Abstract
The enigmatic monocot family Triuridaceae provides a potentially useful model system for studying the effects of an ancient loss of photosynthesis on the plant plastid genome, as all of its members are mycoheterotrophic and achlorophyllous. However, few studies have placed the family in a comparative context, and its phylogenetic placement is only partly resolved. It was also unclear whether any taxa in this family have retained a plastid genome. Here, we used genome survey sequencing to retrieve plastid genome data for Sciaphila densiflora (Triuridaceae) and ten autotrophic relatives in the orders Dioscoreales and Pandanales. We recovered a highly reduced plastome for Sciaphila that is nearly colinear with Carludovica palmata, a photosynthetic relative that belongs to its sister group in Pandanales, Cyclanthaceae–Pandanaceae. This phylogenetic placement is well supported and robust to a broad range of analytical assumptions in maximum-likelihood inference, and is congruent with recent findings based on nuclear and mitochondrial evidence. The 28 genes retained in the S. densiflora plastid genome are involved in translation and other nonphotosynthetic functions, and we demonstrate that nearly all of the 18 protein-coding genes are under strong purifying selection. Our study confirms the utility of whole plastid genome data in phylogenetic studies of highly modified heterotrophic plants, even when they have substantially elevated rates of substitution.
Keywords: dN/dS ratio, gene loss, genome reduction, mycoheterotrophy, Pandanales, plastid genome evolution
Introduction
Mycoheterotrophic plants obtain some or all of their nutrients from soil fungi, typically those involved in mycorrhizal interactions with other plants (e.g., Merckx 2013). Merckx and Freudenstein (2010) counted at least 50 independent origins of full mycoheterotrophy, in which plants have lost the ability to photosynthesize and rely completely on fungal associates. Most of the 400 or so species of full mycoheterotrophs are monocots, a major clade that appears to be particularly prone to this evolutionary transition (Imhof 2010; Merckx and Freudenstein 2010; Merckx, Mennes, et al. 2013). Of these, about 50 species belong to a single monocot family, Triuridaceae, which is exclusively mycoheterotrophic. The family comprises nine extant genera, and has a pantropical distribution (Maas-van de Kamer and Weustenfeld 1998). Triuridaceae are small achlorophyllous, perennial herbs with tiny flowers and reduced scale-like leaves, found mostly in damp and deep-shaded forest habitats (Furness et al. 2002; Merckx, Freudenstein, et al. 2013). There are relatively few collections of this ephemeral and inconspicuous lineage, and their general biology, genomics and evolutionary history remain poorly understood (e.g., Rudall 2003). It seems likely that Triuridaceae experienced an ancient loss of photosynthesis, as a molecular dating analysis indicates that the (nonphotosynthetic) crown clade of the family arose around the Cretaceous or Lower Paleocene (Mennes et al. 2013).
Genome survey sequencing techniques (e.g., Cronn et al. 2008) now allow relatively straightforward retrieval of whole plastid genomes (plastomes) of green and heterotrophic plants (mycoheterotrophs and parasitic plants) for use in comparative analysis. Several studies of whole plastid genomes of heterotrophs have recently investigated their molecular evolution and characterized the structural rearrangements and losses that often occur following the loss of photosynthesis (Krause 2008; Barrett and Davis 2012; Wicke et al. 2013, 2014). Genes encoded in the plastome are also expected to show evidence of degradation due to relaxation or release of purifying selection for photosynthesis-related genes (e.g., Barrett et al. 2014). Seven full circular plastomes of mycoheterotrophic taxa have been published to date, representing the five orchids Corallorhiza striata, Epipogium roseum, Epipogium aphyllum, Neottia nidus-avis and Rhizanthella gardneri (Delannoy et al. 2011; Logacheva et al. 2011; Barrett and Davis 2012; Schelkunov et al. 2015), the monocot Petrosavia stellaris (Logacheva et al. 2014), and the liverwort Aneura mirabilis (Wickett et al. 2008). These plastomes exhibit variation in patterns of gene loss and retention, gene order, and plastome structure. For example, the plastome of Corallorhiza striata (Barrett and Davis 2012) is in a relatively early stage of genome degradation, and has retained a gene order consistent with its green relatives, whereas the plastomes of other mycoheterotrophs (e.g., Petrosavia stellaris, Neottia nidus-avis, R. gardneri) show more complex rearrangements, including substantial reductions in plastome size associated with considerable gene loss.
Based on patterns of gene loss and retention in heterotrophic plants, Barrett and Davis (2012) proposed an ordered trajectory of gene loss in mycoheterotrophs. They hypothesized an initial loss of genes encoding plastid subunits of the NAD(P)H complex, which appears to be involved in responding to photooxidative stress (Martin and Sabater 2010), followed by correlated losses of genes encoding photosynthesis-related protein complexes. Housekeeping genes involved in translation and other nonphotosynthetic functions tend to be retained the longest. Genes retained as open-reading frames are expected to be under purifying selection, if functional. For example, Barrett et al. (2014) found that housekeeping genes retained in fully mycoheterotrophic Corallorhiza are under the same selective regime (i.e., purifying selection) as homologous genes in photosynthetic relatives, consistent with their continued functionality in the plastid, despite the loss of photosynthesis.
Whole plastid genomes retrieved from mycoheterotrophs have also recently been used to determine the phylogenetic placement of several fully mycoheterotrophic lineages with uncertain placement among their photosynthetic relatives. For example, Logacheva et al. (2014) used the 37 protein-coding genes retained in the plastid genome of Petrosavia stellaris (Petrosaviaceae; Petrosaviales) to confirm its placement as the sister group of a photosynthetic taxon, Japonolirion osense. More recently, Mennes et al. (2015) recovered multiple genes from the plastid genomes of two of the three genera of Corsiaceae (16 and 23 protein-coding genes for Arachnitis uniflora and Corsia cf. boridiensis, respectively, and four ribosomal DNA (rDNA) genes from both genera; several transfer RNA (tRNA) genes were also recovered). These plastid genes placed Corsiaceae as the sister group of Campynemataceae in Liliales and supported the family’s monophyly, consistent with nuclear and mitochondrial evidence in the same study. Mennes et al. (2015) showed that all three plant genomes produced a congruent and well-supported picture of phylogenetic relationships of Corsiaceae, which in turn supports the idea that plastid genomes of heterotrophic plants are suitable for large-scale phylogenetic inference, despite extensive rate elevation and gene loss. Mennes et al. (2013) examined the phylogenetic position of Triuridaceae using mitochondrial and nuclear data, and demonstrated that it belongs in the monocot order Pandanales, confirming earlier results based on nuclear 18S rDNA data, mitochondrial atpA data, and morphology (Chase et al. 2000; Davis et al. 2004; Rudall and Bateman 2006). However, the precise relationships of the families within the order are still unclear; they were poorly supported in the analyses of Mennes et al. (2013), for example.
Here, we report on full plastid genomes and plastid gene sets recovered from Sciaphila densiflora (Triuridaceae) and ten related green taxa in Pandanales and Dioscoreales (comprising complete plastid genomes for Sciaphila and a green relative, Carludovica palmata, and plastid gene sets for nine additional relatives). The data from Sciaphila represent the first plastid genome sequences from Triuridaceae. We used these data: 1) To characterize major changes in the plastid genome following the loss of photosynthesis, including gene losses and retentions, and structural rearrangements; 2) to assess whether genes retained in the plastid genome of Sciaphila as open-reading frames are evolving under purifying selection or some other selective regime; and 3) to confirm the placement of Triuridaceae in Pandanales using plastid evidence and to pinpoint its local placement among the four photosynthetic families in the order (Cyclanthaceae, Pandanaceae, Stemonaceae, and Velloziaceae), while exploring the effect of different likelihood approaches on phylogenetic inference.
Materials and Methods
Taxon Sampling
We generated new plastid genome sequences for ten species in Pandanales and one in Dioscoreales (supplementary table S1, Supplementary Material online), and added these to published angiosperm plastome data retrieved from GenBank and from monocot-focused matrices presented in Givnish et al. (2010), Barrett et al. (2013), and Mennes et al. (2015) (supplementary table S1, Supplementary Material online). The 71-taxon matrix included at least one taxon from each of the five families of Pandanales (ten taxa in total), representatives of all major monocot lineages (50 taxa), in addition to representatives of the eudicots, magnoliids and the orders Amborellales, Nymphaeales, and Austrobaileyales (i.e., ANA-grade taxa) as outgroups (11 taxa in total).
DNA Isolation and Library Preparation
We isolated DNA using a modified CTAB protocol (Doyle JJ and Doyle JL 1987; Rai et al. 2003), and prepared whole-genome shotgun sequencing libraries using several library preparation kits. We used Bioo Nextflex DNA sequencing kit (Bioo Scientific Corp., Austin, TX) and KAPA LTP Library Preparation kit (KAPA Biosystems, Boston, MA) when greater than 10 ng of starting DNA was available (we used the Bioo kit for Sciaphila). For lower amounts of initial DNA, we used NuGEN Ovation Ultralow Library System (NuGEN Technologies Inc., San Carlos, CA). We sheared DNAs to 400-bp fragments on a Covaris S220 sonicator (Covaris, Inc., Woburn, MA) for library preparation with all three kits, and size-selected all libraries (550–650 bp fragments). For the Bioo kit we size-selected using a 2% agarose gel, purifying the resulting DNA using a Zymoclean gel recovery kit (Zymo Research, Irving, TX). For the Kapa and NuGEN kits, we used magnetic bead size selection (Agencourt AMPure XP magnetic beads; Beckman Coulter Genomics, Brea, CA). For quality control, we quantified all libraries by Qubit (Qubit fluorometer; ThermoFisher Scientific, Waltham, MA) to ensure a minimum DNA concentration of 0.5 ng/µl. Library fragment sizes were verified by Bioanalyzer (Agilent Technologies, Santa Clara, CA), and concentrations were measured by qPCR on an iQ5 real-time system (Illumina DNA standard kit; KAPA Biosystems; Bio-Rad Laboratories, Inc., Hercules, CA). Individual libraries were multiplexed (Cronn et al. 2008) in several lanes on an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA) and sequenced as 100-bp paired-end reads.
