Abstract
With the advent of quantitative tools for measuring gene expression in single cells, researchers have made the discovery that in many contexts, mRNA and protein levels can vary widely from cell to cell, often due to inherently stochastic events associated with gene expression. The study of this cellular individuality has become a field of study in its own right, characterized by a blend of technological development, theoretical analysis, and more recently, applications to biological phenomena. In this review, we focus on the use of the variability inherent to gene expression as a tool to understand gene regulation. We discuss the use of variability as a natural systems-level perturbation, its use in quantitatively characterizing the biological processes underlying transcription, and its application to the discovery of new gene regulatory interactions. We believe that use of variability can provide new biological insights into different aspects of transcriptional control and can provide a powerful complementary approach to that of existing techniques.
Introduction
Why do cells differ from each other? Traditionally, researchers thought that the differences between cells resulted from genetic differences or environmental differences. Increasingly, however, single cell measurements have revealed that even genetically identical cells in homogeneous conditions can display dramatic fluctuations in their levels of gene expression 1. Such fluctuations manifest at the level of mRNA and proteins, often owing to the inherent randomness of the biochemistry associated with transcription and translation. Experiments in a host of organisms have shown that this phenomenon is widespread, manifesting in bacteria, yeast and in a variety of metazoan cell types, and the field is known for the application of ever more sophisticated methods for making precise measurements of gene expression in single cells2. Next, researchers took to exploring the biological consequences of this variability1,3,4, showing that in some cases, cells exploit noise to generate phenotypic heterogeneity, whereas in other cases, gene regulatory networks appear to be constructed in such a manner as to minimize the effects of noise. Such findings are particularly relevant to fields such as stem cell and cancer biology, where experiments suggest that the widespread cellular heterogeneity observed in these systems may have implications for cell fate specification and disease.
A relatively underexplored topic in the field, however, is the use of variability in gene expression as the basis for characterizing and understanding gene regulation. In principle, the idea is rather simple: a regulatory interaction between two genes would manifest itself as a correlation or anti-correlation in their output. For example, if gene A encoded a transcription factor that repressed the expression of gene B, then one might expect that expression of gene A would be anti-correlated with expression from gene B, meaning that cells high in A would then be low in B and vice versa. This “fluctuation-correlation analysis” can be used to quantitatively characterize such interactions without artificially perturbing the system using drugs or other disruptive agents. Additionally, such single-cell techniques can be used to characterize the biophysics of cellular processes such as transcription5, or alternatively, as a tool for discovering new–and often unexpected–interactions. In this review, we explore some progress researchers in the field have already made along these lines, and some potential directions for future research in this area.
Variability as a natural perturbation
A central goal of systems biology is to take qualitative descriptions of biological processes and determine their quantitative characteristics. For instance, in the case above, even if we know that transcription factor A represses the transcription of gene B, understanding how this interaction fits into the larger context of the cell requires that we know quantitatively how the transcription of gene B depends on the concentration of transcription factor A (Fig 1A,B). The current approach to this problem involves systematically perturbing cells through overexpression or knockdown of a set of genes, among other techniques. However, the naturally occurring cell-to-cell variability in mRNA and protein levels serves as a proxy for these artificial perturbations, and can provide a means by which one can make such quantitative measurements. (It is important to note, however, that single-cell measurements, especially those measuring fluorescence intensity, can be prone to technical noise. It is crucial for investigators to check for systematic errors in their measurements to be sure that the variability measured is of biological relevance and not due to measurement noise.)
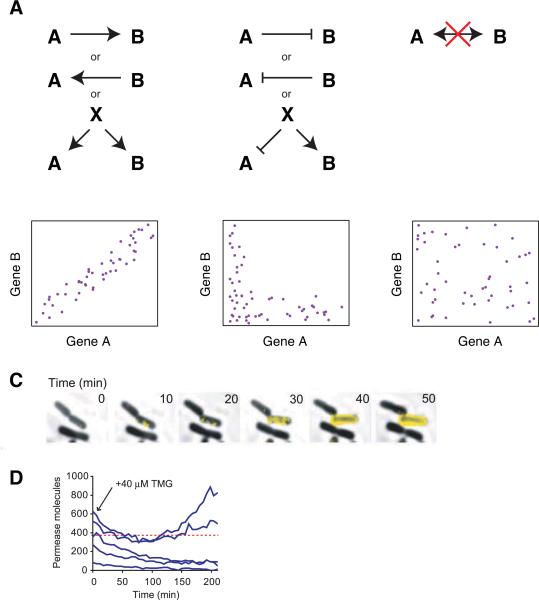
Figure 1.
