Mutant phenotypes reveal a central role for VEG2 in flowering and compound inflorescence formation and suggest that transcription of FT/TFL1 genes is regulated by VEG2-FT protein complexes.
Abstract
As knowledge of the gene networks regulating inflorescence development in Arabidopsis thaliana improves, the current challenge is to characterize this system in different groups of crop species with different inflorescence architecture. Pea (Pisum sativum) has served as a model for development of the compound raceme, characteristic of many legume species, and in this study, we characterize the pea VEGETATIVE2 (VEG2) locus, showing that it is critical for regulation of flowering and inflorescence development and identifying it as a homolog of the bZIP transcription factor FD. Through detailed phenotypic characterizations of veg2 mutants, expression analyses, and the use of protein-protein interaction assays, we find that VEG2 has important roles during each stage of development of the pea compound inflorescence. Our results suggest that VEG2 acts in conjunction with multiple FLOWERING LOCUS T (FT) proteins to regulate expression of downstream target genes, including TERMINAL FLOWER1, LEAFY, and MADS box homologs, and to facilitate cross-regulation within the FT gene family. These findings further extend our understanding of the mechanisms underlying compound inflorescence development in pea and may have wider implications for future manipulation of inflorescence architecture in related legume crop species.
INTRODUCTION
Inflorescences are the shoot structures that bear flowers, and their form and arrangement have important implications for reproductive success and ease of harvest in agricultural systems (Wyatt, 1982). Angiosperm species exhibit incredible diversity in inflorescence form, which derives from complexity and pattern of branching, the number and position of flowers, and the capacity of the inflorescence for continued growth (Weberling, 1992). At the tissue level, this variation can be attributed to differences in the identity and activity of the shoot meristems that produce each component of the inflorescence. Among many genes that have a role in the regulation of flowering, a subset also have a role in specifying the identity of vegetative, inflorescence, or floral meristems, and it is the interaction of these genes that determines how the inflorescence develops. Understanding the genes and regulatory interactions that underlie the development of different inflorescence forms is a crucial step to enable future optimization of inflorescence architecture for maximal crop productivity.
Like most plant processes, inflorescence development is best understood in the model species Arabidopsis thaliana. The Arabidopsis inflorescence is a simple raceme, in which flowers are borne directly on the main stem and the shoot apex remains indeterminate, with organogenesis balanced by self-renewal (Figure 1A). Two key genes, FLOWERING LOCUS T (FT) and TERMINAL FLOWER1 (TFL1), have a major role in generating this inflorescence form, through antagonistic effects on the expression of meristem identity genes (Kardailsky et al., 1999; Kobayashi et al., 1999; McGarry and Ayre, 2012; Jaeger et al., 2013). FT and TFL1 both belong to the phosphatidylethanolamine binding protein family and individually interact with the basic leucine zipper (bZIP) transcription factors FD and FD PARALOG (FDP) to regulate expression of floral target genes within the apex (Abe et al., 2005; Wigge et al., 2005; Hanano and Goto, 2011). TFL1/FD complexes delay flowering and prevent upregulation of floral genes within the shoot apical meristem (SAM) to maintain shoot indeterminacy (Hanano and Goto, 2011). FT/FD complexes promote expression of floral genes, ultimately resulting in the induction of the MADS box gene APETALA1 (AP1) in axillary meristems to specify floral identity (Abe et al., 2005; Wigge et al., 2005). AP1 is also upregulated by the floral integrator and floral identity gene LEAFY (LFY), which acts independently of the FT/FD pathway (Ruiz-García et al., 1997; Abe et al., 2005; Wigge et al., 2005). Within floral meristems, AP1 and LFY directly repress TFL1 expression to maintain determinacy (Wagner et al., 1999; Kaufmann et al., 2010).
Figure 1.
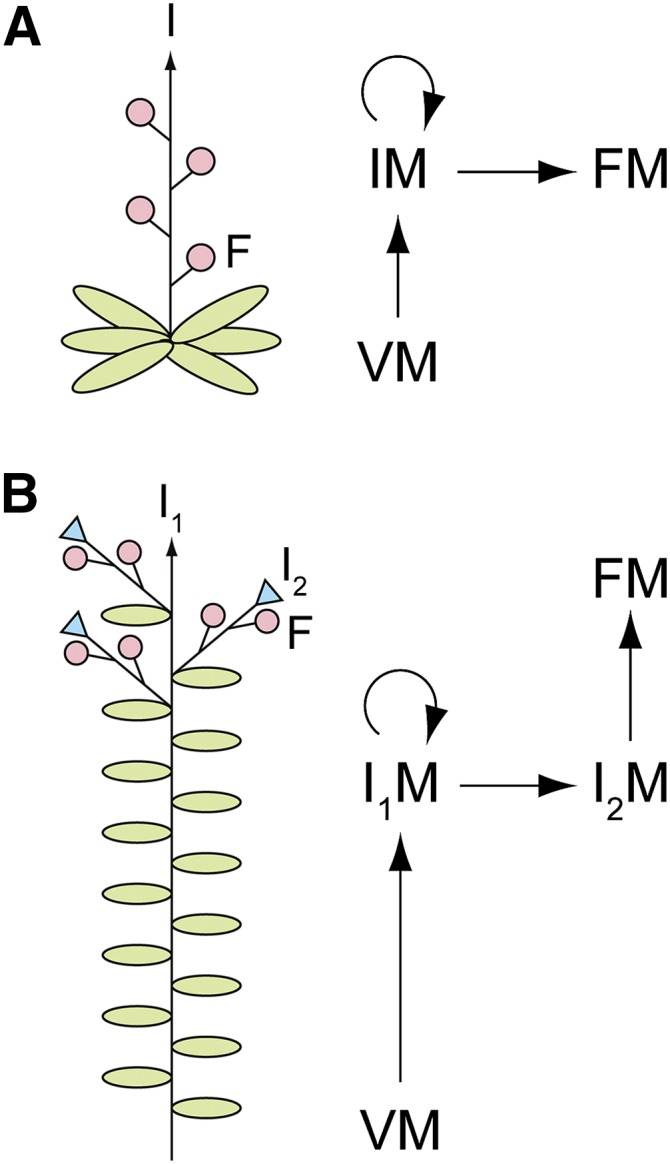
Inflorescence Development in Arabidopsis and Pea.
(A) The simple raceme of Arabidopsis.
(B) The compound raceme of pea.
For each species, a diagram of inflorescence architecture (left) and schematic of the meristem transitions involved in inflorescence development (right) are shown. In diagrams, arrows indicate indeterminate growth of the inflorescence stem (I; I1), green ovals are leaves, pink circles are flowers (F), and blue triangles are stubs terminating each secondary inflorescence (I2) axis. In schematics, straight arrows indicate meristem transitions and products, and circular arrows indicate meristem indeterminacy. Meristem abbreviations are as follows: vegetative meristem (VM), inflorescence meristem (IM), primary inflorescence meristem (I1M), secondary inflorescence meristem (I2M), and floral meristem (FM).
In this study, we investigate genes regulating development of the compound raceme of pea (Pisum sativum). Pea is an important crop plant and is also representative of other agronomically significant legume species within the Papilionoideae, including lentil (Lens culinaris), chickpea (Cicer arietinum), common bean (Phaseolus vulgaris), and soybean (Glycine max), which share similar inflorescence architecture. Relative to the simple raceme of Arabidopsis, the compound raceme of pea has an additional level of branching, such that flowers are not directly borne on the main inflorescence stem but are instead borne on modified lateral branches, termed secondary inflorescences (I2s; Figure 1B). Pea inflorescence development can thus be considered to consist of three distinct stages. During vegetative growth, the SAM has vegetative (V) identity and produces the main stem, bearing alternate leaves with vegetative axillary buds, which normally remain dormant. On commitment to flowering, the pea vegetative SAM undergoes a transition to a primary inflorescence (I1) meristem (Ferguson et al., 1991), which we refer to here as the V/I1 transition, the first stage of inflorescence development. The I1 meristem is similar to the vegetative SAM, in that it remains indeterminate and produces the shoot tissues (stem and leaves) of the main stem. However, the I1 is distinguished by the fact that it bears an axillary I2 at each stem node instead of a vegetative bud (Singer et al., 1999). Specification of I2 meristem identity is the second stage of pea inflorescence development. Each I2 is leafless and determinate, terminating in a hairy stub after bearing several axillary flowers (Hole and Hardwick, 1976). Specification of floral meristem identity is the third and final stage of pea inflorescence development.
A number of key genes that regulate inflorescence development in pea have been identified based on characterization of pea mutants with altered inflorescence form (summarized in Supplemental Figure 1). Mutants of PROLIFERATING INFLORESCENCE MERISTEM (PIM), an AP1 homolog, fail to correctly specify floral meristems (Taylor et al., 2002). In accordance with a conserved role as a floral meristem identity gene, expression of PIM is limited to floral meristems (Berbel et al., 2001). DETERMINATE (DET), a TFL1 homolog (TFL1a), is expressed within the I1 meristem, where it promotes SAM indeterminacy (Foucher et al., 2003; Berbel et al., 2012). det mutants exhibit conversion of the I1 to an I2, which terminates the main stem (Singer et al., 1990). Unlike Arabidopsis TFL1, DET has no influence on flowering time in pea, and this role is instead played by LATE FLOWERING (LF), a second pea TFL1 homolog (TFL1c), which acts to delay flowering (Foucher et al., 2003).
Three other pea loci of particular interest for understanding the regulation of inflorescence development are GIGAS, VEGETATIVE1 (VEG1), and VEG2. Plants carrying severe mutant alleles for any of these loci exhibit normal vegetative development but fail to develop I2 or floral structures under long-day (LD) photoperiods, suggesting that these loci have critical roles in pea inflorescence development (Reid and Murfet, 1984; Murfet and Reid, 1993; Beveridge and Murfet, 1996). GIGAS has been characterized as the FT homolog, FTa1, which is thought to encode a graft-transmissible floral stimulus that can travel from leaf to apex to promote flowering (Beveridge and Murfet, 1996; Hecht et al., 2011), similar to Arabidopsis FT (Corbesier et al., 2007; Mathieu et al., 2007). The most severe gigas mutant, gigas-2, is nonflowering under LD only and is late-flowering with normal I2 and floral morphology under short-day (SD) conditions (Murfet, 1992; Taylor and Murfet, 1994; Hecht et al., 2011). VEG1 has been identified as FULc, a MADS box gene from the AGAMOUS-LIKE79 (AGL79) clade, which appears to have a role in legume compound inflorescence development as a critical I2 identity gene (Berbel et al., 2012). The SAM of the veg1 mutant undergoes the V/I1 transition and acquires I1 identity at the same time as the wild type, but fails to subsequently specify I2 meristems (Berbel et al., 2012). The VEG2 locus has received the least attention of these three pea loci and has not been described in detail. VEG2 is represented by two recessive mutant alleles, veg2-1 and veg2-2, both generated by fast neutron mutagenesis (Murfet, 1992; Murfet and Reid, 1993).
