Abstract
The production of mature eosinophils is a tightly orchestrated process with the aim to sustain normal eosinophil levels in tissues while also maintaining low numbers of these complex and sensitive cells in the blood. To identify regulators of homeostatic eosinophilopoiesis in mice, we took a global approach to identify genome-wide transcriptome and epigenome changes that occur during homeostasis at critical developmental stages, including eosinophil-lineage commitment and lineage maturation. Our analyses revealed a markedly greater number of transcriptome alterations associated with eosinophil maturation (1199 genes) than with eosinophil-lineage commitment (490 genes), highlighting the greater transcriptional investment necessary for differentiation. Eosinophil progenitors (EoPs) were noted to express high levels of granule proteins and contain granules with an ultrastructure distinct from that of mature resting eosinophils. Our analyses also delineated a 976-gene eosinophil-lineage transcriptome that included a repertoire of 56 transcription factors, many of which have never previously been associated with eosinophils. EoPs and eosinophils, but not granulocyte-monocyte progenitors (GMPs) or neutrophils, expressed Helios and Aiolos, members of the Ikaros family of transcription factors, which regulate gene expression via modulation of chromatin structure and DNA accessibility. Epigenetic studies revealed a distinct distribution of active chromatin marks between genes induced with lineage commitment and genes induced with cell maturation during eosinophil development. In addition, Aiolos and Helios binding sites were significantly enriched in genes expressed by EoPs and eosinophils with active chromatin, highlighting a potential novel role for Helios and Aiolos in regulating gene expression during eosinophil development.
Keywords: Hematopoiesis, Eosinophils, Cell Differentiation, Gene Regulation, Transcription Factors, Allergy
Introduction
Eosinophils are predominately tissue-dwelling leukocytes that participate in a variety of homeostatic and pathologic processes primarily through their ability to store and rapidly release a plethora of mediators (1, 2). Eosinophils develop in the bone marrow from an eosinophil lineage–committed progenitor (EoP) that expresses CD34, the receptor for IL-5 (IL-5Rα/CD125), and the transcription factor (TF) GATA-1 (3, 4). In mice, EoPs are derived from the granulocyte-monocyte progenitor (GMP) following eosinophil-lineage commitment (3). Several factors necessitate that the production of mature eosinophils be a tightly regulated process: eosinophils are normally less than five percent of leukocytes in the blood, are incapable of proliferation, and have high rates of spontaneous apoptosis (5, 6), yet need to be regularly replenished during homeostasis in tissues such as the gastrointestinal tract. Murine studies with genetically altered animals provide a wealth of evidence supporting not only a critical role for IL-5 in mediating disease-associated eosinophilia, but also, unexpectedly, that IL-5 is not required for baseline eosinophil production (7–12). The regulatory molecules or networks necessary for optimal homeostatic eosinophil production remain largely unknown.
Gene expression and chromatin analyses in the eosinophil lineage have been limited, likely due to the rarity of the cell populations and the challenges in isolating naïve primary EoPs and eosinophils at sufficient purities and quantities. Eosinophil- and IL-5–dependent changes in gene expression have been reported in experimental models of asthma (13) and infection (14), as well as in ex vivo liquid cultures (15). However, global gene expression analysis or chromatin mapping at specific developmental stages in homeostatic eosinophil production has not been reported. In this study, we took a global approach to identify genome-wide transcriptome and epigenome changes that occur at critical stages in eosinophil development—eosinophil-lineage commitment (EoP compared to GMP) and eosinophil maturation (eosinophil compared to EoP). We performed chromatin immunoprecipitation coupled with massively parallel sequencing (ChIP-seq) and RNA sequencing (RNA-seq) to identify TFs and genetic regulatory elements that are active during homeostatic eosinophil production in the bone marrow. Our analyses revealed that markedly more transcriptome alterations were associated with eosinophil maturation (1199 genes) than eosinophil-lineage commitment (490 genes), highlighting the greater transcriptional investment necessary for differentiation. In addition, we noted high expression levels of granule proteins in EoPs and organized granules with an ultrastructure distinct from that of mature eosinophils. We also report that Siglec-F is expressed by EoPs and, further, can be used as a surface marker for identifying progenitors with eosinophil-lineage potential in murine bone marrow. A 976-gene eosinophil-lineage transcriptome was delineated that included 56 TFs, including the unexpected expression of Helios and Aiolos, members of the Ikaros family of TFs. Epigenomic studies revealed that genes that were specifically induced with eosinophil-lineage commitment in EoPs were “poised” with active chromatin marks in GMPs, despite not being expressed in GMPs. In contrast, the majority of the genes that were highly and specifically induced with maturation in eosinophils was not associated with poised chromatin, suggesting distinct epigenetic regulation for genes induced with lineage commitment compared to genes induced with cell maturation during eosinophil development. We further report significant enrichment of potential binding sites for Helios and Aiolos in the genes expressed by EoPs and eosinophils with active chromatin, supporting a critical role for these TFs in regulating gene expression during eosinophil development.
Collectively, our study reveals that the dynamic changes in gene expression associated with eosinophil development include novel transcriptional regulators, such as Helios and Aiolos, and distinct epigenetic profiles between EoPs and eosinophils. Comprehensive epigenomic and transcriptomic profiling during critical stages in eosinophil development will ultimately define the programming and gene regulatory networks necessary for eosinophil development and will likely lead to novel therapeutic strategies to regulate eosinophil production. In addition, our study highlights that the regulatory mechanisms that direct eosinophil homeostasis are likely to be developmental-stage (EoP vs. eosinophil) specific. These findings have implications for a number of diseases, including allergic and eosinophilia-associated disorders, in which these processes may be manipulated for therapeutic benefit.
Materials and Methods
Mice
BALB/c wild-type mice were analyzed at 6 to 8 weeks of age. All mice were housed under specific pathogen-free conditions and handled under approved protocols (#2E09072) of the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center (CCHMC).
Cell isolation and identification
Bone marrow cells were isolated as previously described (16). The cells were then washed twice in 1X PBS, counted and resuspended at 1–2 × 107 cells per mL in 1X PBS/0.1% BSA. Goat anti-rat IgG magnetic beads (New England BioLabs, Inc.) were prepared according to the manufacturer’s instructions and coated with anti-mouse B220 (clone RA3.3A1/6.1), CD19 (clone 1D3), or Ly6-G (clone 1A8) (BioXCell, 1 μL of beads per μg of antibody). The beads were added to the cell suspension at a 5:1 ratio and incubated at least 1 h at 4°C with gentle agitation on a rotating plate. The cell suspension was placed on an iMag rack (BD Bioscience) for 10 min, and the negative fraction was collected, filtered through a 70-μm cell strainer, and washed twice in 1X PBS. Cells were stained for 15 min at room temperature in FACS buffer (1X PBS, 0.5% FBS, and 2 mM EDTA). To sort GMPs and EoPs, the cells were stained as previously described (16), except for the omission of Fc block prior to staining and the addition of PE-Cy7 anti-mouse CD16/CD32 (clone 93, Biolegend) and Zombie NIR (fixable viability stain, Biolegend®) to the staining panel. FITC anti-mouse CCR3 (clone 83101, R&D), PE anti-mouse Siglec-F (clone E50-2440, BD Biosciences), and Zombie NIR were used to sort eosinophils. Cell sorting was performed on a FACS-Aria II (BD Biosciences) maintained by the Research Flow Cytometry Core at CCHMC.
Progenitor analyses
For colony forming unit (CFU) assays, 1,000 sorted GMPs or EoPs were diluted in 0.4 mL culture media with or without IL-5 (50 ng/mL) and were added to a 3.6 mL of Methocult media (Stem Cell Technologies, #3434). The culture plates were then prepared according to the manufacturer’s instructions (1 mL media with cells per 35-mm plate with 3 plates per condition). After 9 days of incubation, colony types (CFU-G [granulocytes], CFU-GM [granulocytes and monocytes] and CFU-M [monocytes]) were counted on each plate on the basis of morphology. In addition, single colonies were picked, diluted in 0.15 mL of 1X PBS with 0.5% BSA and then cytospun at 500 rpm for 5 min. The cytospin slides were stained Differential Quik Modified Giemsa (Electron Microscopy Sciences). The colonies from 1 plate from each group were pooled and prepared for flow cytometry. The media was dissolved in 10 mL of 1X PBS, centrifuged at 1200 rpm for 5 min, and washed again with 1X PBS. The cells were then resuspended in 0.1 mL of FACS buffer and stained with Siglec-F-PE and Zombie near IR-Live/Dead. The data were acquired on a BD Fortessa cell cytometer and analyzed with FlowJo software.