Characterizing gene regulation using expression variability. A. Depiction of different types of regulation and how they would manifest themselves as correlations or anti-correlations in gene expression at the single cell level (assuming only intrinsic noise in gene expression). B. Time-lapse microscopy of an individual E. coli cell that triggers a positive gene expression feedback loop. C. Using variability to map out the threshold level of expression required to trigger the feedback loop. B and C are reproduced from Choi et al. Science 20086.
Such approaches have in particular shed light on the interplay between cell-to-cell variability and gene regulatory interactions in cell fate specification, often providing striking quantitative insights into long-standing questions in these fields. One particularly remarkable example is a recent analysis of the lactose utilization circuit in E. coli1,6. This gene network exhibits a strong positive feedback in that the production of even a small amount of lactose permease can cause gratuitous inducers (such as IPTG or TMG) to enter the cell, thereby increasing the production of lactose permease, eventually causing the cell to switch to a lactose-utilizing phenotype. However, the exact amount of lactose permease required to trigger the positive feedback has long been unresolved. Using cutting edge microscopy methods that enabled them to count individual lactose permease molecules in single living cells (Fig. 1C), Choi et al. explicitly quantified this threshold by taking cells randomly expressing lactose permease at a range of different levels and seeing which ones triggered the positive feedback when they added inducer to the system (Fig. 1D).
In metazoan developmental systems, researchers have utilized other methods for quantifying gene expression to measure expression variability and its relationship to phenotypic variability. In C. elegans, we used quantitative RNA fluorescence in situ hybridization (RNA FISH)7-9 to demonstrate that mutant organisms can display large variability in the expression of key transcription factors, and that the mutant phenotype itself is dependent on whether the expression of this transcription factor fails to exceed a certain threshold10. (We have also used similar tools to arrive at comparable findings in bacterial differentiation11.) In both of these cases, it would be difficult if not impossible to quantitatively make the perturbations required to quantify these thresholds with conventional means, especially in multicellular systems.
Far from being a mere numerical curiosity, the identification of the exact value of the threshold has important implications for how such systems must be constructed. For instance, were the lactose utilization circuit triggered by the production of even a single lactose permease molecule (as had been hypothesized for some time), the prevention of spontaneous lactose utilization in the cell would require exquisite and absolute control over the expression of lactose permease. Similar arguments could be made with respect to developmental networks in metazoans. We believe that measuring these numerical characteristics of gene regulatory networks will contribute greatly to our understanding of the design principles underlying these crucial processes.
Another important use for such methods is in the quantitative characterization of new and relatively unexplored gene regulatory mechanisms, such as control of gene expression via micro RNA (miRNA), which are thought to alter translation and RNA stability12 or, more recently, long non-coding RNA, which appear to regulate gene expression via a host of mechanisms13,14, including epigenetic. Despite the intense excitement over the potential for new biology in these fields, it has often proven difficult to quantify their behavior via conventional means, especially in the case of long non-coding RNA, whose modes of action often preclude standard perturbations. In such cases, variability in gene expression provides a natural means to measure the relevant interactions. For instance, recent quantitative studies of miRNA15 and small RNA16 function have harnessed natural variability to quantitatively map out the interaction between miRNA and its target mRNA, revealing that miRNA activity can lead to threshold-like gene expression behaviors. In the case of long non-coding RNAs, a few new studies are exploring quantitative aspects of their regulatory function17,18, in one case showing that even just 10 molecules of a long non-coding RNA can lead to very sharp downregulation of the target gene17. It is interesting to note that studies on bulk populations have often found that the effects of microRNA and long non-coding RNA are relatively modest compared to that of more conventional regulatory mechanisms such as transcription factor binding, but it is possible that the effects are actually more striking but are not readily apparent at the population level17.
