Abstract
Azospirillum is a rhizobacterial genus containing plant growth-promoting species associated with different crops worldwide. Azospirillum brasilense strains exhibit a growth-promoting effect by means of phytohormone production and possibly by N2 fixation. However, one of the most important factors for achieving an increase in crop yield by plant growth-promoting rhizobacteria is the survival of the inoculant in the rhizosphere, which is not always achieved. The objective of this study was to develop quantitative PCR protocols for the strain-specific quantification of A. brasilense FP2. A novel approach was applied to identify strain-specific DNA sequences based on a comparison of the genomic sequences within the same species. The draft genome sequences of A. brasilense FP2 and Sp245 were aligned, and FP2-specific regions were filtered and checked for other possible matches in public databases. Strain-specific regions were then selected to design and evaluate strain-specific primer pairs. The primer pairs AzoR2.1, AzoR2.2, AzoR5.1, AzoR5.2, and AzoR5.3 were specific for the A. brasilense FP2 strain. These primer pairs were used to monitor quantitatively the population of A. brasilense in wheat roots under sterile and nonsterile growth conditions. In addition, coinoculations with other plant growth-promoting bacteria in wheat were performed under nonsterile conditions. The results showed that A. brasilense FP2 inoculated into wheat roots is highly competitive and achieves high cell numbers (∼107 CFU/g [fresh weight] of root) in the rhizosphere even under nonsterile conditions and when coinoculated with other rhizobacteria, maintaining the population at rather stable levels for at least up to 13 days after inoculation. The strategy used here can be applied to other organisms whose genome sequences are available.
INTRODUCTION
Azospirillum is one of the most important genera of plant growth-promoting rhizobacteria found worldwide under a variety of environmental and soil conditions (1). The diazotroph Azospirillum brasilense is the best-studied species of the genus, is found in close association with many agriculturally important crops, and exerts beneficial effects on plant growth and productivity (2–4). Nitrogen fixation (5, 6) and the production of the auxin 3-indoleacetic acid (IAA) by many representatives of the genus Azospirillum are related to the growth promotion effects observed in inoculated plants, such as increases in root length and the numbers of root hairs and lateral roots (3).
The biotechnological use of A. brasilense inoculants in Latin American and in Brazil, in particular, has increased in recent years (7). Strain FP2 is a spontaneous mutant of A. brasilense Sp7 (8). Strain Sp7 has been shown to be capable of stimulating the growth of several members of the family Poaceae and increasing the productivities of wheat and maize crops (2). Strain FP2 can also promote the growth of wheat (9) and enhance maize and wheat productivity under field conditions (unpublished data). Most of the A. brasilense inoculants in Brazil contain strains Ab-V5 and Ab-V6, which are also derivatives of strain Sp7. Ab-V5 and Ab-V6 were shown to increase the productivity of maize and wheat under field conditions (10) and were officially authorized for use as inoculants in these crops (10).
However, a major problem related to A. brasilense inoculants is the survival of the inoculated strains in the rhizosphere soil (11, 12), which affects inoculant performance, since the effective colonization of roots is necessary for the successful stimulation of plant growth by Azospirillum (13).
To assess the diversity and taxonomy of crop plant-associated bacteria, many cultivation-dependent and -independent methods are currently in use (14–16). However, most of these methods are not quantitative and are based on the evaluation of the 16S rRNA gene coding sequences. They are able to provide highly confident results only at the genus and species levels and are not specific enough to study the bacterial population dynamics at the strain level, which is necessary for inoculant monitoring. Thus, in many cases, it is not possible to quantitatively associate the failure or success of plant growth promotion achieved with the inoculated bacterial population at a strain-specific resolution, leaving the outcome of the inoculation unexplained (17). Furthermore, the crop response to inoculation under field conditions heavily depends on the combination of the plant genotype and the bacterial strain (18–20), stressing the need for methodologies to evaluate the success of plant colonization accurately at a high resolution. Previously, we used whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis to differentiate species of Azospirillum, including several closely related A. brasilense strains (21). However, this method is not quantitative, requires growth on a culture medium, and is time and labor-intensive.
Quantitative PCR (qPCR) has been the method of choice to quantify rhizosphere populations because it allows high specificity, sensitivity, and speed (17, 22, 23). This technique has successfully been used to quantify several bacteria associated with plants. It was successfully used for the quantification of a functionally specific subgroup of pseudomonads in the rhizosphere (24). The pathogen Xylella fastidiosa was quantified in citrus plants (25), while the endophytic bacterium Methylobacterium mesophilicum was monitored by qPCR during Catharanthus roseus colonization (26). In Brassica oleracea, the plant growth-promoting Enterobacter radicincitans population was monitored by qPCR in association with fluorescence in situ hybridization (FISH) (27), with not only the amount of bacteria in the colonized plants but also their location in the plants being determined. Although these reports showed that qPCR is a valuable technique to quantitatively monitor populations of unlabeled bacteria in greenhouse experiments, none has used strain-specific primers. The application of strain-specific primers is difficult in field experiments, where closely related indigenous bacteria may interfere with the amplification and quantification. For strain-specific molecular monitoring, sequence-characterized amplified region (SCAR) markers obtained from BOX-PCR, enterobacterial repetitive intergenic consensus sequence-PCR, and randomly amplified polymorphic DNA (RAPD)-PCR fragments were recently applied to design primers for the qPCR quantification of A. brasilense and Azospirillum lipoferum at the strain-specific level (17, 22).
The objective of this study was to develop qPCR protocols for the strain-specific quantification of the plant growth-promoting bacterium A. brasilense FP2 on the basis of a comparison of its whole-genome sequence (WGS) with that of the closely related strain Sp245. The designed strain-specific primers were then applied for quantification to monitor the FP2 population in inoculated wheat plants under sterile and nonsterile conditions.
MATERIALS AND METHODS
Bacterial strains.
All Azospirillum strains (Table 1) were routinely grown in NFbHPN medium (28) at 30°C under aeration with shaking at 120 rpm. Strains from other genera were grown in DYGS medium (29) containing, per 1,000 ml, 0.10% glucose, 0.20% yeast extract, 0.15% peptone, 0.50% MgSO4·7H2O, and 0.15% l-glutamic acid at pH 6.0 to 6.5; the cultures were incubated at 30°C under aeration with shaking at 120 rpm. Colony counts of all strains were performed after dilutions were spread on the respective medium plates and incubated for 72 h at 30°C.
TABLE 1.
