SUMMARY
Obesity and metabolic syndrome are associated with an increased risk for lipotoxic cardiomyopathy, which is strongly correlated with excessive accumulation of lipids in the heart. Obesity- and type 2 diabetes-related disorders have been linked to altered expression of the transcriptional cofactor PGC-1α, which regulates the expression of genes involved in energy metabolism. Using Drosophila, we identify PGC-1/spargel (PGC-1/srl) as a key antagonist of high-fat diet (HFD)-induced lipotoxic cardiomyopathy. We find that HFD-induced lipid accumulation and cardiac dysfunction are mimicked by reduced PGC-1/srl function and reversed by PGC-1/srl overexpression. Moreover, HFD feeding lowers PGC-1/srl expression by elevating TOR signaling and inhibiting expression of the Drosophila adipocyte triglyceride lipase (ATGL, Brummer), both of which function as upstream modulators of PGC-1/srl. The lipogenic transcription factor SREBP also contributes to HFD-induced cardiac lipotoxicity, likely in parallel with PGC-1/srl. These results suggest a regulatory network of key metabolic genes that modulates lipotoxic heart dysfunction.
Keywords: heart, contractility, arrhythmia, cardiomyopathy, metabolic syndrome, type 2 diabetes, brummer, FAS, S6 Kinase, SREBP, epistasis, lipolysis, lipogenesis
INTRODUCTION
Heart failure is a major cause of mortality in modern society. The high prevalence of obesity and related diseases, including type 2 diabetes, plays a significant role in the increased incidence of cardiovascular disease, which affects more than a billion people worldwide. It is now recognized that obesity and diabetes are associated with lipotoxic cardiomyopathy, a form of cardiac dysfunction that is caused by excessive lipid accumulation in myocardial cells (Borradale and Schaffer, 2005; Christoffersen et al., 2003). The high incidence of obesity-associated heart dysfunction is aggravated by changes in nutritional habits, such as consumption of high caloric diets, combined with reduced physical activity. A major challenge in developing targeted therapeutics for this type of heart disease is to understand how the molecular and genetic changes induced by high caloric diets lead to metabolic imbalances that culminate in cardiac dysfunction.
Although mammalian model organisms are ultimately essential for identifying the underlying mechanisms of obesity and metabolic syndrome (Kanasaki and Koya, 2011; York and Bouchard, 2000). The genetic and metabolic complexity and gene redundancy in higher organisms complicates dissection of the fundamental genetic relationships involved in these diseases. However, the components and regulatory pathways of core metabolic pathways are generally highly conserved, making genetically simpler invertebrate models, such as Drosophila, well suited for such studies (Baker and Thummel, 2007; Botstein et al., 1997; Broughton et al., 2005; Diop and Bodmer, 2012; Kuehnlein, 2012; Noyes et al., 1995). Drosophila is unique in that it is the only invertebrate model organism with a beating heart for which suitable genetic tools and assays are available to study heart function (Bier and Bodmer, 2004; Ocorr et al., 2007; Wessells et al., 2004; Wolf et al., 2006; Ocorr et al., 2014).
Recent studies with Drosophila models of obesity-associated heart dysfunction have demonstrated that consumption of high-fat or -sugar diets, or genetic manipulation of key metabolic regulators, can lead to excessive fat accumulation accompanied by severe heart defects, including increased frequency of arrhythmias, reduced cardiac output, increased non-contractile myocardial cells, and altered myofibrillar structure and collagen content (Birse et al., 2010; Diop et al., 2012; Lee et al., 2010; Lim et al., 2011; Na et al., 2013). Thus, flies challenged with high caloric diets display metabolic responses and increased risk for heart disease reminiscent of those observed in humans. Importantly, these studies showed that inhibition of signaling through insulin, target of rapamycin (TOR) or the lipogenic transcription factor SREBP, or by increasing triacylglyceride (TAG) lipolysis were effective in counteracting excess lipid accumulation as well as the associated cardiac defects. Although these findings suggested that major metabolic pathways could be targeted for therapeutic intervention in human obesity-induced cardiomyopathy, further work is necessary to obtain a more complete understanding of the essential regulators of these lipogenic pathways and their relationship to cardiac function. In this study, we use the Drosophila model to delineate novel genetic relationships between key metabolic regulators relevant to cardiac lipotoxicity.
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1 (PGC-1) is a transcriptional coactivator first identified as a key regulator of thermogenesis in brown adipose tissue in mammals (Puigserver et al., 1998). Subsequently, additional PGC-1 family members have been shown to modulate gene expression by interacting with a number of nuclear receptors (Finck and Kelly, 2006; Gleyzer and Scarpulla, 2011; Lin et al., 2002). Studies in mice and flies have demonstrated that PGC-1 proteins are induced by environmental stimuli, such as cold exposure, exercise, or starvation, that trigger a number of metabolic adaptations (Arany, 2008; Baar et al., 2002; Goto et al., 2000; LeMoine et al., 2008; Tiefenbock et al., 2010; Tinkerhess et al., 2012; Zechner et al., 2010). PGC-1α is the most studied member of the PGC-1 family and plays key roles in mitochondrial biogenesis and electron transport chain assembly by interacting with nuclear respiratory factor (NRF) and estrogen receptor-related receptor (ERR) transcription factors (Rowe et al., 2010; Russell et al., 2004; Scarpulla, 2011; Tiefenbock et al., 2010). PGC-1α also controls mitochondrial fatty acid oxidation in the muscles and liver by coactivating members of the PPAR and retinoic X receptor families of nuclear factors (Vega et al., 2000; Zechner et al., 2010). The PGC-1α/ERR axis has also been implicated in controlling cardiac metabolism and function (Finck and Kelly, 2006, 2007; Lai et al., 2008; Patten and Arany, 2012; Schilling and Kelly, 2011). Interestingly, decreased cardiac PGC-1α expression, observed in adipocyte triglyceride lipase (ATGL)-deficient mice, is associated with cardiac lipid accumulation and subsequent heart failure (Haemmerle et al., 2011). Of note, sequence variation at the PGC-1α locus has been correlated with obesity in humans (Rankinen et al., 2006). Despite the known involvement of PGC-1 in cardiac function, it is still unclear whether or how PGC-1 protein interacts with other metabolic regulators and signaling pathways in modulating the deleterious effects of high caloric diets on lipid accumulation and cardiomyopathy. In particular, it is not known if TOR and SREBP pathways interact with PGC-1/srl function in these processes.
In this study, we provide evidence that reducing the function of Drosophila PGC-1/srl causes a phenotype of excess fat accumulation and severe heart dysfunction that is remarkably similar to that observed in wildtype flies exposed to a HFD. This phenotype is exacerbated in PGC-1/srl mutant flies that are fed a HFD, whereas systemic or cardiac-specific PGC-1/srl overexpression is protective. Genetic interaction experiments suggest that PGC-1/srl is likely to act downstream of TOR signaling to protect the heart from lipotoxicity. We show that the protective effect of reduced TOR signaling requires elevated expression of PGC-1/srl and ATGL (major TAG lipase encoded by brummer, bmm, in Drosophila), with ATGL/bmm positioned downstream of TOR and upstream of PGC-1/srl in the homeostatic control of fat accumulation and cardiac lipotoxicity. Interestingly, these PGC-1/srl manipulations did not change the expression or processing of SREBP, and reducing SREBP does not significantly ameliorate heart dysfunction of PGC-1/srl heterozygotes, suggesting that PGC-1/srl and SREBP do not act in a strict epistatic relationship. Taken together, our data define a new interaction network of key metabolic regulators that contribute to cardiac lipotoxicity in Drosophila, and thus provide insights into possible mechanisms of HFD-associated cardiomyopathy relevant to humans.
