Summary
Execution of the DNA damage response (DDR) relies upon a dynamic array of protein modifications. Using quantitative proteomics, we have globally profiled ubiquitination, acetylation, and phosphorylation in response to ultraviolet and ionizing radiation. To improve acetylation site profiling, we developed the strategy FACET-IP. Our datasets of 33,500 ubiquitination and 16,740 acetylation sites provide valuable insight into DDR remodeling of the proteome. We find that K6- and K33-linked polyubiquitination undergo bulk increases in response to DNA damage, raising the possibility that these linkages are largely dedicated to DDR function. We also show that Cullin Ring Ligases mediate 10% of DNA damage induced ubiquitination events and that EXO1 is an SCF-Cyclin F substrate in the response to UV radiation. Our extensive datasets uncover additional regulated sites on known DDR players such as PCNA and identify previously unknown DDR targets such as CENPs, underscoring the broad impact of the DDR on cellular physiology.
Introduction
In response to genotoxic stress, cells evoke a complex signaling network known as the DNA damage response (DDR) (Ciccia and Elledge, 2010; Jackson and Bartek, 2009). The DDR serves to protect genomic integrity by coordinating pathways involved in DNA repair, cell cycle arrest, gene transcription, apoptosis, and senescence. Execution of the DDR relies upon a dynamic array of protein modifications, with phosphorylation playing a central role. Using quantitative proteomics to identify over 700 substrates of the pivotal DDR kinases ATM and ATR, we previously demonstrated the extensive breadth of phosphorylation-dependent signaling in the DDR (Matsuoka et al., 2007). Since this initial study, the repertoire of known DDR-regulated phosphorylation events has expanded significantly to more than 1300 proteins (Beli et al., 2012; Bennetzen et al., 2010; Bensimon et al., 2010; Stokes et al., 2007).
In addition to phosphorylation, it is now evident the DDR depends upon other protein modifications such as ubiquitination (Bekker-Jensen and Mailand, 2011; Jackson and Durocher, 2013; Messick and Greenberg, 2009). Ubiquitin-dependent signaling regulates post-replication repair, the Fanconi Anemia (FA) pathway, the double-strand break (DSB) response, nucleotide excision repair (NER), and cell cycle arrest. Ubiquitination in these pathways can function to recruit interacting proteins, to allosterically alter protein function, or to target proteins for proteasomal degradation. These roles are often associated with different polyubiquitin chain linkages. For example, K63 linkages serve as a scaffold for signaling complex assembly during template switching and the DSB response, whereas K48 linkages lead to proteasomal degradation. Notably, the atypical K6 linkage has also been associated with the DDR, as the BRCA1/BARD1 ubiquitin ligase can assemble K6 linkages in vitro and in overexpression systems. However, whether endogenous K6 linkages increase after DNA damage and to what extent is currently unknown. This unresolved issue is critical to determining whether K6 ubiquitination occurs physiologically in the DDR (Kulathu and Komander, 2012).
In addition to ubiquitination, other lysine-linked protein modifications, such as acetylation, participate in the DDR. Acetylation regulates p53 activation (Brooks and Gu, 2011), DNA damage kinase signaling (Altmeyer et al., 2013; Floyd et al., 2013; Kaidi and Jackson, 2013; Sun et al., 2007), homologous recombination (Kaidi et al., 2010; Tang et al., 2013), nucleotide excision repair (Fan and Luo, 2010), non-homologous end joining (Miller et al., 2010), and base excision repair (Hasan et al., 2002). Comparison of ubiquitination and acetylation proteomic datasets has demonstrated that many sites of lysine ubiquitination and acetylation overlap, leading to speculation that in certain conditions, reciprocal regulation of acetylation and ubiquitination may occur at common sites (Bennetzen et al., 2013; Kim et al., 2011; Wagner et al., 2011). However, no proteomic studies have surveyed both DDR ubiquitination and acetylation together nor sequenced acetylation sites deeply enough in cell lines in order to make meaningful conclusions about reciprocal regulation.
To directly compare the relative prevalence of ubiquitination, acetylation, and phosphorylation in the DDR, we have quantitatively profiled all three modifications in response to both ultraviolet and ionizing radiation. For ubiquitination, we employed a proteomic strategy that enriches for ubiquitin-derived di-Glycine remnants on trypsinized peptides (Emanuele et al., 2011; Kim et al., 2011; Povlsen et al., 2012; Udeshi et al., 2013; Wagner et al., 2011). To functionally characterize DDR-regulated sites, we utilized secondary perturbations in 8 proteomic screens, producing a dataset that includes more than 33,500 ubiquitination sites. To detect acetylation sites on a similar scale, we developed an approach (FACET-IP) that allowed us to identify over 10,800 acetylation sites in a single experiment. These integrated datasets provide valuable insight into how the DDR remodels the proteome. We demonstrate that SCF-Cyclin F ubiquitinates EXO1 in response to UV radiation and that K6- and K33-linked polyubiquitination undergo bulk increases in response to DNA damage.
Results
Systematic profiling of ubiquitination in response to ultraviolet and ionizing radiation
To globally examine ubiquitination in the cellular responses to UV and ionizing radiation, we performed ubiquitin remnant profiling and metabolic labeling in cell culture. Heavy isotope-labeled cells were left untreated while light cells were stimulated with either UV or IR. Lysates were combined, digested with trypsin, and peptides containing ubiquitin-derived di-Glycine remnants were enriched with an antibody against di-Glycine. Three biological replicates were performed for each stimulus and 3 sequential immunoprecipitations (IPs) for each replicate, giving a total of 9 IPs per stimulus (Figure 1A). We attained strong reproducibility in quantitation between replicates for both the UV (Figure 1B) and IR screens (Figure 1C). In further support of the quality of this data, we identified proteins known to be ubiquitinated in response to UV and/or IR treatment, including FANCD2, FANCI, PCNA, and XPC (Figure 1D). Moreover, the levels of ubiquitination after UV versus IR stimulation were consistent with immunoblot results (Figure 1D): IR induced less FANCD2 ubiquitination and failed to stimulate PCNA ubiquitination, as previously reported (Ulrich, 2009). Notably, UV radiation regulated more sites than IR, as reflected in the histograms in Figure 1E. This elevated regulation is consistent with UV stimulating ubiquitination pathways not activated by IR, such as those coordinated by the stress-activated kinases p38 and JNK in response to non-genotoxic stimuli (Srinivas et al., 2005; Tsuchiya et al., 2010; Villumsen et al., 2013).
