Abstract
Herein, we report the results of study of Anopheles species in Primorsk and Khabarovsk regions of Russia. Three species of the Anopheles hyrcanus group: An. kleini, An. pullus, and An. lesteri were identified by molecular taxonomic diagnostics for the first time in Russia. Surprisingly, An. sinensis, which earlier was considered the only species of Anopheles in Russian Far East, was not observed. We analyzed nucleotide variation in the 610-bp fragment of the 5′ end of the cytochrome c oxidase subunit I (COI) region. All species possessed a distinctive set of COI sequences. A maximum likelihood phylogenetic tree was constructed for members of the hyrcanus group. The examined Anopheles hyrcanus group members could be divided into two major subgroups: subgroup 1 (An. hyrcanus and An. pullus) and subgroup 2 (An. sinensis, An. kleini, and An. lesteri), which were found to be monophyletic.
Introduction
The presence of numerous sibling species displaying high morphological similarity is a common feature of the genus Anopheles (Diptera, Culicidae).1–3 These species play distinct roles in transmitting malaria pathogens. Identification of potential vectors and a study of their distribution and role in a pathogen transmission are important components of malaria control measures.
The Anopheles hyrcanus group of mosquitoes includes about 30 closely related species distributed throughout the Palearctic and Oriental regions.4 Some species of this group are vectors of malaria parasites, in particular Plasmodium vivax.5 Sibling species are difficult or impossible to distinguish morphologically. The method of identification by polymerase chain reaction (PCR) was developed to differentiate five sibling species in the An. hyrcanus group (An. sinensis Wiedemann, 1828; An. pullus Yamada, 1937; An. lesteri Baisas and Hu, 1936; An. belenrae Rueda, 2005; An. kleini Rueda, 2005).6
In this study, we have investigated species composition of the Anopheles hyrcanus group in the Russian Far East. The results of phylogenetic analysis are provided.
Materials and Methods
Mosquito larvae were collected from eight locations in the Primorsk and Khabarovsk regions of Russia from August 22 to September 05, 2011 and from June 22 to June 26, 2013 (Table 1). An. hyrcanus larvae were collected in the southern Kazakhstan and Zhambul regions of the Republic of Kazakhstan from April 28 to May 18, 2012. The larvae were preserved in 96% ethyl alcohol for subsequent molecular and genetic analyses. In total, we identified and studied 913 mosquitoes of the Anopheles hyrcanus group.
Table 1.
Species composition of malaria mosquitoes (Anopheles) in the Primorsk and Khabarovsk regions
| No. | Collection localities, coordinates, and dates | n | Frequencies (f ± sf, %) | ||
|---|---|---|---|---|---|
| An. kleini | An. pullus | An. lesteri | |||
| 1 | Slavyanka, Primorsk region; 42°51′ N 131°23′ E; August 28, 2011 | 48 | 75.00 ± 6.25 | 16.67 ± 5.38 | 8.33 ± 3.99 |
| 2 | Vladivostok, Sputnik; 43°14′ N 132°02′ E; August 29, 2011 (IV stage) | 24 | 100.0 | 0 | 0 |
| 3 | Artem, Primorsk region; 43°22′ N 132°09′ E; August 27, 2011 (IV stage) | 24 | 79.17 ± 8.29 | 20.83 ± 8.29 | 0 |
| 4 | Artem, Primorsk region; 43°22′ N 132° 9′ E; August 27, 2011 (II stage) | 48 | 79.17 ± 5.86 | 18.75 ± 5.63 | 2.08 ± 2.06 |
| 5 | Ussuriisk, Primorsk region; 43°49′ N 131°57′ E; August 31, 2011 (IV stage) | 24 | 87.50 ± 6.75 | 12.50 ± 6.75 | 0 |
| 6 | Ussuriisk, Primorsk region; 43°49′ N 131°57′ E; August 31, 2011 (II stage) | 24 | 66.67 ± 9.62 | 33.33 ± 9.62 | 0 |
| 7 | Sibirtsevo, Primorsk region; 44°12′ N 132°27′ E; August 30, 2011 | 60 | 100.0 | 0 | 0 |
| 8 | Priamurskii, Khabarovsk region, Amur river floodplain; 48°32′ N 134°54′ E; September 4, 2011 (IV stage) | 30 | 96.67 ± 3.28 | 3.33 ± 3.28 | 0 |
| 9 | Priamurskii, Khabarovsk region, Amur river floodplain; 48°32′ N 134°54′ E; September 4, 2011 (II stage) | 30 | 95.74 ± 2.94 | 4.26 ± 2.94 | 0 |
| 10 | Khabarovsk; 48°30′ N 135°03′ E; September 5, 2011 | 48 | 95.83 ± 2.88 | 4.35 ± 2.88 | 0 |
| 11 | Sovetskaya Gavan', Khabarovsk region; 48°56′ N 140°15′ E; August 22 and 23, 2011 | 66 | 100.0 | 0 | 0 |
| 12 | Priamurskii, Khabarovsk region, Amur river floodplain; 48°32′ N 134°54′ E; June 25, 2013 (IV stage) | 117 | 91.45 ± 2.58 | 8.55 ± 2.58 | 0 |
