Dominant mutations in two nucleus-encoded, paralogous octotricopeptide repeat proteins cause the mutated proteins to recognize and destabilize the chloroplast atpA and petA transcripts.
Abstract
We characterized two spontaneous and dominant nuclear mutations in the unicellular alga Chlamydomonas reinhardtii, ncc1 and ncc2 (for nuclear control of chloroplast gene expression), which affect two octotricopeptide repeat (OPR) proteins encoded in a cluster of paralogous genes on chromosome 15. Both mutations cause a single amino acid substitution in one OPR repeat. As a result, the mutated NCC1 and NCC2 proteins now recognize new targets that we identified in the coding sequences of the chloroplast atpA and petA genes, respectively. Interaction of the mutated proteins with these targets leads to transcript degradation; however, in contrast to the ncc1 mutation, the ncc2 mutation requires on-going translation to promote the decay of the petA mRNA. Thus, these mutants reveal a mechanism by which nuclear factors act on chloroplast mRNAs in Chlamydomonas. They illustrate how diversifying selection can allow cells to adapt the nuclear control of organelle gene expression to environmental changes. We discuss these data in the wider context of the evolution of regulation by helical repeat proteins.
INTRODUCTION
During and after transcription, sequence-specific RNA binding proteins control the processing, transport, localization, translation, and stability of coding and noncoding RNAs. Modular proteins, made up of tandem repeats of simple structural motifs (20 to 50 amino acids in length), most often comprising antiparallel α-helices and thus also termed helical repeat proteins, are particularly well suited to develop interactions with macromolecules, including RNA. Repeated motifs fold independently and stack on each other to form elongated or concave surfaces. While tetratricopeptide repeat (34 amino acids), Huntington, Elongation factor 3, protein phosphatase 2A, and yeast kinase TOR1 (HEAT; 39 amino acids), Armadillo (38 amino acids), Ankirin (33 amino acids), and leucine-rich repeat (23 to 24 amino acids) repeats are involved in protein-protein interactions, Pumilio and fem-3 binding factor (PUF; 36 amino acids), Transcription Activator Like Effector (TALE; 34 amino acids), pentatricopeptide repeat (PPR; 35 amino acids), Half A Tetratricopeptide (HAT; 34 amino acids), and mitochondrial termination factor (mTERF; ∼30 amino acids) motifs mediate protein-nucleic acid interactions (reviewed in Rubinson and Eichman, 2012). Crystallographic structures of PUF (Wang et al., 2001, 2002; Miller et al., 2008), TALE (Deng et al., 2012; Mak et al., 2012), and PPR (Ke et al., 2013; Yin et al., 2013; Gully et al., 2015) proteins in complex with their RNA/DNA targets confirmed that nucleic acids bind in an extended conformation to the inner concave surface of the solenoid, with each nucleotide contacting one, or at most two, consecutive repeats. Thus, repeats act in a modular fashion, with each repeat interacting with one nucleotide. Within a repeat, the side chain of a few residues at specific positions determines the recognized nucleotide, mainly by establishing hydrogen bonds with the nucleotide base. Nucleotide recognition by specific amino acid combinations was recently successfully predicted for PUF (Wang et al., 2002; Cheong and Hall, 2006; Filipovska et al., 2011), TALE (Boch et al., 2009; Moscou and Bogdanove, 2009), and PPR (Barkan et al., 2012; Takenaka et al., 2013; Yagi et al., 2013) proteins. Based on this “recognition code,” recombinant PUF and TALE proteins can be engineered to bind virtually any RNA or DNA target of interest (Christian et al., 2010; Cooke et al., 2011; reviewed in Bogdanove and Voytas, 2011; Filipovska and Rackham, 2012; Yagi et al., 2014). Furthermore, this modular architecture endows helical repeat proteins with a great versatility in vivo, as module reorganization through genetic recombination or substitutions of nucleotide-specifying amino acids will allow recognition of new targets.
Nuclear control of organelle gene expression is a key feature of eukaryotic cells that emerged after endosymbiosis (Choquet and Wollman, 2002; Woodson and Chory, 2008; Barkan, 2011). Indeed, every posttranscriptional step of organelle gene expression, RNA editing, splicing, processing, trimming, or translation activation is controlled in a gene- and, thus, sequence-specific manner by nucleus-encoded RNA binding proteins (denoted ROGEs for regulators of organelle gene expression; reviewed in Barkan and Goldschmidt-Clermont, 2000; Choquet and Wollman, 2002; Schmitz-Linneweber and Small, 2008; Woodson and Chory, 2008; Germain et al., 2013). Predictably, most ROGEs belong to helical repeat protein families, such as PPR, HAT, and mTERF (reviewed in Barkan and Small, 2014; Hammani et al., 2014). The great expansion of ROGEs in photosynthetic organisms contrasts with the poor conservation of ROGEs between different lineages. Despite their common structural organization, the various families of modular proteins do not generally share a common origin as their respective consensuses are not related. This suggests a high flexibility of nucleo-organelle interactions, well suited for rapid adaptation to new environmental constraints or ecological niches. For instance, PPR proteins, predominantly targeted to mitochondria or chloroplasts (Lurin et al., 2004) are particularly numerous in land plants with more than 450 members identified in Arabidopsis thaliana or rice (Oryza sativa). By contrast, the unicellular green alga Chlamydomonas reinhardtii possesses only 14 PPR proteins (Tourasse et al., 2013) but more than 120 members of another family of helical repeat proteins, poorly represented in land plants, the OPR (octotricopeptide repeat) proteins, defined by a degenerate motif of 38 residues (Auchincloss et al., 2002; Merendino et al., 2006; Loiselay, 2007; Eberhard et al., 2011; Rahire et al., 2012; Lefebvre-Legendre et al., 2015). As PPR repeats, OPR repeats are predicted to fold into a pair of antiparallel α-helices. Most OPR proteins are predicted to be targeted to organelles (Loiselay, 2007) where several have been shown to control the posttranscriptional steps of gene expression.
Most mutants affected in ROGEs described to date were screened for photosynthetic or respiratory defects after mutagenesis. They display a recessive phenotype, being defective in a gene product that, in the wild type, binds specifically to a given target transcript, usually in its 5′ untranslated region (5′UTR). In contrast, the two nuclear mutations that we describe below in Chlamydomonas appeared spontaneously and are dominant. They correspond to single amino acid substitutions in two OPR proteins that gain a new function by recognizing a new target in the coding region of two chloroplast transcripts, thus providing insights into the evolution of the nuclear control of organelle gene expression.
RESULTS
Isolation of the ncc2 Mutation, Which Alters the Stability of the petA Transcript
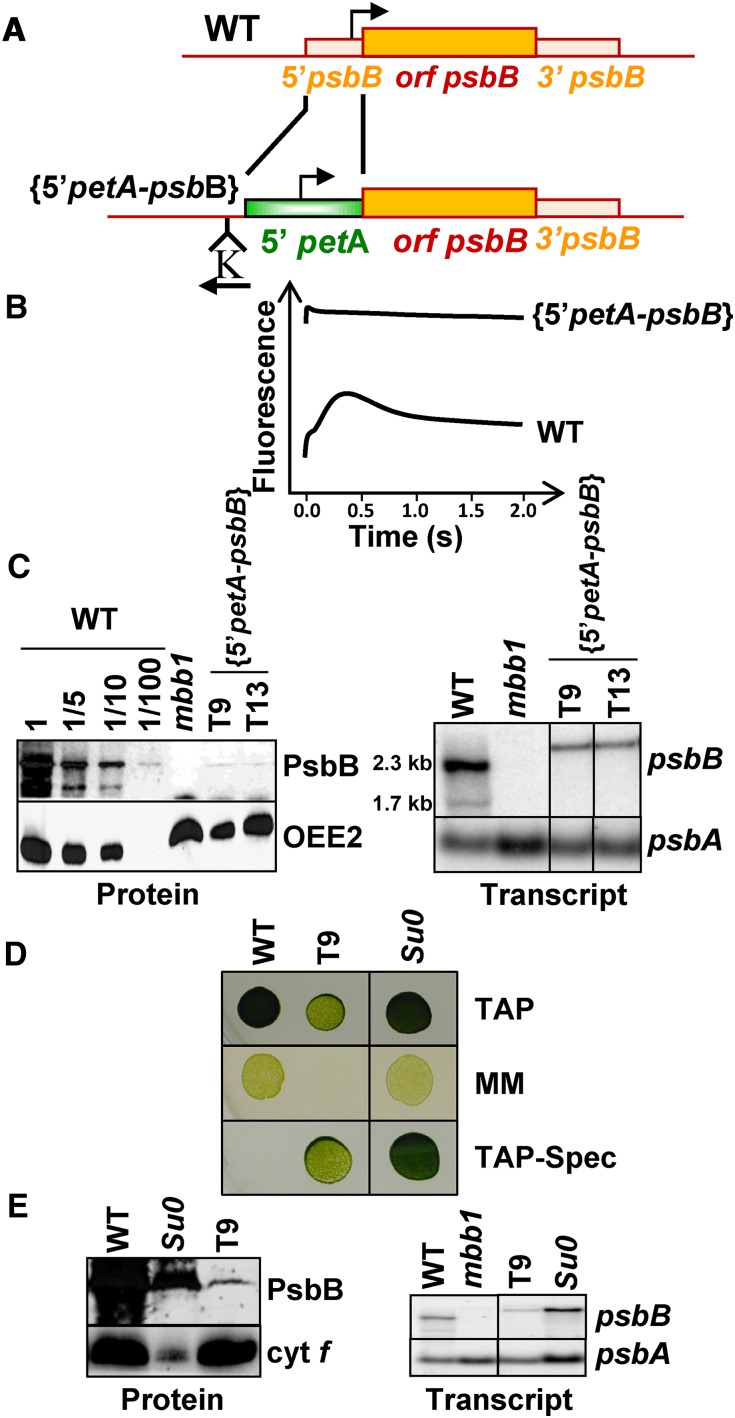
To better understand the regulation of chloroplast gene expression and identify new nuclear mutants, we used a chimeric construct to express a photosystem II (PSII) core protein under the control of the 5′UTR of an unrelated chloroplast gene. This chimera was expressed to reduced levels, insufficient for phototrophic growth, allowing us to isolate strains that had recovered phototropic growth. The 5′petA-psbB chimera is made of the psbB coding sequence (CDS), encoding the PSII core antenna PsbB, translated under the control of the 5′UTR of the chloroplast petA gene, encoding the cytochrome f subunit of the cytochrome b6f complex (Figure 1A). When inserted by chloroplast transformation at the psbB locus (see Table 1 for transformants generated in this work), this chimera only allows a low level of PsbB expression, insufficient to sustain the phototrophic growth of {5′petA-psbB} transformed cells, which display a PSII defective phenotype (Figures 1B to 1D). While plating the T9 {5′petA-psbB} transformant on minimal medium, we found one spontaneous phenotypic revertant, Su0, which had recovered phototrophic growth capability and increased accumulation of PsbB (Figures 1D and 1E). In crosses to a wild-type strain (see Table 2 for crosses performed in this work), all tetrad progeny inherited the 5′petA-psbB chimera, uniparentally transmitted by the mt+ {5′petA-psbB} parent, but the photoautotrophic capability segregated 2:2. This was indicative of a single nuclear suppressor mutation that we called ncc2 for nuclear control of chloroplast gene expression. We then crossed one phototrophic progeny (mt−) to the wild type (mt+) to eliminate the 5′petA-psbB chimera; two tetrad progeny had a wild-type phenotype, while the other two, presumably carrying the ncc2 mutation, although phototrophic, presented fluorescence induction kinetics typical of cytochrome b6f leaky mutants (Figure 2A). Indeed, their ΦPSII (0.2), much lower than that of the wild type (0.59), indicates a decreased electron flow downstream of PSII (Maxwell and Johnson, 2000). This correlates well with the 6-fold reduced abundance in cytochrome f, when compared with the wild type (Figure 2B). In 5-min pulse labeling experiments, cytochrome f synthesis was much reduced and hardly detectable in the ncc2 mutant (Figure 2C). When probing chloroplast transcripts for the major photosynthetic protein subunits by mRNA hybridization, we observed a selective drop in accumulation of the mature petA mRNA (below 5% of the wild-type level; Figure 2D), whereas petA processing intermediates were little affected (Figure 2D, asterisks, top panel). This suggests a preserved transcription but an accelerated degradation of the mature petA transcript in the mutant.
Figure 1.
Isolation of the ncc2 Mutant.
(A) Schematic description of the psbB gene in wild-type and {5′petA-psbB} transformants. Bent arrows indicate promoters, and the position and orientation of the selection cassette (K) are indicated.
(B) Fluorescence induction kinetics of dark-adapted wild-type and {5′petA-psbB} cells. The nearly constant fluorescence intensity over time in strain {5′petA-psbB}, as well as its high initial level, almost similar to the stationary level, is typical of leaky PSII mutants.
