Abstract
Gene regulation in mammals involves a complex interplay between promoters and distal regulatory elements that function in concert to drive precise spatiotemporal gene expression programs. However, the dynamics of the distal gene regulatory landscape and its function in the transcriptional reprogramming that underlies neurogenesis and neuronal activity remain largely unknown. Here, we performed a combinatorial analysis of genome-wide data sets for chromatin accessibility (FAIRE-seq) and the enhancer mark H3K27ac, revealing the highly dynamic nature of distal gene regulation during neurogenesis, which gets progressively restricted to distinct genomic regions as neurons acquire a post-mitotic, terminally differentiated state. We further find that the distal accessible and active regions serve as target sites for distinct transcription factors that function in a stage-specific manner to contribute to the transcriptional program underlying neuronal commitment and maturation. Mature neurons respond to a sustained activity of NMDA receptors by epigenetic reprogramming at a large number of distal regulatory regions as well as dramatic reorganization of super-enhancers. Such massive remodeling of the distal regulatory landscape in turn results in a transcriptome that confers a transient loss of neuronal identity and gain of cellular plasticity. Furthermore, NMDA receptor activity also induces many novel prosurvival genes that function in neuroprotective pathways. Taken together, these findings reveal the dynamics of the distal regulatory landscape during neurogenesis and uncover novel regulatory elements that function in concert with epigenetic mechanisms and transcription factors to generate the transcriptome underlying neuronal development and activity.
The nervous system is the most complex mammalian organ. How this complexity is generated during development remains very poorly understood. The neural plate represents the primordium of the central nervous system (CNS) during development. It consists of a single sheet of neuroepithelial (NE) cells that divide symmetrically with a high rate to allow planar expansion of the neural plate and generate neural tube via the process called neurulation. At mid-gestation, between embryonic day 9 (E9) and 10 (E10) in mice, the first neurons of the nervous system are born, signaling an important transition in the development of neural progenitor cells. Concomitant with the potential to generate neurons, the progenitors acquire the identity of radial glial (RG) cells (Götz and Huttner 2005; Martynoga et al. 2012). In the developing brain, radial glial (RG) cells in the ventricular zone (VZ) undergo asymmetric division, and daughter cells migrate toward the cortical plate (CP), passing through the subventricular zone (SVZ). Our understanding of the transcriptional control of neurogenesis in the cerebral cortex by sequence-specific transcription factors has increased; but despite exciting developments (Feng et al. 2007; Borrelli et al. 2008; Hu et al. 2012; Lister et al. 2013), very little is known about the dynamics of chromatin accessibility and distal epigenetic gene regulation during embryonic neurogenesis and how stage-specific transcription factors utilize differential regulatory regions in driving the stage-specific transcriptional program.
The neuronal signaling via N-methyl-d-aspartate (NMDA) receptors plays a critical role in the development of the CNS and in adult neuroplasticity, learning, and memory (Koenig 1995; Milner et al. 1998). NMDA receptors have also been implicated in the etiology of several neurological disorders (Choi 1992; Lipton and Rosenberg 1994; Hardingham and Bading 2003; Bossy-Wetzel et al. 2004). Excessive stimulation of the NMDA receptor results in disorders involving acute insult to the brain with deprivation of blood supply (e.g., ischemic stroke, traumatic brain injury) (Choi 1988; Kalia et al. 2008). Moreover, neurotoxicity induced via hyper-stimulation of the NMDA receptor also facilitates slow-progressing neurodegenerative diseases (e.g., Huntington's, Alzheimer's, Parkinson's, and amyotrophic lateral sclerosis) (Young et al. 1988; Ulas et al. 1994; Ikonomidou et al. 1996; Blanchet et al. 1997; Snyder et al. 2005; Ahmed et al. 2011; Sgambato-Faure and Cenci 2012; Mehta et al. 2013) and disorders arising from the sensitization of neurons (e.g., epilepsy, neuropathic pain) (Rogawski 1992; Woolf and Mannion 1999). The NMDA receptor has been extensively characterized via electrophysiological assays, and genetic and chemical manipulations. NMDA receptors require the binding of glycine and glutamate in combination with the release of voltage-dependent magnesium blockage. In addition to its role during CNS development, neuronal activity via NMDA receptors also imparts neuronal plasticity, memory formation, and learning that require associated transcriptional changes to mediate physiological responses (Huh et al. 2000; Kandel 2001; Nestler and Landsman 2001; Zhang et al. 2007; Chen et al. 2014). Evidence suggests that NMDA receptor activation leads to strengthening of synapses through long-term potentiation (LTP) and to the weakening of synapses through long-term depression (LTD) (Sanchez-Perez et al. 2005; Massey and Bashir 2007; Zhang et al. 2007; Chen et al. 2014; Connor and Wang 2015). Recent findings have revealed early transcriptome responses following neuronal activity, including via NMDA receptors that further underlie many aspects of neuronal development (Shaywitz and Greenberg 1999; West et al. 2001; Tao et al. 2002; Pokorska et al. 2003; Zhang et al. 2007; Kim et al. 2010; Chen et al. 2014; Malik et al. 2014). Importantly however, the effect of long-term excitation of the NDMA receptor on the gene expression program remains unknown. Furthermore, a systematic genome-wide analysis to delineate whether epigenetic reprogramming underlies the response to long-term NMDA activity has not been conducted.
In eukaryotes, nucleosomes provide a basic layer of transcription repression by reducing access to DNA. A widely emerging concept is that nucleosome positioning and occupancy are organized by the combinatorial action of transcription factors, epigenetic regulators, and DNA sequence to regulate DNA accessibility (Bell et al. 2011). In this regard, open chromatin profiling has been widely used to identify regulatory elements that predict cell-type–specific functional behaviors (Thurman et al. 2012). Active regulatory elements such as promoters and enhancers are accessible and are marked by H3K27ac, thereby allowing a study of the activity state of both proximal and distal regulatory elements (Creyghton et al. 2010; Rada-Iglesias et al. 2011; Bonn et al. 2012). Recently, the term “super-enhancer” was used to describe groups of putative enhancers in close genomic proximity that were further shown to drive expression of genes that define cell identity (Hnisz et al. 2013; Whyte et al. 2013; Pott and Lieb 2014). To understand cell-fate determinants at the level of transcriptional regulation, it is crucial to identify a reliable set of regulatory elements that actively contribute to regulation of gene expression during processes of cell fate commitment. One of the most effective means of discovering these regulatory elements is through the identification of nucleosome-depleted regions (“open chromatin”). The FAIRE assay has emerged as a simple, unbiased but robust, and high-throughput method to identify such functional regulatory regions in a broad range of organisms and cell types (Giresi et al. 2007; Song et al. 2011; Simon et al. 2012; Koohy et al. 2013). Here, we performed a combinatorial analysis of chromatin accessibility using FAIRE assay and the enhancer mark H3K27ac to elucidate the dynamics and function of the distal gene regulatory landscape during neurogenesis and in response to neuronal activity.
Results
FAIRE-seq sensitively identifies accessible chromatin during neurogenesis
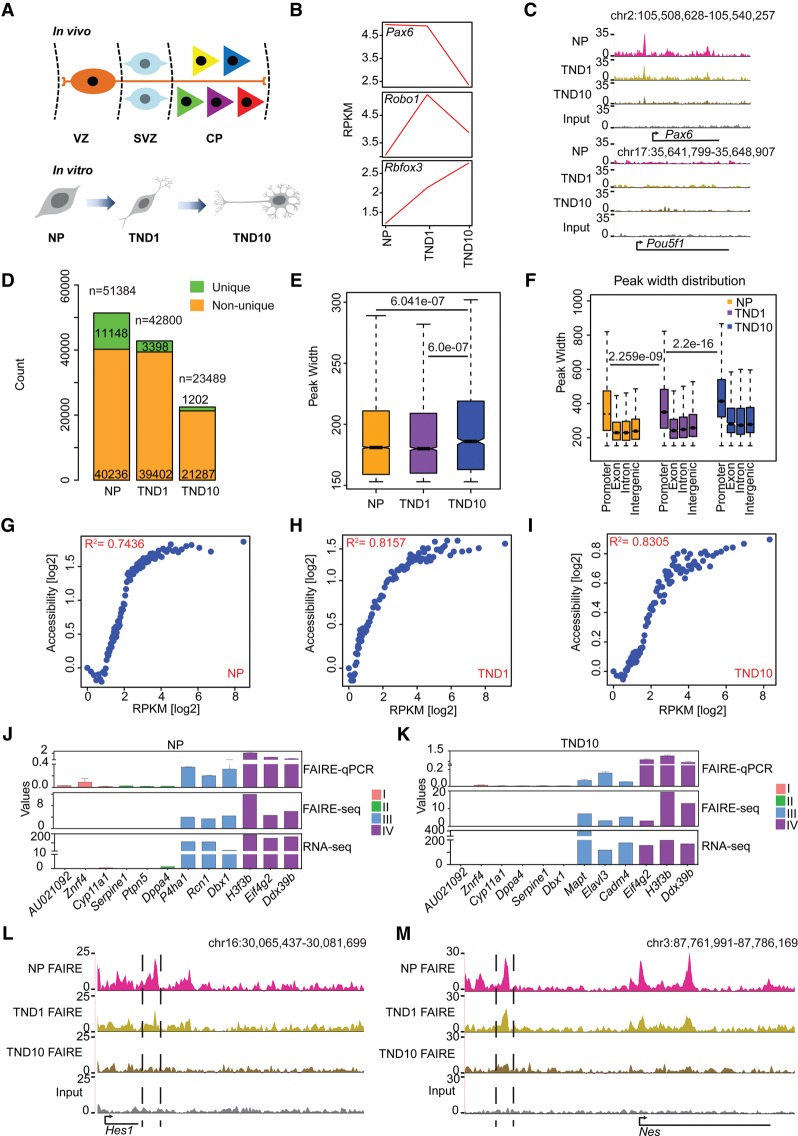
To investigate changes in chromatin accessibility during neurogenesis, we adapted a highly refined system that uses embryonic stem cells (ESC) to generate >95% pure neuronal progenitor (NP) cells (radial glial–like) and subsequently terminally differentiated pyramidal neurons (TN) (Bibel et al. 2004). Previous studies, including our own work, have revealed highly synchronous and reproducible changes in the epigenome and transcriptome during neuronal differentiation in this system that were also in good agreement with mouse primary cortical neurons (Fig. 1A,B; Mohn et al. 2008; Lienert et al. 2011; Stadler et al. 2011; Tiwari et al. 2012a,b; Thakurela et al. 2013). We performed the FAIRE assay at three distinct time points during neuronal differentiation: NP, TN day 1 (TND1) (immediate/early neurons, marking onset of neurogenesis), and TN day 10 (TND10) (late neurons, representing terminally differentiated postmitotic neurons) and subjected the derived material to high-throughput sequencing (FAIRE-seq) (Supplemental Fig. S1A–E; Supplemental Table S1). Exploration of UCSC Genome Browser tracks revealed the expected patterns of accessible (Pax6) and inaccessible (Pou5f1) regions in our FAIRE-seq data (Fig. 1C). Further analysis demonstrated that neuronal differentiation accompanies a progressive reduction in the number of both total and stage-specific (unique) accessible sites (Fig. 1D; Supplemental Fig. S1F). This indicates that post-mitotic differentiated cells have a more compact and defined accessible chromatin state, which may reflect their restricted developmental potential. Genomic distribution of FAIRE sites revealed that a large number of peaks occurred at intergenic regions followed by promoters in NP and TND1; whereas in TND10, promoters exhibited slightly higher accessibility than intergenic regions (Supplemental Fig. S1G,H). Exons in all stages exhibited much less openness, followed by introns (Supplemental Fig. S1G). Interestingly, we also observed that fully differentiated neurons have significantly higher peak widths, and promoters exhibit higher accessibility than other genomic locations at all stages of neuronal differentiation (Fig. 1E,F).
