Abstract
This study aimed at assessing the dynamics of lactic acid bacteria and other Firmicutes associated with durum wheat organs and processed products. 16S rRNA gene-based high-throughput sequencing showed that Lactobacillus, Streptococcus, Enterococcus, and Lactococcus were the main epiphytic and endophytic genera among lactic acid bacteria. Bacillus, Exiguobacterium, Paenibacillus, and Staphylococcus completed the picture of the core genus microbiome. The relative abundance of each lactic acid bacterium genus was affected by cultivars, phenological stages, other Firmicutes genera, environmental temperature, and water activity (aw) of plant organs. Lactobacilli, showing the highest sensitivity to aw, markedly decreased during milk development (Odisseo) and physiological maturity (Saragolla). At these stages, Lactobacillus was mainly replaced by Streptococcus, Lactococcus, and Enterococcus. However, a key sourdough species, Lactobacillus plantarum, was associated with plant organs during the life cycle of Odisseo and Saragolla wheat. The composition of the sourdough microbiota and the overall quality of leavened baked goods are also determined throughout the phenological stages of wheat cultivation, with variations depending on environmental and agronomic factors.
INTRODUCTION
Lactic acid bacteria are ubiquitous microorganisms with potential beneficial effects on crop and livestock production (1). Together with lactic acid bacteria, other Firmicutes (e.g., Bacillus species) are ubiquitous, often found as soil inhabitants and easily transferred from soil to roots. Some strains of Bacillus produce cytokinins, thus acting as plant growth-promoting bacteria (2). Other strains inhibit the pathogenic fungus Fusarium graminearum in durum wheat (3). From the agronomic point of view, wheat is a well-characterized crop, with a worldwide economic relevance, whose associated bacteria could influence plant growth (4). Several plant-associated bacteria contaminate grains, and hence flour, affecting the overall quality of leavened baked goods (5, 6). It is well known that some spore-forming bacteria (e.g., Bacillus species) cause rope spoilage in bread (7).
Some Bacillus and other undesired microorganisms (e.g., molds belonging to Aspergillus and Penicillium) can be inhibited by the use of sourdough, a biological leavening agent containing high numbers of yeasts and, especially, lactic acid bacteria (8, 9). Other advantages that are consistent with the use of sourdough regard the sensory, shelf life, and nutritional features of leavened baked goods. Most of these advantages are attributed to the metabolisms of lactic acid bacteria (10). They are not deliberately added but are naturally present in the dough. Indeed, traditional sourdough originates from multiple steps (backslopping) of fermentation. Since they are not added, the bakery environment (the bakers, insects, and animals) and the flour are the two main routes for contaminating lactic acid bacteria. Although an abundance of the literature has dealt with the characterization of the mature sourdough microbiota (11–16), the origin of lactic acid bacteria has only in part been elucidated. Some lactic acid bacteria circulate throughout the environment of propagation. Lactobacillus sanfranciscensis, which was detected in the air of the storage and work rooms, as well as on benches and dough mixers, dominated in the related traditional sourdoughs (17, 18). Lactobacillus plantarum and Lactobacillus spicheri dominated in the bakery environment and in the sourdough produced in the same bakery (17). Evidence has indicated that sourdough fermentations carried out in the laboratory with the flour as the only nonsterile ingredient were characterized by species and/or strains of lactic acid bacteria different from those in artisan sourdoughs (14, 19–22). This would suggest that the flour microbiota shares very few taxa with that of the corresponding mature sourdough. Evidence has suggested that the flour microbiota was affected by contamination from the bakery environment, especially from humans. For instance, sourdough isolates of Lactobacillus reuteri were human lineage strains (23). Furthermore, many species (e.g., Lactobacillus rossiae, L. plantarum, and L. reuteri) isolated from type II sourdoughs were also identified from rodent and/or swine feces (24–26). Apart from this contamination, the inherent presence of lactic acid bacteria in the flour also has to be considered. Nevertheless, lactic acid bacteria contaminating cereals in the outer layers of kernels are removed during milling and tend to stay in bran-rich fractions (27). Consequently, the lactic acid bacteria contaminating flour or semolina should theoretically be a part of the endophytic microbiota of cereals. Epiphytic lactic acid bacteria were isolated from forage crops (28, 29), and lactobacilli inoculated into Lolium perenne were able to colonize roots at the endophytic level (30). To the best of our knowledge, no data are available for the microbiota of Triticum aestivum or Triticum durum cultivars during plant cultivation.
This study was aimed at assessing, through culture-dependent and -independent approaches, the composition of the wheat plant microbiota during the different phases of growth and to assess the extent of flour contamination by endophytic lactic acid bacteria and other Firmicutes, which from the field become relevant for sourdough fermentation.
MATERIALS AND METHODS
Plant growth and sample collection.
The Odisseo and Saragolla varieties of durum wheat were cultivated in slimy loam soil (36% sand, 40% silt, and 24% clay) in Gravina di Puglia (Bari, Italy). For each cultivar, four comparable allotments (named A, B, C, and D), with individual sizes of 1.5 × 15 m, were established at approximately 500 m apart within the same field in November 2013. Plants were harvested from the following stages of crop growth: tillering, stem elongation, booting, flowering, milk development, and physiological maturity. At each sampling time, a group of 10 plants was carefully removed with intact roots, and the soil adhering to the roots was removed by shaking. Plants were divided by means of a sterile scalpel into hypogeous (roots) and epigeous (leaves or spikes, depending on the phenological stage) organs, which were inserted into Whirl-Pak sterile sampling bags (Nasco, Fort Atkinson, WI), kept at a cool temperature, and processed in the laboratory within 4 h of collection.
Confocal laser scanning microscopy (CLSM).
Small pieces of roots and leaves were taken from wheat and put in isopentane cooled by liquid nitrogen at −150°C, in order to freeze the samples without formation of ice crystals. Twenty-micrometer-thick sections were achieved at −27°C using a cryotome. After drying on a slide, sections were stained by using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Cambridge Bioscience, Cambridge, United Kingdom) according to the manufacturer's instructions. After 15 min, sections were rinsed using distilled water, and an upper lip was sealed on the section using nail polish. Microscopic observations were performed using a Leica confocal microscope (Leica Microsystems, Heidelberg GmbH, Wetzlar, Germany) in epi mode (31).
DNA extraction from plant organs and processed grain samples.
Ten-gram quantities of washed plant organs (roots and leaves or spikes) were immersed in 200 ml of DNA-free 0.1 M potassium phosphate buffer (pH 7.0) in a 500-ml Erlenmeyer flask and treated (7 min at 30°C) in an ultrasonic bath (RK103H Sonorex Super, Ultra Center Europe, Zwolle, The Netherlands) (32). Plant organs were taken out of the flask for further treatment, whereas epiphytic microorganisms released into the buffer were pelleted by centrifugation (20,000 × g for 30 min at 4°C) and then resuspended in 5 ml of potassium phosphate buffer (33). Suspensions of epiphytic microorganisms were used as starting material for extraction of DNA. Plant organs were surface sterilized by soaking in 150 ml of a 15% (vol/vol) H2O2 solution. They were treated for 15 min with gentle rotary shaking, protected from light. Then plant organs were twice washed in sterile demineralized water and finally dried in a laminar flow hood for 1 h (34). Surfaces of plant organs were checked for sterility by blotting them tightly on 1/10-strength tryptic soy agar and incubating plates at 30°C for 48 h (35). No growth was observed. Surface-sterilized plant organs were homogenized (3 min) with 80 ml of 0.1 M potassium phosphate buffer (pH 7.0) in a BagMixer 400P (Interscience, Saint Nom, France) blender. The homogenate, free of plant debris, contained endophytic microorganisms and was used as starting material for extraction of DNA.