Here, we characterize the roles of the VEG2 locus during each stage of inflorescence development in pea through examination of the two veg2 mutant alleles. We identify VEG2 as a pea homolog of FD (FDa) and further investigate the possible mechanisms of VEG2 function. Our findings reveal that VEG2 plays a central role in the regulation of meristem identity, acting in conjunction with multiple FT proteins to regulate expression of FT, TFL1, and LFY homologs and key MADS box genes, VEG1 and PIM, throughout development of the pea compound inflorescence.
RESULTS
VEG2 Acts across All Stages of Inflorescence Development
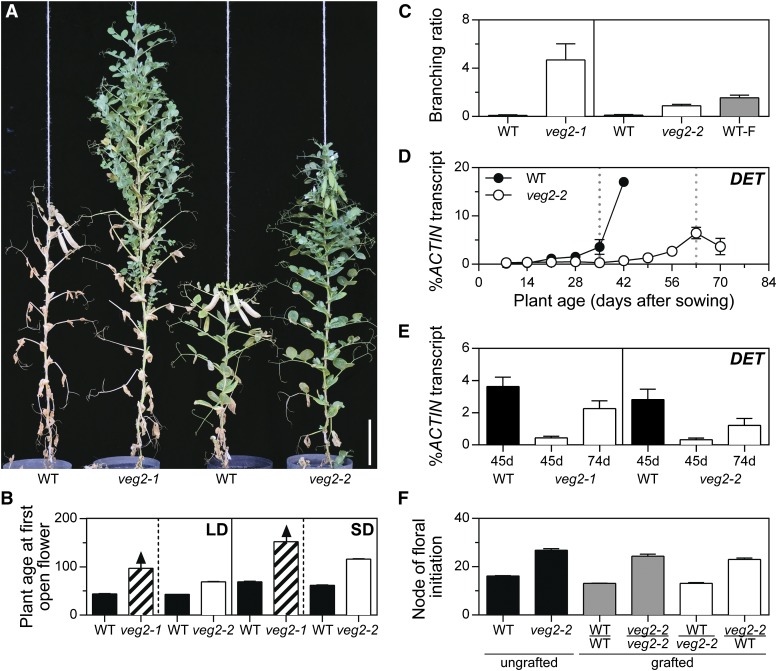
We first examined veg2-1 and veg2-2 mutant phenotypes to investigate the role(s) of VEG2 during compound inflorescence development. Under both LD and SD photoperiods, the veg2-1 mutant remained nonflowering throughout development and the weaker veg2-2 mutant flowered later than wild-type (Figures 2A and 2B). A conspicuous feature of both veg2 mutants was increased aerial branching, with lateral branches occupying aerial nodes in veg2-1, and the aerial nodes prior to the first flowering node in veg2-2 (Figures 2A and 2C; Supplemental Figure 2). This may be linked to the absence of flowers and pods in veg2-1 and their delayed appearance in veg2-2, as increased branching is also observed in wild-type plants when flowers/pods are removed (Figure 2C; Supplemental Figure 2; Lockhart and Gottschall, 1961) and in other nonflowering mutants, veg1 and gigas (Gottschalk, 1979; Beveridge and Murfet, 1996).
Figure 2.
VEG2 Acts Locally in the Apex to Promote Flower Initiation and Inflorescence Development.
(A) Representative veg2-1 and veg2-2 plants and their associated wild-type lines (wild-type siblings of veg2-1 and NGB5839, respectively). Plants are shown 97 d after sowing in LD (18 light/6 dark). Bar = 10 cm.
(B) Plant age at first open flower (days after sowing) in LD (24 light) and SD (8 light/16 dark). Values represent mean ± se for n = 3 to 6 plants. For nonflowering veg2-1 mutants, bars with diagonal hatching and arrow show plant age at the end of the experiment.
(C) Ratio of total branch length to main stem length in intact veg2 mutants, associated wild-type lines, and deflowered wild-type plants (WT-F, line NGB5839, each flower removed after anthesis). Mean values ± se are shown for n = 5 to 6 plants grown in LD (24 light).
(D) and (E) Relative expression of DET transcript as a marker of primary inflorescence (I1) identity in dissected shoot apices at weekly time points throughout development in veg2-2 (D) and specific time points in both veg2 mutants (E). In (D), developing floral buds were first macroscopically visible in the wild type 35 d after sowing and in veg2-2 63 d after sowing (broken lines) in LD (24 light). In (E), time points correspond to early flowering stages in the wild type 45 d after sowing (45d) and veg2-2 plants 74 d after sowing (74d) in LD (18 light/6 dark). Mean values ± se are shown for n = 2 to 3 biological replicates.
(F) Node of floral initiation for graft combinations of the wild type (NGB5839) and veg2-2 created by grafting 7-d-old scions onto 3-week-old stocks, controls comprising grafted plants with stock and scion of identical genotype, and ungrafted control plants. For each graft combination, the genotypes of scion (top) and stock (bottom) are shown, separated by a horizontal line. Values represent mean ± se for n = 5 to 11 plants in LD (18 light/6 dark).
The fact that both veg2 mutations impair the initiation of flowering suggested that they might affect the V/I1 transition (Figure 1B). I1 meristems are characterized by the expression of the TFL1 homolog DET, and DET expression has been used as a developmental marker for I1 meristem identity (Berbel et al., 2012). In nonflowering veg1 and gigas mutants, the timing of DET induction is similar to the wild type, indicating that the V/I1 transition is not affected under LD in these mutants (Hecht et al., 2011; Berbel et al., 2012; Supplemental Figure 3). We first examined DET expression in the late-flowering veg2-2 mutant at weekly time points during development from seedling to flowering adult plant. DET induction was delayed by 3 to 4 weeks in veg2-2 relative to the wild type, comparable to the ∼4-week delay in the appearance of floral buds (Figure 2D). In a second experiment, we also examined DET expression in the more severe allele, veg2-1, but focused on only two time points, in view of limited availability of this sterile genotype. Time points were selected to coincide with the expected peaks in DET expression in the wild type and veg2-2. At 45 d after sowing, when the presence of floral buds indicated that the V/I1 transition had occurred in the wild type, DET expression was 8- to 9-fold lower in the veg2 mutants than respective wild-type plants (Figure 2E). By day 74, when floral buds were first visible in veg2-2 apices and wild-type plants had senesced, DET expression had increased 4- to 5-fold in the veg2 mutants. This suggests that DET induction is also delayed in veg2-1, similar to veg2-2, and that the V/I1 transition is therefore delayed in both mutants. In addition, the fact that DET is eventually expressed in the nonflowering veg2-1 mutant indicates that this mutant does acquire I1 identity, but as no I2 structures subsequently develop, we conclude that the next stage of inflorescence development, I2 meristem specification, must be blocked in this mutant.
Next, we used grafting to investigate whether VEG2 may contribute to the generation of a long-distance flowering signal, as is the case for the GIGAS/FTa1 gene (Beveridge and Murfet, 1996; Hecht et al., 2011). Figure 2F shows that veg2-2 scions grafted onto wild-type stocks flowered as late as self-grafted veg2-2 control plants (P = 0.729), while wild-type scions grafted to veg2-2 stocks flowered at a similar time to self-grafted wild-type plants (P = 1.000). These results indicate that VEG2 cannot influence flowering across a graft union and suggest that VEG2 instead acts locally within the shoot apex.
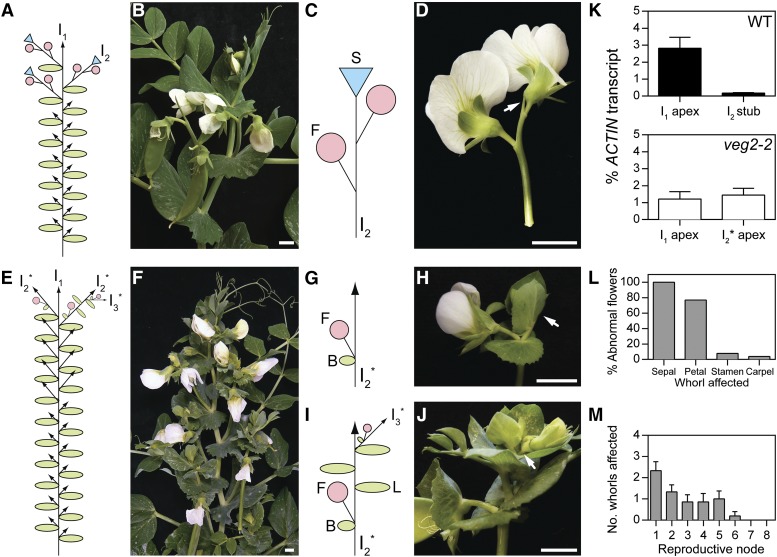
We next examined the weaker veg2-2 mutant phenotype in more detail for insight into the role(s) of VEG2 during the later stages of flowering, I2 and floral development, which do not occur in the nonflowering veg2-1 mutant. In the veg2-2 mutant, I2 morphology was abnormal at all reproductive nodes. Abnormal I2 structures resembled the wild type in that they bore one or more axillary flowers, but unlike wild-type I2 structures, which terminate in a stub, these had a bract subtending each flower and retained an indeterminate apex (Figures 3A to 3J). After producing one or two flowers, subsequent nodes of the veg2-2 I2 bore full compound leaves with vegetative axillary buds or axillary tertiary inflorescence (I3) structures that reiterate the same abnormal I2 pattern (Figures 3I and 3J). This phenotype suggests that I2 identity is initially partly specified in veg2-2, but this identity is not maintained, and reversion to I1 identity occurs. Consistent with this interpretation, DET was expressed in the indeterminate apex of the veg2-2 I2 at a similar level to that in the veg2-2 I1 apex, whereas in the wild type, expression of DET was limited to the I1, and levels in the I2 stub were negligible (Figure 3K). This indicates that VEG2 has an important role not only in specifying but also in maintaining I2 identity.