(C) Phenotypic characterization of {5′petA-psbB} transformed strains. Left: PsbB accumulation probed with a specific antibody in two independent transformants, in a dilution series of the wild type and in the mbb1-222E strain, defective for the accumulation of the psbB mRNA (Monod et al., 1992), as a specificity control. OEE2, whose accumulation is independent of PSII assembly (de Vitry et al., 1989), provides a loading control. Right: psbB mRNA accumulation in the same strains. Because of the larger size of the petA versus psbB 5′UTRs, the chimeric mRNA migrates more slowly than the endogenous psbB transcript.
(D) Growth properties of the wild-type, {5′petA-psbB}, and Su0 strains. Drops of liquid culture (2 × 106 cells⋅mL−1) were spotted on TAP medium and grown under dim light (10 μE⋅m−2⋅s−1, top), on minimal medium (MM) under high light (150 μE⋅m−2⋅s−1, intermediate), or on TAP supplemented with spectinomycin (500 μg⋅mL−1; lower panel). Pictures were taken after 10 d of growth.
(E) Phenotypic characterization of the Su0 strain. Accumulation of PsbB and cytochrome f (left) and of the psbB mRNA (right) in the same strains. psbA was used as a loading control.
Table 1. Strains Generated by Transformation in This Study.
| Recipient Strain | Transforming Plasmid | Transformed Strain | |
|---|---|---|---|
| Wild type | pK5′petA-psbB | {5′petA-psbB} | |
| Wild type | pKΔf::αTr | {Δf::αTr} | |
| Wild type | p5′dAfRK | {dAfR} | |
| Wild type | pf42St | {f42St} | |
| Wild type | pf145St | {f145St} | |
| ncc1 | pKatpAStb | ncc1 {atpASt} | |
| Chloroplast transformationa | Wild type | pKatpAM | {atpAM} |
| ncc1 | pKatpAM | ncc1 {atpAM} | |
| Wild type | pKpetAM | {petAM} | |
| ncc2 | pKpetAM | ncc2 {petAM} | |
| Wild type | pK5′petD::T2 | {5′petD::T2} | |
| ncc1 | pK5′petD::T2 | ncc1 {5′petD::T2} | |
| ncc2 | pK5′petD::T2 | ncc1 {5′petD::T2} | |
| Wild type | pKpetDCod::T2 | {petDCDS::T2} | |
| ncc2 | pKpetDCod::T2 | ncc2 {petDCDS::T2} | |
| Nuclear transformationc | Wild type | pNCC1M-HA | NCC1M-HA |
| Wild type | pNCC2M-HA | NCC2M-HA |
All recipient strains were mt+. Thanks to the uniparental inheritance of the mt+ chloroplast genome, this allowed the transmission of the chimeras introduced by transformation to the whole progeny of crosses. Recipient strains were also spectinomycin sensitive, with all chimera being associated with a spectinomycin resistance cassette for selection of transformants. Transformants were selected for resistance to spectinomycin (100 mg⋅mL−1) under low light (5 μE⋅m−2⋅s−1) and subcloned in darkness until they reached homoplasmy.
Reference: Eberhard et al. (2011).
Transformed strains were selected for resistance to paromomycin (10 mg⋅mL−1) under low light (5 μE⋅m−2⋅s−1).
Table 2. Strains Generated by Crosses in This Study.
| mt+ Parent | mt− Parent | Progeny |
|---|---|---|
| ncc2 {5′petA-psbB} (Su0) | Wild type | ncc2 mt− {5′petA-psbB} |
| Wild type | ncc2 {5′petA-psbB} (Su0) | ncc2 |
| arg2 [1] | ncc2 | ncc2 arg2 |
| arg7 [1] | ncc2 | ncc2 arg7 |
| ncc2 arg7 | ncc2 arg2 | Diploid ncc2/ncc2 |
| arg7 | ncc2 arg2 | Diploid ncc2/NCC2 |
| arg7 | arg2 | Diploid NCC2/NCC2 |
| {Δf::αTr} [2] | ncc2 | ncc2 {Δf::αTr} |
| {dAfR} [2] | ncc2 | ncc2 {dAfR} |
| {petASt} [3] | ncc2 | ncc2 {petASt} |
| {f42St} [2] | ncc2 | ncc2 {f42St} |
| {f145St} [2] | ncc2 | ncc2 {f145St} |
| {FAFA} ncc1 [4] | ncc2 | ncc1 ncc2 {FAFA} |
| {atpASt} [5] | ncc1 | ncc1 {atpASt} |
References: [1] (Ebersold, 1967; Harris, 1989); [2] this work; [3] (Boulouis et al., 2011); [4] (Drapier et al., 2002); [5] (Eberhard et al., 2011). By convention, chloroplast genotypes, when relevant, follow the nuclear genotype and are written between braces.
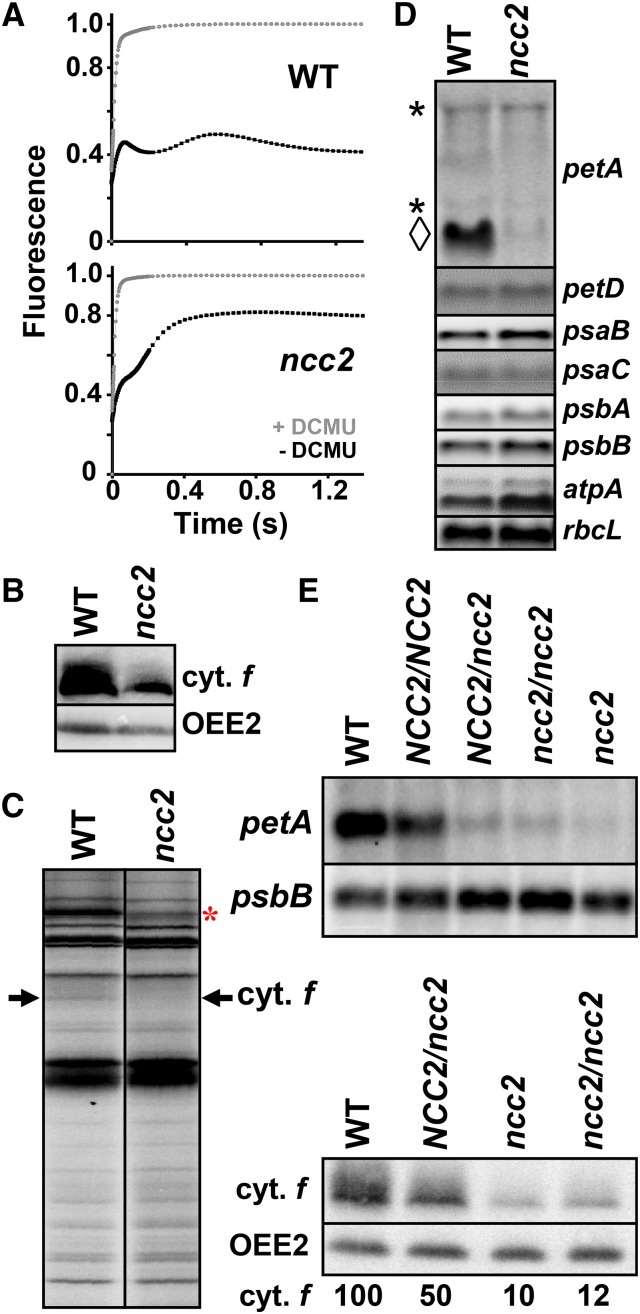
Figure 2.
The Dominant ncc2 Mutation Confers a b6f Leaky Phenotype Due to Reduced Accumulation of the petA mRNA.
(A) Fluorescence induction kinetics of dark-adapted ncc2 and wild-type cells. Black and gray curves show the kinetics recorded in the absence and in the presence, respectively, of DCMU (5 μM), which blocks electron transfer downstream of PSII. Maximal fluorescence levels in the presence of DCMU were normalized to 1.
(B) Cytochrome f accumulation in wild-type and ncc2 strains. OEE2 provides a loading control.
(C) Translation of chloroplast genes determined by 5 min 14C-acetate (5 μCi⋅mL−1) pulse-labeling experiments performed in the presence of cycloheximide (10 μg⋅mL−1) to block cytosolic translation. The arrow indicates the position of cytochrome f. The origin of the reduced rate of synthesis of Rubisco LS (red asterisk) in the ncc2 mutant, despite the unchanged accumulation of the rbcL transcript (D), is unknown.
(D) Accumulation of representative transcripts for chloroplast photosynthesis genes in wild-type and ncc2 strains, assessed by RNA gel blots using the probes indicated on the right of the panel. For the petA gene, the diamond indicates the mature transcript, while asterisks point to precursor RNA species.
(E) Accumulation of petA mRNA and cytochrome f, determined as above, in diploid strains either homozygous or heterozygous for the ncc2 mutation. Wild-type and ncc2 strains are shown for comparison. psbB mRNA and OEE2 provide loading controls in RNA and immunoblots, respectively.
This phenotype readily explains the suppressor effect of the ncc2 mutation on the expression of the 5′petA-psbB chimera. Cytochrome f is a CES (for controlled by epistasy of synthesis) protein, whose rate of synthesis is controlled by its assembly within cytochrome b6f (Choquet et al., 1998, 2003; reviewed in Choquet and Wollman, 2008). In the wild type, a small fraction of cytochrome f remains unassembled and downregulates the translation of the petA mRNA or of any chimera driven by the petA 5′UTR, such as the 5′petA-psbB chimera here, by 3-fold. The ncc2 mutant synthesizes reduced amounts of cytochrome f (Figure 2C), which precludes significant accumulation of unassembled cytochrome f, thereby releasing the translational downregulation of 5′petA-driven genes. The 5′petA-psbB chimera, now expressed 3-fold more in the ncc2 mutant than in the wild type, allows PsbB synthesis at rates sufficient to sustain photoautotrophic growth, even though PsbB levels remain lower than those produced by the endogenous psbB.
We previously identified two nuclear genes, MCA1 and TCA1 (for maturation/stability and translation of the cytochrome b6f complex subunit PetA, respectively), which contribute to the protection of the petA transcript against exonucleolytic degradation (Wostrikoff et al., 2001; Loiselay et al., 2008). However, crosses of ncc2 to either mca1-2 or tca1-2 mutants (Supplemental Table 1) clearly showed genetic independence, indicating that the ncc2 phenotype was not due to altered expression of MCA1 or TCA1.
ncc2 Is a Dominant Mutation
To determine whether the ncc2 mutation was recessive or dominant, we constructed vegetative diploid strains either homozygous or heterozygous for the ncc2 locus, as described in Table 2. In contrast to homozygous NCC2/NCC2 diploids, the heterozygous ncc2/NCC2 and homozygous ncc2/ncc2 diploids displayed the same 20-fold reduced accumulation of the petA mRNA as the haploid ncc2 parental strain (Figure 2E). However, some gene dosage effect partially damped the effect of the ncc2 mutation at the protein level, since cytochrome f abundance only decreased 2-fold in ncc2/NCC2 heterozygotes (Figure 2E). In contrast to almost all mutations in ROGE genes described to date in Chlamydomonas, the ncc2 mutation is not recessive but dominant. It thus strikingly resembles the other dominant nuclear mutation acting on chloroplast transcript isolated in Chlamydomonas, ncc1, which specifically destabilizes the monocistronic transcript of the chloroplast atpA gene encoding the α subunit of ATP synthase (Drapier et al., 2002). Similar to the ncc1 mutation, the ncc2 mutation is unlikely to be a loss-of-function allele, but rather likely modifies some gene product, so that it now acts on the petA mRNA.
The ncc2 Mutation Causes NNC2 to Act on the petA CDS, Similar to the Action of Mutated NCC1 on atpA
In Chlamydomonas, the known ROGEs that control the stability of a chloroplast transcript target its 5′UTR, with the exception of the protein affected by the dominant ncc1 mutation, which targets the atpA CDS (Drapier et al., 2002). We thus tested which part of the petA mRNA was targeted for degradation in the ncc2 mutant, using two distinct petA chimeras.
At the petA locus, we first substituted the sequence coding for mature cytochrome f by that encoding the ATP synthase subunit α, truncated to maintain a similar mRNA length, fused in frame with the sequence coding for the lumen targeting peptide of cytochrome f (Δf::αTr chimera; Figure 3A). All chimeras used in this study were associated with an aadA selection cassette (Table 1), and transformants were selected for resistance to spectinomycin. One of the resulting transformants, {Δf::αTr}, was crossed to the ncc2 mutant. RNA gel blot analysis showed no hybridization with an intragenic petA probe in the parental strain {Δf::αTr} because this sequence had been replaced by that of atpA. An atpA probe detected, in addition to the endogenous atpA transcripts, the Δf::αTr chimeric transcript in the parental transformant and in all progeny (Figure 3B). This was also true for a larger and minor transcript (indicated by an asterisk), resulting from cotranscription of the chimera with the downstream aadA cassette. Strikingly, these two bands accumulated to the same level in all progeny, irrespective of their ncc2 or NCC2 genotypes. Thus, chimeric transcripts lacking the sequence encoding mature cytochrome f are no longer destabilized by the ncc2 mutation.