Figure 1.
FAIRE-seq sensitively identifies proximal and distal accessible regions during neuronal differentiation. (A) Schematic representation of the in vitro differentiation system compared to respective in vivo stages. (B) Line plot showing the expression of established marker genes for different neuronal stages. (C) UCSC Genome Browser tracks indicating the presence of an open region at Pax6 promoter (gene active in NP) and the absence of any active region in the Pou5f1 gene locus (active in ES cells only). (D) Stacked bar plot of the total and unique FAIRE peak count. (E) Box plot showing distribution of peak widths of unique NP, TND1, and TND10 peaks. P-values are calculated using the Wilcoxon test. (F) Peak width distribution in different genomic regions. P-values are calculated using the Wilcoxon test. (G–I) Scatter plot comparing gene expression and promoter accessibility. Genes were binned into percentiles, and the mean expression and mean promoter accessibility of each bin were plotted. The x-axis shows gene expression in reads per kilobase of transcript per million mapped reads (RPKM); the y-axis represents normalized FAIRE enrichment at promoters. (J,K) FAIRE-qPCR validation of promoter accessibility and gene expression. Genes are categorized into four classes: (I) not expressed in any of the three stages; (II) not expressed in the analyzed stage but expressed in at least one other stage; (III) expression specific to the analyzed stage; and (IV) expressed in all the stages. Values on the y-axis represent FAIRE/input normalized to Hspa8 (FAIRE qPCR), normalized enrichment score (FAIRE-seq), and RPKM for normalized expression (RNA-seq). (L) UCSC Genome Browser tracks showing the presence of a distal regulatory element near Hes1 (a NP specific gene). (M) Same as in L for the nestin gene (Nes).
An examination of the relationship between promoter accessibility and transcription revealed a very high positive correlation between promoter openness and gene expression at all three stages (Fig. 1G–I). Surprisingly, such correlation increased following acquisition of the neuronal state compared with neuronal progenitors (Fig. 1G–I). We further validated a number of genes for their promoter accessibility using FAIRE-qPCR, which fully corroborated with the observations based on genome-wide FAIRE-seq and gene expression data (Fig. 1J,K). As recent data suggest that distal regulatory elements are critical players in orchestrating the stage-specific gene expression profile underlying cellular identity during differentiation, we next focused on uncovering distal regulatory elements and their contribution to the transcriptional reprogramming underlying neurogenesis (Kim et al. 2010; Malik et al. 2014). A closer look at UCSC Genome Browser tracks revealed that FAIRE-seq is also highly sensitive and specific for the identification of potential distal regulatory regions (Fig. 1L,M). Overall, these results suggest that FAIRE-seq can sensitively identify both proximal and distal accessible regions during neurogenesis.
Distal open regions marked with H3K27ac define cell-type–specific transcriptional program
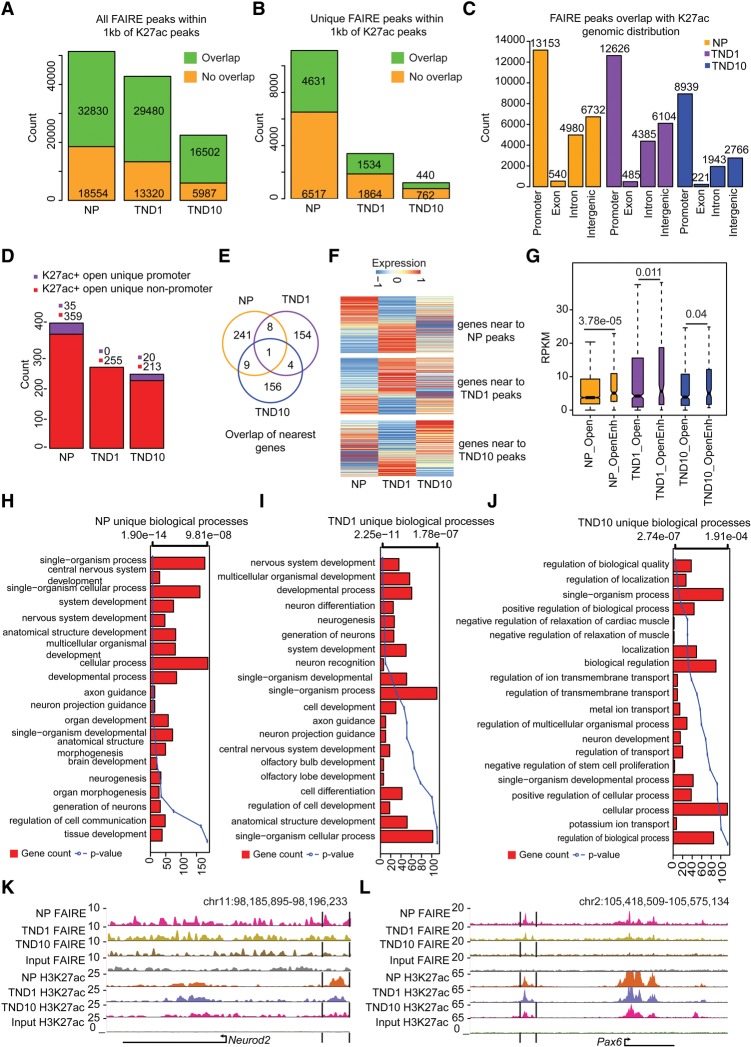
The acetylation of lysine 27 at histone 3 (H3K27ac) has been used as a mark to distinguish active versus inactive distal regulatory elements that function to primarily regulate the expression of proximal genes (Creyghton et al. 2010; Rada-Iglesias et al. 2011; Bonn et al. 2012; Chen et al. 2014). To identify such regulatory elements, we generated H3K27ac ChIP-seq profiles for the three stages of neurogenesis, and extensive quality controls confirmed the high quality of the derived data (Supplemental Fig. S2A–E; Supplemental Table S2). Computational analysis revealed a large number of H3K27ac-enriched regions in each cell type (Supplemental Fig. S2F). Further investigation demonstrated that although the total number of H3K27ac sites remained similar, fully differentiated neurons gained more unique H3K27ac sites compared with NP and TND1 (Supplemental Fig. S2F,G). Furthermore, these unique peaks in neurons displayed significantly higher peak widths (Supplemental Fig. S2H). We also observed a gradual loss of H3K27ac enrichment at promoters (Supplemental Fig. S2I). We speculated that accessible H3K27ac regions that are targeted by regulatory factors could function as sites of high regulatory activity. Interestingly, we observed that the majority of FAIRE-identified open regions harbored H3K27ac within 1 kb for all three cell types, but the fraction of such overlap was higher in differentiated neurons than dividing neuronal progenitors. (Fig. 2A; Supplemental Fig. S2J). Such occurrence of FAIRE positive sites with H3K27ac was also observed at a large number of stage-specific accessible regions (Fig. 2B; Supplemental Fig. S2K). Furthermore, reverse overlap of the H3K27ac sites with FAIRE revealed that many H3K27ac sites were not open and, consequently, may not be functionally active (Supplemental Fig. S2L–O). The genomic distribution of all double-positive sites indicated that a majority were located at promoters, least in exons followed by introns (Fig. 2C; Supplemental Fig. 2P). After promoters, intergenic regions contained the highest number of such sites, likely representing potential distal regulatory elements (Fig. 2C).
Figure 2.
Distal regulatory elements define the cell-type–specific transcriptome during neuronal development. (A) Stacked bar plot showing fraction of total FAIRE peaks overlapping with H3K27ac sites in NP, TND1, and TND10. Peaks were considered to be overlapping if they were within a distance of 1 kb. (B) Same as in A but for unique peaks. (C) Bar plot showing the distribution of FAIRE and H3K27ac overlapping peaks in different genomic regions in NP, TND1, and TND10 stages. (D) Stacked bar plot showing promoter and nonpromoter peaks that are open and uniquely acquire H3K27ac in NP, TND1, and TND10. (E) Venn diagram representing the overlap of genes near the sites identified in D. (F) Heat map showing expression of genes nearest to the peaks that uniquely gain H2K27ac in NP, TND1, or TND10. (G) Box plot showing differences in expression for genes that are near open H3K27ac sites (OpenEnh) and only open sites (Open). P-values are calculated using the Wilcoxon test. (H–J) Bar plots showing enrichment of biological processes for genes near NP, TND1, and TND10 open and unique H3K27ac-positive sites. The bars reflect the number of genes in each category, and the lines represent the multiple testing-corrected P-values, displayed as an alternate x-axis, of the corresponding GO terms. (K,L) Browser tracks for Neurod2 and Pax6 showing potential distal regulatory regions.