Cell morphology
Morphology of eosinophils, EoPs, and GMPs was evaluated by staining cytospins (5 min at 500 rpm) with Differential Quik Modified Giemsa or DAB substrate (Vector Labs) for eosinophil peroxidase activity and counterstaining with Mayer’s haematoxylin (Sigma Aldrich) following manufacturer instructions. For MBP staining, cells were fixed for 5 min in 1X PBS/2% paraformaldehyde, permeabilized with 1X PBS/0.1% BSA/0.01% Triton X-100 for 15 min at room temperature, and then stained with rat anti-mouse MBP (a generous gift of Dr. J.J. Lee, Mayo Clinic, Arizona) at a 1:1000 dilution in permeabilization buffer overnight at 4°C. Cells were washed three times in permeabilization buffer and stained with goat anti-mouse Alexa Fluor 594 (Molecular Probes) at a 1:2000 dilution in permeabilization buffer for 1 h at room temperature. Cells were washed and then counterstained and mounted with DAPI Fluormount G (Southern Biotech). Imaging was performed with an Olympus BX51 microscope at 400X or with an Olympus DP72 digital camera at 1000X (oil immersion) magnification.
Transmission electron microscopy
Sorted EoPs were washed in 1X PBS then fixed in 2.5% glutaraldehyde/0.1M sodium cacodylate (Electron Microscopy Sciences) overnight at 4°C. Post-fixation was performed sequentially in 0.5% osmium tetroxide, 0.8% potassium ferricyanide, 1% tannic acid, and 1% uranyl acetate. Cells were then dehydrated through an ethanol series and embedded in LX-112 resin (Ladd Research Industries). Thin sections were prepared with a Leica EM UC7 ultramicrotome, attached to 200-mesh copper grids (Electron Microscopy Sciences), and stained with 1% uranyl acetate and Reynold lead citrate. Cells were viewed on a Hitachi H-7650 electron microscope at 80 kV using a AMT-600 camera and image capture engine software (Advanced Microscopy Techniques) maintained by the CCHMC Pathology Core Facility.
Gene expression analysis
Total RNA from GMPs, EoPs, and eosinophils sorted from the pooled bone marrow of BALB/c wild-type mice was collected in TriSure reagent (Bioline) and isolated with the Direct-zolTM RNA MiniPrep (Zymo Research) according to the manufacturer’s protocol. RNA-seq libraries were prepared from two independent sorts of each cell type (GMPs, EoPs, or eosinophils) using an unstranded Illumina protocol and sent for sequencing on an Illumina HiSeq 2500 sequencing system at the CCHMC sequencing core facility, resulting in ~10 million reads per sample. The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (17) and are accessible through GEO Series accession number GSE69707 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69707). RNA-seq data analysis was performed using the BioWardrobe Experiment Management System (https://biowardrobe.com/) (18). Briefly, reads were mapped to the mm10 genome and RefSeq-based transcriptome using RNA-STAR (v. 2.4.0c) (19) and assigned to the RefSeq genes using the Wardrobe algorithm. A minimum reads per kilobase per million mapped reads (RPKM) equivalent to 10 was deemed the lower limit of expression on the basis of the genes known to be expressed by EoPs and eosinophils. Differentially expressed genes with DESEQ (v. 1) (20) P value < 0.05 and RPKM > 1 in at least one time point were analyzed. Results were interpreted in the context of biological processes and molecular functions, as well as pathways, through the use of ToppGene Suite (21) for gene list functional enrichment. Heat maps of differentially expressed genes were generated using Genespring version 12.6.1 and BioWardrobe. For independent confirmation of gene expression, cDNA was synthesized from prepared RNA with iScript (BioRad, Hercules CA) using 0.5 μg of RNA template according to manufacturer instructions. Completed cDNA was diluted 1:5 with nuclease-free water, and 5 ng (2 μL) was used as template for quantitative PCR on an ABI7900HT thermal cycler (Life Technologies, Grand Island NY) with FastStart Universal SYBR Green Master + Rox (Roche, Indianapolis IN) and oligonucelotides (IDT, Coralville IA) at 100 nM in a reaction volume of 10 μL. Data were analyzed for fold expression over Gapdh (2−dCt). The following primers were used for quantitative PCR: Ikzf3 (Aiolos, 5′-CTGAATGACTACAGCTTGCCC, 5′-GCTCCGGCTTCATAATGTTCT), Sfpi1 (PU.1, 5′-ATGTAGGAAACCTGGTGACTG, 5′-TTCCCTGAGAACCACTTCAC), Epx (EPX, 5′-ATGGAGACAGATTCTGGTGG, 5′-CCAGTATTGTCGCATACAATCC), and Gapdh (5′-CTGGTATGACAATGAATACGG, 5′-GCAGCGAACTTTATTGATGG).
ChIP-sequencing
Chromatin was cross-linked with the addition of one-tenth of 10X buffer [containing 0.1M NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM Hepes, and 8.8% formaldehyde] to suspensions of sorted GMPs, EoPs, or eosinophils (one independent sort per cell type) and incubated for 8 min on ice. The reaction was stopped with the addition of glycine and incubated at room temperature for 5 min. The cell suspensions were centrifuged for 5 min at 2000 x g at 4°C, and the pellets were washed twice in cold 1X PBS. Pellets were then stored at −80°C. Prior to sonication, the pellets were thawed and resuspended in Tris-EDTA containing 0.1% SDS and proteinase inhibitors. Sonication was performed in a Covaris sonicator for 3 min. A fraction of the chromatin suspension was used to check the quality of the sonication, and glycerol was added to the remaining and stored at −80°C. The chromatin was then pre-cleared via incubation with magnetic beads (Dynal) prior to performing ChIP with Histone H3 lysine 4 trimethyl (H3K4me3) antibody (Millipore) overnight in the SX-8G IP-Star ® ChIP Robot (Diagenode). ChIP-seq data analysis was performed using the BioWardrobe Experiment Management System (https://biowardrobe.com/) (18). Briefly, reads were mapped to the mm10 genome using Bowtie, and islands were detected using MACS. The areas of differential enrichment were identified using MAnorm. The ChIP-seq data have been deposited in NCBI’s Gene Expression Omnibus (17) and are accessible through GEO Series accession number GSE69707 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69707).
TF binding site analysis
TF binding motif enrichment analysis in H3K4me3-marked regions located near genes in our gene sets (e.g., genes expressed specifically in EoPs or eosinophils only) was performed using the HOMER software package with default settings (22). Briefly, HOMER uses a library of TF binding models (in the form of position weight matrices) to scan a set of input sequences for statistical enrichment of each position weight matrix. We supplemented the HOMER motif library with ChIP-seq–derived motifs for Ikaros family TFs (23), as the HOMER library does not include motifs for Aiolos or Helios and these TFs were highly expressed in our RNA-seq data. In addition to the HOMER motif enrichment analysis, we also identified putative Aiolos binding sites in H3K4me3-marked sequences by scanning for matches to the published ChIP-seq–derived motif (23). We scored all sequences using the standard log-likelihood scoring system (24) and a threshold corresponding to 80% of the maximum possible score of the motif.