RESULTS
HFD-Induced Fat Accumulation Requires Reduced PGC-1/srl Expression
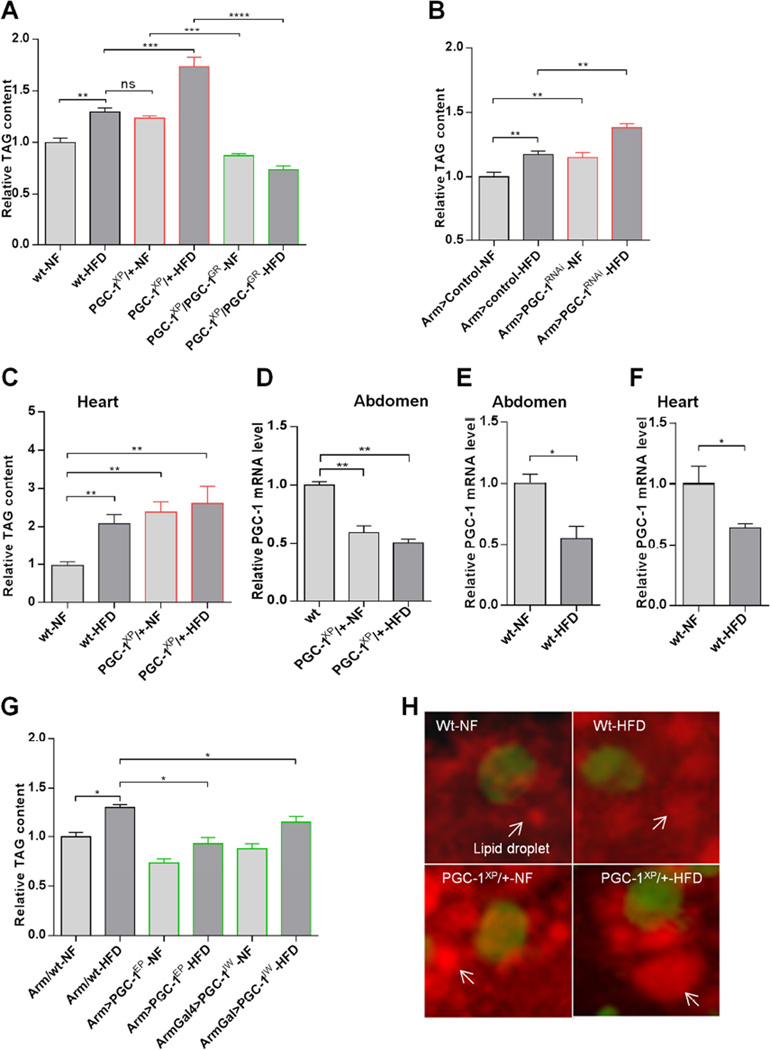
To determine the involvement of Drosophila PGC-1/srl in obesity and heart dysfunction, we first examined the effect of PGC-1/srl modulation on whole body triglyceride (TAG) content in wildtype (w1118) flies, heterozygous PGC-1/srl mutants (PGC-1XP/+ and PGC-1PBAC/+), and flies with systemic RNAi-mediated PGC-1/srl knockdown (KD). Flies were raised on normal food (NF) for 5–10 days and then transferred for a further 5 days to NF or HFD (NF supplemented with 30% coconut oil). We found that flies with reduced PGC-1/srl function had elevated TAG levels compared with wildtype flies, and TAG levels were further increased when the mutant or systemic PGC-1/srl KD flies (Arm>PGC-1RNAi) were fed a HFD (Figures 1A, 1B, S1A). Moreover, TAG levels were increased in the heart and muscle-rich thorax of HFD-fed or PGC-1/srl heterozygous flies (Figures 1C and S1B), consistent with the deposition of TAG in additional organs once the capacity of adipose tissue was exceeded. To further demonstrate that the increased TAG level in PGC-1/srl heterozygous mutant flies was specific to reduced PGC-1/srl function, we used a genomic rescue construct (PGC-1GR; Tiefenbock et al., 2010) in flies carrying the PGC-1XP mutation. We found that these flies (PGC-1XP/PGC-1GR) have a significantly lower TAG level compared to PGC-1 heterozygous flies (PGC-1XP/+) under NF or HFD (Figure 1A). These experiments show that a genomic PGC-1/srl transgene was able to rescue the TAG phenotype of the PGC-1/srl mutant flies in support of the idea that the observed phenotype is specific to loss of PGC-1/srl function. The rescue flies show a somewhat lower TAG level than the wildtype control, which possibly reflects a slightly higher activity by the rescue transgene compared to the wildtype allele. PGC-1/srl KD specifically in adipose tissue (using the lsp-Gal4 driver) or muscle (24B-Gal4 driver) also resulted in systemic fat accumulation in both NF- and HFD-fed flies, supporting an association between reduced PGC-1/srl function and fat accumulation (Figure S1C and S1I).
Figure 1. HFD-Induced Obesity Is Accompanied by a Reduction in PGC-1/srl Function.
(A) TAG content of PGC-1/srl mutant flies. Flies were fed for 5–10 days on normal food (NF) and then transferred to NF or HFD (NF + 30% coconut oil) for a further 5 days. The wildtype (wt) strain is w1118. PGC-1XP/+and PGC-1GR are respectively PGC-1 heterozygous mutant and and PGC-1GR denotes flies that carry a wild type PGC-1 genomic rescue transgene. Results are expressed as the fold difference in whole fly TAG compared with wt-NF flies and are the mean ± SEM of n = at least 27 for all genotypes and conditions (wt-NF replicates were normalized to their average, thus contain error bars here and throughout). **P < 0.01, ***P < 0.001, ****P <0. 0001,one way ANOVA test.
(B) TAG content of PGC-1 knockdown flies. Controls (GD control lines used to make PGC-1 RNAi) are the same genetic background as the PGC-1RNAi line. Results are expressed as the fold difference in whole fly TAG compared with Arm>control-NF flies and are the mean ± SEM of 42 ≤ n ≥ 27. **P < 0.01, one way ANOVA test.
(C) TAG content in the hearts of PGC-1/srl mutant flies. Results are expressed as the fold difference compared with wt-NF flies and are the mean ± SEM of 250 ≤ n ≥ 200. . **P < 0.01, one way ANOVA test.
(D–F) Relative PGC-1 mRNA levels in wt or PGC-1/srl mutant flies. PGC-1 mRNA was measured by qPCR in (D) the abdomen of wt and PGC-1XP/+ heterozygous flies, and (E) the abdomen or (F) the heart of wt flies on NF or HFD. Results are expressed as the fold difference compared with wt flies and are the mean ± SEM of triplicates. *P < 0.05, **P < 0.01, ANOVA and student t-tests.
(G) TAG content of flies overexpressing PGC-1/srl. Results are expressed as the fold difference in whole fly TAG compared with Arm/wt-NF flies and are the mean ± SEM of 36 ≤ n ≥ 27. . *P < 0.05, one way ANOVA test.
(H) Representative images of Nile Red staining of lipid droplets in cardiomyocytes of wt and PGC-1XP/+ flies on NF or HFD. Arrows indicate lipid droplets; cardiomyocyte nuclei are stained green by DAPI. See also Figure S1.
Because wildtype flies on a HFD and flies with reduced PGC-1/srl function on NF had similar levels of TAG, we asked whether PGC-1/srl expression was modulated by HFD feeding. Indeed, qPCR analysis showed that HFD decreased PGC-1/srl mRNA in various tissues to a level comparable to PGC-1/srl heterozygotes, including in the abdomen, the heart and muscle-rich thorax (Figures 1D-1F, S1D and S1H). Conversely, TAG levels were decreased in HFD-fed flies overexpressing PGC-1/srl systemically (Figures 1G, S1G and S1J) or specifically in muscle (Figure S1E) or adipose tissue (Figure S1F). These findings suggest that the HFD-induced reduction in PGC-1/srl function promotes fat accumulation, whereas overexpression of PGC-1/srl blunts the effect.
To further analyze the effects of HFD and reduced PGC-1/srl function on lipid accumulation, we visualized lipid droplets in cardiomyocytes by Nile Red staining. We observed an increase in droplet size and fat content (Figure 1H) in the hearts of wildtype flies on HFD and PGC-1/srl heterozygotes on NF or HFD.
