairs (Figure 3E; Supplemental Figure 7A). In weakly swollen root hairs expressing ROP10:GFP, fluorescence signals spread from the PM at the tip to the PM of the shank (Supplemental Figures 7B and 7C). In strongly ballooning root hairs expressing ROP10:GFP, GFP fluorescence was distributed throughout the PM of the entire root hairs (Figure 3F). The observed phenotypes of root hairs expressing ROP10:GFP were essentially the same as those of root hairs overexpressing ROP10 (Figures 1C and 1D). The short and young root hairs transformed with ROP10:GFP that were comparable to the control root hairs displayed almost normal tip growth, with fluorescence signals in the apical PM of root hair tips, whereas root hairs transformed with ROP10:GFP showing ballooning morphology displayed isotropic growth, with ROP10:GFP fluorescence signals throughout the whole root hair (Figures 3D to 3F). Hence, the ROP10:GFP fluorescence signals at the PM in transformed root hairs that were comparable to root hairs in control roots may reflect the authentic localization of the ROP10 protein in normally growing root hairs. Taken together, these findings suggest that the localization of ROP10 at the apical PM is crucial for the tip growth of root hairs and that the degree of uncontrolled growth of root hairs is associated with ROP10 being distributed at subapical PM parts of the root hairs.
Figure 3.
ROP10:GFP Is Localized at the Apical PM of Root Hairs and the PM of Outgrowths and Curling in Response to NF Treatment and Rhizobial Infection.
(A) and (B) Subcellular localization of free GFP in root epidermal cells (A) and root hairs (B) of control roots expressing GFP alone.
(C) ROP10:GFP is localized at the PM of root epidermal cells close to the root tip.
(D) ROP10:GFP is localized at the PM of root hair bulges, which produce elongating root hairs by tip growth.
(E) ROP10:GFP is localized at the PM in the apex of elongating root hairs close to the root tip.
(F) ROP10:GFP is localized at the PM throughout the ballooning root hairs.
(G) Following treatment with 1 nM NodSm-IV(C16:2, S) for 24 h, ROP10:GFP is localized at the PM of outgrowths (arrowheads) of a swollen root hair.
(H) ROP10:GFP is localized at the PM of outgrowths (arrowheads) of a swollen root hair 7 d after inoculation with S. meliloti.
(I) ROP10:GFP is visible in the curling (green) of the root hair 5 d after inoculation with mCherry-expressing S. meliloti (red; arrowhead).
Bars = 100 μm.
Overexpression of ROP10 and ROP10CA in Root Hairs Results in Aberrant Rhizobial Infection
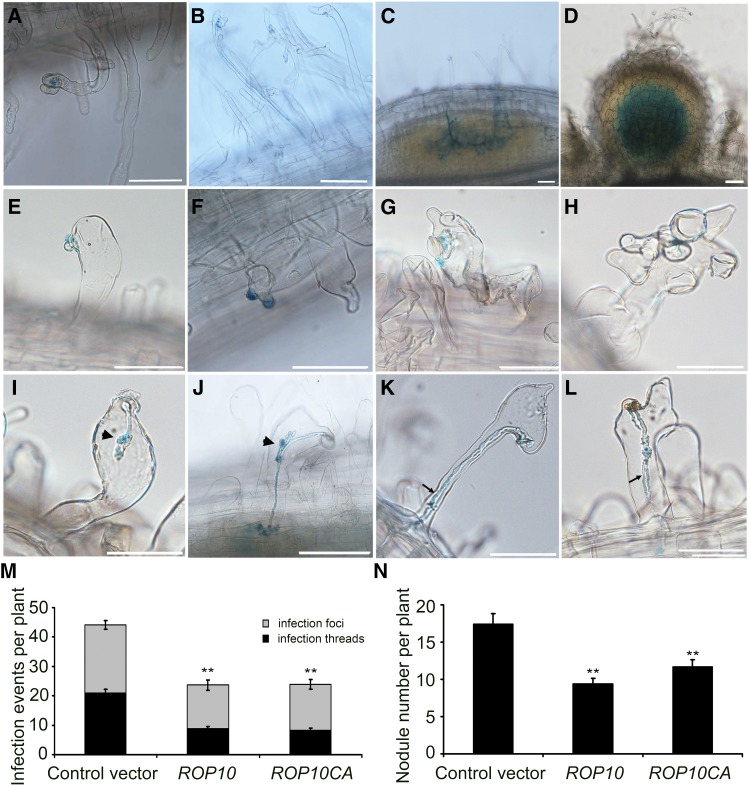
Root hair deformation is directly associated with successful rhizobial entry into root hairs and subsequent infection thread formation. To investigate whether the aberrant root hair morphology caused by the overexpression of ROP10 or ROP10CA affects rhizobial infection, we inoculated transformed roots with S. meliloti 2011 harboring the hemA:lacZ fusion, which allows microscopic visualization of the infection. Following inoculation, control root hairs transformed with the empty vector formed a single curl resulting in both entrapment of bacteria (Figure 4A) and the initiation of an infection thread (Figure 4B). The bacteria then could penetrate the root cortex, and subsequently, the infection threads ramified (branched out) into the nodule primordium and the developing nodule (Figures 4C and 4D). In contrast with the control, root hair infection was abnormal in transformed roots overexpressing ROP10. Some root hairs produced an outgrowth that seemed to be embraced by a single layer of bacterial cells, and no root hair curling was observed (Figure 4E). Most of the ROP10-overexpressing root hairs formed more than one outgrowth from a single root hair tip, and microcolonies apparently developed on multiple outgrowths. However, the bacteria were unable to induce the formation of normal root hair curls, and infection thread initiation was not observed in such root hairs (Figures 4F to 4H). We also noticed that infection threads in root hairs aborted due to the formation of sac-like structures (Figure 4I). Nonetheless, a few of these aberrant infection threads could successfully reach the nodule primordia (Figure 4J). Occasionally, dual infection threads were observed that appeared to be fused together at the base of the root hairs, and new infection threads grew from these fusion points toward the nodule primordia (Figures 4K and 4L). Roots overexpressing ROP10CA exhibited similar aberrant root hair infections (Supplemental Figure 8). To substantiate these findings, we scored quantitatively the total number of infection foci (i.e., the number of microcolonies in root hair tips) as well as the number of infection threads in root hairs and in primordia at 7 d after inoculation. Compared with control roots that were transformed with the empty vector, the numbers of infection foci and infection threads were reduced in ROP10- and ROP10CA-overexpressing roots (Figure 4M). ROP10 transcript levels in transformed roots overexpressing ROP10 or ROP10CA were increased significantly compared with the control roots (Supplemental Figure 9). These observations indicate that root hair deformation and infection thread development were markedly affected by the overexpression of ROP10 or ROP10CA.
Figure 4.
Overexpression of Wild-Type ROP10 and ROP10CA Affects Rhizobial Infection and Nodulation of M. truncatula.
(A) to (D) Control roots transformed with the empty vector showed a normal infection process of root hairs. Rhizobia are present in the infection pocket of a curled root hair tip (A), in the infection thread that extends within a curled root hair (B), in infection threads that ramify into a fine network in the nodule primordium (C), and in a developing nodule (D).
(E) to (L) Roots overexpressing ROP10 showed an aberrant infection process in swollen root hairs. Microcolonies of bacteria were often absent in swollen root hairs (E). Frequently, more than one outgrowth in a single root hair were observed ([F] to [H]) and infection threads aborted due to the formation of a sac-like structure (arrowhead) (I). Occasionally, a new infection thread was initiated from the sac-like structure and reached the root cortex (J). Double infection threads were also observed in a single root hair ([K] and [L]).
Rhizobia were detected by β-galactosidase staining with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside. Photographs were taken 7 d after inoculation with S. meliloti. Bars = 100 μm.
(M) Overexpression of ROP10 and ROP10CA led to a significant reduction of root hair infection. Infection events include observed infection foci and infection threads. The infection events were scored at 7 d after inoculation.
(N) Overexpression of ROP10 and ROP10CA resulted in reduced nodulation. The number of nodules on transgenic roots per plant was scored at 14 d after inoculation.
Statistical significance (**P < 0.01) was evaluated by Student’s t test. Error bars indicate se. Data presented are representative of three independent experiments.
