Abstract
The evasion of anti-growth signaling is an important characteristic of cancer cells. In order to continue to proliferate, cancer cells must somehow uncouple themselves from the many signals that exist to slow down cell growth. Here, we define the anti-growth signaling process, and review several important pathways involved in growth signaling: p53, phosphatase and tensin homolog (PTEN), retinoblastoma protein (Rb), Hippo, growth differentiation factor 15 (GDF15), AT-rich interactive domain 1A (ARID1A), Notch, insulin-like growth factor (IGF), and Krüppel-like factor 5 (KLF5) pathways. Aberrations in these processes in cancer cells involve mutations and thus the suppression of genes that prevent growth, as well as mutation and activation of genes involved in driving cell growth. Using these pathways as examples, we prioritize molecular targets that might be leveraged to promote anti-growth signaling in cancer cells. Interestingly, naturally-occurring phytochemicals found in human diets (either singly or as mixtures) may promote anti-growth signaling, and do so without the potentially adverse effects associated with synthetic chemicals. We review examples of naturally-occurring phytochemicals that may be applied to prevent cancer by antagonizing growth signaling, and propose one phytochemical for each pathway. These are: epigallocatechin-3-gallate (EGCG) for the Rb pathway, luteolin for p53, curcumin for PTEN, porphyrins for Hippo, genistein for GDF15, resveratrol for ARID1A, withaferin A for Notch and diguelin for the IGF1-receptor pathway. The coordination of anti-growth signaling and natural compound studies will provide insight into the future application of these compounds in the clinical setting.
Keywords: Tumor suppressor, anti-growth signaling, reversible and irreversible evasion, cancer prevention, hallmark of cancer
1. Introduction
Carcinogenesis is a complex, stochastic and yet highly coordinated multi-step process in which normal cells progress through hyperplasia to mild, moderate and severe dysplasia to carcinoma in situ, invasive carcinoma, and finally to metastatic disease after initiation by primary carcinogenic insult [1]. Hahn and Weinberg [2] proposed six hallmarks to better define and understand this complex process. They modeled these hallmarks in normal human bronchial epithelial cells and demonstrated immortalization in vitro by targeting tumor suppressor pathways, notably, retinoblastoma (Rb) regulation of cell cycle entry, tumor protein 53 (TP53) regulation of cell cycle progression, human telomerase reverse transcriptase (hTERT) activation, combined with an oncogenic signal using activated Harvey rat sarcoma viral oncogene homolog (hRAS) [3]. As this model shows, and as studies of human tumors progress into the era of high throughput sequencing, it is clear that evasion of anti-growth signaling and loss of tumor suppressors are central hallmarks necessary to the oncogenic process.
Loss of growth control mechanisms allows neoplastic cells to acquire unlimited replicative ability and evade elimination, growth arrest, and senescence by tumor suppressors. In general, tumor suppressor genes block the transformation of normal cells to cancerous cells. Environmental stress factors including ultraviolet (UV), irradiation, and chemicals can induce DNA damage and genetic alteration. These injuries can cause the progression of carcinogenic processes if damage cannot be appropriately repaired and mutated cells continuously proliferate. Dozens of tumor suppressor genes are activated under these circumstances that inhibit the proliferation of damaged/mutated cells by arresting cell cycle progression and inducing apoptosis and other types of programmed cell death, thus their evasion is critical for carcinogenesis. p53 and Rb are typical tumor suppressor genes [4]; they play a key role in determining the fate of cells, i.e whether they proliferate or undergo senescence or apoptotic programs. In solid tumors, the most common genetic changes are losses of tumor suppressor genes. It has been estimated that over 70% of the genetic changes discovered in solid tumors represent evasion of tumor suppressor mechanisms; leading to the suggestion that this leaves us with an un-targetable cancer problem. It would appear necessary to replenish the function associated with the mutated or lost tumor suppressor in every tumor cell, a goal that has so far been unattainable. However, loss of a tumor suppressor usually results in unopposed signaling by a mechanism normally suppressed by the lost tumor suppressor gene. Thus, a viable strategy to overcome the evasion of a tumor suppressor mechanism is to identify and target the unrestrained pathways activated by the loss of tumor suppressors.
This review will briefly discuss how anti-growth signaling mechanisms are inactivated in tumors with emphasis on major tumor suppressor pathways and will explore how these pathways can be targeted for the prevention and treatment of cancer.
2. Dysfunction: mechanism of evasion of tumor suppressors
Tumor cells may evade tumor suppressors by genetic and epigenetic mechanisms. Genetic mechanisms include chromosomal deletion, mutation and inactivation or loss of upstream or downstream effectors. Epigenetic evasion includes DNA methylation, and histone methylation and acetylation. Examples of tumor suppressor losses are abundant in solid tumors. Among the most common are loss, mutation and/or methylation of the cyclin-dependent kinase inhibitor (CDKN) 2A locus on chromosome 9p21, which leads to loss of the CDKN, p16ink4a and often the mouse double minute 2 homolog (hMDM2) inhibitor p14ARF as well. Loss of p16ink4a results in unopposed activation of the cyclin dependent kinases CDK4/6, which phosphorylate the Rb protein thereby activating E2F-mediated transcription of genes involved in entry into the cell cycle. Loss of p14ARF protein results in unopposed MDM2 activity and increased p53 ubiquitination and degradation with effects similar to loss of p53. Mutation, loss or inhibition of TP53 function is also very common, as is loss and/or mutation of phosphatase and tensin homolog (PTEN). Loss of p53 leads to loss of cell cycle checkpoints, the ability of the cell to arrest and effectively repair DNA errors or damage and the accumulation of genetic instability and accumulation of mutations. Additionally, p53 protein has an important role in triggering apoptosis, thus its loss leads to the inappropriate survival of cells with new mutations. Loss of PTEN protein, a phosphatase that dephosphorylates phosphatidylinositol (3,4,5)-trisphosphate (PIP3), leads to unopposed activity of phosphoinositide 3-kinase (PI3K) and protein kinase B (AKT) signaling, which drives tumor growth.
The dysfunctional pathways activated by loss of tumor suppressors provide continuous unopposed tumor growth promoting signals. These pathways have consequently become potential targets for novel anti-cancer compounds. For example, inhibitors of MDM2 are being tested to restore p53 function, mammalian target of rapamycin (mTOR) inhibitors are being tested to overcome PTEN loss, and CDK4/6 inhibitors to restore Rb function from p16ink4a loss of function are entering clinical trials.
3. Prioritized anti-growth signaling pathways
There are hundreds of tumor suppressor genes that possess the ability to stop or slow down the carcinogenesis process. The activation of tumor suppressors is mostly context-dependent and varies by organ site and by molecular and pathological sub-type. The most common and important tumor suppressors, their role in tumorigenesis and approaches to target these genes for cancer treatment and prophylaxis are discussed in the following sections and summarized in Fig. 1.
Figure 1.
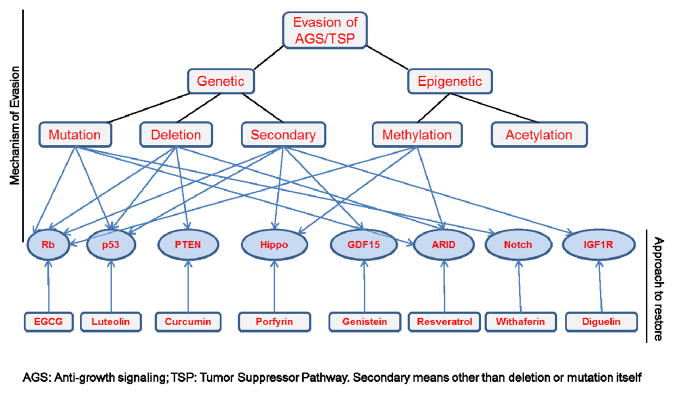
Mechanism of evasion of tumor suppressor pathways and their targeting by natural compounds.
3.1 The Rb pathway
The retinoblastoma (Rb) gene was the first tumor suppressor gene to be described. The development of retinoblastoma was predicted by Alfred Knudsen to involve a “two hit” mechanism, based on the kinetics of appearance of retinoblastoma in the inherited form (single order kinetics) and the sporadic form (second order kinetics). This analysis led to the hypothesis that disease initiation requires two steps involving loss of function of both copies of the affected gene. Thus, Rb was recognized to have a tumor suppressor function long before the gene was identified and demonstrated to be inactivated by mutation of one copy and loss or silencing of the second copy [5]. The Rb protein (pRb) also plays a key role in integrating diverse signals from intra- and extracellular sources and thus driving cell cycle progression. In cells in the G0/G1 phase, pRb, which is in a hypophosphorylated state, binds to the transcription factor E2F family and suppresses E2F-mediated gene transcription. The E2F family encodes a variety of genes involved in cell cycle progression, DNA replication, DNA damage repair, cell cycle checkpoint, and apoptosis [6]. Therefore, inhibition of the E2F family by pRb results in the suppression of cell cycle progression. However, the cyclin D/CDK4 complex phosphorylates pRB and allows E2Fs to bind their target genes by disrupting the formation of the pRB-E2F complex [7]. In addition to controlling cell cycle progression, pRB is also involved in the regulation of DNA replication, differentiation, and apoptosis [7].
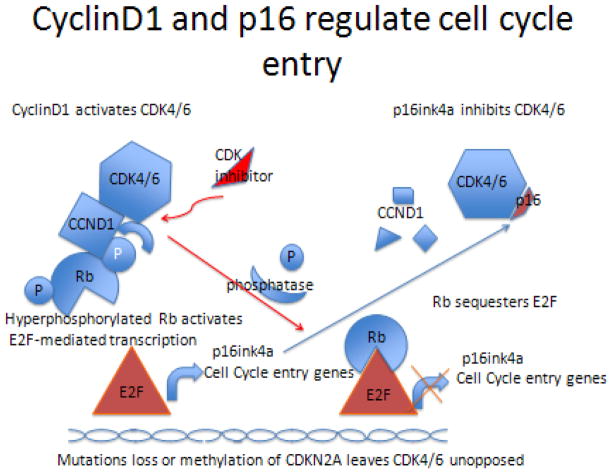
In head and neck squamous cell cancers (HNSCC), loss or mutation of Rb is uncommon [1], but frequent inactivation by loss, mutation and/or methylation of p16 (CDKN2/MTS-1/INK4A) was observed in the majority of tumors [8]. In fact, p16ink4a was first called the multiple tumor suppressor gene-1 (MTS-1) because it is so commonly inactivated in many tumor types (melanoma, breast, head and neck, etc.). Abnormalities in CDKN2A RB1 are found in greater than 70% of lung squamous cancers [9], consistent with the observations in HNSCC. The p16 protein regulates Rb by acting as an inhibitor of the cyclin dependent kinases that phosphorylate Rb, thereby allowing the E2F family of transcription factors to initiate expression of genes involved in entry into the cell cycle. In an important feedback loop, E2F upregulates p16ink4a protein expression, which inactivates the cyclinD1/CDK4/6 complex (Figure 2). With CDK4/6 inactivated, Rb is de-phosphorylated by ubiquitous phosphatases and re-sequesters E2F, preventing cell cycle entry.
Figure 2.
Cyclin D1 and p16 regulate cell cycle entry. When p16 is lost, CDK inhibitors can restore function.
3.2 The p53 pathway
The tumor suppressor p53 family consists of three members, p53, p63 and p73, sharing overlapping anti-growth functions, such as cell cycle arrest, apoptosis and DNA repair. TP53 is very frequently targeted by mutation and loss in cancer. In the recent report of The Cancer Genome Atlas (TCGA) assessment of squamous cell lung cancers, the most common significantly mutated gene was TP53 [9]. Mutation and loss of p53 are very common in cancers related to carcinogens in tobacco smoke, such as lung, head and neck, bladder, etc, but are less common in breast and prostate cancers that have hormonal, genetic and diet-related etiologic factors. In a subset of cancers that retain wild type p53, loss of p14ARF can result in unopposed activity of MDM2, which is a transcriptional target and a negative regulator of p53. MDM2 is a p53-specific E3 ubiquitin ligase that physically interacts with p53, causing its mono-ubiquitination and thus its degradation [10]. Disruption of the p53-MDM2 interaction leads to p53 induction and its biological response such as cell cycle arrest, DNA damage repair and apoptosis. In this subset, MDM2 inhibitors are effective at restoring p53 function leading to growth inhibition and induction of apoptosis [11].
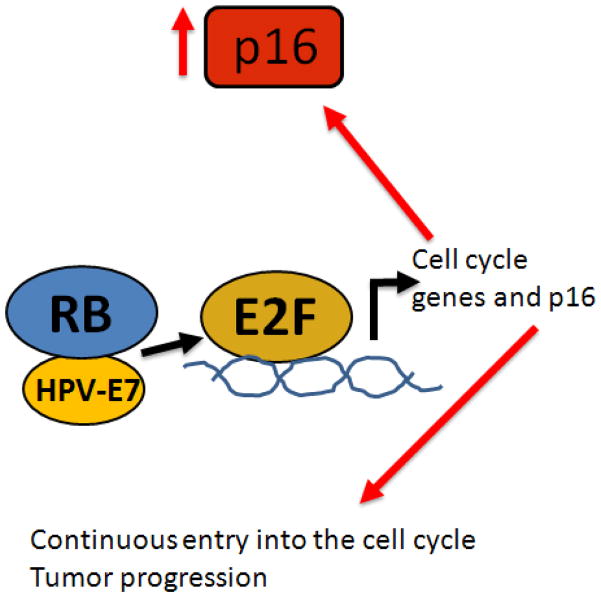
In cancers caused by oncogenic viruses, such as the high risk human papilloma viruses (hrHPV), which are implicated in anogenital and oropharyngeal cancers, p53 and p16ink4a are nearly always wild type. Currently, 70–90% of oropharyngeal cancers diagnosed at the University of Michigan contain hrHPV [12]. The viral oncogenes E6 and E7 target and inactivate p53 and Rb, respectively. E7 binds to Rb protein preventing it from binding to E2F, which then activates continuous transcription of genes involved in cell cycle entry and of p16 (Figure 3). For this reason, overexpression of p16 is a useful surrogate for hrHPV infection. These tumors are also more responsive to therapy, most likely because the p53 and Rb genes are intact, and after treatment can still function if the virus is inactivated by chemotherapy and radiation. However, some HPV-positive tumors are also controlled by surgery [13], suggesting that an immune response to the virus may assist in eliminating the tumor.
Figure 3.
HPV-E7 binds to Rb blocking it from inactivating E2F. E2F upregulates both cell cycle genes and p16 making it a marker for HPV induced cancers.
Mutation and loss of p53 function can be an Achilles heel for cytotoxic therapies such as cisplatin. Cisplatin binds to DNA to cause its cytotoxic effect. When p53 is compromised by mutation it is less effective in causing cell cycle arrest and DNA repair. Thus, the degree of lethal damage to tumor cells caused by cisplatin is often increased when p53 is mutated or inactivated because the cell cannot undergo p53-mediated cell cycle arrest and p53-mediated DNA repair.
DNA damage or stress response mediated by chemicals, UV irradiation, or oncogenic mutation results in the stabilization and accumulation of p53 [14, 15]. Activated p53 can bind to specific DNA sequences in the promoter region of its target genes, including p21, B-cell lymphoma 2 (Bcl2)-associated X protein (Bax), p53 upregulated modulator of apoptosis (PUMA), and growth arrest and DNA damage (GADD45), mediating cell cycle arrest, senescence, and apoptosis [16]. In contrast to these suppressive roles, mutant p53 loses these functions and serves as an oncogene by physically interacting with other proteins and thus modulating their cellular function [17]. For example, the interaction between mutant p53 and p63 results in the decreased tumor suppressive activity of p63 [17]. A recent report suggests that the interaction of mutant p53 with Smad negatively regulates p63, leading to cancer cell metastasis [18]. Mutant p53 is also known to associate with several transcription factors, such as specificity protein 1 (Sp1), Ets-1, and vitamin D receptor (VDR), resulting in the transactivation of their oncogenic target genes [17].
Although p53 is inactivated through deletion or mutation in about half of all cancers, mutation of p63 and p73 are extremely rare in human cancers [19–23], although their expression is frequently dysregulated in human tumors [19]. p73 mRNA and/or protein were shown to be upregulated in breast [24, 25], ovarian [26], hepatocellular [27], neuroblastoma [28], bladder [29], prostate [30], thyroid [31], B-cell chronic leukemias [32] and colorectal [33] cancers, and lost in pancreatic adenocarcinoma [34]. Expression of ΔNp73 isoforms was also increased in head and neck cancers [35]. These members of the p53 family are also targets of many anti-cancer drugs including those frequently used in the clinic such as cisplatin.
3.3 The phosphatase and tensin homolog (PTEN) pathway
PTEN is often lost or inactivated in multiple solid tumor types including prostate, breast, thyroid, and endometrial tumors and others [36]. PTEN is a critical regulator of signaling through the PI3K pathway through its action as a PIP3 phosphatase, thereby negatively regulating the PI3K-AKT-mTOR pathway. In the absence of PTEN, unregulated cell proliferation occurs through activation of a cascade of signals downstream in the AKT pathway. Fortunately, mTOR inhibitors [37] and mitogen/extracellular signal-regulated kinase (MEK) inhibitors [38] are being developed that can target these pathways, providing a strategy to counter this common tumor suppressor loss. For instance, mTOR inhibitors such as everolimus and temsirolimus targeting the PI3/AKT pathway, which is overexpressed as a consequence of PTEN loss, have been shown to improve median overall survival in renal cell carcinomas and pancreatic neuroendocrine carcinomas.
Loss or inactivation of PTEN also inactivates the forkhead box “O” (FOXO) family of tumor suppressors through AKT. The family consists of four members, FOXO1 (also known as FKHR), FOXO3a (also known as FKHRL1), FOXO4 (also known as AFX or MLLT7), and FOXO6. FOXO proteins are well characterized tumor suppressors: simultaneous somatic deletions of FOXO1, 3 and 4 alleles generates thymic lymphomas and systemic hemagiomas in mice (reviewed in [39]). FOXOs are direct substrates of AKT and AKT-dependent phosphorylation sequesters them in the cytoplasm and promotes their degradation through ubiquitylation, thus inactivating them. FOXOs can undergo multiple other post-translational modifications (PTMs) [39], which affect their transcriptional activity and contribute to tumor initiation, progression and drug action. FOXO proteins transcribe genes involved in G0/G1 cell cycle transition (p21, p27, p15, p19), G2/M transition (GADD45, cyclin G2 and Polo-like kinase 1) and apoptosis (BIM, BCL6, PUMA, TNF-related apoptosis-inducing ligand [TRAIL], etc). Since FOXOs are mostly inactivated through activation of PI3K-AKT pathways, these tumor suppressors can be easily activated by targeting PI3K-AKT pathways. Multiple small molecule inhibitors have been developed that target PI3K or AKT and many of them are currently under clinical investigation.
3.4 The Hippo pathway
The Hippo signaling pathway has emerged as a central regulator of growth, connecting processes such as adhesion, cell polarity, and cell density with increases in cell number [40–42]. Overcoming the upstream regulators of the Hippo pathway may be a prerequisite, or hallmark feature, of certain cancer cell types [43].
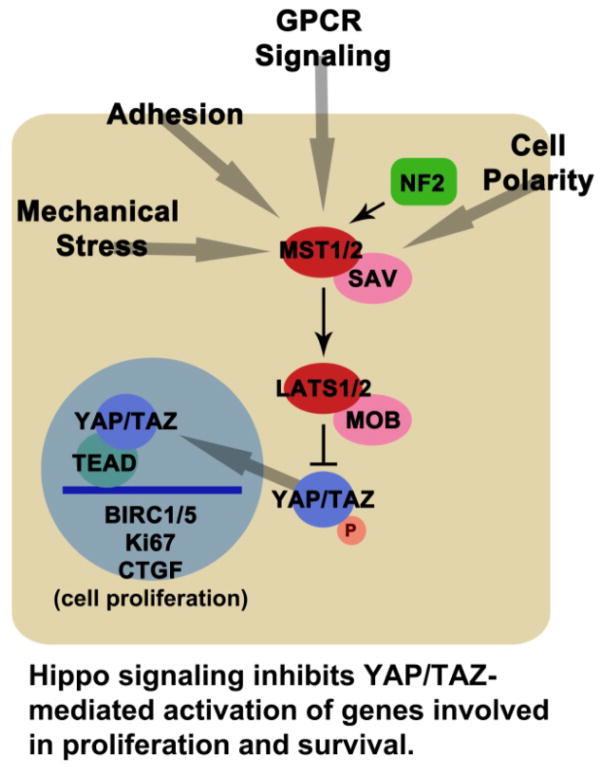
The core Hippo pathway is highly conserved, and functions to restrict growth during development by limiting the output of precursor cells [42]. In this pathway, the Ste20-like kinase MST1/2 (Hippo) works with the adaptor Salvador (SAV) to phosphorylate and activate the nuclear Dbf2-related (NDR)-like kinase large tumor suppressor kinase (LATS1/2). In turn, LATS activity with the adaptor, MOB, results in the phosphorylation of Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) (Figure 4) [41, 42]. This phosphorylation of YAP leads to its accumulation in the cytoplasm where it has been shown to associate with 14-3-3, angiomotin (AMOT), or catenin α1 (CTNNA1) [44, 45]. The phosphorylation and exclusion of YAP from the nucleus is critical because it prevents its activity as a transcriptional coactivator. If it enters the nucleus, YAP increases cell production, both through proliferation and the inhibition of cell death. Hence, the loss of core components such as MST or LATS prevents YAP/TAZ phosphorylation and activates YAP targets. The cytoplasmic association of YAP/TAZ with AMOT and CTNNA1 is also significant, because when these cytoskeletal or cell adhesion scaffolds are disrupted, the cytosolic pool of YAP/TAZ changes. Thus, disruption of adhesion, cytoskeletal, and cell polarity proteins can drive cell proliferation through YAP. There are numerous additional regulators of core pathway members, including: cadherin 1 (CDH1), CD44, FERM domain containing 6 (FRMD6), protein associated with Lin Seven 1 (PALS1), PATJ, AJUBA, Ras-associated family members (RASSF), and G-protein coupled receptors (GPCRs) [40, 41]. The relative contributions of these multiple inputs to the core members is not resolved, although it is likely that several such inputs may exist at any one time in the same cell.
Figure 4.
Hippo signaling inhibits YAP/TAZ-mediated activation of genes involved in proliferation and survival.
Loss of core Hippo pathway components, or hyperactivation of YAP/TAZ, in adult mouse precursor cells has been shown to result in hyperplasia and tumorigenesis [40, 42]. Overall, this suggests that Hippo pathway disruption promotes cancer [40]. Consistent with this notion, YAP has been observed to undergo copy number amplification in both human cancer and mouse cancer models [46, 47]. The Hippo pathway regulator neurofibromatosis 2 (NF2) provides another example of how cancer may result through YAP activation. NF2 is listed in the Catalogue of Somatic Mutations in Cancer (COSMIC) gene database and NF2 loss of function results in the formation of schwanommas [40, 48–50]. Functional assays have shown that NF2 regulates YAP activity, and YAP is required for the overgrowth observed when NF2 is lost in vivo [51].
Genomic analyses by TCGA or COSMIC do not show an enrichment of core Hippo pathway members in cancer [40]. It is unclear why this is the case. Perhaps unidentified components, which link cell adhesion or cytoskeletal proteins with the Hippo pathway, are lost in cancers rather than the core members. Alternatively, epigenetic factors that regulate the expression of core Hippo components could be disrupted in cancer. Regulation of the Hippo pathway by other signaling processes is another means through which cancer cells overcome the growth restrictions normally imposed on tissue cells by the Hippo pathway [40]. For instance, the Wnt signaling pathway component dishevelled segment polarity protein 1 (DVL1) binds to phosphorylated YAP/TAZ, inhibiting Wnt signaling [52, 53]. Thus, loss of Hippo activity could de-phosphorylate YAP and simultaneously activate the Wnt pathway. Hyperactive Wnt itself is thought to drive colorectal cancer through the activation of its target, catenin β1 (CTNNB1) [54], and YAP can bind and synergize with CTNNB1 in some contexts [55]. The disruption of Hippo signaling could be quite significant in cancer by enabling proliferation through multiple other pathways.