Results
Gene expression associated with eosinophil-lineage commitment
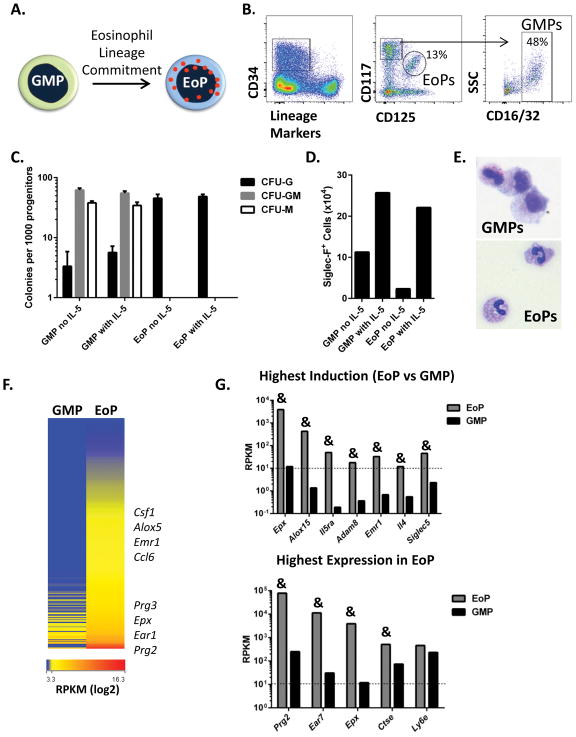
To assess genome-wide changes in gene expression following commitment to the eosinophil lineage (Figure 1A), GMPs and EoPs were identified via flow cytometry based on surface marker expression in naïve murine bone marrow (Figure 1B), sorted, and then subjected to colony-forming assays to confirm lineage potential and to RNA-seq analysis. In GMP and EoP cultures, eosinophil-containing colonies (CFU-G) were observed in the absence and presence of IL-5 (Figure 1C). Progeny of both GMPs and EoPs expressed Siglec-F (Figure 1D) and had cell morphology consistent with eosinophils (Figure 1E). Together, these data indicate that a fraction of GMPs from BALB/c bone marrow have eosinophil-lineage potential, as previously shown in C57BL6 mice (3), and that the frequency of eosinophils increases with IL-5 stimulation. We next investigated the alterations in gene expression that are related to eosinophil development. Eosinophil-lineage commitment was associated with 386 genes that were significantly upregulated (Figure 1F) and with 104 genes that were significantly downregulated in EoPs compared to GMPs (Supplemental Figure S1A–B). The genes with the highest induction in EoPs compared to GMPs (Figure 1G) included known eosinophil-associated genes such as eosinophil peroxidase (Epx), IL-5Rα (Il5ra), IL-4 (Il4), and Siglec-F (Siglec5). The genes with the highest transcript levels in EoPs included genes encoding granule-associated proteins, such as major basic protein (MBP, Prg2) and eosinophil peroxidase (Epx) (Figure 1G). Notably, transcripts for several of these highly expressed genes were also expressed by GMPs (e.g. Ctse and Ly6e), but at lower levels (Figure 1G).
Figure 1. EoPs express high mRNA levels of granule proteins.
(A) Eosinophil-lineage commitment schematic is shown. (B) A representative gating strategy to delineate EoPs and GMPs in whole bone marrow for FACS is shown and starts with pre-gating on viable single cells and then further gating on cells that express CD34, are negative for lineage markers and Sca-1, and co-express either CD117int (c-Kit) and CD125 (IL-5Rα) for EoPs or CD117hi and CD16/32 for GMPs. Percentage of cells in the parent gate is shown. (C) Colony-forming assay from sorted GMPs and EoPs cultured with and without IL-5 is shown (data from a representative of 2 independent experiments is shown with 3 plates per condition). (D) Total number of Siglec-F+ cells in colonies on a single plate from GMPs or EoPs cultured with or without IL-5 is shown (data from a representative of 2 independent experiments is shown). (E) Cytospins from colonies derived from GMPs and EoPs are shown (magnification 1000X). (F) Heat map showing mRNA expression levels for genes that were significantly induced 2-fold or more (P < 0.05) in EoPs compared to GMPs. Expression levels (mean RPKM) for genes that are highly induced in EoPs compared to GMPs (G, upper panel) and for genes that are the most highly expressed in EoPs (G, lower panel) are shown. Minimal expression level was mean RPKM equal to 10 and is indicated with a dashed line. &P < 1e-5.
EoPs express granule proteins and Siglec-F
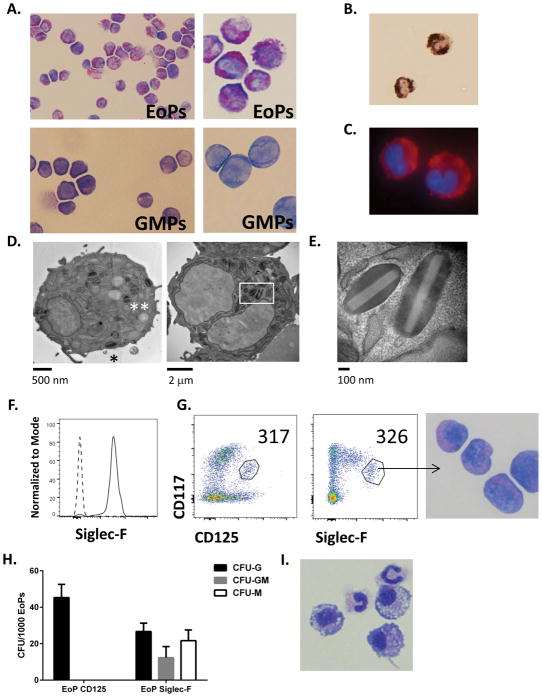
With the unexpected finding of markedly high transcript levels for granule proteins in EoPs, we further investigated EoP morphology and cellular contents. Staining of sorted EoPs revealed significant granularity compared to GMPs (Figure 2A) with high levels of eosinophil peroxidase activity and MBP protein expression in the cytoplasm (Figure 2B–C). Ultrastructural morphology of the granules within EoPs revealed specific granules with a variety of phenotypes (Figure 2D). Some specific granules contained an electron dense core similar to that seen in a resting eosinophil (Figure 2D, asterisk), whereas others had irregular electron-dense regions and clearing of the central matrix (Figures 2D–E), indicating that the specific granules within EoPs are not uniform. As our transcriptome analyses revealed expression of Siglec-F (Siglec5) by EoPs (Figure 1G), we confirmed surface expression of Siglec-F on EoPs by flow cytometry (Figure 2F). Notably, Siglec-F expression was equivalent to IL-5Rα (CD125) expression in identifying EoPs within the CD34-expressing progenitors (Lin−CD34+CD117int) population (Figure 2G). To determine whether these two Siglec-F–expressing populations had similar eosinophil potential, we sorted the two populations, EoP-CD125 (Lin−CD34+CD117intCD125+) and EoP-Siglec-F (Lin−CD34+CD117intSiglec-F+), and subjected the cells to colony-forming assays. Notably, the CD125-expressing EoPs gave rise only to eosinophil-containing colonies, whereas the EoP-Siglec-F cultures yielded colonies containing eosinophils and monocytes (Figures 2H–I), suggesting that Siglec-F is a marker of eosinophil and/or monocyte potential in murine bone marrow progenitors whereas CD125 expression represents eosinophil-lineage commitment.
Figure 2. EoPs express granule proteins and Siglec-F.
(A) Cytospins of isolated EoPs and GMPs are shown (400X magnification on left, 1000X magnification on right). (B) Eosinophil peroxidase activity (brown-black precipitate) in isolated EoPs is shown (400X). (C) Anti-MBP staining (red) in isolated EoPs is shown with nuclei stained with DAPI (1000X). Results shown in panels A–C are representative of 3 experiments. (D) Electron micrographs showing scattered secondary granules with electron-dense core (asterisk) as seen in mature eosinophils, secondary granules with electron dense matrix on one side (double asterisk), and secondary granules with an electron-lucent core (box) are shown. (E) Higher magnification image of secondary granules in EoP with electron-lucent core and electron-dense matrix is shown. (F) Surface expression (solid line) of Siglec-F by EoPs in naïve murine bone marrow compared to isotype control (dashed line) is shown. Pre-gating included live, single cells that were lineage-marker negative but expressed CD34, CD125, and CD117. Results are representative of 3 experiments. (G) EoP identification via surface expression of CD117 and CD125 compared to CD117 and Siglec-F by CD34-expressing, lineage-marker negative bone marrow cells is shown with cell counts within each gate in the upper right corner of the dot blot. Cytospin of EoPs sorted via Siglec-F expression is shown. Results are representative of 2 experiments. (H) Colony-forming assay from EoPs sorted via surface expression of CD125 and Siglec-F is shown (data from a representative of 2 independent experiments is shown with 3 plates per condition). (I) Cytospins from colony-forming assay are shown (magnification 1000X).
Gene expression associated with eosinophil differentiation
To assess genome-wide changes in gene expression that are associated with eosinophil maturation (Figure 3A), eosinophils were identified via flow cytometry based on surface marker expression in naïve murine bone marrow (Figure 3B). We used co-expression of CCR3 and Siglec-F surface expression as a marker of mature eosinophils, as CCR3−Siglec-F+ cells had the morphology of immature eosinophils (Supplemental Figure S1C). Mature eosinophils (Siglec-F+CCR3+) were sorted and then subjected to RNA-seq analysis. Eosinophil maturation was associated with a significant reduction in the expression of 504 genes (eosinophils vs. EoPs, Figure 3C). Notably, transcript levels for genes that encode for granule proteins were significantly lower in eosinophils than EoPs (Figure 3D). Eosinophil maturation was also associated with significant elevation in expression of 695 genes (eosinophils vs. EoPs, Figure 3E). The genes with the greatest induction in mature eosinophils compared to EoPs (Figure 3F) included genes not previously associated with eosinophils, including resistin-like molecule gamma (Retnlg) and ADAM 19 (Adam19), as well as genes encoding proteins important for eosinophil effector functions, such as CCR3 (Ccr3) and the matrix metalloproteinases MMP-8 (Mmp8) and MMP-9 (Mmp9). A majority of the genes with the highest transcript levels in eosinophils were also expressed in EoPs (Figure 3G), including MBP (Prg2), arachidonate 15-lipoxygenase (Alox15), myeloperoxidase (Mpo) and the chemokine CCL6 (Ccl6), suggesting early and sustained expression of a subset of genes associated with effector functions during eosinophil development.