As H3K27ac also marks active promoters, we classified all accessible sites enriched in H3K27ac into two classes: promoters and nonpromoters. Interestingly, sites that are H3K27ac-unique and also accessible were almost exclusively located in nonpromoter regions, arguing that the unique transcription profiles of different cellular stages during neuronal differentiation may be largely determined by distal regulatory elements (Fig. 2D). Moreover, the gene sets located in proximity to these H3K27ac-unique and accessible sites were mostly nonoverlapping (Fig. 2E). Expression analysis of these nearest genes demonstrated that ∼40%–60% of genes were up-regulated compared with the adjacent state (Fig. 2F; Supplemental Fig. S2Q). The genes that do not exhibit induced behavior in association with such distal elements may be wrongly assigned due to computational limitations and that the enhancer-nearest TSS pairing only holds for up to 40% of enhancers (Andersson et al. 2014; Doyle et al. 2014; Samee and Sinha 2014). The correlation of unique distal H3K27ac-accessible sites with nearest gene expression was much higher in TND1 and TND10 than in NPs, further suggesting that distal gene regulation becomes more defined as cells reach a terminally differentiated state. We further observed that the genes that have nearby open and H3K27ac-positive regions exhibit significantly higher expression than genes that only have accessible regions without H3K27ac, suggesting that the presence of H3K27ac at these open sites has an enhancing effect on transcription (Fig. 2G), as previously suggested (Rada-Iglesias et al. 2011; Bonn et al. 2012). GO enrichment analysis showed that these genes are involved in neuronal development for NP (e.g., Pax6 and Neurod2) and TND1 (e.g., Robo1), whereas TND10 (e.g., Nlgn1) genes were involved in more specialized neuronal functions such as ion transport, localization, and regulation of transmembrane transport (Fig. 2H–L). Furthermore, a comparison of TND10 unique H3K27ac sites with similar data from other cell types confirmed that these sites are neuron-specific and not a general consequence of the differentiation process (Supplemental Fig. S2R). These findings suggest that distal accessible regions marked with H3K27ac contribute to the stage-specific transcriptional program during neurogenesis.
Distal regulatory elements recruit distinct transcription factors to regulate gene expression during neurogenesis
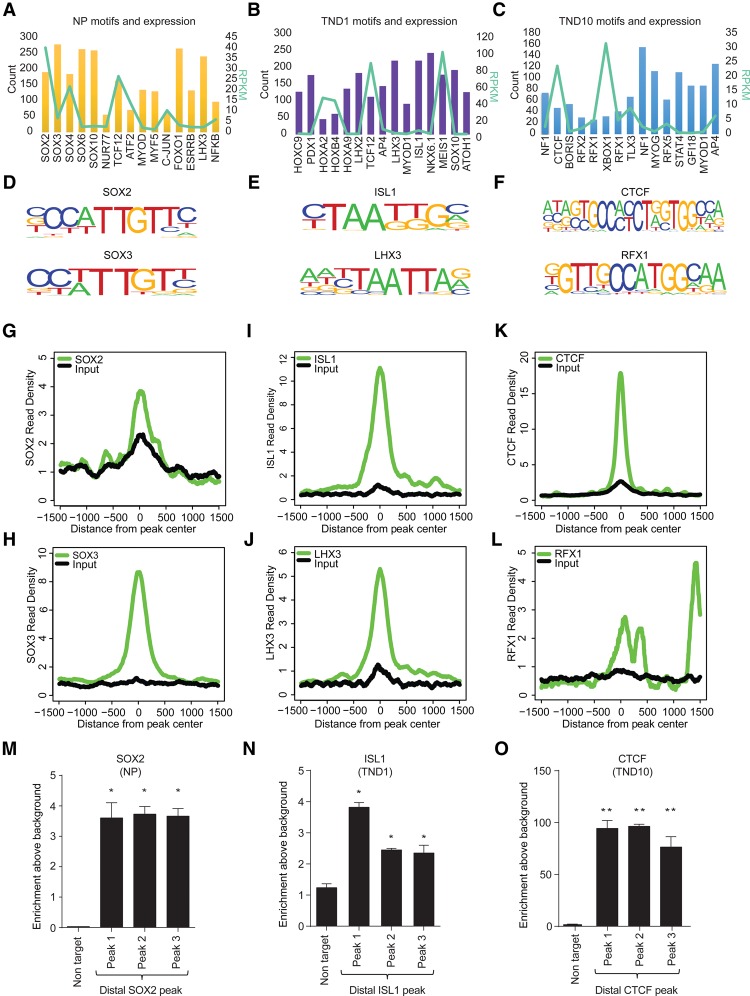
To gain understanding into the action of these distal regulatory elements, we next investigated whether they are enriched for binding sites of regulatory factors. Motif prediction analysis of these open and H3K27ac-positive sites revealed motifs for many known factors, a majority of which were identified in a stage-specific manner (Fig. 3A–C). Furthermore, these transcription factors were expressed in the predicted stage (Fig. 3A–C, line graph). Importantly, a number of these factors have been previously implicated in neuronal development (Supplemental Table S3). To test the targeting of predicted transcription factors at these distal sites, we chose two factors for each stage for which genome-wide binding data sets were publicly available for matching stages of neuronal differentiation (SOX2 and SOX3 for NP; LHX3 and ISL1 for TND1; and CTCF and RFX1 for TND10) and were shown to be critical for the respective stages (Fig. 3D–F; Uwanogho et al. 1995; Sharma et al. 1998; Tanaka et al. 2011; Dekker 2012; Hirayama et al. 2012; Lee et al. 2012, 2013). Comparative analysis of these ChIP-seq data sets with open and H3K27ac-positive sites revealed a high enrichment for these factors at distal regions in the respective stages (Fig. 3G–L). To provide ultimate validation of such transcription factor occupancy within our neuronal differentiation system, we performed ChIP assays for one transcription factor for each stage of neurogenesis (SOX2 for NP; ISL1 for TND1; and CTCF for TND10). These results show that indeed these three factors, viz. SOX2, ISL1, and CTCF, are significantly enriched at the identified distal regulatory elements in neuronal progenitors, early neurons, and mature neurons respectively (Fig. 3 M–O). These findings also argue that the function of distal open sites involves a crosstalk of epigenetic mechanisms and critical transcription factors in defining the cell-type–specific transcriptional profile during neuronal differentiation.
Figure 3.
Distinct transcription factors are targeted to distal regulatory elements in a stage-specific manner during neurogenesis. (A–C) Bar and overlapping line plot showing identified motifs and their counts in NP, TND1, and TND10 at uniquely gained H3K27ac nonpromoter sites (bars, main y-axis) and their expression (line, alternate y-axis, light green). (D–F) Motif sequence of the selected transcription factors from the three stages. (G–L) Plots showing enrichment of the corresponding transcription factors around the center of the uniquely gained open H3K27ac peaks in NP (SOX2 and SOX3) (G,H), TND1 (LHX3 and ISL1) (I,J), and TND10 (CTCF and RFX1) (K,L). The x-axis shows the distance from the peak center; the y-axis represents normalized enrichment of the corresponding transcription factor. (M–O) ChIP-qPCR validation of selected distal regions for their occupancy using specific antibody, SOX2 in NP (M), ISL1 in TND1 (N), and CTCF in TND10 (O). Average enrichments from independent assays are plotted on the y-axis as a ratio of precipitated DNA (bound) to total input DNA and then further normalized to an intergenic region. Error bars represent the SEM of independent biological replicates. (*) P < 0.05; (**) P < 0.01, Student's t-test.
NMDA receptor activation results in massive transcriptional reprogramming in neurons
Our observations suggested a highly dynamic nature of epigenome and transcriptome during neurogenesis that becomes progressively more defined as neurons acquire a terminally differentiated, postmitotic identity. We were next curious to investigate whether the acquired epigenetic and transcriptional profile of neurons could be modulated in response to neuronal function such as neuronal activity. Neuronal activity is mediated via calcium-permeable receptors such as NMDA receptors (NMDARs). To study the effects of a prolonged NMDAR stimulation, we treated differentiated neurons with the established agonist NDMA for 6 h. We confirmed that such treatment did not lead to any cytotoxicity or apoptotic response (Supplemental Fig. S3A–C). Furthermore, although the NMDA treatment led to induction of classical plasticity genes in a manner similar to glutamate, this was effectively blocked by the NMDAR antagonist D-2-amino-5-phosphonovaleric acid (DAPV), suggesting the specificity of the response (Supplemental Fig. S3D). Moreover, the established neuronal activity genes showed a dose-dependent modulation in both ESC-derived neurons and cortical neurons (Supplemental Fig. S3E,F).