De Novo Contig Assembly, Plastid Gene Annotation, and Plastome Reconstruction
Illumina reads were processed with CASAVA 1.8.2. (Illumina, Inc.) to sort the multiplexed data by taxon. To obtain contigs, we performed de novo assemblies for each individual taxon using CLC Genomics Workbench v.6.5.1 (CLC Bio, Aarhus, Denmark) with default settings. We selected all contigs greater than 500 bp in length with greater than 20× coverage, and used a custom Perl script (Daisie Huang, University of British Columbia) to Basic Local Alignment Search Tool (BLAST) contigs against a local database (Altschul et al. 1990) of plastid genes from Dioscorea elephantipes (GenBank accession number NC_009601.1) in order to remove mitochondrial and nuclear contigs. For Cyclanthus, Freycinetia, Sararanga, Croomia, Pentastemona, Stemona, Stichoneuron, Xerophyta and Lophiola, we annotated plastid genes using DOGMA (Wyman et al. 2004), manually inspecting gene and exon boundaries in Sequencher 4.2.2. (Gene Codes Corporation, Ann Arbor, US) using Phoenix dactylifera (NC_013991) and D. elephantipes to annotate start/stop codons and introns for each protein-coding gene. We exported final gene sets (coding regions) as individual FASTA files for each taxon. For Carludovica and Sciaphila, we assembled full circular plastomes, designing primers using Primer3 (Koressaar and Remm 2007; Untergrasser et al. 2007) to bridge gaps between contigs or to verify contig overlap. Amplification of these regions was performed using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific), followed by sequencing using BigDye Terminator v3.1 sequencing chemistry (Applied Biosystems, Inc., Foster City, CA) on an Applied Biosystems 3730S 48-capillary DNA analyzer (Applied Biosystems, Inc.). We used Sequencher to produce a consensus plastome sequence by assembling contigs produced in CLC together with the Sanger-derived sequences, and annotated the consensus sequences in DOGMA, as discussed above. For Sciaphila, we additionally searched all intergenic spacer regions for potential pseudogenes. We used OGDRAW (Lohse et al. 2013) to generate the two plastome maps.
Data Matrix Construction and Sequence Alignment
We added data for ten newly sequenced species in Pandanales and one species in Dioscoreales to published data (supplementary table S1, Supplementary Material online) for 82 plastid genes (78 protein-coding genes and 4 rDNA genes, with 71 taxon terminals per file; missing genes were represented as blanks). We aligned each gene file in Se-Al v.2.0a11 (Rambaut 2002) using criteria laid out in Graham et al. (2000), staggering gene regions that were difficult to align (e.g., Saarela and Graham 2010). We verified that alignments for protein-coding genes were maintained as open-reading frames, and concatenated all individual gene alignments into a single 102,897-bp matrix (derived from 67,506 bp of unaligned plastid sequence data in C. palmata, for reference), including the inverted repeat (IR) regions only once. To check for compilation errors in the final matrix, we exported the concatenated gene sequences for each taxon and used Sequencher to compare them with the original individual taxon files (none was found). We retrieved plastid gene ycf1 for most taxa (the gene is absent in Sciaphila, see below), but did not include it in the final matrix due to difficulties in alignment.
Phylogenetic Inference
We analyzed the data using parsimony and maximum-likelihood (ML) methods. For the parsimony analysis, we ran a heuristic parsimony search for shortest trees in PAUP* v4.0a134 (Swofford 2003) using tree-bisection–reconnection branch swapping and 1,000 random stepwise addition replicates, holding 100 trees at each step, and otherwise using default settings. We estimated branch support with a bootstrap analysis (Felsenstein 1985), using 1,000 replicates, with 100 random addition replicates per bootstrap replicate for the parsimony analysis (for the ML analyses, see below). For all bootstrap analyses performed here, we considered well-supported branches to have at least 95% bootstrap support, and poorly supported branches to have less than 70% support, following Zgurski et al. (2008).
For the ML analyses, we first conducted heuristic searches of the DNA sequence data using nucleotide substitution models with RAxML v.7.4.2 (Stamatakis 2006), using a graphical interface for it (Silvestro and Michalak 2012). We ran three variant analyses, one with all the data unpartitioned, a second with the data partitioned by codon position (a “codon” partitioning scheme, with rDNA genes considered as additional data partitions), and a third with the data partitioned by both gene and codon positions (“G×C,” or gene by codon partitioning); see below for how the final partitioning schemes were set up. We analyzed translated protein-coding genes with amino acid substitution models in RAxML with unpartitioned data, and with the data also partitioned by gene (described below). Finally, we analyzed the unpartitioned DNA sequence data using a codon-based substitution model implemented in Garli 2.0 (Zwickl 2006). For all ML analyses we conducted 20 independent searches for the best tree, and estimated branch support using 500 bootstrap replicates, using GTRGAMMA or PROTGAMMA approximations for the analyses based on nucleotide/codon versus amino acid substitution models, respectively (we used a subset of taxa for the analysis using the codon-based substitution model, because of computational limitations, see below). Each bootstrap analysis used the same substitution models as the searches for best trees.
For the unpartitioned ML analysis of the DNA sequence data, we used jModeltest 2.1.3 (Guindon and Gascuel 2003; Darriba et al. 2012) to find the optimal DNA substitution model using the Bayesian Information Criterion, BIC (Schwarz 1978). This chose GTR (general time reversible)+G as the best model. For the various partitioned analyses we used PartitionFinder v.1.1.1 and PartitionFinderProtein 1.1.0 (Lanfear et al. 2012) to combine partitions that did not have significantly different DNA or amino-acid substitution models, using the hierarchical clustering algorithm and the BIC, and used the final data partitioning schemes for phylogenetic inference. For the codon partitioning scheme, we allocated nucleotides in protein-coding genes according to whether they belong to the first, second or third codon position, and assigned four additional initial partitions for the plastid rDNA genes (for a total of seven initial partitions). PartitionFinder retained four partitions (one for each codon position, and one for all four rDNA genes), with GTR+G identified as the best model in each case (supplementary table S2a, Supplementary Material online). For the G×C (gene by codon) partitioning scheme for the DNA sequence data, we first partitioned the matrix both by gene (treating the trans-spliced exons of 5′-rps12 and 3′-rps12 as two genes, operationally) and by codon position (first, second and third position for the protein-coding genes, leaving the rDNA genes as distinct partitions), for a total of 241 initial partitions. PartitionFinder retained 12 final partitions, with GTR+G or GTR+G+I selected as the best DNA substitution model in all cases (supplementary table S2b, Supplementary Material online). We used the GTR+G model for individual partitions in subsequent phylogenetic analysis, as the I parameter (invariant sites) may be adequately accommodated by the gamma parameter (Yang 2006). For the amino acid data, PartitionFinderProtein retained 71 partitions from the original 80 partitions (partitioned by gene, again considering 5′-rps12 and 3′-rps12 as two genes), and inferred a range of optimal amino acid models that we used in subsequent phylogenetic inference (supplementary table S2c, Supplementary Material online).
We also analyzed the nucleotide sequence data set using an unpartitioned codon-based substitution model. For this analysis we applied the 6-rate (GTR) codon model with F3×4 codon frequencies and one dN/dS parameter, using Garli 2.0 (Zwickl 2006) on the CIPRES Portal (Miller et al. 2010). Because of severe computational constraints for the latter method, we estimated the bootstrap support for two subsets of this matrix, one including only taxa in Pandanales and Dioscoreales (12 taxa), and a second with additional representatives chosen from most major monocot lineages (26 taxa, see below).
Model-Based Tests of Selective Regime in Plastid Genes
We used the CodeML module in PAML4.8 (Yang 2007) to assess changes in selective regime in 18 protein-coding genes retained as open-reading frames in the Sciaphila plastome (table 1). The objective was to test hypotheses of different ω values (ω is the ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site) for Sciaphila (indicated below as “MHT,” an abbreviation for “mycoheterotroph”), compared with photosynthetic (green) outgroups. We built two codon-based “branch” models, which can detect differences in selection regimes in particular lineages (Yang 2007). In the simplest model (M0, one ratio), all branches evolve under one ω-ratio (i.e., ωMHT = ωgreen; see supplementary table S3, Supplementary Material online. In the alternative model (M1, two ratios), Sciaphila was allowed to evolve under a different ω-ratio than the green taxa (i.e., two ratios allowed, ωMHT and ωgreen). We also compared “branch-site” models to survey for positive selection that may affect only a few sites in a prespecified lineage (Yang 2007). For this test, we specified Sciaphila as the foreground lineage and all green taxa as background lineages. For the null model (H0), the foreground branch (ω2) was fixed to ω2 = 1, allowing codons on this branch to evolve neutrally. In the alternative model (H1), ω2 > 1 was estimated, allowing positive selection in the foreground lineage.
Table 1.
