Abstract
Improving enzyme thermostability is of importance for widening the spectrum of application of enzymes. In this study, a structure-based rational design approach was used to improve the thermostability of a highly active, wide-pH-range-adaptable, and stable endopolygalacturonase (PG8fn) from Achaetomium sp. strain Xz8 via the optimization of charge-charge interactions. By using the enzyme thermal stability system (ETSS), two residues—D244 and D299—were inferred to be crucial contributors to thermostability. Single (D244A and D299R) and double (D244A/D299R) mutants were then generated and compared with the wild type. All mutants showed improved thermal properties, in the order D244A < D299R < D244A/D299R. In comparison with PG8fn, D244A/D299R showed the most pronounced shifts in temperature of maximum enzymatic activity (Tmax), temperature at which 50% of the maximal activity of an enzyme is retained (T50), and melting temperature (Tm), of about 10, 17, and 10.2°C upward, respectively, with the half-life (t1/2) extended by 8.4 h at 50°C and 45 min at 55°C. Another distinguishing characteristic of the D244A/D299R mutant was its catalytic activity, which was comparable to that of the wild type (23,000 ± 130 U/mg versus 28,000 ± 293 U/mg); on the other hand, it showed more residual activity (8,400 ± 83 U/mg versus 1,400 ± 57 U/mg) after the feed pelleting process (80°C and 30 min). Molecular dynamics (MD) simulation studies indicated that mutations at sites D244 and D299 lowered the overall root mean square deviation (RMSD) and consequently increased the protein rigidity. This study reveals the importance of charge-charge interactions in protein conformation and provides a viable strategy for enhancing protein stability.
INTRODUCTION
Pectin, the third most abundant polysaccharide of the plant cell wall, is the most structurally complex polysaccharide consisting of covalently linked galacturonic acid (1, 2). Pectinases—microbial enzymes which are capable of depolymerizing pectin—play an important role in wide biotechnological applications, including the fruit juice, vinification, paper, and textile industries, as well as the extraction of oils (3). Polygalacturonase (PG) is the most widely studied pectinase due to its high activity, mesophilic and acidic properties, and satisfactory performance in juice clarification, extraction, viscosity reduction, and yield improvement (4). Based on sequence similarity and action mode (5), PGs are classified into family 28 of glycoside hydrolase (GH) and endo- and exo-types. Most endo-PGs are highly active and stable at 30 to 50°C (6; http://www.brenda-enzymes.org/), which are far from the requirements of thermophilic processing in the feed industry (7).
To obtain a thermostable, highly active PG, either mining new genetic resources of thermophiles (8) or engineering the protein and application environment (9, 10) is the most popular practice. Thermophilic fungi Thielavia arenaria XZ7 (60°C) (7), Thermoascus aurantiacus CBMAI-756 (60 to 65°C) (11), Penicillium sp. strain SPC-F 20 (60°C) (12), and Sclerotium rolfsii (60°C) (13) have been identified as the most excellent microbial sources of thermophilic or thermostable PGs. There have been diverse protein engineering strategies employed. For instance, Dumon et al. (14) used directed evolution to create a hyperthermostable xylanase variant with a melting temperature (Tm) 25°C higher than that of the parent enzyme. Moreover, rational design approaches concerning protein surface electrostatic interactions (15), B-factor values (16), hydrophobic interactions (17), hydrogen bonds (18), ionic bonds (19), cation-π interactions (20), and disulfide bridges (21) have been employed to develop stable proteins. However, most of the above-mentioned methods increased rigidity of domain structures at the cost of catalytic efficiency. Thus, it is important for various applications to investigate how to engineer PGs for both enhanced stability and the absence of activity loss.
Optimization of charge-charge interactions is a structure-based rational design approach that has proven successful in thermostability improvement (22, 23). However, its extensive application has been limited by underdeveloped bioinformatic tools. Tanford and Kirkwood (24) initially set up the Tanford-Kirkwood (TK) model for calculating the contribution of a single charged residue to the overall stability, which was further improved by introducing solvent accessibility (SA) (25), Gibbs free energy (26), and electrostatic interactions (27). Recently, we released a suite of enzyme redesign algorithms, the enzyme thermal stability system (ETSS) (28), in order to refine the protocol of TK-SA model calculation and surface charge-charge interaction analysis. The ETSS is a general protocol for the rational improvement of enzyme thermostability based on the charge of the enzyme. By replacing the negatively charged residues with neutrally or positively charged ones, the thermal stability of enzymes would be improved. In the present study, we employed the ETSS to improve the thermostability of a GH28 endo-PG, namely, PG8fn, without perturbing its enzymatic activity via optimization of charge-charge interactions. PG8fn from Achaetomium sp. strain Xz8 is distinguished by its high specific activity (28,000 ± 293 U/mg) and broad ranges of pH adaptation and stability (pH 3.0 to 8.0) (29). However, its thermal properties are poor: it retains thermostability only at 45°C or below.
MATERIALS AND METHODS
Chemicals, plasmids, and strains.