Figure 1. Quantitative profiling of ubiquitination in response to ultraviolet and ionizing radiation.
(A) Schematic overview of approach to profile ubiquitination regulated by UV or IR. Three biological replicates were performed for each stimulus and 3 sequential IPs for each replicate, giving a total of 9 IPs per stimulus.
(B, C) Experimental reproducibility indicated by Pearson correlation coefficients for Log2(L/H) ratios for 3 biological UV replicates (B) and for 3 biological IR replicates (C).
(D) Log2(L/H) ratios for diGly sites on proteins known to be ubiquitinated in response to UV and/or IR. For both FANCD2 and PCNA, the reduced or absent levels of IR-induced ubiquitination, relative to UV-induced ubiquitination, are consistent between proteomic and immunoblot results.
(E) Histograms depicting Log2(L/H) ratios for all quantified diGly sites in the 3 UV replicates (left) and 3 IR replicates (right).
(F) Log2(L/H) ratios for all quantified sites of PCNA ubiquitination found in both the UV and IR datasets. Error bars in all plots represent the SEM of unique MS1 quantifications for the indicated site. See also Figure S1.
Importance of proteasome inhibition in profiling DNA damage induced ubiquitination
In response to DNA damage, ubiquitination can lead to the proteasomal degradation of substrates such as CDCD25A, CDC25B, SETD8, and EXO1. We failed to detect increased ubiquitination of these proteins upon UV or IR stimulation, likely because of their rapid degradation. We therefore repeated UV and IR screens in the presence of the proteasome inhibitor MG132 (Figure 2A). We again observed strong correlation in quantitation between replicates for both the UV (Figure 2B) and IR screens (Figure 2C). Pre-treatment with MG132 markedly increased the detection of diGly peptides from proteins whose ubiquitination is known to cause proteasomal degradation (Figure 2D). This allowed us to identify the UV-induced ubiquitination of numerous known degraded proteins including CDC25A, CDC25B, SETD8, EXO1, DDB2, and CDC6 (Figure 2E). In the absence of MG132, importantly, many ubiquitinated peptides within these proteins actually decreased upon UV stimulation (Figure 2F). Reversal of these decreases with MG132 (Figure 2F) demonstrated they were due to protein degradation rather than deubiquitination. These results highlight the importance of proteasome inhibition in comprehensive investigation of ubiquitination dynamics. Equally important, however, is ubiquitin profiling in the absence of proteasome inhibition, which can cause ubiquitin pool depletion and thereby diminish the inducibility of non-degradative ubiquitination events. We observed this phenomenon for PCNA and XPC (Figure 2G), whose UV-induced ubiquitination is known to be non-proteolytic (Poulsen et al., 2013; Sugasawa et al., 2005; Ulrich, 2009).
Figure 2. Profiling of UV- and IR-regulated ubiquitination in the presence of proteasome inhibition.
(A) Schematic representation of DDR ubiquitin profiling in the presence of proteasome inhibition. MG132 was added to both damaged (light) and non-damaged (heavy) plates to capture ubiquitination events that lead to proteasomal degradation.
(B, C) Pearson correlation coefficients for Log2(L/H) ratios for 3 biological UV replicates in the presence of MG132 (B) and for 2 biological IR replicates in the presence of MG132 (C).
(D) MG132 increases detection of diGly peptides from proteins whose UV-induced ubiquitination is known to cause proteasomal degradation. Mean diGly TSCs (total spectral counts) depicted in blue are derived from the UV screen (Figure 1A), while mean diGly TSCs in red are from the UV-MG132 screen (Figure 2A). In the absence of MG132, diGly peptides from SETD8, CDC6, and EXO1 are not detected at all.
(E) Quantitation of diGly sites on proteins whose UV-induced ubiquitination is known to result in proteasomal degradation. Log2(L/H) ratios come from the UV-MG132 screen in Figure 2A.
(F) In the absence of MG132 (blue bars), diGly sites on degraded proteins decrease in response to UV stimulation, while pre-treatment with MG132 (red bars) reverses this decrease. Blue bars are from UV screens (Figures 1A or S4A) and red bars from the UV-MG132 screen (Figure 2A).
(G) The UV induction of known non-degradative ubiquitination events is lower in the presence (red bars) than in the absence (blue bars) of MG132. Blue bars are from the UV dataset (Figure 1A) and red bars from the UV-MG132 dataset (Figure 2A). Error bars in all plots represent the SEM of unique MS1 quantifications for the indicated site. See also Figure S1.
Ubiquitin profiling in fractionated nuclear extracts expands the DDR ubiquitinome
In addition to ubiquitin profiling in whole cell lysates, we performed diGly IPs from nuclear extracts. For these experiments, we fractionated nuclear lysates with strong cation exchange chromatography (SCX) prior to diGly enrichment (Figure S4A). IPs were performed from ten fractions for each UV or IR stimulus. We employed this fractionation procedure in order to allow comparison with acetylation and phosphorylation enrichment strategies described below. Combining sites from all of our DNA damage ubiquitin remnant experiments, representing 18 biological replicates and 68 diGly IPs, we identified 33,503 sites and quantified 26,009 sites (Figure S1A). We identified a total of 2197 sites whose abundance increased more than two-fold (Log2(L/H) ratio 1) in response to UV and 741 sites that were upregulated more than two-fold by IR.
Pathways and sites regulated by DDR ubiquitination
We performed gene set enrichment analysis on proteins whose ubiquitination either increased or decreased more than 2-fold following UV or IR radiation. In response to UV, we enriched for multiple DNA repair pathways, including NER and the FA pathway, as well as DNA damage induced checkpoint pathways (Figure 3A). Interestingly, we also enriched for functions related to the mitotic spindle, including centrosome amplification, chromosome alignment, chromatid cohesion, and chromosome segregation (Figure 3A, 3C). Notably, we found that 10 centromere proteins (CENPs) were deubiquitinated in response to UV or ionizing radiation (Figure 3B). This regulation may be related to crosstalk between the DDR and the spindle checkpoint, which was also enriched in our screens (Figure 3A, 3C), or may alternatively involve CENP proteins participating in non-centromere-related DDR functions. Consistent with this idea, the proteins CENP-S and CENP-X are known components of the FA core complex and are necessary for MMC resistance (Singh et al., 2010; Yan et al., 2010), while the proteins CENP-A,-N,-T,-U have been reported to localize to sites of laser microirradiation (Zeitlin et al., 2009). At the other end of the spindle, we identified UV-regulated proteins associated with the centrosome (Figure 3C), including the centriolar satellite proteins AZI1, PCM1, and MIB1, whose UV-induced deubiquitination and centrosomal delocalization was recently reported (Villumsen et al., 2013). We also identified proteins involved in spindle microtubule assembly, including 6 kinesin motor proteins and 4 of 8 subunits from the HAUS complex (Figure 3C), whose depletion results in destabilization of kinetochore mictrotubules (Lawo et al., 2009).