Bacterial strains used in this study
| Microorganism | Reference or source |
|---|---|
| Azospirillum amazonense DSM 2787 | Helmholtz Zentrum München strain collection |
| Azospirillum brasilense FP2 | 8 |
| Azospirillum brasilense NH | Helmholtz Zentrum München strain collection |
| Azospirillum brasilense Sp245 | Helmholtz Zentrum München strain collection |
| Azospirillum brasilense Sp7 | Helmholtz Zentrum München strain collection |
| Azospirillum canadense LMG 23617 | Helmholtz Zentrum München strain collection |
| Azospirillum irakense DSM 11586a | Helmholtz Zentrum München strain collection |
| Azospirillum lipoferum DSM 1691 | Helmholtz Zentrum München strain collection |
| Azospirillum rugosum DSM 19657 | Helmholtz Zentrum München strain collection |
| Burkholderia brasiliensis M171 | Helmholtz Zentrum München strain collection |
| Burkholderia tropica PPe5 | Helmholtz Zentrum München strain collection |
| Gluconacetobacter diazotrophicus DSM 5601 | Helmholtz Zentrum München strain collection |
| Roseomonas genomospecies 6 CCUG 33010 | Helmholtz Zentrum München strain collection |
| Roseomonas fauriae KACC 11694 | Helmholtz Zentrum München strain collection |
Primer design.
To design Azospirillum brasilense FP2 strain-specific primer pairs, the following general strategy was used: (i) the WGS of A. brasilense FP2 from the FASTA genome sequence was fragmented in silico using in-house scripts, producing 500-bp nonoverlapping fragments; (ii) the genome sequence of A. brasilense Sp245 was used to build a local BLAST database, and A. brasilense FP2 sequence fragments were used as queries for a BLASTn similarity search with default parameters; (iii) fragments for which no hits were found were subjected to a second BLASTn (30) search against the NCBI NT database (performed in July 2012; GenBank release 190), using default parameters; and (iv) putative strain-specific sequences, i.e., sequences without any match in the two BLAST sequence analyses, were used to design sets of primer pairs specific for A. brasilense FP2. In order to inspect the selected regions, the draft genome sequence of A. brasilense FP2 was annotated and visually analyzed using the RAST program, version 2.0 (31, 32), and the Unipro UGENE tool kit, version 1.14 (33).
The WGS of Azospirillum brasilense FP2 is publicly available in the NCBI database under accession number APHV00000000 and assembly GCA_000404045.1. Its total sequence length is 6,885,108 bp, it has 413 contigs (N50, 29,432 bp), it has a GC content of 68.1%, and it has a genome coverage of 25 times. The WGS of Azospirillum brasilense Sp245 is available in the NCBI database under accession numbers HE577327 to HE577333 (1 chromosome and 6 plasmids) and assembly GCA_000237365.1. Its total sequence length is 7,530,241 bp (total assembly gap length, 6,000 bp), it has 67 contigs (N50, 186,382 bp), and it has a GC content of 68.6%.
Primer design was performed, using Primer Express software (version 3.0; Applied Biosystems, Foster City, CA), on the basis of (i) an amplicon size inferior to 200 bp and primer lengths ranging from 18 to 22 bp; (ii) a high melting temperature (Tm) for the primers (Tm, approximately 60°C) and a low Tm difference (ΔTm) between primers (ΔTm, <2°C); and (iii) a lack of predicted hairpin loops, duplexes, and primer-dimer formation.
Primer selection and evaluation.
The designed primer pairs were synthesized by Eurofins (Ebersberg, Germany) and qualitatively analyzed by conventional PCR with about 30 ng of genomic DNA, 10 pmol of each primer, 1 U of Taq DNA polymerase (Taq Dream Invitrogen Inc.), Taq DNA polymerase buffer, 200 mmol/μl of desoxyribonucleotide, and sterile ultrapure water to a final volume of 10 μl. The cycling program included a 10-min initial denaturation, incubation at 95°C, 25 cycles consisting of denaturation at 95°C for 15 s and annealing at 60°C for 60 s followed by 72°C for 30 s, and a final elongation of 10 min at 70°C. A primer pair was considered strain specific if (i) successful amplification occurred using the DNA of the target strain as the template; (ii) cross-amplification with nontarget strains was absent; and (iii) amplification in the control tube reaction, to which no DNA was added, was absent. Genomic DNAs from 14 strains of 10 species and 4 genera (Table 1) were used as the templates for the PCRs. A second step was performed under quantitative PCR conditions to check the primer specificity (by the use of melting curves) and amplification efficiency, as described below.
Quantitative PCR conditions.
qPCR was performed in a total reaction volume of 25 μl containing 12.5 μl Power SYBR green PCR master mix (Applied Biosystems), 6.25 μl of a primer mix (final concentration, 1 μmol), and 6.25 μl of 2.5 ng/μl diluted template DNA. A MicroAmp optical 96-well reaction plate (Applied Biosystems) and an ABI Prism 7500 system (Applied Biosystems) were used. The cycling program included a 10-min incubation at 95°C, 40 cycles consisting of 95°C for 15 s and 60°C for 60 s followed by 72°C for 30 s, and an additional incubation at 72°C for 10 min. Amplification specificity was verified by melting curve analysis of the PCR products, performed using the ABI Prism 7500 system sequence detection software (version 1.2.3; Applied Biosystems).
Primer efficiency determination.
Genomic DNA from A. brasilense FP2 was used to prepare 10-fold dilution series (in triplicate). Sterile water was used as a negative control. The cycle threshold (CT) value was automatically determined for each sample by the ABI Prism 7500 system sequence detection software (version 1.2.3; Applied Biosystems). A standard curve was generated by plotting the CT value against the logarithm of the bacterial DNA concentration (data not shown) and used to calculate the amplification efficiency (E) (Table 2).
TABLE 2.
Primer characteristics and parameters evaluated by qPCR
| Primer pair | Orientationa | Sequence | Length (mer) | GC content (%) | R2 | Slope | Efficiencyb |
|
|---|---|---|---|---|---|---|---|---|
| E | % E | |||||||
| 16S rRNA genec | F | TCGCTAGTAATCGCGGATCA | 20 | 50 | 0.9995 | 3.3 | 2.01 | 101.3 |
| R | TGTGACGGGCGGTGTGTA | 18 | 61 | |||||
| Azo-2 | F | GCGCGGGAAGTCCTGAAT | 18 | 61 | 0.9934 | 3.4 | 1.97 | 96.8 |
| R | CCCTTCACCATCCAGTCGAT | 20 | 55 | |||||
| AzoR2.1 | F | CGCCACCATGCGATCAA | 17 | 59 | 0.9980 | 3.3 | 2.01 | 101.3 |
| R | GCATGCCCAGTACTGCAAGTC | 21 | 57 | |||||
| AzoR2.2 | F | CCTTCACCTGGACGGTTCAG | 20 | 60 | 0.9982 | 3.5 | 1.94 | 94.0 |
| R | CGCGGCCAGCAGACTT | 16 | 69 | |||||
| AzoR5.1 | F | GATCACTGGACTCGGCTGTCA | 21 | 57 | 0.9977 | 3.7 | 1.88 | 87.6 |
| R | ATCGACCGTTCTCAGCGTCTA | 21 | 52 | |||||
| AzoR5.2 | F | TCACTGGACTCGGCTGTCAA | 20 | 55 | 0.9996 | 3.6 | 1.89 | 88.8 |
| R | ATATCGACCGTTCTCAGCGTCTA | 23 | 48 | |||||
| AzoR5.3 | F | AATTCTTTCCGTTGGCTTTCAA | 22 | 36 | 0.9995 | 3.4 | 1.97 | 96.8 |
| R | GCTTGCCGACCGGAGTATC | 19 | 63 | |||||
F, forward primer; R, reverse primer.