YAP/TAZ function as transcriptional coactivators of transcription factors such as p73, TEA domain family member 1 (TEAD), SMAD, and possibly others [40, 42]. YAP/TAZ nuclear activity increases the transcription of many genes including baculoviral IAP repeat-containing proteins (BIRC) 2 and 5, marker of proliferation Ki67, connective tissue growth factor (CTGF), and amphiregulin [56–58]. Such targets reveal how YAP activity leads to increased cell production: Ki67 leads to greater proliferation, CTGF and amphiregulin influence further signaling processes, and BIRC2/5 inhibit apoptosis. Moreover, Hippo and Wnt, or transforming growth factor (TGF)-β signaling are integrated [41], and some work on intestinal precursors reveals that the Janus kinase 2 (JAK)/signal transducer and activator of transcription (STAT) and epidermal growth factor (EGF) pathways may also be activated through YAP [59, 60]. The effects of Hippo signaling disruption could therefore influence the output of multiple signaling pathways that regulate the hallmarks of cancer [43].
Since the Hippo pathway suppresses cell growth by inhibiting YAP/TAZ, the desired intervention is to either enhance Hippo signal transduction or to inhibit YAP/TAZ activity. Drugs inhibiting the core kinases are not useful, but it may be possible to develop drugs targeting the phosphorylation sites on YAP/TAZ, which are known [41, 42], or drugs that prevent the activity of YAP/TAZ. For instance, small molecule screening has revealed three porphyrins that prevent the association of YAP with TEAD transcription factors [61]. One of these, verteporfin, is already applied clinically to treat macular degeneration. It is therefore possible that porphyrins could be applied in treating cancers where NF2 function has been lost without impacting normal tissue function.
A recent report has shown that GPCR signaling can both stimulate and repress the core Hippo kinases [62]. The GPCR agonists, lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P), can inhibit the core Hippo pathway member, LATS, which directly phosphorylates and inhibits YAP/TAZ. However, GPCR activation can also produce the opposite outcome. Adrenergic receptors, for instance stimulated by epinephrine, can actually increase the phosphorylation of YAP/TAZ by LATS, meaning that Hippo signal transduction can be enhanced [62]. GPCR signaling is complex in that the involved ligands, GPCRs, and G-α proteins work in a combinatorial and tissue-specific fashion. This specificity presents an opportunity to develop context-specific interventions that inhibit proliferation by agonizing the activity of LATS.
The Hippo pathway is just one of many mechanisms that suppress growth, but its disruption has extremely potent effects on cell proliferation. Future treatments of cancers that evade Hippo pathway growth suppression are likely to involve inhibition of YAP or enhancement of the upstream Hippo components that negatively regulate it.
3.5 Growth differentiation factor 15 (GDF15)
GDF15, also known as nonsteroidal anti-inflammatory drug (NSAID)-activated gene-1 (NAG-1), macrophage inhibitory cytokine 1 (MIC-1), placental TGF-β (PTGF-β), prostate differentiation factor (PDF), and placental bone morphogenetic protein (PLAB), is structurally similar to TGF-β [63]. TGF-β has established roles as both a growth suppressor and growth stimulator in cancer cells. Similarly, GDF15 exhibits growth suppressive and growth-promoting effects, which may depend in part on the stage of disease [64, 65]. In comparison to other cytokines and transcription factors, GDF15 has not been extensively studied in the context of cancer cell growth suppression. However, accumulating data suggest that GDF15 has multiple functions in cancer cells, including roles in the evasion of anti-growth signaling.
The growth-suppressing function of GDF15 may be due in part to the presence of p53 response elements within its promoter. GDF15 is a p53 transcriptional target that mediates G1 cell cycle arrest and apoptosis [66, 67]. In addition to its p53-dependent activity, GDF15 has been shown to reduce cancer cell growth through other mechanisms. The Krüppel-like factor 4 (KLF4) tumor suppressor also binds to the GDF15 promoter and initiates transcription of GDF15 in pancreatic cancer cells [68]. Other transcriptional mediators of GDF15 identified in colorectal cancer cell lines include Sp1, Sp3, and early growth response 1 (EGR1) [69, 70]. Finally, peroxisome proliferator-activated receptor gamma (PPARγ) ligand has been shown to induce GDF15 expression in pancreatic and colon cancers [68, 71, 72]. Thus, there are several major transcriptional regulators of growth suppression that upregulate GDF15. However, similar to TGF-β, GDF15 also exhibits growth-promoting effects. In fact, serum levels of GDF15 are frequently elevated in patients with various types of cancer, including pancreatic, prostate, and ovarian cancers. Therefore, although GDF15 appears to play a role in regulating the growth of cancer cells, its exact functions and mechanisms of action remain incompletely defined and may vary depending on cellular and disease context. In addition, GDF15 is upregulated by various signaling molecules, such as mitogen-activated protein kinase (MAPK) family members. Various chemical compounds, including 12-O-tetradecanoylphorbol-13-acetate (TPA), vitamin E, NSAID agents, and the proteasome inhibitor MG132 induce the transcription of GDF15/NAG-1 in a p38 MAPK-dependent manner [73–76]. Interestingly, MG132 increased GDF15 at the promoter level and stabilized the GDF15 transcript. In addition to being regulated by MAPK signaling, GDF15 expression is regulated by PI3K/Akt/glycogen synthase kinase-3 beta (GSK-3β) signaling [77], with PI3K inhibition inducing GDF15/NAG-1 expression. However, pharmacological inhibition or genetic knockdown of GSK-3β, which is negatively regulated by PI3K/Akt, blocks the upregulation of GDF15/NAG-1 in response to PI3K inhibition. Thus, PI3K signaling may suppress GDF15 expression in some contexts, whereas PI3K/Akt inhibitors induce GDF15 via GSK-3β. This induction of GDF15 may contribute to the growth suppressive or apoptotic effects of pharmacological PI3K inhibitors. Conversely, PI3K/MAPK signaling pathways are also implicated in the growth-promoting effects of GDF15. Stimulation or overexpression of GDF15 induces phosphorylation of Akt, p38 MAPK, and extracellular signal-regulated kinases (ERK1/2) in breast and ovarian cancer cells [78, 79]. These seemingly paradoxical results suggest context-dependent effects and possible feedback pathways that regulate GDF15 expression and/or activity.
GDF15 has potential roles in growth suppression as well as in metastasis and invasion. Increased circulating levels of secreted GDF15 or increased tissue expression are reported in patients with colorectal [80], ovarian [78, 81, 82], prostate [83–85], pancreatic [86, 87], breast [88], and bladder [89] cancers and in the cerebrospinal fluid of patients with glioblastoma [90], suggesting that GDF15 may serve as a cancer biomarker and potential therapeutic target in advanced cancers. However, targeting GDF15 must be carefully considered, given its possible role in the growth suppression of early-stage cancers. Increased metastasis, invasiveness, proliferation, and migration have been observed in the presence of GDF15 overexpression. However, the signaling pathways contributing to these biological events appear to vary between cancer types and remain largely uncharacterized.
GDF15 stimulates proliferation via ERK activation in prostate cancer cell lines [91–93]. GDF15 stimulation has been shown to activate phosphorylation of the receptor tyrosine kinase human epidermal growth factor receptor 2 (erbB2/HER2) [79, 94]. Src inhibition partially disrupts this phosphorylation [79], suggesting that GDF15 may mediate crosstalk to HER2 from another unidentified receptor. GDF15 stimulation results in reduced response to the HER2 monoclonal antibody, trastuzumab [79], and knockdown of GDF15 improves response to trastuzumab [79]. MAPK and PI3K/mTOR signaling pathways are activated by GDF15 [78, 79]; for example, mTOR inhibition by rapamycin overcomes the increased invasive phenotype of GDF15-stably transfected ovarian cancer cells [78]. Further, this GDF15-mediated invasion is matrix metalloproteinase (MMP)-dependent [78]. Increases in gelatinolytic activity and upregulation of the urokinase plasminogen activator (uPA) system are also linked to GDF15-mediated invasiveness in gastric cancer cell lines but in an ERK1/2-dependent manner [95]. The role of TGF-β receptor in this process and the exact signaling mechanisms that mediate resistance and invasion in GDF15-overexpressing breast or ovarian cancer cells remain to be established.
GDF15 serum levels are increased in colon cancer patients treated with non-steroidal anti-inflammatory drugs in association with reduced tumor recurrence [80]. Non-synonymous protein-coding single nucleotide polymorphisms (nsSNPs) that cause a cytosine-to-guanine (C-to-G) substitution at exon 202 of the GDF15 precursor protein have been identified in patients with prostate cancer. This substitution, known as H6D, is associated with a reduced risk of developing prostate cancer [96–98]. However, one study showed that prostate cancer patients with this CG mutation suffered a higher mortality rate versus the CC genotype [97]. Thus, the exact functional changes induced by the mutation and the validity of this genetic change as a predictor of survival or cancer biomarker remain to be determined.
The strongest evidence that GDF15 suppresses tumor growth has been obtained in mouse models in which GDF15 overexpression decreased tumorigenesis in models of lung cancer [99], breast cancer [100], prostate cancer [101], glioblastoma [102], and colon carcinoma [103]. Further, GDF15 transgenic mice developed fewer carcinogen-induced lung [99] or colorectal [103] tumors relative to control mice. Cross-breeding of GDF15 transgenic and transgenic adenocarcinoma of the mouse prostate (TRAMP) mice resulted in significantly fewer tumors, which were lower in grade compared to those in the control TRAMP mice. However, tumors in GDF15/TRAMP mice showed increased metastasis [93]. These results are particularly intriguing given the paradoxical results that have been reported for GDF15 in the literature. The data suggest that GDF15 may function as a growth suppressor in pre-malignant or early stages of cancer, but that a pro-growth/pro-invasive phenotype is stimulated by GDF15 in advanced tumors.
GDF15 also has putative anti-inflammatory activities. GDF15 appears to suppress secretion of inflammatory cytokines from lipopolysaccharide-treated macrophages [63]. Indeed, GDF15 transgenic mice exposed to a carcinogen show reduced lung tissue inflammation than control mice, mediated by p38 MAPK inhibition, and increased caspase activity in lung cancers [99]. The primary receptor for GDF15 remains to be identified. Structural similarity with TGF-β suggests that GDF15 may be a ligand for the TGF-β receptor. Phosphorylation of the TGF-β substrate Smad2 occurs in response to GDF15 stimulation in breast cancer cells [79]; however, direct evidence of the receptor has not yet been provided in the literature. Further, the multiple forms of endogenous versus secreted GDF15 may have differential functions, including as a growth suppressor, mediator of invasion, and anti-inflammatory factor, which complicates drug development efforts targeting GDF15. However, its high circulating levels in metastatic forms of various cancers suggest that it may be a relevant predictor of disease progression and possibly a novel molecular target in metastatic cancers that are refractory to standard therapies.
3.6 AT-rich interactive domain 1A (ARID1A)
The switching defective/sucrose nonfermenting (SWI/SNF) complex is an ATP-dependent, chromatin-remodeling, multiple-subunit enzyme critical for many biological processes including cellular differentiation and proliferation [104, 105]. Several subunits of the complex are crucial for proliferation control and function as tumor suppressors in various cancer types [106]. It is estimated that loss of SWI/SNF complex components is a critical event in carcinogenesis in 10–20% of solid tumors [107]. ARID1A encodes a human homolog of yeast SWI1, which contains a DNA-binding motif (AT-rich interactive domain, ARID) and is an integral member of the SWI/SNF complex.
Mutations in ARID1A have been detected in a wide variety of human cancers, with the highest mutation frequency (~50%) occurring in carcinomas arising from endometriosis or endometrial tissue. ARID1A mutations have also been found in renal, gastric, and breast tumors, medulloblastoma, clear-cell ovarian carcinoma, endometrioid ovarian carcinoma, endometrioid endometrial cancer and transitional cell carcinoma [108–118].
ARID1A is located on chromosome 1p and its protein product is predominantly localized to the nucleus. ARID1A was initially discovered to be a p300/cAMP response element binding protein (CBP)-associated partner that participates in recruitment of the SWI/SNF complex to specific transcription sites [119]. ARID1A contains an AT-rich DNA-interacting domain (ARID) and a glucocorticoid receptor binding domain which stimulates glucocorticoid receptor-dependent transcriptional activation [120].
ARID1A mutation is highly associated with loss of ARID1A expression. However, ARID1A protein expression is also low or absent in some tumors that lack mutations in ARID1A. ARID1A is a nucleo-cytoplasmic protein whose stability depends on its subcellular localization. Nuclear ARID1A is less stable than cytoplasmic ARID1A because ARID1A is rapidly degraded by the ubiquitin-proteasome system in the nucleus [121]. Another study showed that the promoter region of ARID1A is highly methylated in many invasive breast cancers. Promoter hypermethylation correlates with decreased expression of ARID1A in invasive ductal carcinomas of the breast [122]. These results demonstrate that there are multifaceted mechanisms to regulate ARID1A expression that include ubiquitination and promoter hypermethylation.
ARID1A suppresses cancer cell growth through several distinct mechanisms: 1) Inhibition of cancer cell proliferation: restoration of wild-type ARID1A expression suppressed cellular proliferation, colony formation and tumor growth in mice, whereas gene silencing of ARID1A in non-transformed epithelial cells enhanced cellular proliferation and tumorigenicity [110, 121, 123]; 2) Differentiation: ARID1A knockdown disrupted differentiation of cultured osteoblasts. The C-myc promoter is a direct target of mammalian ARID1A containing the SWI/SNF complex during preosteoblast differentiation. ARID1A is required for repression of C-myc during differentiation [124]; 3) Apoptosis: shRNA-mediated ARID1A knockdown inhibited both Fas-induced caspase-8 cleavage and FAS-induced mitochondrial leakage and led to inhibition of Fas-mediated cell death in Jurkat T cells. Knockdown of ARID1A in a leukemia cell line also conferred resistance to Fas-mediated apoptosis [125]; 4) Cell adhesion: The SWI/SNF complex regulates the expression of several important cell-adhesion proteins, such as CD44 and E-cadherin, as well as extracellular matrix proteins including MMPs and integrins [107]. These proteins are known to play important roles in tumor progression and metastasis; 5) DNA repair: SWI/SNF is recruited to double-strand DNA break sites and is required for efficient DNA repair in vivo [105]; and 6) Immune surveillance: Major histocompatibility complex (MHC) class I and II gene expression is regulated by SWI/SNF [126]. Loss of SWI/SNF function may inhibit immune surveillance and decrease detection of tumor cells.
Finally, ARID1A mutation or loss of ARID1A expression has prognostic and predictive value in ovarian clear-cell carcinoma, gastric cancer, breast cancer, and bladder cancer [123] and loss of ARID1A in ovarian clear cell carcinoma correlated with worse prognosis in patients treated with platinum-based chemotherapy [127].
3.7 Notch pathways
Over the past decade, oncologic signaling pathways (K-Ras, Wnt, β-catenin, etc) have emerged as being critical to the development of neoplasia. In addition to conventional pathways, primordial embryonic pathways have recently been thought to be integral to the initiation and maintenance of carcinogenesis. These include the Hedgehog and Notch embryonic pathways. In the process of carcinogenesis, aberrant regulation of these pathways leads to neoplasia. In addition to contributing to the process of carcinogenesis, dysregulation of Notch signaling has been shown to mediate chemotherapy resistance, facilitate epithelial to mesenchymal transition (EMT), and also maintain the cancer stem cell population.
Structurally, Notch signaling involves a transmembrane receptor and a transcription factor, which interacts with other nuclear proteins to control gene expression, thus transmitting growth and proliferation signals to the cell. Notch receptors are represented by four homologs in mammals (Notch1-Notch4), and contain a large extracellular domain and an intracellular signaling domain (NICD). Activation of Notch occurs through ligand binding. Two Notch ligand families, Jagged and Delta, have been described in mammals with five ligands identified to date (Jagged 1, 2, and Delta 1, 3, 4). After ligand binding, two successive proteolytic cleavage steps occur. The first step is mediated extracellularly by ADAM/TACE (a disintegrin and metalloprotease/tumor-necrosis factor (TNF) α converting enzyme) and occurs at the S2 cleavage site. The second step occurs at the S3 cleavage site and is mediated intramembranously by the γ-secretase complex, consisting of a catalytic subunit (presenilin 1 or 2), and accessory subunits (nicastrin, presenilin enhancer 2 [Pen-2], and anterior pharynx-defective 1 [Aph-1]). The resulting active form of NICD translocates to the nucleus where it binds a transcriptional repressor known as C promoter-binding factor (CBF-1, also known as recombination signal binding protein or immunoglobulin kappa J RBPJ), or CBF-1/suppressor of Hairless/Lag1 (CSL), converting it to a transcriptional activator. This NICD-CBF1 complex then activates the transcription of several downstream Notch target genes, such as Myc, p21, and Hes (hairy/enhancer of split) family members (for a review of the Notch signaling pathway and its role in cancer see [128–130]), which in turn act as transcriptional regulators of further downstream genes [131–134]. In the absence of NICD, the CBF-1 protein binds to specific DNA sequences in the regulatory elements of various target genes and represses transcription of these genes by recruiting histone deacetylases and other components to form a co-repressor complex.
In tumor types with activating mutations of Notch, blocking Notch signaling via γ-secretase inhibition produces a slower growing, less transformed phenotype in human cancer cells in vivo. Inappropriate activation of Notch signaling in T-cell acute lymphoblastic leukemia [135, 136], breast cancer [137–139], melanoma [140–142] and lung cancer [143–145] has been shown to result in stimulation of proliferation, restriction of differentiation and prevention of apoptosis. Overexpression of Notch also occurs in other hematologic malignancies, including B-cell malignancies [128]. In contrast, in squamous epithelial tumors such as SCC of the oral mucosa or skin, inactivating mutations of Notch1 are more common and leave the Wnt signaling pathway unopposed, so that continuous growth ensues. In squamous epithelium, Notch1 signals the cell to undergo differentiation at the basal suprabasal junction. The development of benign skin tumors has been observed in patients taking γ-secretase inhibitors because of Notch1 inhibition.
A variety of cancers have been characterized by aberrant Notch signaling, which serves an oncogenic function [146, 147]. This association was first described in T cell acute lymphoblastic leukemia where it was found that point mutations of the Notch receptors lead to constitutive over-activation [147]. More recently, over the past decade, several studies have emerged suggesting that dysregulated Notch activity is also involved in the inception and maintenance of other human cancers such as glioma, melanoma, breast cancer, pancreatic cancer, medulloblastoma, and colorectal carcinomas [131, 148–150]..
More recently, the role of Notch as an important tumor suppressor has been illustrated in the pathogenesis of pancreatic cancer. Pancreatic cancer is a highly aggressive malignancy with a very poor prognosis. Effective treatment options are very limited largely due to the innate chemotherapy and radiotherapy resistance that characterizes this aggressive neoplasm. Interestingly, Notch signaling has been found to be important in the pathogenesis of chemo-resistance and radio-resistance of pancreatic cancer and also in the process of pancreatic carcinogenesis from initiation to cancer formation and maintenance. The study of Notch function in the pancreas, however, has been limited by early embryonic lethality of mice with Notch signaling deficiency, and thus most data exist for early pancreatic organogenesis. It has been shown that activation of Notch 1 prevented exocrine and endocrine differentiation of pancreatic progenitor cells, leaving them in an undifferentiated state [151, 152]. Thus Notch signaling regulates cell fate decisions in both exocrine and endocrine lineages during organogenesis. The role of Notch during later embryonic stages and in adult tissue homeostasis is also being investigated. In the adult pancreas, Notch is normally suppressed except for in the centroacinar cells [153], while Notch target genes and signaling molecules are upregulated in invasive pancreatic cancer in addition to pancreatic precursor lesions from both mice and humans, suggesting that Notch might play an important role in the process of pancreatic carcinogenesis. Moreover, aberrant activation of the Notch signaling pathway has also been demonstrated in several transgenic models of pancreatic cancer [154].
3.8 Evading apoptosis
Evasion of apoptosis is a hallmark of cancer [43]. During proliferation and/or growth, cancer cells encounter a variety of unfavorable conditions, such as an insufficient supply of growth factors, oxygen, and other nutrients. Under these harsh circumstances, cells generally undergo regulatory programs that induce cell cycle arrest, apoptosis, and/or other types of programmed cell death; many of these programs are modulated by the actions of various tumor suppressor genes, such as p53 and Rb. However, cancer cells can circumvent these programs by activating several pathways that promote cell survival and proliferation. Of these bypassing pathways, the insulin-like growth factor receptor (IGF-1R)-mediated signaling cascade, which is regulated by ligands, receptors, and IGF-binding proteins, plays a key role in sustained cell survival and proliferation and the evasion of apoptosis. Accordingly, a number of clinical trials directly targeting IGF-1R have been conducted in various cancer types; however, these have met with only modest or unsustained efficacy through yet to be identified mechanisms.
Recent studies have implicated IGF-1R-mediated signaling as a main player in the control of cancer cell proliferation, growth, and survival. The IGF-1R signaling axis is composed of ligands (IGF1 and IGF2), receptors (IGF-1R, IGF-2R, and IR), and IGF-binding proteins (IGFBPs) [155]. IGFs are polypeptides that are structurally similar to insulin (approximately 50% homology to insulin) [156]. IGFs are mainly produced in the liver upon stimulation of growth hormone (GH) and influence autocrine, paracrine, and endocrine systems [155]. IGF1 displays high affinity for IGF-1R and the IGF-1R/insulin receptor (IR) hybrid receptor, while having a relatively low binding capacity for IGF-2R or IR [155, 157]. In contrast, IGF2 possesses high binding affinity for the IR-A isoform as well as IGF-1R, IGF-2R, and the IGF-1R/IR hybrid receptor [155, 157]. IGF-1R and IGF-2R are glycoproteins located on the cell membrane. IGF-1R is a receptor tyrosine kinase that exists as a tetrameric α2β2 complex and can be associated with IR to form the IGF-1R/IR hybrid receptor [158]. In contrast, IGF-2R is a monomer with no tyrosine kinase activity and the binding of IGF2 to IGF-2R results in the termination of IGFR-mediated signaling activation. Activation of IGF-2R exerts anti-proliferative and pro-apoptotic activities [159] via a variety of mechanisms, such as binding and subsequent internalization and degradation of IGF-2 [158], surface activation of the latent TGF-β [160], or serving as a receptor for retinoic acid [161].
Binding of an IGF to its receptor induces autophosphorylation of the receptor, stimulation of intrinsic tyrosine kinase activity, and phosphorylation of cellular substrates including insulin receptor substrate 1 (IRS-1), leading to gene activation and ultimately to proliferation or differentiation of cells [155]. Two distinct signal transduction pathways for IGF-IR with major roles in transmitting the cellular effects of IGFs have been identified: the Ras/Raf/MAPK and the PI3K/Akt pathways [155, 162]. PI3K, which is also a Ras mediator, phosphorylates the D3 position of phosphatidylinositol on PI4P and PI(4,5)P2 to produce PI(3,4)P2 and PI(3,4,5)P3 [163]. This mechanism is negatively regulated by the PTEN tumor suppressor that dephosphorylates the 3′ sites of PI(3,4)P2 and PI(3,4,5)P3 [163]. PI(3,4,5)P3 and PI(3,4)P2 recruit intracellular proteins containing the pleckstrin homology (PH) domain to the cytoplasmic membrane, which is an essential event in the activation of PI3K-dependent kinases such as pyruvate dehydrogenase kinase (PDK-1) and Akt/PKB [164, 165]. The serine/threonine protein kinase Akt is a direct target of PI3K. Three members of this family, Akt1/PKBα, Akt2/PKBβ, and Akt3/PKBγ, are activated by growth factors, integrins, and signals initiated by the stimulation of GPCR [165–168]. Activated Akt promotes cell survival and blocks apoptosis by phosphorylating substrates such as Bad, caspase-9, human telomerase reverse transcriptase subunit, transcription factor FKHRL1, GSK-3β, and IκB kinases [164, 169]. Activated IGF-1R recruits the adaptor protein Shc, which, in turn, mediates the binding of growth factor receptor-bound protein 2 (Grb2) via its SH2 domain. Grb2 further recruits a guanine nucleotide exchange factor son of sevenless (Sos), which then activates Ras through converting bound GDP to GTP. Activated Ras stimulates the sequential Raf/MEK/ERK cascade, which upregulates the phosphorylation and activation of target transcription factors c-jun and ETS domain-containing protein (Elk-1), eventually leading to the promotion of cell cycle progression through modulating cyclin D1, p21, and p27 expression.