Figure 3. DDR-regulated ubiquitination of mitotic spindle proteins.
(A) Biological pathways and functions significantly enriched among proteins ubiquitinated in response to UV radiation in the UV, UV-MG132, or UV-SCX datasets. Analysis was performed with Ingenuity and significance determined using the right-tailed Fisher's exact test.
(B) Log2(L/H) ratios for diGly sites on centromere proteins (CENPs) deubiquitinated in response to UV and/or IR in the UV-SCX or IR-SCX datasets.
(C) Mitotic spindle proteins ubiquitinated or deubiquitinated in response to UV (rectangles), IR (ovals), or both (rectangular oval). Green proteins (Log2 ratios >1) are ubiquitinated and red proteins (Log2 ratios <−1) deubiquitinated. Data are from the UV, UV-MG132, UV-SCX, IR, IR-MG, or IR-SCX datasets.
In addition to identifying previously uncharacterized DDR targets, we uncovered additional ubiquitination sites on known DDR players. Ubiquitination of PCNA on K164 is a well-known DDR event that leads to the recruitment of translesion-polymerases or the translocase ZRANB3 (Ciccia et al., 2012). We identified UV-induced ubiquitination of PCNA on not only K164 but also the additional site K117 (Figure 1F). Prior reports have demonstrated that ubiquitination of K107 in S. cerevisiae PCNA occurs in response to the failure of Okazaki fragment ligation arising from deletion of DNA ligase I and that this ubiquitination is necessary for RAD53 phosphorylation (Das-Bradoo et al., 2010b). This finding has led to the proposal that different types of DNA damage (nicks in nascent strands versus replication-blocking lesions in template strands) may elicit ubiquitination on different sites in S. cerevisiae PCNA (Das-Bradoo et al., 2010a). Our results suggest that similar regulation may occur in human cells. Notably, depletion of DNA ligase-I in human cells causes PCNA ubiquitination on an unknown site (Das-Bradoo et al., 2010b).
Systematic profiling of acetylation in response to ultraviolet and ionizing radiation
Proteomic studies have demonstrated that many sites of lysine ubiquitination and acetylation overlap, suggesting that reciprocal regulation of acetylation and ubiquitination may occur at common sites (Bennetzen et al., 2013; Kim et al., 2011; Wagner et al., 2011). Importantly no prior study has sequenced acetylation in cell lines deeply enough to make meaningful conclusions about coregulation. To examine such coregulation in the DDR, we sought to identify acetylation sites at a scale similar to that possible for ubiquitination and thus developed the approach FACET-IP (Fractionated ACETylation IP). Peptides from nuclear lysates were separated by strong cation exchange chromatography (SCX) into ten fractions, and acetylated peptides were enriched with a pan-acetyl antibody from each fraction (Figure 4A). SCX improved the number of acetylation sites identified in a single acetylation IP. Whereas a single IP from 20mg of total cellular peptides yielded 1006 sites (Figure 4B, Table S5), IP from a single SCX fraction containing only 1-2 mg of peptides, produced up to 2556 sites (Figure 4B). Over ten SCX fractions, this method generated over 10,800 sites per 20 mg of total cellular peptides (Figure S2A). Including pilot fractionation studies using whole cell lysates, we have identified a total of 16,740 acetylation sites (Table S5), which is the largest collection of sites in a single study to-date (Choudhary et al., 2014).
Figure 4. FACET-IP identifies acetylation sites regulated by ultraviolet or ionizing radiation.
(A) Schematic for proteomic identification of acetylation sites by FACET-IP.
(B) Number of acetylation sites (blue) and acetylated proteins (red) detected by FACET-IP among 10 SCX fractions (range 745-2556 sites, 523-1220 proteins). Elution of fractions 1-5 occurred using a KCl gradient to 91 mM and fractions 6-10 using isocratic 350 mM KCl. Dashed lines represent the number of acetylated sites (1006 sites, blue) and proteins (554 proteins, red) identified in a single IP from 20 mg of nuclear lysates.
(C) Number of acetylated sites and proteins identified, quantified, and upregulated at least 2-fold in response to UV and IR.
(D) Identification of site K382 on TP53 known to be acetylated in response to UV and IR.
(E) Heat map representing Log2(L/H) ratios for ubiquitination (upper) and acetylation (lower) among 237 quantified sites that were both acetylated and ubiquitinated and that underwent a more than 2-fold increase in ubiquitination in response to UV.
(F) Percentage of quantified ubiquitination, acetylation, and phosphorylation sites that are upregulated at least 2-fold in response to UV. All values are derived from proteomic screens (anti-diGly, anti-acK, or IMAC) involving SCX fractionation of nuclear lysates.
(G) Heat map representing the number of UV or IR-inducible acetylation, ubiquitination, and phosphorylation sites on DNA repair proteins (belonging to the Gene Ontology category GO:000628) from six prior proteomic studies, as described in Experimental Procedures. See also Figures S2-S4.
To examine UV- and IR-regulated acetylation, we utilized SILAC in conjunction with FACET-IP (Figure 4A). We identified 12,456 sites and quantitated 10,192 sites for the UV and IR experiments together (Figure 4C). This data set is nearly 5 times larger than that for a previous study of etoposide (10 ıM 24 hours) and IR (10 Gy 1 hour) treatment and 20 times larger when considering acetylation events that occur within 1 hour and thus are less prone to cell cycle or transcriptional effects (Figure S2B) (Beli et al., 2012). The success of our technique is demonstrated by our ability to detect UV and IR-induced p53 acetylation on its known site K382 (Figure 4D). We identified 219 sites whose acetylation increased at least two-fold in response to UV radiation and 107 in response to IR (Figure 4C, Table S5). Increased acetylation occurred on multiple proteins with known roles in the DDR including RPA1, RPA3, ERCC8, ERCC4, and APEX1 (Figure S3A). We also observed de-acetylation of known DDR factors including NBN, RAD50, XRCC1, BLM, CLSPN, EXO1, USP7, and MSH2. These latter proteins may be targets of histone deacetylase inhibitors, possibly explaining how this class of drugs causes DNA damage sensitization, the mechanism for which is currently unclear.