Figure 3.
Inflorescence and Floral Morphology Is Abnormal in the veg2-2 Mutant.
(A) to (D) Wild-type inflorescence structure in pea line NGB5839.
(A) Diagram of plant architecture.
(B) Photograph of reproductive nodes on the main stem.
(C) and (D) Diagram (C) and photograph (D) of the secondary inflorescence (I2) which bears axillary flowers (F) and terminates in a stub (S; arrow).
(E) to (J) Inflorescence structure in veg2-2.
(E) Diagram of plant architecture.
(F) Photograph of reproductive nodes on the main stem.
(G) and (H) Diagram (G) and photograph (H) of a typical veg2-2 I2, which bears an axillary flower with subtending bract (B) and retains an indeterminate apex (arrow).
(I) and (J) Diagram (I) and photograph (J) of an older veg2-2 I2, which has a pod and subtending bract at the first node, three nodes with full compound leaves (L), and a flower on an axillary tertiary inflorescence (I3; arrow in photograph).
Note all veg2-2 I2 structures are indeterminate, similar to (H), and may develop additional nodes after bearing axillary flowers, similar to (J).
(K) Relative expression of DET transcript as an indicator of primary inflorescence (I1) identity in the dissected shoot apex during early flowering stages (I1 apex; 45 and 74 d after sowing in wild-type NGB5839 and veg2-2, respectively) and in the I2 (wild-type I2 stub 59 d after sowing and veg2-2 I2 apex 74 d after sowing) in LD (18 light/6 dark). Mean values ± se are shown for n = 2 to 3 biological replicates.
(L) Proportion of abnormal flowers in veg2-2 defective in each of the four floral whorls for n = 26 flowers grown in LD (24 light).
(M) Number of whorls affected by floral defects at each reproductive node on veg2-2 plants. Values represent mean ± se for n = 7 plants grown in LD (24 light).
In diagrams, arrows indicate the potential for indeterminate growth, circles are flowers, triangles are terminal stubs, ovals are leaves or bracts, and asterisks indicate abnormal nature of structures. In photographs, bars = 1 cm.
We also observed that flowers produced on veg2-2 I2 structures were frequently abnormal. Defects were most common in outer (sepal and petal) whorls (Figure 3L) and included fusion of floral organs to leaf or other floral tissue, a reduction in organ number, and organ displacement or malformation (Supplemental Figure 4). The severity of these floral defects decreased acropetally on the main stem axis, and flowers on I2 structures at higher reproductive nodes showed normal morphology (Figure 3M; Supplemental Figure 4). These observations indicate a further role for VEG2 in specification of floral meristems.
Previous study has shown that PIM, a pea AP1 homolog, has a major role in specification of floral meristems in pea (Taylor et al., 2002). To determine whether VEG2 could affect floral phenotype in the absence of functional PIM, we also examined the phenotype of the pim-2 veg2-2 double mutant. pim-2 mutant plants produce I2 structures but fail to specify floral meristems correctly, and single flowers are typically replaced by groups of abnormal flowers (Taylor et al., 2002; Supplemental Figure 5). By contrast, no discrete units recognizable as flowers were observed in the pim veg2-2 double mutant. Although structures with floral identity did form, these were limited to isolated floral organs that were fused to, or borne in the axils of, leaves or bracts on upper nodes of the main stem or branches (Supplemental Figure 5). This more severe phenotype of pim veg2-2 relative to pim indicates that VEG2 acts at least in part through genes other than PIM to specify floral meristem identity.
The VEG2 Locus Corresponds to an FD Gene
Preliminary mapping indicated that VEG2 was located toward the base of pea LGI (Murfet and McKay, 2012). We exploited the close synteny between pea and Medicago truncatula (Duarte et al., 2014) to search for an appropriate candidate for the VEG2 locus. In the syntenic region of M. truncatula chromosome 5, we identified a homolog of the Arabidopsis bZIP transcription factor FD (Supplemental Figure 6). Mutants for FD genes in other species typically exhibit delayed flowering time (Abe et al., 2005; Wigge et al., 2005; Muszynski et al., 2006; Park et al., 2014), which is one of the features of the veg2-2 mutant (Figure 2B). Isolation and mapping of the full-length coding sequence for the pea ortholog of this gene confirmed its location close to the VEG2 locus (Supplemental Figure 6 and Supplemental Table 1).
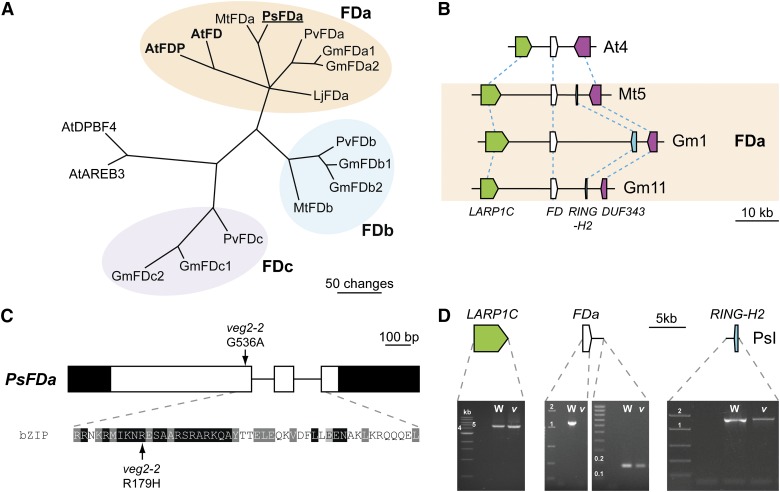
With this gene as a likely candidate for VEG2, we investigated the legume FD family further. We identified FD genes from M. truncatula, soybean, common bean, and Lotus japonicus, excluding homologs of the closely related AREB3/DPBF4 genes and other Group A bZIP transcription factors (Corrêa et al., 2008). Figure 4A shows that this approach identified three subclades of legume FD genes, which we have designated as FDa, FDb, and FDc. The VEG2 candidate was included within the legume FDa subclade, which showed the greatest similarity to Arabidopsis FD and FDP. FDa and FDb subclades were represented in species from both galegoid and phaseoloid legume clades (Figure 4A) but were not apparent in other rosid I orders, including Rosales, Cucurbitales, and Malpighiales (Supplemental Figure 7 and Supplemental Table 2). This suggests that FDa and FDb subclades resulted from a legume-specific duplication event prior to the divergence of galegoid and phaseoloid lineages, ∼54 million years ago (Lavin et al., 2005). Although FDb is present in M. truncatula, BLAST searches of pea transcript databases (Franssen et al., 2011; Kaur et al., 2012) and PCR approaches using degenerate and Mt FDb-specific primers (Supplemental Table 3) provided no evidence for an FDb ortholog in pea. FDc genes were identified only in the phaseoloid species soybean and common bean (Figure 4A). The gene present immediately upstream of FD in Arabidopsis, LA RELATED PROTEIN 1C (LARP1C), was found to be conserved close to all legume FD genes investigated (Figure 4B; Supplemental Figure 8 and Supplemental Table 4). This microsynteny between genomic regions surrounding FD in Arabidopsis and legume homologs supports the probable common origin of Arabidopsis FD and all three legume FD subclades. No microsynteny was apparent between regions containing FDP in Arabidopsis and any legume FD genes.
Figure 4.
The VEG2 Locus Corresponds to FDa.
(A) Phylogram of the legume FD family. Branches with bootstrap values <55% obtained from 10,000 trees have been collapsed. Two related group A bZIP transcription factors from Arabidopsis, DPBF4 and AREB3, are included as an outgroup. Alternative names for previously identified soybean proteins GmFDL02 (GmFDb1), GmFDL04 (GmFDc1), and GmFDL0602 (GmFDc2) are adopted to better reflect wider phylogenetic relationships. The analysis is based on the sequence alignment shown in Supplemental Data Set 1. Sequence details are given in Supplemental Table 2. Ps, Pisum sativum; At, Arabidopsis thaliana; Mt, Medicago truncatula; Lj, Lotus japonicus; Gm, Glycine max; Pv, Phaseolus vulgaris.
(B) Microsynteny between genomic regions containing Arabidopsis FD, legume FDa genes, and flanking genes. Genes are represented as boxes with point showing putative direction of transcription on black lines representing regions of the genome with chromosome number indicated. Between species, corresponding genes are connected by dashed lines. Microsynteny for legume FDb and FDc genes is shown in Supplemental Figure 8. Gene details are given in Supplemental Table 4.
(C) Diagram of the pea FDa gene showing nature and location of the SNP in veg2-2, which affects a conserved amino acid within the functional bZIP domain. Exons are shown as boxes, with coding sequence in white and untranslated regions in black. Shading levels in bZIP domain sequence indicate degree of conservation (black = 100%, dark gray = 80%, and light gray = 60%) from alignment with other FD proteins shown in Supplemental Figure 9. Nucleotide numbering begins at the start of the coding sequence.
(D) Representative PCR results for full-length coding sequence for FDa, its putative 5′ and 3′ flanking genes, and a region immediately downstream of FDa from the wild type (Kaliski) and veg2-1 mutant genomic DNA template. For each gel, lanes containing a DNA ladder and PCR product for a no template negative control, wild-type (W) positive control, and veg2-1 (v) mutant gDNA are shown (from left to right). Band size (kb) is indicated for relevant ladder bands. Above each gel, the genomic regions isolated are shown diagrammatically, as for (B). Regions between gene diagrams are not drawn to scale.
Sequencing of FDa in the veg2-2 mutant revealed a single nucleotide polymorphism (SNP) (G536A) directing a substitution (R179H) within the DNA binding, basic region of the bZIP domain (Figure 4C). This SNP cosegregated perfectly with veg2-2 phenotype in a population of 114 F2 progeny that included 34 veg2-2 mutants (Supplemental Figure 6). An arginine is highly conserved at this position in FD proteins from diverse angiosperm species and in 95% of all Arabidopsis bZIP family proteins (Supplemental Figure 9), which comprise 13 divergent groups with minimal sequence similarity outside of the bZIP domain (Corrêa et al., 2008). This high level of conservation strongly implies that this residue is important for general bZIP transcription factor function. The same arginine-to-histidine substitution was previously reported for the maize FD homolog DLF1 in the inflorescence mutant dlf1-N2461A (Muszynski et al., 2006). Results from 3D modeling indicated that the arginine at this position comes into direct contact with the phosphate groups on target DNA and conversion to a histidine results in distortion of the DNA backbone, which reduces binding strength (Muszynski et al., 2006). The R179H amino acid substitution in the pea veg2-2 mutant is likely to reduce FDa function in a similar manner.