| 13 | Priamurskii, Khabarovsk region, Amur river floodplain; 48°32′ N 134°54′ E; June 25, 2013 (II stage) | 21 | 100.0 | 0 | 0 |
| 14 | Khabarovsk; 48°30′ N 135°03′ E; June 25, 2013 (IV Stage) | 71 | 98.61 ± 1.38 | 1.39 ± 1.38 | 0 |
| 15 | Khabarovsk; 48°30′ N 135°03′ E; June 25, 2013 (I–III stage) | 87 | 100.0 | 0 | 0 |
| 16 | Vladivostok, Sputnik; 43°14′ N 132°02′ E; June 23, 2013 (IV stage) | 23 | 100.0 | 0 | 0 |
| 17 | Vladivostok, Sputnik; 43°14′ N 132°02′ E; June 23, 2013 (II stage) | 96 | 95.83 ± 1.84 | 0 | 4.16 ± 1.84 |
| 18 | Vladivostok, Ugol'naya; 43°16′ N 132°02′ N; June 24, 2013 | 54 | 85.18 ± 3.28 | 0 | 14.81 ± 3.28 |
sf = standard deviation.
DNA was extracted using DNA Invisorb® Spin Tissue Mini Kit (Invitek, Berlin, Germany). Species identification was performed by PCR analysis as described by Li and others.6 The phylogenetic analysis of species from the Anopheles hyrcanus group was carried out on the basis of the comparison of cytochrome c oxidase subunit I (COI) nucleotide sequences. The standard LCO1490 and HCO2198 primers7 were used to amplify the 5′-region of the COI gene.
The 5′-region of the COI gene was sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) with an ABI 3130 sequencer in 5–9 specimens of each species. The sequences of 610-bp fragments for 28 specimens of four mosquito species from the Anopheles hyrcanus group were obtained (GenBank accession numbers KC855638–KC855665). The 25 COI sequences of five other species from the Anopheles hyrcanus group and the COI sequence of An. messeae Falleroni, 1926 (an out-group) were retrieved from the GenBank and were also included in the analysis.
The sequences were edited and analyzed using the Sequencing Analysis 5.2 (Applied Biosystems) and SeqMan™ II (DNASTAR Inc., Madison, WI). For the phylogenetic analysis, the sequences were aligned using ClustalW. A maximum likelihood analysis in MEGA5 was used to examine phylogenetic relationships among taxa.8,9 Pairwise nucleotide sequence divergences were calculated using the Kimura 2-parameter (K2P) model.10
Results and Discussion
An. hyrcanus, a member of the Anopheles hyrcanus group, is believed to be present in the territories of the former Union of Soviet Socialist Republics (USSR).11 Within this species “western” and “eastern” forms were identified on the basis of the morphological features: the coloration of the fourth hindtarsal segment and the wing pattern. In the western specimens the fourth hindtarsal segment is more often pale, wing is more often with sharp pattern, and both costal spots well developed, pattern sometimes diffuse. In the eastern specimens the fourth hindtarsal segment is more often dark with narrow pale apical ring, sometimes with pale basal ring, wing is more often with diffuse pattern, and pale costal spots sometimes partly reduced, one may be nearly or wholly lacking. The western form included An. hyrcanus of the European part of Russia, while the eastern form included An. hyrcanus mosquitoes from the Russian Far East.11 However, it was impossible to identify the taxonomic status of these forms because of a great variability in the morphological characters. On the basis of the egg exochorion and cytogenetic analysis of larval karyotype, the eastern form of An. hyrcanus was identified as An. sinensis; it was stated that it was the only species of the Anopheles genus in the Russian Far East.12 However, more species of the Anopheles hyrcanus group have been described in the nearby countries of the Oriental zone. In particular, the list of hyrcanus species in China comprises 21 species13 and in the Republic of Korea comprises six species,5 five of which (An. sinensis, An. pullus, An. lesteri, An. kleini, and An. belenrae) are extremely difficult to identify morphologically.3,6 The molecular genetic assay used here allowed five species of “hyrcanus” group to be identified: An. sinensis, An. pullus, An. lesteri, An. belenrae, and An. kleini.6 For the first time, we identified three species of the Anopheles hyrcanus group in Russia: An. kleini, An. pullus, and An. lesteri.14 In 2011 and 2013, we found variation in species composition according to different habitats. Table 1 presents the data on the distribution and proportion of species from the Anopheles hyrcanus group in the Primorsk and Khabarovsk regions.