Lastly, we determined the frequency of ubiquitination and acetylation co-regulation at common sites following UV radiation. Among 2276 quantitated sites that were both ubiquitinated and acetylated, 237 underwent a ≥ 2-fold increase in ubiquitination, and only 5 of these sites experienced a reciprocal ≥ 2-fold decrease in acetylation (Figure 4E). Similarly, among 180 sites undergoing ≥ 2-fold deubiquitination, only 5 experienced an increase in acetylation (Figure S3B). These results suggest that reciprocal regulation of ubiquitination and acetylation is not a frequent event in the DNA damage response. It is possible that different cellular populations are ubiquitinated versus acetylated on the same sites.
Comparative analysis of ubiquitination, acetylation, and phosphorylation in the DNA damage response
Given a meaningful number of acetylation sites regulated by DNA damage, we set out to quantify the relative prevalence of ubiquitination, acetylation, and phosphorylation regulation in the DNA damage response. To allow for an accurate comparison among the three modifications, we quantitated ubiquitination and phosphorylation using the same approach we utilized for acetylation: SCX fractionation of nuclear lysates. Nuclear peptides were fractionated by SCX and ubiquitinated peptides were then enriched with diGly antibody and phosphopeptides with IMAC (Figure S4A). We found that the percentage of sites that increased at least 2-fold in response to UV radiation was similar for phosphorylation (8.4%) and ubiquitination (8.5%) and was over 3 times greater than that for acetylation (Figure 4F). When looking at PTM removal, we also found that UV induced deubiquitination and dephosphorylation were more prevalent than deacetylation (Figure S4B). These results suggest that regulation of ubiquitination is as common as phosphorylation in the DNA damage response and that both modifications are more common than acetylation.
We next compared DNA damage induced ubiquitination, acetylation, and phosphorylation on specifically DNA repair proteins (belonging to the Gene Ontology category GO:000628). To depict a heat map summary of inducible sites on DDR proteins, we included two-fold induced sites from this work and from all prior proteomic studies of phosphorylation and acetylation. We found fewer instances of inducible acetylation than inducible ubiquitination or phosphorylation on proteins within the DNA repair category (Figure 4G). Moreover, the number of inducible sites on these proteins was greater for ubiquitination and phosphorylation than for acetylation (Figure 4G). Notably, DDR components were among the most heavily acetylated proteins in the proteome, with DNAPK and PARP1 falling among the top 12 proteins with the most acetylation sites (Figure S3C). Despite the presence of many acetylation sites on DDR proteins, our results collectively suggest that acetylation regulation by DNA damage is significantly less common than for ubiquitination and phosphorylation, consistent with previous findings that compared acetylation and phosphorylation (Beli et al., 2012).
K6- and K33-linked polyubiquitination occur in response to UV radiation
Lysine linkages between ubiquitin monomers within polyubiquitin chains are associated with different physiological roles. K48- and K11-linked polyubiquitination lead to proteasomal degradation, whereas K63-linked polyubiquitination serves as a scaffold for signaling complex assembly. Increases in both K48 and K63 linkages occur on specific proteins in response to IR and UV radiation. K48 linkage is involved in the degradation of cell cycle proteins such as CDC25A and CDT1, while K63-linked polyubiquitination occurs on the scaffolds H2A and PCNA to recruit proteins that promote DNA repair.
Our ubiquitin profiling efforts revealed no changes in bulk K48 or K63 linkages upon IR or UV radiation. These findings are expected given the plethora of biological processes that utilize K48 and K63 linkages outside of DNA damage signaling. Much to our surprise, however, we did detect bulk increases in K6 (3.6-fold) and to a lesser extent K33 (1.8-fold) linkages in response to UV but not ionizing radiation (Figure 5A). Importantly, we observed increases in K6 and K33 linkages in all 7 biological UV replicates and in none of 6 IR replicates (Figures 5B, 5C), supporting the strong reproducibility of our data. For the K6 linkage, these replicates represent a total of 76 unique peptide measurements for the UV data and 68 measurements for the IR data. Remarkably, increases in K6 linkage occurred for all 76 unique peptides in the UV data and none of the 68 peptides in the IR data (Figure 5B). Similarly, increases in K33 linkage occurred in all of 27 unique peptide measurements for the UV data and none of 21 unique peptide measurements for the IR data (Figure 5C). These results demonstrate that endogenous K6- and K33-linkages increase in response to DNA damage, the implications for which are discussed below.
Figure 5. K6- and K33-linked polyubiquitination occurs in response to UV radiation.
(A) Log2(L/H) ratios for ubiquitin diGly sites from cells treated with UV (blue bars) or IR (red bars). Data are from the UV and IR datasets in Figure 1A. Error bars represent the SEM of multiple MS1 quantifications for the indicated site.
(B) Log2(L/H) ratios for ubiquitin peptides modified on Lys-6 with diGly. For UV stimulation, all 76 unique peptide measurements across 7 biological replicates demonstrate upregulation, whereas none of 68 IR peptide measurements across 6 biological replicates are regulated.
(C) Log2(L/H) ratios for ubiquitin peptides modified on Lys-33 with diGly. For UV stimulation, all 27 unique peptide measurements across 7 biological replicates demonstrate upregulation, whereas none of 21 IR peptide measurements across 6 biological replicates are regulated. Gray bars in (B) and (C) represent median and standard deviation of Log2(L/H) values.
Cullin-RING-ligases are responsible for 10% of UV-induced ubiquitination events
Determining E3 ligases upstream of ubiquitination events is important for a systems analysis of DNA damage induced ubiquitination. While few pharmacologic inhibitors of ubiquitin ligases are available for this purpose, Cullin-RING-ligase (CRL) activity can be probed with the drug MLN4924. MLN4924 inhibits CRL neddylation, which is necessary for the activation of all CRL ligases. To identify UV-induced ubiquitination events mediated by CRLs, we performed ubiquitin remnant profiling with UV radiation and MLN4924. Both heavy and light cells were treated with UV radiation and with MG132, while only light cells were treated with MLN4924 (Figure 6A). Di-Glycine IPs were performed in triplicate and yielded strong correlation between replicates (Figure 6B). We identified 206 sites on 116 proteins whose ubiquitination increased at least 2-fold in response to UV radiation and decreased at least 2-fold in response to MLN4924 treatment (Figure 6C). This data suggests that approximately 10% of UV-induced ubiquitination events are mediated by CRLs (Figure 6C). Our approach successfully identified many proteins known to be ubiquitinated by CRLs in response to UV radiation, including CDC25A, SETD8, XPC, DDB2, and CDC25B (Figure 6D), demonstrating the validity of our approach.