Processed grain samples were obtained as follows. Chaff was removed from grain by using a Wintersteiger AG (Ried, Austria) thresher (model Seedmech Classic) and stored for further analyses. The grain was further separated from residual chaff and tempered to a moisture content of 18%. An aliquot of tempered grain (100 g) was used for extraction of epiphytic and endophytic microorganisms. Epiphytic microorganisms were extracted according to the protocol described for plant organs. Endophytic microorganisms were extracted upon grinding of chemically surface-sterilized grain, devoid of epiphytic microorganisms. Ten grams of ground grain was homogenized (3 min) with 90 ml of 0.1 M potassium phosphate buffer (pH 7.0) in a BagMixer 400P blender. The homogenate, free of grain debris, containing endophytic microorganisms, was used as starting material for extraction of DNA. Another aliquot of tempered grain (10 kg) was milled into flour with a Bühler-Miag MLI-204 grinder (Bühler S.p.A., Segrate, Milan, Italy). Chaff, bran, and flour were used for detection of lactic acid bacteria and for DNA extraction. Commercial grains and the related bran and flour were also analyzed as controls.
Genomic DNA was extracted from 0.5 ml of suspensions containing either epiphytic or endophytic microorganisms, using the FastDNA Pro Soil-Direct kit (MP Biomedicals, Santa Ana, CA) coupled to the FastPrep instrument (MP Biomedicals), according to the protocol described by Minervini et al. (36). The quality and concentration of DNA extracts were assayed by spectrophotometric measurements using a NanoDrop ND-1000 (Thermo Fisher Scientific Inc., Marietta, OH).
Illumina MiSeq analysis.
DNA extracted from plant organs collected in each of the four allotments (A, B, C, and D) was pooled and used as the template for Illumina MiSeq diversity analyses, which were carried out at the Research and Testing Laboratory (RTL; Lubbock, TX). Primers Firm350F and Firm814R were used to amplify a fragment of the 16S rRNA gene for analysis of diversity inside the phylum of Firmicutes (37). PCR and sequencing analyses were carried out according to the protocol of the RTL.
The sequenced reads were processed through denoising and chimera detection. Denoising was performed through the following steps: (i) merging together the forward and reverse reads using the PEAR Illumina paired-end read merger (38); (ii) reading the runs through an internally developed quality trimming algorithm that truncates reads having an average quality higher than 25; (iii) grouping reads by using the USEARCH (39) algorithm (prefix dereplication) into clusters (4% dissimilarity among sequences of the same cluster), so that each sequence of shorter length to the centroid sequence must be a 100% match to the centroid sequence for the length of the sequence; and (iv) operational taxonomic unit (OTU) selection by using the UPARSE OTU selection algorithm (40) to classify the large number of clusters into OTUs. Following denoising, the selected OTUs were chimera checked using the UCHIME software executed in de novo mode (41). Each trimmed read was mapped to its corresponding nonchimeric cluster using the USEARCH global alignment algorithm (39). Each sequence in a cluster was then aligned to the consensus sequence. Each sequence was corrected base by base in order to remove noise. Analysis of microbial diversity was finally performed by running the centroid sequence from each cluster against the USEARCH algorithm, using a database of high-quality sequences derived from the NCBI database. Lastly, the output was analyzed using an internally developed python program that assigns taxonomic information to each sequence.
The percentage of each bacterial OTU was analyzed individually for each sample, providing relative abundance information among the samples based on the relative numbers of reads within each (42).
Enumeration and isolation of endophytic lactic acid bacteria from plant organs.
Surface-sterilized plant organs and chemically surface-sterilized grain and flour were used. Ten-gram quantities of roots, leaves, and spikes were immersed in 90 ml of sterile 0.1 M potassium phosphate buffer (pH 7.0) and homogenized for 3 min in a BagMixer 400P. Endophytic microorganisms from 10 g of grain or flour were also analyzed. Lactic acid bacteria were enumerated and isolated from plant organs, grain, and flour by the use of two selective media: de Man, Rogosa, and Sharpe medium (MRS) with maltose (10 g liter−1) and fresh yeast extract (50 ml liter−1), pH 5.6 (modified MRS [mMRS]), and sourdough bacterial medium (SDB) (43). Cycloheximide (0.1 g liter−1) was added to both media in order to prevent fungal growth. One milliliter of suspension containing endophytic microorganisms was serially diluted in sterile saline (NaCl 9 g liter−1) solution and plated onto mMRS and SDB agar. Plates were incubated at 30°C for 48 h. For each medium, a number of colonies equal to the square root of the total number recorded in plates coming from the highest dilution was randomly picked up. Gram-positive, catalase-negative, nonmotile rods and coccal isolates were cultivated in mMRS or SDB at 30°C for 24 h and restreaked onto the same agar medium. All isolates considered for further analyses were able to acidify the culture medium.
Phenotypic identification of endophytic lactic acid bacteria.
Metabolic profiles of endophytic lactic acid bacteria isolated from plant organs, grain, and flour were determined using Biolog AN microplates (Rigel Life Sciences, Rome, Italy). The wells of the microplates were inoculated with 150 μl of bacterial suspensions adjusted to 0.6 ± 0.05 unit of absorbance (at a wavelength of 620 nm). Positive reactions were automatically recorded after incubation (for 24 h at 30°C under anaerobic conditions) using a microplate reader with a 590-nm wavelength filter.
Genotypic characterization of endophytic L. plantarum isolates by RAPD-PCR analysis.
Genomic DNA of L. plantarum isolates was extracted using a DNeasy blood and tissue kit (Qiagen, SA, Courtaboeuf, France) according to the manufacturer's instructions (44). Three oligonucleotides, P4 (5′-CCGCAGCGTT-3′), P7 (5′-AGCAGCGTGG-3′), and M13 (5′-GAGGGTGGCGGTTCT-3′), with arbitrarily chosen sequences were used for biotyping isolates of L. plantarum from leaves, spikes, and processed grain samples of Odisseo and Saragolla wheat. The reaction mixture and PCR conditions for primers P4 and P7 were those described by De Angelis et al. (45). PCR conditions for primer M13 were as described by Siragusa et al. (46). Randomly amplified polymorphic DNA-PCR (RAPD-PCR) profiles were acquired by the MCE-202 MultiNA microchip electrophoresis system (Shimadzu Italia s.r.l., Milan, Italy), using the DNA-12000 reagent kit (100 to 12,000 bp) and the 2-log DNA ladder (0.1 to 10.0 kb) according to the manufacturer's instructions. The reproducibility of the RAPD profiles was assessed by comparing the PCR products obtained with primers P4, P7, and M13 and DNA prepared from three independent cultures of the same strain. Nine strains were studied, and the patterns for the same strain were 95% similar, indicating that the reproducibility of the technique under the conditions used was high.
Statistical analyses.
Data were obtained at least in triplicate. Analysis of variance (ANOVA) was carried out on transformed data, followed by separation of means with Tukey's honestly significant difference (HSD) test, using the statistical software Statistica 7.0 for Windows (StatSoft, Vigonza, Italy). Weighted and unweighted UniFrac distance matrices and OTU tables were used to perform ADONIS and ANOSIM statistical tests through the compare_category.py script of QIIME to verify the microbial populations in the different plant organs. Permutation analysis was also performed for the Illumina MiSeq data. Multiple testing of corrected pairwise Spearman correlations was computed between OTU at the genus level (abundance of >0.1% in at least 5 samples) and environmental conditions. Only significant correlations (false discovery rate [FDR] < 0.050) were considered (47). Cell density of presumptive lactic acid bacteria, lactic acid bacterium OTU (genus level), environmental temperature, and water activity (aw) of the wheat organs were analyzed by principal component analysis (PCA), using Statistica 7.0.
Nucleotide sequence accession number.
The 16S rRNA gene sequences are available in the Sequence Read Archive of NCBI (accession number PRJNA268304).
RESULTS
Phenological stages of durum wheat samples and endophytic bacteria.