Figure 3. Granule protein transcripts are higher in EoPs than eosinophils.
(A) Eosinophil maturation schematic is shown. (B) A representative gating strategy to identify mature eosinophils from naïve murine bone marrow is shown and starts with pre-gating on viable single cells and then further gating on cells that co-express Siglec-F and CCR3. Percentage of parent gate is shown. (C) Heat map showing transcript levels of genes that were significantly reduced 2-fold or more (P < 0.05) in eosinophils (Eos) compared to EoPs is shown. (D) Expression level (mean RPKM) of genes that were significantly reduced in eosinophils compared to EoPs. &P < 1e-5. (E) Heat map showing transcript levels of genes that were significantly induced 2 fold or more (P <0.05) in eosinophils (Eos) compared to EoPs is shown. Expression level (mean RPKM) of genes with the greatest relative induction in eosinophils compared to EoPs (F) or the highest expression (G) in eosinophils (Eos) is shown. &P < 1e-5, *P < 5e-2.
Identification of eosinophil-lineage transcriptome
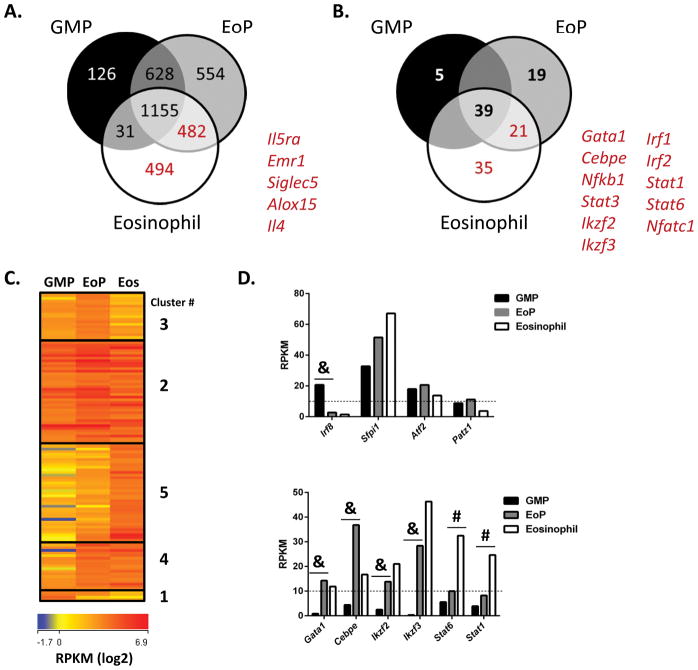
We next compared gene expression between GMPs, EoPs, and eosinophils and noted that the largest grouping of genes were genes that were expressed by all three cell types (Figure 4A). Of the expressed genes that were not common to all three cell types, EoPs shared a significant proportion of this non-communal transcriptome subset with GMPs (628/1664, 38%) and mature eosinophils (482/1664, 29%), which is consistent with a cell early in lineage commitment and differentiation (Figure 4A). Eosinophils shared a much greater proportion of their non-communal transcriptome with EoPs (482/1007, 48%) than GMPs (31/1007, 3%) (Figure 4A). We identified an eosinophil-lineage transcriptome comprising 976 genes that were expressed by eosinophils, but not GMPs (Supplemental Table S1), which included Il5ra, Emr1, Siglec5, Alox15, and Il4 (Figure 4A, numbers in red font). Of note, 49% (482/976) of the eosinophil-lineage transcriptome was also expressed by EoPs (Figure 4A). To identify potential regulators of the eosinophil-lineage transcriptome, we next compared expression of TFs between GMPs, EoPs, and eosinophils. We identified 119 TFs that were expressed by GMPs, EoPs, or eosinophils (Figure 4B, Supplemental Table S2). One-third (39/119) of the TFs were expressed by all three cell types, whereas there was cell-specific expression of 5, 19, and 35 TFs in GMPs, EoPs, and eosinophils, respectively (Figure 4B), suggesting a more diverse repertoire of TF expression is necessary to regulate eosinophil maturation. We identified a subset of 56 TFs that were included in the eosinophil-lineage transcriptome, including Gata1, Cebpe, and Stat3 (Figure 4B). A number of these TFs have not been previously associated with eosinophils, including Helios (Ikzf2) and Aiolos (Ikzf3), members of the Ikaros family of zinc finger TFs. We next looked at the differential expression levels of the 119 TFs expressed by GMPs, EoPs, and eosinophils and identified 5 clusters based on expression pattern (Figure 4C). Cluster 1 included the TFs that were expressed only by GMPs (e.g. Irf8, Figure 4D). Cluster 2 included TFs that were expressed by all three cell types, such as PU.1 (Sfpi1) and ATF2 (Atf2, Figure 4D). Cluster 3 included TFs with higher levels of expression in GMPs and EoPs than in eosinophils (e.g. Patz1, Figure 4D). Cluster 4 included TFs that were expressed by EoPs and eosinophils but not by GMPs (e.g. Gata1, Cebpe, Ikzf2, and Ikzf3, Figure 4D), and Cluster 5 included TFs that were expressed by eosinophils but not by EoPs or GMPs (e.g. Stat6 and Stat1, Figure 4D).
Figure 4. Identification of eosinophil-lineage transcriptome and repertoire of TFs.
(A) Venn diagram with overlap of gene expression between GMPs, EoPs, and eosinophils is shown. The number of genes, and select gene names, included in the eosinophil-lineage transcriptome (EoP and eosinophil shared expression and eosinophil-only expression) appear in red. (B) Venn diagram with overlap of TF expression between GMPs, EoPs, and eosinophils is shown. The number of TF genes, and select TF gene names, included in the eosinophil-lineage transcriptome appear in red. (C) Heap map showing differential TF expression between GMPs, EoPs, and eosinophils (Eos) is shown. Clusters based on differential expression pattern are delineated with boxes and numbered on the right. (D) Expression level (mean RPKM) of TFs in isolated GMPs, EoPs, and eosinophils is shown with lower limit of expression (RPKM equal to 10) depicted with a dashed line. &P < 1e-5, #P < 1e-4.
Chromatin modifications during eosinophil development
To learn more about the regulatory networks controlling differential gene expression during eosinophil development, we next investigated the distribution of a chromatin modification (H3K4me3) associated with active promoters and strong enhancers (25–27). Chromatin marked with H3K4me3 was associated with gene induction, and the marks clustered around the transcriptional start sites (TSSs) of the expressed genes in GMPs, EoPs, and eosinophils (Figure 5A). The majority of the H3K4me3 islands at the TSSs were present in both EoPs and GMPs and in EoPs and eosinophils (Figure 5B), highlighting that EoPs share gene regulatory mechanisms with both GMPs and eosinophils. More than 75% of the genes expressed by EoPs or eosinophils contained H3K4me3 marks in their promoters (Figure 5C), supporting a correlation between the presence of this mark and promoter activity. We next compared H3K4me3 marks between expressed genes (RPKM ≥ 10) and silent genes (RPKM < 2). Nearly all of the expressed genes in EoPs and eosinophils were marked with H3K4me3 (Figure 5D). In contrast, a vast majority of the silent genes in either EoPs or eosinophils was unmarked, but a small proportion of the silent genes had H3K4me3 at their TSS (Figure 5D). Of the silent genes that were marked with H3K4me3 in EoPs, 53 genes were expressed in eosinophils, including those encoding ST2 (Il1rl1), β7 integrin (Itgb7), and PAF receptor (Ptafr), suggesting that a subset of genes associated with eosinophil effector functions may be poised for expression as early as lineage commitment. Gene ontology analysis of the silent genes that were H3K4me3 marked in eosinophils showed that this gene set was enriched for cell cycle genes; thirteen of these genes were expressed in GMPs, and 23 of these genes were expressed in EoPs, suggesting that these genes were marked early in development and remained marked through differentiation despite downregulation of expression in the mature eosinophil.