Figure 6. Screen for CRL-mediated ubiquitination events in the DDR identifies EXO1 as an SCF substrate.
(A) Diagram of approach to identify UV or IR induced ubiquitination events mediated by CRLs. Light cells were treated with 10 ιM MLN4924 and 5 ιM MG132 for 30 minutes while heavy cells were treated with 5 ιM MG132 for 30 minutes. Both heavy and light plates were then irradiated with 40 J/m2 UV or 10 Gy IR and harvested 1 hour later.
(B) Correlation coefficients for Log2(L/H) ratios for 3 biological replicates identifying CRL substrates in the presence of UV and for 2 biological replicates in the presence of IR.
(C) Overlap of sites and proteins ubiquitinated more than 2-fold in response to UV (from UV, UV-MG, and UV-SCX datasets) with sites whose ubiquitination is suppressed more than 2-fold by MLN4924 (from UV-MLN dataset).
(D) Identification of proteins whose ubiquitination is stimulated by UV and inhibited by MLN4924. Blue bars represent Log2(L/H) ratios of diGly sites from UV-MG132 dataset (Figure 2A) while red bars represent ratios from UV-MLN dataset (Figure 6A). EXO1 represents a previously uncharacterized CRL substrate.
(E) (Above) HeLa cells were released from a double-thymidine block and harvested at 2 hour intervals. Two hours prior to each point of harvest, cells were treated with 1 ιg/mL 4NQO. Propidium iodide stained cells from each time point were analyzed by flow cytometry, and lysates blotted for EXO1. (Below) HeLa cells were released from a double thymidine arrest and harvested 6 hours later. At 4 hours prior to harvest, they were treated with 5 ιM MLN4924 for 2 hours followed by 1 ιg/mL 4NQO for 2 hours.
(F) HeLa cells infected with lentiviruses expressing dominant negative Cullins or empty vector (EV) were harvested 2 hours after 1 ιg/mL 4NQO treatment and blotted for EXO1. See also Figure S5.
SCF-Cyclin F mediates EXO1 ubiquitination in response to UV damage
In addition to known CRL substrates, we also identified previously uncharacterized targets of CRLs in the response to UV radiation. For example, the nuclease EXO1 is known to be ubiquitinated and degraded by the proteasome in response to HU-induced replication fork stalling (El-Shemerly et al., 2005). Our proteomic analyses detected EXO1 ubiquitination in response to UV radiation (Figure 2E, Table S2). Consistent with UV-induced ubiquitination mediating proteasomal degradation, we did not detect EXO1 ubiquitination proteomically in the absence of proteasome inhibition (Figure 2D, Table S1), and MG132 inhibited EXO1 degradation detected by immunoblot (Figure S5A). Also consistent with immunoblot results (Figure S5B), our proteomic data indicated that EXO1 ubiquitination was stimulated by UV but not IR (Figure S5B, Table S2). Importantly, the E3 ligase for EXO1 has not been previously identified. Our proteomic data indicated that UV-induced ubiquitination of EXO1 at K796 was inhibited by MLN4924 (Figure 6D). To validate this finding, we demonstrated by immunoblot that EXO1 degradation was maximal in late S / G2 phase of the cell cycle and that this degradation was inhibited by MLN4924 (Figure 6E). To determine which of five Cullins in the CRL family mediate EXO1 ubiquitination, we expressed dominant negative mutants of each Cullin and found that CUL1 was responsible for EXO1 degradation (Figure 6F). These findings establish EXO1 as an SCF substrate in the DDR.
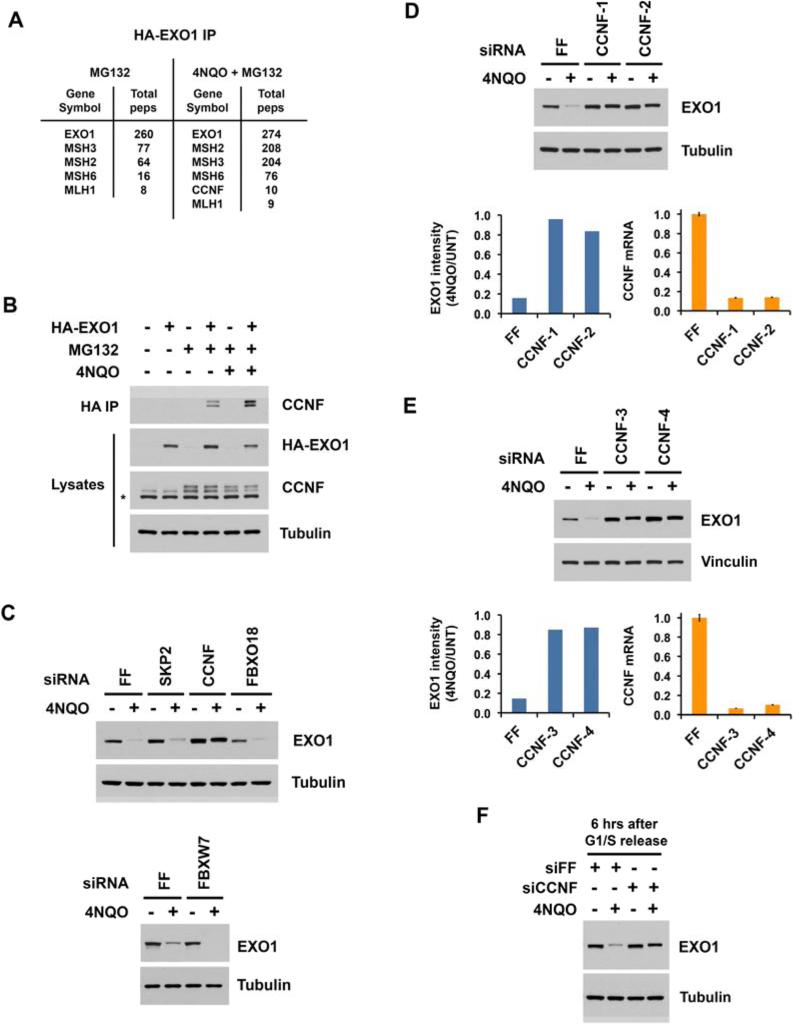
To determine the F-box protein responsible for EXO1 ubiquitination, we sought to identify candidates interacting with EXO1 preferably in a DNA damage-induced fashion. We therefore treated 293T cells with 4NQO (in the presence of MG132) and screened for proteins interacting with HA-tagged EXO1 by mass spectrometry. We found that the F-box protein Cyclin F (CCNF) associated with EXO1 in 4NQO-stimulated but not non-damaged cells (Figure 7A). Importantly, immunoblotting of HA-EXO1 immunoprecipitates with a Cyclin F antibody confirmed the interaction (Figure 7B). MG132 treatment was necessary to detect binding, consistent with degradation of EXO1 occurring upon Cyclin F association (Figure 7B). MG132 additionally increased the levels of Cyclin F, whose own stability is known to be regulated by the proteasome (D'Angiolella et al., 2013). Importantly, stimulation with 4NQO enhanced interaction of the proteins beyond that with MG132 alone (Figure 7B). These results demonstrate that binding of Cyclin F to EXO1 is induced by DNA damage.