How does this approach compare to the standard methods for quantifying gene regulation, such as inducible expression, RNA knockdown and transgenic overexpression? In many ways, we view these approaches as complementary, and indeed, combining intrinsic variability approaches with standard methods can lead to new insights, particularly in higher eukaryotes. The benefits of using variability stem primarily from its “perturbation-less” quality: the abundances of the molecules in question are at their physiologically relevant levels, there are no non-specific effects, and they do not require time-consuming or complex (or intractable) genetic manipulations. Conventionally, in mammalian cells, the standard methods for changing levels of gene expression are RNA knockdown (via siRNA, shRNA, antisense oligonucleotides (ASO), RNAi or morpholino, amongst other methods) and transgenic overexpression (with other new methods under development19,20). The former methods often suffer from the fact that they afford little control over the degree of knockdown (not to mention a lack of specificity), and single cell analyses often reveal that the level of knockdown can vary wildly from cell to cell17. Meanwhile, transgenic overexpression usually results in expression that exceeds endogenous levels by significant margins unless great care is taken to ensure that expression does not exceed normal levels, and even then, transgenes are notoriously variable in their expression from cell to cell. Of course, the benefit of these perturbations is that the results can establish a clear direction of causality: whereas correlations merely indicate that genes co-vary and may potentially regulate each other, knockdowns and overexpression can reveal which gene regulates which. Ultimately, we believe that using the variability inherent to RNA knockdown and transgenic overexpression can actually serve to help elucidate further information from these perturbations. For instance, in a recent study17, ASO-mediated RNA knockdown of a long non-coding RNA in mouse ES cells yielded only marginal effects on a putative target gene at the population level, but a single cell analysis showed that the effects were actually much larger in the specific cells affected. We anticipate that studying knockdown and overexpression at the single molecule level will result in a much more nuanced and detailed interpretation of such experiments.
It is important, however, to note that using variability to characterize gene regulatory interactions may not work in every situation. First and foremost, it is entirely conceivable that the expression of the gene in question does not vary enough to provide sufficient sampling. This is likely the case for many “housekeeping” genes 21, among others; in such situations, there may be enough variability to potentially confirm an interaction, but perhaps not enough to characterize it. Also, it is possible that variability in the upstream component of the system does not transmit to the downstream component, due to noise buffers, as strikingly demonstrated in a recent study22. Finally, we note that much care must be taken in making and interpreting such measurements, in particular controlling for extrinsic global factors in variation such as cell size23-25.
Variability for biophysical characterization
One can also use variability as a means to extract more details about the various biochemical processes involved in gene expression5. An illustrative case study is that of transcription, specifically, transcriptional bursts. One of the main conceptual contributions of the cell-to-cell variability field to our understanding of transcription is the finding that many genes (if not the vast majority in higher eukaryotes) transcribe in a pulsatile fashion, with even “constitutively expressed” housekeeping genes remaining transcriptionally inactive much of the time and only occasionally transcribing several mRNA in rapid succession (constituting a “burst”). Early studies using fluorescent protein reporters hinted at this sort of behavior26, and quantitative RNA imaging studies confirmed the existence of bursts27-31.
Inserting the notion of bursting into the field of transcriptional regulation has led to a more nuanced framework within which to understand how the biochemical underpinnings of transcriptional regulation manifest themselves. Previously, when researchers uncovered a mechanism governing the degree of transcription of a gene (say, binding affinity of a particular transcription factor), the only discernable change would be to the mean level of transcription. Now, one can consider whether change to the binding affinity of said transcription factor changes the mean by altering, say, burst frequency or burst size. Indeed, experiments measuring burst size and frequency via RNA FISH have shown that modulating various types of transcription factors and promoter elements can differentially affect burst size and frequency29,30,32,33. (A particularly striking study of several genes in bacteria suggests that transcriptional bursting scales solely with the mean abundance of the transcript, and is primarily independent of other factors34.) Such results suggest that these different perturbations affect the transcriptional process in different ways, leading one to make more specific biochemical interpretations. For instance, many researchers believe that transcriptional bursts are a consequence of or otherwise related to chromatin remodeling and modification; thus, changes to burst frequency may be related to the degree to which the perturbation in question is involved in chromatin remodeling. Researchers will have to make more progress, however, to make the links between the biochemistry and the more operational characteristics of transcriptional burst frequency and size more explicit.
Biologically, the next step is to link the modulation of transcriptional bursts to actual biological outcome. One study has shown that modulating transcriptional bursts can actually qualitatively affect the behavior of a gene network35, and we expect more such examples in the future; indeed, it is likely that bursts underlie the variability observed in developmental systems10,36. Meanwhile, a host of new methods, including single-nucleotide discrimination for allele-specific expression37 and live imaging methods38,39 are likely to bring a new level of clarity to experimental results.
There are several other aspects of transcription that may benefit from the more detailed measurements that variability in gene expression affords. Good examples include transcriptional pausing, in which a polymerase remains “poised” near the transcriptional start site, which is thought to facilitate rapid transcriptional activation. It is unclear how these pausing mechanisms influence the kinetics of transcription at the level of bursts. Other examples in which measurements of cell-to-cell variability reveal something about the mechanisms include the way in which a transcription factor regulates transcriptional40 and translational bursting41-43.