HFD-Induced Reduction in PGC-1/srl Expression Provokes Cardiac Lipotoxicity
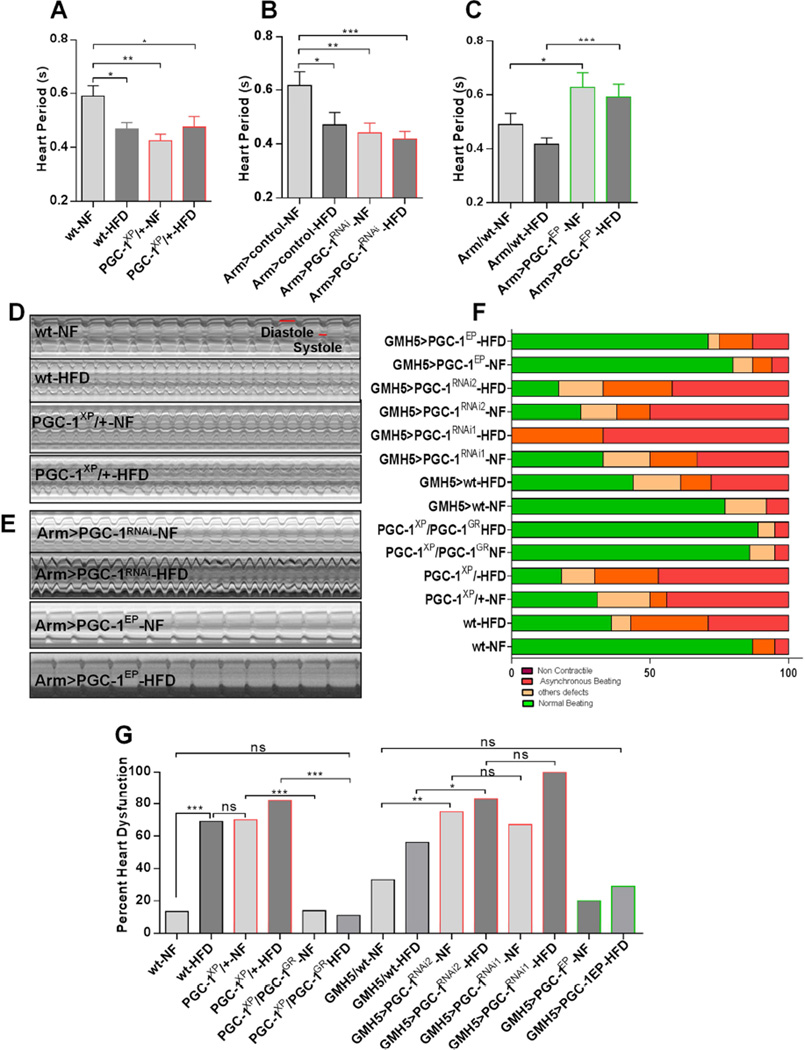
The finding that PGC-1/srl heterozygotes and HFD-fed wildtype flies accumulated fat in the heart (Figures 1C and 1H), together with the reported link between obesity and heart disease in humans and other mammals (Borradaile and Schaffer, 2005; Zhou et al., 2000), prompted us to investigate the effect of PGC-1/srl manipulation on heart function in Drosophila. For this, we used the previously established methods of semi-intact heart preparation, high-speed video recording, and SOHA software analysis (Ocorr et al., 2007; Fink et al., 2009). We observed that wildtype flies on HFD and flies with reduced PGC-1/srl function on either NF or a HFD showed decreased heart period (Figure 2A, 2B and 2D and Movies S1–S4) due to reductions in both diastolic and systolic intervals (Figure S2A, S2B, S2D, and S2E). This is also illustrated in M-mode traces (Figures 2D and 2E). Conversely, systemic PGC-1/srl overexpression increased the heart period even under HFD conditions (PGC-1EP flies; Figures 2C and 2E), which was primarily due to an increased diastolic interval (Figure S2C and S2F). Wildtype flies on HFD also showed reduced fractional shortening (Figure S2G and S2H), as previously noted (Birse et al., 2010), but this parameter was unaffected by reduced PGC-1/srl function (Figure S2G and S2H).
Figure 2. HFD-Induced Cardiac Dysfunction is Modulated by PGC-1/srl Function.
(A) Heart periods of wt and PGC-1 heterozygous flies. Results are the mean ± SEM of 45 ≤ n ≥ 33. *P < 0.05, **P < 0.01, one way ANOVA test.
(B) Heart periods of control and PGC-1 knockdown flies. Results are expressed as the mean ± SEM of 22 ≤ n ≥ 20. *P < 0.05, **P < 0.01, ***P < 0.001, one way ANOVA test.
(C) Heart periods of control and PGC- 1-overexpressing flies. Results are the mean ± SEM of 32 ≤ n ≥ 16. *P < 0.05, ***P < 0.001, one way ANOVA test.
(D–E) Representative M-modes traces (5s) showing heart wall movements (Y-axis) versus time (X-axis). Stills are from high-speed movies of semi-intact fly heart preparations for (D) wt and PGC-1XP/+ flies on NF or HFD, and (E) PGC-1 knockdown (Arm>PGC- 1RNA) and PGC-1-overexpressing (Arm>PGC-1EP) flies on NF or HFD.
(F) Graphical representation of the proportion of PGC-1/srl mutant, RNAi and overexpression flies displaying heart dysfunction phenotypes, classified as non-contractile regions, asynchronous beating, and other defects (dysfunctional ostia, narrowed heart regions, and transmission defects).
(G) Quantification of heart defects shown in (F). Statistical significance was determined using chi-square test 33 ≤ n ≥ 14. wt-NF vs. wt- HFD, x2 = 15; P < 0.001. wt-HFD vs. PGC-1XP/+-NF, x2 = 0.008; ns. PGC-1XP/+-NF vs. PGC-1XP/PGC-1 GR-NF, x2 = 15.78; P < 0.001. PGC- 1XP/+-HFD vs. PGC-1XP/PGC-1GR-HFD, x2 = 23.00; P < 0.001. wt-NF vs. PGC- 1XP/PGC-1GR-HFD, x2 = 0.002; ns. GMH5/wt-NF vs. GMH5>PGC-1RNAi1-NF, x2 = 7.03; P < 0.01. GMH5/wt-HFD vs. GMH5>PGC-1RNAi2-HFD, x2 = 3.33; P < 0.05. GMH5>PGC-1RNAi2-NF vs. GMH5>PGC-1RNAi1-NF, x2 = 0.17; ns. GMH5>PGC-1RNAi2-HFD vs. GMH5>PGC-1RNAi1-HFD, x2 = 2.30; ns. GMH5/wt-NF vs. GMH5>PGC-1EP-HFD x2 = 013; ns. not significant (ns). See also Figure S2 and Movies S1–S7.
PGC-1/srl heterozygotes exhibited striking defects in other aspects of cardiac contraction that were remarkably similar to those observed upon HFD feeding. The defects included heart tubes with regions lacking active contractions (‘non-contractile’), asynchronous anterior versus posterior beating patterns (‘asynchronous beating’), dysfunctional inflow valves (ostia), and occasional localized constriction (‘other defects’) (Movies S5–S7). Quantification of these parameters revealed that cardiac dysfunction was markedly increased in wildtype flies fed a HFD and in flies with reduced PGC-1/srl function (PGC-1xp/+), and was further increased in PGC-1xp/+ flies on a HFD (Figures 2F and 2G). Remarkably, PGC-1/srl heterozygous flies containing a genomic PGC-1/srl transgene (PGC-1XP/PGC-1GR) showed a significant decreased in cardiac dysfunction under NF and HFD (Figures 2F and 2G), suggesting that adding a genomic copy of PGC-1/srl to PGC-1/srl heterozygotes rescued also the heart phenotypes. These data therefore suggested that the HFD-induced reduction in PGC-1/srl function is associated not only with fat accumulation but also with HFD-induced heart dysfunction.
To determine whether cardiac-specific KD of PGC-1/srl was sufficient to cause heart dysfunction, we expressed two different PGC-1/srl RNAi constructs using a cardiomyocyte-specific driver (GMH5). Indeed, flies with PGC-1/srl KD exhibited pronounced hearts defects comparable to those observed in PGC-1/srl heterozygotes or wildtype flies fed a HFD (Figures 2F and 2G), indicating that reducing PGC-1/srl function in the heart alone was sufficient to cause dysfunction. The heart defects tended to be more severe when cardiac PGC-1/srl KD flies were exposed to a HFD, whereas HFD-fed flies with cardiac overexpression of PGC-1/srl (PGC-1EP) showed a much lower incidence of heart defects (Figures 2F and 2G). These data suggest that high levels of PGC-1/srl protected the heart from HFD-induced dysfunction in a cardiac-autonomous fashion. Conversely, downregulation of PGC-1/srl activity correlated with the deleterious effects of HFD on cardiac function, although it cannot be ruled out that reduced PGC-1/srl function also has effects on the heart not related to fat accumulation.
Inhibition of TOR Activity Elevates PGC-1/srl Expression and Protects Against HFD-Induced Heart Dysfunction
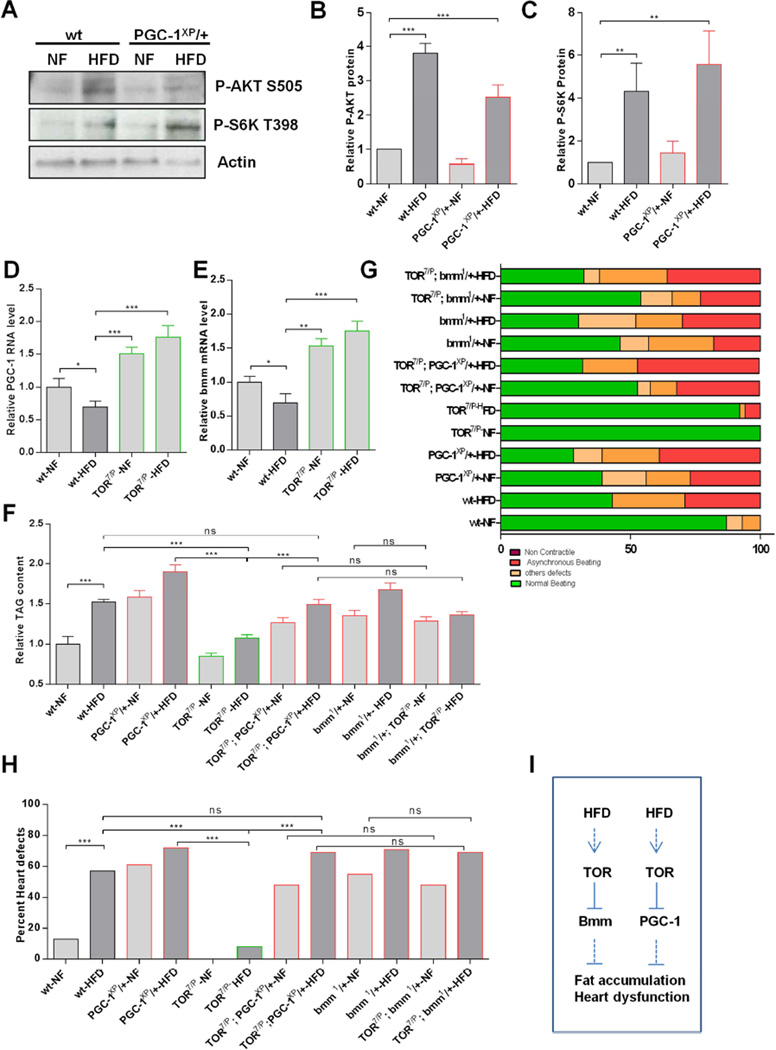
We previously showed that reduced TOR signaling can protect against HFD-induced obesity and heart dysfunction in Drosophila (Birse et al., 2010). Here, we find that HFD feeding of wildtype flies activated TOR signaling, as shown by increased levels of the phosphorylated (i.e. active) forms of AKT (P-AKTS505) and S6K (P-S6KT398) (Figure 3A–C). These observations are consistent with the previously demonstrated protective effect of reduced TOR activity in lipotoxic heart dysfunction (Birse et al., 2010).To determine whether PGC-1/srl and TOR might function through a common pathway, we examined activation of AKT (upstream regulator of TOR) and S6K (downstream effector of TOR) in PGC-1/srl heterozygotes. Like wildtype flies fed HFD, PGC-1 heterozygotes flies show an increase of P-AKTS505 and P-S6KT398 under HFD, however, P-AKT and P-S6K levels did not appear to be altered differently in PGC-1/srl heterozygous flies compared to wildtype (Figure 3A–C), suggesting that PGC-1/srl was unlikely an upstream regulator of TOR signaling in this situation.