Figure 5. Differential epigenetic regulation during eosinophil development.
(A) Heat maps of mRNA levels and distribution of H3K4me3 around transcriptional start sites (TSSs) in isolated GMPs, EoPs, and eosinophils are shown with each horizontal line being the same single gene across the heat and distribution maps. (B) Pie charts representing the presence of H3K4me3 marks in gene promoter regions (+/− 1000 bp from TSS) that are in common (black) or unique for GMPs (green), EoPs (blue), or eosinophils (orange) are shown, with the upper pie chart representing a comparison between GMPs and EoPs and the lower pie chart representing a comparison between EoPs and eosinophils. (C) Pie charts representing the proportion of expressed genes (RPKM ≥ 10) that contain at least one H3K4me3 mark in the promoter region in EoPs (left chart) or eosinophils (right chart) are shown. (D) Heat maps of mRNA levels and H3K4me3 distribution relative to TSS for genes that are expressed (RPKM ≥ 10) or silent (RPKM ≤ 2) in EoPs or eosinophils are shown. (E) Heat maps of mRNA levels and H3K4me3 distribution relative to TSS for genes that are significantly induced 2 fold or more (P ≤ 1e-2) in the EoPs compared to GMPs or in eosinophils (Eos) compared to EoPs are shown.
We next investigated whether the genes that were specifically induced with eosinophil-lineage commitment (i.e. genes expressed in EoPs but not in GMPs) were already marked (or poised) for expression in GMPs. A vast majority of the genes that are expressed in EoPs but not in GMPs do contain H3K4me3 marks in GMPs (Figure 5E), suggesting that they are poised for induction prior to lineage commitment. We next examined whether the genes that were specifically induced with eosinophil maturation (i.e. genes expressed in eosinophils but not in EoPs) were already marked in EoPs. Notably, a majority (87%, 392/451, P = e-92 hypergeometric test) of the genes expressed in eosinophils but silent in EoPs were not H3K4me3 modified (or poised) in EoPs (Figure 5E). These genes included metalloendopeptidases (Mmp25, Adam19, Mmp8, Mmp9) and complement C3a receptor (C3ar1), highlighting that during eosinophil maturation, these genes undergo an additional regulatory step (i.e. chromatin modification) for upregulation of expression.
TF binding sites in H3K4me3-marked promoters
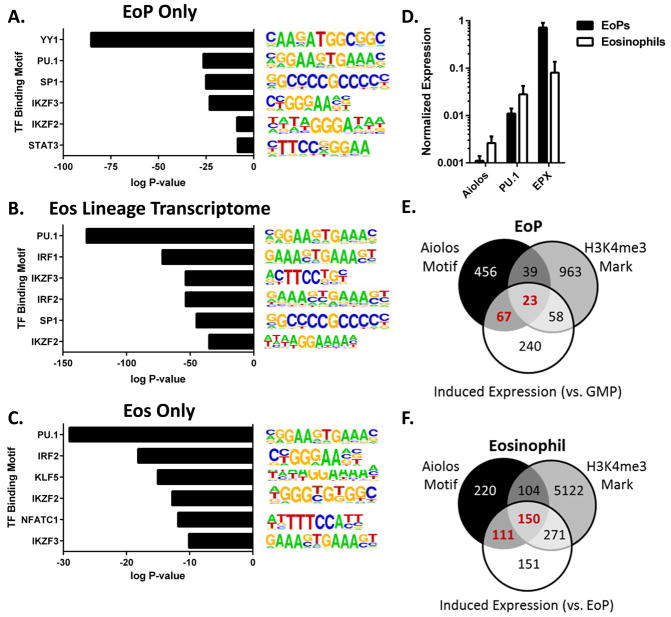
To identify potential novel regulators of eosinophil-lineage commitment and differentiation, we analyzed the sequences marked with H3K4me3 for enrichment of TF binding sites within and near genes expressed by EoPs and/or eosinophils. Notably, potential binding sites for Helios (IKZF2) and Aiolos (IKZF3) were significantly enriched in all 3 subsets of genes analyzed: 1) genes only expressed by EoPs (and not by GMPs or eosinophils, Figure 6A), 2) genes included in the eosinophil-lineage transcriptome (expressed by EoPs and eosinophils but not GMPs, Figure 6B), and 3) genes only expressed by eosinophils (and not by GMPs or EoPs, Figure 6C), supporting a potential regulatory role for Helios and Aiolos in eosinophil development. In genes expressed by EoPs, but not eosinophils or GMPs, potential binding sites for YY1 and STAT3, TFs expressed by both EoPs and eosinophils, were significantly enriched in the sequences marked by H3K4me3 (Figure 6A). In the eosinophil-lineage transcriptome, potential binding sites for IRF1 and IRF2, TFs expressed by eosinophils (and not by GMPs or EoPs), were significantly enriched in the sequences marked by H3K4me3, suggesting a novel role for these TFs in eosinophil maturation (Figure 6B). In the genes expressed by only eosinophils, binding sites for KLF5 and NFATc1 were significantly enriched in the H3K4me3-marked sequences, highlighting potential novel roles for these TFs in regulating gene expression during eosinophil maturation (Figure 6C).
Figure 6. Enrichment of Helios and Aiolos binding motifs in eosinophil-lineage transcriptome.
Highly represented TF motifs (HOMER motif enrichment algorithm) in H3K4me3-marked sequences in genes expressed by EoPs only (not by GMPs or eosinophils) (A), in the eosinophil (Eos)-lineage transcriptome (EoPs and eosinophils but not GMPs) (B), and by eosinophils only (not by GMPs or EoPs) (C) are shown with log-transformed P values on the x-axis. (D) Normalized expression of Aiolos, PU.1, and EPX in EoPs and eosinophils as determined by quantitative PCR is shown (n = 2 independent experiments with 2–6 samples per cell type). Shown are Venn diagrams representing genes with significantly induced expression (white circle, P < 0.01) in EoPs (compared to GMPs) (E) or in eosinophils (compared to EoPs) (F) and whether they contain Aiolos binding motifs (Aiolos Motif) and/or H3K4me3 peaks that are significantly elevated (P < 0.01, H3K4me3 Mark). The number of genes with Aiolos binding motifs and induced expression is in red font.
With the unexpected expression of Helios and Aiolos by EoPs and eosinophils in our RNA-seq data and significant over-representation of potential Helios and Aiolos binding sites in genes expressed by EoPs and/or eosinophils, we further investigated a role for Aiolos in eosinophil production. We confirmed elevated Helios and/or Aiolos expression in mature murine eosinophils compared to neutrophils in data sets assembled by the ImmGen consortium (Supplemental Figure S1D) (28), as well as Aiolos expression in sorted EoPs and eosinophils from independent experiments (Figure 6D). A search of the GEO Profiles database (29) revealed Helios and Aiolos expression in human eosinophils as well (Supplemental Figure S1E; GEO accession GSE28492) (30). Collectively, these data demonstrate conserved Helios and Aiolos expression in the eosinophil lineage in mice and humans.
To further investigate the potential for Aiolos as a positive regulator of gene expression during eosinophil development, we next investigated whether the genes with Aiolos binding sites also contained significantly elevated H3K4me3 peaks (2 fold or more, P < 0.01) and were significantly induced (2 fold or more, P < 0.05) during eosinophil development. During eosinophil commitment (GMP to EoP), 23% (90/388) of the genes with significantly elevated expression in the EoP compared to GMP had Aiolos binding motifs in the gene (P = 0.04, hypergeometric test), and 26% (23/90) of those genes also had significantly higher H3K4me3 peaks (Figure 6D and Supplemental Table S3). Notably, 15 of the induced genes with Aiolos binding motifs are associated with cytoplasmic membrane–bound vesicles (P = 1.9 × 10−6), including SNAP23, a protein involved in exocytosis in human eosinophils (31). During eosinophil differentiation (EoP to eosinophil), 38% (261/683) of the genes with significantly elevated expression in eosinophils compared to EoPs contained Aiolos binding motifs (P = 4.4e-15, hypergeometric test), and 57% (150/261) of those genes also contained elevated H3K4me3 modifications (Figure 6E and Supplemental Table S3). More than 30 of the induced genes that contained both Aiolos binding motifs and elevated H3K4me3 modification are associated with cell migration, including integrins (Itgam, Itgb2, Itgb3, Itgb7) and CCR1, suggesting that Aiolos may regulate a subset of genes important for eosinophil trafficking. Collectively, these data highlight that Aiolos has the potential to regulate a significant portion of the genes that are induced during eosinophil-lineage commitment and eosinophil maturation.