Variability as a tool for discovery
In the preceding examples, we have described situations in which one uses variability to characterize known interactions. However, variability can also aid in the discovery of new interactions, essentially through a process of identifying correlations and anticorrelations in the natural variability of a large number of different molecules. This is a nascent area of investigation, largely owing to the relatively recent development of the tools required for making measuring the expression of hundreds or thousands of genes at the single cell level. Here, we report a few such efforts, primarily organized by methodology.
One of the workhorse tools for studying cell-to-cell variability in gene expression is the use of fluorescent proteins, which provide robust measures of protein levels in single cells. In organisms amenable to facile genetic manipulation such as E. coli and, in particular, yeast, researchers have now constructed libraries of strains in which they have engineered large numbers of genes such that the resultant proteins are fused to particular fluorescent reporters44, and have even measured variability in a genome-wide manner21,45. A particular technique to measure variability involves creating yeast strains with pairs of genes labeled by distinguishable fluorescent reporters and looking for correlations between these genes. Using this method, the El-Samad group revealed several regulatory interactions between sets of genes, showing further that sets of genes with related functions tend to co-fluctuate in “noise regulons”, all without making any perturbations to the cell itself24. The Alon group has also made progress in applying such methods in mammalian cells through an approach they call dynamic proteomics, in which they generate a panel of cell lines46, each of which has GFP fused to a specific gene (akin to a gene trap). In a remarkable example of the utility of their method, they showed that variability in the expression of particular genes is correlated with whether or not certain cells respond to chemotherapeutic drugs47. In this situation, of course, it is impossible to say which genes are actually responsible for the cellular phenotype (as opposed to merely being associated with it), but such an analysis can greatly narrow the scope of genes one may consider for further study.
Another approach to using variability for discovery-based applications is to leverage our increasingly sophisticated genomic and image-based tools for high throughput measurements in single cells. In one incarnation, this involves the sorting of cells into high and low expressing subpopulations of a particular gene and then performing comparative expression profiling in these two cases. This method identifies genes that either correlate or anti-correlate with the gene of interest, potentially revealing previously undiscovered regulatory interactions. Researchers have applied such techniques in stem cells48-50, in which case many believe that the notion of gene expression heterogeneity is central to the biology of the cell. However, one must take care in applying this approach to control for effects of population dynamics, which may account for some of the observed variability50,51. Nevertheless, such methods would enable one to determine all genes in the regulatory pathway associated with a single gene without requiring any perturbations.
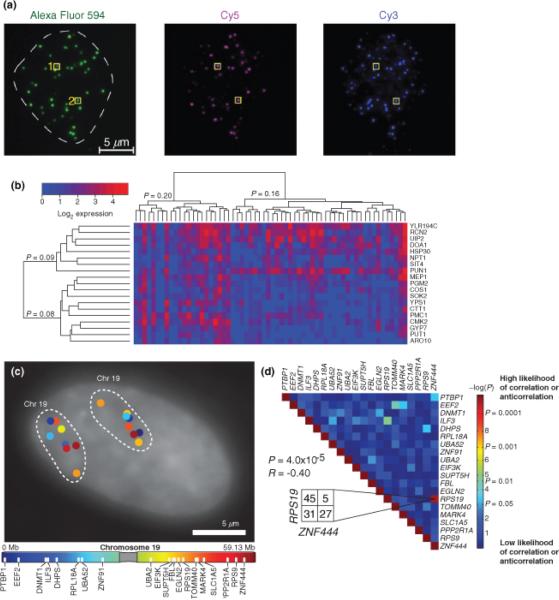
At the same time, the steady march of biotechnology has lead us to the point where we are increasingly able to interrogate large numbers of genes simultaneously within a single cell, potentially greatly expanding the scope of fluctuation-correlation-based regulatory interaction identification. Such methods typically fall into two categories: image-based, which have high resolution and accuracy but are typically limited to 10s to 100s of genes, and single-cell RNA sequencing methods, which provide genomic-level analyses, but are still in their infancy. Image-based methods involve trying to increase multiplexing beyond the standard 3-4 fluorophores one typically uses in fluorescence microscopy, either through color-coding or now barcoding methods. The most successful such methods to date involve RNA FISH7,52, and include transcriptional measurements53 and now single transcripts54, with perhaps the most spectacular example from Lubeck and Cai55, which utilized super resolution microscopy to identify 32 genes simultaneously in single cells (Fig. 2a,b). The latter two studies revealed categories of genes that appeared to be co-regulated just by examining the correlations between the panel of genes in single cells. Another example is a recent study from our lab in which we measured transcription of 20 genes by imaging transcription directly from individual chromosomes56 (Fig. 2c,d). Using this method, we found a striking anti-correlation between two genes (RPS19 and ZNF444) at the single chromosome level, meaning that if a chromosome is transcribing one of these genes, the other gene “knows” to not transcribe. These genes are separated by 14 megabases, revealing the possibility of extremely long-range transcriptional regulation mechanisms. It is important to note that the use of imaging enabled us to assign transcription to particular chromosomes, thus enabling us to unambiguously assert that the interaction was occurring in cis to the chromosome.