Figure 3. TOR is an Upstream Regulator of PGC-1/srl and bmm in HFD-Induced Heart Dysfunction.
(A) Western blots of phospho-AKT and phospho-S6K from wt and PGC-1/srl heterozygous flies on NF and HFD. Actin was used as loading control.
B–C: quantification of relative P (S6K) and P (AKT) protein based on densitometry measurements. Statistics were done using the Kruskal-Wallis test. **P < 0.05, ***P <0.001.
(D–E) Relative PGC-1/srl (D) and bmm (E) mRNA levels in wt and TOR mutant on NF and HFD. Results are expressed as the fold difference compared with wt flies. *P < 0.05, one way ANOVA test.
(F) Relative TAG content in PGC-1/srl, bmm and TOR single and double mutant flies on NF and HFD. Results are expressed as the fold difference in whole fly TAG compared with wt-NF flies and are the mean ± SEM of 66 ≤ n ≥ 18. . **P < 0.01, ***P < 0.001, one way ANOVA test.
(G) Graphical representation of the proportion of PGC-1/sr, bmm and TOR single and double mutant flies displaying heart dysfunction phenotypes, classified as non-contractile regions, asynchronous beating, and other defects (dysfunctional ostia, narrowed heart regions, and transmission defects).
(H) Quantification of heart defects shown in (G). Statistical significance was determined using chi-square test 39 ≤ n ≥ 19. wt-NF vs. wt-HFD, x2 = 12; P < 0.001. wt-HFD vs. TOR7/P-HFD, x2 = 19.33; P <0.001. PGC-1XP/+-HFD vs. TOR7/P-HFD, x2 = 23.58; P < 0.001. TOR7/P-HFD vs. TOR7/P; PGC-1XP/+-HFD, x2 = 23.58; P < 0.001. wt-HFD vs. TOR7/P; PGC-1XP/+-HFD, x2 = 0.46; ns. TOR7/P; PGC-1XP/+-NF vs. TOR7/P; bmm1/+-NFx2 = 0; ns. TOR7/P; PGC-1XP/+-HFD vs. TOR7/P; bmm1/+-HFD, x2 = 0.01; ns. Bmm1/+-NF vs. TOR7/P; bmm1/+-HFDx2 = 106; ns. . ns: not significant.
(I) Schematic of proposed regulatory network. HFD feeding activates TOR signaling, which reduces PGC-1/srl and bmm expression to lead to fat accumulation and heart dysfunction. See also Figure S3.
We next asked whether manipulation of TOR signaling affected PGC-1/srl expression. Indeed, NF-fed flies with reduced TOR signaling (TOR7/P, previously shown to protect from HFD-induced heart dysfunction; Birse et al., 2010) had significantly increased PGC-1/srl mRNA levels (Figure 3D), as did flies with systemic expression of dominant-negative S6K (S6KDN; Figure S3B). Remarkably, the HFD-induced reduction in PGC-1/srl mRNA level was reversed in TOR7/P mutants (Figure 3D). These data therefore suggested that reducing PGC-1/srl expression in TOR7/P mutants might reduce or abrogate the protective effect of reduced TOR signaling on HFD-induced fat accumulation. To test this, we created flies with reduced function of both TOR and PGC-1/srl (TOR7/P;PGC-1xp/+). In keeping with our hypothesis, the double-mutant flies had elevated levels of TAG, similar to those in PGC-1XP/+ flies, and TAG levels were further increased upon HFD feeding (Figure 3F).
Next, we tested whether PGC-1/srl may also function downstream of TOR in regards to heart function. Indeed, we found a high incidence of heart dysfunction in TOR7/P;PGC-1xp/+ double-mutant flies, similar to that observed for single PGC-1XP/+ mutants (Figure 3G and 3H). These data suggested an epistatic relationship between PGC-1/srl and TOR, in which TOR acted as a negative upstream regulator of PGC-1/srl function in controlling fat content and heart function (Figure 3I). The findings further suggested that the previously demonstrated cardioprotective effect of reduced TOR signaling may require upregulation of PGC-1/srl expression.
The ATGL/Bmm Lipase Acts Downstream of TOR and Upstream of PGC-1/srl in Modulating HFD-Induced Heart Dysfunction
Lipolysis, which is the process by which triglycerides are degraded into free fatty acids, is a major component of lipid metabolism. Brummer (bmm), which encodes the Drosophila homologue of mammalian ATGL lipase, also mediates fat hydrolysis in this invertebrate model system (Gronke et al., 2005). We previously showed that bmm overexpression efficiently prevented HFD-induced fat accumulation and heart dysfunction (Birse et al., 2010). Therefore, we asked how Bmm might contribute to the TOR- and PGC-1-regulated effects on lipid accumulation and cardiac function. Interestingly, the phenotype of bmm heterozygous flies was remarkably similar to that of PGC-1/srl heterozygotes. Thus, bmm1/+ flies accumulated excess fat and considerable heart dysfunction on NF, which were further increased by HFD feeding (Figures 3F–H). Conversely, systemic bmm overexpression prevented HFD-induced TAG accumulation (Figure S3A) and heart dysfunction (Birse et al., 2010). Importantly, bmm heterozygotes also exhibited heart dysfunction that was further aggravated by HFD (Figures 3G and 3H). These findings raised the possibility that ATGL/bmm acts in the same pathway as PGC-1/srl and TOR in modulating cardiac lipotoxicity.
To test this idea, we first examined bmm expression in TOR7/P mutant flies. As was observed for PGC-1/srl expression (Figure 3D and S3B), bmm mRNA levels were increased in TOR7/P and S6KDN flies (Figure 3E and S3C). Conversely, bmm mRNA was reduced by HFD feeding, and this was reversed in TOR7/P mutant flies (Figure 3E), suggesting that bmm acts downstream of TOR signaling and may be required for the fat-lowering effect of reduced TOR function. To examine this possibility, we created double-mutant flies with reduced TOR and bmm function (TOR7/P;bmm1/+). Compared to the protective effect of TOR7/P from HFD effects, TOR7/P;bmm1/+ and bmm1/+ mutant flies exhibited similarly increased TAG level and severe degrees of heart dysfunction on both NF and HFD, as wildtype flies on HFD (Figures 3F–H), consistent with bmm loss of function compromising the protection conferred by reduced TOR signaling. These data therefore suggest that like with PGC-1/srl, TOR pathway activation under HFD leads to reduced bmm expression, which in turn promotes fat accumulation and heart dysfunction (Figure 3I).