Discussion
Eosinophils are terminally differentiated cells with a high turnover rate in the circulation and a tissue lifespan of less than a week (32–35). In addition, mature eosinophils do not have the capability to divide and have high rates of spontaneous or intrinsic apoptosis (36), highlighting the need for highly regulated (and perpetual) homeostatic production of new eosinophils. Even though IL-5 has been shown experimentally to be essential for disease-associated eosinophilia (10, 37–39), homeostatic eosinophil production is independent of IL-5, as eosinophil levels are just modestly reduced in IL-5–deficient mice (10). In contrast, regulatory mechanisms involving G-CSF are responsible for both steady-state neutrophil production and disease-associated neutrophilia (40). The regulatory mechanisms that direct homeostatic eosinophilopoiesis are largely unknown. As changes in gene expression are tightly associated with hematopoiesis (22, 41, 42), we took a global approach combining genome-wide transcriptomic and epigenomic analyses at critical stages in homeostatic eosinophil development to delineate the changes in gene expression and characterize the promoters, or genetic regions surrounding the TSS, that drive these transcriptome alterations. Our aim was to begin to molecularly define the regulatory program associated with eosinophil-lineage commitment and eosinophil maturation. Eosinophil maturation from EoPs was associated with markedly greater number of transcriptome alterations, including a larger repertoire of TFs, than eosinophil-lineage commitment. In addition, we reveal distinct epigenetic regulation of the promoters of genes associated with lineage commitment compared to eosinophil maturation. Our study highlights that the regulatory mechanisms responsible for eosinophil homeostasis are likely to be developmental-stage specific. These findings have implications for a number of diseases, including allergic disorders, in which these processes may be manipulated for therapeutic benefit.
In this study, we describe several new EoP characteristics, including surface Siglec-F expression. Notably, our data suggest that Siglec-F expression is a surface marker for bone marrow progenitors with eosinophil-lineage potential, whereas CD125 expression is associated with eosinophil-lineage commitment. Siglec-F is a functional paralog to Siglec-8, a marker for maturing and mature human eosinophils (43, 44). Siglec-F has been shown to be a useful marker to identify mature eosinophils in the bone marrow and spleen of mice (45), and now our data suggest that anti–Siglec-F can be used in the bone marrow, along with antibodies directed against CD34 and c-KIT, to identify cells with eosinophil-lineage potential. EoPs are identified in murine bone marrow via surface expression of CD125 (IL-5Rα) (3). As surface expression of IL-5Rα on eosinophils is substantially influenced by the microenvironment (46–48), Siglec-F may ultimately prove to be a more advantageous marker to identify these rare cells. We also demonstrate abundant expression of granule proteins, especially MBP, by EoPs. As similarly noted in earlier studies (49), the transcript levels for the granule proteins MBP and EPO were higher in the progenitors than in the mature eosinophils, highlighting that synthesis of granule contents occurs early in eosinophil development. Indeed, early synthesis of these granule proteins is critical for normal eosinophilopoiesis, as deficiency in both Mbp and Epx results in impaired eosinophil production (50). We also observed that granules in EoPs exhibit an ultrastructure morphology that differs from that of granules in mature eosinophils. Though the majority of specific granules in mature eosinophils contain an internal, electron-dense crystalline core surrounded by an electron-lucent matrix (12, 51), the granule morphology in EoPs varied, with some specific EoP granules having an electron-lucent core with an electron-dense matrix. Interestingly, fading or clearing of the central core as a result of loss of contents with degranulation has been observed in activated eosinophils (52–54). We observed this seeming reversal of electron density of core and matrix in granules in EoPs sorted from naïve murine bone marrow that also demonstrated high levels of MBP and EPO in their cytoplasm as determined by immunohistochemistry and enzyme activity (Figure 2). Thus, this electron-lucent core may be a baseline EoP-specific granule characteristic and not secondary to activation and release of granule contents. The unexpected observation of high expression levels of granule proteins and the presence of organized granules in murine EoPs, as identified via lack of lineage markers and expression of CD34, IL-5Rα, and intermediate levels of c-KIT, prompts speculation that a less-differentiated progenitor, or preEoP, upstream of the murine EoP may exist. We have studies underway to identify potential preEoPs via Siglec-F expression.
TFs play a key role in activating lineage-specific genetic programs (55). The order of TF expression in myeloid progenitors significantly affects lineage potential (55, 56). For example, induction of GATA-1, in the presence of CCAAT/enhancer binding protein alpha (C/EBP-α) and PU.1, in myeloid progenitors has been shown to result in eosinophil formation. Our data are consistent with this model, as we observed significantly elevated GATA-1 expression in EoPs compared to GMPs and that GMPs also express PU.1 and low levels of C/EBPα (Figure 4 and data not shown). The expression of the granule protein MBP has been shown to be regulated by a combination of TFs, including GATA-1, PU.1, and C/EBP-ε, in eosinophils (57, 58). Notably, we observed co-expression of GATA-1, PU.1, and C/EBP-ε in EoPs and eosinophils (and not in GMPs), which was also associated with high expression levels of transcripts for granule proteins in EoPs and eosinophils. In addition to confirming expression of TFs known to have a role in eosinophil development, we noted expression of a large number of TFs that have not previously been associated with eosinophils. For example, our data demonstrate conserved Helios and Aiolos expression in eosinophil-lineage-committed cells in mice and humans. Helios and Aiolos can act as both activators and repressors of gene transcription (59–61) and mediate their effects through their interaction with other Ikaros family members to anchor protein complexes that regulate chromatin restructuring around target genes (62–64). For example, a change in Aiolos expression results in altered histone modifications that yield more permissive or repressive chromatin environments for gene expression (60). The importance of the chromatin environment (e.g. histone acetylation) in eosinophil differentiation is underscored by studies in which treating a promyelocytic cell line with histone deacetylase inhibitors that prevent the removal of acetate from histone lysine residues results in upregulation of IL5RA expression and in differentiation along the eosinophil lineage (65, 66). Thus, n-butyrate, a histone deacetylase inhibitor that has been used for decades to promote differentiation of human eosinophilic leukemia cell lines (67–69), likely mediates its eosinophil-promoting effects via continuous histone acetylation that results in upregulation in expression of lineage-specific genes.
The Ikaros family of TFs have two zinc finger motifs in the carboxy-terminus that enables them to homodimerize and to dimerize with other members of the family (70). Helios has been shown to heterodimerize with Ikaros and Aiolos (71, 72), and Aiolos has been shown to heterodimerize with Ikaros (73). We observed that EoPs and eosinophils, as well as GMPs and neutrophils, express Ikaros (data not shown). Compared to wild-type mice, Ikaros-deficient mice have increased numbers of basophils, but similar numbers of eosinophils, in the bone marrow at homeostasis (74). Ikaros regulates basophil development via expression of the basophil-promoting TF C/EBP-α (74). We noted Aiolos binding motifs in genes that encode for TFs, including Stat3, Nfatc1, Nfkb1, Irf1 and Irf2, in the eosinophil-lineage transcriptome, suggesting that Aiolos binding may be an important hierarchical step regulating local changes in chromatin states during eosinophil developmental progression. Similarly, we noted that PU.1 binding motifs were highly enriched in genes expressed by both EoPs and eosinophils that also contain H3K4me3 histone modifications. This finding is consistent with studies that demonstrated a majority of PU.1 that was bound to DNA was located near H3K4me3 marks in macrophages and B cells (22). A model has been proposed in which tiers of TFs cooperate to activate regulatory elements required for cell lineage development and function (22, 75, 76); the first tier includes TFs such as PU.1, and perhaps Aiolos, that target potential regulatory elements and create “accessible” DNA structures, and the subsequent second tier of TFs results in fully active regulatory elements and gene expression. Our data support this model, as many of the TFs that were expressed in eosinophils, but not GMPs or EoPs, contained potential Aiolos binding motifs. Future studies are underway to investigate the roles for Helios and Aiolos in regulating the chromatin environment and gene expression during eosinophil development. Delineating the function of the Ikaros family of TFs in eosinophil development will likely lead to clinically relevant findings for eosinophilia-associated disorders, as the mechanism of action of the clinical drug lenalidomide, which is effective in treating multiple myeloma, was determined to be the selective degradation of Aiolos (and Ikaros) in hematopoietic cells (77).