Attempts to amplify FDa by PCR from veg2-1 genomic DNA were unsuccessful, whether using primers within or spanning the FDa coding sequence (Figure 4D), and this failure to amplify FDa clearly distinguished all veg2-1 mutant plants (n = 37) in a segregating population (n = 210; data not shown). The simplest interpretation of these findings is that a deletion encompassing FDa has occurred in this mutant. As the pea genome sequence is not yet available, we again made use of microsynteny to investigate the extent of this apparent deletion. Figure 4B shows that the two genes flanking FDa in M. truncatula, LARP1C and a RING-H2 gene, also have conserved positions flanking FDa in soybean, and in view of the close relationship of pea to M. truncatula, we considered it likely that this arrangement was also preserved in pea. The pea homologs of these genes were isolated, mapped, and found to be closely linked to each other and to FDa, as expected (Supplemental Figure 6), and full-length coding sequence for both genes was found to be intact in the veg2-1 mutant (Figure 4D). In addition, a fragment 1.4 kb downstream of the FDa stop codon was found to be present in veg2-1, revealing that the 3′ boundary for the veg2-1 deletion is close to FDa coding sequence (Figure 4D). Further attempts to define the precise boundaries of the deletion by isolating the region between LARP1C and this fragment in the wild type and veg2-1 mutant were unsuccessful. However, as both the flanking genes predicted by microsynteny were found to be intact in veg2-1, our results suggest that this mutant contains a deletion restricted to FDa. Therefore, based on the evidence of distinct functionally significant mutations that specifically affect FDa in both of the veg2 mutants, and the correlation between the molecular nature of the mutations and the severity of the respective mutant phenotypes, we conclude that the VEG2 locus corresponds to FDa and subsequently refer to FDa as the VEG2 gene.
VEG2 Is Expressed in the Apex throughout Development
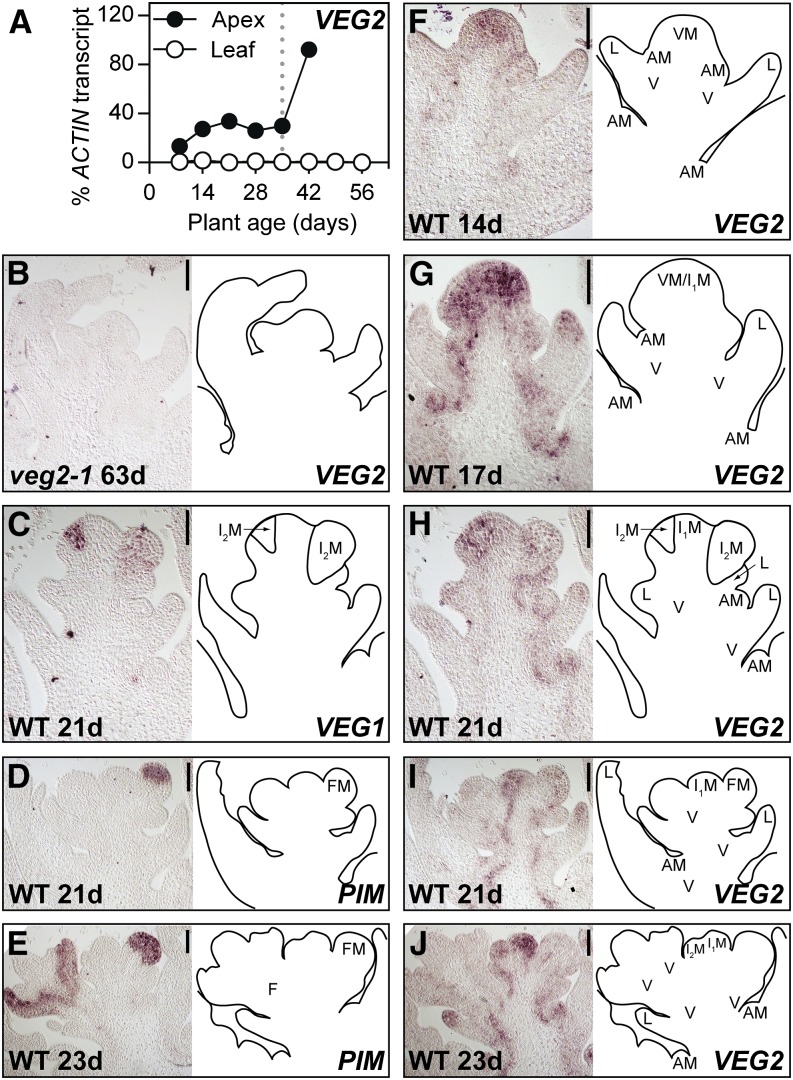
In view of observations that VEG2 is important for multiple stages of inflorescence development, it was of interest to investigate the developmental and spatial pattern of VEG2 expression. We first examined the expression of VEG2 by quantitative RT-PCR (qRT-PCR) in shoot apex and leaf tissues of wild-type plants throughout development from seedling to flowering adult plant in LD. Figure 5A shows that VEG2 was not significantly expressed in expanded leaves but was expressed in the shoot apex throughout development, where VEG2 transcript levels increased during early vegetative growth and showed a further increase during later floral development.
Figure 5.
VEG2 Is Expressed in the Apex throughout Development.
(A) Expression of VEG2 in dissected shoot apices and the uppermost fully expanded leaf of wild-type (NGB5839) plants throughout development. Relative transcript levels were determined by qRT-PCR, normalized to the transcript level of ACTIN, and represent mean ± se for n = 2 biological replicates, each consisting of pooled material from two plants grown in LD (24 light). Developing floral buds were first macroscopically visible in the wild-type apex 35 d after sowing (gray line).
(B) to (J) In situ hybridization results for VEG2 and meristem marker genes.
(B) VEG2 expression in the shoot apex of the veg2-1 deletion mutant, as a negative control for the VEG2 probe.
(C) VEG1 expression domain, as a marker for secondary inflorescence (I2) meristems.
(D) and (E) PIM expression domain, as a marker for floral meristems.
(F) to (J) FDa expression in the vegetative apex (F), the apex at the approximate time of the transition from vegetative to primary inflorescence (I1) meristem identity (G), the I1 apex during early I2 and floral development ([H] and [I]), and the I1 apex during development of floral primordia (J).
Each pair, (C) and (H), (D) and (I), and (E) and (J), represents serial sections from the same apex. Shoot apices shown in photographs in (C) to (J) are from wild-type (NGB5839) plants grown in LD (16 light/8 dark). For each sample, plant age in days after sowing (d) is indicated. Regions of expression are indicated by abbreviations on diagrams as follows: axillary meristem (AM), floral meristem (FM), developing flower (F), leaf primordia (L), primary inflorescence meristem (I1M), secondary inflorescence meristem (I2M), vasculature (V), and vegetative shoot apical meristem (VM).
The VEG2 expression pattern in the wild-type apex was next investigated in more detail by in situ hybridization during the vegetative phase (Figure 5F), the V/I1 transition (Figure 5G), and early flowering stages (Figures 5H to 5J). Apical samples from the veg2-1 mutant were included as negative controls for the VEG2 in situ probe (Figure 5B). Expression patterns for VEG1 (Figure 5C) and PIM (Figures 5D and 5E) were determined on serial sections of the same apices used for VEG2, in order to identify I2 and floral meristem boundaries, respectively (Taylor et al., 2002; Berbel et al., 2012). Consistent with previous reports, expression of VEG1 was observed in I2 meristems, and PIM expression was observed in floral meristems and floral primordia, in the petal region of the petal/stamen common primordia, and in the sepals (Figures 5C to 5E; Taylor et al., 2002; Berbel et al., 2012). VEG2 was expressed in the vegetative SAM, axillary meristems, the I1 meristem, I2 meristems, vasculature, and tips of leaf primordia (Figures 5F to 5J). Expression was also seen in floral meristems during early development (Figure 5I) but was restricted to floral vasculature during later stages of development (Figure 5J). This expression pattern is consistent with roles for VEG2 during the V/I1 transition, in specification and maintenance of I2 meristem identity, and in specification of floral meristem identity.
Flowering and Meristem Identity Genes Are Misregulated in the veg2 Mutants
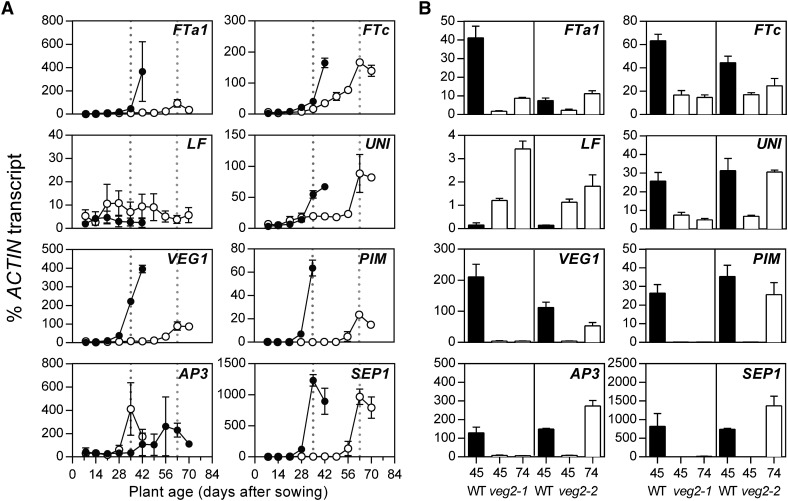
To identify possible regulatory targets of VEG2, we next examined the expression of floral integrator and meristem identity genes in the veg2 mutants, including members of the FT/TFL1 and MADS box gene families and the LFY ortholog UNIFOLIATA (UNI), under LD conditions. Gene expression was investigated in the same experiments described above for DET expression, first in a detailed time course in veg2-2 and then in both veg2 mutants at specific time points chosen to coincide with the appearance of floral buds in wild-type and veg2-2 plants (45 and 74 d after sowing, respectively).