Plants of Odisseo and Saragolla durum wheat were sampled at six phenological stages of crop growth (see Fig. S1 in the supplemental material). Compared to Odisseo, Saragolla is a premature cultivar accelerating the flowering stage of ca. 10 days (see Table S1 in the supplemental material). Consequently, flowering, milk development, and physiological maturity of the two cultivars occur under different climatic conditions (e.g., temperature and rainfall). Preliminarily, the presence of bacteria in plant organs (roots and leaves or spikes) of durum wheat was analyzed by confocal laser scanning microscopy (CLSM). At the sampling times corresponding to tillering, stem elongation, and booting, leaves were analyzed. The samples of booting contained both leaves and spikes. Spikes, instead of leaves, were analyzed in correspondence to flowering, milk development, and physiological maturity. Bacterial cells were visible in all plant organs. An example is given in Fig. S2 in the supplemental material, related to roots and leaves at the stem elongation stage.
Firmicutes diversity through next-generation sequencing.
Total DNA from fractions of roots and leaves or spikes was amplified by Firmicutes-specific primers and subjected to next-generation sequencing. A distinction was made between the surface (epiphytic microbiota) and inside (endophytic microbiota) of plant organs. Total DNA from Odisseo and Saragolla grain (epiphytic and endophytic), chaff, bran, and flour was also analyzed. Commercial durum wheat grain, bran, and related flour were also analyzed. A total of 2,456,502 quality-trimmed sequences of 16S rRNA gene amplicons were obtained. The average number of sequences per sample was 40,942, with an average length of 447 bp. Good's estimated sample coverage (median value of ca. 98%; P > 0.05) indicated that a satisfactory coverage was reached for all the samples analyzed.
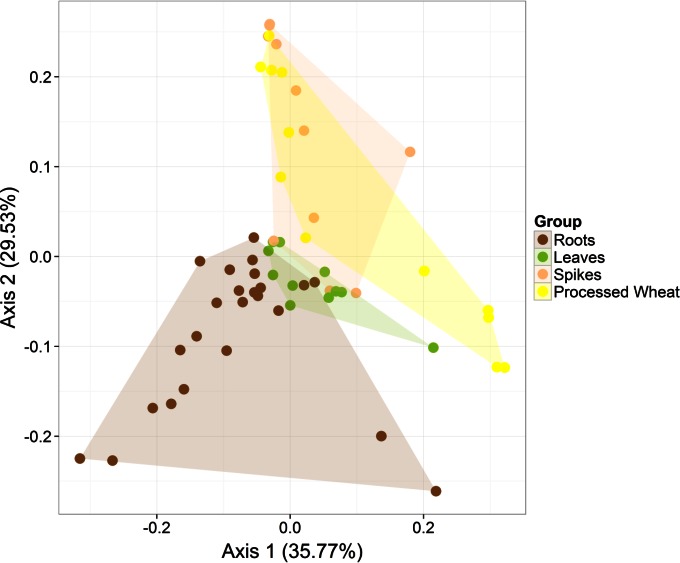
The community of Firmicutes was analyzed for number of operational taxonomic units (OTUs), richness estimator (ACE, Chao1), and diversity index (Shannon and Simpson) (see Table S2 in the supplemental material). The analysis revealed a significantly (P < 0.050) lower microbial diversity of leaves and spikes compared to that of root samples. As shown by principal coordinate analysis (PCoA), roots, leaves, and spikes of Odisseo and Saragolla wheat were clearly differentiated based on unweighted UniFrac distances (Fig. 1). In contrast, spikes and processed wheat samples were not differentiated. In addition, both ADONIS and ANOSIM statistical tests indicated a significant influence of roots and leaves or spikes on the microbial diversity (FDR < 0.050). Leaves did not differ (P > 0.050) from spikes and processed wheat samples.
FIG 1.
Principal coordinate analysis (PCoA) based on unweighted UniFrac analysis of all 16S rRNA gene sequences of epiphytic and endophytic Firmicutes found on hypogeous organs (roots), epigeous organs (leaves and spikes), and processed wheat (grain, chaff, bran, and flour) of Odisseo and Saragolla. Hypogeous and epigeous organs were analyzed at the tillering, stem elongation, booting, flowering, milk development, and physiological maturity stages.
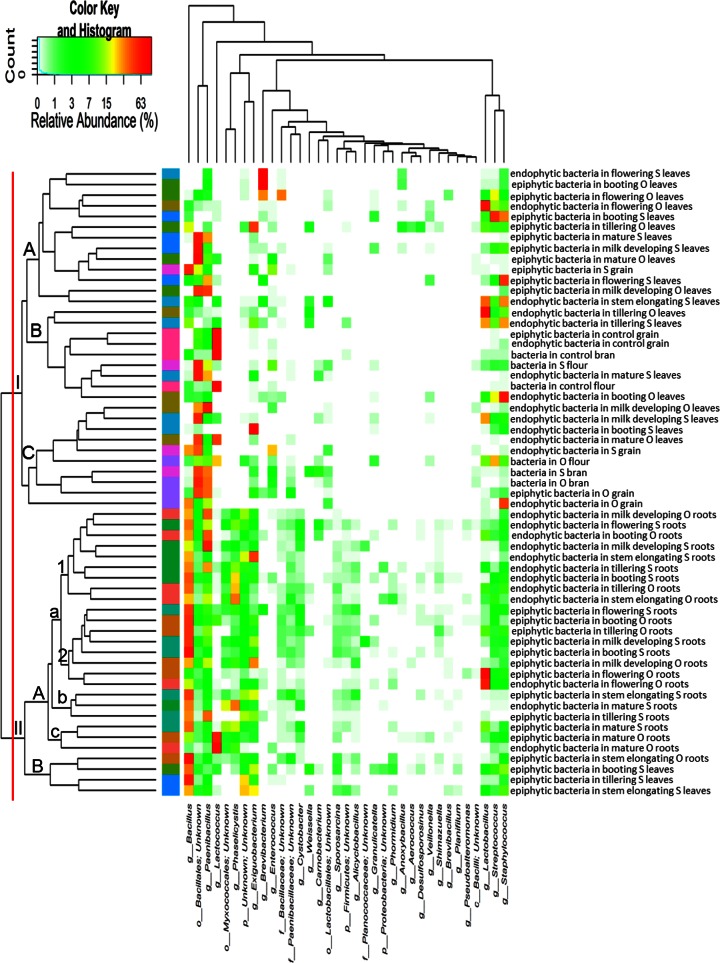
Core Firmicutes microbiome.
According to alpha- and beta-diversity, and considering the 35 most dominant genera of all samples, roots and leaves or spikes were distributed in two clusters (Fig. 2). Exceptions were the epiphytic bacteria belonging to tillering, stem elongation, and booting of Saragolla wheat. With few exceptions, epiphytic and endophytic bacteria grouped in different subclusters (cluster IA versus clusters IB and IC; cluster IIAa1 versus cluster IIAa2). Samples corresponding to endophytic bacteria found in the last phenological stages (milk development and physiological maturity) clustered together with processed wheat samples of both cultivars (cluster IC). A core Firmicutes microbiome, defined at the genus level and occurring in >98% of the samples analyzed was constituted by Bacillus, Exiguobacterium, Lactobacillus, Paenibacillus, Staphylococcus, and Streptococcus. Besides these genera, OTUs belonging to genera Enterococcus and Lactococcus occurred in >85% of the samples.
FIG 2.
Heat map summarizing the relative abundances of the 35 most dominant genera in DNA samples directly extracted from epiphytic and endophytic bacteria found on hypogeous organs (roots), epigeous organs (leaves and spikes), and processed wheat (grain, chaff, bran, and flour) of the Odisseo (O) and Saragolla (S) varieties. Hypogeous and epigeous organs were analyzed at the tillering, stem elongation, booting, flowering, milk development, and physiological maturity stages. Commercial grain, bran, and flour were also analyzed as a control. The color key defines the percentages of OTUs in the samples.