High levels of IGF-1R activation have been observed in early stage lung carcinogenesis [170] and in several sarcoma subtypes [171]. A variety of stimuli, such as platelet-derived growth factors (PDGF) and fibroblast growth factor (FGF), hormones (androgen and estrogen), and various transcription factors, such as Sp1, NF-κB, and estrogen receptor-α (ERα), have been reported to stimulate the promoter activity of the Igfr gene, resulting in increased levels of IGF-1R expression [172–175].
The IGF system is also regulated by six IGFBP family members that bind to IGFs in the extracellular milieu with high affinity and specificity, thus reducing the bioavailability of IGFs [176, 177]. More than 99% of circulating IGF is bound to IGFBPs, and at least 75% of bound IGF is carried as a trimeric complex composed of IGFBP-3 and the acid labile subunit [177]. At the tissue level, IGFBPs interact either with extracellular matrix constituents (IGFBP-2 and IGFBP-5) or directly with cell membranes (IGFBP-1 and IGFBP-3), thereby regulating the interaction between IGFs and IGF-IR [178]. With the exception of IGFBP-6, which binds to IGF-2 with two times greater affinity than it does to IGF-1, the IGFBPs bind to IGF-1 with greater affinity, thereby regulating the mitogenic and anti-apoptotic activity of IGFs [176, 178, 179]. In addition to their modulatory role in IGF action, IGFBP-3 and IGFBP-5 have IGF-independent antiproliferative and pro-apoptotic effects in a variety of cancers [180–182]. Direct functional interactions between IGFBP-3 and the retinoid X receptor (RXR) have been shown to regulate transcriptional signaling and apoptosis [183]. IGF-independent antiangiogenic and antiproliferative effects of IGFBP-3 in human lung and head and neck cancer cells are also possible [184, 185]. IGFBP-3 action is modulated by posttranslational modifications, including proteolysis, phosphorylation, and glycosylation [181]. The cleavage of IGFBPs by a variety of protease families including kallikrein-like serine proteases, cathepsins, and MMPs reduces the affinity for IGFs [179]. However, the role of these posttranslational modification and proteolytic processing events in the regulation of IGFBP stability and function appears to be virtually unexplored.
A number of tumor suppressor genes, such as p53, PTEN, breast cancer gene 1 (BRCA1), Von Hippel–Lindau (VHL), Wilms tumor 1 (WT1), and Klotho, are negative regulators of IGF-1R transcription and/or activation [186–188]. Studies have revealed, however, that these tumor suppressors are frequently mutated in several types of human cancers [189]. An inherited germline mutation in the Rb gene is the first identified tumor suppressor gene implicated in several types of neoplasia, including familial retinoblastoma, small-cell lung cancer, and osteosarcoma [7, 189]. Therefore, loss or mutation of these tumor suppressor genes may contribute to evading growth-inhibitory and/or cell death signals in tumors that succeed in maintaining sustained proliferation and progressing to states of high-grade malignancy.
IGFs regulate cell growth, differentiation, and apoptosis [190]. Imbalance of these diverse processes favors uncontrolled cell proliferation, leading to malignant transformation. In addition, IGFR signaling facilitates angiogenesis and cancer cell metastasis by inducing angiogenic factors and proteases, including hypoxia-inducible factor-1α (HIF-1α) expression, which promotes the expression of vascular endothelial growth factor (VEGF) [191], interleukin-8 (IL-8) [192, 193], MMPs [190, 194] and uPA [190]. In addition, Akt, a downstream protein of IGF-1R-mediated signaling, enhances MDM2-mediated ubiquitination and proteosomal degradation of p53 [195]. A recent report indicates that activated IGF-1R increases the expression of oncogenic initiation of differentiation 2 (Id2) via upregulation of PI3K-Akt activity [196]. These findings indicate a possible oncogenic role of IGFR. IGFR signaling is also involved in intrinsic or acquired resistance to various conventional anticancer chemotherapies, radiation therapy, and recently developed molecularly-targeted anticancer drugs, including tyrosine kinase inhibitors (TKIs) blocking EGFR, HER2, or BRAF [197–200]. Increased IGF2 expression is also found in paclitaxel-resistant ovarian cancer cells [201]. Taken together, these findings implicate IGF-1R as a promising target for anticancer therapy.
3.9 Tumor suppressor gene – context dependent functions of Krüppel-like factor 5 (KLF5)
Deletion of the long arm of human chromosome 13 (13q), especially the region involving 13q21, is the second most frequent chromosomal deletion revealed by comparative genomic hybridization among a large number of different types of human tumors [202]. After analyzing the genomes from hundreds of human prostate and breast cancers, the deletion at 13q21 was mapped to 142-kilobases of the smallest region of overlap, in which the KLF5 gene was the only complete gene and was thus identified as a reasonable candidate for the 13q21 tumor suppressor gene [203, 204]. The majority of KLF5 deletions in cancers are hemizygous, i.e., one of the two KLF5 gene copies is deleted. It has been shown that KLF5 needs both copies to yield sufficient product, and deletion of one copy causes functional insufficiency – haploinsufficiency [205] – so hemizygous deletion impairs KLF5 function during tumorigenesis.
In addition to chromosomal deletion, excess degradation at the protein level has been identified as another common mechanism of KLF5 inactivation during tumorigenesis. KLF5 protein is ubiquitinated and degraded by the ubiquitin proteasome pathway [206], and the WW domain containing E3 ubiquitin protein ligase 1 (WWP1) mediates the ubiquitination of KLF5 [207]. Interestingly, the WWP1 gene is located at a chromosomal region that undergoes frequent copy number gains in human cancers, and WWP1 is indeed often amplified and overexpressed in human cancers, causing excess degradation of KLF5 protein [208, 209]. Therefore, KLF5 is inactivated by two mechanisms: genomic deletion and excess protein degradation, indicating that KLF5 undergoes frequent functional inactivation during tumorigenesis and thus should be considered a tumor suppressor gene. Expression of KLF5 mRNA is frequently reduced or absent in prostate and breast cancer cell lines [203, 204]. In gastric cancer, loss of KLF5 expression occurs more frequently in late stage tumors, in larger tumors, and in tumors with lymph node metastasis [210].
Consistent with a tumor suppressor function, KLF5 has also been found to inhibit cell proliferation and suppress tumorigenesis. Inhibition of cell proliferation has been shown for cancer cell lines from the esophagus, prostate, breast, and epidermis [204, 211, 212], as well as for non-tumorigenic epithelial cells [213]. In prostate cancer cells, KLF5 has been shown to suppress tumorigenesis in xenograft models, and the suppression was suggested to require ERβ and its association with KLF5 and the transcription coactivator CBP on the promoter of FOXO1 and subsequent activation of FOXO1, which induces anoikis in prostate cancer cells thereby suppressing tumor growth.
In contrast, KLF5 can also be tumor promoting, and the reverse of its function can be determined by posttranslational modification, specifically the acetylation of lysine 369 (K369). The oncogenic activity of KLF5 was originally suggested by in vitro studies in which KLF5 was shown to be upregulated in oncogenic H-Ras-transformed NIH3T3 cells [214], and overexpression of KLF5 promoted cell proliferation and induced the transformation of fibroblasts [215] and IEC-6 intestinal epithelial cells [216]. In the TSU-Pr1 bladder cancer cell line, expression of KLF5 promotes cell proliferation and tumorigenesis by inducing the expression of many genes [217]. The tumor promoting function of KLF5 has also been shown in breast cancer cells [218]. Knockdown of KLF5 inhibits multicellular tumor spheroid formation in vitro [219]. In transgenic mice, overexpression of KLF5 promotes the proliferation of basal epithelial cells, but does not produce tumors [220]. In the epidermis, expression of KLF5 is at relatively higher levels in keratinocytes, and overexpression of KLF5 in the basal layer of the epidermis affects epidermal development and disrupts epithelial-mesenchymal interactions necessary for skin adnexae formation [221]. KLF5 promotes cell proliferation through accelerating the G1/S and G2/M cell cycle progression and other mechanisms [216, 217]. Regulation of cell cycle regulators by KLF5 includes the induction of cyclin D1, cyclin B1, Cdc2, Myc, EGFR etc and the inhibition of p27, p15 etc [217, 222, 223].
How KLF5 can be both pro- and anti-tumorigenic is unknown at present. However, a mechanism has been suggested for the reversal of KLF5 functions in gene regulation and cell proliferation control. Using a cultured epidermis epithelial cell line – HaCaT, a model widely used to dissect the TGF-β signaling pathway – it was demonstrated that KLF5 is a cofactor for TGF-β, which inhibits cell proliferation and suppresses early stage tumorigenesis. As reported in several publications [213, 224, 225], without TGF-β, KLF5 forms a transcriptional complex with other transcription factors to repress cell cycle inhibitors such as the p15 CDK inhibitor and induce cell cycle promoting genes such as Myc to promote cell proliferation. When TGF-β is present, KLF5 forms a different transcriptional complex with a reversed function: inducing p15 transcription but repressing Myc transcription, which results in the inhibition of cell proliferation. The molecular basis for the reversal of KLF5 function in gene regulation and cell proliferation control is TGF-β-induced acetylation of KLF5, because interruption of KLF5 acetylation by mutating its acetylation residue prevents the functional reversal of KLF5 [213, 224, 225]. In mouse prostate tissues, unacetylated KLF5 (unAc-KLF5) is expressed in basal or undifferentiated cells, whereas acetylated KLF5 (Ac-KLF5) is expressed primarily in luminal and/or differentiated cells [226], which is consistent with a pro-proliferative function of unAc-KLF5 and anti-proliferative function of acetylated KLF5 in cultured cells.
Based on the available information described above, Ac-KLF5 and unAc-KLF5 have opposing functions in gene regulation and cell proliferation, which could correspond to its opposing functions during tumorigenesis: Ac-KLF5 is responsible for the tumor suppressor function of KLF5 whereas unAc-KLF5 is responsible for the oncogenic function of KLF5 during tumorigenesis. It is well established that the function of TGF-β switches from that of a tumor suppressor in the early stages of tumorigenesis to a tumor promoter in late stage tumor progression [227]. How TGF-β switches function is an intriguing question, and some studies have been published to address this question. It is possible that KLF5 is essential for TGF-β’s tumor suppressor function, and interruption of TGF-β-induced KLF5 acetylation is a key to the reversal of TGF-β function during tumorigenesis. These predictions are currently under investigation.
4. Safety considerations and multi-targeted approach to chemoprevention and therapy with natural compounds
The Latin adage primum non nocere (first, do no harm) that characterizes medical practice over centuries applies perfectly to the prevention/therapy of cancer or any other disease. In particular, recipients of chemopreventive drugs are not cancer patients, but are healthy people who are at high risk for developing cancer such as smokers or those with hepatitis B. Since these essentially healthy people will receive the chemopreventive treatment for a long period of time, the toxicity of the agents severely impacts patient accrual and retention for prevention trials. Toxicity is also a major concern for chemotherapeutic drugs. An ideal compound for both chemoprevention and therapy should be nontoxic, orally active, economical, and easily available. Unlike synthetic compounds, natural compounds have been found safe in long term human consumption in the diet and many of them exhibit potential chemopreventive and anti-tumor effects in preclinical studies [228, 229]. Moreover, the safety and tolerability of many of these natural compounds in pharmacological doses has been established through phase I safety trials.
As described earlier, multiple genetic and epigenetic changes accumulate throughout the carcinogenesis process. While targeting one or two of these pathways using specific agents may not be effective or durable, most natural compounds hit multiple targets [228, 229]. Therefore, long-term use of natural compounds can be an effective and rational strategy for populations at high risk of developing cancers. Clinicians are also paying increasing attention to diet-derived chemopreventive agents as a result of patient preference. Since the recognition of chemoprevention, the National Cancer Institute (NCI) has investigated or sponsored more than 1,000 different potential agents for chemoprevention, of which only about 40 promising agents have been moved to clinical trials including several natural agents http://prevention.cancer.gov/programs-resources/resources/agents. Most preclinical studies using natural compounds were conducted using fully transformed cancer cell lines due to the lack of premalignant cell lines, suggesting that these compounds can also be used for treatment of cancers.
5. Prophylactic and therapeutic potential of targeting anti-growth signaling with natural compounds
Carcinogenesis is a lengthy process, sometimes taking decades for normal cells to transform into invasive cancers. Because of the lengthy transformation process, with several precancerous pathologic stages preceding the change to invasive cancer, there are enormous opportunities to intervene, with the aim to reverse or slow down the transformation process [1, 230]. Such intervention is known as cancer prevention and the use of natural or synthetic agents at pharmacological doses for cancer prevention is called chemoprevention. Chemoprevention is a cost-effective alternative to chemotherapy and its successful implementation may save millions of lives worldwide. An outstanding review article written by Haddad and Shin [231] elegantly describes the general carcinogenesis process with associated genetic and pathologic changes and the article by William et al. [230] distinguishes between chemoprevention and chemotherapy. While the purpose of chemoprevention is to eliminate or slow down the progression of intraepithelial neoplastic or precancerous lesions to invasive cancer, the purpose of chemotherapy is to stop or slow down the growth of fully transformed cells or to eliminate them through activating cell death pathways. Since anti-growth signaling via tumor suppressors challenges tumorigenesis by inhibiting the growth of damaged cells, repairing damage, or eliminating damaged cells through apoptosis or other forms of cell death mechanisms, the reactivation of these pathways using chemical compounds or genetic approaches has high prophylactic and therapeutic potential. Activation (if normal and wild-type) or reactivation (if inactivated via reversible process) of tumor suppressor genes and/or pathways might serve as crucial events for effective chemoprevention and therapy, as discussed later.
5.1 The Rb pathway
The RB-E2F pathway is one of the most important tumor suppressor pathways frequently inactivated or lost in human cancers. As described in section 3.1, Rb and p16ink4a are critical regulators of proliferation. This makes the cyclinD1/CDK4/6 complex an interesting target for chemopreventive and chemotherapeutic drug development. Green tea-derived galloyl polyphenol and epigallocatechin gallate (EGCG) were shown to decrease the phosphorylation of Rb, and as a result, cells were arrested in G1 phase [232]. Several studies also revealed that green tea polyphenols strongly inhibited CDKs or cyclin D1 to exert their chemopreventive (anti-proliferation and cell cycle arrest) effects, which might occur through decreasing the phosphorylation of Rb proteins [233, 234]. Restoration of Rb expression by curcumin in cervical cancer cells [235] and inhibition of the Rb pathway by enhancing CDKN2A/p16 and suppressing phosphorylated Rb in glioblastoma [236] were reported and associated with the chemopreventive potential of curcumin. It has been reported that 1,25-dihydroxivitamin D, the most biologically active metabolite of the micronutrient vitamin D, exerts potent effects on the Rb signaling axis. For example, it suppresses PDGF-induced myocyte proliferation in vitro by inhibiting Rb and checkpoint kinase 1 (Chk1) phosphorylation [237]. In some tumor cells, the natural compound honokiol, derived from trees of the Magnolia genus, activated the production of reactive oxygen, leading to increased phosphorylation of Rb [238].
5.2 The p53 pathway
According to TCGA data available so far, p53 is the most frequently mutated tumor suppressor protein. However, for many tumors p53 is wild-type but inactivated via secondary mechanisms rather than by loss or mutation. Drugs are available and currently under clinical development which inhibit or remove the negative regulator MDM2, thus restoring p53 function leading to growth inhibition and induction of apoptosis. Some of these drugs are DS-3032b, RO5503781, RO5045337, SAR405838 etc. As discussed in section 3.2, viral oncogene-driven tumors also retain wild-type 53 and can be treated with DNA-damaging drugs such as cisplatin, doxorubicin, taxols etc or radiation therapy. Finally, drugs are also under development that reactivate mutant p53, for example, PRIMA-1 and APR-246.
Many natural agents exert their chemopreventive/anti-tumor effects through the induction of cell cycle arrest or apoptosis by activating the p53 pathway. Groups of investigators at Emory University and other institutions have extensively studied these molecular pathways and natural compounds. The green tea component EGCG was shown to activate p53 and its target p21 and Bax in prostate cancer cells with wild-type p53 [239], and Bax in breast carcinoma cells [240]. The vegetable-derived compound luteolin was shown to induce cell cycle arrest and apoptotic effects as well as increased chemosensitization effects associated with the activation of p53 and its targets p21, Bax, and PUMA [241]. Luteolin and a combination of luteolin and EGCG also induced mitochondrial translocation of p53 in lung cancer cell lines [242]. Another extensively investigated dietary chemopreventive agent, curcumin, was reported to activate p53 and its transcriptional target Bax in breast and bladder cancers [243, 244] and induce mitochondrial translocation in prostate cancer [245]. Resveratrol, a component of red wine and grape skin, also activated p53 and its target genes p21, p27, Bax, PUMA, MDM2, and cyclin G [246, 247]. Genistein, an isoflavone and dietary chemopreventive agent from soy also activated p53 and induced G2/M arrest and apoptosis in human malignant glioma cell lines through p21 induction [248]. Activation of p53 was also reported by glycyrrhizic acid in the colon of Wister rats [249], by oleanolic acid in HepG2 transplanted Balb/C mice [250], by amarogentin, a secoiridoid glycoside active component of the medicinal plant Swertia chirata, in a carbon tetrachloride (CCl4)/N-nitrosodiethylamine (NDEA)-induced liver carcinogenesis mouse model [251], by melatonin in cell culture models of MCF-7 and HCT116 [252] and by Kaempferol in A2780/CP70, A2780/wt, and OVCAR-3 ovarian cancer cell lines [253]. An emerging role for p73 activation by natural chemopreventive agents has also been reported. EGCG induced apoptosis by activating p73-dependent expression of a subset of p53 target genes including p21, reprimo, cyclin G1, PERP, MDM2, WIG1, and P53-induced gene 11 (PIG11) in mouse embryonic fibroblasts [254]. Upregulation of p73 was reported in response to EGCG in multiple myeloma cells [255]. Polyphenol-rich Aronia melanocarpa juice induces cell cycle arrest and apoptosis by redox sensitive activation of p73 [256].
Many natural compounds also suppress carcinogenesis by activating upstream or downstream components of the p53 pathway. Curcumin downregulates expression of MDM2 at the transcriptional level in a p53-independent manner, which ultimately upregulates p21 and induces apoptosis in a prostate cancer cell line [257]. Using yeast-based assays, gene reporter assay and computational docking, Leao et al. [258] reported that natural compounds α-mangostin and gambogic acid inhibited p53-MDM2 interaction by binding with MDM2. Disruption of p53-MDM2 interaction is critical for p53 stabilization. Such interaction is inhibited by other natural compounds [259, 260]. Herman-Antosiewicz et al. [261] reported activation of ATR/checkpoint kinase 1-dependent prometaphase checkpoint in cancer cells by the processed garlic constituent diallyl trisulfide. Vitamin D exerts its chemopreventive effects in a chemically-induced carcinogenesis mouse model by promoting expression of the DNA repair genes RAD50 and ataxia-telangiectasia mutated (ATM) and maintaining a positive feedback loop between ATM and vitamin D receptor [262]. The activation of p53 by luteolin also occurs through activation of ATM [242]. The chemopreventive agent selenium also activates DNA damage response by activating ATM [263]. Honokiol is active in cells with both wild type and mutant p53. Studies have shown that tumors with mutant p53 are particularly susceptible to honokiol, especially in the presence of oncogenic Ras. Honokiol decreases the level of Ras in the active GTP configuration [264].
5.3 Epigenetic silencing
Many tumor suppressor proteins are also inactivated via epigenetic silencing. Covalent modifications of histone proteins and DNA hypermethylation of promoters or CpG islands are important epigenetic silencing mechanisms through which tumor cells evade tumor suppressors, and epigenetic intervention is even possible at the precancerous stage [265]. Unlike genetic inactivation, epigenetic inactivation is reversible. DNA methyl transferases (DNMT) are key enzymes regulating DNA methylation and inhibition of such enzymes will reactivate tumor suppressors. Many natural polyphenols act as demethylating agents and reactivate tumor suppressors. In silico docking studies have revealed that curcumin may compete with the cofactor S-adenosyl methionine (SAM) for binding to the catalytic pocket of DNMT and induce global DNA hypomethylation in leukemia cells [266]. Curcumin also decreased p300, histone deacetylase (HDAC) 1 and 3 and p300 histone acetyltransferase (HAT) activity to modify chromatin acetylation [266–268]. Curcumin inhibited the expression of DNMT1 in vitro and in a xenograft model which subsequently reactivated the expression of p15(INK4B) tumor suppressor by promoter demethylation and induced cell cycle arrest and apoptosis [269]. In breast cancer cells, curcumin also reactivated the tumor suppressor RASSF1 through promoter demethylation by inactivating DNMT1 [270]. Human, animal and cell culture studies also revealed that green tea polyphenols are strong demethylating agents and reactivate tumor suppressor genes. Nandakumar et al. [271] reported that EGCG reactivated tumor suppressors p21 and p16INK4a by reducing DNA methylation and increasing histone acetylation. In an azoxymethane-induced APC(Min/+) mouse model of intestinal carcinogenesis, RXR-α is selectively downregulated through CpG hypermethylation, and administration of green tea as the sole source of beverage significantly increased the protein and mRNA levels of RXR-α as well as decreased CpG methylation [272]. In a cell culture model, EGCG was found to reverse the hypermethylation of p16(INK4a), retinoic acid receptor beta (RARβ), O(6)-methylguanine methyltransferase (MGMT), and human mutL homologue 1 (hMLH1) genes, leading to a subsequent increase in their mRNA and protein expression levels [273]. This demethylating activity of EGCG is associated with inhibition of DNMT1 through hydrogen bonding. Resveratrol and genistein also inhibited DNA methylation in cell culture and human intervention studies. Acetylation of STAT3 is elevated in tumors and inhibition of STAT3 by genetic mutation or by resveratrol treatment inhibited tumor growth by reactivating several tumor suppressor genes including ERα, CDKN2A, deleted in lung and esophageal cancer 1 (DLEC1), and STAT1 [274]. In a human intervention study in women with high risk for breast cancer, administration of trans-resveratrol was associated with decreased methylation of tumor suppressor RASSF1a [275].
5.4 The PTEN pathway
Loss of PTEN activates the PI3K-AKT pathway, which is currently an attractive target for drug development. So far, no PI3K inhibitor has obtained FDA approval, but several agents are in clinical trials, including BKM120, BYL719, RP6530, PF04691502, PF-05212384, MK2206 and others. Many natural compounds are also reported to inactivate the PI3K-AKT pathway either by activating PTEN or inactivating oncogenes that drive AKT activation. For example, the chemopreventive agent indole-3-carbinol (I3C), a natural indolecarbinol compound derived from the breakdown of glucobrassicin produced in cruciferous vegetables such as broccoli and Brussels sprouts, induced G1-phase cell-cycle arrest and apoptosis and inhibited the in vivo tumor growth rate by stabilization of PTEN in human melanoma cells that express wild-type PTEN, but not in cells with mutant or null PTEN genotypes [276]. Curcumin treatment significantly resulted in the inhibition of cell proliferation and an increase in the apoptosis rate through the upregulation of PTEN associated with a decrease in DNA methylation level via downregulation of DNMT3b in vivo and in vitro [277]. Many other studies also support that the PTEN-AKT pathway is a major in vitro and in vivo target of curcumin [278]. Reactivation of PTEN by resveratrol has also been reported [279].