FTa1 and FTc are significantly expressed in wild-type apical tissue and are upregulated after commitment to flowering (Hecht et al., 2011; Figure 6A). A comparison of the two time points in Figure 6B suggests that upregulation of FTa1 was delayed in both veg2 mutants; more specifically an ∼3-week delay was apparent in veg2-2 in the detailed time course (Figure 6A). FTc expression was reduced by 4-fold at both time points in veg2-1 and showed delayed induction, by ∼1 week, in veg2-2 (Figure 6). Also, in addition to DET, which exhibits delayed upregulation in both veg2 mutants (Figures 2D and 2E), a second pea TFL1 homolog, LF (TFL1c), was also misregulated, but in an opposite manner. This gene, which inhibits flowering (Foucher et al., 2003), was expressed up to 7-fold higher in the veg2 mutants relative to wild-type levels (Figure 6B).
Figure 6.
Flowering and Meristem Identity Genes Are Misregulated in the veg2 Mutants.
Gene expression in dissected shoot apices at weekly time points throughout development in wild-type (NGB5839; closed circles) and veg2-2 (open circles) (A) and specific time points in both veg2 mutants and associated wild-type lines (wild-type siblings of veg2-1 and NGB5839, respectively) (B). In (A), developing floral buds were first macroscopically visible in the wild type 35 d after sowing and in veg2-2 63 d after sowing (broken lines) in LD (24 light). In (B), time points correspond to early flowering stages in the wild type 45 d after sowing and veg2-2 plants 74 d after sowing in LD (18 light/6 dark). (A) and (B) show results from the same two experiments shown in Figures 2D and 2E, respectively. Mean values ± se are shown for n = 2 to 3 biological replicates.
In Arabidopsis, the floral integrator and floral identity gene LFY defines an FT/FD-independent pathway for AP1 upregulation (Ruiz-García et al., 1997; Abe et al., 2005; Wigge et al., 2005). In wild-type pea, the LFY ortholog UNI is expressed at a low level in the apex during early vegetative development and upregulated at the time of flowering, when FTa1 and FTc are induced (Hecht et al., 2011; Figure 6A). This upregulation was delayed by ∼4 weeks in veg2-2 and UNI expression remained at a low level in veg2-1, more than 3-fold lower than wild-type levels at day 45 (Figure 6), indicating that UNI is downstream of VEG2 in pea.
Several MADS box genes were also misexpressed in the veg2 mutants. The I2 identity gene VEG1 and the floral identity gene PIM have important roles in specifying meristem identity, and consistent with previous reports, these genes were upregulated in the wild-type apex immediately prior to floral development, at a similar time to FTa1 and FTc (Hecht et al., 2011; Berbel et al., 2012; Figure 6). In the veg2-1 mutant, VEG1 and PIM were not expressed, and in veg2-2, these genes showed an ∼4 week delay in induction, roughly corresponding to the delay in flowering time (Figure 6). A similar pattern was also seen for expression of floral organ identity genes AP3 and SEPALATA1 (SEP1), consistent with the absence of flowers in veg2-1 and delayed occurrence of floral development in veg2-2 (Figure 6).
FDa/VEG2 Interacts with All Pea FT Proteins
Studies in diverse species have shown that the physical interaction of FD and FT proteins is widely conserved and functionally significant (Wigge et al., 2005; Taoka et al., 2011; Tsuji et al., 2013). However, in maize (Zea mays), where expansion of the FT family has resulted in functional divergence between family members, there is evidence that these FT proteins differ in their ability to interact with an FD homolog (Danilevskaya et al., 2008; Meng et al., 2011). Differences in expression pattern, differing effects in transgenic Arabidopsis, and inferences from the gigas phenotype all indicate a divergence of function within the pea FT family (Hecht et al., 2011), which could in part be determined by differences in interaction with VEG2/FDa.
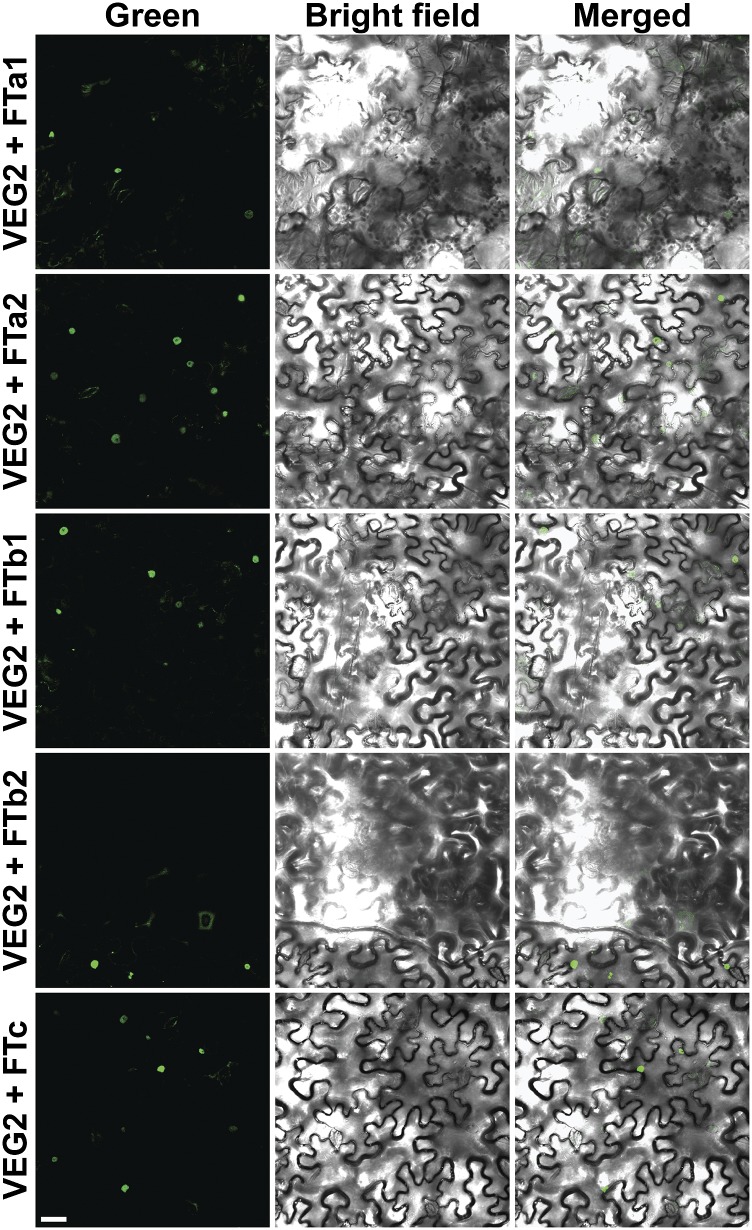
To examine whether this was indeed the case, we tested the interactions of VEG2 with the five pea FT proteins using bimolecular fluorescence complementation analysis in Nicotiana benthamiana leaves. For all five combinations, reconstitution of yellow fluorescent protein (YFP) fluorescence was observed in the nuclei of N. benthamiana leaf epidermal cells (Figure 7), at levels clearly above background (Supplemental Figure 10), indicating that VEG2 can interact with all five pea FT proteins in planta.
Figure 7.
VEG2/FDa Can Interact with Each Pea FT Protein in Planta.
For each interaction, VEG2 fused to the N-terminal half of YFP (YFN) was coexpressed separately with the FT protein fused to the C-terminal half of YFP (YFC). Photographs from left to right comprise the green channel image showing fluorescence of YFP, the bright-field image and the merged YFP fluorescence, and bright-field images. Positive and negative controls are shown in Supplemental Figure 10. Bars = 40 μm.
DISCUSSION
The pea inflorescence is typical of many legumes and is distinguished from the simple Arabidopsis inflorescence by an additional level of branching, with a so-called secondary inflorescence (I2) that displaces flowers from the main inflorescence stem (Figure 1). Three pea loci, GIGAS, VEG1, and VEG2, affect the formation of I2s and thus have the potential to provide insight into the genetic mechanisms that direct compound inflorescence development (Benlloch et al., 2007). GIGAS and VEG1 have been recently characterized as homologs of FT and AGL79, respectively (Hecht et al., 2011; Berbel et al., 2012). Here, we characterized the third of these loci, VEG2, as a pea homolog of the bZIP transcription factor FD and investigated its roles and interactions in the initiation of flowering and throughout inflorescence development.
VEG2 Participates in the Initiation of Flowering
From a developmental perspective, the initiation of flowering in pea is closely associated with the acquisition of I1 identity by the SAM, which is marked by DET/TFL1a expression (Berbel et al., 2012). In veg2 mutants, the induction of DET is delayed relative to the wild type and the other nonflowering mutants, gigas and veg1 (Figures 2D and 2E; Supplemental Figure 3). This indicates that VEG2 has an important role in promoting the V/I1 transition in wild-type plants under LD conditions, whereas GIGAS/FTa1 and VEG1 do not, despite the apparent similarity of their mutant phenotypes. However, the eventual induction of the I1 marker gene DET in the veg2-1 deletion mutant (Figure 2E) does suggest that at least one VEG2-independent pathway also promotes the V/I1 transition in pea.
The majority of work on FD genes in other species has focused on their participation in florigen signaling. Several studies have now shown that FD proteins physically associate with FT proteins and are essential for their flower-promoting function, providing a crucial link between FT proteins and their transcriptional targets (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011). The FD/FT interaction has been examined in most detail in rice (Oryza sativa), where it has been shown that the OsFD1 protein does not bind directly to the FT protein Hd3a, but that their interaction is mediated by 14-3-3 proteins (Taoka et al., 2011; Tsuji et al., 2013). These FT/14-3-3/FD complexes have been referred to as florigen activation complexes (FACs), and it is likely that VEG2 also acts as part of one or more FACs in pea. Pea has five FT genes, and analysis of expression patterns, mutant phenotypes, and activity in transgenic Arabidopsis suggest that these genes may have distinct roles in the flowering process (Hecht et al., 2011). Our results indicate that VEG2 can interact with each of the five pea FT homologs in planta (Figure 7), which suggests that participation in FACs with VEG2 may be important for the function of all pea FT proteins, but appears to rule out differential VEG2 binding as an explanation for differences in their function. Future investigations should instead explore the recent hypothesis that FT functional specificity may in fact derive from interactions with other proteins via a domain distinct from residues required for FAC binding (Ho and Weigel, 2014).