Notably, we observed that a majority of the genes that were induced in the EoP, but silent in the GMP, were already associated with the histone modification H3K4me3 in the GMP, suggesting that these genes were poised for induction in the GMP prior to eosinophil-lineage commitment. Conversely, many of the genes that were specifically induced in the mature eosinophil (and silent in the EoP) did not have the H3K4me3 modification in the EoP, highlighting differences in gene regulation during eosinophil development. These data provide further support for a model of sequential events regulating gene expression during cell maturation. Comprehensive epigenomic and transcriptomic profiling during critical stages in eosinophil development will ultimately define the programming and gene regulatory networks necessary for eosinophil development and will likely lead to novel therapeutic strategies to regulate eosinophil production.
Supplementary Material
Acknowledgments
We wish to thank Shawna Hottinger for her editorial assistance and Drs. Simon Hogan and Marc Rothenberg for critical review of the manuscript.
This work was supported by the NIH grant K08 AI093673 and the Allergy, Asthma, Immunology Education and Research Organization (ARTrust) Faculty Development Award of the American Academy of Allergy, Asthma and Immunology. This project was also supported in part by the NIH grant P30 DK078392 (Gene and Protein Expression and Flow Cytometry Cores) of the Digestive Disease Research Core Center in Cincinnati.
Abbreviations uses in this article
- CCHMC
Cincinnati Children’s Hospital Medical Center
- ChIP
chromatin immunoprecipitation
- EoP
eosinophil-lineage committed progenitor
- Eos
eosinophil
- GMP
granulocyte-monocyte progenitor
- H3K4me3
trimethylation of lysine 4 of histone H3
- preEos
eosinophil precursor
- TF
transcription factor
- TSS
transcriptional start site
References
- 1.Lee J, Rosenberg HF. Eosinophils in health and disease. Elsevier/Academic Press; London; Waltham, MA: 2013. [Google Scholar]
- 2.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nature reviews Immunology. 2013;13:9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. The Journal of experimental medicine. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. The Journal of experimental medicine. 2009;206:183–193. doi: 10.1084/jem.20081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffin R, Leitch AE, Fox S, Haslett C, Rossi AG. Targeting granulocyte apoptosis: mechanisms, models, and therapies. Immunol Rev. 2010;236:28–40. doi: 10.1111/j.1600-065X.2010.00922.x. [DOI] [PubMed] [Google Scholar]
- 6.Simon HU. Molecules involved in the regulation of eosinophil apoptosis. Chemical immunology and allergy. 2006;91:49–58. doi: 10.1159/000090229. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida T, Ikuta K, Sugaya H, Maki K, Takagi M, Kanazawa H, Sunaga S, Kinashi T, Yoshimura K, Miyazaki J, Takaki S, Takatsu K. Defective B-1 cell development and impaired immunity against Angiostrongylus cantonensis in IL-5R alpha-deficient mice. Immunity. 1996;4:483–494. doi: 10.1016/s1074-7613(00)80414-8. [DOI] [PubMed] [Google Scholar]
- 8.Takagi M, Hara T, Ichihara M, Takatsu K, Miyajima A. Multi-colony stimulating activity of interleukin 5 (IL-5) on hematopoietic progenitors from transgenic mice that express IL-5 receptor alpha subunit constitutively. The Journal of experimental medicine. 1995;181:889–899. doi: 10.1084/jem.181.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishinakamura R, Nakayama N, Hirabayashi Y, Inoue T, Aud D, McNeil T, Azuma S, Yoshida S, Toyoda Y, Arai K, et al. Mice deficient for the IL-3/GM-CSF/IL-5 beta c receptor exhibit lung pathology and impaired immune response, while beta IL3 receptor-deficient mice are normal. Immunity. 1995;2:211–222. doi: 10.1016/1074-7613(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 10.Kopf M, Brombacher F, Hodgkin PD, Ramsay AJ, Milbourne EA, Dai WJ, Ovington KS, Behm CA, Kohler G, Young IG, Matthaei KI. IL-5-Deficient mice have a developmental defect in CD5(+) B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity. 1996;4:15–24. doi: 10.1016/s1074-7613(00)80294-0. [DOI] [PubMed] [Google Scholar]
- 11.Dent LA, Strath M, Mellor AL, Sanderson CJ. Eosinophilia in transgenic mice expressing interleukin 5. The Journal of experimental medicine. 1990;172:1425–1431. doi: 10.1084/jem.172.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dent LA, Munro GH, Piper KP, Sanderson CJ, Finlay DA, Dempster RK, Bignold LP, Harkin DG, Hagan P. Eosinophilic interleukin 5 (IL-5) transgenic mice: eosinophil activity and impaired clearance of Schistosoma mansoni. Parasite immunology. 1997;19:291–300. doi: 10.1046/j.1365-3024.1997.d01-210.x. [DOI] [PubMed] [Google Scholar]
- 13.Fulkerson PC, Fischetti CA, McBride ML, Hassman LM, Hogan SP, Rothenberg ME. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc Natl Acad Sci USA. 2006;103:16418–16423. doi: 10.1073/pnas.0607863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bystrom J, Wynn TA, Domachowske JB, Rosenberg HF. Gene microarray analysis reveals interleukin-5-dependent transcriptional targets in mouse bone marrow. Blood. 2004;103:868–877. doi: 10.1182/blood-2003-08-2778. [DOI] [PubMed] [Google Scholar]
- 15.Fulkerson PC, Schollaert KL, Bouffi C, Rothenberg ME. IL-5 Triggers a Cooperative Cytokine Network That Promotes Eosinophil Precursor Maturation. J Immunol. 2014;193:4043–4052. doi: 10.4049/jimmunol.1400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schollaert KL, Stephens MR, Gray JK, Fulkerson PC. Generation of eosinophils from cryopreserved murine bone marrow cells. PloS one. 2014;9:e116141. doi: 10.1371/journal.pone.0116141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kartashov AV, Barski A. BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data. bioRxiv. 2015 doi: 10.1186/s13059-015-0720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids research. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, Petrie HT, Georgopoulos K. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nature immunology. 2012;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 25.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heng TS, Painter MW C. Immunological Genome Project. The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 29.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D’Asaro M, Chiappe A, Sridhar S, Pacheco GD, Burczynski ME, Hochstrasser D, Vonderscher J, Matthes T. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PloS one. 2012;7:e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logan MR, Lacy P, Bablitz B, Moqbel R. Expression of eosinophil target SNAREs as potential cognate receptors for vesicle-associated membrane protein-2 in exocytosis. The Journal of allergy and clinical immunology. 2002;109:299–306. doi: 10.1067/mai.2002.121453. [DOI] [PubMed] [Google Scholar]
- 32.Park YM, Bochner BS. Eosinophil survival and apoptosis in health and disease. Allergy Asthma Immunol Res. 2010;2:87–101. doi: 10.4168/aair.2010.2.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen T, Besse JA, Mingler MK, Fulkerson PC, Rothenberg ME. Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6067–6072. doi: 10.1073/pnas.1220572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farahi N, Singh NR, Heard S, Loutsios C, Summers C, Solanki CK, Solanki K, Balan KK, Ruparelia P, Peters AM, Condliffe AM, Chilvers ER. Use of 111-Indium-labeled autologous eosinophils to establish the in vivo kinetics of human eosinophils in healthy subjects. Blood. 2012;120:4068–4071. doi: 10.1182/blood-2012-07-443424. [DOI] [PubMed] [Google Scholar]
- 35.Ohnmacht C, Pullner A, van Rooijen N, Voehringer D. Analysis of eosinophil turnover in vivo reveals their active recruitment to and prolonged survival in the peritoneal cavity. J Immunol. 2007;179:4766–4774. doi: 10.4049/jimmunol.179.7.4766. [DOI] [PubMed] [Google Scholar]
- 36.Geering B, Stoeckle C, Conus S, Simon HU. Living and dying for inflammation: neutrophils, eosinophils, basophils. Trends Immunol. 2013;34:398–409. doi: 10.1016/j.it.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Tomaki M, Zhao LL, Lundahl J, Sjostrand M, Jordana M, Linden A, O’Byrne P, Lotvall J. Eosinophilopoiesis in a murine model of allergic airway eosinophilia: involvement of bone marrow IL-5 and IL-5 receptor alpha. J Immunol. 2000;165:4040–4050. doi: 10.4049/jimmunol.165.7.4040. [DOI] [PubMed] [Google Scholar]
- 38.Mattes J, Yang M, Mahalingam S, Kuehr J, Webb DC, Simson L, Hogan SP, Koskinen A, McKenzie AN, Dent LA, Rothenberg ME, Matthaei KI, Young IG, Foster PS. Intrinsic defect in T cell production of interleukin (IL)-13 in the absence of both IL-5 and eotaxin precludes the development of eosinophilia and airways hyperreactivity in experimental asthma. The Journal of experimental medicine. 2002;195:1433–1444. doi: 10.1084/jem.20020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogan SP, Koskinen A, Foster PS. Interleukin-5 and eosinophils induce airway damage and bronchial hyperreactivity during allergic airway inflammation in BALB/c mice. Immunology and cell biology. 1997;75:284–288. doi: 10.1038/icb.1997.43. [DOI] [PubMed] [Google Scholar]