Efficiency (E) was calculated using the equation 10−1/slope − 1, and percent efficiency was calculated from the equation (E − 1) × 100.
The forward primer binds the region from 1,267 to 1,286 bp and the reverse primer binds the region from 1,319 to 1,336 bp of the 16S rRNA gene sequence of Azospirillum brasilense Sp7 (GenBank accession number X79739).
Generation of standard curves for qPCR quantification of A. brasilense FP2 in wheat roots.
The standard curves used for the quantification of A. brasilense FP2 in wheat were constructed as described previously (22), with the following modifications. Wheat plants were grown under axenic condition as described below for 7 days, and roots were collected and crushed in liquid nitrogen using a mortar and pestle. A volume of 100 μl of an A. brasilense FP2 culture (dilution range, 102 to 109 CFU) was added to 100 mg of crushed roots, and the components were mixed and incubated for 1 h at room temperature. The whole mixture was used for DNA extraction with a FastDNA spin kit (MP Biomedicals, USA) according to the manufacturer's instructions; qPCR was performed as described above. The standard curve was generated by plotting the CT value versus the number of CFU added to each tube. No bacteria were added to the negative control.
DNA preparation.
Genomic DNA was extracted from the bacterial cultures and wheat roots using a FastDNA spin kit (MP Biomedicals, USA) according to the manufacturer's instructions. DNA concentrations were assessed by measurement of the optical density at 260 nm with a NanoDrop device (NanoDrop Technologies, Wilmington, DE, USA).
qPCR quantification of Azospirillum brasilense FP2 on wheat roots.
For the sterile experiments, seeds of wheat (Triticum aestivum cv. Schöndorfer) were surface sterilized using a protocol described previously (25). Afterward, the seeds were germinated on nutrient agar plates (Analytical Fluka) for 3 days, transferred to glass tubes containing 16 ml of Hoagland solution and quartz beads with a diameter of approximately 3 mm, and then incubated in a greenhouse with a 14-h light/10-h dark cycle at 23°C and a humidity above 50%.
For the experiments performed under nonsterile conditions, seeds were germinated as described above but without surface sterilization in commercial gardener soil (type ED-73; Bayerische Gärtnereigenossenschaft), suspended in Hoagland medium at a final concentration of 1% (wt/vol), and filtered, and this suspension was used as the inoculum in glass tubes containing quartz beads. The negative control consisted of noninoculated plants. Different experiments were conducted to evaluate plants inoculated with A. brasilense FP2 or coinoculated in the presence of other wheat-associated diazotrophs (in the same amount), namely, A. brasilense NH, Herbaspirillum seropedicae Z67, Gluconacetobacter diazotrophicus DSM 5601, and A. lipoferum DSM 1691. The control consisted of A. brasilense FP2-inoculated plants. All microorganisms were grown until the count was about 109 CFU/ml, and the cells were washed once with 1× phosphate-buffered saline buffer (Applichem, Denmark). In all experiments, approximately 107 CFU/plant was inoculated in the plant growth medium and incubated for 14 days. The experiments were performed in biological and technical triplicate, and samples were collected every 2 days.
Determination of number of CFU.
To determine the number of CFU, the roots were crushed using a mortar, serially diluted (10−1 to 10−7) in saline (0.9% NaCl), and plated on NFbHPN medium, and the colonies were counted.
Experimental design and statistical analysis.
The experiments in a growth chamber followed a randomized block design. Colony counts were expressed as the number of CFU per gram (fresh weight) of root, and qPCR quantification data were converted to the equivalent number of CFU per gram (fresh weight) of root. The data were subjected to the Student t test (to compare means of two treatments) or to analysis of variance (to compare many treatments), with means compared by the Tukey test, using the SAEG program (version 8.0; Sistema para Análise Estatísticas, Universidade Federal de Viçosa, Viçosa, Brazil).
RESULTS
Primer design and evaluation of amplification efficiency.
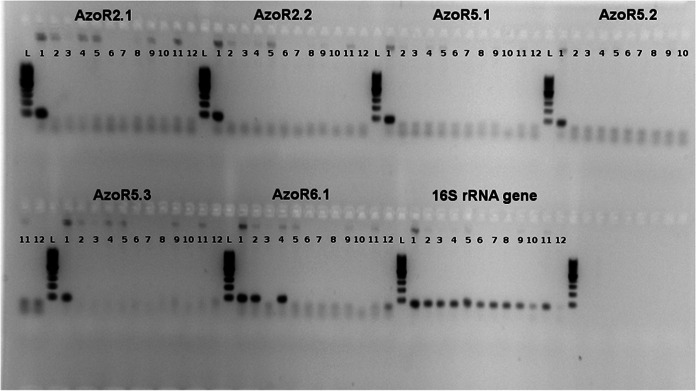
For strain-specific primer design, strain-specific genomic regions were selected after the whole-genome sequences (WGSs) of Azospirillum brasilense FP2 and A. brasilense Sp245, the strain closest to strain FP2 for which a genome sequence is available so far, were compared using the procedures detailed in the Materials and Methods section. The genome sequence comparison was based on BLAST analysis of 500-bp sequence fragments of A. brasilense FP2 against the genome sequence of A. brasilense Sp245 in a local database in the first round and against the sequence in the NCBI NT database in the second round. Although this analysis is database dependent and does not guarantee the selection of strain-specific genomic regions, in practice, comparison of the genomes of two very closely related strains (i.e., strains with very high genomic synteny) allows the selection of genomic regions whose sequences are not likely to match the sequence of a more distantly related organism, as shown by the results of a search of the sequences in a comprehensive database by BLAST analysis. Sequences for which no hits were found in a BLAST analysis against the Sp245 genome sequence also did not show significant hits against the sequences in the NCBI NT database. Using this methodology, six coding and intergenic regions from the A. brasilense FP2 genome were selected, and a total of 10 primer pairs were designed and tested for cross amplification against 13 different bacterial DNAs, including DNAs from four A. brasilense strains, six other Azospirillum spp., and two Roseomonas species strains (Fig. 1 shows the most relevant primer pairs).
FIG 1.