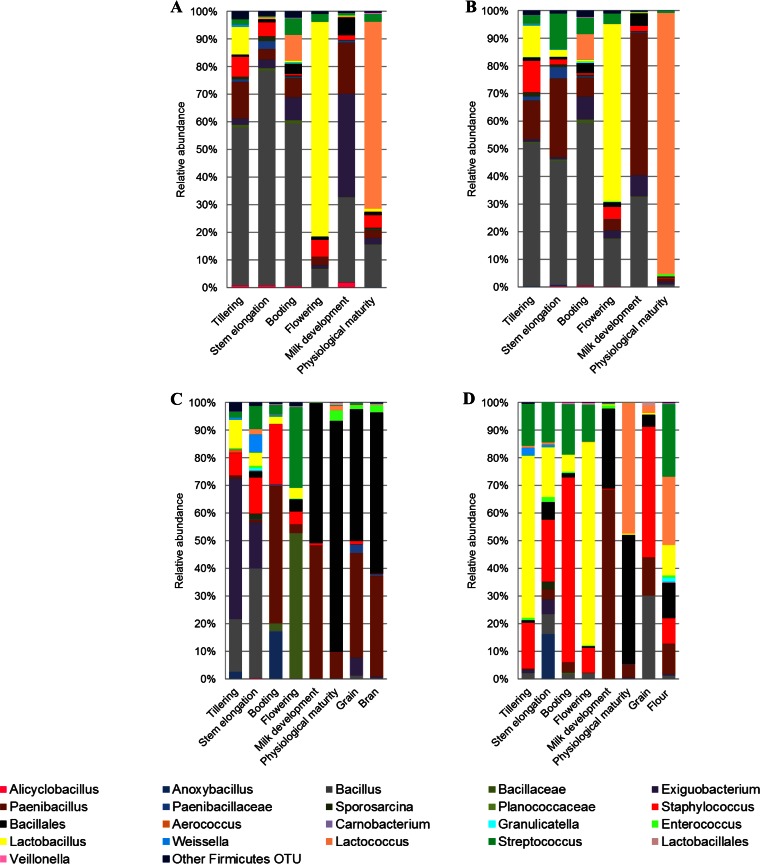
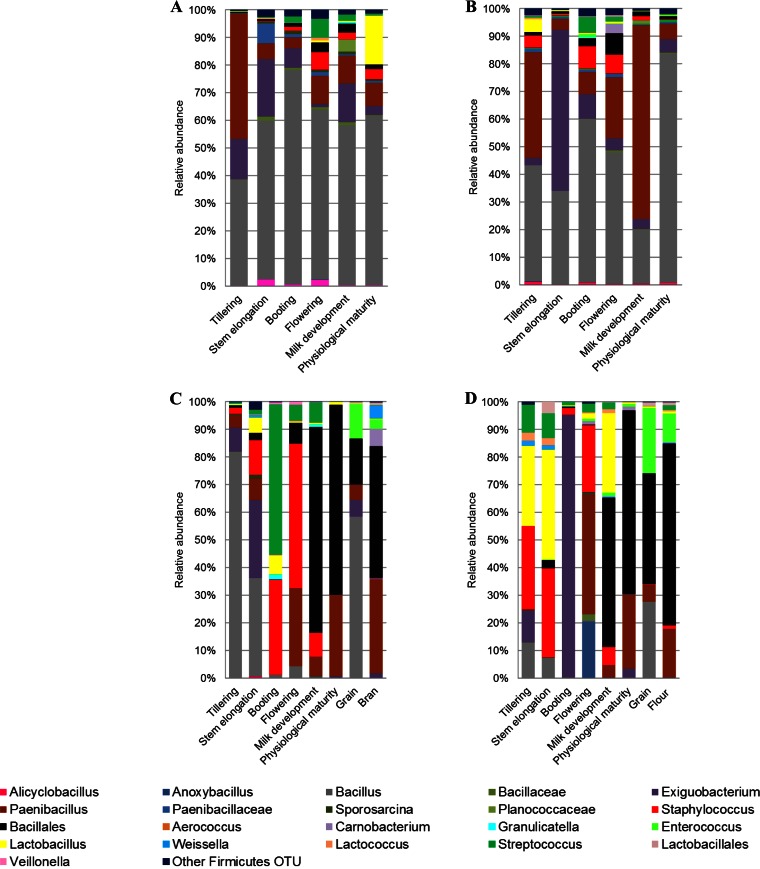
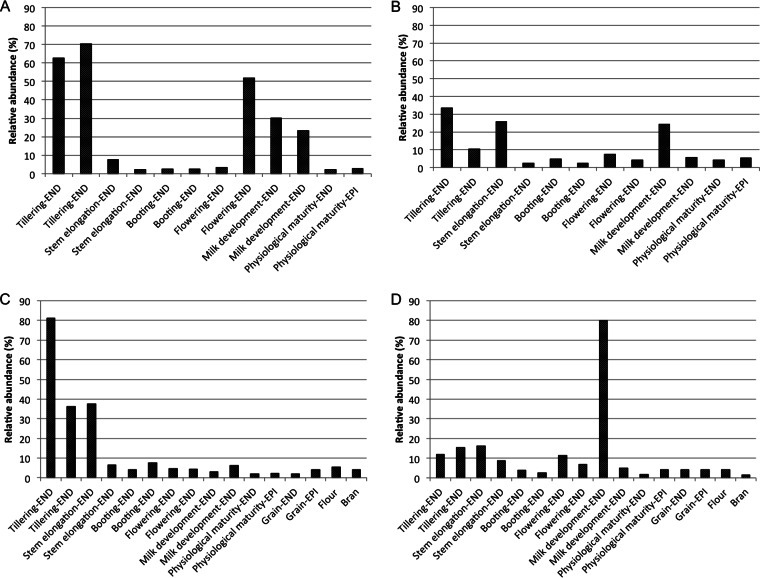
Among Firmicutes, Bacillus was mainly found at the endophytic and, especially, epiphytic levels of roots throughout the wheat life cycle, whereas it was only occasionally detected in leaves and spikes (Fig. 3 and 4). Paenibacillus was found at the endophytic and epiphytic levels of roots regardless of phenological stage. However, the relative abundance of Paenibacillus was lower than that of Bacillus in more than 60% of root samples. In contrast, in samples of leaves and spikes, from flowering to physiological maturity, Paenibacillus was more frequently detected than Bacillus. Exiguobacterium was found in roots throughout the wheat life cycle, whereas it was rarely detected in leaves. However, it was the dominant Firmicutes genus at the epiphytic level of Odisseo leaves at tillering (Fig. 3C) and at the endophytic level of Saragolla leaves at the booting stage (Fig. 4D). Bacillales were among the dominant OTUs at milk development, at physiological maturity, and in processed wheat, especially for Saragolla.
FIG 3.
Relative abundance of epiphytic (A and C) and endophytic (B and D) Firmicutes genera found in hypogeous organs (roots) (A and B), epigeous organs (leaves and spikes), and processed wheat (grain, bran, and flour) (C and D) of Odisseo. Hypogeous and epigeous organs were analyzed at the tillering, stem elongation, booting, flowering, milk development, and physiological maturity stages.
FIG 4.
Relative abundance of epiphytic (A and C) and endophytic (B and D) Firmicutes genera found in hypogeous organs (roots) (A and B), epigeous organs (leaves and spikes), and processed wheat (grain, bran, and flour) (C and D) of Saragolla. Hypogeous and epigeous organs were analyzed at the tillering, stem elongation, booting, flowering, milk development and physiological maturity stages.