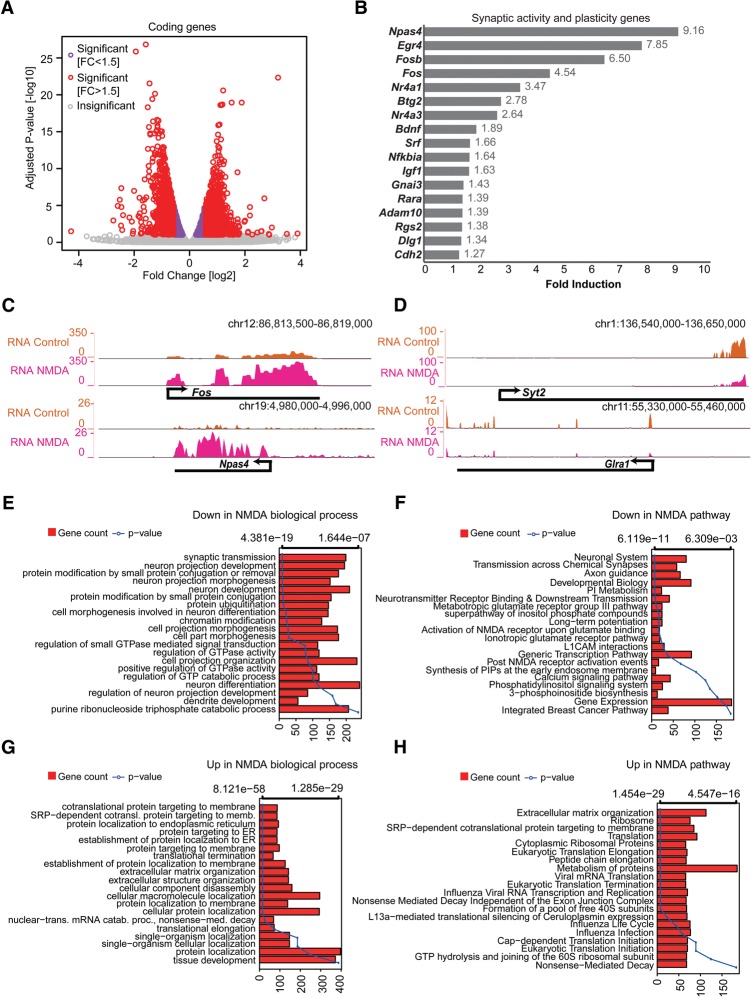
We next profiled the transcriptome (RNA-seq) of NMDA-treated neurons to illuminate the gene expression changes induced by the prolonged activation of NMDA receptors (Supplemental Table S4). Differential gene expression analysis comparing the control and NMDA-treated neurons revealed massive transcriptional reprogramming (Fig. 4A). Furthermore, we observed that many established neuronal activity and neuroplasticity genes were also up-regulated (Fig. 4B,C). Surprisingly, this further accompanied down-regulation of many neuronal genes, particularly those related to synaptic transmission (e.g., Syt2 and Glra1) (Fig. 4D). GO analysis of down-regulated genes revealed a dominant enrichment of genes involved in synaptic transmission and neuronal development (Fig. 4E,F). In contrast, up-regulated genes were enriched for processes related to extracellular matrix reorganization, protein localization, and translation (Fig. 4G,H). Mouse phenotype enrichment analysis demonstrated that the defects in the down-regulated genes were almost exclusively associated with phenotypes such as abnormal learning/memory/conditioning and behavior, whereas those up-regulated were dominantly linked to defects in the development of non-neuronal tissues (Supplemental Fig. S3G,H).
Figure 4.
NMDA receptor signaling causes dramatic transcriptional changes. (A) Volcano plot showing up- and down-regulated genes upon NDMA treatment in TND10 neurons. The x-axis represents the fold change between control and NDMA in log2; the y-axis shows the adjusted P-value in −log10. (B) Bar plot showing fold induction of a few selected known synaptic activity and neuronal plasticity-related genes. The x-axis shows gain of expression fold change upon NMDA stimulation in neurons. (C,D) Browser tracks showing gain (C) and loss (D) of expression for key activity or neuronal-related genes. (E–H) Bar plots showing enrichment of biological processes and pathways in down-regulated (E,F) and up-regulated (G,H) genes upon NDMA stimulation. The bars reflect the number of genes in each category; the lines represent the multiple testing-corrected P-value, displayed as an alternate x-axis, of the corresponding GO term.
The importance of long intergenic noncoding RNAs (lincRNAs) in neuronal development has been increasingly appreciated (Yao and Jin 2014). Our analysis further revealed that NMDA-driven neuronal activity also involves modulation of a large number of lincRNAs (Supplemental Fig. S3I). Furthermore, the expression of the genes nearest to these lincRNAs was also altered under these conditions, suggesting a potential function of these lincRNAs in gene regulation in cis (Supplemental Fig. S3J–L). The genes potentially down-regulated by lincRNAs were enriched for GO terms related to synaptic transmission and neuronal activity, and up-regulated genes were dominantly enriched for many non-neuronal genes (Supplemental Fig. S3M,N).
Induction of NMDA receptor pathway results in neuroplasticity and a neuroprotective response
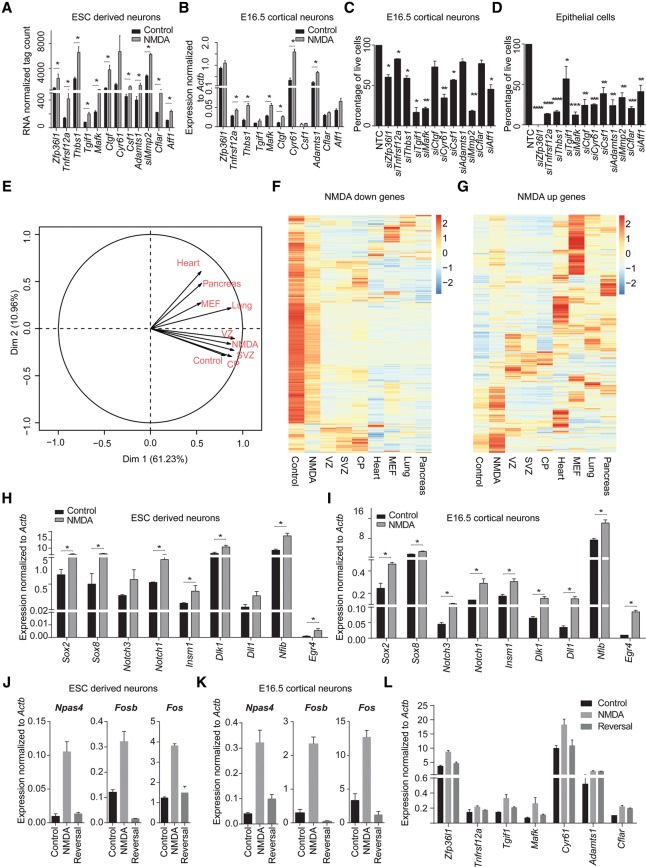
Interestingly, a number of NMDA-induced genes have previously been shown to be important in NMDA-driven neuronal survival (e.g., Btg2, Bdnf, Atf1) (West et al. 2002; Zhang et al. 2007; Zheng et al. 2010; Chen et al. 2014). In addition, this list also contained genes that, based on the literature, could play a role in cell proliferation but have not been implicated in cell survival, especially in the context of neurons (Fig. 5A). Such NMDA-induced expression of these genes was also observed in E16.5 cortical neurons (Fig. 5B). To establish the function of these genes in cell survival, we decided to assess cell death following their siRNA-mediated knockdown in cortical neurons. Interestingly, the depletion of a majority of these genes showed a very strong effect on the viability of cortical neurons (Fig. 5C), which was further extensively validated in mouse epithelial cells (Fig. 5D; Supplemental Fig. S4A–C).
Figure 5.
NMDA receptor stimulation causes induction of non-neuronal and prosurvival genes. (A) Expression values from RNA-seq (normalized tag counts) of selected potential prosurvival candidate genes that are induced upon NMDA. Error bars represent the SEM of independent biological replicates. (B) mRNA levels for selected prosurvival genes (as in A) in NMDA-induced cortical neurons were measured by qRT-PCR relative to Actb and plotted on the y-axis. Error bars represent the SEM of independent biological replicates. (C) Bar plot showing the percentage of live cells upon siRNA-mediated knock down of prosurvival candidates compared with nontargeting control (NTC) in cortical neurons. Error bars represent the SEM of independent biological replicates. (D) Similar analysis as in C but knock down performed in epithelial cells. Error bars represent the SEM of independent biological replicates. (E) Principal component analysis (PCA) plot of control- and NMDA-treated day 10 neurons with ventricular zone (VZ), subventricular zone (SVZ), cortical plate (CP), lung, pancreas, heart, and MEF transcriptomes. (F) Heat map showing the expression of genes down-regulated by NMDA treatment in different tissues from three germ layers (ectoderm: VZ, SVZ, and CP; endoderm: pancreas and lung; mesoderm: heart and MEF). (G) Same as in F but for up-regulated genes. (H,I) Bar plots showing mRNA levels for known neuronal progenitor genes in NMDA-induced ESC-derived neurons (H) and cortical neurons (I) measured by qRT-PCR relative to Actb and plotted on the y-axis. Error bars represent the SEM of independent biological replicates. (J,K) Bar plots showing mRNA levels of key synaptic activity genes upon NMDA treatment and following NMDA withdrawal both in ESC-derived neurons (J) and cortical neurons (K) measured by qRT-PCR relative to Actb and plotted on the y-axis. Error bars represent the SEM of independent biological replicates. (L) Similar analysis as in J was performed in ESC-derived neurons but for prosurvival genes. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001, Student's t-test.
To gain further biological insights in the global gene expression changes induced by NMDA stimulation, we performed a principal component analysis (PCA)-based comparison of the DMSO and NMDA transcriptomes with the transcriptomes of several embryonic tissues from the three germ layers (ectoderm: ventricular zone [VZ], subventricular zone [SVZ], and cortical plate [CP] from mouse cortex; endoderm: lung and pancreas; mesoderm: heart and MEF). This analysis demonstrated that although control neurons were very similar to CP, NMDA treatment of neurons resulted in a transcriptome that was closer to cells from VZ and SVZ but not CP, further confirming a reversal toward the progenitor state (Fig. 5E). Further analysis showed that genes down-regulated upon NMDA treatment were generally highly expressed in control neurons and CP and exhibited the least expression in other tissues (Fig. 5F). In contrast, up-regulated genes were expressed at much higher levels in either VZ/SVZ or in other tissues compared with CP (Fig. 5G). Intrigued by these patterns, a closer gene-wise analysis of up- and down-regulated protein-coding genes further revealed that many established progenitor markers (e.g., Pax6, Nes, Sox2, Notch1, Dll1, and Hes1) were significantly induced, whereas known neuronal genes (e.g., Ncam1, Syn1, Rbfox3, Dscam, Nlgn, and Epha2) were significantly down-regulated (Supplemental Fig. 5A,B). Such NMDA-induced up-regulation of neuronal progenitor genes was further independently validated both in ESC-derived neurons and cortical neurons (Fig. 5H,I).