Specificities of the primer pairs designed to amplify Azospirillum brasilense FP2. Lanes: L, DNA ladder; 1, A. brasilense FP2; 2, A. brasilense NH; 3, A. brasilense Sp7; 4, A. brasilense Sp245; 5, A. lipoferum DSM 1691; 6, Azospirillum rugosum DSM 19657; 7, Azospirillum canadense LMG 23617; 8, Azospirillum amazonense DSM 2787; 9, Azospirillum irakense DSM 1158a; 10, Roseomonas genomospecies 6 CCUG 33010; 11, Roseomonas fauriae KACC 11694; 12, negative control (no template DNA). The primer pair specific for the 16S rRNA-encoding gene was used as a positive amplification control. Primer pairs AzoR2.1, AzoR2.2, AzoR5.1, AzoR5.2, and AzoR5.3 produced amplicons only when A. brasilense FP2 DNA was used as the template and were considered strain-specific primer pairs; primer pair AzoR6.1 produced cross-species amplicons and was not able to amplify all A. brasilense strains tested (i.e., no amplification for strain Sp7 was observed) and so was discarded from further analyses.
Five out of 10 primer pairs were specific for A. brasilense FP2, namely, AzoR2.1, AzoR2.2, AzoR5.1, AzoR5.2, and AzoR5.3 (Table 2). For one of the primer pairs, Azo-2, amplicons were generated for all four strains of Azospirillum brasilense tested (FP2, NH, Sp245, and Sp7), but no amplification was observed for Roseomonas genomospecies 6 CCUG 33010, Roseomonas fauriae KACC 11694, Burkholderia tropica Ppe5, or Burkholderia brasilense M171 (data not shown).
The genome sequences from strains FP2 and Sp245 of A. brasilense share a high degree of synteny; however, strain-specific primer pairs were designed from two FP2 contig sequences that did not align along the chromosome or any plasmid sequences from strain Sp245 (see Fig. S1A in the supplemental material). On the contrary, primer pair Azo-2 was designed from a contig sequence of FP2 that aligns to the Sp245 chromosome sequence (see Fig. S1B in the supplemental material), although the alignment in the region of primer binding did not show a perfect match (data not shown). Automatic annotation of the A. brasilense FP2 draft genome sequence predicted that the amplicon from the Azo-2 primer pair is located at the end of a coding sequence (CDS) for a hypothetical protein. On the other hand, amplicons from strain-specific primer pairs were predicted to be located in a noncoding region (AzoR2.1 and AzoR2.2) or fall into a CDS for the TniQ domain-containing protein (AzoR5.1, AzoR5.2, and AzoR5.3; see Fig. S2 in the supplemental material). Interestingly, the regions surrounding amplicons from strain-specific primer pairs contained some CDSs related to phages and mobile elements.
The efficiency of all strain-specific primer pairs obtained in this study was tested by constructing a standard curve with increasing concentrations of A. brasilense FP2 DNA (Table 2). The primer pairs AzoR5.1 and AzoR5.2 were discarded from further analysis because they had the lowest efficiency rate, and primer pairs AzoR2.1, AzoR2.2, and AzoR5.3 were used to quantify A. brasilense FP2.
qPCR quantification of Azospirillum brasilense FP2 on wheat roots.
In order to test the ability of the strain-specific primer pairs to quantify A. brasilense FP2 in the rhizosphere, a growth chamber experiment was conducted with wheat plants inoculated with A. brasilense FP2 under sterile and nonsterile conditions.
To monitor the population of A. brasilense FP2 in wheat roots, three strain-specific primer pairs with the highest amplification efficiency (AzoR2.1, AzoR2.2, and AzoR5.3) were selected. The primer pair Azo-2 was used to quantify the total A. brasilense population, and a universal 16S rRNA gene-targeted primer pair (Doumit Camilios Neto, personal communication) was used for the quantification of all bacteria present (Table 2).
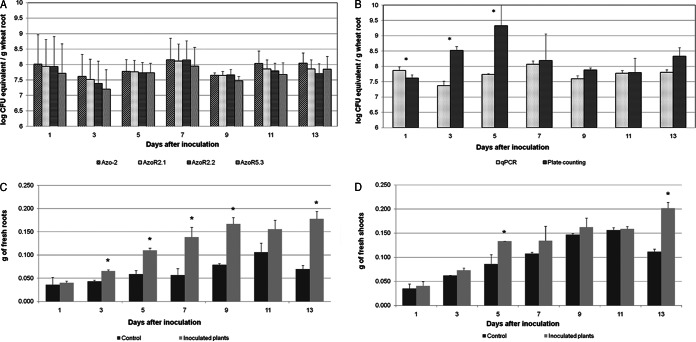
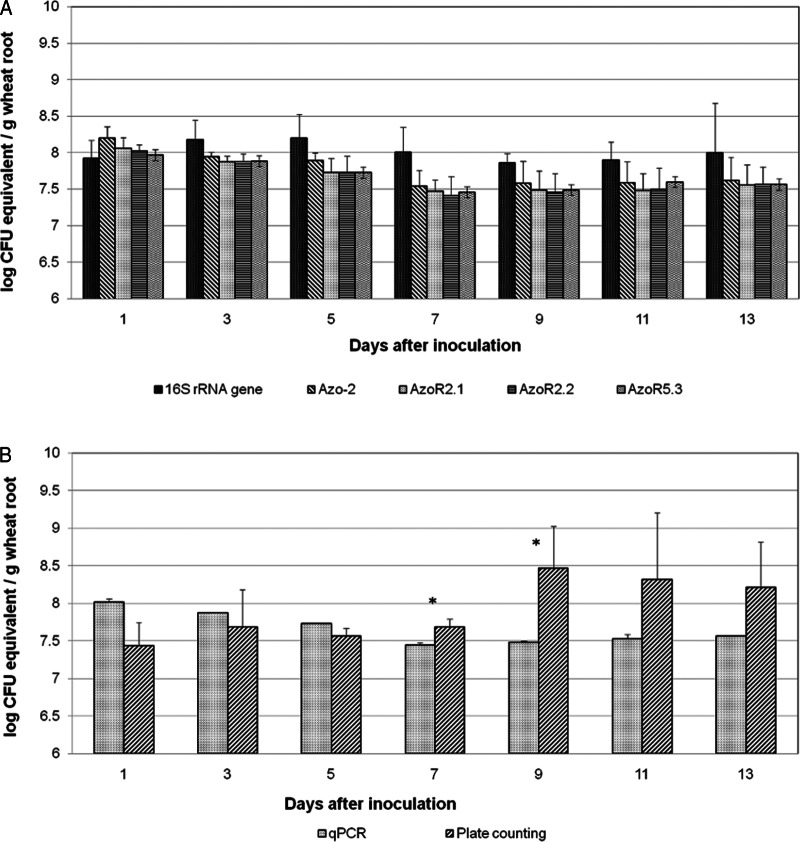
Initially, a standard curve was constructed from a fixed amount of crushed plant root tissues mixed with each sample of serially diluted total DNA of A. brasilense FP2 (see Materials and Methods). The inclusion of plant material during the construction of the standard curve was based on the observation of Couillerot et al. (17) that the presence of root extract decreases the detection limit for the quantification of A. lipoferum CRT1 on maize. The inclusion of root extract allowed conditions including the presence of plant background DNA to be integrated into the technical sensitivity limit of the final standard curve, thereby making the quantification closer to reality. The equation for the qPCR quantification standard curve was used to estimate the amount of bacteria in wheat roots inoculated with A. brasilense FP2. The detection limit of the technique was 104 CFU/g of wheat root (see Fig. S3 in the supplemental material). In the first attempt to monitor the population of A. brasilense FP2, wheat was inoculated and cultivated under sterile conditions. In noninoculated plants, strain A. brasilense FP2 or any other bacteria were not detected by the qPCR or the plate count technique. Figure 2A shows the number of bacteria in wheat inoculated under sterile conditions determined by qPCR using primer pair Azo-2 (specific for Azospirillum spp.) and primer pairs AzoR2.1, AzoR2.2, and AzoR5.3 (specific for strain FP2). There was no statistically significant difference between the measurements obtained using the three strain-specific primer pairs or between those obtained using species- and strain-specific primer pairs. A large number of bacteria were observed in the first days after inoculation (roughly 107 to 108 CFU/g of wheat root; Fig. 2B). The quantification of A. brasilense FP2 was also analyzed by the plate count method in NFbHPN medium. A