We also assessed the nodule number of transgenic hairy roots that overexpress ROP10 or ROP10CA at 2 weeks after inoculation. The morphology of the nodules formed on roots overexpressing ROP10 or ROP10CA was similar to that of nodules formed in control transformed roots (Supplemental Figure 10). However, the number of nodules formed per plant was significantly lower in plants overexpressing ROP10 (9.4 ± 0.8) or ROP10CA (11.6 ± 0.9) than in control plants transformed with the empty vector (17.7 ± 1.3) (Figure 4N). These results are consistent with the reduced infection events observed in these plants.
ROP10 Is Transiently Upregulated in Root Hairs in Response to NFs
A previous study showed that ROP10 is transiently upregulated in M. truncatula roots within 72 h after inoculation with S. meliloti (Liu et al., 2010). Therefore, we wondered whether NF signaling at the preinfection stage affects ROP10 expression. Quantitative RT-PCR (qRT-PCR) analyses were performed on RNA isolated from M. truncatula roots treated for 12 h by a 1 nM solution of NodSm-IV(C16:2, S), an NF purified from S. meliloti. Time-course analyses of ROP10 expression showed that there was no significant change in ROP10 transcript levels in the roots without NF treatment (Supplemental Figure 11A). Following NF treatment, ROP10 expression in the roots appeared to downregulated at 1 h, then upregulated at 3 and 5 h, subsequently returning to the basal expression until 12 h (Supplemental Figure 11B). Further qRT-PCR detection of ROP10 expression in root hairs showed upregulated expression at 3 and 6 h following NF treatment (Supplemental Figure 11C). These results revealed that ROP10 expression in root hairs was transiently stimulated at specific time points by NFs. These data, together with the observations that overexpression of ROP10 and ROP10 led to swollen root hairs, suggest that ROP10 is implicated in NF-induced tip swelling of root hairs.
ROP10:GFP Is Ectopically Localized at the PM of NF-Induced Outgrowths and Infection Sites
Since the extent of PM localization of ROP10 in the root hairs is directly associated with the depolarized growth of root hairs (Figure 2; Supplemental Figure 6) and NFs induce the accumulation of ROP10 transcripts in root hairs (Supplemental Figure 11), we asked whether NF signaling alters the PM localization of ROP10 in the root hairs. Therefore, we examined ROP10:GFP localization in root hairs of M. truncatula treated with NFs or inoculated with rhizobia. Following treatment with 1 nM NodSm-IV(C16:2, S), fluorescence signals in root hairs were visible at the PM of swelling tips, at swelling sites, as well as at NF-induced outgrowths of branched root hairs (Figure 3G; Supplemental Figure 7E). In S. meliloti-inoculated roots, strong ROP10:GFP fluorescence signals were seen at the PM of swelling sites and outgrowths of deformed root hairs (Figure 3H). Moreover, ROP10:GFP was locally and strongly localized at the PM close to infection sites (Figure 3I; Supplemental Figure 7F). Thus, localization of ROP10 at the PM in response to NFs seems to be important for proper root hair curling and bacterial entry into root hairs.
ROP10 Interacts with the Intracellular Kinase Domain of the NF Receptor NFP
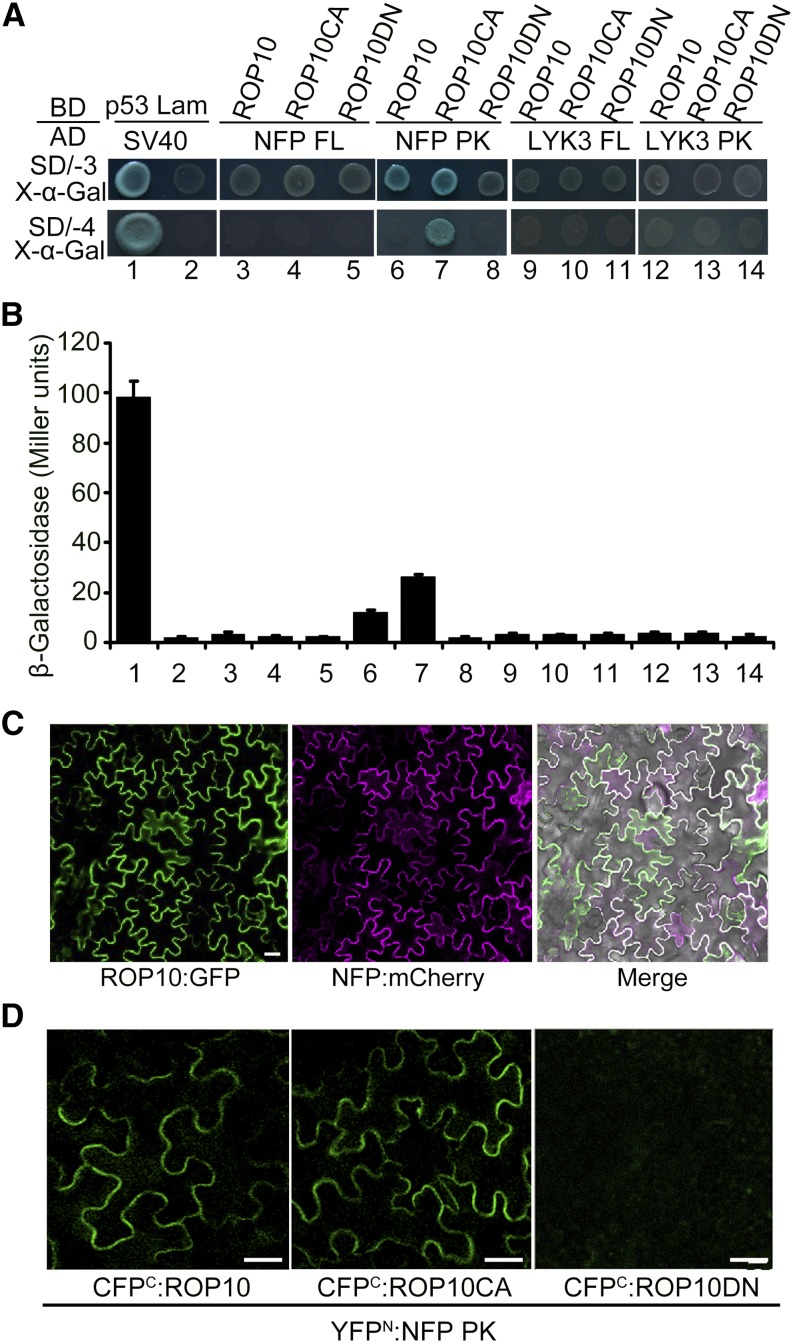
Since ROP10 affects root hair responses that are induced by NFs, we wondered whether ROP10 interacted with the NF receptors implicated in NF signaling. NFP and LYK3 are LysM-containing receptor-like kinases that are localized to the PM, with the LysM domain protruding to the extracellular space perceiving extracellular NF signals and the intracellular kinase domain facing the cytoplasm for the transduction of the signals (Limpens et al., 2003; Smit et al., 2007). Therefore, we tested a possible interaction of ROP10 with NF receptors using the GAL4-based yeast two-hybrid (Y2H) system. We prepared bait proteins that are expressed as fusions of three different ROP10 forms to the GAL4 DNA binding domain (BD) and prey proteins that are expressed as fusions of the full-length NFP and LYK3 (NFP FL and LYK3 FL) and the intracellular kinase domain of NFP and LYK3 (NFP PK and LYK3 PK) fused to the GAL4 activation domain (AD), respectively. In the colony growth assay, either ROP10 or ROP10CA interacted with NFP PK, showing blue colonies on SD/-3/X-α-Gal plates, whereas ROP10DN failed to interact with NFP PK (Figure 5A). Using more stringent SD/-4/X-α-Gal plates, only ROP10CA interacted with NFP PK (Figure 5A). The stronger interaction of ROP10CA with NFP PK was confirmed by the quantitative β-galactosidase activity measurement (Figure 5B). The expression of the fusion proteins in yeast cells was verified by immunoblot analyses (Supplemental Figures 12A and 12B). We also used a LexA-based Y2H system to substantiate these interactions, and similar results were observed (Supplemental Figures 13A and 13B). However, we could not detect the interaction of any forms of ROP10 with full-length NFP in these two systems (Figures 5A and 5B; Supplemental Figures 13A and 13B). These results could be explained by the fact that nuclear import of the full-length NFP, which contains a strongly hydrophobic region (ectotransmembrane), is difficult, and the interaction occurs in the nuclei in these systems. Therefore, using the kinase domain rather than the full-length protein is better for detecting the interaction of different ROP10 forms with NFP. Quantitative and qualitative analyses from GAL4-based and LexA-based Y2H systems showed no interaction of either LYK3 FL or LYK3 PK with any form of ROP10 in the yeast cells (Figures 5A and 5B; Supplemental Figures 13A and 13B).