Next, we wanted to test whether adult-only manipulation of bmm or PGC-1/srl was sufficient to mimic (or protect from) HFD-induced fat accumulation and heart dysfunction. For this purpose, we used the Gene Switch system (GS; Osterwalder et al., 2001; Roman et al., 2001) to conditionally drive - upon RU-486 addition - gene expression in the adult heart, both myocardial and pericardial cells (Hand-GS-Gal4; Monnier et al., 2012). Administration of ethanol vehicle or RU-486 to wildtype flies did not noticeably affect ATGL/bmm or PGC-1/srl expression, fat content or heart function (Figure S3G–K). However, cardiac KD of ATGL/bmm (Hand-GS>bmmRNAi) by RU treatment caused an increase in systemic fat content compared to ethanol vehicle exposure; and conversely, conditional ATGL/bmm or PGC-1/srl overexpression in the adult heart (Hand-GS>UASbmm or Hand-GS>PGC-1JW) decreased fat levels (Figure S3D), suggesting metabolic gene manipulation in cardiac tissue can have a systemic effect (see Grueter CE et al., 2012). Importantly, adult-specific overexpression of PGC-1/srl or ATGL/bmm in the heart of flies treated with RU (Hand-GS>PGC-1JW, Hand-GS>bmm) showed markedly decreased heart dysfunction under HFD, compared to ethanol controls (Figure S3E and S3F). In contrast, Hand-GS>PGC-1RNAi flies treated with RU exhibited significantly increased heart dysfunction under NF and HFD, compared to ethanol controls (Figure S3E and S3F). These data demonstrate that PGC-1/srl and ATGL/bmm manipulation during adult stages is sufficient to modulate heart function under NF and HFD. Together with HFD feeding of adult flies causing increased TOR signaling (Figure 3A) and decreased PGC-1/srl and ATGL/bmm expression (Figure 3D and 3E), these data strongly suggest that PGC-1/srl and ATGL/bmm act as mediators of HFD induced heart dysfunction and effectors of TOR signaling.
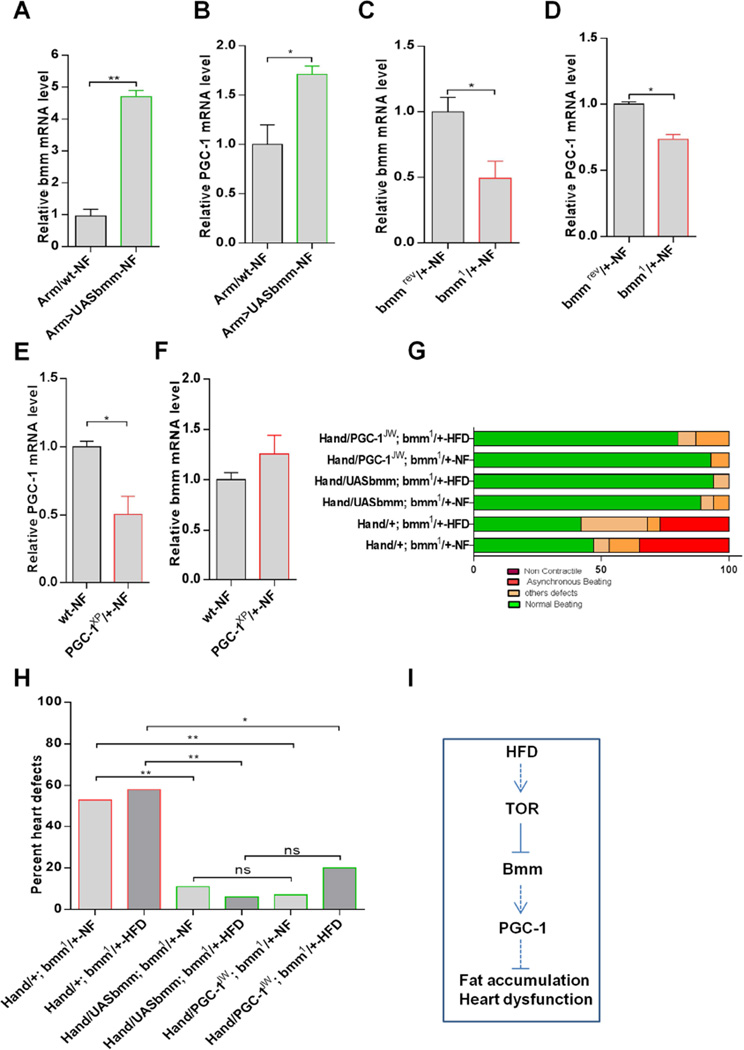
To investigate possible genetic interactions between PGC-1/srl and bmm, we first analyzed PGC-1/srl mRNA levels in bmm mutant flies. We found that PGC-1/srl RNA levels were significantly increased in flies with systemic overexpression of bmm and significantly reduced in bmm heterozygotes (Figures 4A – D). Using the Gene Switch system, we found that induced adult heart expression or KD of ATGL/bmm (Hand-GS>UASbmm-RU or Hand-GS>BmmRNAi) leads to increased or decreased PGC-1/srl expression in the heart, respectively (Figure S4A–D), which is consistent with PGC-1/srl acting downstream of ATGL/bmm in causing (counteracting) HFD-induced heart dysfunction. In PGC-1/srl heterozygous flies on the other hand, bmm mRNA levels were similar to wildtype (Figure 4E–F). Moreover, heart-only induced expression of PGC-1/srl with Hand-GS also did not significantly alter ATGL/bmm expression (Figure S4E–F). Interestingly, however, hand-GS induced cardiac PGC-1/srl KD did exhibit decreased ATGL/bmm levels (Figure S4G–H), suggesting that there may be some feedback regulation from PGC-1/srl to ATGL/bmm, although ATGL/bmm acts predominantly upstream PGC-1/srl in modulating HFD-related cardiac dysfunction (see below and Figures 4G–H and S4I–J).
Figure 4. ATGL/bmm is an Upstream Regulator of PGC-1/srl in HFD-Induced Heart Dysfunction.
(A–F): Relative expression of bmm (A, C, and F) and PGC-1/srl (B, D, and E) mRNA in bmm KD /overexpression flies and in PGC-1/srl mutant flies. *P < 0.05, **P < 0.01, student t-test.
(G) Graphical representation of the proportion of flies with cardiac-specific manipulation of bmm and PGC-1/srl in a bmm mutant background displaying heart dysfunction phenotypes, classified as non-contractile regions, asynchronous beating, and other defects (dysfunctional ostia, narrowed heart regions, and transmission defects).
(H) Quantification of heart defects shown in (G). Statistical significance was determined using chi-square test 19 ≤ n ≥ 14. Hand/+; bmm1/+-NF vs. Hand/UASbmm; bmm1/+-NF\2 = 10.16; P < 0.01. Hand/+; bmm1/+-HFD vs. Hand/UASbmm; bmm1/+-HFDx2 = 10.28; P < 0.01. Hand/+; bmm1/+-NF vs. Hand/PGC-1JW; bmm1/+-NF\2 = 7.36; P < 0.01. Hand/+; bmm1/+-HFD vs. Hand/PGC-1JW; bmm1/+-HFDx2 = 4.96; P < 0.05. Hand/UASbmm; bmm1/+-NF vs. Hand/PGC-1JW; bmm1/+-NFx2 = 0.05; ns. Hand/UASbmm; bmm1/+-HFD vs. Hand/PGC-1JW; bmm1/+-HFDx2 = 1.3; ns.
(I) Schematic of proposed regulatory network. HFD consumption activates TOR signaling, which inhibits the expression of bmm and, subsequently, PGC-1/srl in the same axis, leading to fat accumulation and heart dysfunction. See also Figure S4.
We next examined the relationship between PGC-1/srl and ATGL/bmm in regulating heart function. For this purpose, we examined bmm heterozygous flies with cardiac PGC-1/srl overexpression (Hand>PGC-1JW;bmm1/+). We found that cardiac PGC-1/srl overexpression was able to rescue the heart dysfunction phenotype of bmm heterozygotes under NF and HFD, as well as a control rescue with ATGL/bmm overexpression (Figure 4G and 4H). As expected, we also found that cardiac-specific KD of ATGL/bmm using the myocardial-specific Tin-HE-Gal4 driver induced a high proportion of heart defects in both NF- and HFD-fed flies (Figure S4I–J). However, concomitant cardiac overexpression of PGC-1/srl significantly reduced the deleterious effects of ATGL/bmm KD (Figure S4I–J). Taken together, these data are consistent with ATGL/bmm acting upstream of PGC-1/srl in regulating heart function. These findings suggest that HFD-induced lipid accumulation and cardiac dysfunction are accompanied by the sequential activation of TOR, downregulation of ATGL/bmm and subsequently PGC-1/srl expression, thus leading to cardiac lipotoxicity (Figure 4I).