Summary of Genes Retained in Sciaphila Relative to Carludovica
| Function | Carludovica palmata | Sciaphila densiflora |
|---|---|---|
| Photosynthesis | psaA, psaB, psaC, psaI, psaJ | — |
| psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | — | |
| atpA, atpB, atpE, atpF, atpH, atpI | — | |
| petA, petB, petD, petG, petL, petN | — | |
| rbcL | — | |
| ycf3, ycf4 | — | |
| ndhA, ndhB, ndhC, ndhD, ndhE | — | |
| ndhF, ndhG, ndhH, ndhI, ndhJ | ||
| ndhK | ||
| Ribosomal proteins | rpl2, rpl14, rpl16, rpl20, rpl22, rpl23 | rpl2, rpl14, rpl16, rpl20, rpl36 |
| rpl32, rpl33, rpl36 | ||
| rps2, rps3, rps4, rps7, rps8, rps11 | rps2, rps3, rps4, rps7, rps8, rps11 | |
| rps12, rps14, rps15, rps16, rps18, rps19 | rps12, rps14, rps18, rps19 | |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | — |
| rDNAs | rrn4.5, rrn5, rrn16, rrn23 | rrn4.5, rrn5, rrn16, rrn23 |
| tRNAs | trnA-UGC, trnC-GCA, trnD-GUC | trnC-GCA, trnE-UUC, trnfM-CAU |
| trnE-UUC, trnF-GAA, trnfM-CAU | trnI-CAU, trnQ-UUG, trnW-CCA | |
| trnG-GCC, trnG-UCC, trnH-GUG | ||
| trnI-CAU, trnI-GAU, trnK-UUU | ||
| trnL-CAA, trnL-UAA, trnL-UAG | ||
| trnM-CAU, trnN-GUU, trnP-UGG | ||
| trnQ-UUG, trnR-ACG, trnR-UCU | ||
| trnS-GCU, trnS-GGA, trnS-UGA | ||
| trnT-GGU, trnT-UGU, trnV-GAC | ||
| trnV-UAC, trnW-CCA, trnY-GUA | ||
| Other protein-coding genes | accD, ccsA, cemA, clpP, infA, matK | accD, clpP, matK |
| ycf1, ycf2 |
Note.—Dash (—) indicates the absence of all genes for that protein complex.
To implement both the branch and branch-site models, we removed taxa in alignments lacking sequence data and regions with indels that resulted in missing data for 90% or more of the taxa. We used the 26-taxon best tree inferred from the codon-based substitution model ML analysis (see supplementary fig. S6, Supplementary Material online) as a constraint tree, but with branch lengths generated in PAML, and pruned any taxa missing for individual genes. We ran all models on individual genes using the F3×4 codon frequency model. We used the likelihood ratio test statistic −2(ln L M0/H0 – ln L M1/H1) to compare the fit of M0 versus M1 (branch models) or H0 versus H1 (branch-site models), and calculated P values based on a χ2 test with 1 degree of freedom. We used a Bonferroni correction to account for multiple tests conducted on the same data (Anisimova and Yang 2007), and considered tests significant if the P value was <α/m, where m is the number of branches being tested using the same data (m = 2 for both models). We identified any sites undergoing positive selection in the branch-site model using the Bayes empirical Bayes (BEB) test included in the CodeML package.
Results
Full Circular Plastomes
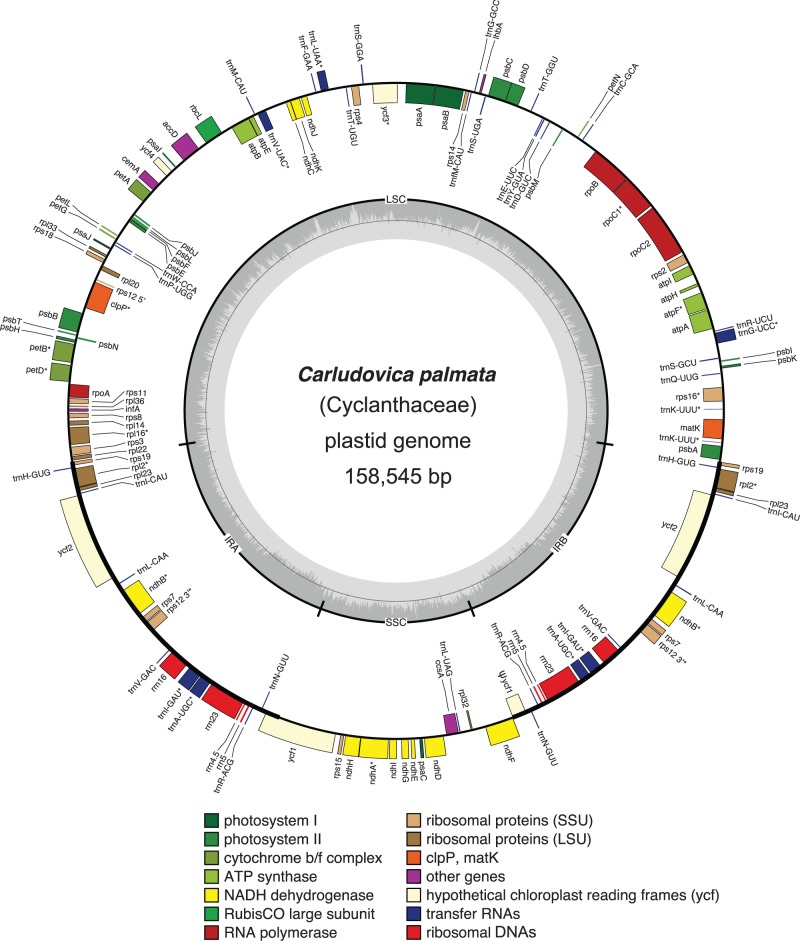
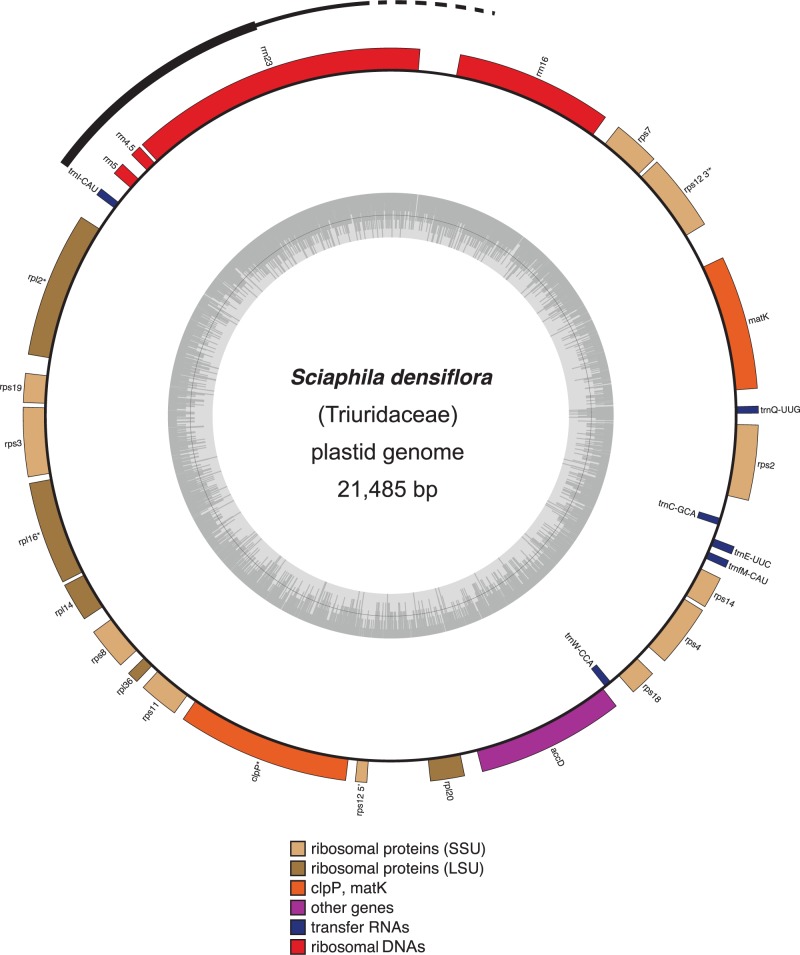
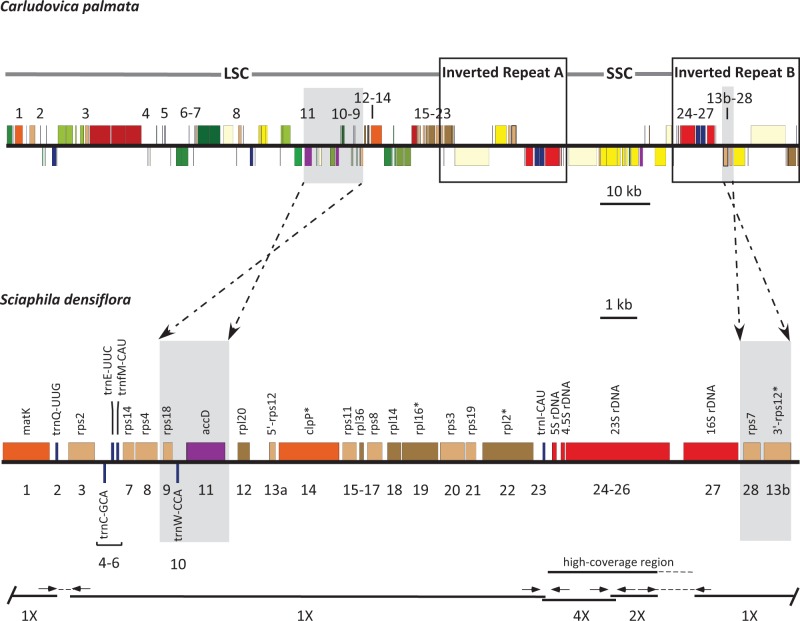
We assembled the plastid genome of C. palmata (GenBank accession number NC026786.1; fig. 1) as a circular sequence of 158,545 bp, with an average of approximately 734.5× coverage from approximately 21.24 million paired-end reads. The Carludovica plastome is comparable to those of other angiosperms in size and organization. It has the typical quadripartite structure of plant plastid genomes, with an 87,041-bp large single copy (LSC) region, an 18,366-bp small single copy (SSC) region, two IR regions of 26,569 bp, and has the same gene order as D. elephantipes (Hansen et al. 2007). We assembled S. densiflora as a circular sequence with a predicted minimum length of 21,485 bp, and an average of approximately 50× coverage (GenBank accession number KR902497.1; fig. 2) from approximately 17.03 million paired-end reads. The DNA extractions for Carludovica and Sciaphila were both done using fresh plant material (Edith Kapinos, Royal Botanic Gardens, Kew, personal communication), and so the order of magnitude lower coverage for the plastome in the mycoheterotroph compared with the autotroph may be consistent with substantially fewer plastid genomes per plant cell for it. Two neighboring sectors of the assembled Sciaphila plastome had substantially higher coverage than the remainder (fig. 3), consistent with them being repeated regions. One sector with approximately 4× the average read depth (214× coverage) includes rrn4.5, rrn5, and part of rrn23; the other includes the remainder of rrn23 and had only approximately twice the average read depth (93× coverage). It is possible that they represent a short series of tandem repeats, but they could also incorporate a reduced and cryptic IR. We were not able to confirm the number or arrangement of these putative repeats because we had a limited amount of DNA for experimental confirmation. No genes from the SSC region that is typical of other angiosperm plastomes (e.g., fig. 1) were recovered. The gene order depicted in figure 3 likely represents the correct order at the ends of any repeated regions, as it is consistent with our ability to connect contigs using direct sequencing (fig. 3; the higher read-depth sectors are also indicated in the Sciaphila genome map). The possibility that high-depth regions instead represent inserts elsewhere (e.g., in the mitochondrial genome) cannot be excluded, although we did not observe obvious sequence variation in these genes that might be indicative of divergent copies in other genomes. The stoichiometry of the coverage levels relative to the remaining plastid contigs is suggestive of replication within the plastid genome rather than elsewhere (i.e., if they are located elsewhere, the coverage depth would not necessarily be near-integer multiples of the rest of the plastome). Also, if there are repeats, we doubt that we have missed additional intervening genes (or pseudogenes), as BLAST-based attempts to recover genes missing from the Sciaphila genome were not successful. Finally, the high degree of colinearity demonstrated here between S. densiflora and its close photosynthetic relative, C. palmata also supports the idea that we have recovered the full complement of retained genes in the mycoheterotrophic species (fig. 3).