Figure 5.
Interactions between Different Forms of ROP10 and the NF Receptor NFP of M. truncatula.
(A) GAL4-based Y2H assays. ROP10 and mutant forms (ROP10CA and ROP10DN) were fused with Gal4 DNA BD in pGBKT7 as baits, and NFP full length (FL), the intracellular kinase domain of NFP (PK), LYK3 PK, and LYK3 FL were fused with the Gal4 AD in pGADT7 as preys. Yeast cells cotransformed with bait and prey constructs were selected on SD medium lacking His, Leu, and Trp supplemented with X-α-Gal (SD/-3/X-α-Gal) and more stringent SD medium lacking His, Ade, Leu, and Trp supplemented with X-α-Gal (SD/-4/X-α-Gal) for 5 d. The results indicate an interaction between NFP PK and ROP10 or ROP10CA. The interaction between mammalian p53 and SV40 served as a positive control, whereas the coexpression of lamin (Lam) and SV40 served as a negative control.
(B) The strength of interaction was quantified by assaying β-galactosidase activities in yeast colonies with chlorophenol red-β-d-galactopyranoside as substrate. Error bars indicate se. Data presented are representative of at least three independent experiments.
(C) Colocalization of ROP10 and NFP at the PM in N. benthamiana leaf epidermal cells. Bars = 50 µm.
(D) BiFC analysis to test interactions between ROP10 or mutant forms and NFP PK in N. benthamiana leaf epidermal cells. Bars = 50 µm.
We further verified the interactions between different ROP10 forms and NFP PK or LYK3 PK using an in vitro protein-protein pull-down assay. Glutathione S-transferase (GST) was fused to different ROP10 forms (GST:ROP10, GST:ROP10CA, and GST:ROP10DN), while a 6xHis tag was added to NFP PK (His:NFP PK) and LYK3 PK (His:LYK3 PK). The results showed that all three GST:ROP10 forms could form a protein complex with NFP PK, whereas the control assay with GST or buffer alone showed no interactions (Supplemental Figure 13C). These findings indicate that ROP10 could not be shifted to the inactive or activated state in the absence of GTP or GDP in vitro pull-down assays, which could not mimic the action of these guanine nucleotides under physiological conditions in yeast cells; thus, all ROP10 forms can interact with NFP PK due to their similar structural conformations (Supplemental Figure 13C). The results from pull-down assays to some extent also confirmed the conclusion from yeast cells that ROP10 interacted with NFP PK in a GTP-dependent manner. Interestingly, even though we did not detect any interactions between ROP10 and LYK3 PK in yeast cells, all three GST-ROP10 forms were able to pull down His:LYK3 PK (Supplemental Figure 13D).
To further define the occurrence of the interactions of ROP10 and NF receptors in planta, we first examined whether ROP10 colocalizes with NF receptors at the PM of N. benthamiana cells. Fluorescent protein constructs (ROP10 fused to the N terminus of GFP; NFP and LYK3 fused to the N terminus of mCherry fluorescent protein) were expressed in N. benthamiana cells under the control of the CaMV 35S promoter. When ROP10:GFP and NFP:mCherry were coexpressed, ROP10:GFP fluorescence colocalized with NFP:mCherry fluorescence at the PM (Figure 5C). However, when ROP10:GFP was coexpressed with LYK3:mCherry, only ROP10:GFP fluorescence but no LYK3:mCherry signals was detected in the PM.
We next used a bimolecular fluorescence complementation (BiFC) approach to detect the direct interactions between different forms of ROP10 and NF receptors. ROP10, ROP10CA, and ROP10DN were fused to the C-terminal part of the cyan fluorescent protein (CFPC), whereas NFP PK and LYK3 PK were fused to the N-terminal part of the yellow fluorescent protein (YFPN). Constructs driven by the CaMV 35S promoter were coexpressed in N. benthamiana cells. The reconstituted strong YFP signals reflecting a protein-protein interaction were observed in the PM for the combinations of CFPC:ROP10 with YFPN:NFP PK and CFPC:ROP10CA with YFPN:NFP PK (Figure 5D). By contrast, no fluorescence signals could be detected for the combination of CFPC:ROP10DN with YFPN:NFP PK (Figure 5D). Coexpression of either CFPC:ROP10 or CFPC:ROP10CA with YFPN:LYK3 PK resulted in weak YFP signals in the PM, whereas no fluorescence was detected for the combination of CFPC:ROP10DN with YFPN:LYK3 (Supplemental Figure 13E). Expression of fusion proteins in N. benthamiana leaf cells was confirmed by immunoblotting using monoclonal His antibodies (Supplemental Figure 14). These data demonstrate that ROP10 physically interacts with NF receptors of M. truncatula in the PM in a GTP-dependent fashion.
Overexpression of ROP10 and ROP10CA Stimulates NF Signaling
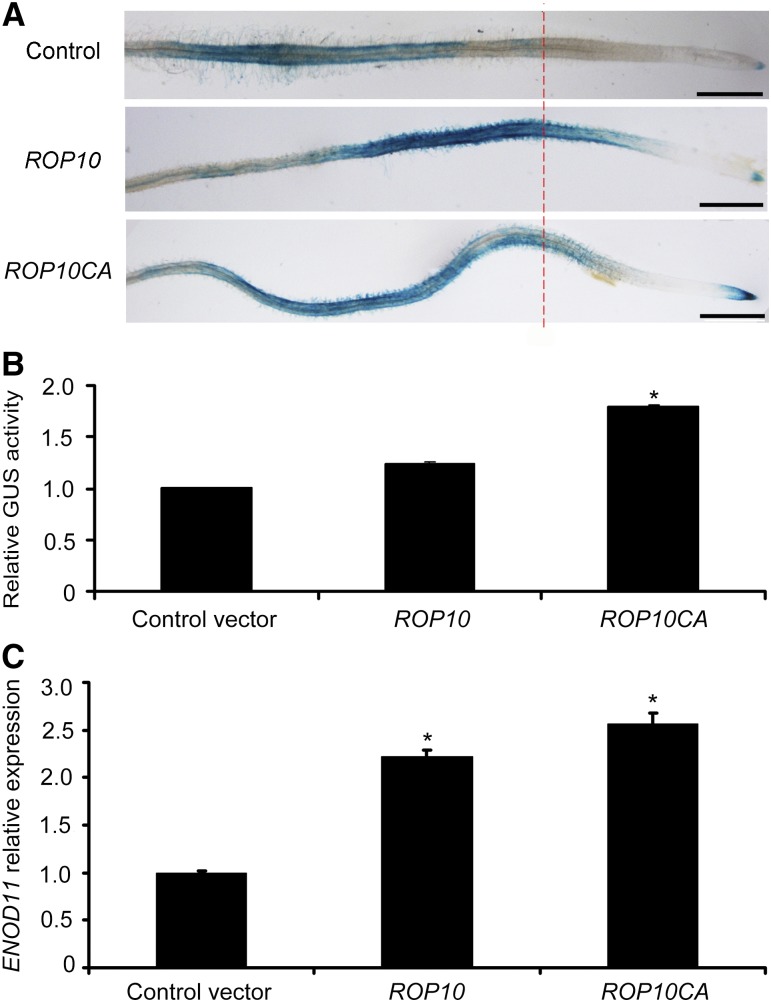
The GUS gene uidA driven by the promoter of the early nodulin gene ENOD11 (ENOD11pro:GUS) is a widely used molecular marker to visualize the activation of NF signaling in the root epidermis of M. truncatula (Andriankaja et al., 2007; Middleton et al., 2007). To substantiate the involvement of ROP10 in the NF signaling pathway, we used stably transformed ENOD11pro:GUS plants (Journet et al., 2001) and generated hairy roots overexpressing ROP10 or ROP10CA under the control of the Ljubq1 promoter. GFP coexpression allowed the visualization of transformed hairy roots. In the absence of NFs, the GUS activity of roots overexpressing ROP10 or ROP10CA was observed only in root cap cells and lateral root primordia. This corresponds to the nonsymbiotic pattern of ENOD11 expression as described previously (Journet et al., 2001). The induction of GUS activity of transformed roots was examined 12 h after treatment with 1 nM NodSm-IV(C16:2, S). In NF-treated control roots transformed with the empty vector, strong GUS activity could be observed in root hairs and epidermal cells ∼1 cm from the growing root tip (Figure 6A). This is the root zone responsive to NFs and S. meliloti infection (Journet et al., 2001; Oldroyd and Long, 2003). By contrast, the area of GUS-stained tissue was greatly expanded close to the root tip in roots overexpressing ROP10 (Figure 6A). A similarly altered GUS staining pattern was also observed in transformed roots overexpressing ROP10CA (Figure 6A). Accordingly, we observed that root hair deformation and rhizobial infection in transformed roots overexpressing ROP10 or ROP10CA occurred close to the root tips (Supplemental Figure 15). Quantitative analysis of GUS activity confirmed that NF-elicited GUS activity was increased in transformed roots overexpressing ROP10 or ROP10CA compared with the control roots (Figure 6B). We also detected by qRT-PCR endogenous transcript levels of ENOD11 in transformed roots overexpressing ROP10 or ROP10CA. Following NF treatment, ENOD11 expression was significantly upregulated in transformed roots overexpressing ROP10 or ROP10CA as compared with the control roots transformed with the empty vector (Figure 6C). As expected, ROP10 transcript levels in these roots were higher than those in control roots (Supplemental Figure 16). These data indicate that elevated levels of ROP10 and ROP10CA in M. truncatula roots resulted in stimulated NF signaling.