Figure 7. SCF-Cyclin F mediates EXO1 ubiquitination in response to UV damage.
(A) HA-tagged EXO1 was immunoprecipitated from 293T cells treated with 5 ιM MG132 for 30 minutes followed by 1 ιg/mL 4NQO for 2 hours. EXO1 complexes were eluted with HA peptide and interacting proteins identified by mass spectrometry.
(B) 293T cells expressing HA-tagged EXO1 were treated with 5 ιM MG132 for 30 minutes followed by 1 ιg/mL 4NQO for 2 hours, and HA immunoprecipitates were blotted for Cyclin F. Asterisk represents a non-specific cross-reacting band.
(C) HeLa cells transfected with siFF or siRNA pools against the F-box proteins SKP2, Cyclin F, FBXO18, or FBXW7 were treated with 1 ιg/mL 4NQO for 2 hours and lysates blotted for EXO1.
(D, E) HeLa cells transfected with siFF or individual siRNAs against Cyclin F were treated with 1 ιg/mL 4NQO for 2 hours and lysates blotted for EXO1. EXO1 band intensities, measured by Image J, were normalized to tubulin or vinculin, and ratios from 4NQO to UNT (untreated) lanes calculated. Cyclin F mRNA depletion was confirmed by RT-qPCR.
(F) HeLa cells transfected with siFF or siCCNF-4 were released from a double thymidine arrest and harvested 6 hours later. At 2 hours prior to harvest, they were treated with 1 ιg/mL 4NQO for 2 hours. See also Figure S6.
We next investigated whether Cyclin F functionally regulates EXO1 stability. We found that depletion of multiple other F-box proteins (ı-TCRP, FBXW7, SKP2, and FBXO18) failed to prevent 4NQO-induced EXO1 degradation (Figures 7C, S6A), while depletion of Cyclin F strongly inhibited its degradation (Figure 7C). We confirmed this result using four individual siRNAs, all of which efficiently depleted Cyclin F and suppressed EXO1 degradation (Figures 7D, 7E). Notably, the degree of Cyclin F depletion correlated with inhibition of EXO1 degradation, as shown for an additional siRNA that was less effective in blocking EXO1 degradation (Figure S6B). These findings reveal that Cyclin F promotes the destabilization of EXO1 in response to DNA damage.
Cyclin F expression oscillates during the cell cycle, with maximal levels occurring in S and G2 (Bai et al., 1994). Because 4NQO-induced EXO1 degradation occurs throughout the cell cycle (Figure 6E), we examined Cyclin F function specifically in S and G2. Similar to MLN4924-induced stabilization of EXO1 in G2 (Figure 6E), depletion of Cyclin F inhibited 4NQO-induced EXO1 degradation at 3 hours (Figure S6C) and 6 hours (Figure 7F) after release from a double thymidine block when cells were in S and G2, respectively. Thus Cyclin F destabilizes EXO1 during the cell cycle phases where its levels are most abundant. This finding along with DNA damage induced interaction of the two proteins identify EXO1 as a substrate of SCF-Cyclin F in the DNA damage response.
Discussion
Systematic orthogonal profiling of ubiquitination in the DDR
We have globally profiled ubiquitination in response to UV and ionizing radiation, utilizing secondary perturbations in 8 orthogonal screens (Figure S1) and producing a dataset of over 33,000 ubiquitination sites. Our work highlights the importance of ubiquitin profiling in the presence of proteasome inhibition, which facilitates detection of proteolytic ubiquitination events. In the absence of proteasome inhibition, we would have failed to detect UV-induced EXO1 ubiquitination (Figure 2D). Profiling in the presence of proteasome inhibition also helps to distinguish inducible deubiquitination from protein degradation (Figure 2F). In a prior proteomic study, UV-induced decrease in diGly sites on the PCNA-binding protein PAF15 was found to result from protein degradation rather than deubiquitination (Povlsen et al., 2012), emphasizing the importance of discriminating between these causes of diGly site reduction. However, ubiquitin profiling should not be performed exclusively in the presence of proteasome inhibition, as it can cause ubiquitin pool depletion and impair the detection of non-degradative ubiquitination events (Figure 2G). Combining results from complementary UV screens performed with MG132 or MLN4924 led to the discovery that EXO1 is a CRL substrate. Integrating orthogonal screens allows for functional site characterization and represents a strength of our datasets.
FACET-IP improves global acetylation profiling in the DDR
In addition to ubiquitination, we profiled acetylation and phosphorylation in response to UV and IR. We found that DDR-regulated ubiquitination is as prevalent as phosphorylation and significantly more common than acetylation. To identify acetylation sites on a scale similar to that for ubiquitination, we developed the approach FACET-IP, which allowed identification of over 10,800 sites in a single experiment. Our improvement in acetylation site yield does not result simply from performing additional IPs per biological replicate. We found that a single IP from 20 mg of total cellular peptides generated 1006 sites. This number was surpassed for 7 of 10 IPs performed from single SCX fractions (range 1240-2556 sites) despite the presence of only 1-2 mg peptides in each fraction (Figure 4B). SCX fractionation prior to acetylpeptide enrichment likely prevents overwhelming of the antibody by highly abundant acetylpeptides such as those arising from histones. Separating these peptides from the bulk population likely enhances access of the antibody to less abundant peptides.