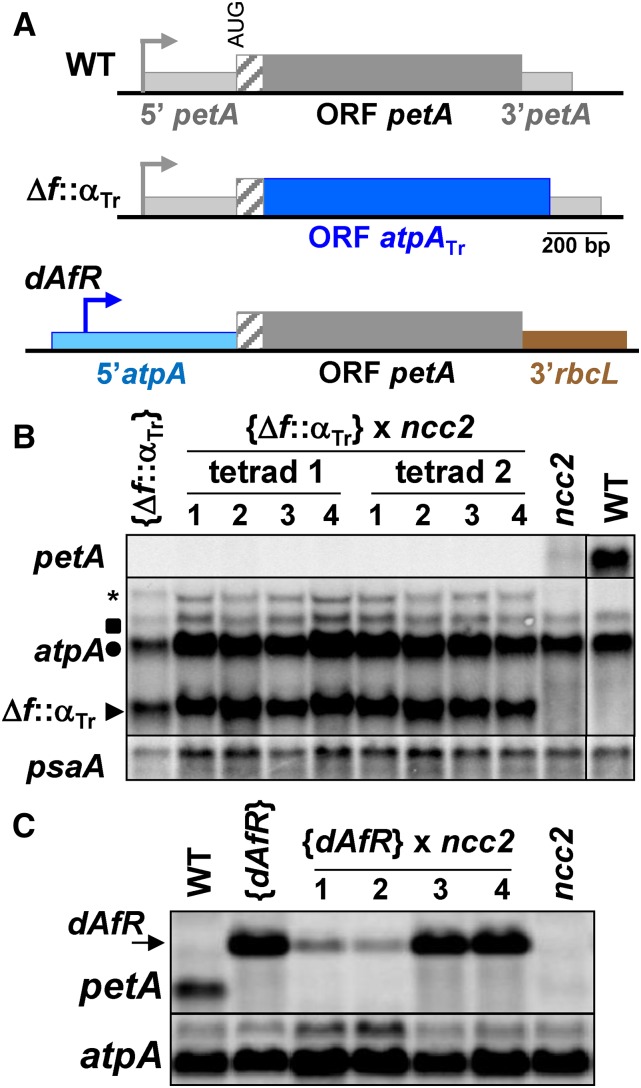
Figure 3.
The ncc2 Mutation Targets the petA CDS.
(A) Scheme of the chimeras. Sequence encoding the mature cytochrome f is shown as a dark-gray rectangle and that encoding the lumen targeting peptide as a hatched rectangle. petA 5′ and 3′UTRs are represented by thinner light-gray rectangles. petA promoter is indicated by a bent arrow. In Δf::αTr chimera, the region encoding mature cytochrome f was replaced by the first 944 nucleotides of the atpA CDS, depicted as a blue rectangle. In dAfR chimera, the petA promoter and 5′UTR regions were replaced by the corresponding atpA regions (pale-blue rectangle), while petA 3′UTR was replaced by that of the rbcL gene (brown rectangle).
(B) Transcript accumulation in tetrad progeny from the cross {Δf::αTr} × ncc2 and in wild-type and parental strains. RNA gel blots were hybridized with probes derived from petA, atpA, and psaA (loading control) CDSs, as indicated on the left. Positions of the endogenous mono- and dicistronic atpA transcripts are indicated by a circle and a square, respectively. Arrow points to the position of the major Δf::αTr chimeric transcript, while the asterisk indicates a minor cotranscript that includes the downstream aadA resistance cassette.
(C) petA and atpA (loading control) transcript accumulation in tetrad progeny from the cross {dAfR} × ncc2 and in wild-type and parental strains. Arrow points to the position the chimeric dAfR transcript.
Symmetrically, the petA coding region, fused to the atpA 5′UTR and rbcL 3′UTR in the dAfR chimera (Figure 3A), was introduced by chloroplast transformation in a wild-type strain. After crossing with ncc2, all tetrad progeny expressed the chimeric petA transcript, larger than the genuine petA transcript. However, two members of each tetrad, likely carrying the ncc2 allele, showed a markedly reduced accumulation of the chimeric transcript, similar to the endogenous petA mRNA in the ncc2 parent (Figure 3C). The petA CDS is thus not only required but also sufficient for transcript degradation in the ncc2 background.
Active Translation of the Target Transcript Is Required for Its Destabilization in ncc2, but Not ncc1, Backgrounds
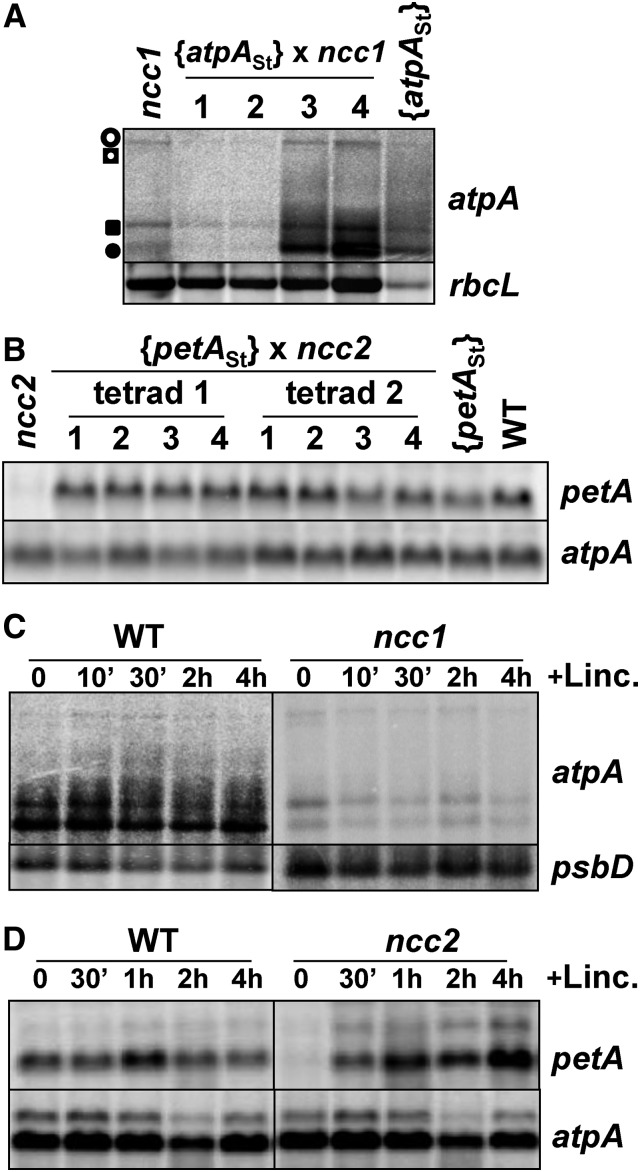
The ncc1 and ncc2 mutations target the CDSs of atpA and petA, respectively; we wondered whether translation of these transcripts was required for their degradation in the ncc1 and ncc2 mutants. To test this, we used {atpASt} and {petASt} strains, which express untranslatable atpA and petA mRNAs that have their initiation codons replaced by a stop codon (Boulouis et al., 2011; Eberhard et al., 2011). We crossed these strains with the ncc1 and ncc2 mutants. In the {atpASt} cells, the atpA transcript pattern is similar to that of the wild-type atpA gene, which comprises four mRNAs transcribed from the tetracistronic atpA gene cluster (Drapier et al., 1998). The ncc1 mutation markedly decreases the amount of monocistronic atpA transcript relative to polycistronic forms (Drapier et al., 2002). We observed a strong reduction in the abundance of monocistronic atpA transcripts in two progeny (1 and 2) from the {atpASt} × ncc1 cross (Figure 4A). Thus, the ncc1 mutation destabilizes the atpA transcript, even when not translated. In contrast, destabilization of the petA transcript by the ncc2 mutation likely depends on active translation, as the accumulation of the petASt transcript remained high in all tetrad progeny (Figure 4B).
Figure 4.
The ncc2, but Not ncc1, Phenotype Is Observed Only When the Target RNA Is Translated.
(A) atpA transcript accumulation in tetrad progeny from the cross {atpASt} × ncc1 and in parental strains. Position of the four transcripts from the atpA tetracistronic transcription unit is indicated. Loading control: rbcL mRNA.
(B) petA transcript accumulation in tetrad progeny from the cross {petASt} × ncc2 and in wild-type and parental strains (loading control: atpA).
(C) and (D) Accumulation of atpA (C) and petA (D) transcripts in ncc1 (C), ncc2 (D), and wild-type strains incubated in the presence of lincomycin for the indicated times. Loading controls: psbD (C) and atpA (D).
Alternatively, the petA AUG initiation codon could be part of the site recognized by the mutated NCC2. We thus studied transcript accumulation over time in cultures treated with lincomycin, an inhibitor of chloroplast translation. The drug did not affect the accumulation of atpA transcripts in ncc1 cells (Figure 4C). By contrast, the accumulation of petA transcripts in the ncc2 mutant increased spectacularly from barely detectable before lincomycin addition, up to wild-type levels after a 4-h treatment (Figure 4D). Thus, active translation of petA transcripts is required for their ncc2-dependent destabilization, whereas the ncc1 mutation destabilizes the atpA transcript independent of translation.
Toward a More Accurate Localization of the Targets of the ncc2 and ncc1 Mutations
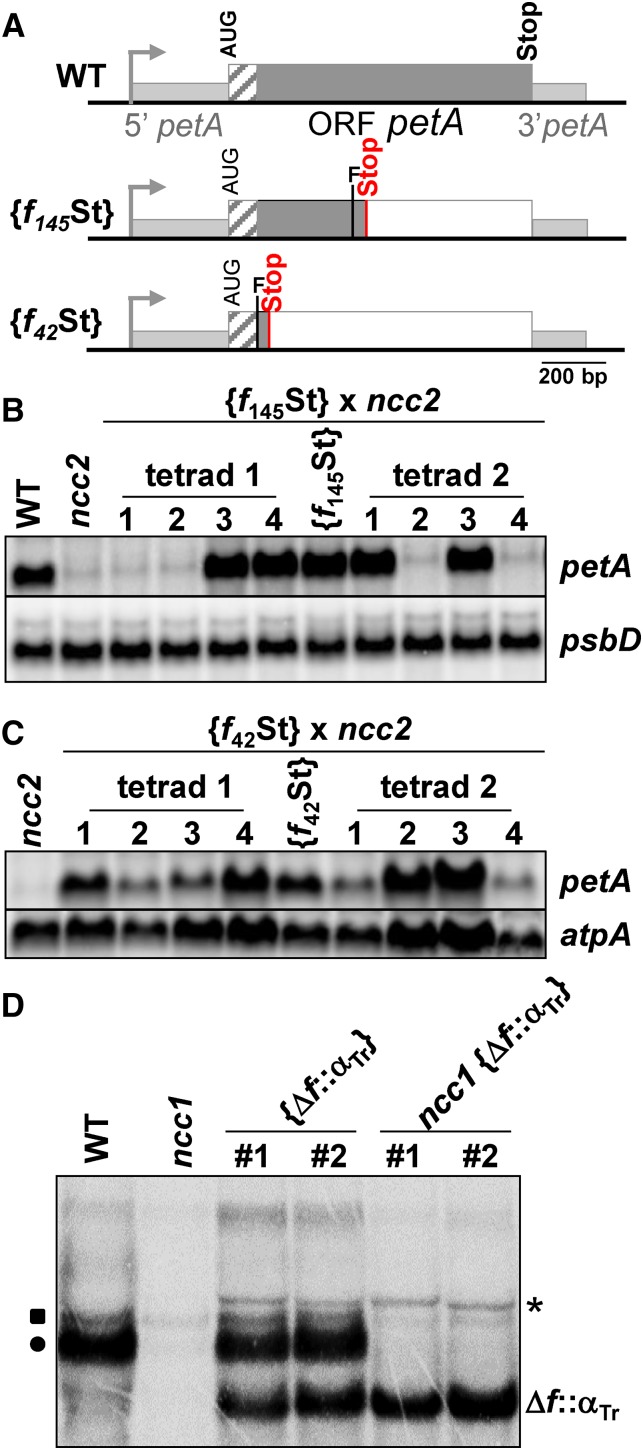
To better understand how translation triggers petA mRNA degradation in strain ncc2, we introduced frame shifts within the petA CDS after nucleotides 93 or 390, causing premature abortion of translation after codons 42 and 145, respectively (Figure 5A). The f42St mutation preserves the translation of the lumen targeting peptide, which is not sufficient to confer ncc2 sensitivity to a translated sequence (Figure 3C). Tetrad analysis after crossing transformants {f42St} and {f145St} to a ncc2 strain showed that the ncc2 mutation still decreased the abundance of the f145St transcript 20-fold (Figure 5B). In contrast, transcripts from the f42St construct were much less sensitive to the ncc2 mutation as their abundance only decreased 2-fold in ncc2 progeny (Figure 5C). Thus, the ncc2-mediated degradation of petA transcripts starts early after the lumen targeting peptide has been synthesized and is completed before translation reaches the 145th codon of cytochrome f.