Fig. 1.—
Circular plastome map of C. palmata (Cyclanthaceae). Genes located inside the circle are transcribed clockwise, those outside are transcribed counterclockwise. The gray circle marks the GC content: The inner circle marks a 50% threshold. Thick branches indicate IR copies. Genes with introns are indicated with asterisks (*). The short pseudogene copy of ycf1 is marked as “ψ”.
Fig. 2.—
Circular plastome map of S. densiflora (Triuridaceae). Genes located inside the circle are transcribed clockwise, those outside are transcribed counterclockwise. The exterior arc is a sector with possible repeats (thicker line indicates higher coverage, see main text and fig. 3; the dotted line indicates Sanger sequence data). The gray circle marks the GC content: The inner circle marks a 50% threshold. Genes with introns are indicated with asterisks (*).
Fig. 3.—
Comparison of linearized plastomes of C. palmata (Cyclanthaceae) and S. densiflora (Triuridaceae). Boxes indicate IR regions (two copies, A and B) in Carludovica. Dashed arrows indicate predicted inversions of the small blocks highlighted in gray. Black lines below the Sciaphila plastome map indicate individual contigs (numbers below the lines indicate the estimated relative depth of coverage, see text). Gaps and contig overlap were, respectively, connected or confirmed using Sanger sequencing with primers at positions indicated with short arrows (primers not to scale; thin dashed lines are sequenced regions not represented in de novo contigs). A sector with higher read depth is indicated (the extent of higher-coverage is uncertain because this sector overlaps with a region produced using Sanger sequencing, indicated with a dotted line; 4×, five times coverage; 2×, two times coverage, see main text). Numbers indicate the 28 genes retained in Sciaphila, 18 of which are protein-coding (note that rps12 is a trans-spliced gene, noted here as 13a and 13b); *Genes with introns. The scale bars indicate relative plastome sizes of Carludovica and Sciaphila (kb, kilobase).
Our current model of the Sciaphila plastid genome provides a minimum size estimate for it (ignoring the possibility that some regions are duplicated), and is approximately 13.6% of the size of Carludovica (discounting duplications in the IR of the latter), with 28 plastid genes retained in total (table 1). Considering unique sequences only, the coding sequences (proteins, rDNA, and tRNA genes) account for 68.7% of the Sciaphila plastome, whereas 58.0% of the Carludovica plastome is composed of coding sequence. The average GC content is also marginally higher in Sciaphila (39.9%) than in Carludovica (36.7%). Most of the 28 retained genes in Sciaphila are involved in protein synthesis; ten code for small ribosomal proteins and five for large ribosomal proteins, all four rDNA loci are retained, along with six tRNA loci (table 1). The remaining loci are accD (which codes for a subunit of acetyl-coA carboxylase or ACCase), clpP (which codes for a proteolytic subunit of the enzyme Clp-protease), and matK (the maturase gene for group-IIA plastid introns); see Wicke et al. (2011) for further details on gene function. All genes except trnC-GCA and trnW-CCA are transcribed on one strand (fig. 2). We did not recover any pseudogenes (or at least all of the genes in table 1 were open-reading frames). Gene order in Sciaphila is nearly colinear with that in Carludovica, although we infer an inversion of a block comprising rps18, trnW-CAA and accD in the LSC of Sciaphila (fig. 3) and a block comprising 3′-rps12 and rps7 in what was the IR, assuming deletion of intervening sequences in the original IR copies (fig. 3). Genes inferred to be lost from Sciaphila include those coding for photosynthesis-related protein subunits (photosystems II and I, cytochrome b6f complex, and ATP synthase), all plastid-encoded RNA polymerase (PEP) loci, the majority of the tRNA loci, several genes involved in protein synthesis (ribosomal proteins rps15, rps16, rpl22, rpl23, rpl32, rpl33, and infA), and two genes with uncertain function (ycf1 and ycf2).
The Phylogenetic Position of Sciaphila (Triuridaceae)
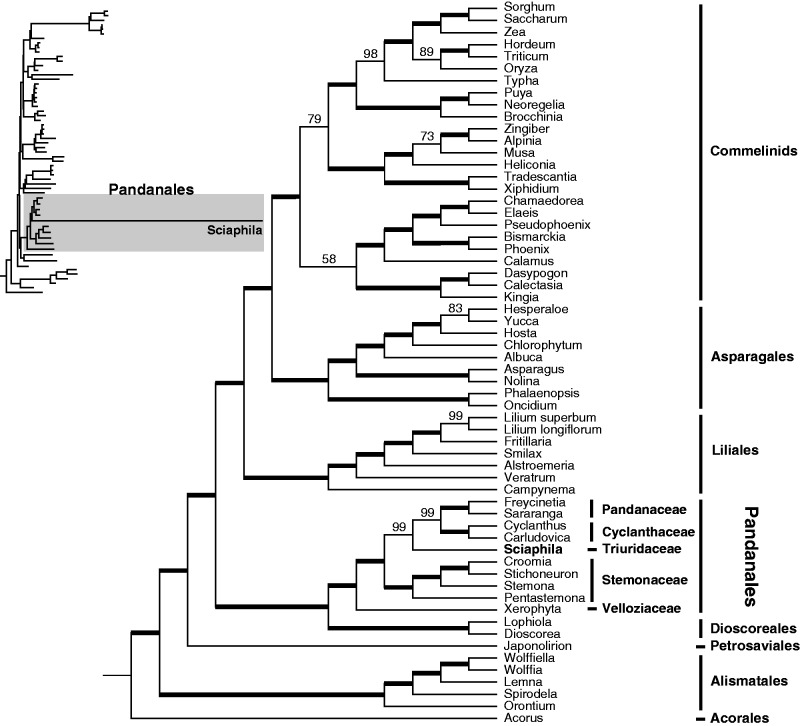
We inferred Sciaphila to be a member of Pandanales in all analyses here, with strong support (fig. 4 and supplementary figs. S1–S7, Supplementary Material online). The monophyly of the order and its sister-group relationship to Dioscoreales were also confirmed with strong support in all likelihood analyses. The position of Sciaphila within Pandanales was completely consistent across all six likelihood analyses, and was also generally strongly supported: A clade comprising Triuridaceae and Cyclanthaceae–Pandanaceae was recovered with 95–99% bootstrap support in the DNA-based ML analyses that used nucleotide substitution models (fig. 4 and supplementary figs. S1–S3, Supplementary Material online), with 88–91% bootstrap support in the amino acid analyses (supplementary figs. S4 and S5, Supplementary Material online), and with 86–87% bootstrap support in the analyses that used a codon-based substitution model (supplementary fig. S6, Supplementary Material online). The clade comprising Cyclanthaceae and Pandanaceae had 88–100% bootstrap support across likelihood analyses (fig. 4 and supplementary figs. S1–S6, Supplementary Material online). Stemonaceae were recovered as the sister group of this clade, and Velloziaceae (represented by Xerophyta) were supported as the sister group of all other Pandanales. The latter relationships all had strong support in all likelihood analyses (97–100%; fig. 4 and supplementary figs. S1–S6, Supplementary Material online; note that one of the analyses shown in supplementary fig. S6, Supplementary Material online, considered only taxa in Dioscoreales and Pandanales).