5.5 The Hippo Pathway
Porphyrin molecules have been shown to inhibit YAP/TEAD activity, suggesting that the negatively-regulated targets of the Hippo pathway can be reduced [61]. Critically, the tumorous overgrowth that occurs when NF2 is conditionally disrupted in mice is inhibited with these molecules in vivo. The ligands glucagon and epinephrine enhance LATS1/2 activity, thus enhancing YAP inhibition through GPCR signaling [62]. This indicates it may be possible to enhance or potentiate the growth suppressive effects of the pathway using GPCR agonists, and that this branch of the GPCR family may be an important potential therapeutic target.
5.6 GDF15
Because of its potential growth suppressive function, GDF15 may inhibit cancer progression [83]. It is possible that endogenous GDF15 mediates the bulk of the growth suppressive activities, and the secreted mature dimer may mediate disease progression. There are multiple cellular forms of GDF15, including pro-GDF15 monomer, pro-GDF15 dimer, pro-peptide N-terminal fragment, and mature dimer [65]. The N-terminal pro-peptide fragment and the mature dimer are rapidly secreted [65, 83]. The mature dimer is believed to be the main bioactive form. However, it remains unclear whether the other forms are biologically active, or whether relative expression levels or ratios of the different forms affect the biological behavior of the cell. Further investigation is needed to establish the relative expression levels of each form in various cancer types and the functional differences between each form. These studies will be important for determining the overall suitability of GDF15 as a target in metastatic cancer.
The anti-cancer flavonoids resveratrol and genistein were shown to induce GDF15 expression in a p53-dependent manner in various types of cancer cell lines, including colorectal, osteosarcoma, and lung cancer cell lines [280, 281]. Another anti-cancer flavonoid, apigenin, was proposed to induce GDF15 expression in a p53-independent manner [282]. Induction of GDF15 in response to anti-cancer drugs and subsequent reductions in MCF-7 breast tumor xenograft volumes correlated with increased ERK1/2 phosphorylation [100]. Thus, MEK signaling and p53 function may be critical mediators of GDF15 expression and growth suppressive function.
GDF15 has been proposed to be a predictor of all-cause mortality [283–285]. Therefore, serum GDF15 levels may indicate the presence of a number of different life-threatening disorders or generally poor health. GDF15 serum evaluation may have a role in general health assessment and blood panels. Routine assessment of GDF15 serum levels may also serve as a minimally invasive way of predicting cancer presence or progression.
5.7 ARID1A
No drugs are currently available that directly target ARID1A. However, ARID1A could interact with several pathways to exert its tumor suppression activities and drug development is underway to target such pathways. These include: 1) PI3K-AKT pathway: ARID1A mutations were also significantly associated with the presence of PIK3CA-activating mutations in tumors, suggesting that ARID1A inactivation may synergize with PIK3CA activation to facilitate cellular transformation [110]. Another study showed that PIK3CA mutations and loss of ARID1A protein expression are early events in the development of cystic ovarian clear cell adenocarcinoma [286]. Liang et al demonstrated that ARID1A siRNA knockdown in endometrial cancer cell lines increased AKT phosphorylation, implicating ARID1A as a novel regulator of PI3K pathway activity [287]. The results indicate that ARID1A and PIK3CA mutations coexist and potentially cooperate in oncogenesis. 2) P21 pathway: p21 is a potent CDK inhibitor (CKI). The p21 (CIP1/WAF1) protein binds to and inhibits the activity of cyclin-CDK2 or -CDK1 complexes, and thus functions as a regulator of cell cycle progression at G1 [288]. p21 is downregulated by ARID1A shRNAs in endometrial cancer cell lines [121]. These findings suggest that ARID1A inhibits cancer cell progression by modulating p21 expression. Since loss of ARID1A upregulates AKT phosphorylation, compounds inhibiting AKT phosphorylation, such as resveratrol could be used for tumors harboring ARID1A loss.
5.8 Notch pathway
Notch inhibition appears to be an attractive target for compounds that might result in chemoprevention and treatment of several solid tumors that have aberrant Notch activation, such as pancreatic cancer, ALL, and brain tumors. Several Notch inhibitors have been evaluated. Since the formation of the NICD following Notch receptor cleavage by γ-secretase is a critical event in Notch signaling, γ-secretase inhibitors (GSI) are actively being investigated as Notch pathway inhibitors. In addition to preclinical studies in mice, several human clinical trials targeting different types of malignancies are currently being conducted. Notch pathways are also targets of many natural compounds. In colon cancer and melanoma cells, honokiol downregulated levels of activated Notch-1, its ligand Jagged-1, and the downstream target gene Hes-1 [289, 290] and can be used to target Notch pathways for therapeutic and preventive purposes. Withaferin A is another natural compound that potentially targets the Notch pathway and inhibits carcinogenesis [291].
5.9 Evading Apoptosis
IGF-1R signaling is a typical prosurvival pathway and is closely associated with hallmarks of cancer, such as sustained activation of growth-promoting signaling and evasion of tumor suppressor-mediated cell death. Accordingly, a number of IGF-1R-targeted agents, mainly anti-IGF-1R monoclonal antibodies (mAbs) and small molecule TKIs, have been evaluated in a variety of preclinical and clinical trials [292, 293]. Indeed, pharmacologic interventions that lead to disruption of the IGF-1R pathway have shown promising activities in inhibiting tumor development and progression as well as sensitizing anticancer therapies in preclinical models [294–296]. In addition, results from phase I or II clinical evaluations of IGF-1R-targeted agents, especially anti-IGF-1R mAbs, demonstrate significant clinical response [297]. However, the therapeutic benefits shown in a subset of patients enrolled in phase II or III clinical trials were unsustained and most of the patients seemed to acquire resistance to the IGF-1R-targeted therapies [297–299]. Furthermore, recent phase III trials of an IGF-1R mAb, figitumumab, were terminated due to an increase in serious adverse events and excess mortality in the experimental arm [297]. However, clinical trials in a number of cancer types are still in progress and the interpretation of previous and current clinical trials remains unclear [292].
Two major categories of drug resistance, de novo and acquired resistance, have been suggested to attenuate the antitumor effects of IGF-1R-targeted agents. Activation of alternative receptor- or non-receptor tyrosine kinases or downstream molecules through various mechanisms have been proposed to bypass IGF-1R signaling, accounting for de novo resistance [300]. Increased expression and/or activation of IR, EGFR, HER2, and VEGF receptor (VEGFR) have been observed in various human cancers and suggested as potential means to bypass the need for the IGF-1R pathway [301–303]. Recent preclinical studies using anti-IGF-1R antibodies suggest that Akt/mTOR is activated through yet to be identified mechanisms, and likely plays a vital role in the primary resistance to an anti-IGF-1R mAb cixutumumab [304]. Overexpression of IGFBPs, activation of heat shock protein 90 (Hsp90), and the EMT phenotype have been also implicated in the resistance to IGF-1R targeted therapies [300].
Given the myriad routes by which cancer cells resist IGF-1R-targeted agents, the clinical utility of IGF-1R-targeted anticancer therapies should be evaluated after the completion of extensive studies on the mechanisms underlying primary and acquired resistance to IGF-1R targeted agents, the discovery of biomarkers to predict the effectiveness of IGF-1R targeted therapies, and the development of effective combinational therapeutic regimens.
5.10 KLF5
KLF5 appears to be a unique tumor suppressor in the sense that its function is context-dependent. Its tumor suppressor function could be dependent upon its acetylation, which can be induced by TGF-β and possibly other signaling pathways. Once experimentally confirmed, how TGF-β and KLF5 become oncogenic will be better understood, and some unique targets could be identified for their potential use in the detection and treatment of malignant cancer.
6. Cross validation of prioritized targets and approaches with other hallmarks of cancer
Given the heterogeneity that is present in most cancers, it is our assumption that the complete arrest of the various subpopulations of immortalized cells in any given cancer will require simultaneous actions on mechanisms that are important for several aspects of cancer biology. We therefore believe that it is important to be able to anticipate synergies that might be achieved by acting on specific targets and with specific approaches (i.e., when contemplating an approach aimed at a broad spectrum of targets). Accordingly, in this review, the prioritized target sites and the approaches that have been identified (as potential methods to reach those targets) were all cross-validated by undertaking a background literature research. A team of researchers consisting of specialists in each cancer hallmark area specifically sought to determine the relevance of these targets and the nominated approaches across a number of important areas of cancer biology. In this regard, targets and approaches that were not only relevant for this area of study, but also relevant for other aspects of cancer biology (i.e., anti-carcinogenic) were noted as having “complementary” effects with symbol “+” in Tables 1 and 2, while those that were found to have pro-carcinogenic actions were noted as having “contrary” effects with symbol “−”. In instances where reports on relevant actions in other aspects of cancer biology were mixed (i.e., reports showing both pro-carcinogenic and anti-carcinogenic potential), the term “controversial” was used with symbol “+/−”. Finally, in instances where no literature support was found to document the relevance of a target site or approach in a particular aspect of cancer biology, we documented this as “no known relationship” with a symbol “0”. These validation results are shown in Tables 1 and 2.
Table 1.
Cross-validation of anti-growth signaling or tumor suppressor pathways with other hallmarks of cancer
| Priority targets for evasion of anti-growth signaling | The Rb pathway: Inactivation of E2F by down-regulation of hyper-phosphorylated Rb or inactivating CDKs. | The p53 pathway: Activation through up-regulation of wild type p53 | PTEN pathway: Activation by inhibiting PI3K-AKT | The Hippo pathway: Activation by inhibiting YAP/TEAD activity | Induction of GDF 15 through p53 activation | Activation of tumor suppressor ARID1A | Blocking NOTCH pathway in association with tumor suppressors | Inhibition of IGF-1R to restore tumor suppressor pathways |
|---|---|---|---|---|---|---|---|---|
| Other cancer hallmarks | ||||||||
| Genomic instability | + [309] | + [310] | + [311, 312] | 0 | − [313] | + [314] | 0 | 0 |
| Sustained proliferative signaling | 0 | + [315, 316] | + [317–320] | + [321–323] | 0 | + [324] | + [325] | + [326–330] |
| Tumor promoting inflammation | 0 [331] | + [332–335] | + [336, 337] | 0 | 0 | 0 | + [338] | + [339, 340] |
| Resistance to apoptosis | + [341] | + [342] | + [343] | + [344] | + [345] | + | + [346] | + [347] |
| Replicative immortality | + [348] | + [349, 350] | + [351] | 0 | 0 | 0 | + [352] | + [353, 354] |
| Dysregulated metabolism | + [355] | + [356–359] | + [360–362] | + [363] | 0 | + [364] | + [365] | 0 |
| Immune system evasion | 0 | + [366] | + [367] | 0 | 0 | 0 | 0 | 0 |
| Angiogenesis | + [368] | 0 [369] | + [370] | 0 | + [371] | 0 | − [372] | −[373] |
| Invasion and metastasis | + [374, 375] | + [376] | + [377] | + [323, 378] | + [379] | − [123, 380] | + [381, 382] | − [383, 384] |
| Interactions in the tumor microenvironment | − [385, 386] | + [387, 388] | + [389, 390] | + [391] | + [392] | 0 | + [393] | + [330] |
Foot notes: “+” indicates complementary; “−” contrary and “0” no known relationship
Table 2.
Cross-validation of potential therapeutic agents targeting anti-growth signaling with other hallmarks of cancer.
| Phytochemical approaches for evasion of anti-growth signaling | EGCG | Luteolin | Curcumin | Genistein | Resveratrol | Curcumin | Withaferin A | Deguelin |
|---|---|---|---|---|---|---|---|---|
| Other cancer hallmarks | ||||||||
| Genomic instability | + [309] | + [310] | + [311, 312] | − [313] | + [314] | + [311, 312, 394] | 0 | 0 |
| Sustained proliferative signaling | + [395, 396] | + [397, 398] | + [399] | +/− [400, 401] | +/− [400, 401] | + [399] | 0 | + [402] |
| Tumor promoting inflammation | + [403–405] | + [406, 407] | + [408–410] | + [411, 412] | + [413, 414] | + [408–410] | + [415] | + [416–418] |
| Resistance to apoptosis | + [419] | + [420] | + [421] | + [422] | + [423] | + [424] | + [425] | + [426] |
| Replicative immortality | + [427–429] | 0 | 0 [430, 431] | + [432–434] | + [435, 436] | + [430, 431] | 0 | 0 |
| Dysregulated metabolism | 0 | + [242, 437, 438] | + [236, 439, 440] | 0 | 0 | 0 | + [441] | 0 |
| Immune system evasion | + [442] | 0 | + [443] | 0 | 0 | + [443] | 0 | 0 |
| Angiogenesis | + [444] | + [445] | + [446] | + [447] | + [448] | + [446] | + [449, 450] | + [451] |
| Invasion and metastasis | + [452, 453] | + [454] | + [455] | + [456, 457] | + [458, 459] | + [460] | + [449, 461] | + [462] |
| Interactions in the tumor microenvironment | + [463, 464] | + [465, 466] | + [467–469] | + [470] | + [471] | + [467–469] | 0 | + [472] |
Foot notes: “+” indicates complimentary; “−” contrary; “+/−” controversial and “0” no known relationship
7. Conclusions and future directions
Anti-growth signaling, mainly mediated by tumor suppressor proteins, and immune surveillance are the two major hurdles that must be overcome for cancer cells to acquire limitless proliferation. Since tumor suppressors function to eliminate damaged cells or cancer cells, their activation is expected for successful cancer therapy. Most cancer drugs currently in use, including recently developed targeted drugs [305], activate anti-growth signaling to prevent cell proliferation. However, the high cost of treatment and the development of resistance to these agents are major challenges for physicians and patients. As discussed, cancer is a multi-factorial and heterogeneous process that involves the accumulation of multiple genetic and epigenetic changes. Resistance arises mainly due to the development of alternative pathways for cell survival and thus multi-targeted approaches with effects on multiple hallmarks of cancer are critical. On the other hand, cost is mostly associated with intellectual issues, investment in development and profit of manufacturing companies. Considering these factors, natural dietary compounds are highly attractive since they are readily available and most are multi-targeted, affecting multiple hallmarks of cancer. Therefore, future directions should explore the development of natural compounds, not only for prevention but also for therapy. Potency and bioavailability of natural compounds are the main barriers to their clinical application, therefore special attention should be given to combinatorial approaches of natural compounds with other natural or synthetic compounds either to achieve synergistic effects or to increase bioavailability. Several such combinations have shown high potential [242, 306]. In parallel, the development of potent analogs of natural compounds and their improved delivery by novel technologies such as nanotechnology are important other directions to be considered in the future. Importantly, proof-of-principle studies demonstrate that the delivery of natural compounds as nanoparticles can significantly improve their efficacy [307, 308]. Approaches to activate anti-growth signaling with synthetic and/or natural compounds promise to open new chapters for cancer prevention and therapy in the near future. Finally, although the “evasion of anti-growth signaling” is a critical component of the hallmarks of cancer, other hallmarks are similarly critical and a more integrative approach is necessary to simultaneously target not only anti-growth signaling but also other properties of cancer to combat this deadly disease.
Acknowledgments
We are thankful to Dr. Anthea Hammond for editorial assistance. The authors also acknowledge the efforts of “Getting to Know Cancer” in organizing and managing this large project.
Footnotes
8. Authors’ Contributions: Dr. Amin contributed to the introduction, p53 pathway, PTEN pathway and prevention/prophylaxis sections. He coordinated with the corresponding author and other contributing authors, and compiled the individual sections. Dr. Karpowicz contributed to the abstract and Hippo pathway sections, and to editing of the entire manuscript. Dr. Carey contributed to the introduction, Rb, p53 and PTEN pathway sections, and to editing of the manuscript. Dr. Arbiser and Dr. Chen contributed to manuscript design and editing. Drs. Nahta, Dong, Kucuk, Khan, Huang, and Lee wrote the GDF15, KLF5, prevention/prophylaxis, Notch, ARID1A, and IGF-1R sections, respectively. Dr. Reichrath compiled information about vitamin D. Drs. Kanya Honoki, Alexandros Georgakilas, Amedeo Amedei, Amr Amin, Bill Helferich, Chandra S. Boosani, Maria Rosa Ciriolo, Sophie Chen, Sulma Mohammed, Asfar S. Azmi, W Nicol Keith, Dipita Bhakta, Dorota Halicka, Elena Niccolai, Hiromasa Fuji, Katia Aquilano, S. Salman Ashraf, Somaira Nowsheen, Xujuan Yang and Alan Bilsland contributed to the cross validation study. Dr. Shin coordinated the whole manuscript writing process with the entire team from the inception of the chapter to the publication as the corresponding author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359(11):1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347(20):1593–603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 3.Lundberg AS, Randell SH, Stewart SA, Elenbaas B, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21(29):4577–86. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 4.Kohno T, Yokota J. How many tumor suppressor genes are involved in human lung carcinogenesis? Carcinogenesis. 1999;20(8):1403–10. doi: 10.1093/carcin/20.8.1403. [DOI] [PubMed] [Google Scholar]
- 5.Knudsen AGJ. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Nat Acad Sci, USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2(12):910–7. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 8.Reed AL, Califano J, Cairns P, Westra WH, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56(16):3630–3. [PubMed] [Google Scholar]
- 9.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1(14):1001–8. [PubMed] [Google Scholar]
- 11.Shangary S, Ding K, Qiu S, Nikolovska-Coleska Z, et al. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol Cancer Ther. 2008;7(6):1533–42. doi: 10.1158/1535-7163.MCT-08-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxwell JH, Kumar B, Feng FY, Worden FP, et al. Tobacco use in human papillomavirus-positive advanced oropharynx cancer patients related to increased risk of distant metastases and tumor recurrence. Clin Cancer Res. 2010;16(4):1226–35. doi: 10.1158/1078-0432.CCR-09-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licitra L, Perrone F, Bossi P, Suardi S, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24(36):5630–6. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 14.Levine AJ, Feng Z, Mak TW, You H, et al. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20(3):267–75. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 15.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15(2):164–71. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 16.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2(8):a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adorno M, Cordenonsi M, Montagner M, Dupont S, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Rufini A, Agostini M, Grespi F, Tomasini R, et al. p73 in Cancer. Genes Cancer. 2011;2(4):491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 21.Dotsch V, Bernassola F, Coutandin D, Candi E, et al. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2(9):a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichimiya S, Nimura Y, Kageyama H, Takada N, et al. p73 at chromosome 1p36.3 is lost in advanced stage neuroblastoma but its mutation is infrequent. Oncogene. 1999;18(4):1061–6. doi: 10.1038/sj.onc.1202390. [DOI] [PubMed] [Google Scholar]
- 23.Douc-Rasy S, Barrois M, Echeynne M, Kaghad M, et al. DeltaN-p73alpha accumulates in human neuroblastic tumors. Am J Pathol. 2002;160(2):631–9. doi: 10.1016/s0002-9440(10)64883-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez G, Garcia JM, Pena C, Silva J, et al. DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol. 2006;24(5):805–15. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- 25.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;59(13):3257–63. [PubMed] [Google Scholar]
- 26.Ng SW, Yiu GK, Liu Y, Huang LW, et al. Analysis of p73 in human borderline and invasive ovarian tumor. Oncogene. 2000;19(15):1885–90. doi: 10.1038/sj.onc.1203512. [DOI] [PubMed] [Google Scholar]
- 27.Tannapfel A, Wasner M, Krause K, Geissler F, et al. Expression of p73 and its relation to histopathology and prognosis in hepatocellular carcinoma. J Natl Cancer Inst. 1999;91(13):1154–8. doi: 10.1093/jnci/91.13.1154. [DOI] [PubMed] [Google Scholar]
- 28.Kovalev S, Marchenko N, Swendeman S, LaQuaglia M, et al. Expression level, allelic origin, and mutation analysis of the p73 gene in neuroblastoma tumors and cell lines. Cell Growth Differ. 1998;9(11):897–903. [PubMed] [Google Scholar]
- 29.Yokomizo A, Mai M, Tindall DJ, Cheng L, et al. Overexpression of the wild type p73 gene in human bladder cancer. Oncogene. 1999;18(8):1629–33. doi: 10.1038/sj.onc.1202474. [DOI] [PubMed] [Google Scholar]
- 30.Yokomizo A, Mai M, Bostwick DG, Tindall DJ, et al. Mutation and expression analysis of the p73 gene in prostate cancer. Prostate. 1999;39(2):94–100. doi: 10.1002/(sici)1097-0045(19990501)39:2<94::aid-pros3>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 31.Malaguarnera R, Vella V, Vigneri R, Frasca F. p53 family proteins in thyroid cancer. Endocr Relat Cancer. 2007;14(1):43–60. doi: 10.1677/erc.1.01223. [DOI] [PubMed] [Google Scholar]
- 32.Leupin N, Luthi A, Novak U, Grob TJ, et al. P73 status in B-cell chronic lymphocytic leukaemia. Leuk Lymphoma. 2004;45(6):1205–7. doi: 10.1080/10298190310001623829. [DOI] [PubMed] [Google Scholar]
- 33.Sunahara M, Ichimiya S, Nimura Y, Takada N, et al. Mutational analysis of the p73 gene localized at chromosome 1p36.3 in colorectal carcinomas. Int J Oncol. 1998;13(2):319–23. [PubMed] [Google Scholar]
- 34.Loukopoulos P, Shibata T, Katoh H, Kokubu A, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98(3):392–400. doi: 10.1111/j.1349-7006.2007.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faridoni-Laurens L, Tourpin S, Alsafadi S, Barrois M, et al. Involvement of N-terminally truncated variants of p73, deltaTAp73, in head and neck squamous cell cancer: a comparison with p53 mutations. Cell Cycle. 2008;7(11):1587–96. doi: 10.4161/cc.7.11.5894. [DOI] [PubMed] [Google Scholar]
- 36.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11(4):289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue Q, Hopkins B, Perruzzi C, Udayakumar D, et al. Palomid 529, a novel small-molecule drug, is a TORC1/TORC2 inhibitor that reduces tumor growth, tumor angiogenesis, and vascular permeability. Cancer Res. 2008;68(22):9551–7. doi: 10.1158/0008-5472.CAN-08-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Garcia M, Banerji U, Albanell J, Bahleda R, et al. First-in-human, phase I dose-escalation study of the safety, pharmacokinetics, and pharmacodynamics of RO5126766, a first-in- class dual MEK/RAF inhibitor in patients with solid tumors. Clin Cancer Res. 2012;18(17):4806–19. doi: 10.1158/1078-0432.CCR-12-0742. [DOI] [PubMed] [Google Scholar]