higher degree of variability was observed by the plate count method than by qPCR in the first days after inoculation. However, no statistically significant differences between sampling points were observed when cells were quantified using either qPCR or the plate count method from day 7 (Fig. 2B). The results also revealed an increase in the fresh weight of roots and shoots of plants inoculated with A. brasilense FP2 (Fig. 2C and D). This stimulation due to inoculation was most evident in the roots. In the second attempt, wheat was inoculated and cultivated under nonsterile conditions. The results showed no statistically significant differences in the results when A. brasilense FP2 was quantified by the qPCR methodology using three different strain-specific primer pairs (AzoR2.1, AzoR2.2, and AzoR5.3). Similar numbers of bacteria were observed when the strain-specific primer pairs and species-specific primer pair Azo-2 were used. As expected, the universal primer pair specific for 16S rRNA-encoding genes (which were used to estimate the total number of bacteria) showed higher numbers of cells per gram of wheat roots, although statistically significant differences were not achieved for any sampling point. Except at day 1, the differences in cell numbers obtained when the results for the universal primer pair specific for 16S rRNA-encoding genes and those for the species- and strain-specific primer pairs were compared were 2- to 5-fold (Fig. 3A). No statistically significant differences were also observed for most sampling points when cell counting techniques were compared, although the plate count method showed a higher degree of variability (Fig. 3B). These results suggest that the population of the inoculated bacteria is high and stable for at least 13 days after inoculation and that the diversity of all bacteria and bacteria of the Azospirillum genus present in the rhizosphere of wheat plants is limited, reflecting the rather low level of diversity of bacteria in the soil used for cultivation. The number of CFU in the soil was evaluated by the plate count method using DYGS medium and reached values of 103 to 104 CFU, confirming the occurrence of a low level of diversity of bacteria in the soil used for the cultivation of wheat and the inoculation experiments.
FIG 2.
Enumeration of Azospirillum brasilense FP2 in inoculated wheat roots under sterile conditions. (A) The primer pair Azo-2 was used for A. brasilense enumeration, and strain-specific primer pairs AzoR2.1, AzoR2.2, and AzoR5.3 were used for A. brasilense FP2 enumeration. (B) Comparison of A. brasilense FP2 enumeration by qPCR and plate counting methods. The values for qPCR are the means from three experiments using strain-specific primer pairs. (C and D) A. brasilense FP2 plant growth promotion effect observed for root fresh weight (C) and shoot fresh weight (D). *, statistically significant difference (P < 0.01).
FIG 3.
Enumeration by qPCR of Azospirillum brasilense FP2 associated with wheat roots under nonsterile conditions. (A) The species-specific primer pair Azo-2 was used for the quantification of A. brasilense; strain-specific primer pairs AzoR2.1, AzoR2.2, and AzoR5.3 were used for the quantification of A. brasilense FP2; and the primer pair specific for the 16S rRNA-encoding gene was used for the quantification of all bacteria present. (B) Quantification of A. brasilense FP2 associated with wheat roots by qPCR and plate count methods. Values for qPCR are means for the three strain-specific primer pairs. *, statistically significant difference (P < 0.01).
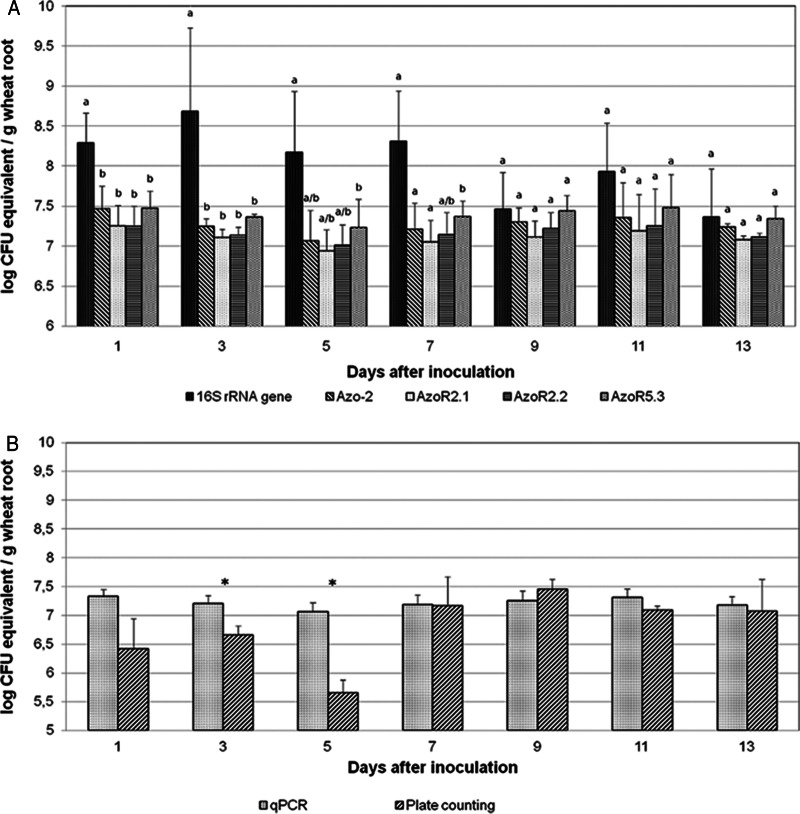
The population of A. brasilense FP2 was stable, even when the rhizobacteria Azospirillum brasilense NH, Herbaspirillum seropedicae Z67, Gluconacetobacter diazotrophicus DSM 5601, and Azospirillum lipoferum DSM 1691 were coinoculated in wheat plants under nonsterile conditions, leaving the FP2 counts above 107 CFU/g (fresh weight) of root. No statistically significant difference in the A. brasilense FP2 numbers obtained by qPCR quantification was achieved when the results obtained with the strain-specific primer pairs and those obtained with the Azo-2 primer pair were compared (Fig. 4A), and no statistically significant differences for most sampling points were observed when the results obtained by qPCR and those obtained by the plate count technique were compared (Fig. 4B). The differences in cell numbers obtained when the universal primer pair for 16S rRNA-encoding genes and species-specific primer pairs were used ranged from 2.2 × 109 (3 days after inoculation) to 3.9 × 107 (13 days after inoculation) (Fig. 4A). However, the total number of bacteria, including the number of bacteria consisting of the other inoculants, was significantly higher by qPCR with the universal primer pair specific for 16S rRNA-encoding genes than qPCR with the species-specific primer pairs until 7 days after inoculation, but after that sampling point the levels dropped and the levels of A. brasilense and strain FP2 were roughly similar by qPCR with both sets of primers. These results reinforce the finding that A. brasilense FP2 maintains a stable population in the rhizosphere/roots of the plants during the period of colonization and further indicate that strain FP2 is highly competitive, a desirable characteristic for inoculant production. When the primer pairs specific for strain FP2 developed in this work (AzoR2.1, AzoR2.2, and AzoR5.3) were used with DNA from plants inoculated with other rhizobacterial strains under nonsterile conditions, there was no amplification product, confirming the specificities of the primers for the detection of A. brasilense FP2 (data not shown).