Figure 6.
NF-Induced ENOD11 Expression Is Enhanced in Transformed M. truncatula Roots Overexpressing ROP10 or ROP10CA.
(A) Transgenic roots overexpressing ROP10 or ROP10CA were generated by A. rhizogenes-mediated transformation of M. truncatula line 416 harboring an ENOD11pro:GUS fusion. Roots were stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid after treatment for 12 h with 1 nM NodSm-IV(C16:2, S). Control roots transformed with the empty vector had a typical NF-induced ENOD11 expression pattern, whereas roots overexpressing ROP10 or ROP10CA showed more extended and stronger GUS staining. Bars = 5 mm.
(B) and (C) Fluorometric analysis of GUS activity (B) and qRT-PCR analysis of ENOD11 expression (C) in transformed roots overexpressing ROP10 or ROP10CA 12 h after treatment with 1 nM NodSm-IV(C16:2, S). Asterisks indicate significant increases relative to the empty vector control (P < 0.05) as evaluated using Student’s t test. Error bars indicate se. Data presented are representative of three independent experiments.
DISCUSSION
Root hair development arises from tip growth, in which all growth is focused on a single specialized region, the apex of the tip-growing cell. In this work, we found that ROP10, a type II ROP GTPase of M. truncatula, regulated the polarized tip growth of root hairs. Overexpression of ROP10 and ROP10CA induced the depolarization of root hair growth, whereas overexpression of ROP10DN inhibited root hair elongation. So far, the roles of type I ROP GTPases of Arabidopsis (namely ROP2, ROP4, and ROP6) in root hair tip growth have been well characterized (Molendijk et al., 2001; Jones et al., 2002). However, type I ROP GTPases seem to regulate root hair development in a different manner from that of ROP10 in M. truncatula. For example, Arabidopsis ROP2 regulates multiple stages of root hair development, including selection of a polar site, establishment of polarity, and maintenance of polar growth (Jones et al., 2002). Overexpression of ROP10 or ROP10CA only induced the isotropic growth of root hairs, suggesting that ROP10 only regulates polarity establishment and the maintenance of root hair tip growth. In addition, the rop10 mutant has no obvious defect in root hair tip growth, suggesting functional redundancy of ROP GTPases in the regulation of root hair tip growth. This notion is supported by the observation that overexpression of constitutively active forms of ROP3, ROP5, and ROP6 causes weak depolarized growth of root hairs in M. truncatula (Riely et al., 2011).
Type I GTPases are associated with the apical PM of elongating root hairs and pollen tubes, which participate in tip growth as well as the generation and maintenance of the apical growing region (Kost et al., 1999; Li et al., 1999; Molendijk et al., 2001; Jones et al., 2002). Consistent with these observations, ROP10 was localized at the PM of the apex in elongating root hairs. GFP-tagged protein localization showed that all ROP10:GFP, ROP10CA:GFP, and ROP10DN:GFP are localized exclusively at the PM, and such specific PM localization is different from the localization of type I ROP GTPases in Arabidopsis. Type I ROP proteins are localized predominantly to the PM, but a fraction are localized in the cytoplasm (Kost et al.,1999; Li et al., 1999; Molendijk et al., 2001; Jones et al., 2002). These data suggest that the mechanisms of ROP10 and type I ROP protein localization at the PM of apical growing regions during tip growth are different. Our data also revealed that ROP10 is attached to the PM by virtue of S-acylation on two conserved C residues in the GC-CG box. The ROP10 mutants with mutated C residues exhibited partial protein detachment from the PM and accumulation of fluorescence in the nuclei and cytoplasm, and the altered PM localization of ROP10 affected the degree of depolarized growth of M. truncatula root hairs. Moreover, the extent of depolarized growth of root hairs expressing ROP10CA was higher than that of root hairs expressing ROP10CAC197+203S. In addition, ROP10CA overexpression caused much stronger depolarized growth than similar or higher levels of ROP10. These data indicate that active GTP-bound ROP10 promotes the isotropic growth of root hairs and that the intensity of depolarized growth of root hairs depends on the levels of activated ROP10 at the PM. Alternatively, active ROP10 localization at the apical PM of growing root hairs is required for root hair tip growth.
Root hair deformation or curling is a unique developmental process that occurs in response to NFs or rhizobial inoculation, which includes root hair tip swelling, selection and initiation of new growth sites, and reestablishment of polarized tip growth (de Ruijter et al., 1998; Esseling et al., 2003). Overexpression of ROP10 and ROP10CA in M. truncatula resulted in short swollen or ballooning root hairs. This is reminiscent of the swelling of root hair tips in response to NFs or rhizobial inoculation. Following rhizobial inoculation, most of the swollen and ballooning root hairs induced by the overexpression of ROP10 or ROP10CA produced extensive deformations. However, such root hairs were unable to undergo normal root hair curling. Typically, sac-like infection structures filled with rhizobia were formed. Consistent with such an aberrant infection process, the total number of infection events and nodules was significantly reduced. Therefore, alterations in normal tip growth of root hairs caused by the overexpression of ROP10 or ROP10CA disturbed the infection process, suggesting that tip growth of root hairs regulated by ROP10 is required for the efficient invasion of bacteria. In general, rhizobial infection or NF treatment induces a transient inhibition of polar tip growth of root hairs, leading to local tip swelling, and then initiates new tip growth near the root hair tip. The resulting root hair curling ensures that bacterial entry and infection thread initiation occur only at one site of the root hair cell (Esseling et al., 2003). However, in swollen root hairs overexpressing ROP10 or ROP10CA, root hair development appears to be permanently arrested by the inhibition of polar tip growth. The swollen root hairs were highly deformed and formed multiple outgrowths in response to bacterial infection or NF treatments. This suggests that swollen root hairs caused by the overexpression of ROP10 or ROP10CA enhance root hair branching and also increase the responsiveness of root hairs to NFs. In fact, ENOD11 promoter activation reflecting nodulation signaling was increased in NF-treated roots overexpressing ROP10 or ROP10CA. Interestingly, GUS staining indicating that ENOD11 promoter activity in these transgenic roots was extended to the root apex. These data indicate that high levels of ROP10 or ROP10CA promote NF-triggered nodulation signaling.