- 39.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13(7):482–95. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 40.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–57. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 41.Varelas X, Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 2012;22(2):88–96. doi: 10.1016/j.tcb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–71. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 44.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144(5):782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B, Li L, Lu Q, Wang LH, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25(1):51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overholtzer M, Zhang J, Smolen GA, Muir B, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103(33):12405–10. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zender L, Spector MS, Xue W, Flemming P, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125(7):1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans DG. Neurofibromatosis 2 [Bilateral acoustic neurofibromatosis, central neurofibromatosis, NF2, neurofibromatosis type II] Genet Med. 2009;11(9):599–610. doi: 10.1097/GIM.0b013e3181ac9a27. [DOI] [PubMed] [Google Scholar]
- 49.Giovannini M, Robanus-Maandag E, van der Valk M, Niwa-Kawakita M, et al. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;14(13):1617–30. [PMC free article] [PubMed] [Google Scholar]
- 50.McClatchey AI, Giovannini M. Membrane organization and tumorigenesis--the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19(19):2265–77. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 51.Zhang N, Bai H, David KK, Dong J, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19(1):27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barry ER, Morikawa T, Butler BL, Shrestha K, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–10. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varelas X, Miller BW, Sopko R, Song S, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18(4):579–91. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Medema JP, Vermeulen L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature. 2011;474(7351):318–26. doi: 10.1038/nature10212. [DOI] [PubMed] [Google Scholar]
- 55.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong J, Feldmann G, Huang J, Wu S, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130(6):1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71(7):2728–38. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 58.Zhao B, Ye X, Yu J, Li L, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137(24):4135–45. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren F, Wang B, Yue T, Yun EY, et al. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci U S A. 2010;107(49):21064–9. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu FX, Zhao B, Panupinthu N, Jewell JL, et al. Regulation of the Hippo-YAP pathway by Gprotein- coupled receptor signaling. Cell. 2012;150(4):780–91. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci U S A. 1997;94(21):11514–9. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mimeault M, Batra SK. Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J Cell Physiol. 2010;224(3):626–35. doi: 10.1002/jcp.22196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Baek SJ, Eling TE. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem Pharmacol. 2013;85(5):597–606. doi: 10.1016/j.bcp.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osada M, Park HL, Park MJ, Liu JW, et al. A p53-type response element in the GDF15 promoter confers high specificity for p53 activation. Biochem Biophys Res Commun. 2007;354(4):913–8. doi: 10.1016/j.bbrc.2007.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan M, Wang Y, Guan K, Sun Y. PTGF-beta, a type beta transforming growth factor (TGF-beta) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-beta signaling pathway. Proc Natl Acad Sci U S A. 2000;97(1):109–14. doi: 10.1073/pnas.97.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Min KW, Zhang X, Imchen T, Baek SJ. A peroxisome proliferator-activated receptor ligand MCC-555 imparts anti-proliferative response in pancreatic cancer cells by PPARgamma-independent up-regulation of KLF4. Toxicol Appl Pharmacol. 2012;263(2):225–32. doi: 10.1016/j.taap.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baek SJ, Horowitz JM, Eling TE. Molecular cloning and characterization of human nonsteroidal anti-inflammatory drug-activated gene promoter. Basal transcription is mediated by Sp1 and Sp3. J Biol Chem. 2001;276(36):33384–92. doi: 10.1074/jbc.M101814200. [DOI] [PubMed] [Google Scholar]
- 70.Baek SJ, Kim JS, Moore SM, Lee SH, et al. Cyclooxygenase inhibitors induce the expression of the tumor suppressor gene EGR-1, which results in the up-regulation of NAG-1, an antitumorigenic protein. Mol Pharmacol. 2005;67(2):356–64. doi: 10.1124/mol.104.005108. [DOI] [PubMed] [Google Scholar]
- 71.Chintharlapalli S, Papineni S, Liu S, Jutooru I, et al. 2-cyano-lup-1-en-3-oxo-20-oic acid, a cyano derivative of betulinic acid, activates peroxisome proliferator-activated receptor gamma in colon and pancreatic cancer cells. Carcinogenesis. 2007;28(11):2337–46. doi: 10.1093/carcin/bgm189. [DOI] [PubMed] [Google Scholar]
- 72.Youns M, Efferth T, Hoheisel JD. Transcript profiling identifies novel key players mediating the growth inhibitory effect of NS-398 on human pancreatic cancer cells. Eur J Pharmacol. 2011;650(1):170–7. doi: 10.1016/j.ejphar.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 73.Shim M, Eling TE. Protein kinase C-dependent regulation of NAG-1/placental bone morphogenic protein/MIC-1 expression in LNCaP prostate carcinoma cells. J Biol Chem. 2005;280(19):18636–42. doi: 10.1074/jbc.M414613200. [DOI] [PubMed] [Google Scholar]
- 74.Shim M, Eling TE. Vitamin E succinate induces NAG-1 expression in a p38 kinase-dependent mechanism. Mol Cancer Ther. 2008;7(4):961–71. doi: 10.1158/1535-7163.MCT-07-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimizu S, Kadowaki M, Yoshioka H, Kambe A, et al. Proteasome inhibitor MG132 induces NAG-1/GDF15 expression through the p38 MAPK pathway in glioblastoma cells. Biochem Biophys Res Commun. 2013;430(4):1277–82. doi: 10.1016/j.bbrc.2012.11.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wynne S, Djakiew D. NSAID inhibition of prostate cancer cell migration is mediated by Nag-1 Induction via the p38 MAPK-p75(NTR) pathway. Mol Cancer Res. 2010;8(12):1656–64. doi: 10.1158/1541-7786.MCR-10-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamaguchi K, Lee SH, Eling TE, Baek SJ. Identification of nonsteroidal anti-inflammatory drug-activated gene (NAG-1) as a novel downstream target of phosphatidylinositol 3-kinase/AKT/GSK-3beta pathway. J Biol Chem. 2004;279(48):49617–23. doi: 10.1074/jbc.M408796200. [DOI] [PubMed] [Google Scholar]
- 78.Griner SE, Joshi JP, Nahta R. Growth differentiation factor 15 stimulates rapamycin-sensitive ovarian cancer cell growth and invasion. Biochem Pharmacol. 2013;85(1):46–58. doi: 10.1016/j.bcp.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joshi JP, Brown NE, Griner SE, Nahta R. Growth differentiation factor 15 (GDF15)-mediated HER2 phosphorylation reduces trastuzumab sensitivity of HER2-overexpressing breast cancer cells. Biochem Pharmacol. 2011;82(9):1090–9. doi: 10.1016/j.bcp.2011.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown DA, Ward RL, Buckhaults P, Liu T, et al. MIC-1 serum level and genotype: associations with progress and prognosis of colorectal carcinoma. Clin Cancer Res. 2003;9(7):2642–50. [PubMed] [Google Scholar]
- 81.Bock AJ, Stavnes HT, Kempf T, Trope CG, et al. Expression and clinical role of growth differentiation factor-15 in ovarian carcinoma effusions. Int J Gynecol Cancer. 2010;20(9):1448–55. doi: 10.1111/IGC.0b013e3181f7d6be. [DOI] [PubMed] [Google Scholar]
- 82.Staff AC, Bock AJ, Becker C, Kempf T, et al. Growth differentiation factor-15 as a prognostic biomarker in ovarian cancer. Gynecol Oncol. 2010;118(3):237–43. doi: 10.1016/j.ygyno.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 83.Bauskin AR, Brown DA, Junankar S, Rasiah KK, et al. The propeptide mediates formation of stromal stores of PROMIC-1: role in determining prostate cancer outcome. Cancer Res. 2005;65(6):2330–6. doi: 10.1158/0008-5472.CAN-04-3827. [DOI] [PubMed] [Google Scholar]
- 84.Brown DA, Stephan C, Ward RL, Law M, et al. Measurement of serum levels of macrophage inhibitory cytokine 1 combined with prostate-specific antigen improves prostate cancer diagnosis. Clin Cancer Res. 2006;12(1):89–96. doi: 10.1158/1078-0432.CCR-05-1331. [DOI] [PubMed] [Google Scholar]
- 85.Selander KS, Brown DA, Sequeiros GB, Hunter M, et al. Serum macrophage inhibitory cytokine-1 concentrations correlate with the presence of prostate cancer bone metastases. Cancer Epidemiol Biomarkers Prev. 2007;16(3):532–7. doi: 10.1158/1055-9965.EPI-06-0841. [DOI] [PubMed] [Google Scholar]
- 86.Koopmann J, Buckhaults P, Brown DA, Zahurak ML, et al. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin Cancer Res. 2004;10(7):2386–92. doi: 10.1158/1078-0432.ccr-03-0165. [DOI] [PubMed] [Google Scholar]
- 87.Koopmann J, Rosenzweig CN, Zhang Z, Canto MI, et al. Serum markers in patients with resectable pancreatic adenocarcinoma: macrophage inhibitory cytokine 1 versus CA19-9. Clin Cancer Res. 2006;12(2):442–6. doi: 10.1158/1078-0432.CCR-05-0564. [DOI] [PubMed] [Google Scholar]
- 88.Welsh JB, Sapinoso LM, Kern SG, Brown DA, et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci U S A. 2003;100(6):3410–5. doi: 10.1073/pnas.0530278100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16(23):5842–51. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 90.Shnaper S, Desbaillets I, Brown DA, Murat A, et al. Elevated levels of MIC-1/GDF15 in the cerebrospinal fluid of patients are associated with glioblastoma and worse outcome. Int J Cancer. 2009;125(11):2624–30. doi: 10.1002/ijc.24639. [DOI] [PubMed] [Google Scholar]
- 91.Chen SJ, Karan D, Johansson SL, Lin FF, et al. Prostate-derived factor as a paracrine and autocrine factor for the proliferation of androgen receptor-positive human prostate cancer cells. Prostate. 2007;67(5):557–71. doi: 10.1002/pros.20551. [DOI] [PubMed] [Google Scholar]
- 92.Senapati S, Rachagani S, Chaudhary K, Johansson SL, et al. Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene. 2010;29(9):1293–302. doi: 10.1038/onc.2009.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Husaini Y, Qiu MR, Lockwood GP, Luo XW, et al. Macrophage inhibitory cytokine-1 (MIC-1/GDF15) slows cancer development but increases metastases in TRAMP prostate cancer prone mice. PLoS One. 2012;7(8):e43833. doi: 10.1371/journal.pone.0043833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim KK, Lee JJ, Yang Y, You KH, et al. Macrophage inhibitory cytokine-1 activates AKT and ERK-1/2 via the transactivation of ErbB2 in human breast and gastric cancer cells. Carcinogenesis. 2008;29(4):704–12. doi: 10.1093/carcin/bgn031. [DOI] [PubMed] [Google Scholar]
- 95.Lee DH, Yang Y, Lee SJ, Kim KY, et al. Macrophage inhibitory cytokine-1 induces the invasiveness of gastric cancer cells by up-regulating the urokinase-type plasminogen activator system. Cancer Res. 2003;63(15):4648–55. [PubMed] [Google Scholar]
- 96.Cheng I, Krumroy LM, Plummer SJ, Casey G, et al. MIC1 and IL1RN genetic variation and advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1309–11. doi: 10.1158/1055-9965.EPI-07-0165. [DOI] [PubMed] [Google Scholar]
- 97.Hayes VM, Severi G, Southey MC, Padilla EJ, et al. Macrophage inhibitory cytokine-1 H6D polymorphism, prostate cancer risk, and survival. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1223–5. doi: 10.1158/1055-9965.EPI-06-0063. [DOI] [PubMed] [Google Scholar]
- 98.Lindmark F, Zheng SL, Wiklund F, Bensen J, et al. H6D polymorphism in macrophage-inhibitory cytokine-1 gene associated with prostate cancer. J Natl Cancer Inst. 2004;96(16):1248–54. doi: 10.1093/jnci/djh227. [DOI] [PubMed] [Google Scholar]
- 99.Cekanova M, Lee SH, Donnell RL, Sukhthankar M, et al. Nonsteroidal anti-inflammatory drug-activated gene-1 expression inhibits urethane-induced pulmonary tumorigenesis in transgenic mice. Cancer Prev Res (Phila) 2009;2(5):450–8. doi: 10.1158/1940-6207.CAPR-09-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinez JM, Sali T, Okazaki R, Anna C, et al. Drug-induced expression of nonsteroidal anti-inflammatory drug-activated gene/macrophage inhibitory cytokine-1/prostate-derived factor, a putative tumor suppressor, inhibits tumor growth. J Pharmacol Exp Ther. 2006;318(2):899–906. doi: 10.1124/jpet.105.100081. [DOI] [PubMed] [Google Scholar]
- 101.Lambert JR, Kelly JA, Shim M, Huffer WE, et al. Prostate derived factor in human prostate cancer cells: gene induction by vitamin D via a p53-dependent mechanism and inhibition of prostate cancer cell growth. J Cell Physiol. 2006;208(3):566–74. doi: 10.1002/jcp.20692. [DOI] [PubMed] [Google Scholar]
- 102.Albertoni M, Shaw PH, Nozaki M, Godard S, et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21(27):4212–9. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- 103.Baek SJ, Okazaki R, Lee SH, Martinez J, et al. Nonsteroidal anti-inflammatory drug-activated gene-1 over expression in transgenic mice suppresses intestinal neoplasia. Gastroenterology. 2006;131(5):1553–60. doi: 10.1053/j.gastro.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 104.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, et al. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186(1):136–45. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 105.Park JH, Park EJ, Lee HS, Kim SJ, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25(17):3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3(1):35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28(14):1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 108.Lichner Z, Scorilas A, White NM, Girgis AH, et al. The chromatin remodeling gene ARID1A is a new prognostic marker in clear cell renal cell carcinoma. Am J Pathol. 2013;182(4):1163–70. doi: 10.1016/j.ajpath.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 109.Wang K, Kan J, Yuen ST, Shi ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 110.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 111.Cornen S, Adelaide J, Bertucci F, Finetti P, et al. Mutations and deletions of ARID1A in breast tumors. Oncogene. 2012;31(38):4255–6. doi: 10.1038/onc.2011.598. [DOI] [PubMed] [Google Scholar]
- 112.Mamo A, Cavallone L, Tuzmen S, Chabot C, et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene. 2012;31(16):2090–100. doi: 10.1038/onc.2011.386. [DOI] [PubMed] [Google Scholar]
- 113.Sausen M, Leary RJ, Jones S, Wu J, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45(1):12–7. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gui Y, Guo G, Huang Y, Hu X, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jones S, Wang TL, Shih Ie M, Mao TL, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guan B, Mao TL, Panuganti PK, Kuhn E, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35(5):625–32. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dallas PB, Cheney IW, Liao DW, Bowrin V, et al. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol. 1998;18(6):3596–603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.King HA, Trotter KW, Archer TK. Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim Biophys Acta. 2012;1819(7):716–26. doi: 10.1016/j.bbagrm.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71(21):6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang X, Sun Q, Shan M, Niu M, et al. Promoter hypermethylation of ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers. PLoS One. 2013;8(1):e53931. doi: 10.1371/journal.pone.0053931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang DD, Chen YB, Pan K, Wang W, et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS One. 2012;7(7):e40364. doi: 10.1371/journal.pone.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, et al. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66(3):1289–93. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 125.Luo B, Cheung HW, Subramanian A, Sharifnia T, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(51):20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Choi NM, Majumder P, Boss JM. Regulation of major histocompatibility complex class II genes. Curr Opin Immunol. 2011;23(1):81–7. doi: 10.1016/j.coi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Katagiri A, Nakayama K, Rahman MT, Rahman M, et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25(2):282–8. doi: 10.1038/modpathol.2011.161. [DOI] [PubMed] [Google Scholar]
- 128.Nefedova Y, Gabrilovich D. Mechanisms and clinical prospects of Notch inhibitors in the therapy of hematological malignancies. Drug Resist Updat. 2008;11(6):210–8. doi: 10.1016/j.drup.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rizzo P, Osipo C, Foreman K, Golde T, et al. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27(38):5124–31. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 130.Shih Ie M, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67(5):1879–82. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 131.Stockhausen MT, Sjolund J, Axelson H. Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res. 2005;310(1):218–28. doi: 10.1016/j.yexcr.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 132.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 133.Bray SJ, Takada S, Harrison E, Shen SC, et al. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev Biol. 2008;8:11. doi: 10.1186/1471-213X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ehebauer M, Hayward P, Arias AM. Notch, a universal arbiter of cell fate decisions. Science. 2006;314(5804):1414–5. doi: 10.1126/science.1134042. [DOI] [PubMed] [Google Scholar]
- 135.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Staal FJ, Langerak AW. Signaling pathways involved in the development of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93(4):493–7. doi: 10.3324/haematol.12917. [DOI] [PubMed] [Google Scholar]
- 137.Dickson BC, Mulligan AM, Zhang H, Lockwood G, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20(6):685–93. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 138.Reedijk M, Odorcic S, Chang L, Zhang H, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65(18):8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 139.Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, et al. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;111(3):439–48. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 140.Massi D, Tarantini F, Franchi A, Paglierani M, et al. Evidence for differential expression of Notch receptors and their ligands in melanocytic nevi and cutaneous malignant melanoma. Mod Pathol. 2006;19(2):246–54. doi: 10.1038/modpathol.3800526. [DOI] [PubMed] [Google Scholar]
- 141.Okuyama R, Tagami H, Aiba S. Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases. J Dermatol Sci. 2008;49(3):187–94. doi: 10.1016/j.jdermsci.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 142.Pinnix CC, Herlyn M. The many faces of Notch signaling in skin-derived cells. Pigment Cell Res. 2007;20(6):458–65. doi: 10.1111/j.1600-0749.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- 143.Chen Y, De Marco MA, Graziani I, Gazdar AF, et al. Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Res. 2007;67(17):7954–9. doi: 10.1158/0008-5472.CAN-07-1229. [DOI] [PubMed] [Google Scholar]
- 144.Konishi J, Kawaguchi KS, Vo H, Haruki N, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67(17):8051–7. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 145.Peacock CD, Watkins DN. Cancer stem cells and the ontogeny of lung cancer. J Clin Oncol. 2008;26(17):2883–9. doi: 10.1200/JCO.2007.15.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bolos V, Blanco M, Medina V, Aparicio G, et al. Notch signalling in cancer stem cells. Clin Transl Oncol. 2009;11(1):11–9. doi: 10.1007/s12094-009-0305-2. [DOI] [PubMed] [Google Scholar]
- 147.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28(3):339–63. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 148.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104(6):1696–702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 149.Miele L, Miao H, Nickoloff BJ. NOTCH signaling as a novel cancer therapeutic target. Curr Cancer Drug Targets. 2006;6(4):313–23. doi: 10.2174/156800906777441771. [DOI] [PubMed] [Google Scholar]
- 150.Sjolund J, Manetopoulos C, Stockhausen MT, Axelson H. The Notch pathway in cancer: differentiation gone awry. Eur J Cancer. 2005;41(17):2620–9. doi: 10.1016/j.ejca.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 151.Hald J, Hjorth JP, German MS, Madsen OD, et al. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260(2):426–37. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 152.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100(25):14920–5. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Apelqvist A, Li H, Sommer L, Beatus P, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400(6747):877–81. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 154.Miyamoto Y, Maitra A, Ghosh B, Zechner U, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3(6):565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 155.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4(7):505–18. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 156.Humbel RE. Insulin-like growth factors I and II. Eur J Biochem. 1990;190(3):445–62. doi: 10.1111/j.1432-1033.1990.tb15595.x. [DOI] [PubMed] [Google Scholar]
- 157.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195(2):127–37. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 158.Riedemann J, Macaulay VM. IGF1R signalling and its inhibition. Endocr Relat Cancer. 2006;13(Suppl 1):S33–43. doi: 10.1677/erc.1.01280. [DOI] [PubMed] [Google Scholar]
- 159.Hebert E. Mannose-6-phosphate/insulin-like growth factor II receptor expression and tumor development. Biosci Rep. 2006;26(1):7–17. doi: 10.1007/s10540-006-9002-3. [DOI] [PubMed] [Google Scholar]
- 160.Dennis PA, Rifkin DB. Cellular activation of latent transforming growth factor beta requires binding to the cation-independent mannose 6-phosphate/insulin-like growth factor type II receptor. Proc Natl Acad Sci U S A. 1991;88(2):580–4. doi: 10.1073/pnas.88.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kang JX, Bell J, Beard RL, Chandraratna RA. Mannose 6-phosphate/insulin-like growth factor II receptor mediates the growth-inhibitory effects of retinoids. Cell Growth Differ. 1999;10(8):591–600. [PubMed] [Google Scholar]
- 162.Ryan PD, Goss PE. The emerging role of the insulin-like growth factor pathway as a therapeutic target in cancer. Oncologist. 2008;13(1):16–24. doi: 10.1634/theoncologist.2007-0199. [DOI] [PubMed] [Google Scholar]
- 163.Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010;10(5):342–52. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 164.Hennessy BT, Smith DL, Ram PT, Lu Y, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 165.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 166.Pankov R, Cukierman E, Clark K, Matsumoto K, et al. Specific beta1 integrin site selectively regulates Akt/protein kinase B signaling via local activation of protein phosphatase 2A. J Biol Chem. 2003;278(20):18671–81. doi: 10.1074/jbc.M300879200. [DOI] [PubMed] [Google Scholar]
- 167.Stupack DG, Cheresh DA. Get a ligand, get a life: integrins, signaling and cell survival. J Cell Sci. 2002;115(Pt 19):3729–38. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- 168.Murga C, Laguinge L, Wetzker R, Cuadrado A, et al. Activation of Akt/protein kinase B by G protein-coupled receptors. A role for alpha and beta gamma subunits of heterotrimeric G proteins acting through phosphatidylinositol-3-OH kinasegamma. J Biol Chem. 1998;273(30):19080–5. doi: 10.1074/jbc.273.30.19080. [DOI] [PubMed] [Google Scholar]