Among a total of 913 mosquitoes from the Anopheles hyrcanus group were identified by PCR, An. kleini was the most commonly collected species; it was found in all samples with a frequency from 66.67% to 100%. An. pullus is subdominant and was identified in 7 habitats with the frequency from 1.39% to 33.33%, followed by An. lesteri (from 2.08% to 14.81%), which was collected in Vladivostok, Slavyanka, and Artem (Primorsk region) (Table 1). The identified species composition is not constant and is subject to a seasonal dynamics, as has been suggested by the data from the Republic of Korea.3,15–17 Of special importance is that An. sinensis has not been detected in the examined region, although it has been regarded as the only representative of the genus Anopheles in the Russian Far East.12
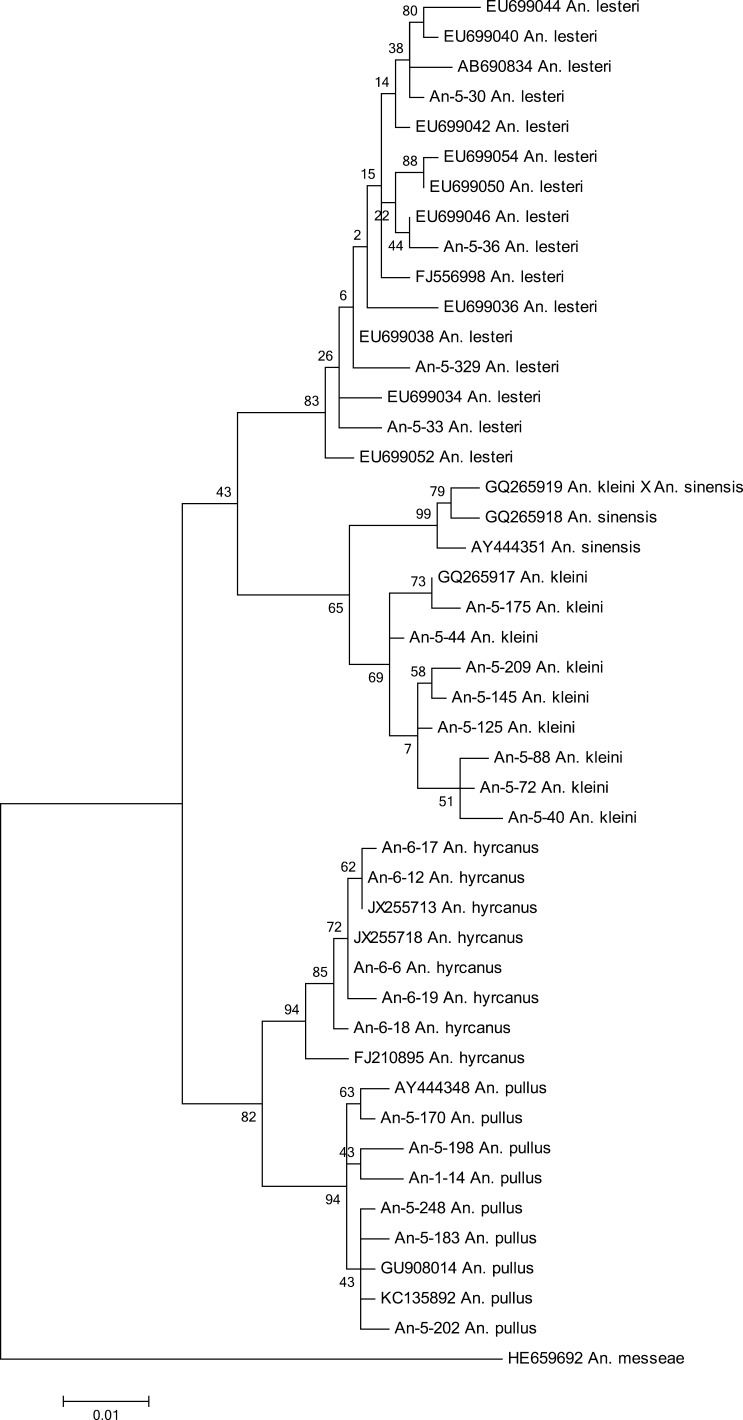
A phylogenetic analysis of the Anopheles hyrcanus group species was carried out on the basis of the comparison of the COI nucleotide sequences.