At the endophytic level, Lactobacillus was the dominant Firmicutes genus at the flowering stage of Odisseo, both in roots and in spikes (Fig. 3B and D). Lactobacillus dominated also in roots at the epiphytic level during flowering (Fig. 3A). Lactococcus dominated in roots (endophytic and epiphytic levels) and spikes (endophytic level) at physiological maturity. Lactococcus was the lactic acid bacterium mostly present in grain at the endophytic level and dominated in flour together with Streptococcus. The genus Lactobacillus was found at a lower level in Saragolla than in Odisseo wheat (Fig. 4). At the flowering stage, Streptococcus was the dominant lactic acid bacterium in roots and spikes. Lactobacillus was the dominant lactic acid bacterial genus only at the endophytic level of spikes during milk development (Fig. 4D) and at the epiphytic level of roots during physiological maturity (Fig. 4A). Enterococcus was the dominant genus of lactic acid bacteria in grain at the endophytic level and dominated in flour. Lactococcus dominated in commercial grain and in the related bran and flour samples (see Table S3 in the supplemental material). At the species level, L. plantarum was the dominant species of Lactobacillus in almost all samples (Fig. 5; see also Table S3). Other lactic acid bacteria (Aerococcus viridans, Carnobacterium maltaromaticum, Enterococcus faecalis, Lactobacillus brevis, Lactobacillus fermentum, Lactobacillus gasseri, Lactobacillus iners, Lactococcus garviae, and Lactococcus lactis) were variously identified.
FIG 5.
Relative abundance of endophytic (END) and epiphytic (EPI) Lactobacillus plantarum within the OTUs belonging to lactic acid bacteria found on hypogeous organs (roots) (A and B), epigeous organs (leaves and spikes), and processed wheat (grain, bran, and flour) (C and D) of Odisseo (A and C) and Saragolla (B and D). Hypogeous and epigeous organs were analyzed at the tillering, stem elongation, booting, flowering, milk development, and physiological maturity stages.
Cell density of endophytic presumptive lactic acid bacteria.
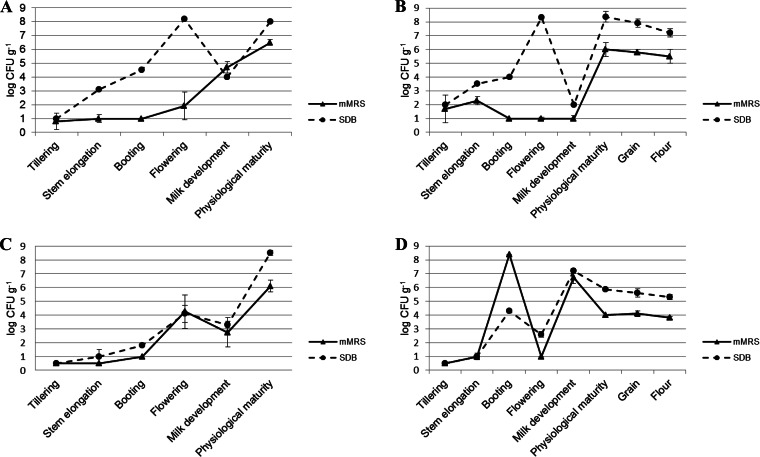
Selective media (mMRS and SDB) were used to detect endophytic presumptive lactic acid bacteria (Fig. 6). Lactic acid bacteria were detected in all plant organs of Odisseo and Saragolla durum wheat at different phenological stages, with cell densities varying from ca. 0.5 to 8.5 log CFU g−1. SDB was the medium showing the highest cell densities. Roots of Odisseo showed a higher cell density of presumptive lactic acid bacteria during tillering, stem elongation, and booting stages than did Saragolla. Roots showed the highest cell density of presumptive lactic acid bacteria during the flowering and physiological maturity stages of Odisseo and Saragolla durum wheat, respectively. Leaves and spikes showed the highest cell density during booting for Saragolla and flowering and physiological maturity for Odisseo.
FIG 6.
Cell numbers of endophytic presumptive lactic acid bacteria enumerated on mMRS and SDB in hypogeous (roots) and epigeous (leaves and spikes) organs of Odisseo (A and B, respectively) and Saragolla (C and D, respectively) durum wheat during the life cycle from tillering to flour.
Isolation and phenotypic identification of endophytic lactic acid bacteria.
Presumptive lactic acid bacteria were isolated (10 colonies for roots or leaves/spikes in each phenological stage) and tested for the ability to acidify the culture medium. All Gram-positive, catalase-negative, acidifying rods or cocci were identified by Biolog AN microplates. L. plantarum was identified in all plant organs at all different phenological stages. Other lactic acid bacteria (Lactobacillus helveticus, Lactobacillus coryniformis, Lactobacillus delbrueckii, L. sanfranciscensis, Weissella confusa, Weissella minor, Streptococcus anginosus, Leuconostoc citreum, and Pediococcus parvulus) were variously identified (data not shown). Within lactobacilli, L. plantarum dominated in roots and samples of leaves and spikes together with L. coryniformis until the flowering stage. L. plantarum, L. helveticus, and L. delbrueckii dominated in both roots and spikes during the milk development and physiological maturity stages of Odisseo and Saragolla wheat (data not shown).
Genotypic characterization of endophytic L. plantarum strains.
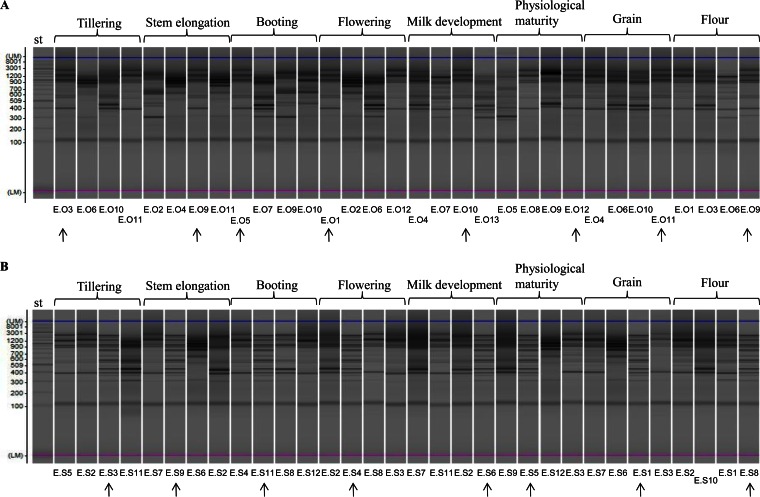
Since L. plantarum was the only lactic acid bacterial species identified at all phenological stages, RAPD-PCR analysis was applied to track the strains of this species. All isolates identified as L. plantarum were characterized by using single primer P4, P7, or M13. Several isolates of L. plantarum at different phenological stages of growth of Odisseo (Fig. 7A; see also Fig. S3A and S4A in the supplemental material) and Saragolla (Fig. 7B; see also Fig. S3B and S4B) wheat as well as in the processed wheat samples (grain and flour) (Fig. 7; see also Fig. S3 and S4) showed the same profile.
FIG 7.
Representative randomly amplified polymorphic DNA-PCR (RAPD-PCR) profiles of Lactobacillus plantarum isolated at the endophytic level from leaves and spikes, grain, or flour of Odisseo (A) and Saragolla (B) durum wheat. Primer M13 was used for RAPD-PCR analysis. A 2-log DNA ladder (0.1 to 10.0 kb) was used as a molecular size standard (st). Capillary electrophoretic profiles were singly acquired by MultiNA. Strains isolated from different phenological stages and processed wheat and showing similar RAPD-PCR profiles are indicated by arrows.
Correlation between lactic acid bacteria, environmental temperature, and wheat organ aw.