Following NF treatment, ROP10 was transiently upregulated in root hairs. This finding demonstrates a feedback response (i.e., the activation of NF signaling stimulates ROP10 expression). As ROP10 or ROP10CA overexpression caused swollen and ballooning root hairs, it is reasonable to assume that upregulation of ROP10 modulates root hair tip swelling induced by NFs. NF treatment and rhizobial infection caused ROP10:GFP to be ectopically accumulated at the PM of outgrowths and curls, revealing that NFs or rhizobial infection spatially determine the PM localization of ROP10 in root hairs. This finding is supported by the fact that NFs and rhizobial infection induced the formation of multiple outgrowths of swollen root hairs in transgenic roots overexpressing ROP10 or ROP10CA. The ectopic PM localization of ROP10 in response to NF treatment may depend on the cooperation of ROP10 with other proteins, such as GAPs, GDIs, and GEFs. In M. truncatula, overexpression of the N-terminal part of RopGEF2 results in short swollen root hairs, and RopGEF2 could interact with ROP10 in yeast cells, suggesting that RopGEF2 is an activator of ROP10 activity (Riely et al., 2011). Similarly, it was reported that the PM-associated chitin receptor CERK1 in rice (Oryza sativa) can phosphorylate RacGEF1 and that the phosphorylated RacGEF1 functions as an activator of Rac1, which represents an essential element of chitin-induced defense gene activation (Akamatsu et al., 2013). Therefore, we speculate that RopGEF2 activates ROP10 of M. truncatula in response to NFs in a similar manner to how RacGEF1 activates Rac1 in chitin-treated rice.
In yeast cells, the active ROP10CA but not the inactive ROP10DN interacts with the NF receptor NFP PK. This result was further verified in plant cells, where the interaction of ROP10 with NFP PK at the PM occurs in a GTP-dependent manner. Although we did not detect the interaction of ROP10 with the NF receptor LYK3 PK in yeast cells, we detected the interaction of ROP10 with LYK3 PK in planta by BiFC. Recently, the interaction of two NF receptors, NFP and LYK3, has been reported. One study indicates that the interaction of NFP and LYK3 was detected in the heterologous N. benthamiana system and that their coexpression causes cell death (Pietraszewska-Bogiel et al., 2013), and another study reveals that NFP and LYK3 form heteromeric complexes at the cell periphery in nodules (Moling et al., 2014). Therefore, it is probable that the GTP-bound ROP10 forms a protein complex with NFP and LYK3. Interestingly, both ROP10CA and ROP10DN are exclusively PM-localized, but only ROP10CA interacted with NFP and LYK3 in planta. This could be because ROP10CA and ROP10DN are possibly localized at different membrane microdomains in the PM. In fact, GTP-bound type I ROP6 of Arabidopsis is partitioned exclusively to lipid rafts by S-acylation to stabilize the protein’s interaction with the PM (Sorek et al., 2007, 2010). Similarly, the activated type II ROP11 (RAC10) of Arabidopsis, one of the closest homologs of ROP10, was detected in lipid rafts (Bloch et al., 2005). Therefore, it is plausible that the active GTP-bound ROP10 is partitioned into lipid rafts of the PM. Current evidence also supports the association of NF receptors with lipid rafts, such as the colocalization of LYK3 and the lipid raft-localized flotillin-like proteins FLOT2 and FLOT4 (Haney et al., 2011) and the interaction of a symbiotic remorin protein SYMREM1 with NFP and LYK3 in the raft during infection (Lefebvre et al., 2010). Thus, we hypothesize that GTP-bound ROP10 is also recruited to lipid rafts during infection, where it forms a protein complex with NF receptors and associated proteins.
Based on our data, we propose a model that NFs temporally and spatially regulate ROP10 localization and activity in the PM of root hair tips to induce root hair deformation (Supplemental Figure 17). For normal development of root hairs to occur, ROP10 is localized and activated at the apical PM of the root hair tip, where it generates and maintains tip growth (Supplemental Figure 17A). In response to NFs, ROP10 is transiently upregulated in root hairs, which causes ectopic localization of ROP10 at the PM in the tip. Tip-localized GEF2 rapidly activates ROP10, and the activated GTP-bound ROP10 likely interacts with the NF receptors NFP and LYK3 in lipid rafts to modulate cellular processes important for cell polarity modification, including actin rearrangement, reactive oxygen species production, and Ca2+ influx (Supplemental Figure 17A). On the other hand, overexpression of ROP10 or ROP10CA in root hairs results in ectopic localization of activated ROP10 throughout the PM, which perhaps promotes a permanent interaction between activated ROP10 and the NF receptors. Therefore, this interaction generates multiple polar growth sites at the PM and extensive root hair deformations (Supplemental Figure 17B). This model offers a possible explanation for the change from normal polarized root hair tip growth to NF-elicited root hair deformation. Since our model presupposes that ROP GTPases cycle between GDP-bound and GTP-bound forms in the root hair, future investigations will focus on how this cycling may happen during root hair deformation.
METHODS
Plant Material and Bacterial Strains
Medicago truncatula ecotypes Jemalong A17 and R108 were used in this study. The NF2968 (rop10 mutant) allele was isolated from a tobacco (Nicotiana tabacum) Tnt1 retrotransposon-tagged mutant collection of M. truncatula (Tadege et al., 2008). Plants were cultivated under a 16-h/8-h light/dark regime with 200 µE m−2 s−1 light irradiance at 22°C and 60% relative humidity. Sinorhizobium meliloti 2011 harboring a hemA:lacZ construct and strain 1021 harboring the mCherry-expressing construct were used for inoculation. Agrobacterium rhizogenes MSU440 was used for hairy root transformation, and Agrobacterium tumefaciens EHA105 was used for transient expression in Nicotiana benthamiana.
DNA Manipulation and Plasmid Construction
Point mutations of ROP10 were introduced into plasmids pMD18-T (Takara) by PCR amplification using primers containing the indicated mutations and the Takara MutanBEST Kit (Takara). To create expression constructs for the analysis of M. truncatula root hairs, pENTR ROP10, ROP10CA, ROP10DN, and ROP10CAC197+203S were used for LR recombination reactions with pUB-GW-GFP (Maekawa et al., 2008), generating pUbi:ROP10, pUbi:ROP10CA, pUbi:ROP10DN, and pUbi:ROP10CAC197+203S plasmids, respectively.
To create ROP10:GFP, ROP10CA:GFP, ROP10DN:GFP, ROP10C203S:GFP, ROP10C197S:GFP, and ROP10C197+203S:GFP constructs, the region containing the CaMV 35S promoter and GFP was released from plasmid pA7-GFP (Voelker et al., 2006) by HindIII and EcoRI and inserted into pCAMBIA 2300S to obtain the control vector 2300S-GFP. Then, ROP10 and derivatives were amplified by PCR and inserted into the 2300S-GFP vector using SalI and SpeI.
To create expression constructs for BiFC analysis, ROP10, ROP10CA, and ROP10DN and the kinase domains of NFP (NFP PK; amino acids 312 to 582) and LYK3 (LYK3 PK; amino acids 323 to 620) were amplified by PCR, inserted into pENTR through TOPO reaction (Invitrogen), and then used for the LR recombination reaction (Invitrogen) with either pCCFP-X or pNYFP-Y (Bai et al., 2007) to obtain CFPC:ROP10, CFPC:ROP10CA, CFPC:ROP10DN, YFPN:NFP PK, and YFPN:LYK3 PK constructs, respectively.
To create expression constructs for colocalization in planta, NFP and LYK3 were fused to the mCherry gene (Clontech; pmCherry Vector) by overlap extension PCR and then inserted into pA7-GFP using XhoI and SmaI. Then, the HindIII and EcoRI fragment was inserted into pCAMBIA 2300S to obtain the NFP:mCherry and LYK3:mCherry constructs, respectively.
To create BD and AD constructs for GAL4-based Y2H assays, the sequences of ROP10, ROP10CA, and ROP10DN were amplified by PCR and cloned with the help of EcoRI and XhoI into the pGBKT7 DNA BD vector (Clontech). NFP full length (NFP FL), NFP kinase domain (NFP PK), LYK3 full length (LYK3 FL), and LYK3 kinase domain (LYK3 PK) were amplified by PCR and cloned with EcoRI and XhoI into the pGADT7 AD vector (Clontech).