Fig. 4.—
Phylogenetic relationships in Pandanales in the context of overall monocot phylogeny, based on plastid genome data (82 plastid genes in photosynthetic taxa; 22 in Sciaphila). The data matrix was partitioned using a G × C (gene by codon) partitioning scheme and analyzed using corresponding nucleotide substitution models (see text and supplementary table S2b, Supplementary Material online, for details). Thick lines indicate 100% bootstrap support; branches with lower support are indicated. The scale bar indicates estimated substitutions per site. This is a subset of a larger angiosperm-wide sampling (supplementary fig. S3, Supplementary Material online, shown as an inset phylogram here; the shaded portion represents Pandanales).
The sole analysis that yielded a different topology concerning the placement of Sciaphila was the parsimony analysis, which recovered it as sister to Xerophyta, but with poor bootstrap support (65%; supplementary fig. S7, Supplementary Material online). The long branch typical of Sciaphila in the likelihood analyses was notably not evident in the parsimony analysis (cf. supplementary figs. S1–S6; fig. S7, Supplementary Material online). This analysis also supported the monophyly of Pandanales, but with only moderate support (71%), suggesting that Sciaphila destabilizes the support for relationships when included in parsimony analysis. We tested this by excluding Sciaphila and rerunning the parsimony analysis; the underlying relationships were not affected, but support values within Dioscoreales and Pandanales improved dramatically (supplementary fig. S7, Supplementary Material online). There were no major differences in monocot relationships across the various likelihood and parsimony analyses.
Tests of Selection
A ω value greater than 1 is interpreted as evidence for positive selection, ω value less than 1 suggests purifying (negative) selection, and ω ≈ 1 indicates neutral evolution (Zhang et al. 2005). Under the branch models (comparing the one-ratio model M0 and the two-ratio model M1), the M0 model fit the data better for 15 of the 18 genes tested (trans-spliced rps12 was treated as two genes, operationally; these portions are listed separately in supplementary table S3, Supplementary Material online, but are lumped as one gene in the discussion below) indicating that these retained genes in Sciaphila are evolving at the same ω rate as in the green outgroups (supplementary table S3, Supplementary Material online). These 15 genes appear to be highly conserved and under purifying selection in the analyzed taxa (0.096 < ω < 0.368). The two-ratio model (M1) was a better fit for clpP, rpl14, and rps7 after Bonferroni correction, suggesting that these genes are under a significantly different selective regime in Sciaphila than in the green taxa. The rps7 locus of Sciaphila (ωMHT = 0.611) approached the expectations for neutral evolution (ω ≈ 1), compared with evidence of strong purifying selection (ω = 0.203) in green taxa. Although we detected significant differences in ω rates for clpP and rpl14, these two genes are still predicted to be experiencing purifying selection in Sciaphila, although this may have also been relaxed (ωMHT = 0.288 and ωMHT = 0.240, respectively, for clpP and rpl14 in Sciaphila; the corresponding values for green taxa are: ωgreen = 0.154 and ωgreen = 0.096).
In the branch-site tests, the null model of neutral evolution (H0), which allows no sites to be under positive selection, appeared to fit the data better for 15 of the 18 genes tested (supplementary table S4, Supplementary Material online). An alternative model of positive evolution (H1), which allows some sites to be under positive selection, was a better fit for accD, rpl20 and rps18, although the result was not significant for rps18 after Bonferroni correction. The BEB test found two positively selected sites in each of the three genes. We located these sites in the alignments and speculate that this result is due to alignment difficulties for these parts of the genes, which are quite variable in Sciaphila. After staggering these hard-to-align sections in a revised alignment (effectively removing them from consideration) and rerunning the PAML tests, we found no evidence of positive selection elsewhere in these genes (supplementary table S4, Supplementary Material online). We therefore suspect that the positive selection results are artifacts. To ensure that these realignments did not affect the phylogenetic placement of Sciaphila, we substituted the realigned versions of these three genes in the original data matrix and reran two ML phylogenetic analyses for the nucleotide data, using nucleotide-substitution models, one with unpartitioned data and the second with the G×C partitioning scheme (see supplementary table S2d, Supplementary Material online, for partitioning scheme), repeating the phylogenetic procedures described above. These minor alterations in the alignment did not affect the placement or support for the placement of Sciaphila (<5% difference in support values; cf. supplementary figs. S1 vs. S8; S3 vs. S9, Supplementary Material online).
Discussion
Gene Loss and Retention in Sciaphila (Triuridaceae)
The retention of only 28 genes in total (18 protein-coding genes, 4 rDNA genes, and 6 tRNA genes) makes the S. densiflora plastid genome one of the smallest ones known in land plants, at least in terms of the number of genes (tables 1 and 2). Sciaphila therefore appears to be in the late stages of plastome reduction, and may be well on its way to full gene loss (Wicke et al. 2013). Photosynthetic land plants have a remarkable degree of conservation of gene content, and considering the nonduplicated genes in the IR, angiosperms typically have 79 protein-coding genes, 4 rDNA genes, and 30 tRNA genes (e.g., Wicke et al. 2011). Carludovica palmata exemplifies a typical angiosperm plastome arrangement (fig. 1; table 1). In contrast, heterotrophic plants may show extensive gene loss, reflecting relaxed evolutionary constraints following the loss of photosynthesis (Krause 2008; Wicke et al. 2011; Barrett and Davis 2012). For example, the mycoheterotrophic liverwort A. mirabilis has retained 125 genes, including duplicates in its IR region, and the holoparasitic species Epifagus virginiana and the mycoheterotrophic orchid R. gardneri (table 2) have retained only 55 genes and 37 genes, respectively (Wolfe et al. 1992; Delannoy et al. 2011). Although Rafflesia lagascae may have lost its plastome entirely (Molina et al. 2014), as in multiple lineages of secondarily heterotrophic unicellular eukaryotes (e.g., Abrahamsen et al. 2004; Smith and Lee 2014; Janouškovec et al. 2015), plastome loss remains to be definitively demonstrated in any land plant. Outside the land plants, the parasitic green alga Helicosporidium sp. has 54 genes (de Koning and Keeling 2006), and the malarial parasite Plasmodium falciparum has 68 genes (Wilson et al. 1996).
Table 2.
Summary of Genes Retained in Published Circular Plastid Genomes of Mycoheterotrophic Species
| Sciaphila densiflora (Triuridaceae) | Aneura mirabilis (Aneuraceae) | Corallorhiza striata (Orchidaceae) | Neottia nidus-avis (Orchidaceae) | Rhizanthella gardneri (Orchidaceae) | Epipogium aphyllum (Orchidaceae) | Epipogium roseum (Orchidaceae) | Petrosavia stellaris (Petrosaviaceae) | |
|---|---|---|---|---|---|---|---|---|
| Photosynthesis | ||||||||
| Photosystem I | – | psaC, I, J, M | psaI, J | – | – | – | – | – |
| ycf4 | ||||||||
| Photosystem II | – | psbA, F, H, I | psbH, I, K, M, | – | – | – | – | psbI, Z |
| J, L, M, N, T, Z | N, T, Z | |||||||
| ATP synthase | – | atpA, B, E, F | atpA, B, E, F | – | – | – | – | atpA, B, E, F |
| H, I | H, I | H, I | ||||||
| Cytochrome b6f | – | petD, G, L, N | petG, L | – | – | – | – | petG |
| Rubisco | – | rbcL | – | – | – | – | – | rbcL |
| NAD(P)H | – | – | ndhJ | – | – | – | – | – |
| dehydrogenase | ||||||||
| Gene expression | ||||||||
| Ribosomal protein | rpl2, 14, 16 | rpl2, 14, 16 | rpl2, 14, 16 | rpl2, 14, 16 | rpl2, 14, 16, 20 | rpl2, 14, 16 | rpl2, 14, 16 | rpl2, 14, 16 |
| 20, 36 | 20, 21, 22, | 20, 22, 23 | 20, 22, 23 | 23, 36 | 36 | 20, 36 | 20, 22, 23 | |
| 23, 32, 33, 36 | 32, 33, 36 | 32, 33, 36 | 32, 33, 36 | |||||
| rps2, 3, 4, 7 | rps2, 3, 4, 7 | rps2, 3, 4, 7 | rps2, 3, 4, 7 | rps2, 3, 4, 7 | rps2, 3, 4, 7 | rps2, 3, 4, 7, | rps2, 3, 4, 7 | |
| 8, 11, 12, 14 | 8, 11, 12, 14 | 8, 11, 12, 14 | 8, 11, 12, 14 | 8, 11, 14, 18 | 8, 11, 12, 14 | 8, 11, 12, 14 | 8, 11, 12, 14 | |
| 18, 19 | 15, 18, 19 | 15, 16, 18, 19 | 15, 19 | 19 | 18, 19 | 18, 19 | 15, 16, 18, 19 | |
| RNA polymerase | – | rpoA, B, C1, C2 | – | – | – | – | – | – |
| rDNAs | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 | rrn4.5, 5, 16, 23 |
| tRNAs | CGCA, EUUC, ICAU | AUGC, CGCA, DGUC | AUGC, CGCA, DGUC | AUGC, CGCA, DGUC | CGCA, DGUC, EUUC | CGCA, EUUC, FGAA | CGCA, EUUC, FGAA | AUGC, CGCA, DGUC |
| fMCAU, QUUG | EUUC, FGAA, GGCC | EUUC, FGAA, GGCC | EUUC, FGAA, GGCC | FGAA, ICAU, fMCAU | ICAU, fMCAU, YGUA | ICAU, fMCAU, QUUG | EUUC, FGAA, GGCC | |
| WCCA | GUCC, HGUG, ICAU | GUCC, HGUG, ICAU | GUCC, HGUG, ICAU | QUGG, WCCA, YGUA | YGUA | GUCC, HGUG, ICAU | ||
| IGAU, KUUU, LCAA, | IGAU, KUUU, LCAA | KUUU, LCAA, LUAA | IGAU, KUUU, LCAA | |||||
| LUAA, LUAG, fMCAU | LUAA, LUAG | LUAG, fMCAU MCAU | LUAA, LUAG, fMCAU | |||||
| MCAU, NGUU, PUGG | fMCAU, MCAU | NGUU, QUUG, RACG | MCAU, NGUU, PUGG | |||||
| QUUG, RUCU, RACG, | NGUU, PUGG, QUUG | RUCU, SGCU, SUGA | QUUG, RACG, RUCU | |||||
| RCCG, SGCU, SGGA | RACG, RUCU, SGCU, | TGGU, TUGU, WCAA | SGCU, SGGA, SUGA | |||||
| SUGA, TUGU, TGGU | SGGA SUGU, TUGU | VGAC, YGUA | VGAC, VUAC WCCA | |||||
| VGCA, VUAC, TUGU | VGAC, VUAC, WCAA | WCCA, YGUA | ||||||
| YGUA | YGUA | |||||||
| Other protein-coding genes | accD, clpP, matK | accD, cemA, clpP | accD, clpP, infA | accD, clpP, infA | accD, clpP, infA | accD, clpP, infA | accD, clpP, infA | accD, clpP, infA |
| infA, matK, chlB | matK, ycf1&2 | ycf1&2 | ycf1&2 | matK, ycf1&2 | ||||
| chlN, chlL, ycf1&2 |
Note.— For protein-coding genes, only those found as “open-reading frames” are included. Genes that are found in all species listed above are shown in bold.