FIG 4.
Quantification of Azospirillum brasilense FP2 associated with wheat roots under nonsterile conditions coinoculated with Azospirillum brasilense NH, Herbaspirillum seropedicae Z67, Gluconacetobacter diazotrophicus DSM 5601, and Azospirillum lipoferum DSM 1691 by the qPCR method. (A) The species-specific primer pair Azo-2 was used for A. brasilense quantification; strain-specific primer pairs AzoR2.1, AzoR2.2, and AzoR5.3 were used for A. brasilense FP2 quantification; and a primer pair specific for the 16S rRNA-encoding gene was used for the quantification of all bacteria present. For each day, different letters indicate statistically significant differences (P < 0.01). (B) Quantification of Azospirillum brasilense FP2 associated with wheat roots by the qPCR and plate count methods. Values for qPCR are the means obtained with the three strain-specific primer pairs. *, statistically significant difference (P < 0.01).
Taken together, the results from all inoculation experiments show that the number of A. brasilense FP2 cells was stable and not below 107 CFU/g (fresh weight) of root, indicating that this bacterium is competitive, maintaining its population at a high level even in the presence of competing rhizobacteria (see Fig. S4 in the supplemental material).
DISCUSSION
Inoculants containing Azospirillum spp. have been tested under field conditions with important crops. A. brasilense strains, including strain FP2, have been recognized to be very effective in promoting plant growth, and some of them have been authorized for use for the production of commercial inoculants in Brazil (10). Despite the importance of these plant growth-promoting bacteria (PGPB), no rapid method has been available to monitor this strain during experiments.
A nested PCR method for the detection of Azospirillum lipoferum CRT1 in the rhizosphere was proposed by Baudoin et al. (34). However, the primers, designed from 16S and 23S rRNA intergenic region-encoding gene fragments, proved not to be specific enough for the development of a strain-specific qPCR quantification method. Several optimizations regarding specificity and efficiency were then applied to design strain-specific qPCR primers to detect bacterial strains on the basis of sequence-characterized amplified region (SCAR) markers (17, 34). In this study, we developed a strain-specific qPCR protocol based on comparative genome analysis to quantify Azospirillum brasilense strain FP2, a plant growth-promoting bacterium, inoculated into the roots of wheat plants. To achieve this goal, we designed strain-specific primers by in silico comparison of 500-bp fragments of a draft A. brasilense FP2 genome with the sequence of the A. brasilense Sp245 genome. Unique strain FP2 fragments were also used to search the NCBI nonredundant database for similar sequences. The strain-specific fragments identified so far were used for primer design.
Many authors have reported on the use of different methods, usually based on experimental approaches, to design taxon-specific primers. Konstantinov et al. (35) isolated specific genomic fragments from the type strain and related strains by digesting the genomic DNA with a restriction enzyme and then making a subtractive hybridization with the closest strains to eliminate shared DNA fragments. The unique fragments were extracted from the gel, cloned, sequenced, and used to design specific primers to detect Lactobacillus sobrius 001T. Fujimoto et al. (36) and Maruo et al. (37) developed a PCR-based method for the identification and quantification of Lactobacillus casei strain Shirota and Lactobacillus lactis subsp. cremoris FC, respectively, using strain-specific primers derived from RAPD analysis. The authors evaluated the survival of these strains through the gastrointestinal tract by qPCR with the strain-specific primers, where they monitored the cell numbers before and after the administration of fermented milk containing this strain. Pereira et al. (38) developed a qPCR method for the quantification of the plant growth-promoting bacterium H. seropedicae in the rhizosphere of maize seedlings. Primer pairs were designed from the genome sequence of H. seropedicae SmR1 (39) and tested against 12 different species. Although the selected genome regions did not match any other sequences in the NCBI database, the primers were not evaluated against the sequences of other H. seropedicae strains, which did not allow any conclusion about their strain specificities to be made.
In the past decade, whole-genome sequencing has become a rapid and cost-effective way to provide comprehensive information about an organism (40). Although the achievement of a complete genome is a demanding process, a draft genome sequence with a wide breadth of coverage can be obtained. In the present work, we have shown that the direct comparison of the genomic sequences of closely related organisms is a rapid and reliable approach to detect specific DNA regions to be used as strain-specific genetic markers for the quantitative detection of bacterial strains colonizing roots and the rhizosphere. The rationale for this approach relies on three facts: (i) the genome sequence provides genetic information to the highest resolution; (ii) regions that diverge between the genome sequences of very closely related organisms (i.e., strains of the same species) are most likely to diverge from the genome sequences of more distant taxa; and (iii) the absence of sequence similarity between possible strain-specific genomic regions and sequences in large public databases covering most of the taxa from different environments can be broadly accepted as the absence of these regions in other organisms.
The strain-specific primers developed in this work to monitor the population of A. brasilense FP2 inoculated into wheat showed that the bacteria colonize the roots of the plant at 107 to 108 CFU/g of root in the first days after inoculation. The population is maintained at relatively stable levels until 13 days after inoculation, at which time the rhizobacteria exert a plant growth promotion effect. Although for some experiments the plant growth promotion effect was not evident during the period analyzed, this effect is frequently observed at later stages of plant development (41). Couillerot et al. (17) also observed a high number of bacteria (105 to 107 CFU/g of maize root) by either qPCR or the plate count method 1 to 3 days after inoculation of A. brasilense UAP-154 and CFN-535 into maize.