To create AD and BD constructs for LexA-based Y2H assays, the sequences of ROP10, ROP10CA, and ROP10DN were amplified by PCR and cloned with the help of EcoRI and XhoI into the pB42 AD vector (Clontech). NFP full length (NFP FL), NFP kinase domain (NFP PK), and LYK3 kinase domain (LYK3 PK) were amplified by PCR and cloned with EcoRI and XhoI into the pLexA DNA BD vector (Clontech).
To create expression constructs for pull-down assays, the sequences of ROP10, ROP10CA, and ROP10DN were amplified by PCR and cloned with EcoRI and XhoI into the pGEX-6P-1 vector (GE Healthcare), which was then used for GST-tagged protein expression in Escherichia coli. Similarly, the NFP kinase domain (NFP PK) and LYK3 kinase domain (LYK3 PK) were amplified by PCR from pENTR/SD/D-NFP PK and pENTR/SD/D-LYK3 PK and cloned with EcoRI and XhoI into pET-28a(+) (Novagen) for His-tagged protein expression.
All PCR products and point mutations were verified by DNA sequencing. The generation of the different constructs is specified in Supplemental Table 1, and primer sequences for plasmid construction are given in Supplemental Table 2.
NF Purification
S. meliloti strain 1021 (pEK327) (Schultze et al., 1992) was used for HPLC purification of NodSm-IV(C16:2, S) as described (Tian et al., 2013).
Actin Staining
M. truncatula roots were harvested and placed in actin-stabilizing buffer (ASB) containing 100 mM PIPES, 10 mM EGTA, and 5 mM MgSO4, pH 6.8, supplemented with 1.5% glycerol, 0.1% Triton X-100, and 2 units of AlexaFluor 488-conjugated phalloidin (Invitrogen), which was added immediately prior to root staining. The roots were incubated in ASB in the dark overnight at room temperature, washed three times in ASB without phalloidin, and mounted in ASB on a glass slide for confocal fluorescence microscopy (Olympus FV1000) analysis. The AlexaFluor 488 dye was excited with the 488-nm line of the argon laser, and emission was captured with a 505- to 530-nm band-pass filter.
Identification of the Tnt1 Insertion Site in the ROP10 Gene
Genomic DNA samples were prepared from wild-type M. truncatula R108 and the rop10 (NF2968) mutant using a standard protocol. PCR was performed using either the Tnt1 primers or ROP10-specific primers listed in Supplemental Table 2. PCR amplification produced one band (∼500 bp) from the wild type, one band (∼1100 bp) from homozygotes of rop10, and two bands (∼500 and ∼1100 bp) from heterozygotes of rop10. These PCR products were fully sequenced.
Transient Expression in N. benthamiana Leaves
A. tumefaciens EHA105 cells carrying a given plasmid were inoculated into 5 mL of Luria-Bertani medium supplemented with 10 mM MES, pH 5.6, 2 μL of 100 mM acetosyringone, and appropriate antibiotics. Cells were grown at 28°C for 12 to 16 h. Then, the bacteria were pelleted, resuspended in 10 mL of 10 mM MgCl2 supplemented with 20 μL of 100 mM acetosyringone, and kept at room temperature for at least 3 h. Infiltration was performed using a 2-mL syringe on expanded leaves of N. benthamiana, which were placed in the dark beforehand. One to 2 d later, transient expression of the proteins was observed with a Zeiss LSM 510 META confocal microscope. For detection of GFP and YFP in the protein localization and BiFC experiments, fluorescence was excited by the 488-nm line of an Ar laser, and signals were captured with a 505- to 530-nm band-pass filter. The same setting was applied for GFP visualization in colocalization studies, where an additional mCherry channel was used (fluorescence excited with the 543-nm line of the Ar laser and emission captured with a 560- to 615-nm band-pass filter).
Protein Gel Blotting of Fusion Proteins in N. benthamiana Leaves
Fusion proteins were extracted from N. benthamiana expressed with ROP10:GFP, ROP10CA:GFP, ROP10DN:GFP, and free GFP by homogenization in buffer 1 (50 mM HEPES-KOH, pH 7.5, 10% sucrose, 50 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, protease inhibitor mix, and 2 mM phenylmethanesulfonyl flouride). The homogenates were centrifuged for 10 min at 4°C at 25,000g, and the supernatants and the pellets were collected. The supernatants were centrifuged again at 100,000g for 1 h at 4°C, and the resulting supernatants were collected as “supernatant samples.” The collected pellets were incubated at 4°C for 30 min in buffer 2 (50 mM HEPES-KOH, pH 7.5, 10% sucrose, 50 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, 0.5% SDS, 1% Triton X-100, plant protease inhibitor mix, and 2 mM phenylmethanesulfonyl flouride) and centrifuged at 25,000g for 15 min at 4°C. The supernatants were collected as “pellet samples.” The supernatant and pellet samples were analyzed by SDS-PAGE and immunoblotted using a GFP monoclonal antibody. GFP antibody was used at 1:2000 (TransGen), and secondary antibody was used at 1:8000 (Goat Anti-Mouse IgG-HRP; Abmart). Protein gel blotting of fusion proteins in the BiFC experiment is described in the Supplemental Methods.
Hairy Root Transformation
M. truncatula seeds were surfaced-sterilized with H2SO4 for 5 min and then with 30 to 50 mL of 10% (v/v) commercial bleach (containing a few drops of Tween 20) for 8 min. The seeds were then imbibed in sterile water for 3 to 4 h (five to six changes), placed on 1% agar plates at 4°C for 24 h, and then incubated at 25°C for 12 h in the dark. Transgenic hairy roots were induced by A. rhizogenes MSU440 as described (Limpens et al., 2004).
Examination of Infection Events and Nodulation Assay
The transformed roots, which were identified by GFP fluorescence using a Nikon SMZ1500 microscope, were transferred to pots filled with a vermiculite and perlite mixture (3:1). After N starvation for 3 d, the roots were incubated with S. meliloti 2011 harboring a hemA:lacZ construct. At 7 d after inoculation, the roots were collected, fixed, and stained with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside as described (Li et al., 2014). The infection events were observed using bright-field microscopy. Nodules were assessed at 14 d after inoculation.
Gene Expression Analysis
Surface-sterilized seeds were germinated and grown on modified Fahraeus medium agar plates for 7 d and then treated with 1 nM NodSm-IV(C16:2, S). Samples for ROP10 expression analysis were harvested at different time points (up to 12 h after NF treatment). At each time point, ∼10 roots were collected for ROP10 expression analysis. Root hairs treated with NodSm-IV(C16:2, S) were collected as described (Wang et al., 2014). Total RNA was isolated from roots and root hairs, and 1 µg of RNA was used for cDNA synthesis with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). qRT-PCR was conducted with the Real-time PCR Master Mix (SYBR Green I; Toyobo) using the StepOne Real-Time System (Applied Biosytems). All reactions were done in triplicate and averaged. Amplification conditions consisted of an initial denaturation step at 95°C for 1 min, 40 cycles at 95°C for 15 s and 60°C for 1 min, followed by a melting curve stage of 95°C for 15 s and a gradient from 65 to 95°C at a rate of 1°C/s. The expression data were normalized against Mt-ACTINB. The primers used for qRT-PCR analysis are listed in Supplemental Table 3.
Y2H Assay
AD and BD constructs based on the GAL4 system were cotransformed into yeast strain AH109 using an LiAc-mediated yeast transformation protocol according to the manufacturer’s instructions (Clontech). At least 10 cotransformed yeast colonies were then plated on SD-His/Leu/Trp and SD-His/Ade/Leu/Trp plates supplemented with X-α-Gal (40 mg/L). The plates were incubated for 3 d at 30°C to test nutritional marker (His and Ade) gene expression and α-galactosidase activity of the MEL1 reporter protein. Quantitative β-galactosidase analysis was performed using a liquid culture assay with chlorophenol red-β-d-galactopyranoside as substrate as described in the Yeast Protocols Handbook (Clontech). Yeast protein extraction is described in the Supplemental Methods. The BD and AD samples were analyzed by SDS-PAGE and immunoblotted using GAL4 BD monoclonal antibody (Sigma) and hemagglutinin monoclonal antibody (CW BIO), respectively. GAL4 BD antibody was used at 1:1000, hemagglutinin antibody was used at 1:8000, and secondary antibody was used at 1:8000 (Goat Anti-Mouse IgG-HRP; Abmart). The Y2H assay based on the LexA system is described in the Supplemental Methods.