Among the five pea FT genes, only three (FTa1, FTb2, and FTc) show clear developmental regulation, consistent with roles in initiation of flowering and/or inflorescence development (Hecht et al., 2011). Because the V/I1 transition is the first stage of inflorescence development, it is likely that the role of VEG2 in this process involves interaction with the FT protein/s that act as florigens and move from leaf to apex following perception of appropriate environmental signals. Grafting experiments with gigas mutants suggest that GIGAS/FTa1 may function as one such mobile signal (Beveridge and Murfet, 1996; Hecht et al., 2011). However, the fact that induction of the I1 marker DET is not affected by the gigas mutation shows that the FAC involving FTa1 is probably not important for initiating the V/I1 transition. This is also consistent with the fact that induction of FTa1 in leaves is delayed relative to floral commitment and with the fact that the gigas phenotype indicates a role later in inflorescence development, promoting I2 specification rather than the V/I1 transition. A second FT gene, FTb2, is also a strong candidate for a florigen signal, as it is induced in leaves within the time frame of the physiological commitment to flowering in LD and is severely misregulated in photoperiod response mutants (Hecht et al., 2011). Furthermore, FTb2 is expressed normally in the gigas mutant in LD (Hecht et al., 2011), which could account for the fact that the V/I1 transition is not affected in the gigas mutant under these conditions. Functional analysis of FTb2, and in particular whether it regulates expression of DET, will be important to clarify its involvement in the V/I1 transition.
VEG2 Is Essential for Secondary Inflorescence Development
We recently showed that expression of the MADS box gene VEG1 is crucial for the formation of the I2 and proposed that I2 meristem identity is specified by VEG1 (Berbel et al., 2012). The nonflowering veg2-1 and gigas mutants are unable to form I2 structures and do not show VEG1 expression (Berbel et al., 2012; Figure 6), which indicates that VEG2/FDa and GIGAS/FTa1 are both required for induction of VEG1 under LD conditions. In addition, our data indicate that the VEG2 and FTa1 proteins can interact (Figure 7), implying that they participate in a FAC that acts to initiate VEG1 expression and specify I2 meristem identity. The fact that the abnormal I2s in the weaker veg2-2 mutant are indeterminate and revert to I1 identity (Figure 3) shows that VEG2/FDa also has a role not only in initial specification of I2 identity, but also in maintenance of this identity. The incomplete specification of I2 identity in veg2-2 is accompanied by a reduction in VEG1 expression levels relative to the wild type (Figure 6), which is consistent with the idea that VEG1 expression is a critical limiting factor in I2 development.
We previously proposed a model for inflorescence development in pea, which elaborates on the simple TFL1/AP1 negative feedback loop described in Arabidopsis (Ratcliffe et al., 1999; Kaufmann et al., 2010). In this model, DET prevents upregulation of VEG1 and PIM in the I1 meristem, and VEG1 prevents upregulation of DET in the I2 meristem, allowing expression of PIM in axillary floral meristems (Berbel et al., 2012). The incomplete and transient specification of I2 identity in the hypomorphic veg2-2 mutant illustrates how disruption of a regulatory loop can destabilize a sharp developmental transition (Sablowski, 2007). This role for VEG2/FDa in the maintenance of I2 identity is comparable to the role recently described for FT in Arabidopsis, in stabilizing inflorescence meristem identity after flowering to prevent floral reversion (Liu et al., 2014; Müller-Xing et al., 2014).
One significant point of contrast between VEG2 and FD genes in other species lies in the severity of its null mutant phenotype. Whereas the veg2-1 mutant is completely unable to flower, FD mutants in both Arabidopsis and maize are merely late-flowering (Koornneef et al., 1991; Abe et al., 2005; Wigge et al., 2005; Muszynski et al., 2006). Even when FD/FT function is completely absent in Arabidopsis, in fd fdp or ft tsf double mutants, flowering will still occur, albeit considerably later than in any single mutant (Jang et al., 2009; Jaeger et al., 2013). However, a nonflowering phenotype is seen in Arabidopsis when fd or ft mutations are combined with lfy, indicating that LFY acts in parallel with FD and FT genes to upregulate AP1 for specification of floral meristems (Ruiz-García et al., 1997; Abe et al., 2005; Wigge et al., 2005). In pea, the LFY ortholog UNI is expressed at a low level in vegetative seedlings where it has a role in leaf development (Hofer et al., 1997), but is upregulated at the time of flowering, and this upregulation is dependent on FTa1 (Hecht et al., 2011). Our results show that the upregulation of UNI is also dependent on VEG2 (Figure 6), which suggests that UNI is acting downstream of VEG2 and FTa1 in pea, and not in parallel, as is seen for LFY in other systems.
A second factor that may contribute to the severity of the VEG2 null phenotype is the existence of I2 specification as an intermediate step in pea inflorescence development and the essential role of VEG1 in this process. All three nonflowering mutants (gigas, veg1, and veg2-1) show a correlation between the absence of VEG1 expression and failure to express PIM (Figure 6B; Hecht et al., 2011; Berbel et al., 2012), suggesting that VEG1 expression is an absolute requirement for PIM expression to occur in secondary inflorescences in the presence of functional DET. The fact that PIM is expressed in the veg1 det double mutant (Berbel et al., 2012) suggests that lack of PIM expression in veg1 may reflect a strong suppression of PIM by DET that is relieved through repression of DET by VEG1. The delay in PIM induction observed in the veg2-2 mutant (Figure 6) suggests that VEG2 promotes PIM expression, either directly, or indirectly through repression of the floral repressor LF/TFL1c.
VEG2 Activity in Floral Meristems Implies a Role for FT Genes in Flower Development
The veg2-2 mutant phenotype also reveals that VEG2 has a role in correct specification of floral identity, especially for sepal and petal whorls (Figure 3L). The observed acropetal decrease in severity of floral defects (Figure 3M; Supplemental Figure 4) indicates that VEG2 is especially important for correct floral development at early reproductive nodes, but this importance decreases with plant age. This could be due either to slow accumulation of downstream targets through partial VEG2 function in the veg2-2 mutant or their activation via an alternative, age-related pathway. The floral abnormalities seen in veg2-2 are similar to those seen in mutants for PIM, which is misregulated in the veg2-2 mutant. Like veg2-2, pim mutants exhibit replacement of sepals with leafy bracts, second and third whorl organs are missing or mosaic, and severity of floral morphology defects decreases acropetally (Singer et al., 1999; Taylor et al., 2002). However, the severity of the pim veg2-2 double mutant phenotype indicates that VEG2 probably has other targets in addition to PIM, most likely other MADS box genes, such as AP3 and SEP1, which are also misregulated in the veg2-2 mutant (Figure 6). MADS box transcription factors are known to act in a combinatorial fashion to guide different stages of the flower initiation and development process (Smaczniak et al., 2012), and the persistent effects of VEG2 might reflect the participation of a limited number of direct VEG2 targets in MADS tetramers throughout reproductive development. Alternatively, VEG2 may be required for direct induction of flower-specific MADS genes, which would also imply an extended postflowering role for FT genes and associated FACs. Our results here and in the previous study of Hecht et al. (2011) show that expression of FTa1 and FTc genes is induced in shoot apical tissue only after the initial commitment to flowering and continues well beyond this transition, consistent with a role in later reproductive development.
VEG2 and the Integration of FT/TFL1 Signaling
In a previous study, we presented evidence for potential cross-regulation among FT genes, specifically, for the positive regulation of FTc at the shoot apex by FTa1 and FTb2 (Hecht et al., 2011). Results from this study show that FTc is also misregulated in the veg2 mutants (Figure 6), providing support for the conclusion that one or more other FT genes may be required for full activation of FTc. In addition, FTa1 expression in the shoot apex is also altered in the veg2 mutants (Figure 6), implying that it too is dependent on one or more FT genes acting together with VEG2. This is consistent with a scenario in which both FTa1 and FTc are transcriptional targets of FT proteins that arrive to the shoot apex, a conclusion also supported by the fact that induction of FTa1 and FTc in the apex occurs only after the plant becomes committed to flower.
One interpretation is that these genes may act as functional integrators of mobile FT signals. However, the induction of DET, which marks the V/I1 transition, occurs with very similar timing to that of FTa1 and FTc in the shoot apex, with no evidence of the delay that might be expected if it was dependent on the expression of these genes. GIGAS/FTa1 is clearly required for VEG1 induction and I2 specification, but the role of FTc is less clear. FTc is still expressed in gigas, albeit at a lower level than the wild type, and the fact that gigas plants do not flower under LD implies that FTc alone cannot substitute for FTa1 in the initiation of I2 formation under these conditions. One explanation is that FTc may play a subsidiary role to FTa1, reinforcing its expression and/or activity and ensuring a clear induction of VEG1 and a sharp developmental transition from vegetative branch to I2.
The observation that VEG2 regulates DET expression provides intriguing evidence for transcriptional regulation of a TFL1 gene by an FD gene. Given the mechanism for FD action, this also implies the involvement of FT family members, and some evidence for this does exist. In strawberry (Fragaria vesca), the FT gene FT1 has been shown to activate TFL1 expression indirectly via SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, to repress flowering and allow production of vegetative shoots under LD conditions in spring and summer (Mouhu et al., 2013). Similarly in Arabidopsis, TFL1 is strongly upregulated in the SAM at the onset of flowering and is expressed in proportion to FT (Jaeger et al., 2013). Furthermore, a computational model designed to simulate flowering in Arabidopsis required the inclusion of a term for activation of TFL1 by FT/FD proteins to correctly model the maintenance of SAM indeterminacy during flowering (Jaeger et al., 2013). Collectively, these findings suggest that direct or indirect transcriptional regulation of TFL1 homologs by FT and FD homologs is also likely to occur more widely, and the functional significance of this and the mechanisms by which it is achieved will be of interest to determine.
In contrast to DET, the expression of the related LF/TFL1c gene was increased in both veg2 mutants relative to the wild type (Figure 6). The tissue specificity of LF expression is not yet known, but the fact that lf mutants are early flowering without any other apparent defects can be interpreted to indicate that LF acts to prevent the acquisition of I2 identity in lateral meristems. In this respect, it is similar to DET, which also prevents the acquisition of I2 identity, but in the SAM. The finding that VEG2 regulates these two TFL1 homologs in opposite ways (i.e., promoting induction of DET and repressing expression of LF) is consistent with the meristem identity changes that occur during the transition to flowering, as DET expression is positively associated with inflorescence development, while LF expression is not. It also implies that one or more FT proteins may act via FACs to relieve suppression of I2 identity by LF in lateral meristems. In other angiosperms, including eudicot and monocot species, FD proteins have been found to also interact with TFL1 proteins, in complexes that inhibit the transcription of floral target genes (Pnueli et al., 2001; Danilevskaya et al., 2010; Hanano and Goto, 2011). The possibility that VEG2/FDa may interact with DET/TFL1a and/or LF/TFL1c in a similar manner in pea remains to be investigated.