The common gene set retained across mycoheterotrophs includes ribosomal proteins (rpl2, 14, 16, and 36; rps2, 3, 4, 7, 8, 11, 14, and 19; the sequence for rps18 in Neottia has a single in-frame internal stop codon which may be RNA edited, so this may also be consistent with a retention of rps18), other protein-coding genes (accD and clpP), all four rDNA loci (rrn4.5, 5, 16, and 23), and four tRNAs (trnC-GCA, E-UUC, I-CAU and fM-CAU). A slightly smaller set of genes is retained in heterotrophic plants in general, that is, including holoparasitic plants (Li et al. 2013; Wicke et al. 2013; Barrett et al. 2014); rps3, rps19 and trnC-GCA are not part of this broader list because they have been lost in some taxa. Sciaphila is evidently near the end of the degradation trajectory proposed by Barrett and Davis (2012) and Barrett et al. (2014), in which the only genes retained are those involved in housekeeping activities.
Commonly retained plastid genes in heterotrophs whose gene products are not involved in photosynthesis or translation include accD, clpP, and matK. However, these loci have been lost individually from the plastid genome of at least one heterotrophic lineage (e.g., Delannoy et al. 2011; Logacheva et al. 2011; Wicke et al. 2013), and in some autotrophs; accD has been lost in several lineages of photosynthetic angiosperms (Jansen et al. 2007), and clpP in several lineages of photosynthetic eudicots (see Straub et al. 2011). Losses in photosynthetic lineages could be explained by functional transfer of the gene to the nuclear genome, which likely occurred for accD in Campanulaceae (Rousseau-Gueutin et al. 2013), for example, or by replacement of the plastid function by a distinct nuclear gene product with similar function, as with replacement of PEPs with nuclear-encoded RNA polymerases (e.g., Zhelyazkova et al. 2012).
The loss of the majority of the plastid tRNA genes in Sciaphila may indicate extensive modification in the functioning of its plastome translation apparatus. Plastid tRNAs may be replaced over evolutionary time by tRNAs imported from the cytosol (e.g., Alkatib et al. 2012), or may be functionally replaced by other tRNAs through the “superwobbling” effect (see Rogalski et al. 2008). One tRNA gene, trnE-UUC, has been found to be retained in the plastid genomes of all heterotrophic plants to date (see table 2 for mycoheterotrophs). Barbrook et al. (2006) hypothesized that this would be the last gene to be retained in the plastid genome of any heterotrophic plant, because of its essential additional role in heme biosynthesis. The precursor of heme, aminolevulinic acid (ALA), is synthesized in land-plant plastids through the C5-pathway, which begins with the ligation of plastid tRNAGlu to glutamate. Secondarily heterotrophic eukaryotes that lack plastid genomes (unicellular eukaryotes: Abrahamsen et al. 2004; Smith and Lee 2014; Janouškovec et al. 2015; possibly the holoparasite Rafflesia: Molina et al. 2014) may either import a viable nuclear or mitochondrial tRNAGlu into the plastid, or instead synthesize ALA through the Shemin pathway in mitochondria (Oborník and Green 2005; Barbrook et al. 2006; Smith and Lee 2014).
General Retention of Colinearity despite Genome Reduction in Sciaphila
Sciaphila exhibits relatively few changes in gene order, despite extensive gene loss (table 2; figs. 2 and 3). Our minimum size estimate of the Sciaphila plastome (21,485 bp) is smaller than most previously published heterotrophic plant genomes, with the exception of the orchid E. roseum, which has a genome size of 19,047 bp (Schelkunov et al. 2015), although undocumented repeats may add to its size, as noted above. The nonrepeated content of the Sciaphila plastid genome is smaller than that of the parasitic green alga Helicosporidium, with a genome size of 37,454 bp (de Koning and Keeling 2006) and the malarial parasite P. falciparum, with a genome size of 34,682 bp (Wilson et al. 1996), although the latter genome includes an IR.
Heterotrophic plant lineages often exhibit extensive changes in their plastomes in terms of gene order, compared with the extensively conserved genomes of photosynthetic land plants (Palmer and Stein 1986). Relaxed selective constraints (e.g., relaxation of selection against repetitive elements that can trigger rearrangements) may contribute to plastid genome rearrangements in heterotrophic lineages (e.g., Wicke et al. 2013). Rearrangements may also be exacerbated by extensive modification or loss of the IR, which may have occurred in Sciaphila (figs. 2 and 3). The IR is thought to act as a stabilizing factor during recombination-dependent replication of the plastome (e.g., Magee et al. 2010; Maréchal and Brisson 2010; Wicke et al. 2011; Sabir et al. 2014). A range of structural alterations have been observed in mycoheterotrophic monocots, including those that are apparently in the early stages of genome reduction, such as Neottia and Corallorhiza (Logacheva et al. 2011; Barrett and Davis 2012), which mostly show only gene loss, to those with more extensive and large-scale rearrangements, such as E. aphyllum, which has lost its SSC region (Schelkunov et al. 2015), and Petrosavia, which has multiple major rearrangements (Logacheva et al. 2014). In contrast, Sciaphila is largely colinear with green angiosperms, such as its close relative Carludovica in Cyclanthaceae (fig. 3). Almost all of the differences can be explained by gene loss events; retained genes are shown in the figure (as numbered labels). The substantial colinearity observed here between Sciaphila and photosynthetic relatives (fig. 3), ignoring gene losses, might point to retention of a cryptic IR (see above) as a stabilizer of genome structure.
Model-Based Tests of Selective Regime in Plastid Genes
Generally relaxed functional constraints resulting from the loss of photosynthesis may also affect plastid-encoded housekeeping genes (e.g., Young and dePamphilis 2005; McNeal et al. 2009). We detected little evidence of this effect here (supplementary table S3, Supplementary Material online), as most retained genes in the Sciaphila plastome are inferred to be under strong purifying selection. Barrett et al. (2014) also found that housekeeping genes retained in the plastome of the fully mycoheterotrophic orchid Corallorhiza were under purifying selection, and the ω-ratios they observed were not significantly different from those of homologous genes in green relatives. Plastid ribosomal protein and tRNA genes are likely retained in the long term because of the general retention of accD and clpP (two nonphotosynthesis-related genes) in land-plant plastid genomes (e.g., Delannoy et al. 2011), which occurs regardless of autotrophy or heterotrophy status. As persistence of any essential plastid encoded-genes requires a functional apparatus for translation, translation apparatus genes would in turn be under strong purifying selection to be retained. Delannoy et al. (2011) hypothesized that plastid-encoded accD and clpP are required for essential plastid-mediated regulation of the production of their respective multisubunit complexes. ClpP is part of the multisubunit Clp protease, and accD codes for the β-carboxyltransferase subunit of ACCase; this subunit regulates fatty-acid biosynthesis in the plastid (see also Bungard 2004, who hypothesized a similar regulatory role for accD). AccD and clpP have both been lost from the plastid genomes of several plant lineages (see above), but these losses appear to be unrelated to the loss of photosynthesis, as they all occurred in green lineages.
Although the branch models test indicates elevated rates of nucleotide substitution in Sciaphila compared with homologous genes in green relatives (data not shown, but also evident in our ML phylograms, e.g., supplementary fig. S3, Supplementary Material online), there appear to have been a proportional increase in both nonsynonymous and synonymous substitution rates, given that the ω-values of most retained genes are consistent with their photosynthetic relatives. Delannoy et al. (2011) also observed this pattern in the mycoheterotrophic orchid Rhizanthella. Although our findings support the continued functionality of all or most retained genes in Sciaphila, recent losses of function cannot be completely ruled out, as there may be a lag between the loss of function/loss of purifying selection and our ability to detect it through pseudogenization, etc. (e.g., Leebens-Mack and dePamphilis 2002). A possible example of this phenomenon concerns the ribosomal protein rps7, which is retained here (table 2) and in all heterotrophic plant plastomes sequenced to date (Li et al. 2013; Wicke et al. 2013; Barrett et al. 2014). This locus may be in the early stages of degradation in Sciaphila, as it has an ω-rate three times that of green taxa (rps7, ω = 0.611; supplementary table S3, Supplementary Material online), approaching ω ≈ 1, the rate expected under neutral evolution. The final expected fate of genes no longer under selective retention is the accumulation of stop codons and indels, leading eventually to complete deletion from the plastome (e.g., Barrett and Davis 2012).