Since NMDA-dependent neuronal activity has been implicated to cause neuroplasticity (Coyle and Tsai 2004), we next probed whether our experimental setup recapitulates similar phenomenon. To investigate this, we performed an NMDA withdrawal experiment on ESC-derived neurons as well as cortical neurons and measured the expression of classical neuronal plasticity genes. This analysis indeed showed that although NMDA stimulation results in a strong up-regulation of neuronal plasticity genes (Npas4, Fosb, and Fos), they were completely reverted back after NMDA withdrawal (Fig. 5J,K). Furthermore, similar behavior was observed for many neuroprotective genes (Fig. 5L). Altogether, these observations indeed establish that NMDA stimulation involves induction of neuroplasticity and neuroprotective response. These findings further imply that under prolonged neuronal activity via NMDA receptors, neurons transiently lose their identity and exhibit greater developmental potential, which may be essential to allow neuronal plasticity.
NMDA-induced neuronal activity results in a massive epigenetic reprogramming of distal regulatory elements
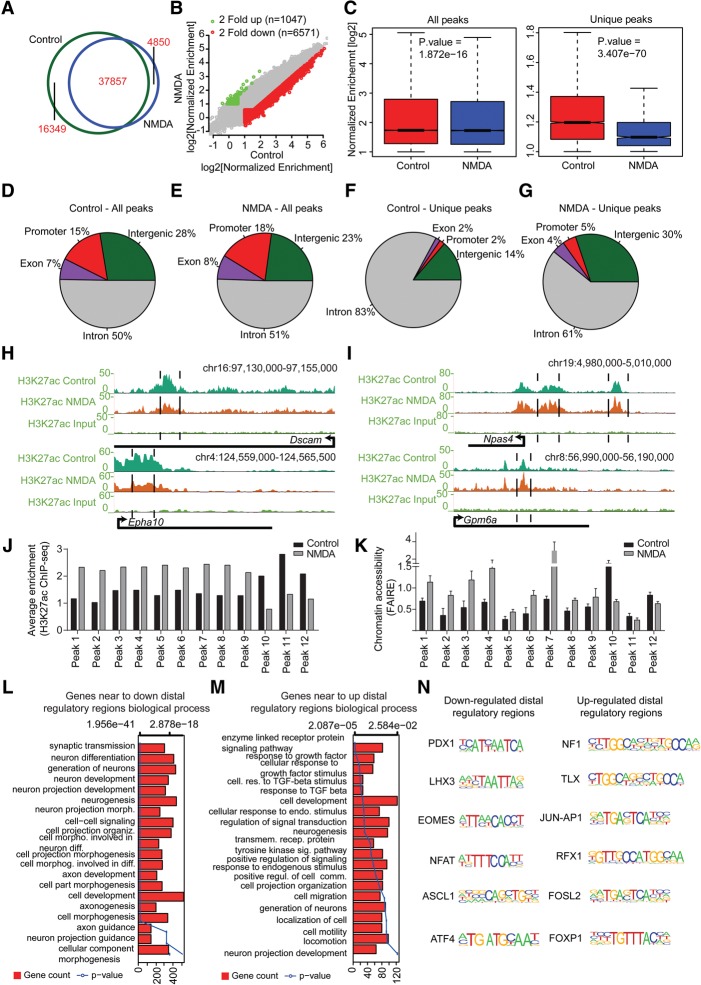
We had previously observed that in differentiated neurons, stage-specific distal regulatory regions marked with H3K27ac regulate the expression of genes involved in ion transport, localization, and the regulation of transmembrane transport—the functions that are known to be involved in neuronal activity (Fig. 2D,J). Furthermore, the gain of stage-specific active regulatory regions was found to occur exclusively at distal sites, which in turn defines cell-type–unique transcriptional programs during neuronal development (Fig. 2D). Although researchers have touched upon the distal regulation of basal neuronal activity, there are no reports studying how distal gene regulation is involved in transcriptional responses following prolonged neuronal activity via NMDA receptors (Kim et al. 2010; Malik et al. 2014). To examine NMDA-dependent transcriptional changes, we performed a ChIP assay using H3K27ac-specific antibody with control and NMDA-treated neurons and performed next-generation sequencing (ChIP-seq) (Supplemental Table S5). Our analysis revealed that NMDA treatment led to a substantial loss of H3K27ac peaks (n = 16,349) and a gain of lesser new peaks (n = 4850) (Fig. 6A). Using stringent enrichment criteria, we further shortlisted genomic sites that strongly lost or gained H3K27ac enrichment (n = 6571, down-regulated; n = 1047, up-regulated) (Fig. 6B). Although decrease in enrichment was significant among all peaks, a dramatic reduction was observed at unique peaks (Fig. 6C). Deeper analysis did not reveal major changes in the percentage of H3K27ac sites distributed across promoters, exons, introns, and intergenic regions following NMDA treatment (Fig. 6D,E). Interestingly, we observed that, upon NMDA stimulation, introns and intergenic regions exhibited the most prominent reprogramming of the H3K27ac mark (Fig. 6F,G). The nearest genes to these sites included known neuronal and synaptic activity-modulated genes (e.g., Dscam and Epha10, which lose H3K27ac and expression upon NMDA treatment; and Npas4 and Gpm6a, which gain H3K27ac and transcription upon NMDA treatment) (Fig. 6H,I). Comparison of epigenome and transcriptome dynamics between neuronal activity versus neurogenesis (NP to TND10) revealed that decreased H3K27ac peaks or down-regulated genes during neuronal activity had more overlap with increased H3K27ac peaks or up-regulated genes during neurogenesis and vice versa (Supplemental Fig. 6A–C). GO enrichment analysis of these overlapping sets of genes further supported our earlier observations that NMDA-stimulated neurons transiently lose their identity (Supplemental Table S6). We also found that a large fraction of these activity-modulated H3K27ac-positive distal regulatory regions were also active in vivo during murine brain development (Supplemental Fig. S6D,E; Nord et al. 2013).
Figure 6.
Prolonged NMDA activity results in a massive modulation of the distal regulatory landscape. (A) Venn diagram showing overlap of H3K27ac peaks identified in control- and NDMA-treated day 10 neurons. (B) Scatter plot showing the gain and loss of H3K27ac sites. Peaks marked in green are either only present in NMDA or at least twofold up-regulated compared with control; peaks marked in red represent the opposite. The x- and y-axes represent the normalized enrichment of H3K27ac in control and NDMA treated neurons. (C) Box plots showing the loss of H3K27ac upon NMDA treatment compared with control at all and unique peaks. P-values are calculated using the Wilcoxon test. (D–G) Pie charts showing the distribution of control or NMDA peaks in different genomic regions for all (D,E) and unique (F,G) peaks. (H) Browser plots showing loss of H3K27ac at potential distal regulatory regions of neuronal genes (Dscam and Epha10). (I) Same as in H but showing gain of H3K27ac at potential distal regulatory regions of activity-related genes. (J) Bar plot showing normalized H3K27ac enrichment of selected distal peaks in control and NMDA-treated neurons as derived from the ChIP-seq data set. (K) FAIRE assay was performed in cells treated with NMDA, and qRT-PCRs were performed for the same distal regulatory regions as in J. Average enrichments from independent assays are plotted on the y-axis as a ratio of precipitated DNA (bound) to total input DNA and then further normalized to an intergenic region. Error bars represent the SEM of independent biological replicates. (L,M) Bar plots showing the enrichment of biological processes for genes near down-regulated (L) and up-regulated (M) distal regulatory regions. The bars reflect the number of genes in each category; the lines represent the multiple testing-corrected P-value, displayed on alternate x-axis, of the corresponding GO term. (N) Representative motifs identified in down- and up-regulated distal regulatory regions.
In line with our previous observations that the distal accessible chromatin sites are largely enriched with H3K27ac in neurons (Fig. 2A), we found that all tested distal regions that gained H3K27ac following NMDA stimulation also gained chromatin accessibility and vice versa (Fig. 6J,K). A GO analysis of genes in proximity to distal regulatory sites modulated via NMDA treatment showed that the genes nearest to those losing enrichment were dominantly enriched for synaptic transmission and neurogenesis (Fig. 6L), and genes in the closest proximity to sites gaining H3K27ac were involved in signaling pathways and cell motility (Fig. 6M). Furthermore, a motif enrichment analysis of these regions revealed their enrichment for binding sites for a number of transcription factors that are known to be critical for neurogenesis (Fig. 6N; Supplemental Fig. 6F,G). Overall, these findings establish that prolonged NMDA activity involves dramatic epigenetic reprogramming at distal regulatory elements.
NMDA-mediated epigenetic reprogramming of distal regulatory elements functions in gene regulation
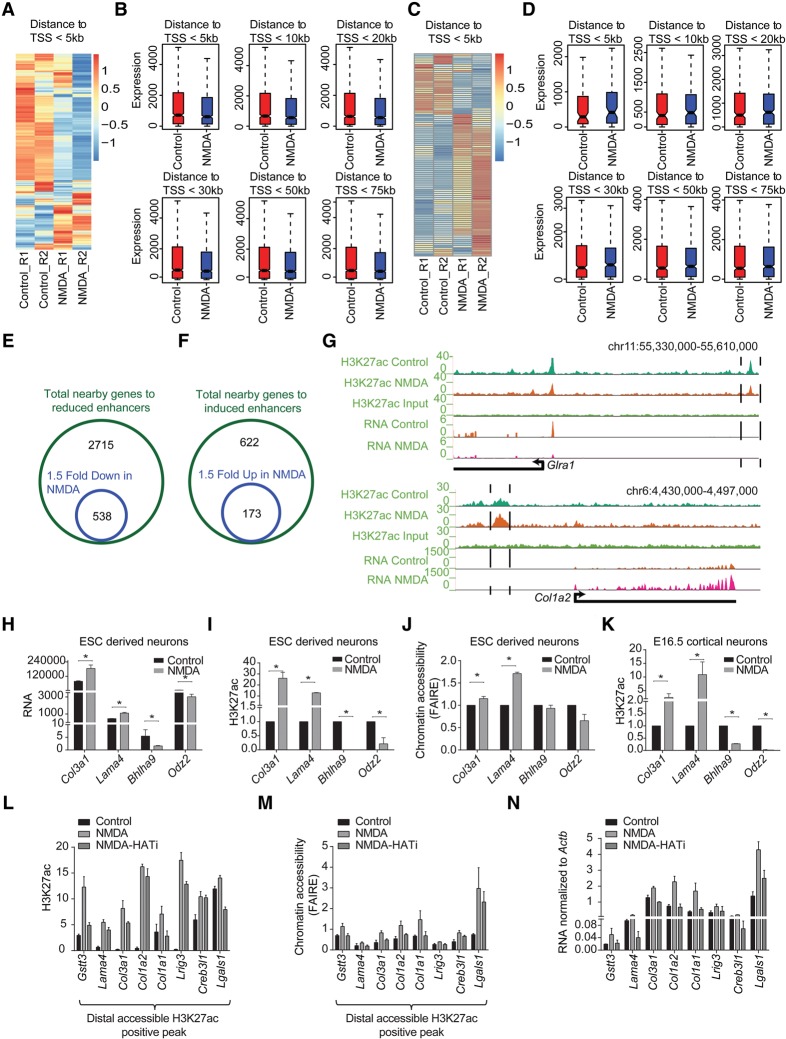
We next attempted to determine whether distal regions that exhibit changes in H3K27ac in response to NMDA activity correlate with expression changes in the nearest genes. To address this question, we created bins of genes based on the distance from these distal regions and plotted their expression levels under control and NMDA-treatment conditions. In line with previous observations, nearby genes displayed a greater decrease or increase in expression when distal regions exhibited a loss or gain of H3K27ac, respectively (Fig. 7A–D; Supplemental Fig. S7A,B). Importantly, these genes included a number of genes that were differentially expressed by at least 1.5-fold upon NMDA treatment (n = 538 genes, down-regulated; n = 173, up-regulated) (Fig. 7E–G). These down-regulated genes were again enriched for synaptic transmission and ion transport, whereas up-regulated genes were enriched for extracellular matrix reorganization (Supplemental Fig. S7C,D). We further independently validated that chromatin changes at distal regulatory elements accompany changes in the expression levels of associated genes both in ESC-derived neurons and cortical neurons derived from E16.5 brain (Fig. 7H–K; Supplemental Fig. S7E).