In conclusion, five primer pairs, AzoR2.1, AzoR2.2, AzoR5.1, AzoR5.2, and AzoR5.3, specific for A. brasilense strain FP2 were successfully designed and tested to monitor the fluctuation of the population of this strain after inoculation into wheat roots under sterile and nonsterile conditions. We demonstrated that A. brasilense FP2 maintained a high number of cells in association with the plant roots within 2 weeks after inoculation. Thus, in our work we showed that the strain-specific primer pairs designed by using available genome sequence information can be effectively applied to quantitatively monitor the population of PGPB in the rhizosphere of the inoculated plants. The strategy for the design of strain-specific primers described here may theoretically be used for any microorganism for which the whole-genome sequence is available in a database. The qPCR methodology developed in this work is a generally applicable tool that may be used to monitor the population dynamics of bacteria inoculated into crop plants and is potentially applicable in field experiments. Furthermore, this technique could also be applied to the quality control of commercially available inoculants, where rigid controls for contamination and the number of inoculant cells have to guarantee the efficiency of the final product.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Roseli Prado, Valter Baura, and Marilza Lamour for technical assistance.
This work was supported by CNPq, INCT da Fixação de Nitrogênio/MCT, Fundação Araucária, and CAPES.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01351-15.
REFERENCES
- 1.Bashan Y, Holguin G, de-Bashan LE. 2004. Azospirillum-plant relationships: physiological, molecular, agricultural, and environmental advances (1997–2003). Can J Microbiol 50:521–577. doi: 10.1139/w04-035. [DOI] [PubMed] [Google Scholar]
- 2.Fibach-Paldi S, Burdman S, Okon Y. 2012. Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol Lett 326:99–108. doi: 10.1111/j.1574-6968.2011.02407.x. [DOI] [PubMed] [Google Scholar]
- 3.Dobbelaere S, Vanderleyden J, Okon Y. 2003. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22:107–149. doi: 10.1080/713610853. [DOI] [Google Scholar]
- 4.Okon Y, Labandera-Gonzalez CA. 1994. Agronomic applications of Azospirillum: an evaluation of 20 years worldwide field inoculation. Soil Biol Biochem 26:1591–1601. doi: 10.1016/0038-0717(94)90311-5. [DOI] [Google Scholar]
- 5.Sant'Anna FH, Almeida LG, Cecagno R, Reolon LA, Siqueira FM, Machado MR, Vasconcelos AT, Schrank IS. 2011. Genomic insights into the versatility of the plant growth-promoting bacterium Azospirillum amazonense. BMC Genomics 12:409. doi: 10.1186/1471-2164-12-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert B, Weber OB, Kirchhof G, Halbritter A, Stoffels M, Hartmann A. 2001. Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int J Syst Evol Microbiol 51:17–26. [DOI] [PubMed] [Google Scholar]
- 7.Castro-Sowinski S, Herschkovitz Y, Okon Y, Jurkevitch E. 2007. Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol Lett 276:1–11. doi: 10.1111/j.1574-6968.2007.00878.x. [DOI] [PubMed] [Google Scholar]
- 8.Pedrosa FO, Yates MG. 1984. Regulation of nitrogen fixation (nif) genes of Azospirillum brasilense by nifA and ntr (gln) type gene products. FEMS Microbiol Lett 23:95–101. doi: 10.1111/j.1574-6968.1984.tb01042.x. [DOI] [Google Scholar]
- 9.Camilios-Neto D, Bonato P, Wassem R, Tadra-Sfeir MZ, Brusamarello-Santos LC, Valdameri G, Donatti L, Faoro H, Weiss VA, Chubatsu LS, Pedrosa FO, Souza EM. 2014. Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15:378. doi: 10.1186/1471-2164-15-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hungria M, Campo RJ, Souza EM, Pedrosa FO. 2010. Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331:413–425. doi: 10.1007/s11104-009-0262-0. [DOI] [Google Scholar]
- 11.Bashan Y. 1999. Interactions of Azospirillum spp. in soils: a review. Biol Fertil Soils 29:246–256. doi: 10.1007/s003740050549. [DOI] [Google Scholar]
- 12.Bashan Y, Puente ME, Rodriguez-Mendoza MN, Toledo G, Holguin G, Ferrera-Cerrato R, Pedrin S. 1995. Survival of Azospirillum brasilense in the bulk soil and rhizosphere of 23 soil types. Appl Environ Microbiol 61:1938–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobbelaere S, Croonenborghs A, Thys A, Ptacek D, Okon Y, Vanderleyden J. 2002. Effect of inoculation with wild type Azospirillum brasilense and A. irakense strains on development and nitrogen uptake of spring wheat and grain maize. Biol Fertil Soils 36:284–297. doi: 10.1007/s00374-002-0534-9. [DOI] [Google Scholar]
- 14.Magnani GS, Cruz LM, Weber H, Bespalhok JC, Daros E, Baura V, Yates MG, Monteiro RA, Faoro H, Pedrosa FO, Souza EM. 2013. Culture-independent analysis of endophytic bacterial communities associated with Brazilian sugarcane. Genet Mol Res 12:4549–4558. doi: 10.4238/2013.October.15.3. [DOI] [PubMed] [Google Scholar]
- 15.Magnani GS, Didonet CM, Cruz LM, Picheth CF, Pedrosa FO, Souza EM. 2010. Diversity of endophytic bacteria in Brazilian sugarcane. Genet Mol Res 9:250–258. doi: 10.4238/vol9-1gmr703. [DOI] [PubMed] [Google Scholar]
- 16.Pisa G, Magnani GS, Weber H, Souza EM, Faoro H, Monteiro RA, Daros E, Baura V, Bespalhok JP, Pedrosa FO, Cruz LM. 2011. Diversity of 16S rRNA genes from bacteria of sugarcane rhizosphere soil. Braz J Med Biol Res 44:1215–1221. doi: 10.1590/S0100-879X2011007500148. [DOI] [PubMed] [Google Scholar]
- 17.Couillerot O, Bouffaud M-L, Baudoin E, Muller D, Caballero-Mellado J, Moënne-Loccoz Y. 2010. Development of a real-time PCR method to quantify the PGPR strain Azospirillum lipoferum CRT1 on maize seedlings. Soil Biol Biochem 42:2298-2305. doi: 10.1016/j.soilbio.2010.09.003. [DOI] [Google Scholar]
- 18.Sánchez AC, Gutiérrez RT, Santana RC, Urrutia AR, Fauvart M, Michiels J, Vanderleyden J. 2014. Effects of co-inoculation of native Rhizobium and Pseudomonas strains on growth parameters and yield of two contrasting Phaseolus vulgaris L. genotypes under Cuban soil conditions. Eur J Soil Biol 62:105–112. doi: 10.1016/j.ejsobi.2014.03.004. [DOI] [Google Scholar]