- 169.Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14(2):342–6. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 170.Kim WY, Jin Q, Oh SH, Kim ES, et al. Elevated epithelial insulin-like growth factor expression is a risk factor for lung cancer development. Cancer Res. 2009;69(18):7439–48. doi: 10.1158/0008-5472.CAN-08-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim SY, Wan X, Helman LJ. Targeting IGF-1R in the treatment of sarcomas: past, present and future. Bull Cancer. 2009;96(7):E52–60. doi: 10.1684/bdc.2009.0915. [DOI] [PubMed] [Google Scholar]
- 172.Werner H, Maor S. The insulin-like growth factor-I receptor gene: a downstream target for oncogene and tumor suppressor action. Trends Endocrinol Metab. 2006;17(6):236–42. doi: 10.1016/j.tem.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 173.Maor S, Mayer D, Yarden RI, Lee AV, et al. Estrogen receptor regulates insulin-like growth factor-I receptor gene expression in breast tumor cells: involvement of transcription factor Sp1. J Endocrinol. 2006;191(3):605–12. doi: 10.1677/joe.1.07016. [DOI] [PubMed] [Google Scholar]
- 174.Ma Y, Zhang L, Peng T, Cheng J, et al. Angiotensin II stimulates transcription of insulin-like growth factor I receptor in vascular smooth muscle cells: role of nuclear factor-kappaB. Endocrinology. 2006;147(3):1256–63. doi: 10.1210/en.2005-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kahlert S, Nuedling S, van Eickels M, Vetter H, et al. Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J Biol Chem. 2000;275(24):18447–53. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- 176.Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20(6):761–87. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 177.McGrath M, Lee IM, Buring J, De Vivo I. Common genetic variation within IGFI, IGFII, IGFBP-1, and IGFBP-3 and endometrial cancer risk. Gynecol Oncol. 2011;120(2):174–8. doi: 10.1016/j.ygyno.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jerome L, Shiry L, Leyland-Jones B. Deregulation of the IGF axis in cancer: epidemiological evidence and potential therapeutic interventions. Endocr Relat Cancer. 2003;10(4):561–78. doi: 10.1677/erc.0.0100561. [DOI] [PubMed] [Google Scholar]
- 179.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23(6):824–54. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 180.Santer FR, Bacher N, Moser B, Morandell D, et al. Nuclear insulin-like growth factor binding protein-3 induces apoptosis and is targeted to ubiquitin/proteasome-dependent proteolysis. Cancer Res. 2006;66(6):3024–33. doi: 10.1158/0008-5472.CAN-05-2013. [DOI] [PubMed] [Google Scholar]
- 181.Jogie-Brahim S, Feldman D, Oh Y. Unraveling insulin-like growth factor binding protein-3 actions in human disease. Endocr Rev. 2009;30(5):417–37. doi: 10.1210/er.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yin P, Xu Q, Duan C. Paradoxical actions of endogenous and exogenous insulin-like growth factor-binding protein-5 revealed by RNA interference analysis. J Biol Chem. 2004;279(31):32660–6. doi: 10.1074/jbc.M401378200. [DOI] [PubMed] [Google Scholar]
- 183.Liu B, Lee HY, Weinzimer SA, Powell DR, et al. Direct functional interactions between insulin-like growth factor-binding protein-3 and retinoid X receptor-alpha regulate transcriptional signaling and apoptosis. J Biol Chem. 2000;275(43):33607–13. doi: 10.1074/jbc.M002547200. [DOI] [PubMed] [Google Scholar]
- 184.Kim JH, Choi DS, Lee OH, Oh SH, et al. Antiangiogenic antitumor activities of IGFBP-3 are mediated by IGF-independent suppression of Erk1/2 activation and Egr-1-mediated transcriptional events. Blood. 2011;118(9):2622–31. doi: 10.1182/blood-2010-08-299784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Oh SH, Kim WY, Lee OH, Kang JH, et al. Insulin-like growth factor binding protein-3 suppresses vascular endothelial growth factor expression and tumor angiogenesis in head and neck squamous cell carcinoma. Cancer Sci. 2012;103(7):1259–66. doi: 10.1111/j.1349-7006.2012.02301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Werner H. Tumor suppressors govern insulin-like growth factor signaling pathways: implications in metabolism and cancer. Oncogene. 2012;31(22):2703–14. doi: 10.1038/onc.2011.447. [DOI] [PubMed] [Google Scholar]
- 187.Kurosu H, Yamamoto M, Clark JD, Pastor JV, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309(5742):1829–33. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27(56):7094–105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 189.Weinberg RA. Tumor suppressor genes. Science. 1991;254(5035):1138–46. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 190.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28(1):20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 191.Stoeltzing O, Liu W, Reinmuth N, Fan F, et al. Regulation of hypoxia-inducible factor-1alpha, vascular endothelial growth factor, and angiogenesis by an insulin-like growth factor-I receptor autocrine loop in human pancreatic cancer. Am J Pathol. 2003;163(3):1001–11. doi: 10.1016/s0002-9440(10)63460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kooijman R, Coppens A, Hooghe-Peters E. IGF-I stimulates IL-8 production in the promyelocytic cell line HL-60 through activation of extracellular signal-regulated protein kinase. Cell Signal. 2003;15(12):1091–8. doi: 10.1016/s0898-6568(03)00069-x. [DOI] [PubMed] [Google Scholar]
- 193.Zhao D, Bakirtzi K, Zhan Y, Zeng H, et al. Insulin-like growth factor-1 receptor transactivation modulates the inflammatory and proliferative responses of neurotensin in human colonic epithelial cells. J Biol Chem. 2011;286(8):6092–9. doi: 10.1074/jbc.M110.192534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang D, Bar-Eli M, Meloche S, Brodt P. Dual regulation of MMP-2 expression by the type 1 insulin-like growth factor receptor: the phosphatidylinositol 3-kinase/Akt and Raf/ERK pathways transmit opposing signals. J Biol Chem. 2004;279(19):19683–90. doi: 10.1074/jbc.M313145200. [DOI] [PubMed] [Google Scholar]
- 195.Ogawara Y, Kishishita S, Obata T, Isazawa Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277(24):21843–50. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 196.Belletti B, Prisco M, Morrione A, Valentinis B, et al. Regulation of Id2 gene expression by the insulin-like growth factor I receptor requires signaling by phosphatidylinositol 3-kinase. J Biol Chem. 2001;276(17):13867–74. doi: 10.1074/jbc.M010509200. [DOI] [PubMed] [Google Scholar]
- 197.van der Veeken J, Oliveira S, Schiffelers RM, Storm G, et al. Crosstalk between epidermal growth factor receptor- and insulin-like growth factor-1 receptor signaling: implications for cancer therapy. Curr Cancer Drug Targets. 2009;9(6):748–60. doi: 10.2174/156800909789271495. [DOI] [PubMed] [Google Scholar]
- 198.Morgillo F, Kim WY, Kim ES, Ciardiello F, et al. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13(9):2795–803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 199.Nahta R, Esteva FJ. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 2006;8(6):215. doi: 10.1186/bcr1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Villanueva J, Vultur A, Lee JT, Somasundaram R, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18(6):683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Huang GS, Brouwer-Visser J, Ramirez MJ, Kim CH, et al. Insulin-like growth factor 2 expression modulates Taxol resistance and is a candidate biomarker for reduced disease-free survival in ovarian cancer. Clin Cancer Res. 2010;16(11):2999–3010. doi: 10.1158/1078-0432.CCR-09-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, et al. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155(3):683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 204.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 205.Shindo T, Manabe I, Fukushima Y, Tobe K, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 206.Chen C, Sun X, Ran Q, Wilkinson KD, et al. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24(20):3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 207.Chen C, Sun X, Guo P, Dong XY, et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 208.Chen C, Zhou Z, Ross JS, Zhou W, et al. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121(1):80–87. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 209.Chen C, Sun X, Guo P, Dong XY, et al. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26(16):2386–94. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 210.Kwak MK, Lee HJ, Hur K, Park do J, et al. Expression of Kruppel-like factor 5 in human gastric carcinomas. J Cancer Res Clin Oncol. 2008;134(2):163–7. doi: 10.1007/s00432-007-0265-2. [DOI] [PubMed] [Google Scholar]
- 211.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 Regulate Proliferation, Apoptosis and Invasion in Esophageal Cancer Cells. Cancer Biol Ther. 2005;4(11):1216–21. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 212.Bateman NW, Tan D, Pestell RG, Black JD, et al. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279(13):12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 213.Guo P, Dong XY, Zhang X, Zhao KW, et al. Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification. J Biol Chem. 2009;284(10):6071–6078. doi: 10.1074/jbc.M806270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nandan MO, Yoon HS, Zhao W, Ouko LA, et al. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23(19):3404–13. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sun R, Chen X, Yang VW. Intestinal-enriched kruppel-like factor (kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276(10):6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, et al. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134(1):120–30. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen C, Benjamin MS, Sun X, Otto KB, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulation in the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118(6):1346–55. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- 218.Zheng HQ, Zhou Z, Huang J, Chaudhury L, et al. Kruppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene. 2009;28(42):3702–13. doi: 10.1038/onc.2009.235. [DOI] [PubMed] [Google Scholar]
- 219.Dardousis K, Voolstra C, Roengvoraphoj M, Sekandarzad A, et al. Identification of Differentially Expressed Genes Involved in the Formation of Multicellular Tumor Spheroids by HT-29 Colon Carcinoma Cells. Mol Ther. 2007;15(1):94–102. doi: 10.1038/sj.mt.6300003. [DOI] [PubMed] [Google Scholar]
- 220.Goldstein BG, Chao HH, Yang Y, Yermolina YA, et al. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. 2007;292(6):G1784–92. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- 221.Sur I, Rozell B, Jaks V, Bergstrom A, et al. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci. 2006;119(Pt 17):3593–601. doi: 10.1242/jcs.03070. [DOI] [PubMed] [Google Scholar]
- 222.Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579(21):4757–62. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yang Y, Goldstein BG, Nakagawa H, Katz JP. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. Faseb J. 2007;21(2):543–50. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 224.Guo P, Dong XY, Zhao KW, Sun X, et al. Opposing effects of KLF5 on the transcription of MYC in epithelial proliferation in the context of transforming growth factor beta. J Biol Chem. 2009;284(41):28243–52. doi: 10.1074/jbc.M109.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Guo P, Zhao KW, Dong XY, Sun X, et al. Acetylation of KLF5 alters the assembly of p15 transcription factors in transforming growth factor-beta-mediated induction in epithelial cells. J Biol Chem. 2009;284(27):18184–93. doi: 10.1074/jbc.M109.007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Xing C, Fu X, Sun X, Guo P, et al. Different expression patterns and functions of acetylated and unacetylated klf5 in the proliferation and differentiation of prostatic epithelial cells. PLoS ONE. 2013;8(6):e65538. doi: 10.1371/journal.pone.0065538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Akhurst RJ, Derynck R. TGF-beta signaling in cancer-a double-edged sword. Trends Cell Biol. 2001;11(11):S44–51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 228.Amin AR, Kucuk O, Khuri FR, Shin DM. Perspectives for cancer prevention with natural compounds. J Clin Oncol. 2009;27(16):2712–25. doi: 10.1200/JCO.2008.20.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gullett NP, Ruhul Amin AR, Bayraktar S, Pezzuto JM, et al. Cancer prevention with natural compounds. Semin Oncol. 2010;37(3):258–81. doi: 10.1053/j.seminoncol.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 230.William WN, Jr, Heymach JV, Kim ES, Lippman SM. Molecular targets for cancer chemoprevention. Nat Rev Drug Discov. 2009;8(3):213–25. doi: 10.1038/nrd2663. [DOI] [PubMed] [Google Scholar]
- 231.Haddad RI, Shin DM. Recent advances in head and neck cancer. New England Journal of Medicine. 2008;359(11):1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 232.Kennedy DO, Kojima A, Moffatt J, Yamagiwa H, et al. Cellular thiol status-dependent inhibition of tumor cell growth via modulation of retinoblastoma protein phosphorylation by (−)-epigallocatechin. Cancer Lett. 2002;179(1):25–32. doi: 10.1016/s0304-3835(01)00856-4. [DOI] [PubMed] [Google Scholar]
- 233.Gupta S, Hussain T, Mukhtar H. Molecular pathway for (−)-epigallocatechin-3-gallate-induced cell cycle arrest and apoptosis of human prostate carcinoma cells. Arch Biochem Biophys. 2003;410(1):177–85. doi: 10.1016/s0003-9861(02)00668-9. [DOI] [PubMed] [Google Scholar]
- 234.Nihal M, Roelke CT, Wood GS. Anti-melanoma effects of vorinostat in combination with polyphenolic antioxidant (−)-epigallocatechin-3-gallate (EGCG) Pharm Res. 2010;27(6):1103–14. doi: 10.1007/s11095-010-0054-5. [DOI] [PubMed] [Google Scholar]
- 235.Maher DM, Bell MC, O’Donnell EA, Gupta BK, et al. Curcumin suppresses human papillomavirus oncoproteins, restores p53, Rb, and PTPN13 proteins and inhibits benzo[a]pyrene-induced upregulation of HPV E7. Mol Carcinog. 2011;50(1):47–57. doi: 10.1002/mc.20695. [DOI] [PubMed] [Google Scholar]
- 236.Su CC, Wang MJ, Chiu TL. The anti-cancer efficacy of curcumin scrutinized through core signaling pathways in glioblastoma. Int J Mol Med. 2010;26(2):217–24. doi: 10.3892/ijmm_00000455. [DOI] [PubMed] [Google Scholar]
- 237.Damera G, Fogle HW, Lim P, Goncharova EA, et al. Vitamin D inhibits growth of human airway smooth muscle cells through growth factor-induced phosphorylation of retinoblastoma protein and checkpoint kinase 1. Br J Pharmacol. 2009;158(6):1429–41. doi: 10.1111/j.1476-5381.2009.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hahm ER, Singh SV. Honokiol causes G0-G1 phase cell cycle arrest in human prostate cancer cells in association with suppression of retinoblastoma protein level/phosphorylation and inhibition of E2F1 transcriptional activity. Mol Cancer Ther. 2007;6(10):2686–95. doi: 10.1158/1535-7163.MCT-07-0217. [DOI] [PubMed] [Google Scholar]
- 239.Hastak K, Agarwal MK, Mukhtar H, Agarwal ML. Ablation of either p21 or Bax prevents p53-dependent apoptosis induced by green tea polyphenol epigallocatechin-3-gallate. FASEB J. 2005;19(7):789–91. doi: 10.1096/fj.04-2226fje. [DOI] [PubMed] [Google Scholar]
- 240.Roy AM, Baliga MS, Katiyar SK. Epigallocatechin-3-gallate induces apoptosis in estrogen receptor-negative human breast carcinoma cells via modulation in protein expression of p53 and Bax and caspase-3 activation. Mol Cancer Ther. 2005;4(1):81–90. [PubMed] [Google Scholar]
- 241.Wu B, Zhang Q, Shen W, Zhu J. Anti-proliferative and chemosensitizing effects of luteolin on human gastric cancer AGS cell line. Mol Cell Biochem. 2008;313(1–2):125–32. doi: 10.1007/s11010-008-9749-x. [DOI] [PubMed] [Google Scholar]
- 242.Amin AR, Wang D, Zhang H, Peng S, et al. Enhanced anti-tumor activity by the combination of the natural compounds (−)-epigallocatechin-3-gallate and luteolin: potential role of p53. J Biol Chem. 2010;285(45):34557–65. doi: 10.1074/jbc.M110.141135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Choudhuri T, Pal S, Agwarwal ML, Das T, et al. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002;512(1–3):334–40. doi: 10.1016/s0014-5793(02)02292-5. [DOI] [PubMed] [Google Scholar]
- 244.Tian B, Wang Z, Zhao Y, Wang D, et al. Effects of curcumin on bladder cancer cells and development of urothelial tumors in a rat bladder carcinogenesis model. Cancer Lett. 2008;264(2):299–308. doi: 10.1016/j.canlet.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 245.Shankar S, Srivastava RK. Involvement of Bcl-2 family members, phosphatidylinositol 3′-kinase/AKT and mitochondrial p53 in curcumin (diferulolylmethane)-induced apoptosis in prostate cancer. Int J Oncol. 2007;30(4):905–18. [PubMed] [Google Scholar]
- 246.Huang C, Ma WY, Goranson A, Dong Z. Resveratrol suppresses cell transformation and induces apoptosis through a p53-dependent pathway. Carcinogenesis. 1999;20(2):237–42. doi: 10.1093/carcin/20.2.237. [DOI] [PubMed] [Google Scholar]
- 247.Shankar S, Siddiqui I, Srivastava RK. Molecular mechanisms of resveratrol (3,4,5- trihydroxy-trans-stilbene) and its interaction with TNF-related apoptosis inducing ligand (TRAIL) in androgen-insensitive prostate cancer cells. Mol Cell Biochem. 2007;304(1–2):273–85. doi: 10.1007/s11010-007-9510-x. [DOI] [PubMed] [Google Scholar]
- 248.Schmidt F, Knobbe CB, Frank B, Wolburg H, et al. The topoisomerase II inhibitor, genistein, induces G2/M arrest and apoptosis in human malignant glioma cell lines. Oncol Rep. 2008;19(4):1061–6. [PubMed] [Google Scholar]
- 249.Khan R, Khan AQ, Lateef A, Rehman MU, et al. Glycyrrhizic acid suppresses the development of precancerous lesions via regulating the hyperproliferation, inflammation, angiogenesis and apoptosis in the colon of Wistar rats. PLoS One. 2013;8(2):e56020. doi: 10.1371/journal.pone.0056020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wang X, Bai H, Zhang X, Liu J, et al. Inhibitory effect of oleanolic acid on hepatocellular carcinoma via ERK-p53-mediated cell cycle arrest and mitochondrial-dependent apoptosis. Carcinogenesis. 2013;34(6):1323–30. doi: 10.1093/carcin/bgt058. [DOI] [PubMed] [Google Scholar]
- 251.Pal D, Sur S, Mandal S, Das A, et al. Prevention of liver carcinogenesis by amarogentin through modulation of G1/S cell cycle check point and induction of apoptosis. Carcinogenesis. 2012;33(12):2424–31. doi: 10.1093/carcin/bgs276. [DOI] [PubMed] [Google Scholar]
- 252.Santoro R, Marani M, Blandino G, Muti P, et al. Melatonin triggers p53Ser phosphorylation and prevents DNA damage accumulation. Oncogene. 2012;31(24):2931–42. doi: 10.1038/onc.2011.469. [DOI] [PubMed] [Google Scholar]
- 253.Luo H, Rankin GO, Li Z, Depriest L, et al. Kaempferol induces apoptosis in ovarian cancer cells through activating p53 in the intrinsic pathway. Food Chem. 2011;128(2):513–519. doi: 10.1016/j.foodchem.2011.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Amin AR, Thakur VS, Paul RK, Feng GS, et al. SHP-2 tyrosine phosphatase inhibits p73- dependent apoptosis and expression of a subset of p53 target genes induced by EGCG. Proc Natl Acad Sci U S A. 2007;104(13):5419–24. doi: 10.1073/pnas.0700642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Shammas MA, Neri P, Koley H, Batchu RB, et al. Specific killing of multiple myeloma cells by (−)-epigallocatechin-3-gallate extracted from green tea: biologic activity and therapeutic implications. Blood. 2006;108(8):2804–10. doi: 10.1182/blood-2006-05-022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sharif T, Alhosin M, Auger C, Minker C, et al. Aronia melanocarpa juice induces a redox-sensitive p73-related caspase 3-dependent apoptosis in human leukemia cells. PLoS One. 2012;7(3):e32526. doi: 10.1371/journal.pone.0032526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Li M, Zhang Z, Hill DL, Wang H, et al. Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res. 2007;67(5):1988–96. doi: 10.1158/0008-5472.CAN-06-3066. [DOI] [PubMed] [Google Scholar]
- 258.Leao M, Gomes S, Pedraza-Chaverri J, Machado N, et al. alpha-Mangostin and Gambogic Acid as Potential Inhibitors of the p53-MDM2 Interaction Revealed by a Yeast Approach. J Nat Prod. 2013;76(4):774–778. doi: 10.1021/np400049j. [DOI] [PubMed] [Google Scholar]
- 259.Sasiela CA, Stewart DH, Kitagaki J, Safiran YJ, et al. Identification of inhibitors for MDM2 ubiquitin ligase activity from natural product extracts by a novel high-throughput electrochemiluminescent screen. J Biomol Screen. 2008;13(3):229–37. doi: 10.1177/1087057108315038. [DOI] [PubMed] [Google Scholar]
- 260.Hardcastle IR, Ahmed SU, Atkins H, Farnie G, et al. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction based on an isoindolinone scaffold. J Med Chem. 2006;49(21):6209–21. doi: 10.1021/jm0601194. [DOI] [PubMed] [Google Scholar]
- 261.Herman-Antosiewicz A, Stan SD, Hahm ER, Xiao D, et al. Activation of a novel ataxia-telangiectasia mutated and Rad3 related/checkpoint kinase 1-dependent prometaphase checkpoint in cancer cells by diallyl trisulfide, a promising cancer chemopreventive constituent of processed garlic. Mol Cancer Ther. 2007;6(4):1249–61. doi: 10.1158/1535-7163.MCT-06-0477. [DOI] [PubMed] [Google Scholar]
- 262.Ting HJ, Yasmin-Karim S, Yan SJ, Hsu JW, et al. A positive feedback signaling loop between ATM and the vitamin D receptor is critical for cancer chemoprevention by vitamin D. Cancer Res. 2012;72(4):958–68. doi: 10.1158/0008-5472.CAN-11-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wu M, Kang MM, Schoene NW, Cheng WH. Selenium compounds activate early barriers of tumorigenesis. J Biol Chem. 2010;285(16):12055–62. doi: 10.1074/jbc.M109.088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Garcia A, Zheng Y, Zhao C, Toschi A, et al. Honokiol suppresses survival signals mediated by Ras-dependent phospholipase D activity in human cancer cells. Clin Cancer Res. 2008;14(13):4267–74. doi: 10.1158/1078-0432.CCR-08-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kopelovich L, Crowell JA, Fay JR. The epigenome as a target for cancer chemoprevention. J Natl Cancer Inst. 2003;95(23):1747–57. doi: 10.1093/jnci/dig109. [DOI] [PubMed] [Google Scholar]
- 266.Liu Z, Xie Z, Jones W, Pavlovicz RE, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009;19(3):706–9. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 267.Chen Y, Shu W, Chen W, Wu Q, et al. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factor kappa B and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007;101(6):427–33. doi: 10.1111/j.1742-7843.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- 268.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279(49):51163–71. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 269.Yu J, Peng Y, Wu LC, Xie Z, et al. Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS One. 2013;8(2):e55934. doi: 10.1371/journal.pone.0055934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Du L, Xie Z, Wu LC, Chiu M, et al. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr Cancer. 2012;64(8):1228–35. doi: 10.1080/01635581.2012.717682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32(4):537–44. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Volate SR, Muga SJ, Issa AY, Nitcheva D, et al. Epigenetic modulation of the retinoid X receptor alpha by green tea in the azoxymethane-Apc Min/+ mouse model of intestinal cancer. Mol Carcinog. 2009;48(10):920–33. doi: 10.1002/mc.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Fang MZ, Wang Y, Ai N, Hou Z, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63(22):7563–70. [PubMed] [Google Scholar]