Figure 5.
Narrowing Down the Target Sequences of the ncc1 and ncc2 Mutations.
(A) Schematic representation of the mutant petA genes, presented as in Figure 3A. The “F” indicates the position of the introduced frame shifts, while gray and white rectangles show translated and untranslated petA sequences, respectively.
(B) and (C) petA transcript accumulation in tetrad progeny from crosses {f145St} × ncc2 (B) and {f42St} × ncc2 (C). Loading controls: psbD (B) and atpA (C).
(D) Accumulation of atpA-hybridizing transcripts in wild-type and ncc1 strains transformed with the Δf::αTr chimera (Figure 3A). The probe hybridizes with the chimeric transcript, either alone (Δf::αTr) or cotranscribed with the aadA cassette (asterisk) and with endogenous mono- (circle) and dicistronic (square) atpA mRNA.
The target of the NCC1 mutated factor was previously localized in the last 1360 bases of the atpA mRNA (Drapier et al., 2002). We narrowed down this region by transforming the chloroplast of the ncc1 strain with the Δf::αTr construct, which contains the atpA CDS deprived of its last 579 nucleotides (Figure 3A). In contrast to the endogenous atpA transcript, the chimeric Δf::αTr mRNA was not destabilized in the ncc1 background (Figure 5D), suggesting that the ncc1 mutation targets the last 579 nucleotides of the atpA transcript.
Identification of the ncc2 and ncc1 Mutations
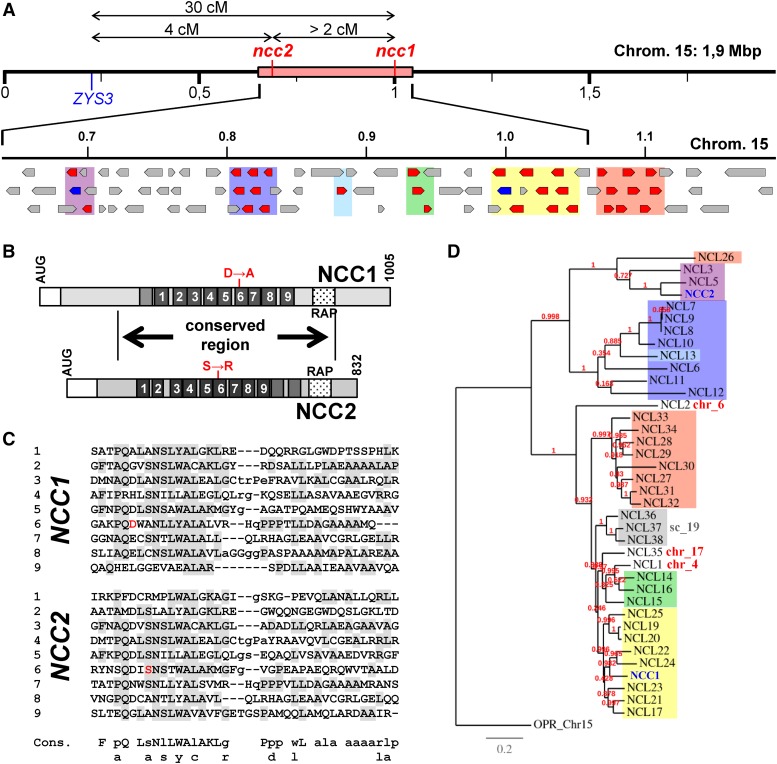
Because the ncc1 and ncc2 mutations are dominant and allow photoautotrophic growth, we could not clone the genes by complementation. Thus, we mapped the mutations onto the Chlamydomonas nuclear genome by crossing the two mutants with the S1D2 strain, which shows a profusion of polymorphisms compared with laboratory strains (Gross et al., 1988; Kathir et al., 2003; Rymarquis et al., 2005). We harvested ∼50 tetrads from each cross and picked one mutant per tetrad. ncc2 progeny were identified based on their fluorescence phenotype. To identify ncc1 progeny, we used for the cross the nonphotosynthetic strain ncc1 {FAFA}, bearing the CDS of atpA fused to the 5′ and 3′UTRs of the petA gene. This chimera is expressed at a low level in a wild-type background and, when combined with the ncc1 mutation, prevents phototrophic growth (Drapier et al., 2002). By PCR-based mapping with diagnostic markers on all chromosome arms (Kathir et al., 2003), we found linkage for both mutations to the ZYS3 marker on the long arm of chromosome 15 (96 and 71% for the ncc2 and ncc1 mutations, respectively; Figure 6A). Using the new markers listed in Supplemental Table 2, the ncc2 mutation was localized between nucleotides 659,176 and 1,063,367 (Chlamydomonas genome v5.5, available on Phytozome at http://phytozome.jgi.doe.gov/; Merchant et al., 2007; Blaby et al., 2014).
Figure 6.
The NCC1 and NCC2 Genes.
(A) Top: Genetic and molecular map of the ncc1 and ncc2 loci. Locations on chromosome 15 of the ZYS3 marker and ncc1 and ncc2 mutations are shown, along with genetic distances. The origin of the discrepancy between genetic distances determined in the three point test has not been investigated but likely originates from the poor fluorescence identification of some double mutants. The pink rectangle represents the molecular region containing the ncc2 mutation, as determined by map-based cloning. Bottom: Physical map of the NCL gene cluster on chromosome 15. NCL genes are drawn in red, NCC1 and NCC2 in blue, and non-OPR genes in gray.
(B) Schematic representation of the NCC1 and NCC2 proteins. White rectangles depict the chloroplast transit peptide predicted by the ChloroP program. Dark-gray boxes represent the OPR repeats. Punctuated rectangles show the position of the RAP domains. The positions of the two substitutions in the ncc1 and ncc2 strains are shown in red. A highly conserved region (57% identity and 69% similarity) between the two proteins is indicated.
(C) Alignment of OPR repeats in the NCC1 (top) and NCC2 (bottom) proteins, with residues corresponding to the consensus (bottom) shaded in gray. Mutated amino acids in ncc1 and ncc2 strains are shown in red.
(D) Phylogeny of NCL proteins. Maximum likelihood tree of the NCL proteins using Chlre_OPR68 as an outgroup. Branch length represents the estimated rate of amino acid substitution. Colored boxes indicate the genomic location of the corresponding NCL genes, as indicated in (A). NCC1 and NCC2 are written in blue.
We then crossed strains ncc2 and ncc1 {FAFA} and recovered a few double mutants among 80 tetrads, identified by their ncc2-like fluorescence induction kinetics and their poor phototrophic growth, due to the ncc1 {FAFA} combination. They showed decreased levels of both atpA and petA transcripts, which were comparable to those in the respective parental ncc1 {FAFA} and ncc2 strains. Accordingly, the accumulation of subunit α and cytochrome f was decreased, as shown in Supplemental Figure 1 for one typical double mutant. Its genome was sequenced using the Illumina platform (2 × 100 bp). After eliminating synonymous and intergenic polymorphisms and those also present in a collection of quasi-isogenic photosynthetic mutants carrying neither ncc1 nor ncc2 mutation, we were left with two point mutations in the interval determined by molecular mapping: an A → C substitution at position 693,478 and a double substitution AT → GG at position 1,001,095/6. These mutations cause a single amino acid substitution in two distinct genes encoding OPR proteins: Cre15.g638950.t1 and Cre15.g640400.t1, whose gene models are fully supported by EST evidence displayed at http://genomes.mcdb.ucla.edu/Cre454/.
Direct sequencing of PCR products amplified from ncc1 and ncc2 strains (for primers, see Supplemental Table 3) localized the A → C substitution in the genome of the ncc2 mutant but not in that of the ncc1 mutant. It leads to a S431→R substitution in Cre15.g640400.t1, therefore named NCC2. Conversely, strain ncc1, but not strain ncc2, carried the AT → GG mutation, leading to the D568 → A substitution in Cre15.g638950.t1, hereafter called NCC1. The NCC1 and NCC2 proteins both contain nine OPR repeats, the mutations changing the 6th residue of the 6th OPR repeat of NCC1 and the 8th residue of the 6th OPR repeat of NCC2 (Figures 6B and 6C). In addition, both proteins also contain a RAP domain (RNA Binding Abundant in Apicomplexa; Lee and Hong, 2004) at their C terminus.
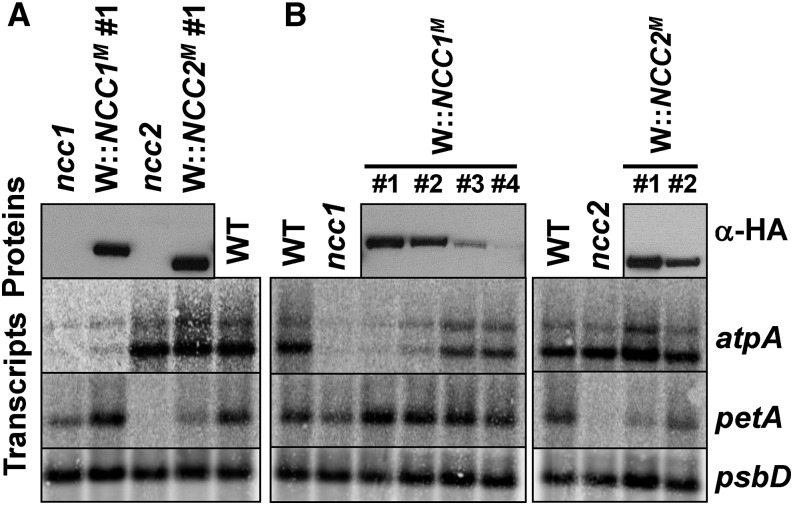
Transgenic Expression of the Mutated NCC1 or NCC2 Protein Confers the ncc1 or ncc2 Phenotype to Transformed Strains
Since the ncc1 and ncc2 mutations are dominant, mutated copies of the NCC1/NCC2 genes, introduced in the wild type, should confer the ncc1/ncc2 phenotypes to transformants. The genomic sequences coding for the mutated NCC1 and NCC2 proteins (hereafter referred to as NCC1M and NCC2M) were fused to a triple HA tag to allow their immunodetection and introduced into Chlamydomonas by transformation, using a vector carrying the aphVIII gene (Sizova et al., 1996). Clones, selected for paromomycin resistance, were screened with an HA-specific antibody for expression of NCC1M or NCC2M. As predicted from their respective molecular mass, NCC2M migrates slightly faster than NCC1M (Figure 7A). When assessing the accumulation of the atpA/petA transcripts by RNA gel blots, transformants expressing NCC1M showed a marked decrease in the accumulation of the atpA monocistronic transcript. In independent transformants, the higher the accumulation of NCC1M, the less atpA monocistronic transcripts (Figure 7B), supporting an NCC1M-mediated degradation of the atpA mRNA. Similarly, the accumulation of the petA mRNA was strongly reduced in transformants expressing NCC2M (Figure 7A), even though still higher than in the ncc2 strain, probably because of insufficient expression level of the transgenic NCC2M. In two transformants expressing NCC2M, the extent of petA mRNA destabilization correlated with the abundance of the protein (Figure 7B).
Figure 7.
Expression of NCC1M (/NCC2M) Confers the ncc1 (/ncc2) Phenotype to Transformed Strains.
(A) Accumulation of the petA and atpA mRNAs in wild-type, ncc1, and ncc2 strains and in two transformed strains expressing NCC1M and NCC2M, as shown with an antibody against the HA tag (upper panel).
(B) Left: Decreasing accumulation of atpA mRNA in a series of transformants accumulating increasing amounts of NCC1M. Right: Two transformants illustrating the negative correlation between accumulations of petA mRNA and NCC2M.
Identification of the NCC1M and NCC2M Targets
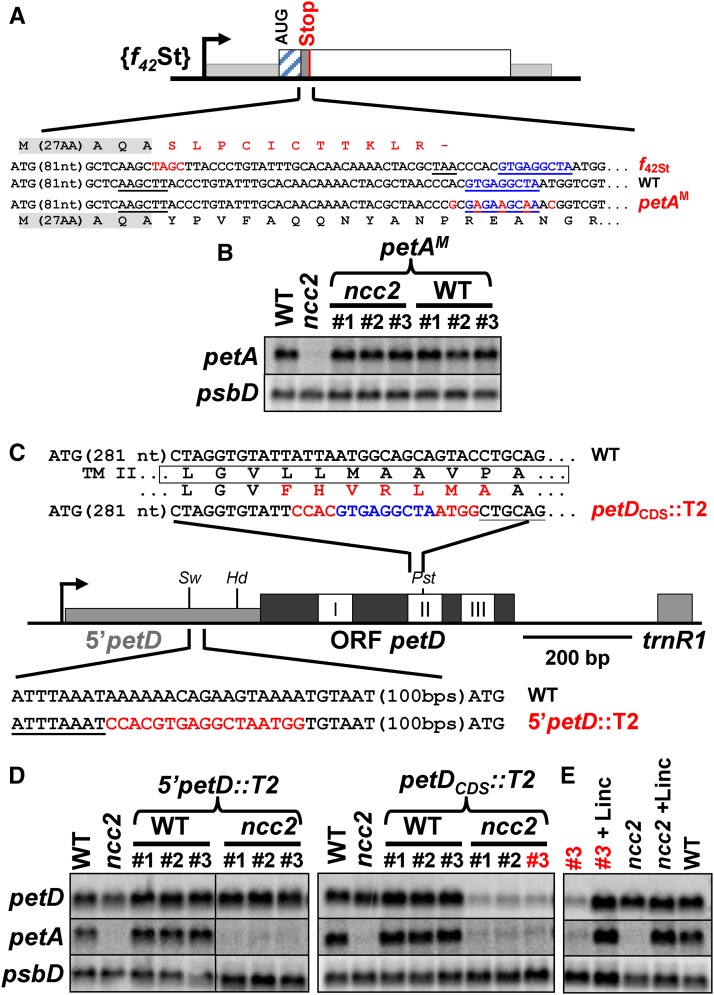
Based on a preliminary version of the code for nucleotide recognition by OPR repeats (Y. Choquet, unpublished results), we predicted that NCC1M and NCC2M should target the NAGNGATTA and GTGAGGNTA sequences, respectively, found at positions 1066 to 1075 and 130 to 139 of the atpA and petA CDS (Figures 8A and 9A). The NCC2M target is located 5 bp downstream of the premature stop codon in the chimera f42St, a region critical for the degradation of the petA transcript in the ncc2 mutant (Figures 5C and 9A).