- 19.Araújo AEDS, Baldani VLD, Galisa PDS, Pereira JA, Baldani JI. 2013. Response of traditional upland rice varieties to inoculation with selected diazotrophic bacteria isolated from rice cropped at the northeast region of Brazil. Appl Soil Ecol 64:49–55. doi: 10.1016/j.apsoil.2012.10.004. [DOI] [Google Scholar]
- 20.Sasaki K, Ikeda S, Eda S, Mitsui H, Hanzawa E, Kisara C, Kazama Y, Kushida A, Shinano T, Minamisawa K, Sato T. 2010. Impact of plant genotype and nitrogen level on rice growth response to inoculation with Azospirillum sp. strain B510 under paddy field conditions. Soil Sci Plant Nutr 56:636–644. doi: 10.1111/j.1747-0765.2010.00499.x. [DOI] [Google Scholar]
- 21.Stets MI, Pinto AS Jr, Huergo LF, de Souza EM, Guimarães VF, Alves AC, Steffens MBR, Monteiro RA, Pedrosa FDO, Cruz LM. 2013. Rapid identification of bacterial isolates from wheat roots by high resolution whole cell MALDI-TOF MS analysis. J Biotechnol 165:167–174. doi: 10.1016/j.jbiotec.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Couillerot O, Poirier M-A, Prigent-Combaret C, Mavingui P, Caballero-Mellado J, Moënne-Loccoz Y. 2010. Assessment of SCAR markers to design real-time PCR primers for rhizosphere quantification of Azospirillum brasilense phytostimulatory inoculants of maize. J Appl Microbiol 109:528–538. doi: 10.1111/j.1365-2672.2010.04673.x. [DOI] [PubMed] [Google Scholar]
- 23.Sørensen J, Haubjerg Nicolaisen M, Ron E, Simonet P. 2009. Molecular tools in rhizosphere microbiology—from single-cell to whole-community analysis. Plant Soil 321:483–512. doi: 10.1007/s11104-009-9946-8. [DOI] [Google Scholar]
- 24.Mavrodi OV, Mavrodi DV, Thomashow LS, Weller DM. 2007. Quantification of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains in the plant rhizosphere by real-time PCR. Appl Environ Microbiol 73:5531–5538. doi: 10.1128/AEM.00925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinheiro RDO, Boddey LH, James EK, Sprent JI, Boddey RM. 2002. Adsorption and anchoring of Azospirillum strains to roots of wheat seedlings. Plant Soil 246:151–166. doi: 10.1023/A:1020645203084. [DOI] [Google Scholar]
- 26.Lacava PT, Li WB, Araújo WL, Azevedo JL, Hartung JS. 2006. Rapid, specific and quantitative assays for the detection of the endophytic bacterium Methylobacterium mesophilicum in plants. J Microbiol Methods 65:535–541. doi: 10.1016/j.mimet.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Ruppel S, Rühlmann J, Merbach W. 2006. Quantification and localization of bacteria in plant tissues using quantitative real-time PCR and online emission fingerprinting. Plant Soil 286:21–35. doi: 10.1007/s11104-006-9023-5. [DOI] [Google Scholar]
- 28.Machado HB, Funayama S, Rigo LU, Pedrosa FO. 1991. Excretion of ammonium by Azospirillum brasilense mutants resistant to ethylenediamine. Can J Microbiol 37:549–553. doi: 10.1139/m91-092. [DOI] [Google Scholar]
- 29.Rodrigues J III, Malavolta VA Jr, Victor O. 1986. Meio simples para o isolamento e cultivo de Xanthomonas campestris pv. citri tipo B Summa. Phytopathology 12:32. [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okonechnikov K, Golosova O, Fursov M, the UGENE team . 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 34.Baudoin E, Couillerot O, Spaepen S, Moënne-Loccoz Y, Nazaret S. 2010. Applicability of the 16S-23S rDNA internal spacer for PCR detection of the phytostimulatory PGPR inoculant Azospirillum lipoferum CRT1 in field soil. J Appl Microbiol 108:25-38. doi: 10.1111/j.1365-2672.2009.04393.x. [DOI] [PubMed] [Google Scholar]
- 35.Konstantinov SR, Smidt H, de Vos WM. 2005. Representational difference analysis and real-time PCR for strain-specific quantification of Lactobacillus sobrius sp. nov. Appl Environ Microbiol 71:7578–7581. doi: 10.1128/AEM.71.11.7578-7581.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujimoto J, Matsuki T, Sasamoto M, Tomii Y, Watanabe K. 2008. Identification and quantification of Lactobacillus casei strain Shirota in human feces with strain-specific primers derived from randomly amplified polymorphic DNA. Int J Food Microbiol 126:210–215. doi: 10.1016/j.ijfoodmicro.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Maruo T, Sakamoto M, Toda T, Benno Y. 2006. Monitoring the cell number of Lactococcus lactis subsp. cremoris FC in human feces by real-time PCR with strain-specific primers designed using the RAPD technique. Int J Food Microbiol 110:69–76. doi: 10.1016/j.ijfoodmicro.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 38.Pereira TP, do Amaral FP, Dall'Asta P, Brod FCA, Arisi ACM. 2014. Real-time PCR quantification of the plant growth promoting bacteria Herbaspirillum seropedicae strain SmR1 in maize roots. Mol Biotechnol 56:660–670. doi: 10.1007/s12033-014-9742-4. [DOI] [PubMed] [Google Scholar]
- 39.Pedrosa FO, Monteiro RA, Wassem R, Cruz LM, Ayub RA, Colauto NB, Fernandez MA, Fungaro MHP, Grisard EC, Hungria M, Madeira HMF, Nodari RO, Osaku CA, Petzl-Erler ML, Terenzi H, Vieira LGE, Steffens MBR, Weiss VA, Pereira LFP, Almeida MIM, Alves LR, Marin A, Araujo LM, Balsanelli E, Baura VA, Chubatsu LS, Faoro H, Favetti A, Friedermann G, Glienke C, Karp S, Kava-Cordeiro V, Raittz RT, Ramos HJO, Ribeiro EMSF, Rigo LU, Rocha SN, Schwab S, Silva AG, Souza EM, Tadra-Sfeir MZ, Torres RA, Dabul ANG, Soares MAM, Gasques LS, Gimenes CCT, Valle JS, Ciferri RR, Correa LC, Murace NK, et al. . 2011. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet 7:e1002064. doi: 10.1371/journal.pgen.1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havlak P, Chen R, Durbin KJ, Egan A, Ren Y, Song X-Z, Weinstock GM, Gibbs RA. 2004. The Atlas genome assembly system. Genome Res 14:721–732. doi: 10.1101/gr.2264004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloepper JW, Schroth MN, Miller TD. 1980. Effects of rhizosphere colonization by plant growth-promoting rhizobacteria on potato plant development and yield. Phytopathology 70:1078–1082. doi: 10.1094/Phyto-70-1078. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.