PGC-1/srl and SREBP Likely Act in Parallel to Modulate HFD-Induced Heart Dysfunction
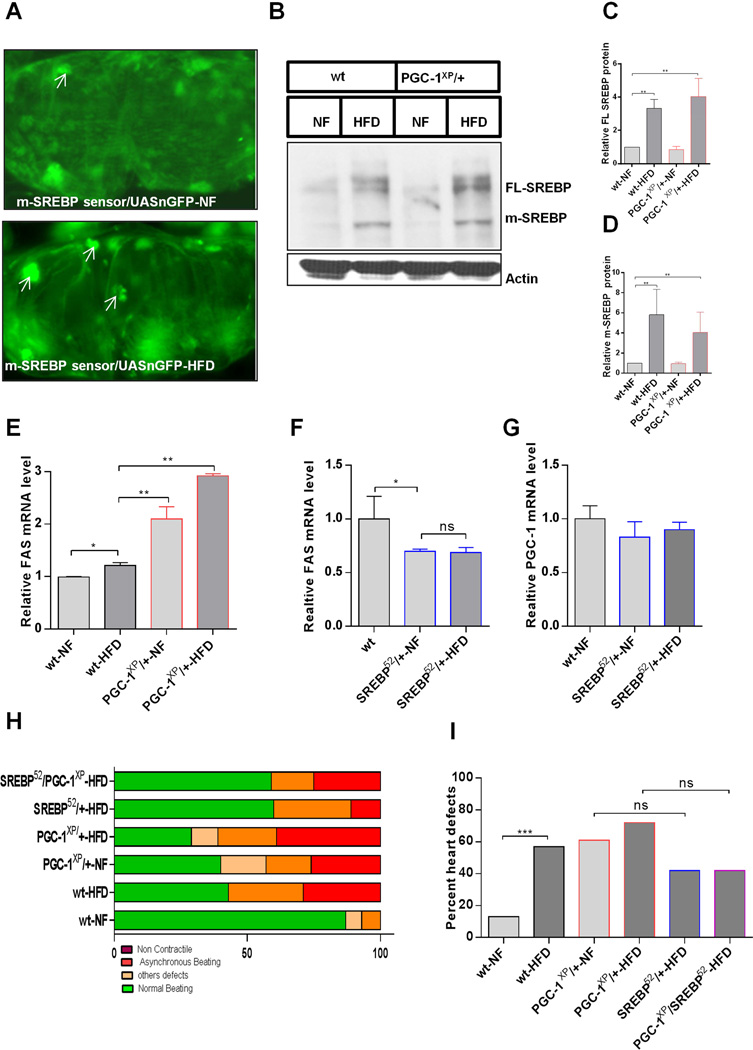
SREBP is a conserved transcriptional regulator of lipogenic gene expression (Dobrosotskaya et al., 2002; Osborne and Espenshade, 2009; Seegmiller et al., 2002). Activation of SREBP or its lipogenic targets, including fatty acid synthase (FAS), increases fat accumulation and causes heart dysfunction in Drosophila (Lim et al., 2011), similar to the effects of reduced PGC-1/srl function (Figures 1 and Figure 2). To determine the effect of HFD on processing of full-length (FL) SREBP to the transcriptionally active mature form (m-SREBP), we created flies carrying a sensor for m-SREBP–mediated transcriptional activation (Kunte et al., 2006) together with a UAS-nGFP reporter. In these flies, activation of m-SREBP is indicated by an increase in GFP fluorescence. We observed that flies fed a HFD showed higher levels of nuclear GFP in the heart (Figure 5A) and thoracic muscle (Figure S5A) than NF-fed flies, indicating that SREBP activity was induced by the HFD. In keeping with this, expression of the SREBP target gene FAS was also elevated in HFD-fed compared to NF-fed flies (Figure 5E).
Figure 5. SREBP and PGC-1/srl Function through Parallel Pathways in HFD-Induced Heart Dysfunction.
(A) Representative micrographs of sections of hearts of flies carrying a UAS-GFP/SREBP sensor and fed NF (top) or HFD (bottom). Sections were stained with anti-GFP antibody (green). Arrowheads indicate increased GFP staining (SREBP activation) in the nuclei of hearts from HFD-fed flies.
(B–D): Western blots of SREBP (FL: full length, m: mature isoform) in whole fly extracts of wt and PGC-1XP/+ flies on NF or HFD (A). Actin was included as a loading control. C and D are quantification of relative FL SREBP protein (C) and m-SREBP from westerns in B (D). Kruskal Wallis test was used for the statistics. **P < 0.01.
(E) Relative expression of FAS mRNA in wt and PGC-1XP/+ flies on NF or HFD. Results are mean ± SD of n = 30 *P < 0.05, **P < 0.01, one way ANOVA test.
(F–G) Relative expression of (F) FAS and (G) PGC-1/srl mRNA in wt and SREBP mutant flies on NF or HFD. Results are mean ± SD of n = 30 *P < 0.05. ns: not significant, one way ANOVA test.
(H) Graphical representation of the proportion of SREBP and PGC-1/srl mutant flies displaying heart dysfunction phenotypes, classified as non-contractile regions, asynchronous beating, and other defects (dysfunctional ostia, narrowed heart regions, and transmission defects).
(I) Quantification of heart defects shown in (F). Statistical significance was determined using chi-square test, 26 ≤ n ≥ 14. wt-NF vs. wt-HFD, x2 = 12; P < 0.001. PGC-1XP/+-NF vs. SREBP52/+- NF, x2 = 0.3; ns. PGC-1XP/+- HFD vs. SREBP52/+- HFD, x2 = 3; ns. SREBP52/+-NF vs. SREBP52/PGC-1 XP-NF, x2 = 1.5; ns. SREBP52/+-HFD vs. SREBP52/PGC-1XP-HFD, x2 = 0.01; ns: not significant. See also Figure S5.
Interestingly, FAS expression was also increased in PGC-1/srl heterozygotes compared to wildtype flies on both NF and HFD (Figures 5E). Also, bmm overexpression and mutant flies have decreased and increased FAS levels, respectively (Figure S5B and S5C). Therefore, we asked whether SREBP and PGC-1/srl interacted to induce fat accumulation and heart dysfunction in response to the HFD. For this purpose, processing of FL-SREBP to m-SREBP was analyzed by western blot analysis. Wildtype flies fed a HFD showed increased levels of both FL-SREBP and m-SREBP compared with flies on NF (Figure 5B–D); however, SREBP expression in PGC-1/srl heterozygotes, which contain similar levels of TAG on NF as do wildtype flies on HFD, was not significantly different from wildtype flies in either NF or HFD conditions (Figure 5B–D). Similarly, SREBP heterozygotes expressed reduced levels of FAS mRNA (Figure 5F), but not of PGC-1/srl mRNA (Figure 5G). Therefore, these data suggest that PGC-1/srl and SREBP do not affect each other’s expression in their modulation of fat accumulation, and may occur through distinct mechanisms.
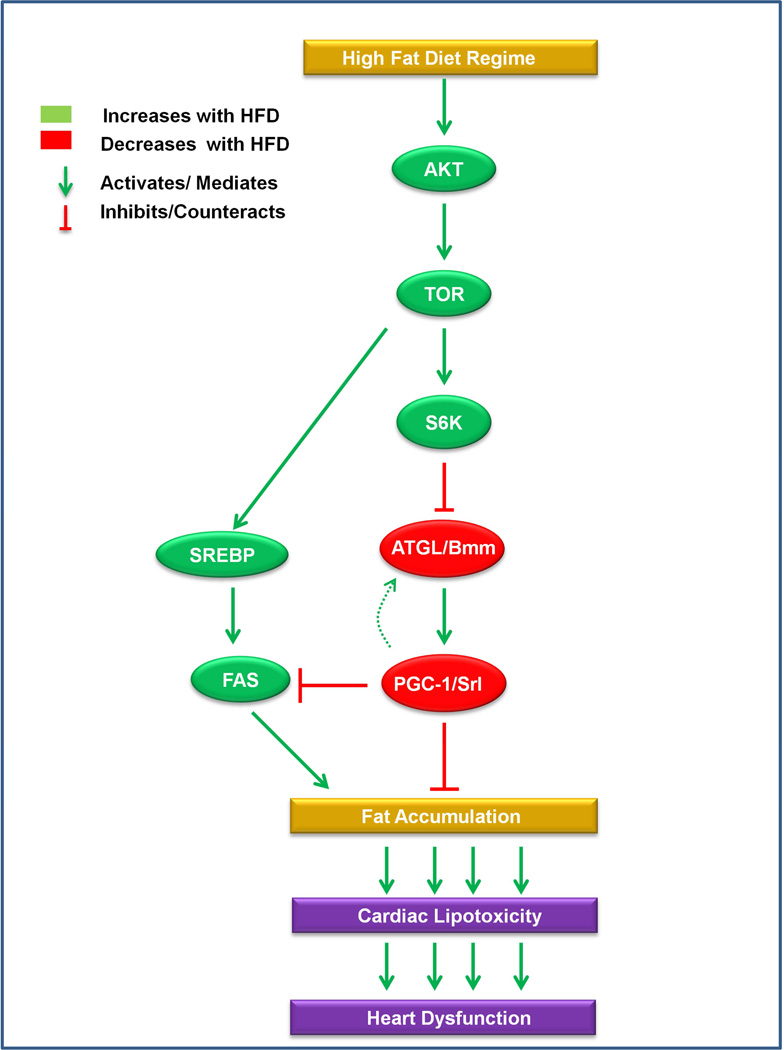
Finally, we examined directly the relationship between SREBP and PGC-1/srl in modulating heart function. Cardiac dysfunction was slightly less prevalent in HFD-fed SREBP heterozygotes than in wildtype flies (Figure 5H and 5I), suggesting that SREBP partially mediate the deleterious effect of HFD on the heart., SREBP and PGC-1/srl double-heterozygous flies exposed to a HFD showed a heart phenotype strikingly similar to that of SREBP heterozygotes (Figures 5H and 5I). In addition, we found that PGC-1/srl and FAS double KD in the heart produced only a slight reduction of heart dysfunction compared to PGC-1/srl single KD (Figure S5D–E), which would be consistent with SREBP being the dominant regulator of FAS in the heart (see Lim et al., 2011) and PGC-1/srl also acting through other effectors. Overall, the data suggest that SREBP may at least in part act in parallel to PGC-1/srl, but it cannot be ruled out that PGC-1/srl also acts downstream (or vice versa), through FAS and possibly other effectors, in regulating heart function (illustrated in Fig. 6). Collectively, these findings are consistent with a model in which SREBP and Bmm-PGC-1/Srl function downstream of TOR and in parallel to each other, interconnected via FAS, to control fat accumulation and heart function. In conclusion, we found that HFD causes excess fat accumulation and cardiac dysfunction via TOR-dependent activation of two parallel pathways, TOR–ATGL/Bmm–PGC-1/Srl and TOR–SREBP (Figure 6).