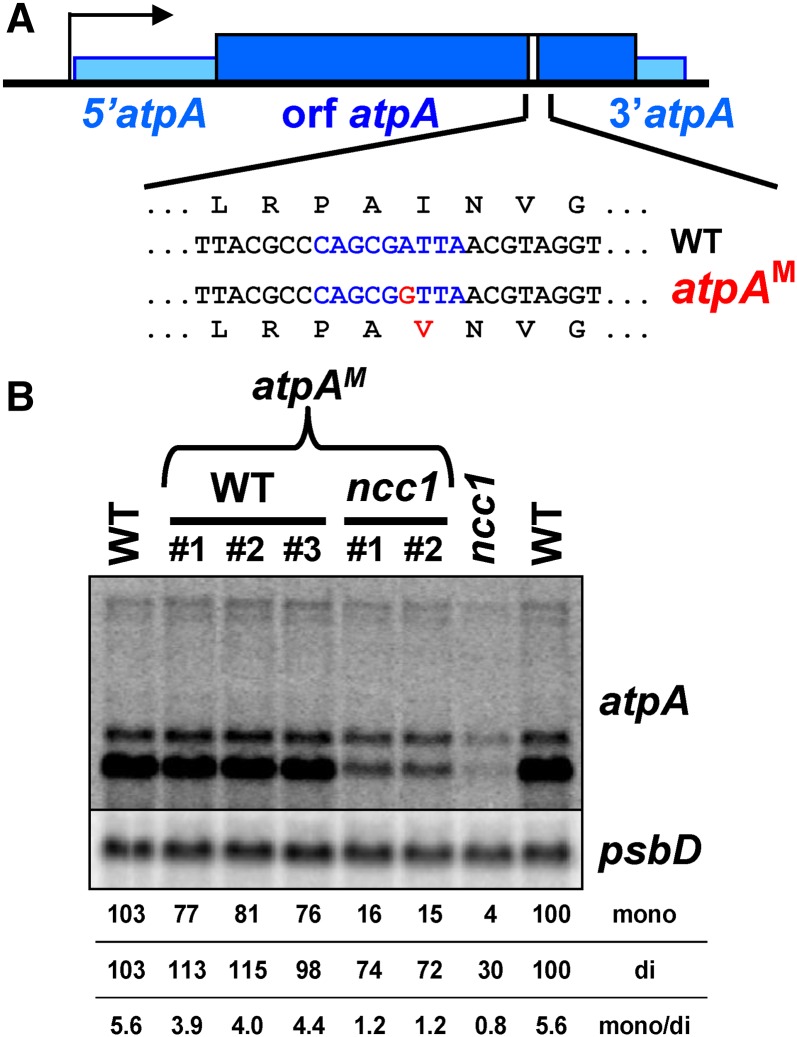
Figure 8.
Identification of the Target of NCC1M.
(A) Location of the target of NCC1M, written in blue along the atpA gene. The mutation introduced in the atpAM construct is shown in red.
(B) Accumulation of the atpA transcript in wild-type and ncc1 strains transformed with the atpAM construct. Independent transformants are presented for each background. Untransformed wild-type and ncc1 strains are shown for comparison. Loading control: psbD.
Figure 9.
Identification of the Target of NCC2M.
(A) Location of the target of NCC2M, written in blue along the petA gene. Silent mutations introduced in the petAM construct are written in red. Residual translation of petA in the f42St mutant, downstream of the frame shift, is shown. The HindIII site used to introduce the frame shift indicated in red is underlined.
(B) Accumulation of the petA transcript in wild-type and ncc2 strains transformed with the petAM construct. Three independent transformants (#1 to #3) are presented for each background. Untransformed wild-type and ncc2 strains are shown for comparison. Loading control: psbD.
(C) Insertion sites of the NCC2M target within the petD gene. Schematic representation of the petD gene with the 5′UTR and CDS drawn as thin light-gray and thick dark-gray rectangles, respectively, while the three white boxes represent transmembrane helices. Relevant restriction sites (SwaI, HindIII, and PstI) are indicated. Nucleotide regions surrounding the NCC2M target are shown with restriction sites underlined.
(D) Accumulation of petD transcript in wild-type and ncc2 strains transformed with the 5′petD::T2 (left) and petDCod::T2 (right) constructs. Three independent transformants (#1 to #3) are presented for each background. Untransformed wild-type and ncc2 strains are shown for comparison. A petA probe reveals ncc2 background. Loading control: psbD.
(E) petD and petA mRNAs accumulation in strains ncc2 and ncc2 {petDCod::T2} #3, before and after a 4-h incubation with lincomycin. The wild type is shown for comparison, and psbD was the loading control.
To confirm these target sequences, we substituted the nucleotide presumably recognized by the mutated OPR repeat in the predicted target of NCC1M, leading to the conservative mutation I391→V (Figure 8A). After transformation of wild-type and ncc1 strains, the mutated atpA gene, atpAM, replaced the endogenous atpA gene. In the wild-type background, this mutation slightly decreased (∼20%) the accumulation of the monocistronic atpA transcript and slightly increased that of the dicistronic atpA transcript (Figure 8B). In contrast, in the ncc1 background, the mutation increased the level of atpA transcripts, with a 4-fold increase in monocistronic transcript and a 2-fold increase in dicistronic transcript. Thus, the ability of NCC1M to destabilize the atpA transcripts was reduced, but not abolished, by a point mutation in the predicted target.
The partial suppression of the NCC1M effect, when only one nucleotide was substituted in its target, prompted us to introduce several silent mutations when assessing the target sequence of NCC2M, converted from GTGAGGCTA to GAGAAGCAA (petAM; Figure 9A). Following transformation, the mutation petAM had no effect on the petA mRNA steady state level in the wild type. In stark contrast, it completely abolished the ncc2 phenotype as it led to a spectacular restoration of the accumulation of petA mRNA in the ncc2 strain, up to wild-type level (Figure 9B), demonstrating that we indeed identified the target of NCC2M.
Wondering whether the target of NCC2M would still be recognized when placed in a different nucleotide context, we inserted it either in the petD 5′UTR (5′petD::T2) or within the petD CDS (petDCDS::T2) (Figure 9C). We used these two insertion sites to assess whether active translation would still be required for the destabilization of the petD transcript. When transformed with the 5′petD::T2 construct, both wild-type and ncc2 strains remained phototrophic and the mutated petD transcript accumulated to the same level as does the original petD transcript in the wild type (Figure 9D). In contrast, chloroplast transformants expressing the petDCDS::T2 chimera were incapable of phototrophic growth, in wild-type or in ncc2 backgrounds, as expected from the probable impairment of transmembrane helix II insertion within the membrane. In the ncc2 background, the accumulation of the chimeric petD mRNA was largely prevented, whereas it remained unaffected in the wild type, when compared with that of the wild-type petD transcript (Figure 9D). Moreover, a 4-h lincomycin treatment of the ncc2 {petDCDS::T2} strain restored the accumulation of the chimeric petD mRNA to a wild-type level, confirming the dependence of the ncc2 phenotype on the translation of its target (Figure 9E).
NCC1 and NCC2 Belong to the NCL Subfamily of Paralogs Encoding OPR-RAP Proteins
In BLAST searches against the Chlamydomonas proteome, NCC1 and NCC2 hit each other with low E-values (< 10−100), along with a set of 36 closely related proteins (Supplemental Table 4). The similarity between these proteins was much higher than with any other protein in the nonredundant Protein Database (http://www.ncbi.nlm.nih.gov/protein/), including those of the closely related alga Volvox carteri (E > 10−30). Thus, NCC1 and NCC2 are part of a Chlamydomonas-specific group of 38 highly similar paralogs that will hereafter be called NCL, for NCC-Like (described in Supplemental Data Set 1). As shown in the alignment (Supplemental Figure 2A), NCL proteins comprise a highly conserved central region, containing 7 to 12 OPR repeats and an ∼60-residue C-terminal RAP domain. By contrast, N- and C-terminal extensions, upstream and downstream of this conserved block, are more divergent.
Strikingly, 32 of these 38 NCL genes are clustered on the long arm of chromosome 15 between positions 686,690 and 1,113,927 (Figure 6A), while three are found on the as yet unassembled scaffold 19 and 3 isolated genes lie on chromosomes 4, 6, and 17.
At variance with the bulk of OPR-encoding genes, which have an average number of 12 introns regularly scattered along the CDS, most NCL CDSs contain a single intron, at a conserved position with respect to the protein sequence (Supplemental Data Set 1 and Supplemental Figure 2). Most probably this intron was already present in the ancestor gene that gave rise to the NCL family by duplications. Phylogeny of NCL proteins was studied using maximum likelihood inference (Figure 6D) and was well correlated with the position and orientation of NCL genes along the cluster (Figures 6A and 6D), suggesting that local tandem gene duplications played a major role in the expansion of the NCL family. This evolution is probably still going on: the adjacent NCL7, NCL8, and NCL9 genes probably originated from very recent duplication events, as NCL7 and NCL8 only differ from NCL9 by 6 and 15 bases, respectively, along the 2277-bp CDS. This leads to three amino acid substitutions in NCL8 and to very limited changes in the very C-terminal end of NCL7. Conversely, the NCL family contains at least two inactivated genes, NCL2 and NCL21, whose sequences, although similar to that of other NCL genes over 598 and 1171 codons, respectively, are interrupted by premature stop codons at positions 200 and 305. According to expression data available on Phytozome, NCL2 is probably not transcribed. Some other NCL genes, such as NCL12 and NCL16, encode truncated proteins comprising only the N-terminal extension and the first four OPR repeats, which explains their higher BLAST E-value (Supplemental Table 4).
NCL Genes Evolved under Diversifying Selection Pressure
Within the conserved block, successive OPR repeats, although all obeying the OPR consensus, significantly differ one from another (Supplemental Figure 2B). By contrast, the sequence of a given repeat is remarkably conserved between the different NCL proteins (Supplemental Figure 2A), except at some variable positions (e.g., 3 and 6). This, together with the spontaneous appearance of the ncc1 and ncc2 mutations, suggests that the NCL proteins may be under diversifying selection, i.e., that nonsynonymous substitutions are selected at some sites, probably because they provide enhanced fitness. We tested this hypothesis by computing the nonsynonymous versus synonymous substitution ratio (ω = dN/dS) for NCL proteins, using the program suite PAML, which reconstitutes the evolution of codons based on an alignment. dN/dS ratios were compared with predictions of evolution models allowing the presence of sites with ω > 1 (diversifying selection) or not (nearly neutral selection) (Yang and Swanson, 2002; Yang, 2007). Likelihood ratio tests shown in Table 3 clearly show that models allowing classes with ω > 1 much better fit the observed values, as judged by the P values of χ2 tests. In the alignment, 26 sites were considered to be under diversifying selection at 99% confidence (49 at 95%), most of them within OPR repeats.
Table 3. Comparison of Codon Evolution Models in NCL Genes.
| Likelihooda | ||||||
|---|---|---|---|---|---|---|
| Nearly Neutral | Positive Selection | 2Δlb | P Valuec | pd | ωe | |
| A | M1 | M2 | (M2 vs. M1) | |||
| −43839.38 | −43673.49 | −331.78 | 1.33 E−72 | 0.093 | 3.05 | |
| B | M7 | M8 | (M7 vs. M8) | |||
| −43806.28 | −43600.42 | −411.72 | 5.66 E−90 | 0.147 | 2.35 | |
Part A is a comparison of model M1 versus M2. M1 allows two classes of codons (ω = 0, negative selection; ω = 1, neutral selection) and M2 an additional class under positive selection (ω > 1). Part B is a comparison of model M7 versus M8. M7 allows a continuous β-distribution of 0 < ω < 1, with an additional class (ω > 1) in M8.