As the genes retained in Sciaphila are mostly housekeeping genes involved in basic plastid processes, we did not expect to find substantial evidence of positive selection. Our initial findings for evidence of positive selection using the branch-site model for three genes (accD, rpl20, and rps18) are probably artifacts of alignment difficulties in highly variable regions, highlighting the sensitivity of this test to slight misalignment. Previous studies have identified sites under positive selection in plastid genes of heterotrophic plants using branch-site models. For example, Barrett et al. (2014) found evidence of positive selection in atp genes retained in fully mycoheterotrophic Corallorhiza. The atp gene complex plays a critical role in photosynthesis, and changes in selective regime may be due to genes having additional or modified plastid functions (Wicke et al. 2013; Barrett et al. 2014). McNeal et al. (2009) found that positive selection may be acting on a codon of gene matK retained in the plastome of the parasitic plant Cuscuta nitida. The gene product of this locus is likely involved in splicing seven plastid group IIA introns (Zoschke et al. 2010); Cu. nitida has lost six of the seven, and may be undergoing positive selection in the matK X-domain (a putative RNA binding domain) in response to this (McNeal et al. 2009). Of the three group IIA introns retained in Sciaphila (one each in clpP, rpl2, 3′-rps12; fig. 2), only rpl2 and 3′-rps12 are targets of matK (Zoschke et al. 2010). However, we found no signs of positive selection in matK in Sciaphila (supplementary table S4, Supplementary Material online). A fourth intron is present in rpl16 (fig. 3).
Resolution of the Phylogenetic Position of Triuridaceae in Pandanales
Until recently, most phylogenetic studies of mycoheterotrophs focused on mitochondrial and nuclear genes for phylogenetic inference (e.g., Neyland and Hennigan 2003; Davis et al. 2004; Merckx et al. 2006, 2009; Mennes et al. 2013, 2015), as rate elevation and the loss of mycoheterotroph plastid genes were thought to make them problematic for phylogenetic inference (e.g., Cronquist 1988, p. 467; Merckx, Mennes, et al. 2013). Molecular data have been scarce for Triuridaceae (Mennes et al. 2013), and previous attempts to amplify plastid genes from the family were unsuccessful (Chase et al. 2000; Caddick et al. 2002). The only purported plastid marker available for Triuridaceae on GenBank is an unpublished rbcL sequence of Sciaphila sp. (FN870930.1), which is a probable contaminant (it has 97% BLAST match to Commelinaceae, and the gene is not retained in the species of Sciaphila we sequenced).
The phylogenetic affinities of members of Triuridaceae have proved to be elusive since the first species was described by Miers (1842). Previous studies suggested relationships with other mycoheterotrophic taxa, such as Petrosaviaceae (Cronquist 1988; Takhtajan 1997). In an early phylogenetic study based on morphology, Dahlgren and Rasmussen (1983) placed the family within Alismatales (as the sister group of the core alismatid families). Dahlgren et al. (1985) later considered its phylogenetic relationship to other families and even its placement within the monocots to be unclear, see also the overview of Triuridaceae systematics in Mennes et al. (2013). Chase et al. (2000) generated the first molecular data for this family (a nuclear 18S rDNA sequence of Sciaphila), placing it in Pandanales, a small order of monocots that includes the four photosynthetic families Cyclanthaceae, Pandanaceae, Stemonaceae, and Velloziaceae (APG 2009). Additional studies using one or a few mitochondrial and nuclear sequences (Caddick et al. 2002; Davis et al. 2004; Mennes et al. 2013) and morphological characters (Caddick et al. 2002; Rudall and Bateman 2006) added support for the inclusion of the family in Pandanales. Triuridaceae were therefore assigned to Pandanales in the most recent version of the Angiosperm Phylogeny Group classification system (APG 2009). However, the family’s precise position within Pandanales has remained uncertain or poorly supported. Different studies have placed it with weak support as the sister group of Cyclanthaceae and Pandanaceae based on 18S rDNA (Chase et al. 2000), as the sister group of Velloziaceae or of a clade comprising Cyclanthaceae, Pandanaceae, and Stemonaceae based on mitochondrial atpA (Davis et al. 2004), or even embedded within Stemonaceae based on morphological data (Rudall and Bateman 2006). More recently, Mennes et al. (2013) found it to be the sister group of Cyclanthaceae, Pandanaceae, and Stemonaceae using nuclear and mitochondrial data (18S rDNA and three mitochondrial genes). The support for the latter relationship was weak, however, although their analyses strongly rejected a close relationship between Triuridaceae and Velloziaceae, or an origin of Triuridaceae within Stemonaceae. Mennes et al. (2013) also found strong support for the monophyly of Triuridaceae, considering five sampled genera.
Broad inferences of angiosperm phylogeny have relied extensively on a few plastid genes until relatively recently (e.g., APG 2009). It would therefore be valuable to integrate mycoheterotrophs into this large body of evidence. Here, we demonstrated the utility of whole plastid genome data for the higher-order phylogenetic placement of Triuridaceae. We confirmed its inclusion in Pandanales (fig. 4), as proposed in previous studies (Chase et al. 2000; Caddick et al. 2002; Davis et al. 2004; Rudall and Bateman 2006; Mennes et al. 2013). We also inferred a more confident placement of Triuridaceae among the four green families in Pandanales, as we found strong evidence that it is the sister group of a clade comprising Cyclanthaceae and Pandanaceae (fig. 4). Stemonaceae are the sister group of this clade, and Velloziaceae are the sister group of all remaining families in the order. Whole plastome data may also be useful for inferring relationships among the nine genera in Triuridaceae, several of which have not been sampled to date (e.g., Mennes et al. 2013). However, we have not been able to retrieve plastid genes to date from two of the genera, Kupea and Seychellaria.
Elevated rates of substitution are generally observed across all three genomes of mycoheterotrophs compared with their green relatives (e.g., Merckx et al. 2006, 2009), but may be particularly acute in the plastid genome. The resulting long branches may cause misinference in phylogenetic analyses, especially when taxon sampling is sparse (Felsenstein 1978). This may result in incorrect placements of heterotrophic lineages, and destabilizing effects on phylogenetic inference for neighboring clades. We demonstrated the latter effect here for parsimony by comparing the results with Sciaphila included versus excluded from consideration in the analysis (supplementary fig. S7, Supplementary Material online). Fortunately, long-branch effects are expected to be less acute for model-based methods such as ML (e.g., Felsenstein 1988; Yang 1996; Huelsenbeck 1997, 1998; Swofford et al. 2001; Yang and Rannala 2012), and may also be minimized by using a dense sampling of species and by using the most realistic model of sequence evolution (Philippe et al. 2011). Here we used model-testing, different data partitioning schemes and examined substitution models that operate at the level of nucleotides, amino acid residues or codons to explore this issue. Nucleotide and amino acid models are commonly used and well-characterized methods for phylogenetic inference (for comprehensive reviews, see Swofford et al. 1996; Lío and Goldman 1998). Codon-based substitution models are less frequently implemented, as they consider sequence change at the level of codons, rather than nucleotides or amino acid residues, and are considerably more computationally intensive. However, they may be more reliable and biologically meaningful than other methods (Goldman and Yang 1994), and may accommodate both closely related and highly divergent sequences in phylogenetic inference (Miyazawa 2013). Here, the codon-based method recovered parallel results to the nucleotide and amino acid-based substitution models. This supports the idea that our phylogenetic inferences and the identification of the closest living green relatives of Triuridaceae are robust to these diverse analytical assumptions.
Supplementary Material
Supplementary figures S1–S9 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Ruth Stockey, Gar Rothwell, Timothy Gallaher, Royal Botanic Gardens, Kew, and Kirstenbosch National Botanical Garden for assistance in obtaining material. They also thank Greg Ross for help in retrieving plastid genomes, Craig Barrett for providing data and advice on plastome assembly and selection tests, Wesley Gerelle for advice on selection tests, David Tack and Daisie Huang for bioinformatics support, Carl Rothfels for advice on phylogenetic inference using codon substitution models, and two anonymous reviewers for their comments. This work was supported by an NSERC (Natural Sciences and Engineering Research Council of Canada) Discovery grant to S.W.G., and by NSERC postgraduate fellowships to V.K.Y.L. and M.S.G. and a UBC (University of British Columbia) Four-Year Fellowship to V.K.Y.L.