Figure 7.
Neuronal activity-dependent transcriptome changes are driven via distal regulatory elements. (A) Heat map showing the expression of genes (within a 5-kb window) near down-regulated H3K27ac-positive distal regulatory regions in the control and NMDA treatments. (B) Box plots showing differences in the expression of genes near down-regulated H3K27ac-positive distal regulatory regions after the genes were binned based on their distance (5, 10, 20, 30, 50, and 75 kb) from these sites. P-values for the differences between control and NMDA conditions for distances 5 kb (n = 911), 10 kb (n = 1076), 20 kb (n = 1367), 30 kb (n = 1621), 50 kb (n = 1970), and 75 kb (n = 2252) are 1.5 × 10−54, 5.7 × 10−56, 3.4 × 10−51, 1.4 × 10−55, 1.5 × 10−54, and 4.4 × 10−56, respectively. P-values are calculated using the Wilcoxon test. (C) Same as in A but for up-regulated H3K27ac-positive distal regulatory regions. (D) Same as in B but for up-regulated H3K27ac-positive distal regulatory regions. P-values for the differences between control and NMDA conditions for distances 5 kb (n = 97), 10 kb (n = 171), 20 kb (310), 30 kb (406), 50 kb (528), and 75 kb (n = 608) are 0.0005, 0.05, 0.04, 0.13, 0.56, and 0.91, respectively. (E,F) Venn diagram showing overlap of genes near down-regulated distal regulatory regions (green circle) with genes at least 1.5-fold down-regulated upon NMDA treatment (blue circle) (E) and up-regulated distal regulatory regions (green circle) with genes at least 1.5-fold up-regulated (blue circle) (F). (G) Browser plots showing concomitant loss (Glra1) or gain (Col1a2) of distal H3K27ac and expression. (H) Normalized tag counts from RNA-seq data of selected genes that show changes in H3K27ac at their distal regions upon NMDA treatment. (I) ChIP-qPCR validation of NMDA-dependent H3K27ac changes for the distal regions for genes shown in (H). (J) FAIRE-qPCRs to asses changes in chromatin accessibility at the same regions shown in (I) upon NMDA treatment. (K) ChIP-qPCR validation of NMDA-dependent H3K27ac changes for the distal regions of genes shown in H in mouse cortical neurons treated with NMDA. (L–N) Bar plots showing analysis of H3K27ac (by ChIP-qPCRs) (L), chromatin accessibility (by FAIRE-qPCRs) (M), of selected distal regions and expression of nearest genes (by RT-qPCRs) (N), following NMDA stimulation in the presence and absence HAT inhibitor (C646). All error bars represent the SEM of independent biological replicates. (*) P < 0.05, Student's t-test.
We next attempted to assess the downstream epigenetic machinery involved in altering chromatin landscape and gene expression in response to NMDA stimulation. To address this, we used C646, a specific inhibitor of the histone acetyltransferase (HAT) EP300 (also known as p300)/CREBBP (also known as CBP) (Bowers et al. 2010) and analyzed distal regulatory elements for certain genes that showed an increase in H3K27ac upon NMDA treatment following treatment with NMDA in the presence and absence of C646. These results reveal that for NMDA-induced distal H3K27ac peaks, the histone acetyltransferase (HAT) EP300/CREBBP activity is required for a gain of H3K27ac enrichment in response to NMDA treatment (Fig. 7L). In addition, such depletion of H3K27ac enrichment further accompanied loss of chromatin accessibility at all measured distal regulatory elements (Fig. 7M). Furthermore, this also accompanied down-regulation in transcription of genes nearest to these distal sites (Fig. 7N). Taken together, these results suggest that the NMDA pathway uses distinct epigenetic players such as EP300/CREBBP at distal regulatory elements to govern epigenetic state and transcriptional dynamics following NMDA receptor activation.
NMDA-induced neural activity involves a dramatic reorganization of super-enhancers
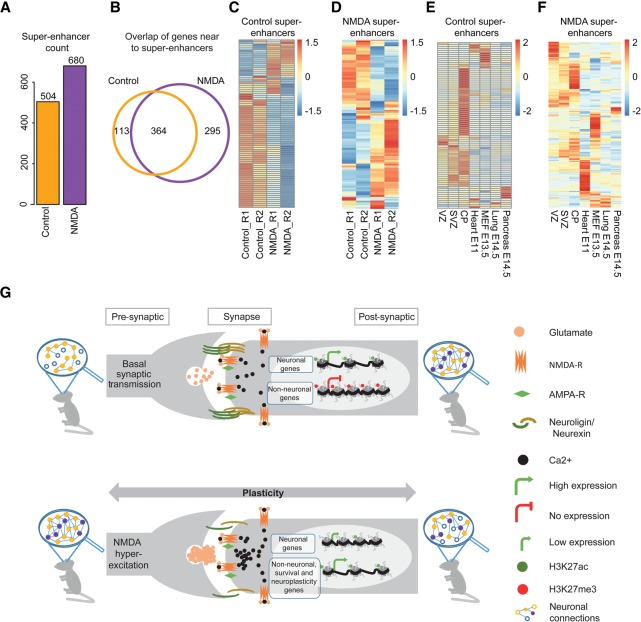
Recent studies have demonstrated that super-enhancers are large clusters of transcriptional enhancers that drive the expression of genes defining cell identity (Hnisz et al. 2013; Whyte et al. 2013). Prompted by our observations that NMDA-mediated gene expression responses involve epigenetic reprogramming of distal regulatory elements, we next investigated whether the NMDA response involves the reorganization of super-enhancers. Surprisingly, despite the massive loss of classical H3K27ac-marked distal regulatory regions, we observed a substantial increase in the number of super-enhancers upon NMDA treatment (Fig. 8A). Furthermore, when we assigned these super-enhancers to nearby genes, we observed that few genes lost super-enhancers (n = 113), whereas a larger number of genes gained super-enhancers (n = 295) following NMDA treatment (Fig. 8B). Among the genes in proximity to control- or NMDA-specific super-enhancers were a number of genes that are known to be crucial for neuronal identity (e.g., Ncam1, Mapt, Rbfox3, control super-enhancers) or neuronal activity (Fos, Per1, and Ephb2, NMDA super-enhancers), respectively (Supplemental Table S7). Comparing the loss and gain of super-enhancers with the transcription changes following NMDA treatment demonstrated that the substantial number of control- or NMDA-specific super-enhancers were associated with higher expression levels of the associated genes in the respective states (Fig. 8C,D; Supplemental Fig. S9A,B). Considering the expression of nearby genes in the three layers of the embryonic cortex and representative tissues from all three lineages, we observed that the majority of genes associated with super-enhancers that were present in control-treated neurons but lost after NMDA treatment were almost exclusively expressed in the cortical plate (CP), where mature neurons reside (Fig. 8E). Interestingly, although many genes associated with super-enhancers acquired exclusively following NMDA treatment were expressed in the cortex, a major fraction of these genes were also expressed in other tissues (Fig. 8F). Accordingly, super-enhancers identified in control neurons were enriched with genes related to neurogenesis (e.g., Ncam1, Mapt, Rbfox3) (Supplemental Fig. 8A), whereas genes associated with NMDA-induced super-enhancers were related to general transcription or metabolic regulation and included many established neuronal activity up-regulated genes (e.g., Fos, Per1, and Ephb2) (Supplemental Fig. 8B; Supplemental Table S7). These findings suggest that prolonged NMDA activity causes a massive reorganization of super-enhancers, which then mediate changes in the expression of critical genes.
Figure 8.
Neuronal activity results in a global reorganization of super-enhancers at key responsive genes. (A) Bar plot showing the number of super-enhancers in the control and NMDA-treated neurons. (B) Venn diagram showing the overlap of genes near control and NMDA super-enhancer. (C,D) Expression of genes near control (C) and NDMA (D) nonpromoter super-enhancers in control- and NDMA-treated neurons (enlarged heat maps along with gene names are provided as Supplemental Fig. S9). (E,F) Heat map showing expression of the same genes as in C and D in tissues from three germ layers. (G) Schematic representation of a model showing epigenome and transcriptome changes in response to sustained NMDA receptor activity. Neurons exhibiting basal synaptic activity express neuronal genes that are marked by an active chromatin state while non-neuronal genes are kept silent in heterochromatin. A prolonged NMDA activity results in a dramatic epigenetic reprogramming, including at many distal regulatory elements and super-enhancers, resulting in the down-regulation of neuronal genes (loss of H3K27ac) and activation of neuronal plasticity, neuroprotective, and several non-neuronal genes (gain of H3K27ac).