Log likelihood values for nearly neutral (M1 and M7) or positive selection (M2 and M8) models.
Likelihood ratio between the two models.
Evaluated from χ2 distribution (df = 2).
Proportion of sites under positive selection.
Mean ω value for sites under positive selection.
In conclusion, NCL genes are still under dynamic evolution and undergo a “birth, diversification, and death” process that is driven by diversifying selection pressure and likely generates new RNA targets via mutations in OPR repeats.
DISCUSSION
ncc1 and ncc2, Dominant and Atypical Mutations in Two OPR Proteins, Lead to RNA Degradation Rather Than RNA Protection
All mutations affecting ROGEs characterized so far in Chlamydomonas are recessive, as they inactivate a protein that binds specifically to a given target transcript, usually in its 5′UTR. Thus, ROGEs likely coevolved with the 5′UTR of their target mRNA. By contrast, the ncc1 and ncc2 mutations described here represent a different category of ROGE mutations in Chlamydomonas. Both appeared spontaneously in our laboratory strains, are dominant, and act on the CDS of their target mRNA. Their unusual dominant nature results from their molecular basis: a single amino acid substitution in one OPR repeat of two different OPR proteins. By introducing a mutant copy of either NCC1 or NCC2 in the wild type, we show that these single substitutions are sufficient to destabilize the atpA or petA transcripts, respectively. They would change the recognized nucleotide and create new targets for the mutated proteins that fortuitously lie within CDSs. As observed for PPR proteins and in agreement with our preliminary version of the OPR code, well-conserved positions (e.g., 4, 5, and 7 to 15; Supplemental Figure 2) within the first antiparallel α-helix of the OPR repeats should mostly contribute to protein scaffolding, whereas more variable positions (e.g., 3 and 6) would be involved in nucleotide recognition. The ncc1 mutation, that changes the variable 6th residue of the repeat, was thus expected to alter nucleotide recognition. Surprisingly, the ncc2 mutation changing the quite conserved 8th residue nevertheless leads to the recognition of a new target.
ROGEs that bind the 5′UTR of their target transcript either activate its translation or protect it from exonucleolytic degradation. In contrast, the interaction of NCC1M and NCC2M with the atpA and petA CDSs leads to the degradation of the atpA and petA mRNAs by a mechanism that remains to be studied: NCC1M and NCC2M may recruit an endonuclease or may themselves carry an endonucleolytic activity, which may be carried by the RAP domain found at the C terminus of both proteins. Indeed, structural modeling of the RAP domains of NCC1 and NCC2 by the I-TASSER software (Zhang, 2008) used four endonucleases as major templates (Supplemental Figures 3 and 4).
A Link with Translation
Strikingly, NCC2M only degrades its target transcript upon translation. This was observed in two different sequence contexts, within the petA and petD transcripts, in which the target is involved in widely different secondary structures. It is therefore unlikely that NCC2M binding to its target site requires the ribosome-mediated unfolding of RNA secondary structures. Rather, the degradation of the transcript would depend on a tight contact between the ribosome and NCC2M, as supported by the limited decrease in petA mRNA observed upon early translation termination, a few nucleotides upstream of the NCC2M target. This interaction would change the conformation of NCC2M itself or of its interacting endonuclease, thereby activating nucleolytic activity, as shown for ribosome associated YoeB, RelE, and RegB RNases (Odaert et al., 2007; Neubauer et al., 2009; Feng et al., 2013).
By contrast, the degradation of the atpA transcript by NCC1M does not require ongoing atpA translation. However, the monocistronic atpA transcript is by far more destabilized than polycistronic atpA transcripts. Possibly, the target of NCC1M, localized 460 bp before the end of the atpA CDS, is fully accessible in the monocistronic atpA transcript, but trapped within secondary structures with downstream sequences in polycistronic transcripts.
NCC1 and NCC2 Belong to the NCL Family of Paralogous OPR-RAP Proteins
NCC1 and NCC2 belong to the NCL family of highly similar OPR paralogs that differ in many respects from the bulk of genes encoding OPR proteins in Chlamydomonas (hereafter “OPR genes”). Unlike most OPR genes, for which orthologs can be easily found in V. carteri, probably because of a conserved role in organelle biogenesis, NCL genes appeared after the separation between the V. carteri and Chlamydomonas lineages. They share a single intron at a fixed position and show high sequence similarity, indicating recent appearance by gene duplication. Their genomic organization is also striking. Whereas OPR genes are randomly dispersed throughout the Chlamydomonas or V. carteri genomes, most NCL genes are clustered on chromosome 15. The rapid evolution of this family likely rests on tandem duplications, giving rise to seven subclusters of closely related genes (Figures 6A and 6D). This tandem organization would favor unequal crossovers, leading to gene duplication/loss and to repeat duplications. Interallelic recombination and gene conversion could also participate in repeat shuffling and, thus, to the diversification of binding sites. Indeed, NCL genes evolve under diversifying selection pressure, with specific positions showing a high dN/dS ratio. The spontaneous appearance of ncc1 and ncc2 mutations in laboratory conditions suggests that this diversification is still active. NCL genes also can decay, as evidenced by the presence of inactivated and truncated genes. Thanks to this vigorous “birth-diversification-death” process, NCL genes represent a constant source of RNA binding proteins with new target specificities.
The NCL family strikingly resembles two other rapidly evolving gene families in plants. Restorer of Fertility-Like (RFL)-PPR proteins are distributed throughout higher plants and include Restorers of fertility (Rfs) proteins characterized in radish (Raphanus sativus), petunia (Petunia hybrida), or rice (Fujii et al., 2011; reviewed in Dahan and Mireau, 2013). Rfs repress the expression of mitochondrion-encoded chimeric open reading frames that are generated by recombination between different copies of the mitochondrial genome and cause cytoplasmic male sterility (CMS) in various crop species. RFL proteins are considered as a reservoir for the evolution of Rfs counteracting the expression of new CMS genes. As NCL genes, most RFL genes are clustered (in two regions of chromosome 1 in Arabidopsis or on chromosome 10 in rice), favoring unequal crossovers and local duplications, which likely contribute to the expansion of the RFL family (Hernandez Mora et al., 2010). RFL genes are under diversifying selection, especially those residues of the PPR motifs that are involved in nucleotide recognition (Geddy and Brown, 2007; Fujii et al., 2011), thus favoring the appearance of new RNA targets. They evolve rapidly, with large divergence between species or even between different accessions (Jonietz et al., 2010).
Similarly, pathogen resistance (R) genes, which activate plant defense reactions upon recognition of specific pathogen effectors, evolve rapidly in an “arms race” against the constantly evolving plant pathogens. R genes predominantly belong to the large and highly dynamic family of nucleotide binding site leucine-rich repeat (NLR) proteins (reviewed in DeYoung and Innes, 2006; Ye and Ting, 2008; Qi and Innes, 2013). They are often clustered on the genome, a situation that, again, favors the expansion and evolution of the gene family (Michelmore and Meyers, 1998), and are under diversifying selection that targets mainly the solvent-exposed residues involved in protein-protein interaction (Wulff et al., 2009; Seeholzer et al., 2010). It is of note that the new alleles created by these diversifying selection processes in RFL and NLR families confer a dominant phenotype of fertility or pathogen resistance to their host organism (Elkonin, 2005; Song et al., 2006; Moffett, 2009).
The Elusive Physiological Function of NCL Proteins
In the above examples, the evolutionary drives for the expansion of gene families are genome warfare and biotic stress. However, the selective pressure that led to the expansion of NCL genes in Chlamydomonas is not known. To address this question, some knowledge of the physiological function of the wild-type NCC1 and NCC2 proteins would be of interest.
We fail to find additional phenotypes conferred by the ncc1 or ncc2 mutations. Either the gain of function provided by the substitutions within NCC1M and NCC2M preserves the original function of the wild-type proteins or these functions are subtle and escaped our phenotypic analysis. In Arabidopsis, inactivation of the RFL genes RPF1 or RPF2, while modifying the processing of a few mitochondrial transcripts (nda4 or cox3 and nad9, respectively), do not alter the accumulation of their gene products nor lead to any obvious phenotype (Jonietz et al., 2010; Hölzle et al., 2011). Furthermore the RPF2 gene seems inactivated in some accessions (Forner et al., 2008; Jonietz et al., 2010). Similarly, would the gene duplication be too recent to have elicited the recruitment of NCC1 and NCC2 for some physiological process, these proteins could have no function, in contrast to their mutant allele. However, their transcription seems regulated: Cursory examination of their expression profiles on the Phytozome browser suggests a transient repression of NCC2 during nitrogen starvation (GSE34585) and a repression of NCC1 by H2O2 treatment (GSE34826).
The spontaneous appearance of the ncc1 and ncc2 mutations likely illustrates a “trial and error” process operating on genes whose physiological function is not essential or redundant. It may ultimately lead to the recognition of new RNA targets, which could provide a selective advantage in specific environmental conditions. This is illustrated by the experimental conditions in which we recovered the ncc2 strain. It appeared when plating a nonphototrophic chimeric strain in phototrophic conditions (Figure 1) but shows a leaky cytochrome b6f-defective phenotype and would have been counterselected in cells with a wild-type chloroplast genome kept in phototrophic conditions.
METHODS
Strains and Growth Conditions
Wild-type, mutant, and transformed Chlamydomonas reinhardtii strains, all derived from 137c, were grown at 25°C in Tris-acetate-phosphate (TAP) medium, pH 7.2 (Harris, 1989), under continuous light (5 to 10 μE m−2 s−1). Strains ncc1 and ncc1 {FAFA} were previously described as mda1-ncc1 and mda1-ncc1{FAFA} by Drapier et al. (2002). Crosses were performed according to Harris (1989). Vegetative diploids were selected on arginine-free plates from crosses between strains carrying the arg2 and arg7 mutations (Ebersold, 1967; Harris, 1989). After 12 d in low light, dark-green colonies comprised of large cells were checked by PCR for the presence of both mt loci (Werner and Mergenhagen, 1998).
Constructs and Nucleic Acid Manipulations
Standard nucleic acid manipulations were performed according to Sambrook et al. (1989). Primers used in this study are listed in Supplemental Table 3.
5′petA-psbB Chimera
The petA 5′UTR and promoter regions were excised with EcoRV and NcoI from plasmid p5F (Choquet et al., 1998) and cloned into the p38A.NcoI vector (Vaistij et al., 2000) digested with StuI and NcoI to yield plasmid p5′petA-psbB. The spectinomycin resistance cassette, excised from plasmid pUC-ATPX-AAD (Goldschmidt-Clermont, 1991) by SmaI and EcoRV, was inserted in the Klenow-treated AvrII site, in reverse orientation with respect to the psbB gene, yielding plasmid pK5′petA-psbB.
pΔf::αTr
A truncated version of the atpA CDS, fused the petA lumen targeting peptide, replaced the sequence encoding mature cytochrome f. Truncated atpA CDS was amplified in two steps from plasmid patpA2 (Ketchner et al., 1995). A 254-bp fragment amplified with primers atpAFusFW/atpAFusRV1 was digested with HindIII and PstI and cloned into HindIII-PstI digested pf::H6 vector (Choquet et al., 2003) to create plasmid p5f-int. A 965-bp fragment amplified with primers atpAFW/atpAFusRV2 was digested with EcoRI and PstI and cloned into vector p5f-int, digested with the same enzymes to create plasmid pΔf::αTr. The aadA cassette was then inserted at the HincII site, upstream and in reverse orientation with respect to the petA CDS, to yield the plasmid pKΔf::αTr.
p5′dAfR
Plasmid p5′dAfR, comprising the petA CDS expressed under the control of the atpA 5′UTR and the rbcL 3′UTR, was created from plasmid pFADBE1 (Kuras and Wollman, 1994), which encompasses the petA genomic region, but where the whole petA gene was replaced by the aadA cassette. The petA CDS, amplified from plasmid pWF (Kuras and Wollman, 1994) using primers AFRF_FW and AFRF_RV and digested by NcoI and PstI, replaced the aadA CDS, excised with NcoI and PstI. The 5′psaA-aadA-3′rbcL selection cassette (Wostrikoff et al., 2004), excised with SmaI and EcoRV, was introduced at the HincII site upstream and in reverse orientation of the chimeric petA gene.
Frame-Shifted petA Genes: pf31St and pf130St
To create frame shifts after the 31st and 130th codons of the petA gene, plasmid pWF was digested by HindIII and BstEII, Klenow-treated, and religated on itself to yield plasmids pf42St and pf145St. The 5′psaA-driven selection cassette was then inserted at the HincII site, upstream and in reverse orientation with respect to the petA CDS, to yield plasmids pKf42St and pKf145St.