Figure 6. Model of genetic network in HFD-induced cardiac lipotoxicity.
HFD feeding causes increased TOR signaling that leads to decreased PGC-1/srl expression via decreased ATGL/bmm function mediating obesity and cardiac lipotoxicity. There may be some feedback from PGC-1/srl to ATGL/bmm in the heart itself, but in regards to modulation of HFD-dependent heart dysfunction PGC-1/srl acts downstream of ATGL/bmm. SREBP is also activated by TOR and is interconnected to the ATGL/bmm-PGC-1/srl axis via FAS to contribute to the regulation of HFD-inflicted cardiac lipotoxicity.
DISCUSSION
It is well established that lipid accumulation in cardiomyocytes can lead to cardiac dysfunction (e.g. Zhang and Ren, 2011), but the genetic and metabolic regulatory networks that mediate cardiac lipotoxicity have yet to be fully understood. Here, we used the Drosophila model to investigate the genetic relationships between PGC-1/srl and other metabolic regulators to elucidate the mechanisms of HFD-induced obesity and cardiac dysfunction. We found that reducing PGC-1/srl function was sufficient to induce TAG accumulation and cardiac lipotoxicity, a phenotype remarkably similar to that observed in wildtype HFD-fed flies. Indeed, HFD feeding reduced PGC-1/srl expression wildtype flies. Conversely, PGC-1/srl overexpression countered both the fat accumulation and associated heart defects induced by HFD, indicating that PGC-1/srl is both necessary and sufficient for counteracting obesity and associated cardiac lipotoxicity. In humans, genome-wide association studies conducted with various ethnic and geographical cohorts have suggested a possible link between obesity and variants in PGC-1 genes (Muller el al., 2003; Ayra et al., 2004). Moreover, reduced PGC-1 gene expression was observed in patients with diabetes and those developing insulin resistance, but no clear mechanism of action has been established (Mootha et al., 2003; Patti et al., 2003). In mammals, PGC-1 genes are known to control cardiac metabolism and function (Schilling and Kelly, 2011; Lai et al., 2008; Russel et al., 2004; Rowe et al., 2010), and reduced PGC-1α has been linked to heart failure and hypertrophy in rodent models of heart disease (Lehman and Kelly, 2002; Arany et al., 2006; Huss et al., 2006). Thus, our demonstration of a close association between PGC-1/srl function, excess fat accumulation, and cardiac lipotoxicity further supports a critical role for this coactivator in diabetic cardiomyopathy. In addition, we demonstrate for the first time that reduction PGC-1/srl function is essential in mediating HFD-induced heart dysfunction.
We demonstrated that HFD feeding activates TOR signaling (increased P-AKT and P-S6K), which in turn leads to downregulation of PGC-1/srl expression. In contrast, a reduction in TOR function, which protects against obesity and heart dysfunction, is associated with increased PGC-1/srl levels. Furthermore, reducing both PGC-1/srl and TOR function (TOR7/P;PGC-1xp/+) abrogated the protective effects of TOR reduction against HFD-induced obesity and heart dysfunction. These findings therefore suggest that PGC-1/srl is a negative downstream effector of TOR. We suggest that persistent activation of TOR in response to a HFD reduces PGC-1/srl expression, which in turn leads to lipid accumulation in the myocardium and provokes heart dysfunction.
We also identified ATGL/Bmm lipase as a downstream effector of TOR in HFD-induced obesity and heart dysfunction, and the effects of a HFD were similar in bmm and PGC-1/srl mutant flies. Importantly, bmm RNA levels were increased in TOR mutant flies and – as observed with PGC-1/srl – reduction of bmm function in TOR mutant flies (TOR7/P;bmm/+) eliminated the beneficial effects of TOR. Because modulation of bmm dominantly induced changes in PGC-1/srl mRNA levels and cardiac-specific increase in PGC-1/srl levels in bmm KD flies protected against heart dysfunction, we conclude that PGC-1/srl acts downstream of bmm and TOR, thus delineating a genetic pathway from TOR to PGC-1/srl via ATGL/bmm.
Interestingly, a recent study showed that mammalian ATGL controls murine heart dysfunction via PPARs (Haemmerle et al., 2011). Here, we demonstrated that this relationship also appears to exist in the fly, and importantly, provides strong evidence that under HFD conditions ATGL/bmm and PGC-1/srl act downstream of TOR in mediating cardiac lipotoxicity. Interestingly, in the above study cardiac PGC-1 gene expression is also reported to be reduced with ATGL deficiency (Haemmerle et al., 2011), thus placing PGC-1 gene function downstream of ATGL in both the fly and the mouse heart. Although molecules of equivalent function to mammalian PPARs have not yet been conclusively identified in Drosophila, our findings are consistent with the presence of a fundamentally conserved epistatic relationship between TOR, bmm, and PGC-1/srl in the metabolic control of cardiac lipotoxicity.
Previous studies have suggested that TOR stimulates lipogenesis via a SREBP-dependent increase in FAS expression (Porstmann et al., 2008; Hagiwara et al., 2012), and we have recently shown that SREBP lipogenic activity strongly influences heart function in Drosophila (Lim et al., 2011). In this study, we demonstrate that under HFD conditions, PGC-1/Srl and SREBP do not seem to directly regulate each other and may act independently to regulate fat accumulation and heart function, which implies that HFD activation of TOR signaling triggers parallel but partially interdependent pathways to regulate lipogenesis (through FAS). This does not preclude that PGC-1/srl and SREBP (as well as bmm) also have FAS-unrelated functions in modulating fat accumulation and cardiac function. In our model, we propose that HFD activation of the nutrient-sensing TOR pathway leads to downregulation of the bmm–PGC-1/srl axis and to upregulation of the SREBP axis, both resulting, at least in part, in FAS upregulation. Such a mechanism would have two deleterious consequences: increased circulating fatty acid levels and increased storage of fat in non-adipose tissues, leading to lipotoxicity and cardiac dysfunction. Interestingly, rodent models of obesity also show increased TAG deposition in the heart and subsequent development of cardiac lipotoxicity (Borradale and Schaffer, 2005; Zhou et al., 1989; Christoffersen et al., 2003). These findings suggest that HFD feeding perturbs homeostatic pathways that are tightly regulated when fat intake is normal.
In humans, type-2 diabetes and associated diseases of the metabolic syndrome are aggravated with age. A recent study in Drosophila by Rera et al. (2011) have shown that PGC-1/srl expression decreases with age in the intestine and PGC-1/srl overexpression was sufficient to delay tissue aging, at least in part by increasing mitochondrial activity. These data are consistent with our findings that PGC-1/srl overexpression protects against obesity and cardiac dysfunction. Further studies will be required to determine our proposed network of gene regulation is also operational in regards to PGC-1/srl’s role in cardiac and overall aging. Conversely, it will be of interest to determine whether increasing certain aspects of mitochondrial function in the heart is sufficient to counteract the cardiac lipotoxicity of HFD feeding.
In a recent report TOR signaling was found to function as a positive regulator of PGC-1/srl in growth process during larval development of Drosophila (Mukherjee and Duttaroy, 2013), which is consistent with previous findings that PGC-1/srl-deficient Drosophila larvae exhibit dramatic growth impairment (Tiefenbock et al., 2010). In contrast, however, we provided strong evidence in this study that during metabolic homeostasis in the adult animal, PGC-1/srl is a negative effector of HFD-induced augmentation of TOR signaling. Thus, during organismal and tissue growth, PGC-1/srl mediates TOR signaling. But in the adult upon a metabolic or dietary insult, activation of TOR function and downstream ATGL/bmm lipase inhibition, prevents PGC-1/srl function, establishing a new genetic relationship between TOR and PGC-1/srl.
In this study, we have identified an integrated genetic network of metabolic regulators in the control cardiac lipotoxicity, in particular, PGC-1/srl acting as a negative effector of TOR. The network connects insulin–TOR, ATGL/Bmm, PGC-1/Srl, SREBP, and FAS, all of which have been associated with obesity in humans (Rankinen et al., 2006; Yang et al., 2007), but their relationships has not been comprehensively elucidated. Our work thus identifies a prototypical gene network with significant relevance for understanding lipotoxic cardiomyopathy in humans, and further emphasizes the utility of the Drosophila heart model in unraveling complex metabolic processes in higher organisms.