Figure 2.
Multiplex imaging-based approaches for measuring the expression of multiple genes at the same time in single cells. A. Color-coding and bar-coding approaches currently enable the measurement of up to 32 genes simultaneously in yeast. B. Expression profiles of 32 genes in single cells cluster into distinct categories of genes. A and B are reproduced from Lubeck and Cai, Nature Methods 201255. C. Simultaneous measurement of transcriptional activity of 20 genes on chromosome 19 in primary human cells. D. Correlating the transcriptional activity of the 20 genes on a per-chromosome basis revealed a strong anti-correlation between two genes separated by roughly 14 megabases. C and D are reproduced from Levesque and Raj, Nature Methods 201356.
One limitation of single molecule methods, however, is that they are generally rather laborious and technically difficult to perform, typically precluding the ability to interrogate the entire genome in such a manner. Other without such limitations typically involve scaling down conventional genome-scale methods for expression profiling like RNA-sequencing57 and multiplex real-time PCR (RT-PCR)58-60 to the point where one can perform them on single cells. Such methods can provide a picture of the genome in single cells, greatly enhancing the scope of the correlations one can measure. It is still an ongoing process to determine the accuracy and sensitivity of these methods, especially because they typically involve amplification steps, but they have the potential for producing comprehensive data that can reveal interactions at a very broad scale. With the rapid decrease in sequencing costs, we believe this will prove to be a hugely useful tool in extending the power of fluctuation-based methods of analysis.
It is worth noting that most of these methods thus far focus on quantifying nucleic acids, specifically RNA, to quantify gene expression. Currently, most comprehensive efforts21,43,46 utilize libraries of cell lines, each with a single gene labeled. Such a scheme makes it difficult to measure single-cell correlations between genes. Moreover, such methods only work in organisms in which genetic manipulations are relatively facile. The development of single cell proteomics would mark a huge advance in the field.
What are the advantages and limitations of fluctuation-correlation analysis for discovering regulatory interactions compared to conventional methods? The primary advantage is that it is generally non-perturbative, and the main disadvantage is that it cannot define any arrow of causality. In mammalian systems, one of the most common ways to check for downstream targets of a regulatory gene is to knock it down via siRNA or the equivalent. One of the main issues with such treatments, however, is that they often have many off-target effects, and so if one pursues a genomic-scale analysis after knockdown, one will obtain a large number of false-positive interactions. We believe that the use of fluctuation-correlation analysis can nicely complement the use of knockdown methods to help identify and characterize interacting genes. Firstly, one can cross-check all the genes identified via fluctuation-correlation analysis and those identified by knockdown to eliminate those genes that are likely off-target effects of the knockdown treatment. Secondly, knockdown treatments will generally affect genes downstream of the gene of interest, whereas fluctuation-correlation analysis identifies both upstream and downstream (and co-regulated) genes. Thus, in a coherently fluctuating set of genes, one could potentially begin to characterize where in the gene network the gene of interest resides.
Meanwhile, as the ability to profile expression in single cells increases, so will our ability to identify not just the networks associated with a particular gene, but rather entire regulatory clusters of genes in an unbiased way24. Currently, the methodology people employ is akin to “guilt by association”61, in which researchers accumulate several different large-scale sets of gene expression data across a large number of perturbations or cell types and look for co-fluctuating genes. In a sense, this is similar to fluctuation-correlation analysis, but it is likely that the interactions identified by fluctuation-correlation analysis will be more specific; for example, single cell analysis can reveal regulatory interactions that are in fact opposite to those detected at the population level upon global perturbations17. As with knockdown experiments, though, we anticipate that combining perturbation-based guilt by association methods with fluctuation-correlation analysis23 has the potential to greatly enhance our understanding of gene regulatory networks.