Literature Cited
- Abrahamsen MS, et al. 2004. Complete genome sequencing of the apicocomplexan, Cryptosporidium parvum. Science 304:441–445. [DOI] [PubMed] [Google Scholar]
- Alkatib S, Fleischmann TT, Scharff LB, Bock R. 2012. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res. 40:6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121. [Google Scholar]
- Anisimova A, Yang Z. 2007. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol Biol Evol. 24:1219–1228. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. 2006. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci.. 11:102–108. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99:1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI, Leebens-Mack J, Conran JG, Stevenson DW. 2013. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics 29:65–87. [DOI] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in early-transitional heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31:3095–3112. [DOI] [PubMed] [Google Scholar]
- Bungard RA. 2004. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays 26:235–247. [DOI] [PubMed] [Google Scholar]
- Caddick LR, Rudall PJ, Wilkin P, Hedderson TAJ, Chase MW. 2002. Phylogenetics of Dioscoreales based on analyses of morphological and molecular data. Bot J Linn Soc. 138:123–144. [Google Scholar]
- Chase MW, et al. 2000. Higher-level systematics of the monocotyledons: an assessment of current knowledge and a new classification. In: Wilson KL, Morisson DA, editors. Monocots: systematics and evolution. Melbourne (Vic.): CSIRO Publishing; p. 3–16. [Google Scholar]
- Cronn R, et al. 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronquist A. 1988. The evolution and classification of flowering plants. 2nd ed. Bronx (NY): The New York Botanical Garden. [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. 1985. The families of the monocotyledons. structure, evolution, and taxonomy. Berlin (Germany): Springer. [Google Scholar]
- Dahlgren RMT, Rasmussen FN. 1983. Monocotyledon evolution: characters and phylogenetic estimation. Evol Biol. 16:255–395. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JI, et al. 2004. A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values. Syst Bot. 29:467–510. [Google Scholar]
- de Koning AP, Keeling PJ. 2006. The complete plastid genome of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, des Francs-Small CC, Brundrett M, Small I. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28:2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 27:401–410. [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. Am Nat. 125:1–15. [Google Scholar]
- Felsenstein J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 22:521–565. [DOI] [PubMed] [Google Scholar]
- Furness CA, Rudall PJ, Eastman A. 2002. Contribution of pollen and tapetal characters to the systematics of Triuridaceae. Plant Syst Evol. 235:209–218. [Google Scholar]
- Givnish TJ, et al. 2010. Assembling the tree of the monocotyledons: plastome sequence phylogeny and evolution of Poales. Ann Mo Bot Gard. 97:584–616. [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Graham SW, Reeves PA, Burns AC, Olmstead RG. 2000. Microstructural changes in noncoding chloroplast DNA: interpretation, evolution, and utility of indels and inversions in basal angiosperm phylogenetic inference. Int J Plant Sci. 161:S83–S96. [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hansen DR, et al. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 45:547–563. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP. 1997. Is the Felsenstein zone a fly trap? Syst Biol.. 46:69–74. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP. 1998. Systematic bias in phylogenetic analysis: is the Strepiptera problem solved? Syst Biol.. 47:519–537. [PubMed] [Google Scholar]
- Imhof S. 2010. Are monocots particularly suited to develop mycoheterotrophy? In: Seberg O, Peterson G, Barfod AS, Davis JI, editors.. Diversity, phylogeny, and evolution in the monocotyledons. Aarhus (Denmark): Aarhus University Press; p. 11–23. [Google Scholar]
- Janouškovec J, et al. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1423790112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104:19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. [DOI] [PubMed] [Google Scholar]
- Krause K. 2008. From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet. 54:111–121. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack JH, dePamphilis CW. 2002. Power analysis of tests for loss of selective constraint in cave crayfish and nonphotosynthetic plant lineages. Mol Biol Evol. 19:1292–1302. [DOI] [PubMed] [Google Scholar]
- Li X, et al. 2013. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One 8:e58747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lío P, Goldman N. 1998. Models of molecular evolution and phylogeny. Genome Res. 8:1233–1244. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. 2014. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 6:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA. 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis . Genome Biol Evol. 3:1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas-van de Kamer H, Weustenfeld T. 1998. Triuridaceae. In: Kubitzki K, editor. The families and genera of vascular plants. III. Flowering plants: Monocotyledons. Berlin (Germany): Springer; p. 452–458. [Google Scholar]
- Magee AM, et al. 2010. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20:1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186:299–317. [DOI] [PubMed] [Google Scholar]
- Martin M, Sabater B. 2010. Plastid ndh genes in plant evolution. Plant Physiol Biochem. 48:636–645. [DOI] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. 2009. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS One 4:e5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennes CB, et al. 2015. Ancient Gondwana break-up explains the distribution of the mycoheterotrophic family Corsiaceae (Liliales). J Biogeogr. 42:1123–1136. [Google Scholar]
- Mennes CB, Smets EF, Moses SN, Merckx VSFT. 2013. New insights in the long-debated evolutionary history of Triuridaceae (Pandanales). Mol Phylogenet Evol. 69:994–1004. [DOI] [PubMed] [Google Scholar]
- Merckx V, Bakker FT, Huysman S, Smets E. 2009. Bias and conflict in phylogenetic inference of myco-heterotrophic plants: a case study in Thismiaceae. Cladistics 25:64–77. [DOI] [PubMed] [Google Scholar]
- Merckx V, et al. 2006. Phylogeny and evolution of Burmanniaceae (Dioscoreales) based on nuclear and mitochondrial data. Am J Bot. 93:1684–1698. [DOI] [PubMed] [Google Scholar]
- Merckx V, Freudenstein JV. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytol. 185:605–609. [DOI] [PubMed] [Google Scholar]
- Merckx VSFT, editor. 2013. Mycoheterotrophy: an introduction. In: Mycoheterotrophy. The biology of plants living on fungi. New York: Springer-Verlag; p. 2–13. [Google Scholar]
- Merckx VSFT, Freudenstein JV, et al. 2013. Taxonomy and classification. In: Merckx V, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer-Verlag; p. 49–50. [Google Scholar]
- Merckx VSFT, Mennes CB, Peay KG, Geml J. 2013. Evolution and diversification. In: Merckx V, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer-Verlag; p. 222–226. [Google Scholar]
- Miers J. 1842. Description of a new genus of plants from Brazil. Trans Linn Soc Lond. 19:77–80. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE); 2010 Nov 14; New Orleans, LA. p. 1-8. [Google Scholar]
- Miyazawa S. 2013. Superiority of a mechanistic codon substitution model even for protein sequences in phylogenetic analysis. BMC Evol Biol. 13:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J, et al. 2014. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol Biol Evol. 31:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyland R, Hennigan M. 2003. A phylogenetic analysis of large-subunit (26S) ribosome DNA sequences suggests that the Corsiaceae are polyphyletic. N Z J Bot. 41:1–11. [Google Scholar]
- Oborník M, Green BR. 2005. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol. 22:2343–2353. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Stein DB. 1986. Conservation of chloroplast genome structure. Curr Genet. 10:823–833. [Google Scholar]
- Philippe H, et al. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai HS, O’Brien H, Reeves PA, Olmstead RG, Graham SW. 2003. Inference of higher-order relationships in the cycads from a large chloroplast data set. Mol Phylogenet Evol. 29:350–359. [DOI] [PubMed] [Google Scholar]
- Rambaut A. 2002. Se-Al v. 2.0a11: Sequence alignment program. Available from: http://tree.bio.ed.ac.uk/software/seal/. [Google Scholar]
- Rogalski M, Karcher D, Bock R. 2008. Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol. 15:192–198. [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, et al. 2013. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 161:1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall PJ. 2003. Monocot pseudanthia revisited: floral structure of the mycoheterotrophic family Triuridaceae. Int J Plant Sci. 164:307–320. [Google Scholar]
- Rudall PJ, Bateman RM. 2006. Morphological phylogenetic analysis of Pandanales: testing contrasting hypotheses of floral evolution. Syst Bot. 31:223–238. [Google Scholar]
- Saarela JMS, Graham SW. 2010. Inference of phylogenetic relationships among the subfamilies of grasses (Poaceae: Poales) using meso-scale exemplar-based sampling of the plastid genome. Botany 88:65–84. [Google Scholar]
- Sabir J, et al. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12:743–754. [DOI] [PubMed] [Google Scholar]
- Schelkunov MI, et al. 2015. Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 7:1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann Stat. 6:461–464. [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RaxML. Org Divers Evol. 12:335–337. [Google Scholar]
- Smith DR, Lee RW. 2014. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 164:1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RaxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Straub SCK, et al. 2011. Building a model: developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics 12:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4 . Sunderland (MA): Sinauer Associates. [Google Scholar]
- Swofford DL, et al. 2001. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst Biol. 50:525–539. [PubMed] [Google Scholar]
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. 1996. Phylogenetic inference. In: Hillis DM, Moritz D, Mable BK, editors. Molecular systematics. Sunderland (MA): Sinauer Associates; p. 407–514. [Google Scholar]
- Takhtajan AL. 1997. Diversity and classification of flowering plants. New York: Columbia University Press. [Google Scholar]
- Untergrasser A, et al. 2007. Primer3Plus, an enhanced web interface to Primer 3. Nucleic Acids Res. 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25:3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schăferhoff B, dePamphilis CW, Müller KF. 2014. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol Bio Evol. 31:529545. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, et al. 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 25:393–401. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, et al. 1996. Complete gene map of the plastid-like DNA of malaria parasite Plasmodium falciparum . J Mol Biol. 261:155–172. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. 1992. Function and evolution of a minimal parasitic plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 89:10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1996. Phylogenetic analysis using parsimony and likelihood methods. J Mol Evol. 42:294–307. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2006. Computational molecular evolution. Oxford: Oxford University Press. [Google Scholar]
- Yang Z. 2007. PAML4: phylogenetic analysis by maximum likelihood. Mol Bio Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. 2012. Molecular phylogenetics: principles and practice. Nat Rev Genet. 13:303–314. [DOI] [PubMed] [Google Scholar]
- Young ND, dePamphilis CW. 2005. Rate variation in parasitic plants: correlated and uncorrelated patterns among plastid genes of different function. BMC Evol Biol. 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurski JM, et al. 2008. How well do we understand the overall backbone of cycad phylogeny? New insights from a large, multigene plastid data set. Mol Phylogenet Evol. 47:1232–1237. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evolution of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhelyazkova P, et al. 2012. The primary transcriptome of barley chloroplasts: noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, et al. 2010. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A. 107:3245–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [Ph.D. dissertation]. The University of Texas at Austin [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.