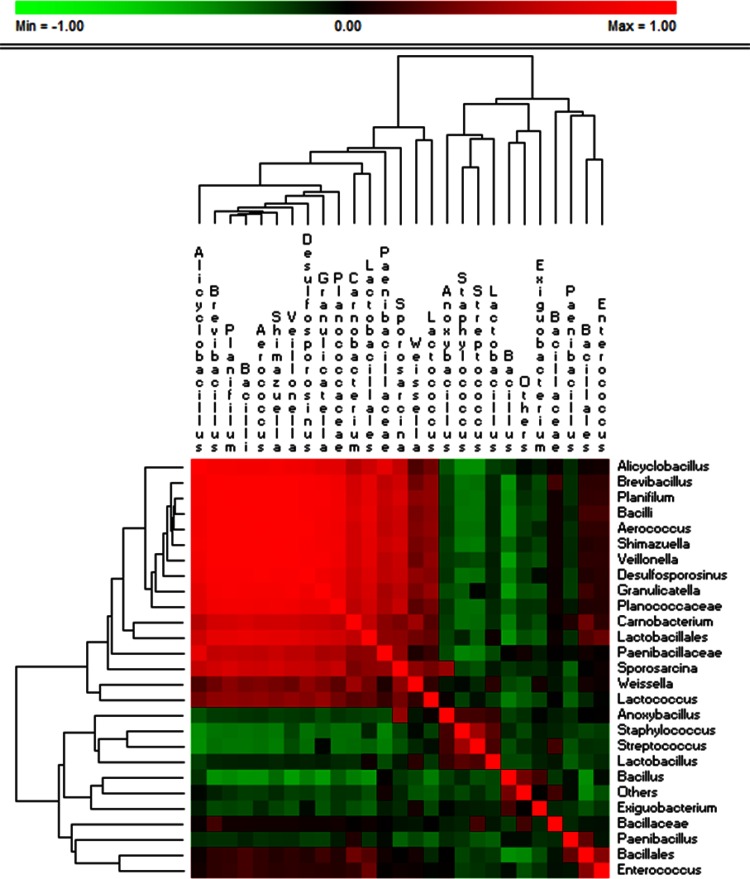
OTU correlation was investigated considering Firmicutes genus level (Fig. 8) taxonomic assignments and significant correlations at an FDR of <0.050. Lactobacillus showed positive correlation with Anoxybacillus, Staphylococcus, Streptococcus, and Weissella. Lactobacillus was negatively correlated mainly with Bacillus and Paenibacillus. Weissella, Lactococcus, and Enterococcus showed positive correlation with many genera, including other lactic acid bacteria (Aerococcus, Granulicatella, and Carnobacterium). Cell density of presumptive lactic acid bacteria, relative abundance of OTU belonging to lactic acid bacterial genera, environmental temperature, and wheat organ aw data were analyzed using PCA. The discrimination of samples between tillering, stem elongation, and booting and milk development, physiological maturity, grain, chaff, flour, and bran was evident (see Fig. S5 in the supplemental material). The cell density of presumptive lactic acid bacteria, Lactococcus and Enterococcus, was positively correlated (r > 0.5) with environmental temperature. Lactobacillus showed a higher positive correlation (r = 0.72) with aw than did Lactococcus and Enterococcus.
FIG 8.
Significant correlations between lactic acid bacterial OTUs (genus level) found on hypogeous organs (roots), epigeous organs (leaves and spikes), and processed wheat (grain, chaff, bran, and flour) of Odisseo and Saragolla. Hypogeous and epigeous organs were analyzed at the tillering, stem elongation, booting, flowering, milk development, and physiological maturity stages. The colors of the scale bar denote the nature of the correlation, with 1 indicating a perfectly positive correlation (red) and −1 indicating a perfectly negative correlation (green) between two microbial orders, families, or genera. Only significant correlations (FDR < 0.05) are shown.
DISCUSSION
This study used the 16S rRNA gene-based high-throughput sequencing approach targeting DNA to describe the ecology dynamics of Firmicutes and, especially, lactic acid bacteria during growth of durum wheat plants. Epiphytic and endophytic Firmicutes were analyzed to track the main source of contaminating lactic acid bacteria from plant to flour. Previously, lactic acid bacteria were found to be components of the epiphytic and/or endophytic microbiota of forage crops and roots of Lolium perenne (28–30). The biodiversity of Firmicutes was higher in roots than in leaves and spikes. At the contact zone with soil, plants host a distinctive root-associated bacterial microbiota, which is believed to have a positive role in plant nutrition and health (48). Diverse bacterial populations were described for the plant rhizosphere and phyllosphere. Beta-diversity indices showed that the composition of Firmicutes in durum wheat spikes was similar to that of processed grains. Bacillus, Exiguobacterium, Paenibacillus, Staphylococcus, Lactobacillus, Streptococcus, Enterococcus, and Lactococcus were the core genera of wheat plant and processed samples. All the above-mentioned Firmicutes were previously identified in wheat flour (16, 18, 49), strengthening the hypothesis that the wheat plant microbiota affected the microbial composition of the flour. Based on the results of this study, the spike or grain microbiota strongly contaminated the related flour. When flour is used as the base for producing sourdough, the most adapted microorganisms (mainly consisting of lactobacilli) are selected, leading to mature sourdough, whose microbiota greatly differs from that of flour (18). Dominant lactic acid bacteria and spore-forming bacteria in wheat flour could affect the quality of bread and other baked goods.
Within the core microbiota of wheat plant and processed samples, the relative abundance of each genus was affected by the plant organs, the cultivars, and the phenological stages. Previously, it was shown that microorganisms can randomly emerge from neighboring environmental ecosystems, but their survival is regulated by the plant (50). Overall, Bacillus was the most abundant genus in roots, especially at the epiphytic level. In this study, Bacillales were found as dominant OTUs in the Saragolla flour. Besides Bacillus, other Firmicutes commonly found in the rhizosphere, such as Paenibacillus and Exiguobacterium, promote plant growth (51). Exiguobacterium acetylicum, inoculated on wheat seeds, positively affected plant growth during the first phenological stages (52). In this study, Exiguobacterium was found as the dominant genus at the endophytic level of roots and leaves, at the stem elongation and booting stages, respectively, of Saragolla wheat.
Recently, a relatively high number and diversity of lactic acid bacteria was demonstrated in agricultural soils (53). Culture-dependent, denaturing gradient gel electrophoresis (DGGE), and 16S rRNA gene sequencing analyses revealed the following main genera: Lactococcus, Leuconostoc, Weissella, Lactobacillus, and Enterococcus (53, 54). Saragolla showed a lower relative abundance of OTUs belonging to Lactobacillus in roots, leaves, and spikes than did Odisseo durum wheat. Saragolla had the fastest increase of OTUs belonging to lactic acid bacteria (highest abundance for Streptococcus) in roots and, especially, at epiphytic levels (leaves and spikes) during booting. At the endophytic level (spikes), Saragolla showed the highest number of OTUs belonging to Lactobacillus during milk development. However, the relative abundance of Lactobacillus markedly decreased at physiological maturity. The influence of pesticides, farming practice, and atmosphere also on the composition of the microbial communities should not be excluded. Since the accelerated flowering stage of Saragolla, the environmental conditions differed between the phenological stages of the two cultivars. The lactic acid bacterial composition of spikes directly affected the bacterial community of processed wheat samples. As hypothesized, the OTUs found as endophytic bacteria of grains (Lactobacillus, Lactococcus, Enterococcus, and Streptococcus) were also found in the flour. L. plantarum was the only species identified in both cultivars at all the phenological stages. Commercial processed wheat grain (used as the control) harbored a high relative abundance of endophytic Lactococcus, which reflected on the related flour. Some strains of Lactococcus lactis showed significant biocontrol activity and/or direct plant growth promotion (55). Recently, it was shown that the inoculum of wheat roots with L. plantarum decreased the oxidative stress (56). Lactobacilli were shown to be effective against biotic stresses, due to their high antagonistic activity against plant pathogens (57). With the exception of L. plantarum, which is known to be a member of the sourdough microbiota, other lactic acid bacteria are present only as intermediate organisms in spontaneous sourdough until more characteristic microorganisms become predominant (16).