Conclusion
The past decade has seen an explosion of interest in single cell analysis, and it is now broadly accepted that superficially similar cells can display dramatic cell-to-cell differences in the expression of key genes. We believe that this variability affords an opportunity to not only characterize known interactions and properties of gene expression, but also to discover new interaction networks and mechanisms of gene regulation in ways that complement existing methods. As we develop ever more sophisticated measurement techniques, single-cell methods using variability will no doubt become increasingly important in the toolkit of molecular biologists in the coming years.
Footnotes
Conflict of interest: AR receives royalties from patents related to RNA FISH methods described in the article, as well as consulting income.
Contributor Information
Olivia Padovan-Merhar, Department of Physics, University of Pennsylvania.
Arjun Raj, Department of Bioengineering, University of Pennsylvania, 210 S. 33rd St. Philadelphia PA 19104..
References
- 1.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annual review of biophysics. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balázsi G, van Oudenaarden A, Collins JJ. Cellular Decision Making and Biological Noise: From Microbes to Mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013 doi: 10.1038/nrg3484. doi:10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi PJ, Cai L, Frieda K, Xie XS. A Stochastic Single-Molecule Event Triggers Phenotype Switching of a Bacterial Cell. Science. 2008;322:442–446. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raj A, Tyagi S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Meth Enzymol. 2010;472:365–386. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- 9.Nair G, Walton T, Murray JI, Raj A. Gene transcription is coordinated with, but not dependent on, cell divisions during C. elegans embryonic fate specification. Development. 2013 doi: 10.1242/dev.098012. doi:10.1242/dev.098012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raj A, Rifkin SA, Andersen EC, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 15.Mukherji S, et al. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative Characteristics of Gene Regulation by Small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes & development. 2013;27:1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bumgarner SL, et al. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013 doi: 10.1016/j.cell.2013.06.044. doi:10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nevozhay D, Zal T, Balázsi G. Transferring a synthetic gene circuit from yeast to mammalian cells. Nat Commun. 2013;4:1451. doi: 10.1038/ncomms2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman JRS, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 22.Shah K, Tyagi S. Consequences of stochastic mRNA synthesis in a gene regulatory pathway. Molecular Systems Biology [Google Scholar]
- 23.Rinott R, Jaimovich A, Friedman N. Exploring transcription regulation through cell-to-cell variability. Proc Natl Acad Sci USA. 2011;108:6329–6334. doi: 10.1073/pnas.1013148108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol. Cell. 2012;45:483–493. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volfson D, et al. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–864. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- 26.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 28.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suter DM, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 31.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blake WJ, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Octavio LM, Gedeon K, Maheshri N. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS Genet. 2009;5:e1000673. doi: 10.1371/journal.pgen.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So L-H, et al. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.To T-L, Maheshri N. Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science. 2010;327:1142–1145. doi: 10.1126/science.1178962. [DOI] [PubMed] [Google Scholar]
- 36.Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levesque MJ, Ginart P, Wei Y, Raj A. Single cell allele-specific expression via single nucleotide variant detection in situ. Nature Methods. doi: 10.1038/nmeth.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nature Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 39.Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nature Methods. 2012;10:119–121. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elf J, Li G-W, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 42.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 45.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 46.Sigal A, et al. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nature Methods. 2006;3:525–531. doi: 10.1038/nmeth892. [DOI] [PubMed] [Google Scholar]
- 47.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 48.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marks H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pina C, et al. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14:287–294. doi: 10.1038/ncb2442. [DOI] [PubMed] [Google Scholar]
- 51.Nevozhay D, Adams RM, Van Itallie E, Bennett MR, Balázsi G. Mapping the environmental fitness landscape of a synthetic gene circuit. PLoS Comput Biol. 2012;8:e1002480. doi: 10.1371/journal.pcbi.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 53.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 54.Jakt LM, Moriwaki S, Nishikawa S. A continuum of transcriptional identities visualized by combinatorial fluorescent in situ hybridization. Development. 2013;140:216–225. doi: 10.1242/dev.086975. [DOI] [PubMed] [Google Scholar]
- 55.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nature Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levesque MJ, Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nature Methods. 2013 doi: 10.1038/nmeth.2372. doi:10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nature Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 58.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan DWM, et al. Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development. 2013;140:1433–1444. doi: 10.1242/dev.087551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]