- 274.Lee H, Zhang P, Herrmann A, Yang C, et al. Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci U S A. 2012;109(20):7765–9. doi: 10.1073/pnas.1205132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhu W, Qin W, Zhang K, Rottinghaus GE, et al. Trans-resveratrol alters mammary promoter hypermethylation in women at increased risk for breast cancer. Nutr Cancer. 2012;64(3):393–400. doi: 10.1080/01635581.2012.654926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Aronchik I, Kundu A, Quirit JG, Firestone GL. The antiproliferative response of indole-3- carbinol in human melanoma cells is triggered by an interaction with NEDD4-1 and disruption of wild-type PTEN degradation. Mol Cancer Res. 2014;12(11):1621–34. doi: 10.1158/1541-7786.MCR-14-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zheng J, Wu C, Lin Z, Guo Y, et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation--a novel mechanism suppressing liver fibrosis. FEBS J. 2014;281(1):88–103. doi: 10.1111/febs.12574. [DOI] [PubMed] [Google Scholar]
- 278.Park W, Amin AR, Chen ZG, Shin DM. New perspectives of curcumin in cancer prevention. Cancer Prev Res (Phila) 2013;6(5):387–400. doi: 10.1158/1940-6207.CAPR-12-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Dhar S, Kumar A, Li K, Tzivion G, et al. Resveratrol regulates PTEN/Akt pathway through inhibition of MTA1/HDAC unit of the NuRD complex in prostate cancer. Biochim Biophys Acta. 2015;1853(2):265–75. doi: 10.1016/j.bbamcr.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 280.Baek SJ, Wilson LC, Eling TE. Resveratrol enhances the expression of non-steroidal anti-inflammatory drug-activated gene (NAG-1) by increasing the expression of p53. Carcinogenesis. 2002;23(3):425–34. doi: 10.1093/carcin/23.3.425. [DOI] [PubMed] [Google Scholar]
- 281.Wilson LC, Baek SJ, Call A, Eling TE. Nonsteroidal anti-inflammatory drug-activated gene (NAG-1) is induced by genistein through the expression of p53 in colorectal cancer cells. Int J Cancer. 2003;105(6):747–53. doi: 10.1002/ijc.11173. [DOI] [PubMed] [Google Scholar]
- 282.Zhong Y, Krisanapun C, Lee SH, Nualsanit T, et al. Molecular targets of apigenin in colorectal cancer cells: involvement of p21, NAG-1 and p53. Eur J Cancer. 2010;46(18):3365–74. doi: 10.1016/j.ejca.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Brown DA, Lindmark F, Stattin P, Balter K, et al. Macrophage inhibitory cytokine 1: a new prognostic marker in prostate cancer. Clin Cancer Res. 2009;15(21):6658–64. doi: 10.1158/1078-0432.CCR-08-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Daniels LB, Clopton P, Laughlin GA, Maisel AS, et al. Growth-differentiation factor-15 is a robust, independent predictor of 11-year mortality risk in community-dwelling older adults: the Rancho Bernardo Study. Circulation. 2011;123(19):2101–10. doi: 10.1161/CIRCULATIONAHA.110.979740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wiklund FE, Bennet AM, Magnusson PK, Eriksson UK, et al. Macrophage inhibitory cytokine-1 (MIC-1/GDF15): a new marker of all-cause mortality. Aging Cell. 2010;9(6):1057–64. doi: 10.1111/j.1474-9726.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Yamamoto S, Tsuda H, Takano M, Tamai S, et al. PIK3CA mutations and loss of ARID1A protein expression are early events in the development of cystic ovarian clear cell adenocarcinoma. Virchows Arch. 2012;460(1):77–87. doi: 10.1007/s00428-011-1169-8. [DOI] [PubMed] [Google Scholar]
- 287.Liang H, Cheung LW, Li J, Ju Z, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22(11):2120–9. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Harper JW, Adami GR, Wei N, Keyomarsi K, et al. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 289.Ponnurangam S, Mammen JM, Ramalingam S, He Z, et al. Honokiol in combination with radiation targets notch signaling to inhibit colon cancer stem cells. Mol Cancer Ther. 2012;11(4):963–72. doi: 10.1158/1535-7163.MCT-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Kaushik G, Venugopal A, Ramamoorthy P, Standing D, et al. Honokiol inhibits melanoma stem cells by targeting notch signaling. Mol Carcinog. 2014 doi: 10.1002/mc.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Koduru S, Kumar R, Srinivasan S, Evers MB, et al. Notch-1 inhibition by Withaferin-A: a therapeutic target against colon carcinogenesis. Mol Cancer Ther. 2010;9(1):202–10. doi: 10.1158/1535-7163.MCT-09-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer. 2012;12(3):159–69. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 293.Gualberto A, Pollak M. Clinical development of inhibitors of the insulin-like growth factor receptor in oncology. Curr Drug Targets. 2009;10(10):923–36. doi: 10.2174/138945009789577945. [DOI] [PubMed] [Google Scholar]
- 294.Hartog H, Van Der Graaf WT, Boezen HM, Wesseling J. Treatment of breast cancer cells by IGF1R tyrosine kinase inhibitor combined with conventional systemic drugs. Anticancer Res. 2012;32(4):1309–18. [PubMed] [Google Scholar]
- 295.Yavari K, Taghikhani M, Maragheh MG, Mesbah-Namin SA, et al. SiRNA-mediated IGF-1R inhibition sensitizes human colon cancer SW480 cells to radiation. Acta Oncol. 2010;49(1):70–5. doi: 10.3109/02841860903334429. [DOI] [PubMed] [Google Scholar]
- 296.Wang YH, Wang ZX, Qiu Y, Xiong J, et al. Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1 receptor inhibits growth, reduces invasion, and enhances radiosensitivity in human osteosarcoma cells. Mol Cell Biochem. 2009;327(1–2):257–66. doi: 10.1007/s11010-009-0064-y. [DOI] [PubMed] [Google Scholar]
- 297.Yee D. Insulin-like growth factor receptor inhibitors: baby or the bathwater? J Natl Cancer Inst. 2012;104(13):975–81. doi: 10.1093/jnci/djs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Reidy DL, Vakiani E, Fakih MG, Saif MW, et al. Randomized, phase II study of the insulin-like growth factor-1 receptor inhibitor IMC-A12, with or without cetuximab, in patients with cetuximab- or panitumumab-refractory metastatic colorectal cancer. J Clin Oncol. 2010;28(27):4240–6. doi: 10.1200/JCO.2010.30.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Guha M. Anticancer IGF1R classes take more knocks. Nat Rev Drug Discov. 2013;12(4):250. doi: 10.1038/nrd3992. [DOI] [PubMed] [Google Scholar]
- 300.Hendrickson AW, Haluska P. Resistance pathways relevant to insulin-like growth factor-1 receptor-targeted therapy. Curr Opin Investig Drugs. 2009;10(10):1032–40. [PubMed] [Google Scholar]
- 301.Buck E, Gokhale PC, Koujak S, Brown E, et al. Compensatory insulin receptor (IR) activation on inhibition of insulin-like growth factor-1 receptor (IGF-1R): rationale for cotargeting IGF-1R and IR in cancer. Mol Cancer Ther. 2010;9(10):2652–64. doi: 10.1158/1535-7163.MCT-10-0318. [DOI] [PubMed] [Google Scholar]
- 302.Ulanet DB, Ludwig DL, Kahn CR, Hanahan D. Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy. Proc Natl Acad Sci U S A. 2010;107(24):10791–8. doi: 10.1073/pnas.0914076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Abraham J, Prajapati SI, Nishijo K, Schaffer BS, et al. Evasion mechanisms to Igf1r inhibition in rhabdomyosarcoma. Mol Cancer Ther. 2011;10(4):697–707. doi: 10.1158/1535-7163.MCT-10-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Shin DH, Min HY, El-Naggar AK, Lippman SM, et al. Akt/mTOR counteract the antitumor activities of cixutumumab, an anti-insulin-like growth factor I receptor monoclonal antibody. Mol Cancer Ther. 2011;10(12):2437–48. doi: 10.1158/1535-7163.MCT-11-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Bean GR, Ganesan YT, Dong Y, Takeda S, et al. PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci Signal. 2013;6(268):ra20. doi: 10.1126/scisignal.2003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.D’Angelo L, Piazzi G, Pacilli A, Prossomariti A, et al. A combination of eicosapentaenoic acid-free fatty acid, epigallocatechin-3-gallate and proanthocyanidins has a strong effect on mTOR signaling in colorectal cancer cells. Carcinogenesis. 2014;35(10):2314–20. doi: 10.1093/carcin/bgu173. [DOI] [PubMed] [Google Scholar]
- 307.Majumdar D, Jung KH, Zhang H, Nannapaneni S, et al. Luteolin nanoparticle in chemoprevention: in vitro and in vivo anticancer activity. Cancer Prev Res (Phila) 2014;7(1):65–73. doi: 10.1158/1940-6207.CAPR-13-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Siddiqui IA, Adhami VM, Bharali DJ, Hafeez BB, et al. Introducing nanochemoprevention as a novel approach for cancer control: proof of principle with green tea polyphenol epigallocatechin-3-gallate. Cancer Res. 2009;69(5):1712–6. doi: 10.1158/0008-5472.CAN-08-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Dickinson D, Yu H, Ohno S, Thomas C, et al. Epigallocatechin-3-gallate prevents autoimmune-associated down- regulation of p21 in salivary gland cells through a p53-independent pathway. Inflamm Allergy Drug Targets. 2014;13(1):15–24. doi: 10.2174/1871528112666131211102500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Cantero G, Campanella C, Mateos S, Cortes F. Topoisomerase II inhibition and high yield of endoreduplication induced by the flavonoids luteolin and quercetin. Mutagenesis. 2006;21(5):321–5. doi: 10.1093/mutage/gel033. [DOI] [PubMed] [Google Scholar]
- 311.Holy JM. Curcumin disrupts mitotic spindle structure and induces micronucleation in MCF-7 breast cancer cells. Mutat Res. 2002;518(1):71–84. doi: 10.1016/s1383-5718(02)00076-1. [DOI] [PubMed] [Google Scholar]
- 312.Chen L, Li WF, Wang HX, Zhao HN, et al. Curcumin cytotoxicity is enhanced by PTEN disruption in colorectal cancer cells. World J Gastroenterol. 2013;19(40):6814–24. doi: 10.3748/wjg.v19.i40.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kim YM, Yang S, Xu W, Li S, et al. Continuous in vitro exposure to low-dose genistein induces genomic instability in breast epithelial cells. Cancer Genet Cytogenet. 2008;186(2):78–84. doi: 10.1016/j.cancergencyto.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Denissova NG, Nasello CM, Yeung PL, Tischfield JA, et al. Resveratrol protects mouse embryonic stem cells from ionizing radiation by accelerating recovery from DNA strand breakage. Carcinogenesis. 2012;33(1):149–55. doi: 10.1093/carcin/bgr236. [DOI] [PubMed] [Google Scholar]
- 315.Jacque E, Billot K, Authier H, Bordereaux D, et al. RelB inhibits cell proliferation and tumor growth through p53 transcriptional activation. Oncogene. 2013;32(21):2661–9. doi: 10.1038/onc.2012.282. [DOI] [PubMed] [Google Scholar]
- 316.Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587(15):2346–51. doi: 10.1016/j.febslet.2013.05.059. [DOI] [PubMed] [Google Scholar]
- 317.Zhang J, Yang Y, Zhang Z, He Y, et al. Gankyrin plays an essential role in estrogen-driven and GPR30-mediated endometrial carcinoma cell proliferation via the PTEN/PI3K/AKT signaling pathway. Cancer Lett. 2013;339(2):279–87. doi: 10.1016/j.canlet.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 318.Lin C, Liang Y, Zhu H, Zhang J, et al. R280T mutation of p53 gene promotes proliferation of human glioma cells through GSK-3beta/PTEN pathway. Neurosci Lett. 2012;529(1):60–5. doi: 10.1016/j.neulet.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 319.Kong L, Schafer G, Bu H, Zhang Y, et al. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33(4):751–9. doi: 10.1093/carcin/bgs022. [DOI] [PubMed] [Google Scholar]
- 320.Moon SH, Kim DK, Cha Y, Jeon I, et al. PI3K/Akt and Stat3 signaling regulated by PTEN control of the cancer stem cell population, proliferation and senescence in a glioblastoma cell line. Int J Oncol. 2013;42(3):921–8. doi: 10.3892/ijo.2013.1765. [DOI] [PubMed] [Google Scholar]
- 321.Nallet-Staub F, Marsaud V, Li L, Gilbert C, et al. Pro-invasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J Invest Dermatol. 2014;134(1):123–32. doi: 10.1038/jid.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mizuno T, Murakami H, Fujii M, Ishiguro F, et al. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31(49):5117–22. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 323.Lamar JM, Stern P, Liu H, Schindler JW, et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109(37):E2441–50. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Zhang Y, Xu X, Zhang M, Bai X, et al. ARID1A is downregulated in non-small cell lung cancer and regulates cell proliferation and apoptosis. Tumour Biol. 2014;35(6):5701–7. doi: 10.1007/s13277-014-1755-x. [DOI] [PubMed] [Google Scholar]
- 325.Li Y, Zhang J, Zhang L, Si M, et al. Diallyl trisulfide inhibits proliferation, invasion and angiogenesis of osteosarcoma cells by switching on suppressor microRNAs and inactivating of Notch-1 signaling. Carcinogenesis. 2013;34(7):1601–10. doi: 10.1093/carcin/bgt065. [DOI] [PubMed] [Google Scholar]
- 326.Jia CY, Li HH, Zhu XC, Dong YW, et al. MiR-223 suppresses cell proliferation by targeting IGF-1R. PLoS One. 2011;6(11):e27008. doi: 10.1371/journal.pone.0027008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wang B, Wang H, Yang Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS One. 2012;7(10):e47053. doi: 10.1371/journal.pone.0047053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Nian W, Ao X, Wu Y, Huang Y, et al. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclindependent kinase 2. Oncol Lett. 2013;6(2):359–366. doi: 10.3892/ol.2013.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zhang Y, Chen X, Lian H, Liu J, et al. MicroRNA-503 acts as a tumor suppressor in glioblastoma for multiple antitumor effects by targeting IGF-1R. Oncol Rep. 2014;31(3):1445–52. doi: 10.3892/or.2013.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Shi ZM, Wang XF, Qian X, Tao T, et al. MiRNA-181b suppresses IGF-1R and functions as a tumor suppressor gene in gliomas. RNA. 2013;19(4):552–60. doi: 10.1261/rna.035972.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Ying L, Hofseth AB, Browning DD, Nagarkatti M, et al. Nitric oxide inactivates the retinoblastoma pathway in chronic inflammation. Cancer Res. 2007;67(19):9286–93. doi: 10.1158/0008-5472.CAN-07-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hudson JD, Shoaibi MA, Maestro R, Carnero A, et al. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190(10):1375–82. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Gudkov AV, Gurova KV, Komarova EA. Inflammation and p53: A Tale of Two Stresses. Genes Cancer. 2011;2(4):503–16. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Shrimali D, Shanmugam MK, Kumar AP, Zhang J, et al. Targeted abrogation of diverse signal transduction cascades by emodin for the treatment of inflammatory disorders and cancer. Cancer Lett. 2013;341(2):139–49. doi: 10.1016/j.canlet.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 335.Rogler G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014;345(2):235–41. doi: 10.1016/j.canlet.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 336.Covey TM, Edes K, Coombs GS, Virshup DM, et al. Alkylation of the tumor suppressor PTEN activates Akt and beta-catenin signaling: a mechanism linking inflammation and oxidative stress with cancer. PLoS One. 2010;5(10):e13545. doi: 10.1371/journal.pone.0013545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Miyake K, Tanonaka K, Minematsu R, Inoue K, et al. Possible therapeutic effect of naftidrofuryl oxalate on brain energy metabolism after microsphere-induced cerebral embolism. Br J Pharmacol. 1989;98(2):389–96. doi: 10.1111/j.1476-5381.1989.tb12609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Debeb BG, Cohen EN, Boley K, Freiter EM, et al. Pre-clinical studies of Notch signaling inhibitor RO4929097 in inflammatory breast cancer cells. Breast Cancer Res Treat. 2012;134(2):495–510. doi: 10.1007/s10549-012-2075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Durfort T, Tkach M, Meschaninova MI, Rivas MA, et al. Small interfering RNA targeted to IGF-IR delays tumor growth and induces proinflammatory cytokines in a mouse breast cancer model. PLoS One. 2012;7(1):e29213. doi: 10.1371/journal.pone.0029213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Garofalo C, Mancarella C, Grilli A, Manara MC, et al. Identification of common and distinctive mechanisms of resistance to different anti-IGF-IR agents in Ewing’s sarcoma. Mol Endocrinol. 2012;26(9):1603–16. doi: 10.1210/me.2012-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Indovina P, Marcelli E, Casini N, Rizzo V, et al. Emerging roles of RB family: new defense mechanisms against tumor progression. J Cell Physiol. 2013;228(3):525–35. doi: 10.1002/jcp.24170. [DOI] [PubMed] [Google Scholar]
- 342.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1(5):a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Wang Q, Ding H, Liu B, Li SH, et al. Addition of the Akt inhibitor triciribine overcomes antibody resistance in cells from ErbB2/Neu-positive/PTEN-deficient mammary tumors. Int J Oncol. 2014;44(4):1277–83. doi: 10.3892/ijo.2014.2271. [DOI] [PubMed] [Google Scholar]
- 344.Enderle L, McNeill H. Hippo gains weight: added insights and complexity to pathway control. Sci Signal. 2013;6(296):re7. doi: 10.1126/scisignal.2004208. [DOI] [PubMed] [Google Scholar]
- 345.Li J, Yang L, Qin W, Zhang G, et al. Adaptive induction of growth differentiation factor 15 attenuates endothelial cell apoptosis in response to high glucose stimulus. PLoS One. 2013;8(6):e65549. doi: 10.1371/journal.pone.0065549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wang Z, Azmi AS, Ahmad A, Banerjee S, et al. TW-37, a small-molecule inhibitor of Bcl-2, inhibits cell growth and induces apoptosis in pancreatic cancer: involvement of Notch-1 signaling pathway. Cancer Res. 2009;69(7):2757–65. doi: 10.1158/0008-5472.CAN-08-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Negi A, Ramarao P, Kumar R. Recent advancements in small molecule inhibitors of insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase as anticancer agents. Mini Rev Med Chem. 2013;13(5):653–81. doi: 10.2174/1389557511313050004. [DOI] [PubMed] [Google Scholar]
- 348.Kan CY, Petti C, Bracken L, Maritz M, et al. Up-regulation of survivin during immortalization of human myofibroblasts is linked to repression of tumor suppressor p16(INK4a) protein and confers resistance to oxidative stress. J Biol Chem. 2013;288(17):12032–41. doi: 10.1074/jbc.M112.447821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Liu TM, Ng WM, Tan HS, Vinitha D, et al. Molecular basis of immortalization of human mesenchymal stem cells by combination of p53 knockdown and human telomerase reverse transcriptase overexpression. Stem Cells Dev. 2013;22(2):268–78. doi: 10.1089/scd.2012.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Yasaei H, Gilham E, Pickles JC, Roberts TP, et al. Carcinogen-specific mutational and epigenetic alterations in INK4A, INK4B and p53 tumour-suppressor genes drive induced senescence bypass in normal diploid mammalian cells. Oncogene. 2013;32(2):171–9. doi: 10.1038/onc.2012.45. [DOI] [PubMed] [Google Scholar]
- 351.Ulbricht U, Sommer A, Beckmann G, Lutzenberger M, et al. Isogenic human mammary epithelial cell lines: novel tools for target identification and validation. Comprehensive characterization of an isogenic human mammary epithelial cell model provides evidence for epithelial-mesenchymal transition. Breast Cancer Res Treat. 2013;138(2):437–56. doi: 10.1007/s10549-013-2472-7. [DOI] [PubMed] [Google Scholar]
- 352.Cui H, Kong Y, Xu M, Zhang H. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer Res. 2013;73(11):3451–9. doi: 10.1158/0008-5472.CAN-12-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Mendivil A, Zhou C, Cantrell LA, Gehrig PA, et al. AMG 479, a novel IGF-1-R antibody, inhibits endometrial cancer cell proliferation through disruption of the PI3K/Akt and MAPK pathways. Reprod Sci. 2011;18(9):832–41. doi: 10.1177/1933719111398501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Wetterau LA, Francis MJ, Ma L, Cohen P. Insulin-like growth factor I stimulates telomerase activity in prostate cancer cells. J Clin Endocrinol Metab. 2003;88(7):3354–9. doi: 10.1210/jc.2002-021326. [DOI] [PubMed] [Google Scholar]
- 355.Zhang L, Yang Z, Ma A, Qu Y, et al. Growth arrest and DNA damage 45G down-regulation contributes to Janus kinase/signal transducer and activator of transcription 3 activation and cellular senescence evasion in hepatocellular carcinoma. Hepatology. 2014;59(1):178–89. doi: 10.1002/hep.26628. [DOI] [PubMed] [Google Scholar]
- 356.Lamonte G, Tang X, Chen JL, Wu J, et al. Acidosis induces reprogramming of cellular metabolism to mitigate oxidative stress. Cancer Metab. 2013;1(1):23. doi: 10.1186/2049-3002-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 358.Jiang P, Du W, Yang X. A critical role of glucose-6-phosphate dehydrogenase in TAp73- mediated cell proliferation. Cell Cycle. 2013;12(24):3720–6. doi: 10.4161/cc.27267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Fox MM, Phoenix KN, Kopsiaftis SG, Claffey KP. AMP-Activated Protein Kinase alpha 2 Isoform Suppression in Primary Breast Cancer Alters AMPK Growth Control and Apoptotic Signaling. Genes Cancer. 2013;4(1–2):3–14. doi: 10.1177/1947601913486346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Cordero-Espinoza L, Hagen T. Increased concentrations of fructose 2,6-bisphosphate contribute to the Warburg effect in phosphatase and tensin homolog (PTEN)-deficient cells. J Biol Chem. 2013;288(50):36020–8. doi: 10.1074/jbc.M113.510289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Chen WT, Zhu G, Pfaffenbach K, Kanel G, et al. GRP78 as a regulator of liver steatosis and cancer progression mediated by loss of the tumor suppressor PTEN. Oncogene. 2013 doi: 10.1038/onc.2013.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Clem BF, O’Neal J, Tapolsky G, Clem AL, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12(8):1461–70. doi: 10.1158/1535-7163.MCT-13-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Wang Z, Wu Y, Wang H, Zhang Y, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci U S A. 2014;111(1):E89–98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Yonekura S, Itoh M, Okuhashi Y, Takahashi Y, et al. Effects of the HIF1 inhibitor, echinomycin, on growth and NOTCH signalling in leukaemia cells. Anticancer Res. 2013;33(8):3099–103. [PubMed] [Google Scholar]
- 366.Menendez D, Shatz M, Resnick MA. Interactions between the tumor suppressor p53 and immune responses. Curr Opin Oncol. 2013;25(1):85–92. doi: 10.1097/CCO.0b013e32835b6386. [DOI] [PubMed] [Google Scholar]
- 367.Bian Y, Hall B, Sun ZJ, Molinolo A, et al. Loss of TGF-beta signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 2012;31(28):3322–32. doi: 10.1038/onc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Gabellini C, Del Bufalo D, Zupi G. Involvement of RB gene family in tumor angiogenesis. Oncogene. 2006;25(38):5326–32. doi: 10.1038/sj.onc.1209631. [DOI] [PubMed] [Google Scholar]
- 369.Dameron KM, Volpert OV, Tainsky MA, Bouck N. The p53 tumor suppressor gene inhibits angiogenesis by stimulating the production of thrombospondin. Cold Spring Harb Symp Quant Biol. 1994;59:483–9. doi: 10.1101/sqb.1994.059.01.053. [DOI] [PubMed] [Google Scholar]
- 370.Wen S, Stolarov J, Myers MP, Su JD, et al. PTEN controls tumor-induced angiogenesis. Proc Natl Acad Sci U S A. 2001;98(8):4622–7. doi: 10.1073/pnas.081063798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Whitson RJ, Lucia MS, Lambert JR. Growth differentiation factor-15 (GDF-15) suppresses in vitro angiogenesis through a novel interaction with connective tissue growth factor (CCN2) J Cell Biochem. 2013;114(6):1424–33. doi: 10.1002/jcb.24484. [DOI] [PubMed] [Google Scholar]
- 372.Li JL, Harris AL. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer Cell. 2005;8(1):1–3. doi: 10.1016/j.ccr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 373.Reinmuth N, Liu W, Fan F, Jung YD, et al. Blockade of insulin-like growth factor I receptor function inhibits growth and angiogenesis of colon cancer. Clin Cancer Res. 2002;8(10):3259–69. [PubMed] [Google Scholar]