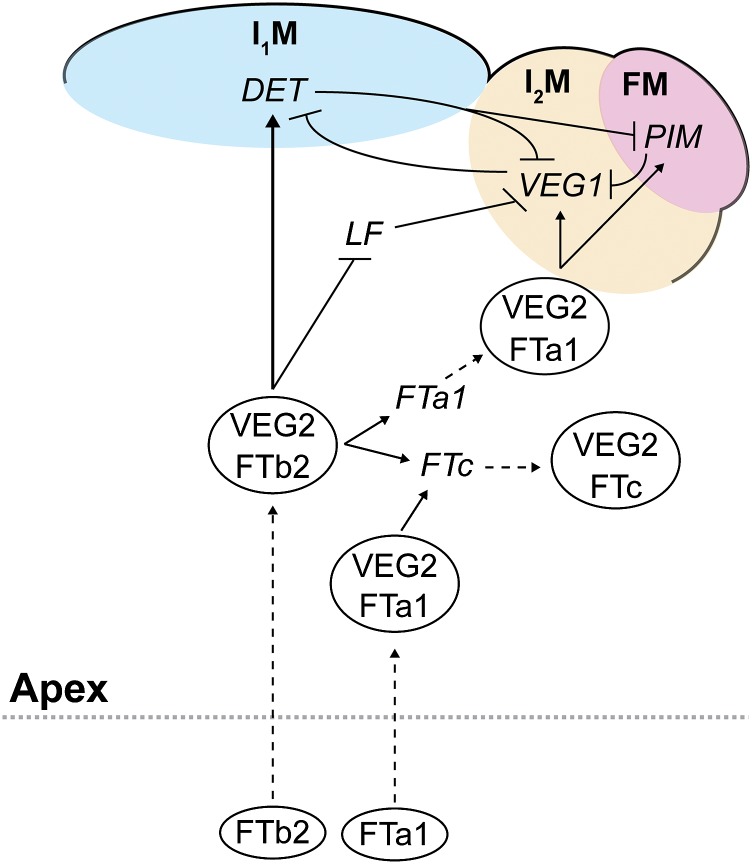
The observations and hypotheses resulting from this study are summarized in the model in Figure 8, which suggests that FTb2 arriving at the shoot apex in LD may form a FAC with VEG2 and regulate a number of other FT/TFL1 genes in the SAM and lateral meristems. The collective action of these secondary FACs may then allow specific expression of VEG1 and formation of I2s in lateral meristems, where LF has been downregulated, and promote upregulation of DET in the SAM to maintain SAM indeterminacy. Future work to test these ideas should include a detailed analysis of expression dynamics of FT and LF genes within the SAM, an examination of the interactions between VEG2 and TFL1 proteins, and characterization of FTb2 and FTc mutants.
Figure 8.
A Model for the Roles and Interactions of VEG2 during Pea Inflorescence Development.
This model summarizes the main hypotheses derived from the major results of this study and previous studies. We propose that FTb2, the best candidate for the pea florigen signal, travels from the leaf to the shoot apex under LD and interacts within a FAC with VEG2/FDa in the apex to promote primary inflorescence meristem (I1M) identity, through upregulation of DET, GIGAS/FTa1, and FTc and repression of the floral repressor LF. FTa1 is expressed in the leaf, encodes a graft-transmissible floral stimulus, and is also expressed in the apex. We infer that FTa1 protein acts in a FAC with VEG2 in the apex to promote expression of FTc and induce VEG1 for specification of secondary inflorescence meristem (I2M) identity. Repressive interactions between DET, VEG1, and PIM are based on a previous model (Berbel et al., 2012). Proteins and protein complexes are shown as ovals, and genes are shown in italics. Unbroken lines indicate inferred roles for genes/proteins as promoting (arrows) or repressing (blunt ends) expression of other genes, either directly or indirectly. Broken lines indicate movement of proteins or translation of genes into proteins. Colored zones indicate specific meristems boundaries. Genes/proteins shown outside colored meristem zones are not intended to represent spatial patterns of expression within the apex.
Overall, the findings from this study have extended our previous work to make a significant contribution to understanding of how differences in inflorescence architecture are generated. They will also assist the investigation of this process in a range of other important legume crop species that share similar inflorescence architecture. Understanding the complex network of genes controlling inflorescence development may ultimately contribute to crop improvement through optimization of inflorescence architecture for efficient harvest and maximal yield.
METHODS
Plant Material and Growth Conditions
The origins of pea (Pisum sativum) veg2-1, veg2-2, pim-2, gigas-2, and veg1 mutants have been described previously (Gottschalk, 1979; Murfet and Reid, 1993; Taylor et al., 2002; Hecht et al., 2011). Molecular characterization of the veg2-1 allele compared the mutant line in the original cv Kaliski background with cv Kaliski. All other experiments used mutant lines derived from multiple backcrosses in the dwarf NGB5839 background, as previously described (Hecht et al., 2007). In the case of nonflowering veg1 and veg2-1 mutants, wild-type siblings were used as controls. Plants for the qRT-PCR experiments shown in Figures 2D, 5A and 6A were grown in growth cabinets at 20°C, and plants for all other experiments were grown in the Phytotron. Growth media, light sources, Phytotron conditions, and grafting protocols have been described previously (Weller et al., 1997; Hecht et al., 2007). For branching data shown in Figure 2C, plants were measured 97 d after sowing and measurements include all vegetative laterals 5 mm or longer in length.
Gene Isolation
Partial length Ps FDa was isolated from a cDNA library screen of 1,000,000 clones from a 5′ Stretch Plus λgt11 cDNA library (Clontech) from pea apical buds (Lester et al., 1997) using partial Mt FDa as a probe. Partial Ps FDa sequence was extended using rapid amplification of cDNA ends (SMART RACE cDNA amplification kit; Clontech), genome walking (GenomeWalker Universal kit; Clontech), and standard PCR techniques to obtain full-length coding sequence. Putative FDa flanking genes and molecular marker genes for mapping of FDa/VEG2 were isolated using primers designed from either pea sequence, where available, or conserved regions of Medicago truncatula orthologs. Primer details are given in Supplemental Table 3. PCR fragments were purified and sequenced directly or cloned in pGEM-T Easy (Promega) and then sequenced at the Australian Genome Research Facility (Brisbane, Australia) or Macrogen (Seoul, Korea).
Phylogenetic Analysis
FD genes from other species were identified by performing BLAST searches using Arabidopsis thaliana FD protein sequence as a query and identity was confirmed by reciprocal BLAST search against Arabidopsis at TAIR (www.arabidopsis.org) and preliminary phylogenetic analysis (data not shown). For full sequence details, including source, see Supplemental Table 2. For each alignment (Supplemental Data Sets 1 and 2), full-length amino acid sequence was aligned using ClustalX (Thompson et al., 1997) and adjusted manually, where necessary, using GeneDoc (version 2.7.000; Nicholas and Nicholas, 1997; http://www.psc.edu/biomed/genedoc). Using these alignments, distance-based methods were used for phylogenetic analyses in PAUP* 4.0b10 (http://paup.csit.fsu.edu/).
Gene Expression Studies
For qRT-PCR, harvested tissue for each sample consisted of both leaflets from the uppermost fully expanded leaf or dissected apical buds (∼2 mm) from two plants. Samples were frozen in liquid nitrogen and total RNA extracted using the SV Total RNA isolation system (Promega). RNA concentrations were determined using a NanoDrop 8000 (Thermo Scientific). Reverse transcription was conducted in 20 μL with 1 μg total RNA using the Tetro cDNA synthesis kit (Bioline) according to the manufacturer’s instructions. RT-negative (no enzyme) controls were performed to monitor for contamination with genomic DNA. First-strand cDNA was diluted five times, and 2 μL was used in each real-time PCR. Reactions using SYBR green chemistry (Sensimix; Bioline) were set up with a CAS-1200N robotic liquid handling system (Corbett Research) and run for 50 cycles in a Rotor-Gene RG3000 (Corbett Research). Two technical replicates and at least two biological replicates were performed for each sample. All primer details are given in Supplemental Table 3.
RNA in situ hybridization with digoxigenin-labeled probes was performed as previously described (Ferrándiz et al., 2000). Probes used for VEG1 and PIM have been described previously (Berbel et al., 2001; Berbel et al., 2012). Primer details for the VEG2/FDa probe are given in Supplemental Table 3.
Bimolecular Fluorescence Complementation Assay
Full-length coding sequences of VEG2/FDa, FTa1, FTa2, FTb1, FTb2, and FTc were amplified from NGB5839 cDNA, cloned in-frame into the pCR8/GW/TOPO entry vector (Invitrogen), and transferred by Gateway LR reaction (Invitrogen) into pYFPN43 and pYFPC43 destination vectors. Primer details are given in Supplemental Table 3. Constructs for the positive control interaction between Arabidopsis proteins AKIN10 and AKINβ2 (Supplemental Figure 10) have been described previously (Belda-Palazón et al., 2012). Constructs were introduced into Agrobacterium tumefaciens C58C1 (pGV2260) and used to infiltrate young fully expanded leaves of 4-week-old tobacco (Nicotiana benthamiana) plants as previously described (Scacchi et al., 2009). Leaves were examined after 3 to 4 d with a Leica TCS-SL confocal microscope and a confocal laser scanning imaging system.
Statistical Analysis
Statistical analysis was conducted using IBM SPSS Statistics (version 21), using a significance level of 0.05. Levene’s test for homogeneity of variance was applied, and one-way ANOVA (with Tukey’s HSD post-hoc test) or Welch’s test for ANOVA (with Games-Howell post-hoc test) were conducted, as appropriate.
Accession Numbers
Please refer to Supplemental Table 2 for details of sequences used in phylogenetic analyses and to Supplemental Table 4 for details of FD and flanking genes in Arabidopsis and legume species. GenBank accession numbers for other pea genes are as follows: AP3 (JN412098), DET (AY340579), FTa1 (HQ538822), FTa2 (HQ538823), FTb1 (HQ538824), FTb2 (HQ538825), FTc (HQ538826), LARP1C (JI919144, JI924790, and JR963915), LF (AY343326), PIM (AJ291298), RING-H2 (KP739948), SEP1 (AY884290), UNI (AF010190), VEG1 (JN974184), and VEG2/FDa (KP739949 and KP739950).