Pull-Down Assay
The GST- and His-tagged proteins were expressed in E. coli strain BL21. A 100-mL culture containing the appropriate plasmid was grown at 37°C to a density of OD600 = 0.4. Isopropylthio-β-galactoside was added to reach a concentration of 0.5 mM, and cells continued to grow for 5 h at 30°C. GST- and His-tagged cells were pelleted by centrifugation and resuspended in 3 to 5 mL of ice-cold lysis buffer (PBS; 8 g/L NaCl, 0.2 g/L KCl, 3.58 g/L Na2HPO4·12H2O, and 0.27 g/L KH2PO4, pH 7.4) and pull-down buffer (20 mM Tris, 100 mM NaCl, 0.1 mM EDTA, and 0.2% Triton X-100, pH 7.4) supplemented with protease inhibitors (Roche) and 1 mM benzamidine, respectively. Cells were then homogenized by sonication. The debris were pelleted by centrifugation at 12,000 rpm for 10 min at 4°C. The soluble fraction containing GST-tagged protein was incubated with Glutathione Sepharose 4B (GE Healthcare) (prewashed with PBS) at 4°C for 1.5 h. The beads with bound GST-tagged proteins were washed with ice-cold PBS and pull-down buffer prior to incubation with His-tagged proteins. The binding reaction was performed at 4°C for 1 h. Then the bound proteins were eluted with low-salt elution buffer (20 mM Tris, 100 mM NaCl, 0.1 mM EDTA, and 0.5% Nonidet P-40, pH 7.4) and high-salt elution buffer (20 mM Tris, 300 mM NaCl, 0.1 mM EDTA, and 0.5% Nonidet P-40, pH 7.4). Eluted proteins were analyzed by SDS-PAGE and protein gel blotting using a His-tag monoclonal antibody (Proteintech). His antibody was used at 1:8000, and secondary antibody was used at 1:8000 (Goat Anti-Mouse IgG-HRP, Abmart) for protein detection.
Histochemical Localization of GUS Activity and Quantitative GUS Activity Assay
The pUbipro:ROP10 and pUbipro:ROP10CA constructs were introduced into stably transformed M. truncatula line L416 harboring the ENOD11pro:GUS fusion by hairy root transformation. Transformed roots (identified by GFP fluorescence using a Nikon SMZ1500 microscope) were transferred to modified Fahraeus medium plates (for N starvation) for 3 d. The roots were incubated for 12 h with 1 nM NodSm-IV(C16:2, S). Roots were then collected for GUS staining, GUS activity measurement, or qRT-PCR analysis. GUS staining was performed as described using 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (Li et al., 2014). For quantitative GUS activity determination, transformed roots were ground in liquid nitrogen and homogenized in GUS extraction buffer. Enzymatic reactions were performed using 1 mg of total protein extract with 4-methylumbelliferyl-β-d-glucuronide as substrate (Sigma-Aldrich) using a microtiter fluorimeter.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU625287 (ROP10) and AJ297721 (ENOD11).
Supplemental Data
Supplemental Figure 1. Structure Analysis of M. truncatula ROP10.
Supplemental Figure 2. Identification and Phenotypic Characterization of the rop10 Mutant.
Supplemental Figure 3. Disorganized Actin Cytoskeleton in Root Hairs of M. truncatula Transformed Roots Overexpressing ROP10 or ROP10 Mutants.
Supplemental Figure 4. Addition of 10% (v/v) DMSO Had No Effect on PM Localization of Different GFP-Fused ROP10 Forms Expressed in N. benthamiana Leaf Cells.
Supplemental Figure 5. Immunoblot Analyses of ROP10 and ROP10CA Proteins with Mutations of C Residues in the GC-CG Box.
Supplemental Figure 6. Root Hair Phenotypes of M. truncatula Roots Overexpressing ROP10 and ROP10CA Mutants with Mutations of C Residues in the GC-CG Box.
Supplemental Figure 7. ROP10:GFP Is Localized at the Apical PM of Root Hair Tip and Localized at the PM of Swelling Tips and Curling after NF Treatment and Rhizobial Infection.
Supplemental Figure 8. Root Hairs of M. truncatula Expressing ROP10CA Showed Aberrant Rhizobial Infection.
Supplemental Figure 9. qRT-PCR Analysis of ROP10 Transcripts in M. truncatula Roots Overexpressing ROP10 or ROP10CA.
Supplemental Figure 10. Nodule Formation on Transformed M. truncatula Roots Overexpressing ROP10 and ROP10CA.
Supplemental Figure 11. ROP10 Is Transiently Upregulated in Response to NF Treatment.
Supplemental Figure 12. Immunoblot Analysis of Protein Expression in Yeast Cells.
Supplemental Figure 13. Interactions between ROP10 and NF Receptors NFP and LYK3 of M. truncatula.
Supplemental Figure 14. Immunoblot Analysis of Expressed Fusion Proteins of N. benthamiana for BIFC Assays.
Supplemental Figure 15. Root Hair Deformation and Rhizobial Infection in M. truncatula Roots Overexpressing ROP10 or ROP10CA.
Supplemental Figure 16. qRT-PCR Analysis of ROP10 Transcripts in M. truncatula roots Overexpressing ROP10 or ROP10CA Treated with NFs.
Supplemental Figure 17. Proposed Model for NFs Temporal and Spatial Regulation of ROP10 PM Localization and Activity to Induce Root Hair Deformation.
Supplemental Table 1. Constructs Used in Various Assays.
Supplemental Table 2. Primers Used in Various Constructs.
Supplemental Table 3. Primers Used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Weihua Tang for experimental support, David Barker for providing stable transgenic ENOD11pro:GUS seeds, Hongquan Yang for the MATCHMAKER LexA two-hybrid system, Sharon Long for the mCherry rhizobium strain, Eva Kondorosi for S. meliloti strain 1021 (pEK327), and the National BioResource Project Biological Resource Center for pUB plasmids. We also thank Ertao Wang and Lin Xu for valuable discussion and Christian Wagner for helpful comments on the manuscript. The M. truncatula Tnt1 mutant line NF2968, jointly owned by the Centre National de la Recherche Scientifique and the Noble Foundation, was created through research funded by the National Science Foundation (Grant 0703285). This work was supported by the National Basic Research Program of China (973 Program Grant 2010CB126501) and the National Natural Science Foundation of China (Grants 31071065 and 31270292).
AUTHOR CONTRIBUTIONS
Y.-Z.W. conceived and designed the experiments. M.-J.L., Q.W., X.L., A.C., and Y.X. performed the experiments. C.S. and Z.-P.X. contributed NFs. K.S.M. and J.W. provided the Tnt1 mutant NF2968. Y.-Z.W. and M.-J.L. analyzed the data. Y.-Z.W., C.S., and M.-J.L. wrote the article with the assistance of L.L., D.L., and G.L.