Chimeric petD Genes
To generate the plasmid ppetD, which contains a single SwaI site, plasmid pWQ (Kuras and Wollman, 1994) was digested with SacI and AflII, Klenow-treated, and religated on itself. To insert the target of NCC2M within the petD 5′UTR or CDS, a PCR fragment of 462 bp was amplified using primers petD5::T2_FW and petDCod::T2_RV and plasmid ppetD as a template. This amplicon was digested with SwaI and HindIII and the 88-bp fragment was cloned into the SwaI-HindIII-digested vector ppetD to create plasmid p5′petD::T2 or with HindIII and PstI and the 358-bp fragment was cloned into ppetD digested by the same enzymes to create plasmid ppetDCod::T2.
These two plasmids were then digested by AvrII, Klenow-treated, and ligated to a recycling 5′psaA-driven spectinomycin resistance cassette. Because the ncc1 mutation targets the atpA mRNA, we avoided using the 5′atpA-driven recycling cassette (Fischer et al., 1996) as a selectable marker and constructed a 5′psaA-aadA-3′rbcL recycling cassette. The atpA 5′UTR, excised from plasmid paadA485 (Fischer et al., 1996) by NcoI and NruI, was replaced by the psaA 5′UTR, itself excised from plasmid pfaAK (Wostrikoff et al., 2004) by EcoRV and NcoI, yielding plasmid p5′aA-aadA485. The recycling cassette was excised from this vector by digestion with SacI and KpnI and Klenow treatment. In the final plasmids pKr5′petD::T2 and pKrpetDCod::T2, the aadA cassette is transcribed in reverse orientation with respect to the petD gene.
Mutation of the NCC1M Target
We mutated the target of NCC1M by two-step megaprime PCR (Higuchi, 1990): Primers atpAExtFW/atpAMT1RV and atpAMT1FW/atpAExtRV allowed the amplification from plasmid patpA2 of two partially overlapping amplicons that were mixed and used as templates in a third PCR with the external primers atpAExtFW/atpAExtRV. The final amplicon, digested by MfeI and PacI, two restriction sites on both sides of the mutation, was cloned into plasmid pKratpA300St (Drapier et al., 2007) digested with the same enzymes, yielding plasmid pKratpAM.
Mutation of the NCC2M Target
We destroyed the target of NCC2M by the same two-step PCR procedure using the mutagenic primers petAMT2FW and petAMT2RV and the external primers petAExtFW and petAExtRV to amplify from plasmid template pWF a 1053-bp fragment. This amplicon, digested by BglII and AccI, was cloned into BglII/AccI-digested plasmid pKWFStop (Boulouis et al., 2011) to create plasmid pKpetAMutT2.
Transformation Vector for Expression of the Mutated NCC1 and NCC2 Proteins
Because of the high percentage of similarity between the paralogous genes of the NCL cluster, designing specific primers to amplify the mutant ncc1 and ncc2 genes turned out to be difficult. We thus ordered the synthetic DNA sequences shown in the supplemental data section (Genscript). They were digested by EcoRI and BglII (ncc1-HA) or EcoRI and BamHI (ncc2-HA) and cloned into the vector pJHL (kindly provided by Jae-Hyeok Lee, University of British Columbia) digested by EcoRI and BamHI.
All DNA constructs were sequenced before transformation in Chlamydomonas. RNA gel blot analyses were performed as described by Drapier et al. (2002) using 33P-labeled probes described by Eberhard et al. (2002). Transcript accumulation was quantified from phosphor imager scans of the blots, as described by Choquet et al. (2003).
Chloroplast translation was arrested by supplementing cells grown in TAP medium (2 × 106 cells mL−1) with lincomycin (final concentration 500 μg mL−1) at t = 0. Aliquots, taken at the indicated time points, were briefly chilled on ice before RNA extraction.
Map-Based Localization of the ncc2 Mutation
To localize the ncc2 mutation, a first set of ∼50 ncc2 progeny (based on their fluorescence phenotype) was selected out of independent meiosis from the cross ncc2 × S1-D2. After Chelex-based DNA extraction (Werner and Mergenhagen, 1998), amplified fragment length polymorphism markers (Kathir et al., 2003; Rymarquis et al., 2005) allowed us to determine the proportion of each parental version of the marker on each chromosome arm by PCR. Once linkage (96%) to the ZYS3 marker was established, new amplified fragment length polymorphism markers, based on identified polymorphisms in S1-D2 ESTs or on putative differences in tandem repeat copy numbers, were designed along chromosome 15 (Supplemental Table 2) to define the region containing the ncc2 mutation. To observe rare crossing-over events, this analysis was performed on 500 independent meioses and allowed us to restrict the location of the mutation to a 405-kb region.
Genomic DNA Preparation, Whole-Genome Sequencing, and Data Analysis
DNA from wild-type and {FAFA} ncc1 ncc2 double mutant strains was extracted with the DNAeasy Plant Maxi Kit (Qiagen), according to the manufacturer’s protocol, starting from 100 mL of stationary culture.
Genome sequencing was performed using the high-throughput, short-read, Illumina technology. Sequencing was done at the Tufts University Core Facility, Boston, MA, on a HiSequation 2000 instrument in paired-end mode, 2 × 100 nucleotides. Libraries had insert sizes of ∼300 bp. About 180 million read pairs were generated. Reads were mapped simultaneously onto the nuclear, chloroplast, and mitochondrial genomes of Chlamydomonas using BWA 0.6.0-r85 (Li and Durbin, 2009). The organelle genomes were taken from GenBank (accession numbers FJ423446 and CRU03843), and Phytozome v.5 was used for the nuclear genome. The BWA “aln” and “sampe” commands were run with default parameter values, except for the following options: “-q -1 -R 10 -o 2 -e 0 -l 30 -t 20” for “aln” and “-a 600 -n 9999 -N 9999” for “sampe.” The alignment files created by BWA in SAM format were then converted to BAM format and indexed, and duplicate read pairs were removed using Samtools 0.1.16 (Li et al., 2009). Read mapping data were visualized in IGV 2.0 (Robinson et al., 2011; Thorvaldsdóttir et al., 2013).
The duplicate-filtered alignment files were then fed to the SVMerge 1.1r32 pipeline (Wong et al., 2010) to detect single nucleotide polymorphisms, short insertions and deletions (indels), and large structural variants. SVMerge included the following tools: breakdancer 1.1 (Chen et al., 2009), pindel 0.2.4q (Ye et al., 2009), cnD 1.3 (Simpson et al., 2010), and SECluster (bundled with SVMerge). The results from InGAP-sv 2.8.1 (Qi et al., 2010; Qi and Zhao, 2011) were also integrated in the SVMerge pipeline. Breakdancer was run with the following options: “-q 35 -c 3 -r 3 -y 40 -m 10000000”; pindel was run with the following options: “-T 1 -x 9 -e 0.02 -u 0.05 -a 1 -m 3 -n 50 -v 50 -d 30 -A 35 -M 6”; cnD was run with the following options: “-threshold=0.5-window=5”; SECluster was run with the following options: “-q 35 -m 6 -c 6 -r {1} -x 10000”; inGAP-sv was run with default parameters, but a maximum coverage of 1000. In the SVMerge configuration file, the following cutoff scores were set: “BDscore=40,” “BDrs=3,” and “PDscore=200.” Velvet 1.1.05 (Zerbino and Birney, 2008) and exonerate 2.2.0 (Slater and Birney, 2005) were run for local assemblies of structural variants in SVMerge. The following Velvet parameters were specifically set: “hashlen=25, exp_cov=auto, cov_cutoff=3.” SVMerge requires the BEDtools package (Quinlan and Hall, 2010); version 2.16.2 was used.
Based on version 5.0 of the gene predictions (obtained from Phytozome), single nucleotide polymorphisms and insertions and deletions were annotated using SHOREmap_annotate from the SHOREmap 1.2 package (Schneeberger et al., 2009) to determine in which features they were located (gene, UTR, CDS, and intron) and whether they would cause synonymous or nonsynonymous changes.
The phylogenic tree was constructed on the Phylogeny.fr platform (Dereeper et al., 2008), including the following steps: Sequences were aligned with MUSCLE (v3.7) (Edgar, 2004) configured for highest accuracy (MUSCLE with default settings; see Supplemental Data Set 2 for the alignment) and cured with Gblocks (v0.91b), using relaxed parameters. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v3.0) (Guindon et al., 2010) with reliability for internal branch assessed using the aLRT test (SH-Like) (Anisimova and Gascuel, 2006) and visualized using TreeDyn (v198.3) (Chevenet et al., 2006).
Bioinformatic Analysis of Positive (Diversifying) Selection
Multiple sequence alignment of the NCL proteins was generated with Muscle (Edgar, 2004) and manually refined to optimize conservation of the repeats. Pal2NAL v.14 was used to align the CDS based on this alignment (Suyama et al., 2006) and PhyML to generate the phylogenetic tree using maximum likelihood. A text file of the alignment is provided in Supplemental Data Set 2. Diversifying selection was analyzed using PAML v4.7a (routine codml) as described (Yang, 2007), using the graphic interface in PAMLX (Xu and Yang, 2013). The likelihood of neutral selection models (M1 and M7) was compared with that of models allowing an additional class with ω > 1 (M2 and M8, positive selection). Model 1 allows two classes of codon with ω = 0 (negative selection) and ω = 1 (neutral selection), while model 7 allows a continuous β-distribution of ω values <1 (Yang and Swanson, 2002). Posterior probability of positive selection at each site was calculated the using Bayes-Empirical Bayes method (Anisimova et al., 2002).
Transformation Experiments
Chloroplast transformation by tungsten particle bombardment (Boynton et al., 1988) was conducted as described (Kuras and Wollman, 1994). Transformants were selected on TAP-Spec (100 mg⋅mL−1) and subcloned on TAP-Spec (500 μg⋅mL−1) until they reached homoplasmy, assessed by restriction fragment length polymorphism analysis. At least three independent transformants were analyzed for each transformation. Phenotypic variations between independent transformants proved negligible.
Nuclear transformation of the wild type was performed by electroporation, as described by Raynaud et al. (2007), with the following parameters: 25 μF and 1000 V cm−1. Transformants were selected on plates supplemented with paromomycin (5 μg⋅mL−1).
Protein Analysis
Pulse-labeling experiments, protein electrophoresis, and immunoblots were performed on exponentially growing cells (2 × 106 cells⋅mL−1) according to Kuras and Wollman (1994). Cell extracts, loaded on equal chlorophyll basis, were analyzed by SDS-PAGE (12 to 18% acrylamide and 8 M urea). Anti-OEE2 and -cytochrome f antibodies, used for [135I]protein A detection, were raised in the laboratory against proteins isolated from Chlamydomonas and have been described previously (de Vitry et al., 1989; Kuras and Wollman, 1994). PsbB and HA-tagged NCC1M and NCC2M proteins were detected by ECL using the monoclonal antibody anti HA.11 (Covance) and horseradish peroxidase-conjugated antibody against mouse IgG (Promega).
Fluorescence Measurements
Fluorescence measurements were performed on dark-adapted liquid cultures using a home-built spectrofluorimeter according to Zito et al. (1997).
Accession Numbers
Sequence data for NCC1, NCC2, and NCL genes from this article can be found in the Phytozome database, as indicated in Supplemental Data Set 1. Other sequence data used in this article can be found in the GenBank/EMBL databases under the following accession numbers: petA (cytochrome f), FJ423446.1; petD, X72919.1; psaB, X05848.1; psaC, U43964.1; psbA, CAA25670; psbB (CP47), X64066.1; psbD, X04147.1; OEE2, M15187.1; atpA, X60298.1; and rbcL, J01399.1.
Supplemental Data
Supplemental Figure 1. Phenotype of ncc1 ncc2 {FAFA} double mutants.
Supplemental Figure 2. Alignment of NCL proteins.
Supplemental Figure 3. Comparison of NCC1 RAP domain model with known structures of endonucleases.
Supplemental Figure 4. I-TASSER alignment used for threading of the NCC1 RAP domain.
Supplemental Table 1. Genetic independence of the ncc2 mutation from MCA1 and TCA1 genes.
Supplemental Table 2. Markers designed to map the ncc2 mutation.
Supplemental Table 3. Oligonucleotides used in this study.
Supplemental Table 4. BLAST search-based identification of NCL proteins in Phytozome v5.5.
Supplemental Data Set 1. Description of NCL proteins, improved gene models for NCL7, 8, 21, 30, and 35, and intracellular targeting of NCL proteins.
Supplemental Data Set 2. Text file of alignment corresponding to the phylogenetic analysis in Figure 6D.
Supplementary Material
Acknowledgments
We thank Jae-Hyeok Lee (University of British Columbia) for his kind gift of the pJHL Chlamydomonas transformation vector, D.B. Stern for providing markers for molecular mapping prior to publication (Rymarquis et al., 2005), and all members of UMR7141 for stimulating discussions and/or critical reading of the article. This work was supported by Unité Mixte de Recherche 7141, CNRS, and Université Pierre et Marie Curie, Paris 06, by the European Community (“SunBioPath” Contract FP7-KBBE-2009-3-02; GIAVAP Contract FP7-KBBE.2010.3; GA No. 266401), by Agence Nationale de la Recherche (ChloroMitoCES: BLAN-NT09_451610 and ChloroRNP: ANR-13-BSV7-0001-01), and by the “Initiative d’Excellence” program (Grant “DYNAMO,” ANR-11-LABX-0011-01). A.B. was “Attachée Temporaire d’Enseignement et de Recherche” at Université Pierre et Marie Curie. N.J.T. was supported by GIAVAP and Dynamo.