Supplemental Data
Supplemental Figure 1. Phenotypes for key inflorescence mutants in pea.
Supplemental Figure 2. Branching in the veg2 mutants.
Supplemental Figure 3. DET expression in nonflowering gigas and veg1 mutants.
Supplemental Figure 4. Floral morphology in the veg2-2 mutant.
Supplemental Figure 5. The pim-2 veg2-2 double mutant phenotype.
Supplemental Figure 6. Comparative map for pea and M. truncatula showing relative locations of VEG2/FDa and surrounding genes.
Supplemental Figure 7. Phylogram of the angiosperm FD family.
Supplemental Figure 8. Microsynteny between genomic regions containing FD and flanking genes in Arabidopsis and legume species.
Supplemental Figure 9. Conserved nature of the amino acid altered by the veg2-2 SNP.
Supplemental Figure 10. Positive and negative BiFC controls.
Supplemental Table 1. Mapping loci details.
Supplemental Table 2. Details of sequences for FD proteins and related bZIP transcription factors used for phylogenetic analyses and alignments.
Supplemental Table 3. Primer details.
Supplemental Table 4. Details for FD and flanking genes in Arabidopsis and legume species.
Supplemental Data Set 1. Alignment used for Figure 4A.
Supplemental Data Set 2. Alignment used for Supplemental Figure 7.
Supplementary Material
Acknowledgments
We thank Ian Cummings, Tracey Winterbottom, and Michelle Lang for help with plant husbandry and Ian Murfet for providing seed of the original veg2-1 and veg2-2 mutant lines. We also thank Alejandro Ferrando for kindly sharing pYFPN43 and pYFPC43 vectors, and YFN-AKIN10 and YFC-AKINβ2 constructs, and Julie Hofer for sharing unpublished primer sequences for isolation of the FENR1 marker gene. We acknowledge the use of facilities administered by the University of Tasmania Central Science Laboratory. This work was supported by the Australian Research Council (J.L.W.) and the Spanish Ministerio de Ciencia e Innovación (F.M.).
AUTHOR CONTRIBUTIONS
F.C.S., J.L.W., V.H., F.M., and C.F. designed the research. F.C.S., A.B., and J.K.V.S. performed the research. F.C.S. and J.L.W. wrote the article.
Glossary
- bZIP
basic leucine zipper
- SAM
shoot apical meristem
- LD
long-day
- SD
short-day
- SNP
single nucleotide polymorphism
- qRT-PCR
quantitative RT-PCR
- FAC
florigen activation complex
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B., Ruiz L., Martí E., Tárraga S., Tiburcio A.F., Culiáñez F., Farràs R., Carrasco P., Ferrando A. (2012). Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS ONE 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R., Berbel A., Serrano-Mislata A., Madueño F. (2007). Floral initiation and inflorescence architecture: a comparative view. Ann. Bot. (Lond.) 100: 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A., Ferrándiz C., Hecht V., Dalmais M., Lund O.S., Sussmilch F.C., Taylor S.A., Bendahmane A., Ellis T.H.N., Beltrán J.P., Weller J.L., Madueño F. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 3: 797. [DOI] [PubMed] [Google Scholar]
- Berbel A., Navarro C., Ferrándiz C., Cañas L.A., Madueño F., Beltrán J.P. (2001). Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J. 25: 441–451. [DOI] [PubMed] [Google Scholar]
- Beveridge C.A., Murfet I.C. (1996). The gigas mutant in pea is deficient in the floral stimulus. Physiol. Plant. 96: 637–645. [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Corrêa L.G.G., Riaño-Pachón D.M., Schrago C.G., dos Santos R.V., Mueller-Roeber B., Vincentz M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Ananiev E.V. (2010). Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol. 153: 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Meng X., Hou Z., Ananiev E.V., Simmons C.R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146: 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J., Rivière N., Baranger A., Aubert G., Burstin J., Cornet L., Lavaud C., Lejeune-Hénaut I., Martinant J.-P., Pichon J.-P., Pilet-Nayel M.-L., Boutet G. (2014). Transcriptome sequencing for high throughput SNP development and genetic mapping in pea. BMC Genomics 15: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C.J., Huber S.C., Hong P.H., Singer S.R. (1991). Determination for inflorescence development is a stable state, separable from determination for flower development in Pisum sativum L. buds. Planta 185: 518–522. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Gu Q., Martienssen R., Yanofsky M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734. [DOI] [PubMed] [Google Scholar]
- Foucher F., Morin J., Courtiade J., Cadioux S., Ellis N., Banfield M.J., Rameau C. (2003). DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15: 2742–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen S.U., Shrestha R.P., Bräutigam A., Bornberg-Bauer E., Weber A.P. (2011). Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk W. (1979). A Pisum gene preventing transition from the vegetative to reproductive stage. Pisum Newsletter 11: 10–11. [Google Scholar]
- Hanano S., Goto K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Knowles C.L., Vander Schoor J.K., Liew L.C., Jones S.E., Lambert M.J.M., Weller J.L. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144: 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Laurie R.E., Vander Schoor J.K., Ridge S., Knowles C.L., Liew L.C., Sussmilch F.C., Murfet I.C., Macknight R.C., Weller J.L. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W.W.H., Weigel D. (2014). Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., Ellis N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7: 581–587. [DOI] [PubMed] [Google Scholar]
- Hole C.C., Hardwick R.C. (1976). Development and control of the number of flowers per node in Pisum sativum L. Ann. Bot. (Lond.) 40: 707–722. [Google Scholar]
- Jaeger K.E., Pullen N., Lamzin S., Morris R.J., Wigge P.A. (2013). Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25: 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Torti S., Coupland G. (2009). Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60: 614–625. [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kaur S., Pembleton L.W., Cogan N.O., Savin K.W., Leonforte T., Paull J., Materne M., Forster J.W. (2012). Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66. [DOI] [PubMed] [Google Scholar]
- Lavin M., Herendeen P.S., Wojciechowski M.F. (2005). Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54: 575–594. [DOI] [PubMed] [Google Scholar]
- Lester D.R., Ross J.J., Davies P.J., Reid J.B. (1997). Mendel’s stem length gene (Le) encodes a gibberellin 3 β-hydroxylase. Plant Cell 9: 1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Farrona S., Klemme S., Turck F.K. (2014). Post-fertilization expression of FLOWERING LOCUS T suppresses reproductive reversion. Front. Plant Sci. 5: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart J.A., Gottschall V. (1961). Fruit-induced and apical senescence in Pisum sativum L. Plant Physiol. 36: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- McGarry R.C., Ayre B.G. (2012). Manipulating plant architecture with members of the CETS gene family. Plant Sci. 188-189: 71–81. [DOI] [PubMed] [Google Scholar]
- Meng X., Muszynski M.G., Danilevskaya O.N. (2011). The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhu K., Kurokura T., Koskela E.A., Albert V.A., Elomaa P., Hytönen T. (2013). The Fragaria vesca homolog of suppressor of overexpression of constans1 represses flowering and promotes vegetative growth. Plant Cell 25: 3296–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Xing R., Clarenz O., Pokorny L., Goodrich J., Schubert D. (2014). Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell 26: 2457–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfet I.C. (1992). Garden pea and allies - an update from Hobart. Flowering Newsletter 13: 10–20. [Google Scholar]
- Murfet I.C., Reid J.B. (1993). Developmental mutants. In Peas: Genetics, Molecular Biology and Biotechnology, Casey R., Davies D.R., eds (Cambridge, UK: CAB International; ), pp. 165–216. [Google Scholar]
- Murfet I.C., McKay M.J. (2012). Evidence of linkage between VEG2 and I in pea LG I. Pisum Genet. 44: 7–8. [Google Scholar]
- Muszynski M.G., Dam T., Li B., Shirbroun D.M., Hou Z., Bruggemann E., Archibald R., Ananiev E.V., Danilevskaya O.N. (2006). delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142: 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K.B., and Nicholas, H.B. (1997). GeneDoc: a tool for editing and annotating multiple sequence alignments (distributed by the author), http://www.psc.edu/biomed/genedoc.
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Gutfinger T., Hareven D., Ben-Naim O., Ron N., Adir N., Lifschitz E. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13: 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Bradley D.J., Coen E.S. (1999). Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120. [DOI] [PubMed] [Google Scholar]
- Reid J.B., Murfet I.C. (1984). Flowering in Pisum: A fifth locus, VEG. Ann. Bot. (Lond.) 53: 369–382. [Google Scholar]
- Ruiz-García L., Madueño F., Wilkinson M., Haughn G., Salinas J., Martínez-Zapater J.M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9: 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. (2007). Flowering and determinacy in Arabidopsis. J. Exp. Bot. 58: 899–907. [DOI] [PubMed] [Google Scholar]
- Scacchi E., Osmont K.S., Beuchat J., Salinas P., Navarrete-Gómez M., Trigueros M., Ferrándiz C., Hardtke C.S. (2009). Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136: 2059–2067. [DOI] [PubMed] [Google Scholar]
- Singer S., Sollinger J., Maki S., Fishbach J., Short B., Reinke C., Fick J., Cox L., McCall A., Mullen H. (1999). Inflorescence architecture: A developmental genetics approach. Bot. Rev. 65: 385–410. [Google Scholar]
- Singer S.R., Hsiung L.P., Huber S.C. (1990). Determinate (det) mutant of Pisum sativum (Leguminosae: Papilionoideae) exhibits an indeterminate growth pattern. Am. J. Bot. 77: 1330–1335. [Google Scholar]
- Smaczniak C., Immink R.G.H., Angenent G.C., Kaufmann K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098. [DOI] [PubMed] [Google Scholar]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- Taylor S.A., Murfet I.C. (1994). A short day mutant in pea is deficient in the floral stimulus. Flowering Newsletter 18: 39–43. [Google Scholar]
- Taylor S.A., Hofer J.M.I., Murfet I.C., Sollinger J.D., Singer S.R., Knox M.R., Ellis T.H.N. (2002). PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol. 129: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Nakamura H., Taoka K., Shimamoto K. (2013). Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 54: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Sablowski R.W.M., Meyerowitz E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584. [DOI] [PubMed] [Google Scholar]
- Weberling, F. (1992). Morphology of Flowers and Inflorescences. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Weller J.L., Murfet I.C., Reid J.B. (1997). Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 114: 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wyatt R. (1982). Inflorescence architecture - how flower number, arrangement, and phenology affect pollination and fruit-set. Am. J. Bot. 69: 585–594. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.