As shown by cultivation on mMRS and SDB, the cell density of presumptive lactic acid bacteria differed depending on the cultivar and/or the phenological stage. Saragolla durum wheat, which had an accelerated flowering stage, showed a faster increase of the cell density of presumptive lactic acid bacteria in leaves and spikes during booting than did Odisseo wheat. According to the 16S rRNA gene-based high-throughput sequencing, L. plantarum was the endophytic lactic acid bacterial species found in both cultivars at all the phenological stages, grain, and flour. This finding reinforced the concept that L. plantarum has a strong ecological or metabolic adaptability to different habitats (58, 59). Mesophilic lactobacilli (e.g., L. coryniformis) were mainly isolated in the first phenological stages. In contrast, thermophilic lactobacilli (L. helveticus and L. delbrueckii) were the highest in the milk development and physiological maturity stages.
Overall, the relative abundance of lactic acid bacterial genera was affected by other Firmicutes genera, environmental temperature, and aw of plant organs. Lactobacilli showed the highest positive correlation with aw. This finding could explain the strong decrease of OTU belonging to Lactobacillus during milk development (Odisseo) and physiological maturity (Saragolla). At these stages, Lactobacillus was mainly replaced by Streptococcus, Lactococcus, and Enterococcus. With the goal of decreasing chemical treatments (e.g., fertilizers, herbicides, and fungicides), microorganism-based commercial products were used to promote plant growth and to combat biotic disease (50, 60).
The results of this study highlight that the wheat microbiota differed from those of the grains and flour. Only a few microorganisms identified in wheat plants play a role in sourdough. Further experimental approaches, e.g., experiments involving competition of wheat isolates with sourdough isolates, are necessary to understand whether the L. plantarum strains from wheat grains dominate in mature sourdoughs.
Supplementary Material
ACKNOWLEDGMENT
This work was partially funded by the Istituto Agronomico Mediterraneo (IAM) Bari, project FOODING.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01852-15.
REFERENCES
- 1.Ikeda DM, Weinert E, Chang KCS, McGinn JM, Miller SA, Keliihoomalu C, DuPonte MW. August 2013. Natural farming: lactic acid bacteria. Sustain Agric 2013:1–4. [Google Scholar]
- 2.Arkhipova TN, Anokhina NL. 2009. Effects of wheat plant inoculation with cytokinin-producing microorganisms on plant growth at increasing level of mineral nutrition. Russ J Plant Physiol 56:814–819. doi: 10.1134/S1021443709060119. [DOI] [Google Scholar]
- 3.Zhao Y, Selvaraj JN, Xing F, Zhou L, Wang Y, Song H, Tan X, Sun L, Sangare L, Folly YM, Liu Y. 2014. Antagonistic action of Bacillus subtilis strain SG6 on Fusarium graminearum. PLoS One 9:e92486. doi: 10.1371/journal.pone.0092486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 5.De Vuyst L, Vrancken G, Ravyts F, Rimaux T, Weckx S. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol 26:666–675. doi: 10.1016/j.fm.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Gobbetti M, Rizzello CG, Di Cagno R, De Angelis M. 2014. How the sourdough may affect the functional features of leavened baked goods. Food Microbiol 37:30–40. doi: 10.1016/j.fm.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Valerio F, De Bellis P, Di Biase M, Lonigro SL, Giussani B, Visconti A, Lavermicocca P, Sisto A. 2012. Diversity of spore-forming bacteria and identification of Bacillus amyloliquefaciens as a species frequently associated with the ropy spoilage of bread. Int J Food Microbiol 156:278–285. doi: 10.1016/j.ijfoodmicro.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Gobbetti M. 1998. Interactions between lactic acid bacteria and yeasts in sourdoughs. Trends Food Sci Technol 9:267–274. doi: 10.1016/S0924-2244(98)00053-3. [DOI] [Google Scholar]
- 9.Vogel RF, Knorr R, Müller MRA, Steudel U, Gänzle MG, Ehrmann M. 1999. Non dairy lactic fermentations: the cereal world. Antonie Van Leeuwenhoek 76:403–411. doi: 10.1023/A:1002089515177. [DOI] [PubMed] [Google Scholar]
- 10.Gobbetti M, De Angelis M, Corsetti A, Di Cagno R. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci Technol 16:57–69. doi: 10.1016/j.tifs.2004.02.013. [DOI] [Google Scholar]
- 11.Gobbetti M, Corsetti A, Rossi J. 1994. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of amino acids. World J Microbiol Biotechnol 10:275–279. [DOI] [PubMed] [Google Scholar]
- 12.De Vuyst L, Schrijvers V, Paramithiotis S, Hoste B, Vancanneyt M, Swings J, Kalantzopoulos G, Tsakalidou E, Winy M. 2002. The biodiversity of lactic acid bacteria in Greek traditional wheat sourdoughs is reflected in both composition and metabolite formation. Appl Environ Microbiol 68:6059–6069. doi: 10.1128/AEM.68.12.6059-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:475–482. doi: 10.1128/AEM.69.1.475-482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheirlinck I, Van der Meulen R, Van Schoor A, Vancanneyt M, De Vuyst L, Vandamme P, Huys G. 2007. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl Environ Microbiol 73:6262–6269. doi: 10.1128/AEM.00894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minervini F, Di Cagno R, Lattanzi A, De Angelis M, Antonielli L, Cardinali G, Cappelle S, Gobbetti M. 2012. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: interactions between ingredients and microbial species diversity. Appl Environ Microbiol 78:1251–1264. doi: 10.1128/AEM.07721-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ercolini D, Pontonio E, De Filippis F, Minervini F, La Storia A, Gobbetti M, Di Cagno R. 2013. Microbial ecology dynamics during rye and wheat sourdough fermentation. Appl Environ Microbiol 79:7827–7836. doi: 10.1128/AEM.02955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheirlinck I, Van der Meulen R, De Vuyst L, Vandamme P, Huys G. 2009. Molecular source tracking of predominant lactic acid bacteria in traditional Belgian sourdoughs and their production environments. J Appl Microbiol 106:1081–1092. doi: 10.1111/j.1365-2672.2008.04094.x. [DOI] [PubMed] [Google Scholar]
- 18.Minervini F, Lattanzi A, De Angelis M, Celano G, Gobbetti M. 2015. House microbiotas as sources of lactic acid bacteria and yeasts in traditional Italian sourdoughs. Food Microbiol 52:66–76. doi: 10.1016/j.fm.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Van der Meulen R, Scheirlinck I, Van Schoor A, Huys G, Vancanneyt M, Vandamme P, De Vuyst L. 2007. Population dynamics and metabolite target analysis during laboratory fermentations of wheat and spelt sourdoughs. Appl Environ Microbiol 73:4741–4750. doi: 10.1128/AEM.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheirlinck I, Van der Meulen R, Van Schoor A, Vancanneyt M, De Vuyst L, Vandamme P, Huys G. 2008. Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems as assessed by culture and population fingerprinting. Appl Environ Microbiol 74:2414–2423. doi: 10.1128/AEM.02771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weckx S, Van der Meulen R, Allemeersch J, Huys G, Vandamme P, Van Hummelen P, De Vuyst L. 2010. Community dynamics of bacteria in sourdough fermentations as revealed by their metatranscriptome. Appl Environ Microbiol 76:5402–5408. doi: 10.1128/AEM.00570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weckx S, Van der Meulen R, Maes D, Scheirlinck I, Huys G, Vandamme P, De Vuyst L. 2010. Lactic acid bacteria community dynamics and metabolite production of rye sourdough fermentations share characteristics of wheat and spelt sourdough fermentations. Food Microbiol 27:1000–1008. doi: 10.1016/j.fm.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Su MSW, Oh PL, Walter J, Gänzle MG. 2012. Intestinal origin of sourdough Lactobacillus reuteri isolates as revealed by phylogenetic, genetic, and physiological analysis. Appl Environ Microbiol 78:6777–6780. doi: 10.1128/AEM.01678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Toit M, Dicks LMT, Holzapfel WH. 2003. Identification of heterofermentative lactobacilli isolated from pig faeces by numerical analysis of total soluble cell protein and RAPD patterns. Lett Appl Microbiol 37:12–16. doi: 10.1046/j.1472-765X.2003.01334.x. [DOI] [PubMed] [Google Scholar]