- 40.Wirths S, Bugl S, Kopp HG. Neutrophil homeostasis and its regulation by danger signaling. Blood. 2014;123:3563–3566. doi: 10.1182/blood-2013-11-516260. [DOI] [PubMed] [Google Scholar]
- 41.May G, Soneji S, Tipping AJ, Teles J, McGowan SJ, Wu M, Guo Y, Fugazza C, Brown J, Karlsson G, Pina C, Olariu V, Taylor S, Tenen DG, Peterson C, Enver T. Dynamic analysis of gene expression and genome-wide transcription factor binding during lineage specification of multipotent progenitors. Cell stem cell. 2013;13:754–768. doi: 10.1016/j.stem.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laslo P, Pongubala JM, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Seminars in immunology. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Bochner BS. Siglec-8 on human eosinophils and mast cells, and Siglec-F on murine eosinophils, are functionally related inhibitory receptors. Clin Exp Allergy. 2009;39:317–324. doi: 10.1111/j.1365-2222.2008.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hudson SA, Herrmann H, Du J, Cox P, el Haddad B, Butler B, Crocker PR, Ackerman SJ, Valent P, Bochner BS. Developmental, malignancy-related, and cross-species analysis of eosinophil, mast cell, and basophil siglec-8 expression. Journal of clinical immunology. 2011;31:1045–1053. doi: 10.1007/s10875-011-9589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyer KD, Garcia-Crespo KE, Killoran KE, Rosenberg HF. Antigen profiles for the quantitative assessment of eosinophils in mouse tissues by flow cytometry. Journal of immunological methods. 2011;369:91–97. doi: 10.1016/j.jim.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei JT, Martinez-Moczygemba M. Separate endocytic pathways regulate IL-5 receptor internalization and signaling. J Leukoc Biol. 2008;84:499–509. doi: 10.1189/jlb.1207828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hellman C, Hallden G, Hylander B, Lundahl J. Regulation of the interleukin-5 receptor alpha-subunit on peripheral blood eosinophils from healthy subjects. Clin Exp Immunol. 2003;131:75–81. doi: 10.1046/j.1365-2249.2003.02051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory B, Kirchem A, Phipps S, Gevaert P, Pridgeon C, Rankin SM, Robinson DS. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor alpha-chain expression by cytokines: IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor alpha expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor alpha expression. J Immunol. 2003;170:5359–5366. doi: 10.4049/jimmunol.170.11.5359. [DOI] [PubMed] [Google Scholar]
- 49.Gruart V, Truong MJ, Plumas J, Zandecki M, Kusnierz JP, Prin L, Vinatier D, Capron A, Capron M. Decreased expression of eosinophil peroxidase and major basic protein messenger RNAs during eosinophil maturation. Blood. 1992;79:2592–2597. [PubMed] [Google Scholar]
- 50.Doyle AD, Jacobsen EA, Ochkur SI, McGarry MP, Shim KG, Nguyen DT, Protheroe C, Colbert D, Kloeber J, Neely J, Shim KP, Dyer KD, Rosenberg HF, Lee JJ, Lee NA. Expression of the secondary granule proteins major basic protein 1 (MBP-1) and eosinophil peroxidase (EPX) is required for eosinophilopoiesis in mice. Blood. 2013;122:781–790. doi: 10.1182/blood-2013-01-473405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melo RC, Spencer LA, Dvorak AM, Weller PF. Mechanisms of eosinophil secretion: large vesiculotubular carriers mediate transport and release of granule-derived cytokines and other proteins. J Leukoc Biol. 2008;83:229–236. doi: 10.1189/jlb.0707503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, Foster PS, Matthaei KI. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood. 2007;110:1578–1586. doi: 10.1182/blood-2007-01-071340. [DOI] [PubMed] [Google Scholar]
- 53.Malm-Erjefalt M, Persson CG, Erjefalt JS. Degranulation status of airway tissue eosinophils in mouse models of allergic airway inflammation. American journal of respiratory cell and molecular biology. 2001;24:352–359. doi: 10.1165/ajrcmb.24.3.4357. [DOI] [PubMed] [Google Scholar]
- 54.Spencer LA, Bonjour K, Melo RC, Weller PF. Eosinophil secretion of granule-derived cytokines. Frontiers in immunology. 2014;5:496. doi: 10.3389/fimmu.2014.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes & development. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McNagny K, Graf T. Making eosinophils through subtle shifts in transcription factor expression. The Journal of experimental medicine. 2002;195:F43–F47. doi: 10.1084/jem.20020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, Ackerman SJ. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. Journal of Biological Chemistry. 2002;277:43481–43494. doi: 10.1074/jbc.M204777200. [DOI] [PubMed] [Google Scholar]
- 58.Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP epsilon and PU.1. Blood. 2003;101:3265–3273. doi: 10.1182/blood-2002-04-1039. [DOI] [PubMed] [Google Scholar]
- 59.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nature immunology. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, Rangachari M, Zhu C, Xiao S, Seavitt J, Georgopoulos K, Kuchroo VK. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nature immunology. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans HG, Roostalu U, Walter GJ, Gullick NJ, Frederiksen KS, Roberts CA, Sumner J, Baeten DL, Gerwien JG, Cope AP, Geissmann F, Kirkham BW, Taams LS. TNF-alpha blockade induces IL-10 expression in human CD4+ T cells. Nature communications. 2014;5:3199. doi: 10.1038/ncomms4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 63.Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, Smale ST, Fisher AG, Merkenschlager M. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 64.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, Dickins RA, Zuber J, Jaritz M, Busslinger M. Stage-specific control of early B cell development by the transcription factor Ikaros. Nature immunology. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tagari P, Pecheur EI, Scheid M, Brown P, Ford-Hutchinson AW, Nicholson D. Activation of human eosinophils and differentiated HL-60 cells by interleukin-5. International archives of allergy and immunology. 1993;101:227–233. doi: 10.1159/000236450. [DOI] [PubMed] [Google Scholar]
- 66.Ishihara K, Hong J, Zee O, Ohuchi K. Possible mechanism of action of the histone deacetylase inhibitors for the induction of differentiation of HL-60 clone 15 cells into eosinophils. British journal of pharmacology. 2004;142:1020–1030. doi: 10.1038/sj.bjp.0705869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fischkoff SA. Graded increase in probability of eosinophilic differentiation of HL-60 promyelocytic leukemia cells induced by culture under alkaline conditions. Leukemia research. 1988;12:679–686. doi: 10.1016/0145-2126(88)90103-8. [DOI] [PubMed] [Google Scholar]
- 68.Fischkoff SA, Condon ME. Switch in differentiative response to maturation inducers of human promyelocytic leukemia cells by prior exposure to alkaline conditions. Cancer research. 1985;45:2065–2069. [PubMed] [Google Scholar]
- 69.Zimmermann N, Daugherty BL, Stark JM, Rothenberg ME. Molecular analysis of CCR-3 events in eosinophilic cells. J Immunol. 2000;164:1055–1064. doi: 10.4049/jimmunol.164.2.1055. [DOI] [PubMed] [Google Scholar]
- 70.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Molecular immunology. 2011;48:1272–1278. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 71.Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, Morgan BA. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Current biology : CB. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 72.Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, Akashi K, Weissman IL, Fisher AG, Smale ST. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes & development. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. The EMBO journal. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rao KN, Smuda C, Gregory GD, Min B, Brown MA. Ikaros limits basophil development by suppressing C/EBP-alpha expression. Blood. 2013;122:2572–2581. doi: 10.1182/blood-2013-04-494625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heinz S, Glass CK. Roles of lineage-determining transcription factors in establishing open chromatin: lessons from high-throughput studies. Current topics in microbiology and immunology. 2012;356:1–15. doi: 10.1007/82_2011_142. [DOI] [PubMed] [Google Scholar]
- 76.Rothenberg EV. Epigenetic mechanisms and developmental choice hierarchies in T-lymphocyte development. Briefings in functional genomics. 2013;12:512–524. doi: 10.1093/bfgp/elt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, Ciarlo C, Hartman E, Munshi N, Schenone M, Schreiber SL, Carr SA, Ebert BL. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.