Ubiquitin regulation of mitotic spindle proteins
We detected DDR-regulated ubiquitination sites on kinetochore and other mitotic spindle proteins (Figure 3C). Regulation of kinetochore-microtubule attachment and chromosome alignment by DDR factors has been previously reported (Rozier et al., 2013). Our data suggests that ubiquitin signaling may mediate DDR regulation of the kinetochore. We found that 10 centromeric proteins (CENPs) were deubiquitinated in response to UV and/or IR. This regulation may represent the DDR interfacing with the spindle assembly checkpoint or alternatively the involvement of CENPs in non-centromere-related DDR functions that require CENP localization to DNA damage sites as described (Singh et al., 2010; Yan et al., 2010; Zeitlin et al., 2009). An additional possibility is that CENP ubiquitination regulates chromosome positional stability (Soutoglou et al., 2007) or enhanced chromosome mobility (Dimitrova et al., 2008) that has been observed in different DNA damage settings. Yet another possibility is that ubiquitination of CENPs may regulate recombination between repetitive DNA sequences found at centromeres. Future studies to elucidate the function of CENP deubiquitination are likely to yield important insights into the DDR.
K6- and K33-linked polyubiquitination in response to UV radiation
K6-linked polyubiquitination has been associated with the DDR through the ubiquitin ligase BRCA1. Prior studies have shown that BRCA1 can assemble K6-linked polyubiquitin chains in vitro and in cells when overexpressed (see references in Kulathu and Komander, 2012; Christensen et al., 2007). However, these studies did not examine whether DNA damage can actually induce K6 polyubiquitination nor did they assay for endogenous K6 linkages. Here, we demonstrate that UV, but not IR, increases endogenous K6-linked polyubiquitination. BRCA1 localizes to UV-induced foci, functions in post-replication repair, and stabilizes stalled replication forks (Pathania et al., 2011; Schlacher et al., 2012). It therefore has known roles in the response to replication stress and represents a candidate for regulating UV-induced K6 ubiquitination. Involvement of BRCA1's ubiquitin ligase in this response would provide another function for the ligase, which is dispensable for homologous recombination repair of DSBs (Reid et al., 2008).
In addition to K6-linked polyubiquitination, we discovered a smaller UV-induced increase in K33 ubiquitination. Little is known about K33 ubiquitination, and it has not been previously linked to the DDR. For both K6 and K33 linkages, it is significant they underwent bulk increases in response to UV radiation. It is surprising that ubiquitin linkages of any type would undergo UV-induced aggregate changes detected by mass spectrometry, as this requires that the linkage is produced in such high quantity that it overwhelms levels resulting from all other concomitant cellular processes. For example, it is well established that K63- and K48-linked ubiquitination are induced by DNA damage, yet we observed little or no bulk increases in these linkages upon UV or ionizing radiation, likely because of their significant involvement in other processes. Thus, there exists a strong and unique relationship between UV radiation and polyubiquitination involving K6 and K33 linkages that raises the question as to whether these linkages are largely dedicated to the DNA damage response.
EXO1 is an SCF-Cyclin F substrate in the response to replication stress
EXO1 is a member of the RAD2 family of structure-specific nucleases with 5’-3’ processive exonuclease activity and 5’-flap endonuclease activity. It functions in multiple DNA metabolic processes, including mismatch repair, DSB resection during homologous recombination, and Okazaki fragment processing (Tran et al., 2004). We demonstrate that EXO1 is ubiquitinated and degraded by the proteasome in response to UV radiation and 4NQO, both of which cause replication stress. Prior reports have shown that EXO1 deletion suppresses replication fork instability caused by RAD53 deletion mutants in S. cerevisiae (Segurado and Diffley, 2008). Furthermore, EXO1 mediates the resection and degeneration of stalled replication bubbles in RAD53 mutant cells (Cotta-Ramusino et al., 2005). EXO1 is present at replication forks due to its roles in mismatch repair and lagging strand synthesis. The purpose of its degradation during replication stress may therefore be to prevent unwanted resection of stalled replication forks. Notably, 4NQO-induced EXO1 destabilization also occurs during G2, where its degradation may serve to prevent excessive DSB resection during homologous recombination. Unchecked nucleases can threaten genomic stability, so limiting their activity through negative feedback circuitry is important for avoiding unintended toxicity (Ciccia and Elledge, 2010). Importantly, we show that EXO1 ubiquitination is mediated by SCF-Cyclin F. Increase in basal EXO1 upon Cyclin F depletion (Figures 7C-E) is reminiscent of CDC25A elevation upon ı-TCRP depletion (Figure S6A) and may result from basal suppression of EXO1 levels by endogenous DNA damage or may indicate an additional role for SCF-Cyclin F ubiquitination of EXO1 in the absence of damage. Although Cyclin F was the founding member of the F-box protein family (Bai et al., 1996), relatively few of its substrates have been identified to-date (D'Angiolella et al., 2013), and EXO1 represents an example that expands this important list.
Our findings on SCF-Cyclin F mediated EXO1 ubiquitination and K6 and K33-linked polyubiquitination demonstrate the utility of our proteomic datasets, which are likely to yield additional valuable insights into DDR pathways.
Experimental Procedures
SILAC sample preparation
HeLa cells were cultured in lysine- and arginine-free DMEM containing 10% dialyzed FBS. Light media was supplemented with 50 mg/mL L-lysine and 40 mg/ml L-arginine, while heavy was supplemented with 50 mg/ml L-lysine-U-13C6-15N2 and 40 mg/ml L-arginine-U-13C6-15N4(Cambridge Isotope Labs). For all SILAC screens (except those described in Figure 6), heavy cells were not damaged while light cells were treated with 40 J/m2 UV or 10 Gy IR and then harvested 1 hour later. For the MG132 screens in Figure 2, both light and heavy cells were treated with 5 ıM MG132 for 30 minutes prior to UV or IR treatment of the light cells. For the MLN4924 screens in Figure 6, light cells were treated with 10 ıM MLN4924 and 5 ıM MG132 for 30 minutes while heavy cells were treated with 5 M MG132 for 30 minutes. Both light and heavy cells were then irradiated with 40 J/m2 UV or 10 Gy IR and harvested 1 hour later. For all screens, cells were harvested, washed with PBS, and lysed in denaturing buffer consisting of 8M urea, 20 mM HEPES pH 7.5, 1 mM ı-glycerophosphate, 2.5 mM sodium pyrophosphate, and 1 mM Na3VO4 with sonication. Heavy and light lysates were combined in a 1:1 ratio, and proteins (20 mg) were reduced with 4 mM DTT, alkylated with 5.5 mM chloroacetamide, and digested with trypsin overnight at room temperature. The solution was acidified with trifluoroacetic acid (TFA) and clarified by centrifugation. The supernatant was then desalted on a Sep-Pak C18 column (Waters), and peptides were lyophilized.