Discussion
Differentiation is characterized by sequential changes in the cellular state specified by transcriptional reprogramming that is believed to be generated by a combinatorial action of epigenetic mechanisms and transcription factors. Despite progress, we lack a comprehensive understanding of how specific genes are precisely turned on and off during neuronal development and how regulatory sequences contribute to this process. Recent evidence has indicated a crucial role for distal regulatory elements in defining a cell-type–specific gene expression program during development. Using the FAIRE-seq assay, here we have revealed genome-wide remodeling of chromatin accessibility during successive stages of neurogenesis. These data reveal that a distinct set of H3K27ac-marked distal regulatory regions are uniquely gained in each stage of differentiation and function to drive the expression of genes that define cell identity. We further demonstrated that in addition to chromatin competence with respect to the presence of the H3K27ac marker, these distal elements recruit distinct transcription factors that are known to be critical for the respective stages of neuronal differentiation. These findings further highlight a complex crosstalk among epigenetic mechanisms, DNA sequence, and transcription factors in determining the potential of distal regulatory elements that in turn define the transcriptome underlying cellular identity. Furthermore, such combinatorial analysis of chromatin accessibility (FAIRE-seq) and H3K27ac (ChIP-seq) has led to the first high-sensitivity map of regulatory elements for progressive stages of neuronal differentiation.
The identification of open chromatin regions using FAIRE-seq revealed that accessible chromatin is rapidly remodeled during neurogenesis. We observed that dividing neuronal progenitors have more open chromatin than post-mitotic neurons, reflecting genome compaction upon differentiation. This result also suggests that a large number of accessible sites from differentiated neurons may preexist in neuronal progenitors, arguing for an “epigenetic priming”–like phenomenon that later gains transcriptional competence upon the induction of neurogenesis. This suggestion is further supported by our observations that promoter accessibility correlates more strongly with gene expression in post-mitotic neurons than dividing neuronal progenitors. Alternatively, a certain fraction of the openness detected in dividing neuronal progenitor cells may be contributed by DNA replication; whereas in post-mitotic neurons, the open chromatin state may be exclusively related to transcriptional activity. These observations were further extended to distal regulatory elements because the fraction of open distal regions enriched with the active mark H3K27ac was much higher in post-mitotic cells than dividing neuronal progenitors. Overall, these observations suggest that neurogenesis accompanies establishment of a more structured landscape of regulatory elements and chromatin accessibility that function primarily in gene regulation in post-mitotic, terminally differentiated neurons.
Enhancers are distal regulatory elements that function to activate gene expression and are primary drivers of tissue-specific gene expression programs. Using our high-resolution map of chromatin accessibility and H3K27ac, which marks active enhancers, we illustrated the dynamic changes in usage of regulatory elements during neurogenesis. Interestingly, genes in close proximity to stage-specific nonpromoter accessible H3K27ac sites were nonoverlapping between the consecutive stages of neuronal differentiation (NP, TND1, and TND10) and gained expression in the stage associated with H3K27ac. A GO enrichment analysis for these genes revealed involvement in biological processes and pathways that reflects the cellular identity of each of the three cell types. These findings further argue for a critical importance of distal gene regulation in defining cell fates during neurogenesis.
We were next tempted to ask whether the acquired epigenetic and transcriptional state in neurons is subject to modulation during neuronal function such as neuronal activity. The activity-dependent plasticity of vertebrate neurons allows the brain to respond to external stimuli. Neuronal activity-dependent LTP and LTD impart activity-driven neuronal plasticity. Basal neuronal activity or LTP results in the formation of synapses and thus creates memories; but after prolonged neuronal activity, LTD is required to rewire old synapses to allow the acquisition of new learning. This study unravels many novel aspects of neuronal activity induced by NMDA receptors and demonstrates that prolonged NMDA activity results in significant epigenetic remodeling of the distal regulatory elements to mediate the required gene expression responses.
Here, we provide the first comprehensive map of all coding and noncoding genes that are transcriptionally modulated in response to sustained NMDA receptor activity. The genes down-regulated following NMDA receptor stimulation included many neuron-specific genes, and those induced contained extracellular matrix genes and many other genes expressed in non-neuronal lineages in addition to the classical neuronal activity genes (Fig. 8G). This result is also in agreement with a previous study demonstrating that the NDMA response leads to a major reorganization of the extracellular matrix (Chen et al. 2014). Interestingly, such an NMDA receptor activity–driven transcriptome resembled neuronal progenitors more closely than neurons and showed induction of classical neuronal progenitor genes (e.g., Pax6, Sox2) and down-regulation of hallmark neuronal genes (e.g., Mapt, Rbfox3). Importantly, transcription of neuronal plasticity genes was completely reversed soon after the NMDA withdrawal, suggesting a true neuronal plasticity in response to neuronal activity. These findings suggested that sustained activity of an NMDA receptor results in a transient loss of neuronal identity, and we propose that this may be essential to allow cellular and chromatin plasticity. Future studies should aim to investigate the relevance of this phenomenon in the in vivo contexts of such neuronal activity. Furthermore, given the generation of such a “plastic” cellular state in response to neuronal activity, it would be interesting to assess whether this condition predisposes neurons for reprogramming to other cell types.
Furthermore, this study also discovered for the first time a number of novel genes that were transcriptionally induced by NMDA and function in neuronal survival (Fig. 8G). In addition, NMDA-induced genes also included several genes established to be important in NMDA-driven neuronal survival (e.g., Btg2, Bdnf, Atf1) (West et al. 2002; Zhang et al. 2007; Zheng et al. 2010; Chen et al. 2014). Such induction of prosurvival genes upon neuronal activity may reflect a neuroprotective response (Fig. 8G) and envisage the possible protective effects of neuronal activity against neurodegeneration. In the future, it would be interesting to explore whether these novel prosurvival genes could offer therapeutic avenues against neurodegenerative diseases such as Parkinson's, Alzheimer's, or Huntington's disease.
Current literature suggests that basal neuronal activity results in a gain of H3K27ac at distal sites and assists memory formation (Deisseroth et al. 1998; West et al. 2002; Feng et al. 2007; Kim et al. 2010; Zheng et al. 2010; Chen et al. 2014; Lopez-Atalaya and Barco 2014). Therefore, any LTD event that results in a selective loss of synaptic strength may also involve the erasure of memory stored on chromatin during basal activity. We observed that prolonged NMDA activity results in a massive loss of potential distal regulatory regions, whereas only a limited set of potential distal regulatory regions were gained. The genes in close proximity to these lost distal regulatory regions were dominantly enriched in neuronal genes such as those involved in neuronal transmission and neuronal development (Fig. 8G). Furthermore, such NMDA-induced lost and gained distal H3K27ac sites showed opposite dynamics during neurogenesis (upon transition from NP to TND10), further suggesting that sustained neuronal activity also generates an epigenome that is closer to the neuronal progenitor state and reflects a partial de-differentiation.
In contrast to these classical H3K27ac-marked distal regulatory regions that were mostly lost upon NMDA stimulation, such treatment led to a massive reorganization of super-enhancers that were both lost and gained. Furthermore, genes gaining super-enhancers following NMDA treatment included key neuronal activity genes, and genes that lose super-enhancers contained many neuronal genes (Fig. 8G). These observations also imply that NMDA receptor signaling also functions to reprogram critical regulatory regions such as super-enhancers, which have been shown to drive expression of genes that define cell identity (Hnisz et al. 2013; Whyte et al. 2013; Pott and Lieb 2014) and thus may have a profound effect on the cell fate. We also observed that the activation of the NMDA pathway results in the recruitment of histone acetyltransferase EP300/CREBBP at distal regulatory elements that then acetylates Histone H3 at lysine 27 to result in the observed chromatin accessibility of these distal sites as well as transcriptional activation of the nearest genes.
Taken together, our findings reveal the dynamics of distal regulatory elements and a vast trove of potential distal regulatory regions and super-enhancers that function in determining the transcriptome underlying stage-specific cellular identity during neurogenesis and in response to NMDA-induced neuronal activity. It is very exciting to uncover how the distal regulatory landscape is actively remodeled during neuronal development as well as upon activation of the NMDA receptor pathway to mediate neuroplasticity and neuroprotective responses (Fig. 8G). Future work should aim to unravel the mechanistic details of the collaborative partnership between transcription factors and epigenetic mechanisms at distal regulatory regions and how this partnership contributes to the gene expression program that underlies neuronal development and activity.
Methods
Cell culture
Wild-type embryonic stem cells derived from blastocysts (3.5 post coitum) from a mixed 129-C57BL/6 background (called 159.2) were cultured and differentiated as previously described (Bibel et al. 2004). A subclone of NMuMG cells has been described previously and was grown in DMEM supplemented with 10% FBS, 2 mM L-Glutamine, and 1× nonessential amino acids (Maeda et al. 2005). All cells were grown in 7% CO2 at 37°C.
Cortical neuron culture
Cortical neurons were cultured as described previously (Kim et al. 2010). Please refer to Supplemental Methods for more details.
FAIRE assay
FAIRE-assay was performed as described previously (Giresi et al. 2007). Please refer to Supplemental Methods for more details.
ChIP assay
ChIP experiments were performed as described previously (Stadler et al. 2011), starting with 60 μg chromatin and 5 μg antibodies. ChIP-qPCR was performed using SYBR Green chemistry (ABI). Primers used for ChIP-qPCRs are listed in Supplemental Table S8.
Neuronal activation
Neurons were washed three times with BSS (balanced salt solution: 25 mM Tris, 120 mM NaCl, 15 mM glucose, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2, pH 7). After treating the neurons with 200 µM NMDA (Sigma) or control only for 6 h, cells were processed for RT-qPCR, ChIP, and FAIRE analysis or washed three times with BSS and fresh neuronal medium added for other downstream experiments. For recovery, cells were incubated 72 h in neuronal medium without NMDA.