Glossary
- PM
plasma membrane
- NF
nodulation factor
- CaMV
cauliflower mosaic virus
- qRT-PCR
quantitative RT-PCR
- Y2H
yeast two-hybrid
- BD
binding domain
- AD
activation domain
- BiFC
bimolecular fluorescence complementation
- ASB
actin-stabilizing buffer
References
- Akamatsu A., Wong H.L., Fujiwara M., Okuda J., Nishide K., Uno K., Imai K., Umemura K., Kawasaki T., Kawano Y., Shimamoto K. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476. [DOI] [PubMed] [Google Scholar]
- Amor B.B., Shaw S.L., Oldroyd G.E., Maillet F., Penmetsa R.V., Cook D., Long S.R., Dénarié J., Gough C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34: 495–506. [DOI] [PubMed] [Google Scholar]
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D., Lavy M., Efrat Y., Efroni I., Bracha-Drori K., Abu-Abied M., Sadot E., Yalovsky S. (2005). Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol. Biol. Cell 16: 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L., Vidali L., Dominguez J., Pérez H., Sánchez F., Hepler P.K., Quinto C. (1998). Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol. 116: 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol R.J., Takeda S., Linstead P., Durrant M.C., Kakesova H., Derbyshire P., Drea S., Zarsky V., Dolan L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016. [DOI] [PubMed] [Google Scholar]
- Catoira R., Timmers A.C., Maillet F., Galera C., Penmetsa R.V., Cook D., Dénarié J., Gough C. (2001). The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518. [DOI] [PubMed] [Google Scholar]
- Craddock C., Lavagi I., Yang Z. (2012). New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 22: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter N.C.A., Rook M.B., Bisseling T., Emons A.M.C. (1998). Lipochito-oligosaccharides re-initiate root hair tip growth in Vicia sativa with high calcium and spectrin-like antigen at the tip. Plant J. 13: 341–350. [Google Scholar]
- Esseling J.J., Lhuissier F.G., Emons A.M. (2003). Nod factor-induced root hair curling: Continuous polar growth towards the point of nod factor application. Plant Physiol. 132: 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseling J.J., Lhuissier F.G., Emons A.M. (2004). A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: Implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell 16: 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352. [DOI] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Li S., Lord E.M., Yang Z. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C.H., Riely B.K., Tricoli D.M., Cook D.R., Ehrhardt D.W., Long S.R. (2011). Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23: 2774–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R., Geurts R., Franssen H., Spaink H.P., Van Kammen A., Bisseling T. (1994). Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 105: 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Vernoud V., Szumlanski A., Nielsen E., Yang Z. (2008). A tip-localized RhoGAP controls cell polarity by globally inhibiting Rho GTPase at the cell apex. Curr. Biol. 18: 1907–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Wu G., Yan A., Lee Y.J., Grierson C.S., Yang Z. (2010). Pollen-tube tip growth requires a balance of lateral propagation and global inhibition of Rho-family GTPase activity. J. Cell Sci. 123: 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.M., Kobayashi H., Davies B.W., Taga M.E., Walker G.C. (2007). How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Shen J.J., Fu Y., Li H., Yang Z., Grierson C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E.P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D.G., Gianinazzi-Pearson V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14: 737–748. [DOI] [PubMed] [Google Scholar]
- Kaló P., Gleason C., Edwards A., Marsh J., Mitra R.M., Hirsch S., Jakab J., Sims S., Long S.R., Rogers J. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Ke D., Fang Q., Chen C., Zhu H., Chen T., Chang X., Yuan S., Kang H., Ma L., Hong Z., Zhang Z. (2012). The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol. 159: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiirika L.M., Bergmann H.F., Schikowsky C., Wimmer D., Korte J., Schmitz U., Niehaus K., Colditz F. (2012). Silencing of the Rac1 GTPase MtROP9 in Medicago truncatula stimulates early mycorrhizal and oomycete root colonizations but negatively affects rhizobial infection. Plant Physiol. 159: 501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Kost B. (2006). Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/Rop to the apex of pollen tubes. Plant Cell 18: 3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Becker C., Schmitt A.C., Kost B. (2006). Nt-RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. Plant J. 46: 1018–1031. [DOI] [PubMed] [Google Scholar]
- Kost B. (2008). Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 18: 119–127. [DOI] [PubMed] [Google Scholar]
- Kost B., Lemichez E., Spielhofer P., Hong Y., Tolias K., Carpenter C., Chua N.H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Yalovsky S. (2006). Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J. 46: 934–947. [DOI] [PubMed] [Google Scholar]
- Lavy M., Bracha-Drori K., Sternberg H., Yalovsky S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA 107: 2343–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lei M., Yan Z., Wang Q., Chen A., Sun J., Luo D., Wang Y. (2014). The REL3-mediated TAS3 ta-siRNA pathway integrates auxin and ethylene signaling to regulate nodulation in Lotus japonicus. New Phytol. 201: 531–544. [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633. [DOI] [PubMed] [Google Scholar]
- Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55: 983–992. [DOI] [PubMed] [Google Scholar]
- Liu W., Chen A.M., Luo L., Sun J., Cao L.P., Yu G.Q., Zhu J.B., Wang Y.Z. (2010). Characterization and expression analysis of Medicago truncatula ROP GTPase family during the early stage of symbiosis. J. Integr. Plant Biol. 52: 639–652. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kusakabe M., Shimoda Y., Sato S., Tabata S., Murooka Y., Hayashi M. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21: 375–382. [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E., Mun J.H., Yeun L.H., Jayaraman D., Rougé P., Barre A., Lougnon G., Schornack S., Bono J.J., Cook D.R., Ané J.M. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 20: 912–921. [DOI] [PubMed] [Google Scholar]
- Middleton P.H., Jakab J., Penmetsa R.V., Starker C.G., Doll J., Kaló P., Prabhu R., Marsh J.F., Mitra R.M., Kereszt A. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E., Long S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk A.J., Bischoff F., Rajendrakumar C.S., Friml J., Braun M., Gilroy S., Palme K. (2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20: 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moling S., Pietraszewska-Bogiel A., Postma M., Fedorova E., Hink M.A., Limpens E., Gadella T.W., Bisseling T. (2014). Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26: 4188–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C., Wu H.M., Cheung A.Y. (2006). RAC/ROP GTPases: ‘Hubs’ for signal integration and diversification in plants. Trends Plant Sci. 11: 309–315. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E.D., Long S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol. 131: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietraszewska-Bogiel A., Lefebvre B., Koini M.A., Klaus-Heisen D., Takken F.L., Geurts R., Cullimore J.V., Gadella T.W. (2013). Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS ONE 8: e65055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Riely B.K., He H., Venkateshwaran M., Sarma B., Schraiber J., Ané J.M., Cook D.R. (2011). Identification of legume RopGEF gene families and characterization of a Medicago truncatula RopGEF mediating polar growth of root hairs. Plant J. 65: 230–243. [DOI] [PubMed] [Google Scholar]
- Riely B.K., Lougnon G., Ané J.M., Cook D.R. (2007). The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49: 208–216. [DOI] [PubMed] [Google Scholar]
- Schultze M., Quiclet-Sire B., Kondorosi E., Virelizer H., Glushka J.N., Endre G., Géro S.D., Kondorosi A. (1992). Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc. Natl. Acad. Sci. USA 89: 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152. [DOI] [PubMed] [Google Scholar]
- Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Sorek N., Gutman O., Bar E., Abu-Abied M., Feng X., Running M.P., Lewinsohn E., Ori N., Sadot E., Henis Y.I., Yalovsky S. (2011). Differential effects of prenylation and S-acylation on type I and II ROPS membrane interaction and function. Plant Physiol. 155: 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N., Poraty L., Sternberg H., Bar E., Lewinsohn E., Yalovsky S. (2007). Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol. Cell. Biol. 27: 2144–2154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sorek N., Segev O., Gutman O., Bar E., Richter S., Poraty L., Hirsch J.A., Henis Y.I., Lewinsohn E., Jürgens G., Yalovsky S. (2010). An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr. Biol. 20: 914–920. [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347. [DOI] [PubMed] [Google Scholar]
- Tian Y., Liu W., Cai J., Zhang L.Y., Wong K.B., Feddermann N., Boller T., Xie Z.P., Staehelin C. (2013). The nodulation factor hydrolase of Medicago truncatula: Characterization of an enzyme specifically cleaving rhizobial nodulation signals. Plant Physiol. 163: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., Truchet G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628. [DOI] [PubMed] [Google Scholar]
- Voelker C., Schmidt D., Mueller-Roeber B., Czempinski K. (2006). Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J. 48: 296–306. [DOI] [PubMed] [Google Scholar]
- Wang Q., Lei M.J., Chen A.M., Wang R.G., Li G.J., Wang Y.Z. (2014). MtROP8 is involved in root hair development and the establishment of symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti. Chin. Sci. Bull. 59: 4289–4297. [Google Scholar]
- Yalovsky S., Bloch D., Sorek N., Kost B. (2008). Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147: 1527–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Fu Y. (2007). ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 10: 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18830–18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.