AUTHOR CONTRIBUTIONS
A.B., D.D., F.-A.W., and Y.C. designed research. A.B., D.D., H.R., K.W., J.G.-B., and Y.C. performed research. N.J.T., K.P., and O.V. contributed to analytic tools and bioinformatics analysis. A.B., D.D., F.-A.W., and Y.C. analyzed data. A.B., D.D., F.-A.W., and Y.C. wrote the article.
Glossary
- ROGE
regulator of organelle gene expression
- UTR
untranslated region
- TAP
Tris-acetate-phosphate
- CDS
coding sequence
- PSII
photosystem II
References
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552. [DOI] [PubMed] [Google Scholar]
- Anisimova M., Bielawski J.P., Yang Z. (2002). Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 19: 950–958. [DOI] [PubMed] [Google Scholar]
- Auchincloss A.H., Zerges W., Perron K., Girard-Bascou J., Rochaix J.D. (2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157: 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011). Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572. [DOI] [PubMed] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby I.K., et al. (2014). The Chlamydomonas genome project: a decade on. Trends Plant Sci. 19: 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bogdanove A.J., Voytas D.F. (2011). TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846. [DOI] [PubMed] [Google Scholar]
- Boulouis A., Raynaud C., Bujaldon S., Aznar A., Wollman F.A., Choquet Y. (2011). The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., et al. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538. [DOI] [PubMed] [Google Scholar]
- Chen K., et al. (2009). BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C.G., Hall T.M. (2006). Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. USA 103: 13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Bañuls A.L., Jacq B., Christen R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529: 39–42. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.-A. (2009). The CES process. In Chlamydomonas Source Book, 2nd ed, Vol. 2, E.E. Harris, D.B. Stern, and G.B. Witman, eds (New York, London, Amsterdam: Academic Press, Elsevier), pp. 1027–1064. [Google Scholar]
- Choquet Y., Zito F., Wostrikoff K., Wollman F.A. (2003). Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Stern D.B., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95: 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A., Prigge A., Opperman L., Wickens M. (2011). Targeted translational regulation using the PUF protein family scaffold. Proc. Natl. Acad. Sci. USA 108: 15870–15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan J., Mireau H. (2013). The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 10: 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.K., Shi Y., Yan N. (2012). Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C., Olive J., Drapier D., Recouvreur M., Wollman F.A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109: 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B.J., Innes R.W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7: 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Stern D.B., Wollman F.A. (2002). A dominant nuclear mutation in Chlamydomonas identifies a factor controlling chloroplast mRNA stability by acting on the coding region of the atpA transcript. Plant J. 31: 687–697. [DOI] [PubMed] [Google Scholar]
- Drapier D., Rimbault B., Vallon O., Wollman F.A., Choquet Y. (2007). Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 26: 3581–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D., Suzuki H., Levy H., Rimbault B., Kindle K.L., Stern D.B., Wollman F.A. (1998). The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Functional analysis of a polycistronic transcription unit. Plant Physiol. 117: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160. [DOI] [PubMed] [Google Scholar]
- Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., Wollman F.A. (2011). Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 67: 1055–1066. [DOI] [PubMed] [Google Scholar]
- Ebersold W.T. (1967). Chlamydomonas reinhardtii: heterozygous diploid strains. Science 157: 447–449. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkonin L.A. (2005). Dominant male sterility in sorghum: effect of nuclear background on inheritance of tissue-culture-induced mutation. Theor. Appl. Genet. 111: 1377–1384. [DOI] [PubMed] [Google Scholar]
- Feng S., Chen Y., Kamada K., Wang H., Tang K., Wang M., Gao Y.G. (2013). YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 41: 9549–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A., Rackham O. (2012). Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol. Biosyst. 8: 699–708. [DOI] [PubMed] [Google Scholar]
- Filipovska A., Razif M.F., Nygård K.K., Rackham O. (2011). A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 7: 425–427. [DOI] [PubMed] [Google Scholar]
- Fischer N., Stampacchia O., Redding K., Rochaix J.D. (1996). Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251: 373–380. [DOI] [PubMed] [Google Scholar]
- Forner J., Hölzle A., Jonietz C., Thuss S., Schwarzländer M., Weber B., Meyer R.C., Binder S. (2008). Mitochondrial mRNA polymorphisms in different Arabidopsis accessions. Plant Physiol. 148: 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Bond C.S., Small I.D. (2011). Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. USA 108: 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddy R., Brown G.G. (2007). Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genomics 8: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain A., Hotto A.M., Barkan A., Stern D.B. (2013). RNA processing and decay in plastids. Wiley Interdiscip Rev RNA 4: 295–316. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19: 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.H., Ranum L.P., Lefebvre P.A. (1988). Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13: 503–508. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Gully B.S., Cowieson N., Stanley W.A., Shearston K., Small I.D., Barkan A., Bond C.S. (2015). The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 43: 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Bonnard G., Bouchoucha A., Gobert A., Pinker F., Salinas T., Giegé P. (2014). Helical repeats modular proteins are major players for organelle gene expression. Biochimie 100: 141–150. [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press; ). [Google Scholar]
- Hernandez Mora J.R., Rivals E., Mireau H., Budar F. (2010). Sequence analysis of two alleles reveals that intra-and intergenic recombination played a role in the evolution of the radish fertility restorer (Rfo). BMC Plant Biol. 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. (1990). Recombinant PCR. In PCR Protocols: A Guide to Methods and Applications, D.H. Gelfand, M.A. Innis, J.J. Sninsky, and T.J. White, eds (New York/London/Amsterdam: Academic Press, Elsevier), pp. 177–183. [Google Scholar]
- Hölzle A., Jonietz C., Törjek O., Altmann T., Binder S., Forner J. (2011). A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 65: 737–744. [DOI] [PubMed] [Google Scholar]
- Jonietz C., Forner J., Hölzle A., Thuss S., Binder S. (2010). RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell 22: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P., LaVoie M., Brazelton W.J., Haas N.A., Lefebvre P.A., Silflow C.D. (2003). Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2: 362–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J., Chen R.Z., Ban T., Zhou X.E., Gu X., Tan M.H., Chen C., Kang Y., Brunzelle J.S., Zhu J.K., Melcher K., Xu H.E. (2013). Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 20: 1377–1382. [DOI] [PubMed] [Google Scholar]
- Ketchner S.L., Drapier D., Olive J., Gaudriault S., Girard-Bascou J., Wollman F.A. (1995). Chloroplasts can accommodate inclusion bodies. Evidence from a mutant of Chlamydomonas reinhardtii defective in the assembly of the chloroplast ATP synthase. J. Biol. Chem. 270: 15299–15306. [DOI] [PubMed] [Google Scholar]
- Kuras R., Wollman F.A. (1994). The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13: 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Hong W. (2004). RAP—a putative RNA-binding domain. Trends Biochem. Sci. 29: 567–570. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L., Choquet Y., Kuras R., Loubéry S., Douchi D., Goldschmidt-Clermont M. (2015). A nucleus-encoded helical-repeat protein which is regulated by iron availability controls chloroplast psaA mRNA expression in Chlamydomonas. Plant Physiol., 10.1104/pp.114.253906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselay, C. (2007). Importance of Nucleus-Encoded Factors for the Expression of the Chloroplast Genome: The Example of the Chloroplast petA Gene in Chlamydomonas reinhardtii. PhD dissertaion (Paris: UFR de Biologie, UPMC). [Google Scholar]
- Loiselay C., Gumpel N.J., Girard-Bascou J., Watson A.T., Purton S., Wollman F.A., Choquet Y. (2008). Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 28: 5529–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A.N., Bradley P., Cernadas R.A., Bogdanove A.J., Stoddard B.L. (2012). The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—a practical guide. J. Exp. Bot. 51: 659–668. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L., Perron K., Rahire M., Howald I., Rochaix J.D., Goldschmidt-Clermont M. (2006). A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 34: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R.W., Meyers B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Miller M.T., Higgin J.J., Hall T.M. (2008). Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 15: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P. (2009). Mechanisms of recognition in dominant R gene mediated resistance. Adv. Virus Res. 75: 1–33. [DOI] [PubMed] [Google Scholar]
- Monod C., Goldschmidt-Clermont M., Rochaix J.D. (1992). Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol. Gen. Genet. 231: 449–459. [DOI] [PubMed] [Google Scholar]
- Moscou M.J., Bogdanove A.J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Neubauer C., Gao Y.G., Andersen K.R., Dunham C.M., Kelley A.C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D.E. (2009). The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139: 1084–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaert B., Saïda F., Aliprandi P., Durand S., Créchet J.B., Guerois R., Laalami S., Uzan M., Bontems F. (2007). Structural and functional studies of RegB, a new member of a family of sequence-specific ribonucleases involved in mRNA inactivation on the ribosome. J. Biol. Chem. 282: 2019–2028. [DOI] [PubMed] [Google Scholar]
- Qi D., Innes R.W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Zhao F. (2011). inGAP-sv: a novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res. 39: W567–W575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Zhao F., Buboltz A., Schuster S.C. (2010). inGAP: an integrated next-generation genome analysis pipeline. Bioinformatics 26: 127–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahire M., Laroche F., Cerutti L., Rochaix J.D. (2012). Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 72: 652–661. [DOI] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A., Choquet Y. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 104: 9093–9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson E.H., Eichman B.F. (2012). Nucleic acid recognition by tandem helical repeats. Curr. Opin. Struct. Biol. 22: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis L.A., Handley J.M., Thomas M., Stern D.B. (2005). Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 137: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13: 663–670. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.E., Weigel D., Andersen S.U. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Seeholzer S., Tsuchimatsu T., Jordan T., Bieri S., Pajonk S., Yang W., Jahoor A., Shimizu K.K., Keller B., Schulze-Lefert P. (2010). Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact. 23: 497–509. [DOI] [PubMed] [Google Scholar]
- Simpson J.T., McIntyre R.E., Adams D.J., Durbin R. (2010). Copy number variant detection in inbred strains from short read sequence data. Bioinformatics 26: 565–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I.A., Lapina T.V., Frolova O.N., Alexandrova N.N., Akopiants K.E., Danilenko V.N. (1996). Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene 181: 13–18. [DOI] [PubMed] [Google Scholar]
- Slater G.S., Birney E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L.Q., Fu T.D., Tu J.X., Ma C.Z., Yang G.S. (2006). Molecular validation of multiple allele inheritance for dominant genic male sterility gene in Brassica napus L. Theor. Appl. Genet. 113: 55–62. [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Brennicke A., Graichen K. (2013). Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8: e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse N.J., Choquet Y., Vallon O. (2013). PPR proteins of green algae. RNA Biol. 10: 1526–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij F.E., Goldschmidt-Clermont M., Wostrikoff K., Rochaix J.D. (2000). Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 21: 469–482. [DOI] [PubMed] [Google Scholar]
- Wang X., Zamore P.D., Hall T.M. (2001). Crystal structure of a Pumilio homology domain. Mol. Cell 7: 855–865. [DOI] [PubMed] [Google Scholar]
- Wang X., McLachlan J., Zamore P.D., Hall T.M. (2002). Modular recognition of RNA by a human pumilio-homology domain. Cell 110: 501–512. [DOI] [PubMed] [Google Scholar]
- Werner R., Mergenhagen D. (1998). Mating type determination of Chlamydomonas reinhardtii by PCR. Plant Mol. Biol. Rep. 16: 295–299. [Google Scholar]
- Wong K., Keane T.M., Stalker J., Adams D.J. (2010). Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 11: R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Choquet Y., Wollman F.A., Girard-Bascou J. (2001). TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Girard-Bascou J., Wollman F.A., Choquet Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23: 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B.B., Heese A., Tomlinson-Buhot L., Jones D.A., de la Peña M., Jones J.D. (2009). The major specificity-determining amino acids of the tomato Cf-9 disease resistance protein are at hypervariable solvent-exposed positions in the central leucine-rich repeats. Mol. Plant Microbe Interact. 22: 1203–1213. [DOI] [PubMed] [Google Scholar]
- Xu B., Yang Z. (2013). PAMLX: a graphical user interface for PAML. Mol. Biol. Evol. 30: 2723–2724. [DOI] [PubMed] [Google Scholar]
- Yagi Y., Nakamura T., Small I. (2014). The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78: 772–782. [DOI] [PubMed] [Google Scholar]
- Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. (2013). Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 8: e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Swanson W.J. (2002). Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19: 49–57. [DOI] [PubMed] [Google Scholar]
- Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. (2009). Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Ting J.P. (2008). NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr. Opin. Immunol. 20: 3–9. [DOI] [PubMed] [Google Scholar]
- Yin P., et al. (2013). Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504: 168–171. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F., Kuras R., Choquet Y., Kössel H., Wollman F.A. (1997). Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 33: 79–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.