Inhibition of the histone acetyltransferase (HAT) EP300/CREBBP
Neurons were pretreated with the EP300/CREBBP inhibitor (10 µM, C646, Sigma) for 30 min; then NMDA treatment was performed as described above but in the presence of C646.
Next-generation sequencing data analysis
Details of genomics data analysis including RNA-seq, ChIP-seq, and FAIRE-seq are provided as Supplemental Methods.
Data access
FAIRE-seq, ChIP-seq, and RNA-seq data generated in this study data have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE65713.
Supplementary Material
Acknowledgments
We thank members of the Tiwari laboratory for support and critical feedback during the project. Support by the Core Facilities of the Institute of Molecular Biology (IMB), Mainz, is gratefully acknowledged, particularly the Genomics Core Facility and the Bioinformatics Core Facility. We thank Prof. Jason Lieb (The University of Chicago) for providing protocols and critical comments during the study. Research in the laboratory of V.K.T. is supported by the Deutsche Forschungsgemeinschaft (DFG) Grant TI 799/1-1, Marie Curie CIG 322210, EpiGeneSys RISE1 program, and Wilhelm Sander Stiftung 2012.009.2.
Author contributions: S.T. analyzed data and wrote the manuscript. S.K.S. performed experiments, analyzed data, and wrote the manuscript. A.G. performed experiments. V.K.T. designed the study, analyzed data, and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.190926.115.
Freely available online through the Genome Research Open Access option.
References
- Ahmed I, Bose SK, Pavese N, Ramlackhansingh A, Turkheimer F, Hotton G, Hammers A, Brooks DJ. 2011. Glutamate NMDA receptor dysregulation in Parkinson's disease with dyskinesias. Brain 134(Pt 4): 979–986. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thomä NH, Schübeler D. 2011. Determinants and dynamics of genome accessibility. Nat Rev Genet 12: 554–564. [DOI] [PubMed] [Google Scholar]
- Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, Korte M, Goetz M, Barde YA. 2004. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci 7: 1003–1009. [DOI] [PubMed] [Google Scholar]
- Blanchet PJ, Papa SM, Metman LV, Mouradian MM, Chase TN. 1997. Modulation of levodopa-induced motor response complications by NMDA antagonists in Parkinson's disease. Neurosci Biobehav Rev 21: 447–453. [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EE. 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. 2008. Decoding the epigenetic language of neuronal plasticity. Neuron 60: 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. 2004. Molecular pathways to neurodegeneration. Nat Med 10(Suppl: S2–S9. [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. 2010. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol 17: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Modrusan Z, Sheng M, Kaminker JS. 2014. Regulation of neuronal gene expression and survival by basal NMDA receptor activity: a role for histone deacetylase 4. J Neurosci 34: 15327–15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW. 1988. Glutamate neurotoxicity and diseases of the nervous system. Neuron 1: 623–634. [DOI] [PubMed] [Google Scholar]
- Choi DW. 1992. Excitotoxic cell death. J Neurobiol 23: 1261–1276. [DOI] [PubMed] [Google Scholar]
- Connor SA, Wang YT. 2015. A place at the table: LTD as a mediator of memory genesis. Neuroscientist 10.1177/1073858415588498. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Tsai G. 2004. NMDA receptor function, neuroplasticity, and the pathophysiology of schizophrenia. Int Rev Neurobiol 59: 491–515. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW. 1998. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature 392: 198–202. [DOI] [PubMed] [Google Scholar]
- Dekker J. 2012. CTCF and cohesin help neurons raise their self-awareness. Proc Natl Acad Sci 109: 8799–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny LA. 2014. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput Biol 10: e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. 2007. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res 61(5 Pt 2): 58R–63R. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. 2005. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. 2003. The Yin and Yang of NMDA receptor signalling. Trends Neurosci 26: 81–89. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. 2012. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep 2: 345–357. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XL, Wang Y, Shen Q. 2012. Epigenetic control on cell fate choice in neural stem cells. Protein Cell 3: 278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. 2000. Functional requirement for class I MHC in CNS development and plasticity. Science 290: 2155–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidou C, Qin Qin Y, Labruyere J, Olney JW. 1996. Motor neuron degeneration induced by excitotoxin agonists has features in common with those seen in the SOD-1 transgenic mouse model of amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 55: 211–224. [DOI] [PubMed] [Google Scholar]
- Kalia LV, Kalia SK, Salter MW. 2008. NMDA receptors in clinical neurology: excitatory times ahead. Lancet Neurol 7: 742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294: 1030–1038. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig O. 1995. [Perception and memory: illustration of an approach of the study of visual mechanisms in cognitive neuroscience]. Rev Neurol 151: 451–456. [PubMed] [Google Scholar]
- Koohy H, Down TA, Hubbard TJ. 2013. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS One 8: e69853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cuvillier JM, Lee B, Shen R, Lee JW, Lee SK. 2012. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc Natl Acad Sci 109: 3383–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Shen R, Cho HH, Kwon RJ, Seo SY, Lee JW, Lee SK. 2013. STAT3 promotes motor neuron differentiation by collaborating with motor neuron-specific LIM complex. Proc Natl Acad Sci 110: 11445–11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB, Schübeler D. 2011. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet 7: e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton SA, Rosenberg PA. 1994. Excitatory amino acids as a final common pathway for neurologic disorders. N Engl J Med 330: 613–622. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Barco A. 2014. Can changes in histone acetylation contribute to memory formation? Trends Genet 30: 529–539. [DOI] [PubMed] [Google Scholar]
- Maeda M, Johnson KR, Wheelock MJ. 2005. Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci 118(Pt 5): 873–887. [DOI] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, et al. 2014. Genome-wide identification and characterization of functional neuronal activity–dependent enhancers. Nat Neurosci 17: 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Drechsel D, Guillemot F. 2012. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb Perspect Biol 4: a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey PV, Bashir ZI. 2007. Long-term depression: multiple forms and implications for brain function. Trends Neurosci 30: 176–184. [DOI] [PubMed] [Google Scholar]
- Mehta A, Prabhakar M, Kumar P, Deshmukh R, Sharma PL. 2013. Excitotoxicity: bridge to various triggers in neurodegenerative disorders. Eur J Pharmacol 698: 6–18. [DOI] [PubMed] [Google Scholar]
- Milner B, Squire LR, Kandel ER. 1998. Cognitive neuroscience and the study of memory. Neuron 20: 445–468. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Landsman D. 2001. Learning about addiction from the genome. Nature 409: 834–835. [DOI] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. 2013. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorska A, Vanhoutte P, Arnold FJ, Silvagno F, Hardingham GE, Bading H. 2003. Synaptic activity induces signalling to CREB without increasing global levels of cAMP in hippocampal neurons. J Neurochem 84: 447–452. [DOI] [PubMed] [Google Scholar]
- Pott S, Lieb JD. 2014. What are super-enhancers? Nat Genet 47: 8–12. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogawski MA. 1992. The NMDA receptor, NMDA antagonists and epilepsy therapy. A status report. Drugs 44: 279–292. [DOI] [PubMed] [Google Scholar]
- Samee MA, Sinha S. 2014. Quantitative modeling of a gene's expression from its intergenic sequence. PLoS Comput Biol 10: e1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez A, Llansola M, Cauli O, Felipo V. 2005. Modulation of NMDA receptors in the cerebellum. II. Signaling pathways and physiological modulators regulating NMDA receptor function. Cerebellum 4: 162–170. [DOI] [PubMed] [Google Scholar]
- Sgambato-Faure V, Cenci MA. 2012. Glutamatergic mechanisms in the dyskinesias induced by pharmacological dopamine replacement and deep brain stimulation for the treatment of Parkinson's disease. Prog Neurobiol 96: 69–86. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. 1998. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell 95: 817–828. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 68: 821–861. [DOI] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD. 2012. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY, Nairn AC, Salter MW, Lombroso PJ, Gouras GK, et al. 2005. Regulation of NMDA receptor trafficking by amyloid-β. Nat Neurosci 8: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. 2011. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21: 1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Nojima Y, Shoji W, Sato M, Nakayama R, Ohshima T, Okamoto H. 2011. Islet1 selectively promotes peripheral axon outgrowth in Rohon-Beard primary sensory neurons. Dev Dyn 240: 9–22. [DOI] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME. 2002. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron 33: 383–395. [DOI] [PubMed] [Google Scholar]
- Thakurela S, Garding A, Jung J, Schübeler D, Burger L, Tiwari VK. 2013. Gene regulation and priming by topoisomerase IIα in embryonic stem cells. Nat Commun 4: 2478. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Burger L, Nikoletopoulou V, Deogracias R, Thakurela S, Wirbelauer C, Kaut J, Terranova R, Hoerner L, Mielke C, et al. 2012a. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc Natl Acad Sci 109: E934–E943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schübeler D, Beisel C. 2012b. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet 44: 94–100. [DOI] [PubMed] [Google Scholar]
- Ulas J, Weihmuller FB, Brunner LC, Joyce JN, Marshall JF, Cotman CW. 1994. Selective increase of NMDA-sensitive glutamate binding in the striatum of Parkinson's disease, Alzheimer's disease, and mixed Parkinson's disease/Alzheimer's disease patients: an autoradiographic study. J Neurosci 14(11 Pt 1): 6317–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. 1995. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev 49: 23–36. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. 2001. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci 98: 11024–11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. 2002. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci 3: 921–931. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf CJ, Mannion RJ. 1999. Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet 353: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Yao B, Jin P. 2014. Unlocking epigenetic codes in neurogenesis. Genes Dev 28: 1253–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AB, Greenamyre JT, Hollingsworth Z, Albin R, D'Amato C, Shoulson I, Penney JB. 1988. NMDA receptor losses in putamen from patients with Huntington's disease. Science 241: 981–983. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Steijaert MN, Lau D, Schütz G, Delucinge-Vivier C, Descombes P, Bading H. 2007. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53: 549–562. [DOI] [PubMed] [Google Scholar]
- Zheng S, Eacker SM, Hong SJ, Gronostajski RM, Dawson TM, Dawson VL. 2010. NMDA-induced neuronal survival is mediated through nuclear factor I-A in mice. J Clin Invest 120: 2446–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.