The conspecific K2P divergence averaged 0.73% (range 0.36–1.09%), and it was higher than the conspecific K2P divergence reported earlier for mosquitoes from China (0.39%), Canada mosquitoes (0.55%) and Aedes and Ochlerotatus genera from Tomsk and Kemerovo regions (0.57%).18–20 The sequence divergences between species averaged 3.71% (range 2.34–4.50%), which is higher than the threshold of the intra- and interspecific differences (2%).21
Individuals of a single species always occurred on a common clade, regardless of where they were collected (Figure 1). The phylogenetic tree divides the Anopheles hyrcanus group into two major subgroups: the first subgroup comprises An. hyrcanus and An. pullus; the second subgroup consists of An. sinensis, An. kleini, and An. lesteri (Figure 1). These patterns correlate with those found using the ribosomal internal transcribed spacer.22,23 The observed low interspecific divergence is characteristic of An. hyrcanus and An. pullus, the K2P distance (2.34%) only slightly exceeds the interspecific variability. In addition, the lowest intraspecific divergence (K2P, 0.36%) occurred in An. hyrcanus. The interspecies divergence was the highest for An. hyrcanus and An. kleini (K2P, 4.50%). The maximal intraspecies variability (K2P, 1.09%) was observed in An. lesteri. At the same time, probably, An. lesteri species may consist of three forms.23
Figure 1.
Maximum likelihood phylogenetic tree of Kimura 2-parameter (K2P) of cytochrome c oxidase subunit I (COI) Hyrcanus group members sequences. The numbers at the branches bases indicate the bootstrapping percentages (1,000 replicates).
Thus, we obtained new data on species composition and phylogenetic relationships for malaria mosquitoes in the Russian Far East. The list of mosquito species in Russia is now supplemented by three additional species, An. kleini, An. pullus, and An. lesteri. More comprehensive and detailed morphological, cytogenetic, and genetic research studies of the Anopheles hyrcanus group species are still required to better understand the malaria vector system in the Russian Far East.
Footnotes
Financial support: This study was supported by the Russian Fund of Basic Research (grant no. 11-04-00716-а “The genetic mechanisms of adaptation of sibling-species of malaria mosquitoes of Anopheles hyrcanus complex in Russia,” 2011–2013).
Authors' addresses: Natalia V. Khrabrova, Yulia V. Andreeva, Anuarbek K. Sibataev, and Svetlana S. Alekseeva, Tomsk State University, Tomsk, Russian Federation, E-mails: hrabrova@yandex.ru, andreeva_y@mail.200.ru, anuar@res.tsu.ru, and culex@res.tsu.ru. Perizat A. Esenbekova, Institute of Zoology, Committee of Science, Ministry of Education and Science of Republic of Kazakhstan, Almaty, Republic of Kazakhstan, E-mail: chronometer2012@yandex.ru.