Enrichment of diGly, acetylated, and phosphorylated peptides
For diGly and acetylation proteomics, lyophilized peptides were dissolved in IP buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM NaCl) and then enriched by overnight incubation with protein A agarose beads conjugated to a monoclonal diGly antibody (Cell Signaling Technology, Inc) or polyclonal acetyl-lysine antibody (ImmuneChem, Inc). Beads were washed with IP buffer followed by water, and enriched diGly-modified or acetylated peptides were eluted with 0.15% TFA. For diGly screens in Figures 1, 2, and 6, three sequential immunoprecipitations were performed. Eluates were desalted by Stage tip chromatography and lyophilized before analysis by LC-MS/MS.
For phosphorylation proteomics, one-half of each lyophilized SCX peptide fraction was dissolved in IMAC buffer (250 mM acetic acid, 30% acetonitrile) and then enriched by 90 minute incubation with precharged IMAC resin (Phos-Select iron affinity gel, Sigma–Aldrich). Beads were washed with IMAC buffer and bound peptides eluted with 50 mM Tris, 300 mM NH4OH, pH 10. The remainder of each lyophilized SCX fraction was dissolved in TiO2 buffer (2 M dihydroxybenzoic acid, 50% acetonitrile, 0.1% TFA) and enriched by 90 minute incubation with Titansphere TiO2 beads (GL Sciences). Beads were washed with TiO2 buffer followed by 50% acetonitrile, 0.1% TFA, and bound peptides were eluted as for IMAC. Eluted peptides from both IMAC and TiO2 enrichment were desalted by Stage tip chromatography and lyophilized before MS analysis.
Mass spectrometry
Lyophilized peptides enriched by diGly IP, FACET-IP, or IMAC/TiO2 were dissolved in 5% acetonitrile / 5%formic acid. Using a Famos autosampler (LC Packings), they were loaded onto a reversed phase microcapillary column (100 mm I.D.) packed first with 5 mm of Magic C4 resin (5 mm, 100 Ao, Michrom Bioresources) followed by 20 cm of Maccel C18AQ resin (3 mm, 200 Ao, The Nest Group, Inc.). Ubiquitin-derived diGly and acetylated peptides were separated using a gradient of 5%–27% acetonitrile in 0.125% formic acid over 180 minutes and detected in a LTQ-Orbitrap Velos Pro mass spectrometer (ThermoFisher). IMAC/TiO2-enriched phosphorylated peptides were separated using a 95 minute gradient and detected in an LTQ-Orbitrap XL mass spectrometer (ThermoFisher).
Supplementary Material
Highlights.
Proteomic discovery of DNA damage regulated ubiquitination and acetylation sites
FACET-IP approach improves acetylation site detection
Atypical K6- and K33-polyubiquitin linkages increase in response to UV radiation
SCF-Cyclin F mediates EXO1 ubiquitination in response to UV radiation
Acknowledgments
We thank members of the Elledge and Gygi labs for helpful discussions. We also thank W. Harper for the Cul3 and Cul4 dominant-negative clones, and M. Rape for pCS2 His-ubiquitin plasmids. A.E.H.E is supported by a Burroughs Wellcome Fund CAMS Award and K12 Paul Calabresi Award for Oncology. I.H. is a recipient of an EMBO long-term fellowship. This work was supported by NIH grants to S.J.E. and S.P.G. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altmeyer M, Toledo L, Gudjonsson T, Grofte M, Rask MB, Lukas C, Akimov V, Blagoev B, Bartek J, Lukas J. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol Cell. 2013;52:206–220. doi: 10.1016/j.molcel.2013.08.025. [DOI] [PubMed] [Google Scholar]
- Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Dinant C, Watanabe S, Bartek J, Lukas J, Andersen JS. Acetylation dynamics of human nuclear proteins during the ionizing radiation-induced DNA damage response. Cell Cycle. 2013;12:1688–1695. doi: 10.4161/cc.24758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal. 2010;3:rs3. doi: 10.1126/scisignal.2001034. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2:456–462. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- D'Angiolella V, Esencay M, Pagano M. A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis. Trends Cell Biol. 2013;23:135–140. doi: 10.1016/j.tcb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo S, Nguyen HD, Bielinsky AK. Damage-specific modification of PCNA. Cell Cycle. 2010a;9:3674–3679. doi: 10.4161/cc.9.18.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo S, Nguyen HD, Wood JL, Ricke RM, Haworth JC, Bielinsky AK. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat Cell Biol. 2010b;12:74–79. doi: 10.1038/ncb2007. sup pp 71-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes nonhomologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 2005;65:3604–3609. doi: 10.1158/0008-5472.CAN-04-4069. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S, El-Andaloussi N, Hardeland U, Hassa PO, Burki C, Imhof R, Schar P, Hottiger MO. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol Cell. 2002;10:1213–1222. doi: 10.1016/s1097-2765(02)00745-1. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–74. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, Feunteun J, Livingston DM. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell. 2011;44:235–251. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA, et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol. 2013;201:797–807. doi: 10.1083/jcb.201212075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povlsen LK, Beli P, Wagner SA, Poulsen SL, Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N, Choudhary C. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol. 2012;14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, Baer R, Ludwig T. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc Natl Acad Sci U S A. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozier L, Guo Y, Peterson S, Sato M, Baer R, Gautier J, Mao Y. The MRN-CtIP pathway is required for metaphase chromosome alignment. Mol Cell. 2013;49:1097–1107. doi: 10.1016/j.molcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Diffley JF. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008;22:1816–1827. doi: 10.1101/gad.477208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas H, Juroske DM, Kalyankrishna S, Cody DD, Price RE, Xu XC, Narayanan R, Weigel NL, Kurie JM. c-Jun N-terminal kinase contributes to aberrant retinoid signaling in lung cancer cells by phosphorylating and inducing proteasomal degradation of retinoic acid receptor alpha. Mol Cell Biol. 2005;25:1054–1069. doi: 10.1128/MCB.25.3.1054-1069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Asano T, Nakayama K, Kato T, Jr., Karin M, Kamata H. Nuclear IKKbeta is an adaptor protein for IkappaBalpha ubiquitination and degradation in UV-induced NF-kappaB activation. Mol Cell. 2010;39:570–582. doi: 10.1016/j.molcel.2010.07.030. [DOI] [PubMed] [Google Scholar]
- Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc. 2013;8:1950–1960. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, Yang YG, Choudhary C, Nielsen ML, Mailand N, et al. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013;32:3029–3040. doi: 10.1038/emboj.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.