EXPERIMENTAL PROCEDURES
HFD Feeding Regimen
Flies were aged for 5 days after eclosion in tubes (25 flies/tube) containing normal fly food (NF) and then transferred to NF or HFD (NF + 30% coconut oil) for 5 days, as previously described (Birse et al., 2010). Identical groups of flies were either weighed and frozen at −80°C for determination of TAG content (Birse et al., 2010) or analyzed for heart function abnormalities (see below).
Semi-Intact Drosophila Heart Preparation and Digital High-Speed Movie Analysis
All dissection steps were performed in artificial hemolymph saline, as previously described (Ocorr et al., 2007). Flies were anesthetized with fly-nap (Carolina Biochem.) and transferred to a Vaseline-coated Petri dish for dissection as described (Vogler and Ocorr, 2009). The submerged dissected hearts were oxygenated for 15 min at room temperature to equilibrate. High-speed digital movies were made using a Leica DM-LFSA microscope with a 10× water immersion lens, a Hamamatsu EM-CCD camera, and HCI image capture software (Hamamatsu, Inc.). Movies were analyzed with custom designed software (Fink et al., 2009; Ocorr et al., 2007). See also supplemental experimental procedures and references.
Supplementary Material
HIGHLIGHTS.
PGC-1/srl mutations mimic HFD-induced cardiac lipoxicity
HFD-induced cardiac lipotoxicity requires PGC-1/srl inhibition
The effect of TOR on PGC-1/srl is mediated by ATGL/bmm inhibition
PGC-1/srl and SREBP act via parallel pathways to control cardiac lipotoxicity
ACKNOWLEDGEMENTS
We would like to thank Leanne Jones, Ronald Kuehnlein, Tom Neufeld, Christian Frei, Hugo Stocker and Robert Rawson for reagents and fly stocks, and Daniel Kelly and Timothy Osborne for critical reading of the manuscript. We are grateful to Bloomington Stock Center, DGRC (Japan) and VDRC for fly strains. We are also grateful for excellent technical assistance by Cherie Celeste and Joan Choi. This work was funded by grants from the National Institutes of Health (P01 HL098053, P01 AG033561 and R01 HL054732) to R.B., an NHLBI postdoctoral research supplement (R01 HL085481) and a postdoctoral fellowship from the American Association of University Women (AAUW) to S.B.D., and grants from the American Heart Association to K.O. and to R.T.B. S.B.D designed and performed most of the experiments, analyzed all data, and wrote the paper; J.B. performed experiments; K.O. developed analytical tools, analyzed data, and helped in writing the paper; R.T.B and S.O. analyzed parts of the data, and R.B. supervised the project, designed experiments, analyzed data, and wrote the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Duggirala R, Jenkinson CP, Almasy L, Blangero J, O'Connell P, Stern MP. Evidence of a novel quantitative-trait locus for obesity on chromosome 4p in Mexican Americans. Am J Hum Genet. 2004;74:272–28. doi: 10.1086/381717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. Diabetic larvae and obese flies - emerging studies of metabolism in Drosophila . Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342:1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, Ocorr K, Bodmer R, Oldham S. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010;12:533–544. doi: 10.1016/j.cmet.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradaile NM, Schaffer JE. Lipotoxicity in the heart. Curr Hypertens Rep. 2005;7:412–417. doi: 10.1007/s11906-005-0035-y. [DOI] [PubMed] [Google Scholar]
- Botstein D, Chervitz SA, Cherry JM. Yeast as a model organism. Science. 1997;277:1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen C, Bollano E, Lindegaard ML, Bartels ED, Goetze JP, Andersen CB, Nielsen LB. Cardiac lipid accumulation associated with diastolic dysfunction in obese mice. Endocrinology. 2003;144:3483–3490. doi: 10.1210/en.2003-0242. [DOI] [PubMed] [Google Scholar]
- Diop SB, Bodmer R. Drosophila as a model to study the genetic mechanisms of obesity-associated heart dysfunction. J Cell Mol Med. 2012;16:966–971. doi: 10.1111/j.1582-4934.2012.01522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila . Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) regulatory cascade in cardiac physiology and disease. Circulation. 2007;115:2540–2548. doi: 10.1161/CIRCULATIONAHA.107.670588. [DOI] [PubMed] [Google Scholar]
- Gleyzer N, Scarpulla RC. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. J Biol Chem. 2011;286:39715–39725. doi: 10.1074/jbc.M111.291575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasaki K, Koya D. Biology of obesity: lessons from animal models of obesity. J Biomed Biotechnol. 2011;2011:197636. doi: 10.1155/2011/197636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnlein RP. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res. 2012;53:1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Bodmer R, Bier E, Karin M. Sestrins at the crossroad between stress and aging. Aging (Albany NY) 2010;2:369–374. doi: 10.18632/aging.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- LeMoine CM, Genge CE, Moyes CD. Role of the PGC-1 family in the metabolic adaptation of goldfish to diet and temperature. J Exp Biol. 2008;211:1448–1455. doi: 10.1242/jeb.014951. [DOI] [PubMed] [Google Scholar]
- Lim HY, Wang W, Wessells RJ, Ocorr K, Bodmer R. Phospholipid homeostasis regulates lipid metabolism and cardiac function through SREBP signaling in Drosophila. Genes Dev. 2011;25:189–200. doi: 10.1101/gad.1992411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Monnier V, Iche-Torres M, Rera M, Contremoulins V, Guichard C, Lalevee N, Tricoire H, Perrin L. dJun and Vri/dNFIL3 are major regulators of cardiac aging in Drosophila. PLoS Genet. 2012;8:e1003081. doi: 10.1371/journal.pgen.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Duttaroy A. Spargel/ dPGC-1 is a new downstream effector in the Insulin-TOR Signaling pathway in Drosophila . Genetics. 2013;195:433–441. doi: 10.1534/genetics.113.154583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes. 2003;52:895–898. doi: 10.2337/diabetes.52.3.895. [DOI] [PubMed] [Google Scholar]
- Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, Cagan R. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 2013;9:e1003175. doi: 10.1371/journal.pgen.1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes BE, Katz FN, Schaffer MH. Identification and expression of the Drosophila adipokinetic hormone gene. Mol Cell Endocrinol. 1995;109:133–141. doi: 10.1016/0303-7207(95)03492-p. [DOI] [PubMed] [Google Scholar]
- Ocorr K, Vogler G, Bodmer R. Methods to assess Drosophila heart development, function and aging. Methods. 2014;68:265–272. doi: 10.1016/j.ymeth.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Fink M, Cammarato A, Bernstein S, Bodmer R. Semi-automated Optical Heartbeat Analysis of small hearts. JoVE. 2009:31. doi: 10.3791/1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007a;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten IS, Arany Z. PGC-1 coactivators in the cardiovascular system. Trends Endocrinol Metab. 2012;23:90–97. doi: 10.1016/j.tem.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Bray MS, Hagberg JM, Perusse L, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2005 update. Med Sci Sports Exerc. 2006;38:1863–1888. doi: 10.1249/01.mss.0000233789.01164.4f. [DOI] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling J, Kelly DP. The PGC-1 cascade as a therapeutic target for heart failure. J Mol Cell Cardiol. 2011;51:578–583. doi: 10.1016/j.yjmcc.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Tiefenbock SK, Baltzer C, Egli NA, Frei C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 2010;29:171–183. doi: 10.1038/emboj.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkerhess MJ, Healy L, Morgan M, Sujkowski A, Matthys E, Zheng L, Wessells RJ. The Drosophila PGC-1alpha homolog spargel modulates the physiological effects of endurance exercise. PLoS One. 2012;7:e31633. doi: 10.1371/journal.pone.0031633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Kelly T, He J. Genetic epidemiology of obesity. Epidemiol Rev. 2007;29:49–61. doi: 10.1093/epirev/mxm004. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- York D, Bouchard C. How obesity develops: insights from the new biology. Endocrine. 2000;13:143–154. doi: 10.1385/ENDO:13:2:143. [DOI] [PubMed] [Google Scholar]
- Zechner C, Lai L, Zechner JF, Geng T, Yan Z, Rumsey JW, Collia D, Chen Z, Wozniak DF, Leone TC, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ren J. Role of cardiac steatosis and lipotoxicity in obesity cardiomyopathy. Hypertension. 2011;57:148–150. doi: 10.1161/HYPERTENSIONAHA.110.164178. [DOI] [PubMed] [Google Scholar]
- Zhou YT, Grayburn P, Karim A, Shimabukuro M, Higa M, Baetens D, Orci L, Unger RH. Lipotoxic heart disease in obese rats: implications for human obesity. Proc Natl Acad Sci U S A. 2000;97:1784–1789. doi: 10.1073/pnas.97.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.