References
- 1.Stegnii VN. Population Genetic and Evolution of Malaria Mosquitoes. Tomsk, Russian Federation: Tomsk State University; 1991. p. 136. [Google Scholar]
- 2.Collins FH, Paskewitz SM. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol Biol. 1996;5:1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- 3.Jeong KY, Un S, Lee J, Lee IY, Yong TS, Ree HI. Population dynamics of five Anopheles species of the hyrcanus group in northern Gyeonggi-do, Korea. Korean J Parasitol. 2010;48:351–353. doi: 10.3347/kjp.2010.48.4.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harbach RE. Review of the internal classification of the genus Anopheles (Diptera: Culicidae): the foundation for comparative systematics and phylogenetic research. Bull Entomol Res. 1994;84:331–342. [Google Scholar]
- 5.Rueda LM, Kim HC, Klein TA, Pecor JE, Li C, Sithiprasasna R, Debboun M, Wilkerson R. Distribution and larval habitat characteristics of Anopheles hyrcanus group and related mosquito species (Diptera: Culicidae) in South Korea. J Vector Ecol. 2006;31:198–205. doi: 10.3376/1081-1710(2006)31[198:dalhco]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Lee JS, Groebner JL, Kim HC, Klein TA, O'Guinn ML, Wilkerson RC. A newly recognized species in the Anopheles hyrcanus group and molecular identification of related species from the Republic of South Korea (Diptera: Culicidae) Zootaxa. 2005;939:1–8. [Google Scholar]
- 7.Folmer M, Black W, Hoeh R, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 8.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 11.Gutsevich AV. The polytypic mosquito species. 1. Anopheles hyrcanus (Pallas, 1771) Parazitologiya. 1976;10:148–153. [PubMed] [Google Scholar]
- 12.Gordeev MI, Klein SV. The cytogenetic analysis of the Anopheles hyrcanus group mosquitoes. In: Stegnii VN, editor. The Problems of Evolution Cytogenetic, Selection, and Introduction. Tomsk, Russian Federation: Tomsk State University; 1997. pp. 21–24. [Google Scholar]
- 13.Rueda LM, Zhao T, Ma Ya, Gao Q, Ding ZG, Khuntirat B, Sattabongkot J, Wilkerson RC. Updated distribution records of the Anopheles (Anopheles) hyrcanus species-group (Diptera: Culicidae) in China. Zootaxa. 2007;1407:43–55. [Google Scholar]
- 14.Khrabrova NV, Perevozkin VP, Andreeva YuV, Sibataev AK, Stegniy VN. Species composition of the mosquito Anopheles hyrcanus (Diptera, Culicidae) group in the Russian Far East. J Vector Ecol. 2012;37:450–452. doi: 10.1111/j.1948-7134.2012.00249.x. [DOI] [PubMed] [Google Scholar]
- 15.Rueda LM, Brown TL, Kim HC, Chong S-T, Klein TA, Foley DH, Anyamba A, Smith M, Pak EP, Wilkerson RC. Species composition, larval habitats, seasonal occurrence and distribution of potential malaria vectors and associated species of Anopheles (Diptera: Culicidae) from the Republic of Korea. Malar J. 2010;9:55. doi: 10.1186/1475-2875-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HC, Rueda LM, Wilkerson RC, Foley DH, Sames WJ, Chong ST, Nunn PV, Klein TA. Distribution and larval habitats of Anopheles species in northern Gyeonggi province, Republic of Korea. J Vector Ecol. 2011;36:124–134. doi: 10.1111/j.1948-7134.2011.00149.x. [DOI] [PubMed] [Google Scholar]
- 17.Foley DH, Klein TA, Kim HC, Kim M-S, Wilkerson RC, Harrison G, Rueda LM, Lee W-J. Synchronous peaks in trap catches of malaria-infected mosquito species at Daeseongdong, a border village between North and South Korea. J Vector Ecol. 2011;37:29–36. doi: 10.1111/j.1948-7134.2012.00197.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Li C, Guo X, Xing D, Ya Dong, Wang Z, Zhang Y, Liu M, Zheng Z, Zhang H, Zhu X, Wu Z, Zhao T. Identifying the main mosquito species in China based on DNA barcoding. PLoS One. 2012;7:e47051. doi: 10.1371/journal.pone.0047051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cywinska A, Hunter FF, Hebert PDN. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- 20.Khrabrova NV, Andreeva YuV, Vaulin OV, Alekseeva SS, Sibataev AK. Variability of the mitochondrial cytochrome oxydase subunit I gene sequence in species of the genera Aedes and Ochlerotatus (Diptera: Culicidae) Russ J Genet Appl Res. 2013;3:279–286. [Google Scholar]
- 21.Hebert PDN, Cywinska A, Ball S, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Ya, Xu A. The hyrcanus group of Anopheles (Anopheles) in China (Diptera: Culicidae): species discrimination and phylogenetic relationships inferred by ribosomal DNA internal transcribed spacer 2 sequences. J Med Entomol. 2005;42:610–619. [PubMed] [Google Scholar]
- 23.Hwang UW. Revisited ITS2 phylogeny of Anopheles (Anopheles) hyrcanus group mosquitoes: reexamination of unidentified and misidentified ITS2 sequences. Parasitol Res. 2007;101:885–894. doi: 10.1007/s00436-007-0553-4. [DOI] [PubMed] [Google Scholar]