- 374.Singh S, Johnson J, Chellappan S. Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochim Biophys Acta. 2010;1799(10–12):788–94. doi: 10.1016/j.bbagrm.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Kim KJ, Godarova A, Seedle K, Kim MH, et al. Rb suppresses collective invasion, circulation and metastasis of breast cancer cells in CD44-dependent manner. PLoS One. 2013;8(12):e80590. doi: 10.1371/journal.pone.0080590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Tan EH, Morton JP, Timpson P, Tucci P, et al. Functions of TAp63 and p53 in restraining the development of metastatic cancer. Oncogene. 2014;33(25):3325–33. doi: 10.1038/onc.2013.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Zhang LL, Liu J, Lei S, Zhang J, et al. PTEN inhibits the invasion and metastasis of gastric cancer via downregulation of FAK expression. Cell Signal. 2014;26(5):1011–20. doi: 10.1016/j.cellsig.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 378.Wang L, Shi S, Guo Z, Zhang X, et al. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8(6):e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 379.Wallin U, Glimelius B, Jirstrom K, Darmanis S, et al. Growth differentiation factor 15: a prognostic marker for recurrence in colorectal cancer. Br J Cancer. 2011;104(10):1619–27. doi: 10.1038/bjc.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Yan HB, Wang XF, Zhang Q, Tang ZQ, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis. 2014;35(4):867–76. doi: 10.1093/carcin/bgt398. [DOI] [PubMed] [Google Scholar]
- 381.Wang Z, Li Y, Banerjee S, Kong D, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109(4):726–36. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 382.Wang Z, Banerjee S, Li Y, Rahman KM, et al. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66(5):2778–84. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 383.Christofori G. New signals from the invasive front. Nature. 2006;441(7092):444–50. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 384.Bahr C, Groner B. The IGF-1 receptor and its contributions to metastatic tumor growth-novel approaches to the inhibition of IGF-1R function. Growth Factors. 2005;23(1):1–14. doi: 10.1080/08977190400020229. [DOI] [PubMed] [Google Scholar]
- 385.Mankame TP, Lingen MW. The RB tumor suppressor positively regulates transcription of the anti-angiogenic protein NOL7. Neoplasia. 2012;14(12):1213–22. doi: 10.1593/neo.121422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Johnson JL, Pillai S, Pernazza D, Sebti SM, et al. Regulation of matrix metalloproteinase genes by E2F transcription factors: Rb-Raf-1 interaction as a novel target for metastatic disease. Cancer Res. 2012;72(2):516–26. doi: 10.1158/0008-5472.CAN-11-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Guo G, Marrero L, Rodriguez P, Del Valle L, et al. Trp53 inactivation in the tumor microenvironment promotes tumor progression by expanding the immunosuppressive lymphoid-like stromal network. Cancer Res. 2013;73(6):1668–75. doi: 10.1158/0008-5472.CAN-12-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Lin SY, Dolfi SC, Amiri S, Li J, et al. P53 regulates the migration of mesenchymal stromal cells in response to the tumor microenvironment through both CXCL12-dependent and -independent mechanisms. Int J Oncol. 2013;43(6):1817–23. doi: 10.3892/ijo.2013.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461(7267):1084–91. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Wallace JA, Li F, Leone G, Ostrowski MC. Pten in the breast tumor microenvironment: modeling tumor-stroma coevolution. Cancer Res. 2011;71(4):1203–7. doi: 10.1158/0008-5472.CAN-10-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Serrano I, McDonald PC, Lock F, Muller WJ, et al. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Vanhara P, Lincova E, Kozubik A, Jurdic P, et al. Growth/differentiation factor-15 inhibits differentiation into osteoclasts--a novel factor involved in control of osteoclast differentiation. Differentiation. 2009;78(4):213–22. doi: 10.1016/j.diff.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 393.Ishida T, Hijioka H, Kume K, Miyawaki A, et al. Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol Lett. 2013;6(5):1201–1206. doi: 10.3892/ol.2013.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Wong TS, Chan WS, Li CH, Liu RW, et al. Curcumin alters the migratory phenotype of nasopharyngeal carcinoma cells through up-regulation of E-cadherin. Anticancer Res. 2010;30(7):2851–6. [PubMed] [Google Scholar]
- 395.Deng YT, Lin JK. EGCG inhibits the invasion of highly invasive CL1–5 lung cancer cells through suppressing MMP-2 expression via JNK signaling and induces G2/M arrest. J Agric Food Chem. 2011;59(24):13318–27. doi: 10.1021/jf204149c. [DOI] [PubMed] [Google Scholar]
- 396.Wang H, Bian S, Yang CS. Green tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1alpha. Carcinogenesis. 2011;32(12):1881–9. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Xie F, Lang Q, Zhou M, Zhang H, et al. The dietary flavonoid luteolin inhibits Aurora B kinase activity and blocks proliferation of cancer cells. Eur J Pharm Sci. 2012;46(5):388–96. doi: 10.1016/j.ejps.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 398.Tsui KH, Chung LC, Feng TH, Chang PL, et al. Upregulation of prostate-derived Ets factor by luteolin causes inhibition of cell proliferation and cell invasion in prostate carcinoma cells. Int J Cancer. 2012;130(12):2812–23. doi: 10.1002/ijc.26284. [DOI] [PubMed] [Google Scholar]
- 399.Ono M, Higuchi T, Takeshima M, Chen C, et al. Antiproliferative and apoptosis-inducing activity of curcumin against human gallbladder adenocarcinoma cells. Anticancer Res. 2013;33(5):1861–6. [PubMed] [Google Scholar]
- 400.Moore AB, Castro L, Yu L, Zheng X, et al. Stimulatory and inhibitory effects of genistein on human uterine leiomyoma cell proliferation are influenced by the concentration. Hum Reprod. 2007;22(10):2623–31. doi: 10.1093/humrep/dem185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Di X, Yu L, Moore AB, Castro L, et al. A low concentration of genistein induces estrogen receptor-alpha and insulin-like growth factor-I receptor interactions and proliferation in uterine leiomyoma cells. Hum Reprod. 2008;23(8):1873–83. doi: 10.1093/humrep/den087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Murillo G, Peng X, Torres KE, Mehta RG. Deguelin inhibits growth of breast cancer cells by modulating the expression of key members of the Wnt signaling pathway. Cancer Prev Res (Phila) 2009;2(11):942–50. doi: 10.1158/1940-6207.CAPR-08-0232. [DOI] [PubMed] [Google Scholar]
- 403.Ahmad N, Adhami VM, Gupta S, Cheng P, et al. Role of the retinoblastoma (pRb)-E2F/DP pathway in cancer chemopreventive effects of green tea polyphenol epigallocatechin-3-gallate. Arch Biochem Biophys. 2002;398(1):125–31. doi: 10.1006/abbi.2001.2704. [DOI] [PubMed] [Google Scholar]
- 404.Hardtner C, Multhoff G, Falk W, Radons J. (−)-Epigallocatechin-3-gallate, a green tea-derived catechin, synergizes with celecoxib to inhibit IL-1-induced tumorigenic mediators by human pancreatic adenocarcinoma cells Colo357. Eur J Pharmacol. 2012;684(1–3):36–43. doi: 10.1016/j.ejphar.2012.03.039. [DOI] [PubMed] [Google Scholar]
- 405.Hoffmann J, Junker H, Schmieder A, Venz S, et al. EGCG downregulates IL-1RI expression and suppresses IL-1-induced tumorigenic factors in human pancreatic adenocarcinoma cells. Biochem Pharmacol. 2011;82(9):1153–62. doi: 10.1016/j.bcp.2011.07.063. [DOI] [PubMed] [Google Scholar]
- 406.Lin Y, Shi R, Wang X, Shen HM. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr Cancer Drug Targets. 2008;8(7):634–46. doi: 10.2174/156800908786241050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Pratheeshkumar P, Son YO, Budhraja A, Wang X, et al. Luteolin inhibits human prostate tumor growth by suppressing vascular endothelial growth factor receptor 2-mediated angiogenesis. PLoS One. 2012;7(12):e52279. doi: 10.1371/journal.pone.0052279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Fang LY, Izumi K, Lai KP, Liang L, et al. Infiltrating macrophages promote prostate tumorigenesis via modulating androgen receptor-mediated CCL4-STAT3 signaling. Cancer Res. 2013;73(18):5633–46. doi: 10.1158/0008-5472.CAN-12-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Dai XZ, Yin HT, Sun LF, Hu X, et al. Potential therapeutic efficacy of curcumin in liver cancer. Asian Pac J Cancer Prev. 2013;14(6):3855–9. doi: 10.7314/apjcp.2013.14.6.3855. [DOI] [PubMed] [Google Scholar]
- 410.Subbaramaiah K, Sue E, Bhardwaj P, Du B, et al. Dietary polyphenols suppress elevated levels of proinflammatory mediators and aromatase in the mammary gland of obese mice. Cancer Prev Res (Phila) 2013;6(9):886–97. doi: 10.1158/1940-6207.CAPR-13-0140. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 411.Zhang Y, Li Q, Chen H. DNA methylation and histone modifications of Wnt genes by genistein during colon cancer development. Carcinogenesis. 2013;34(8):1756–63. doi: 10.1093/carcin/bgt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Chen FP, Chien MH. Phytoestrogens induce apoptosis through a mitochondria/caspase pathway in human breast cancer cells. Climacteric. 2014;17(4):385–92. doi: 10.3109/13697137.2013.869671. [DOI] [PubMed] [Google Scholar]
- 413.Fernandez D, Bolli P, Snedden W, Vasdev S, et al. Modulation of left ventricular hypertrophy by dietary salt and inhibition of angiotensin converting enzyme. J Hypertens Suppl. 1988;6(4):S145–7. doi: 10.1097/00004872-198812040-00042. [DOI] [PubMed] [Google Scholar]
- 414.Fischbach F, Thurmayr R, Kolben M, Graeff H. Possible prediction of neonatal and puerperal infections by intrapartum determination of polymorphonuclear granulocyte elastase levels in maternal blood. Eur J Clin Microbiol Infect Dis. 1988;7(4):588–9. doi: 10.1007/BF01962627. [DOI] [PubMed] [Google Scholar]
- 415.Heyninck K, Lahtela-Kakkonen M, Van der Veken P, Haegeman G, et al. Withaferin A inhibits NF-kappaB activation by targeting cysteine 179 in IKKbeta. Biochem Pharmacol. 2014;91(4):501–9. doi: 10.1016/j.bcp.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 416.Dell’Eva R, Ambrosini C, Minghelli S, Noonan DM, et al. The Akt inhibitor deguelin, is an angiopreventive agent also acting on the NF-kappaB pathway. Carcinogenesis. 2007;28(2):404–13. doi: 10.1093/carcin/bgl162. [DOI] [PubMed] [Google Scholar]
- 417.Suh YA, Kim JH, Sung MA, Boo HJ, et al. A novel antitumor activity of deguelin targeting the insulin-like growth factor (IGF) receptor pathway via up-regulation of IGF-binding protein-3 expression in breast cancer. Cancer Lett. 2013;332(1):102–9. doi: 10.1016/j.canlet.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Oh SH, Woo JK, Jin Q, Kang HJ, et al. Identification of novel antiangiogenic anticancer activities of deguelin targeting hypoxia-inducible factor-1 alpha. Int J Cancer. 2008;122(1):5–14. doi: 10.1002/ijc.23075. [DOI] [PubMed] [Google Scholar]
- 419.Sarkar FH, Li Y, Wang Z, Kong D. Cellular signaling perturbation by natural products. Cell Signal. 2009;21(11):1541–7. doi: 10.1016/j.cellsig.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Ding S, Hu A, Hu Y, Ma J, et al. Anti-hepatoma cells function of luteolin through inducing apoptosis and cell cycle arrest. Tumour Biol. 2014;35(4):3053–60. doi: 10.1007/s13277-013-1396-5. [DOI] [PubMed] [Google Scholar]
- 421.Premkumar DR, Jane EP, DiDomenico JD, Vukmer NA, et al. ABT-737 synergizes with bortezomib to induce apoptosis, mediated by Bid cleavage, Bax activation, and mitochondrial dysfunction in an Akt-dependent context in malignant human glioma cell lines. J Pharmacol Exp Ther. 2012;341(3):859–72. doi: 10.1124/jpet.112.191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Banerjee S, Li Y, Wang Z, Sarkar FH. Multi-targeted therapy of cancer by genistein. Cancer Lett. 2008;269(2):226–42. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Wesolowska O, Wisniewski J, Bielawska-Pohl A, Paprocka M, et al. Stilbenes as multidrug resistance modulators and apoptosis inducers in human adenocarcinoma cells. Anticancer Res. 2010;30(11):4587–93. [PubMed] [Google Scholar]
- 424.Yu LL, Wu JG, Dai N, Yu HG, et al. Curcumin reverses chemoresistance of human gastric cancer cells by downregulating the NF-kappaB transcription factor. Oncol Rep. 2011;26(5):1197–203. doi: 10.3892/or.2011.1410. [DOI] [PubMed] [Google Scholar]
- 425.Grogan PT, Sarkaria JN, Timmermann BN, Cohen MS. Oxidative cytotoxic agent withaferin A resensitizes temozolomide-resistant glioblastomas via MGMT depletion and induces apoptosis through Akt/mTOR pathway inhibitory modulation. Invest New Drugs. 2014;32(4):604–17. doi: 10.1007/s10637-014-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Yang YL, Ji C, Bi ZG, Lu CC, et al. Deguelin induces both apoptosis and autophagy in cultured head and neck squamous cell carcinoma cells. PLoS One. 2013;8(1):e54736. doi: 10.1371/journal.pone.0054736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Wang X, Hao MW, Dong K, Lin F, et al. Apoptosis induction effects of EGCG in laryngeal squamous cell carcinoma cells through telomerase repression. Arch Pharm Res. 2009;32(9):1263–9. doi: 10.1007/s12272-009-1912-8. [DOI] [PubMed] [Google Scholar]
- 428.Sadava D, Whitlock E, Kane SE. The green tea polyphenol, epigallocatechin-3-gallate inhibits telomerase and induces apoptosis in drug-resistant lung cancer cells. Biochem Biophys Res Commun. 2007;360(1):233–7. doi: 10.1016/j.bbrc.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 429.Berletch JB, Liu C, Love WK, Andrews LG, et al. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103(2):509–19. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Khaw AK, Hande MP, Kalthur G, Hande MP. Curcumin inhibits telomerase and induces telomere shortening and apoptosis in brain tumour cells. J Cell Biochem. 2013;114(6):1257–70. doi: 10.1002/jcb.24466. [DOI] [PubMed] [Google Scholar]
- 431.Nasiri M, Zarghami N, Koshki KN, Mollazadeh M, et al. Curcumin and silibinin inhibit telomerase expression in T47D human breast cancer cells. Asian Pac J Cancer Prev. 2013;14(6):3449–53. doi: 10.7314/apjcp.2013.14.6.3449. [DOI] [PubMed] [Google Scholar]
- 432.Khaw AK, Yong JW, Kalthur G, Hande MP. Genistein induces growth arrest and suppresses telomerase activity in brain tumor cells. Genes Chromosomes Cancer. 2012;51(10):961–74. doi: 10.1002/gcc.21979. [DOI] [PubMed] [Google Scholar]
- 433.Li Y, Liu L, Andrews LG, Tollefsbol TO. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009;125(2):286–96. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Ouchi H, Ishiguro H, Ikeda N, Hori M, et al. Genistein induces cell growth inhibition in prostate cancer through the suppression of telomerase activity. Int J Urol. 2005;12(1):73–80. doi: 10.1111/j.1442-2042.2004.00973.x. [DOI] [PubMed] [Google Scholar]
- 435.Fuggetta MP, Lanzilli G, Tricarico M, Cottarelli A, et al. Effect of resveratrol on proliferation and telomerase activity of human colon cancer cells in vitro. J Exp Clin Cancer Res. 2006;25(2):189–93. [PubMed] [Google Scholar]
- 436.Lanzilli G, Fuggetta MP, Tricarico M, Cottarelli A, et al. Resveratrol down-regulates the growth and telomerase activity of breast cancer cells in vitro. Int J Oncol. 2006;28(3):641–8. [PubMed] [Google Scholar]
- 437.Wang TT, Wang SK, Huang GL, Sun GJ. Luteolin induced-growth inhibition and apoptosis of human esophageal squamous carcinoma cell line Eca109 cells in vitro. Asian Pac J Cancer Prev. 2012;13(11):5455–61. doi: 10.7314/apjcp.2012.13.11.5455. [DOI] [PubMed] [Google Scholar]
- 438.Rao PS, Satelli A, Moridani M, Jenkins M, et al. Luteolin induces apoptosis in multidrug resistant cancer cells without affecting the drug transporter function: involvement of cell line-specific apoptotic mechanisms. Int J Cancer. 2012;130(11):2703–14. doi: 10.1002/ijc.26308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Zanotto-Filho A, Braganhol E, Edelweiss MI, Behr GA, et al. The curry spice curcumin selectively inhibits cancer cells growth in vitro and in preclinical model of glioblastoma. J Nutr Biochem. 2012;23(6):591–601. doi: 10.1016/j.jnutbio.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 440.Hossain M, Banik NL, Ray SK. Synergistic anti-cancer mechanisms of curcumin and paclitaxel for growth inhibition of human brain tumor stem cells and LN18 and U138MG cells. Neurochem Int. 2012;61(7):1102–13. doi: 10.1016/j.neuint.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Hahm ER, Lee J, Kim SH, Sehrawat A, et al. Metabolic alterations in mammary cancer prevention by withaferin A in a clinically relevant mouse model. J Natl Cancer Inst. 2013;105(15):1111–22. doi: 10.1093/jnci/djt153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Huang AC, Cheng HY, Lin TS, Chen WH, et al. Epigallocatechin gallate (EGCG), influences a murine WEHI-3 leukemia model in vivo through enhancing phagocytosis of macrophages and populations of T- and B-cells. In Vivo. 2013;27(5):627–34. [PubMed] [Google Scholar]
- 443.Bhattacharyya S, Md Sakib Hossain D, Mohanty S, Sankar Sen G, et al. Curcumin reverses T cell-mediated adaptive immune dysfunctions in tumor-bearing hosts. Cell Mol Immunol. 2010;7(4):306–15. doi: 10.1038/cmi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Jung YD, Ellis LM. Inhibition of tumour invasion and angiogenesis by epigallocatechin gallate (EGCG), a major component of green tea. Int J Exp Pathol. 2001;82(6):309–16. doi: 10.1046/j.1365-2613.2001.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Bagli E, Stefaniotou M, Morbidelli L, Ziche M, et al. Luteolin inhibits vascular endothelial growth factor-induced angiogenesis; inhibition of endothelial cell survival and proliferation by targeting phosphatidylinositol 3′-kinase activity. Cancer Res. 2004;64(21):7936–46. doi: 10.1158/0008-5472.CAN-03-3104. [DOI] [PubMed] [Google Scholar]
- 446.Arbiser JL, Klauber N, Rohan R, van Leeuwen R, et al. Curcumin is an in vivo inhibitor of angiogenesis. Mol Med. 1998;4(6):376–83. [PMC free article] [PubMed] [Google Scholar]
- 447.Fotsis T, Pepper M, Adlercreutz H, Fleischmann G, et al. Genistein, a dietary-derived inhibitor of in vitro angiogenesis. Proc Natl Acad Sci U S A. 1993;90(7):2690–4. doi: 10.1073/pnas.90.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Chen Y, Tseng SH. Review. Pro- and anti-angiogenesis effects of resveratrol. In Vivo. 2007;21(2):365–70. [PubMed] [Google Scholar]
- 449.Gao R, Shah N, Lee JS, Katiyar SP, et al. Withanone Rich Combination of Ashwagandha Withanolides Restricts Metastasis And Angiogenesis Through hnRNP-K. Mol Cancer Ther. 2014 doi: 10.1158/1535-7163.MCT-14-0324. [DOI] [PubMed] [Google Scholar]
- 450.Mohan R, Hammers HJ, Bargagna-Mohan P, Zhan XH, et al. Withaferin A is a potent inhibitor of angiogenesis. Angiogenesis. 2004;7(2):115–22. doi: 10.1007/s10456-004-1026-3. [DOI] [PubMed] [Google Scholar]
- 451.Lee H, Lee JH, Jung KH, Hong SS. Deguelin promotes apoptosis and inhibits angiogenesis of gastric cancer. Oncol Rep. 2010;24(4):957–63. doi: 10.3892/or.2010.957. [DOI] [PubMed] [Google Scholar]
- 452.Sharma C, Nusri Qel A, Begum S, Javed E, et al. (−)-Epigallocatechin-3-gallate induces apoptosis and inhibits invasion and migration of human cervical cancer cells. Asian Pac J Cancer Prev. 2012;13(9):4815–22. doi: 10.7314/apjcp.2012.13.9.4815. [DOI] [PubMed] [Google Scholar]
- 453.Farabegoli F, Papi A, Orlandi M. (−)-Epigallocatechin-3-gallate down-regulates EGFR, MMP-2, MMP-9 and EMMPRIN and inhibits the invasion of MCF-7 tamoxifen-resistant cells. Biosci Rep. 2011;31(2):99–108. doi: 10.1042/BSR20090143. [DOI] [PubMed] [Google Scholar]
- 454.Weng CJ, Yen GC. Flavonoids, a ubiquitous dietary phenolic subclass, exert extensive in vitro anti-invasive and in vivo anti-metastatic activities. Cancer Metastasis Rev. 2012;31(1–2):323–51. doi: 10.1007/s10555-012-9347-y. [DOI] [PubMed] [Google Scholar]
- 455.Mitra A, Chakrabarti J, Banerji A, Chatterjee A, et al. Curcumin, a potential inhibitor of MMP- 2 in human laryngeal squamous carcinoma cells HEp2. J Environ Pathol Toxicol Oncol. 2006;25(4):679–90. doi: 10.1615/jenvironpatholtoxicoloncol.v25.i4.70. [DOI] [PubMed] [Google Scholar]
- 456.Hussain A, Harish G, Prabhu SA, Mohsin J, et al. Inhibitory effect of genistein on the invasive potential of human cervical cancer cells via modulation of matrix metalloproteinase-9 and tissue inhibitors of matrix metalloproteinase-1 expression. Cancer Epidemiol. 2012;36(6):e387–93. doi: 10.1016/j.canep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 457.Li Y, Kucuk O, Hussain M, Abrams J, et al. Antitumor and antimetastatic activities of docetaxel are enhanced by genistein through regulation of osteoprotegerin/receptor activator of nuclear factor-kappaB (RANK)/RANK ligand/MMP-9 signaling in prostate cancer. Cancer Res. 2006;66(9):4816–25. doi: 10.1158/0008-5472.CAN-05-3752. [DOI] [PubMed] [Google Scholar]
- 458.Ji Q, Liu X, Fu X, Zhang L, et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLoS One. 2013;8(11):e78700. doi: 10.1371/journal.pone.0078700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Athar M, Back JH, Kopelovich L, Bickers DR, et al. Multiple molecular targets of resveratrol: Anti-carcinogenic mechanisms. Arch Biochem Biophys. 2009;486(2):95–102. doi: 10.1016/j.abb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269(2):199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 461.Thaiparambil JT, Bender L, Ganesh T, Kline E, et al. Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. Int J Cancer. 2011;129(11):2744–55. doi: 10.1002/ijc.25938. [DOI] [PubMed] [Google Scholar]
- 462.Thamilselvan V, Menon M, Thamilselvan S. Anticancer efficacy of deguelin in human prostate cancer cells targeting glycogen synthase kinase-3 beta/beta-catenin pathway. Int J Cancer. 2011;129(12):2916–27. doi: 10.1002/ijc.25949. [DOI] [PubMed] [Google Scholar]
- 463.Zgheib A, Lamy S, Annabi B. Epigallocatechin gallate targeting of membrane type 1 matrix metalloproteinase-mediated Src and Janus kinase/signal transducers and activators of transcription 3 signaling inhibits transcription of colony-stimulating factors 2 and 3 in mesenchymal stromal cells. J Biol Chem. 2013;288(19):13378–86. doi: 10.1074/jbc.M113.456533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Jang JY, Lee JK, Jeon YK, Kim CW. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. 2013;13:421. doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Chen SS, Michael A, Butler-Manuel SA. Advances in the treatment of ovarian cancer: a potential role of antiinflammatory phytochemicals. Discov Med. 2012;13(68):7–17. [PubMed] [Google Scholar]
- 466.Chen KC, Chen CY, Lin CR, Yang TY, et al. Luteolin attenuates TGF-beta1-induced epithelial-mesenchymal transition of lung cancer cells by interfering in the PI3K/Akt-NF-kappaB-Snail pathway. Life Sci. 2013;93(24):924–33. doi: 10.1016/j.lfs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 467.Xu Y, Zhang J, Han J, Pan X, et al. Curcumin inhibits tumor proliferation induced by neutrophil elastase through the upregulation of alpha1-antitrypsin in lung cancer. Mol Oncol. 2012;6(4):405–17. doi: 10.1016/j.molonc.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Dudas J, Fullar A, Romani A, Pritz C, et al. Curcumin targets fibroblast-tumor cell interactions in oral squamous cell carcinoma. Exp Cell Res. 2013;319(6):800–9. doi: 10.1016/j.yexcr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Zlotogorski A, Dayan A, Dayan D, Chaushu G, et al. Nutraceuticals as new treatment approaches for oral cancer--I: Curcumin. Oral Oncol. 2013;49(3):187–91. doi: 10.1016/j.oraloncology.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 470.Dai W, Wang F, He L, Lin C, et al. Genistein inhibits hepatocellular carcinoma cell migration by reversing the epithelial-mesenchymal transition: Partial mediation by the transcription factor NFAT. Mol Carcinog. 2013 doi: 10.1002/mc.22100. [DOI] [PubMed] [Google Scholar]
- 471.Shamim U, Hanif S, Albanyan A, Beck FW, et al. Resveratrol-induced apoptosis is enhanced in low pH environments associated with cancer. J Cell Physiol. 2012;227(4):1493–500. doi: 10.1002/jcp.22865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Nair AS, Shishodia S, Ahn KS, Kunnumakkara AB, et al. Deguelin, an Akt inhibitor, suppresses IkappaBalpha kinase activation leading to suppression of NF-kappaB-regulated gene expression, potentiation of apoptosis, and inhibition of cellular invasion. J Immunol. 2006;177(8):5612–22. doi: 10.4049/jimmunol.177.8.5612. [DOI] [PubMed] [Google Scholar]