- 25.De Angelis M, Siragusa S, Berloco M, Caputo L, Settanni L, Alfonsi G, Amerio M, Grandi A, Ragni A, Gobbetti M. 2006. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res Microbiol 157:792–801. doi: 10.1016/j.resmic.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Groenewald WH, Van Reenen CA, Todorov SD, Du Toit M, Corli Witthuhn RC, Holzapfel WH, Dicks LMT. 2006. Identification of lactic acid bacteria from vinegar flies based on phenotypic and genotypic characteristics. Am J Enol Vit 57:519–525. [Google Scholar]
- 27.Alfonzo A, Ventimiglia G, Corona O, Di Gerlando R, Gaglio R, Francesca N, Moschetti G, Settanni L. 2013. Diversity and technological potential of lactic acid bacteria of wheat flours. Food Microbiol 36:343–354. doi: 10.1016/j.fm.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JG, Cai Y, Kobayashi R, Kumai S. 2000. Characteristics of lactic acid bacteria isolated from forage crops and their effects on silage fermentation. J Sci Food Agric 80:1455–1460. doi:. [DOI] [Google Scholar]
- 29.Pang H, Tan Z, Qin G, Wang Y, Li Z, Jin Q, Cai Y. 2012. Phenotypic and phylogenetic analysis of lactic acid bacteria isolated from forage crops and grasses in the Tibetan Plateau. J Microbiol 50:63–71. doi: 10.1007/s12275-012-1284-5. [DOI] [PubMed] [Google Scholar]
- 30.Gaggìa F, Baffoni L, Di Gioia D, Accorsi M, Bosi S, Marotti I, Biavati B, Dinelli G. 2013. Inoculation with microorganisms of Lolium perenne L.: evaluation of plant growth parameters and endophytic colonization of roots. N Biotechnol 30:695–704. doi: 10.1016/j.nbt.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Herbert S, Bouchet B, Riaublanc A, Dufour E, Gallant DJ. 1999. Multiple fluorescence labelling of proteins, lipids and whey in dairy products using confocal microscopy. Lait 79:567–575. doi: 10.1051/lait:1999646. [DOI] [Google Scholar]
- 32.Kinkel LL, Wilson M, Lindow SE. 1996. Utility of microcosm studies for predicting phylloplane bacterium population sizes in the field. Appl Environ Microbiol 62:3413–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suda W, Nagasaki A, Shishido M. 2009. Powdery mildew-infection changes bacterial community composition in the phyllosphere. Microbes Environ 24:217–223. doi: 10.1264/jsme2.ME09114. [DOI] [PubMed] [Google Scholar]
- 34.Stock JD, Hirano SS. 1991. Location of phyllosphere bacteria on or within leaves of field-grown snap bean plants. Phytopathology 81:1222. [Google Scholar]
- 35.Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A. 2004. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl Environ Microbiol 70:2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minervini F, De Angelis M, Di Cagno R, Pinto D, Siragusa S, Rizzello CG, Gobbetti M. 2010. Robustness of Lactobacillus plantarum starters during daily propagation of wheat flour sourdough type I. Food Microbiol 27:897–908. [DOI] [PubMed] [Google Scholar]
- 37.Muḧling M, Woolven-Allen J, Murrell JC, Joint I. 2008. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J 2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 40.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 41.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreotti R, Perez de Leon AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol 11:6. doi: 10.1186/1471-2180-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kline L, Sugihara TF. 1971. Microorganisms of the San Francisco sour dough bread process. II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl Microbiol 21:459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed W, Sawant S, Huygens F, Goonetilleke A, Gardner T. 2009. Prevalence and occurrence of zoonotic bacterial pathogens in surface waters determined by quantitative PCR. Water Res 43:4918–4928. doi: 10.1016/j.watres.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 45.De Angelis M, Corsetti A, Tosti N, Rossi J, Corbo MR, Gobbetti M. 2001. Characterization of non-starter lactic acid bacteria from Italian ewe cheeses based on phenotypic, genotypic and cell wall protein analyses. Appl Environ Microbiol 67:2011–2020. doi: 10.1128/AEM.67.5.2011-2020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siragusa S, Di Cagno R, Ercolini D, Minervini F, Gobbetti M, De Angelis M. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl Environ Microbiol 75:1099–1109. doi: 10.1128/AEM.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suchodolski JS, Dowd SE, Wilke V, Steiner JM, Jergens AE. 2012. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One 7:e39333. doi: 10.1371/journal.pone.0039333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlaeppi K, Dombrowski N, Oter RG, van Themaat EVL, Schulze-Lefert P. 2014. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A 111:585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rizzello CG, Cavoski I, Turk J, Ercolini D, Nionelli L, Pontonio E, De Angelis M, De Filippis F, Gobbetti M, Di Cagno R. 2015. Organic cultivation of Triticum turgidum subsp. durum is reflected in the flour-sourdough fermentation-bread axis. Appl Environ Microbiol 81:3192–3204. doi: 10.1128/AEM.04161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berlec A. 2012. Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci 193–194:96–102. [DOI] [PubMed] [Google Scholar]
- 51.Vessey JK. 2003. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255:571–586. doi: 10.1023/A:1026037216893. [DOI] [Google Scholar]
- 52.Selvakumar G, Joshi P, Nazim S, Mishra PK, Kundu S, Gupta HS. 2009. Exiguobacterium acetylicum strain 1P (MTCC 8707) a novel bacterial antagonist from the North Western Indian Himalayas. World J Microbiol Biotechnol 25:131–137. doi: 10.1007/s11274-008-9874-4. [DOI] [Google Scholar]
- 53.Somers E, Amke A, Croonenborghs A, van Overbeek LS, Vanderleyden J. 2007. Lactic acid bacteria in organic agricultural soils, abstr 367827. Abstr Plant Res Int, Montpellier, France. [Google Scholar]
- 54.Yang L, Deng Y, Zhang H, Diao Q. 2012. Isolation and characterization of an Enterococcus strain from Tibetan alpine meadow soil. Wei Sheng Wu Xue Bao 52:1421–1426. [PubMed] [Google Scholar]
- 55.Shrestha A, Kim EC, Lim CK, Cho S, Hur JH, Park DH. 2009. Biological control of soft rot on chinese cabbage using beneficial bacterial agents in greenhouse and field. Korean J Pestic Sci 13:325–331. [Google Scholar]
- 56.Yarullina DR, Asafova EV, Kartunova JE, Ziyatdinova GK, Ilinskaya ON. 2014. Probiotics for plants: NO-producing lactobacilli protect plants from drought. Appl Biochem Microbiol 50:166–168. doi: 10.1134/S0003683814020197. [DOI] [PubMed] [Google Scholar]
- 57.Stoyanova LG, Ustyugova EA, Netrusov AI. 2012. Antibacterial metabolites of lactic acid bacteria: their diversity and properties. Appl Biochem Microbiol 48:229–243. doi: 10.1134/S0003683812030143. [DOI] [PubMed] [Google Scholar]
- 58.Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, van Hylckama Vlieg JE. 2010. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12:758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- 59.de Melo Pereira GV, Magalhàes KT, Lorenzetti ER, Souza TP, Schwan RF. 2012. A multiphasic approach for the identification of endophytic bacterial [sic] in strawberry fruit and their potential for plant growth promotion. Microb Ecol 63:405–417. doi: 10.1007/s00248-011-9919-3. [DOI] [PubMed] [Google Scholar]
- 60.Wani ZA, Ashraf N, Mohiuddin T, Riyaz-Ul-Hassan S. 2015. Plant-endophyte symbiosis, an ecological perspective. Appl Microbiol Biotechnol 99:2955–2965. doi: 10.1007/s00253-015-6487-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.