All chemicals were of analytical grade and commercially available. FastPfu DNA polymerase from TransGen (Beijing, China) and restriction endonucleases and T4 DNA ligase from New England BioLabs (Hitchin, United Kingdom) were purchased. The substrate polygalacturonic acid was purchased from Sigma-Aldrich (St. Louis, MO). The recombinant plasmid pPIC9-pg8fn harbors the GH28 endo-PG gene pg8fn from Achaetomium sp. strain Xz8 (CGMCC6545 of the China General Microbiological Culture Collection Center, Beijing, China) (29). Escherichia coli Trans I-T1 and the pEASY-T3 vector (TransGen) were used for plasmid amplification and construction, respectively. Pichia pastoris GS115 from Invitrogen (Carlsbad, CA) was used for heterologous expression.
Selection of the mutagenesis site.
The initial homology model of wild-type PG8fn was built with Discovery Studio 4.1 (Accelrys, San Diego, CA) using the crystal structure of CluPG1 from Colletotrichum lupini (PDB code 2IQ7_A; 79.6% identity) as the template. The total interaction energy between charged amino acids of points i and j (Eij) of wild-type PG8fn was calculated using the ETSS. A negative Eij value of a given residue signifies a favorable interaction, and vice versa. Hence, the modification of residues with positive Eij values may enhance PG8fn thermostability. Four criteria were used to redesign PG8fn, as follows: (i) four disulfide bridges conserved in Aspergillus niger endo-PGs (30) were retained, (ii) residues with high positive Eij values were taken as priorities, (iii) residues far from the catalytic center were selected to retain the activity, and (iv) multiple-sequence alignment was performed to determine the mutation sites. When oppositely charged residues were located at the same position, negatively charged residues were mutated to positively charged ones. Otherwise, the similarly charged residues were mutated to neutral Ala. The three-dimensional structure was performed by PyMOL (Delano Scientific, Portland, OR).
Site-directed mutagenesis.
The expression primers (PGS-F and PGN-R) for amplifying the gene pg8fn were reported in our previous work (29). Site-directed mutagenesis was performed by overlap extension PCR (31) with pg8fn as the DNA template and the primers listed in Table 1. The first degenerate PCR fragment was amplified by introducing the mutation site into the oligonucleotide primers, and the second fragment was amplified by annealing the overlapping ends of the first fragments. Final products were ligated into the pEASY-T3 vector and sequenced.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| D7A-F | CTCCCCGCCAAGCGCGCGAGCTGCACCTTTACCGCTGCTGCCT |
| D25A-F | CATCACCCTCAAGGCTATCACCGTCCCCG |
| D25A-R | GGGGACGGTGATAGCCTTGAGGGTGATGGT |
| E73A-F | CATCCACGTCGCTGGCGCCCCCGGCCATGT |
| E73A-R | CGGGGGCGCCAGCGACGTGGATGTTGTT |
| E132A-F | TCAACGGCGCTGCTAACCTCGGAGTGTAC |
| E132A-R | CCGAGGTTAGCAGCGCCGTTGATGCTGAAG |
| D138A-F | CTCGGAGTGTACGCTGTCCACCTCGACAAC |
| D138A-R | TCGAGGTGGACAGCGTACACTCCGAGGTTT |
| D244A-F | CTCCGTGTCGGCTGTCGAGTACTCGGGCA |
| D244A-R | GAGTACTCGACAGCCGACACGGAGCCCTTG |
| E246K-F | TGTCGGACGTCAAGTACTCGGGCATCACAC |
| E246K-R | ATGCCCGAGTACTTGACGTCCGACACGGAG |
| E267K-F | AGCAGGACTACAAGAACGGCTCGCCCACCG |
| E267K-R | GGCGAGCCGTTCTTGTAGTCCTGCTCGATG |
| D299R-F | CGGGCGCCACCAGAGTCTACATCCTCTGCG |
| D299R-R | AGGATGTAGACTCTGGTGGCGCCCGACTTG |
Mutation sites are underlined.
Overexpression and purification of wild-type PG8fn and its mutants.
To overproduce PG8fn and its mutants, the cDNA fragments of pg8fn and its nine mutants without the signal peptide-coding sequence were cloned into the vector pPIC9 (Invitrogen). The recombinant plasmids were then linearized by BglII and transformed into P. pastoris GS115 competent cells by electroporation (Bio-Rad, Boston, MA). The positive transformants were screened on the basis of their enzymatic activities in shake tubes, and those exhibiting the highest activities were selected for fermentation in 1-liter conical flasks. Minimal methanol (MM) or minimal dextrose medium agar plates for selection of His+ transformants, buffered glycerol complex medium (BMGY), and buffered methanol complex medium (BMMY) were prepared and used for P. pastoris growth and enzyme induction according to the manual for the Pichia expression kit (Invitrogen).
The enzymes were purified by following previously described procedures (29), and their molecular weights were determined using a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 12% (wt/vol) gel. The concentrations of the purified proteins were determined using the Bio-Rad protein assay kit. Enzymatic deglycosylation of purified recombinant proteins was performed with 200 U of endo-β-N-acetylglucosaminidase H (endo H) at 37°C for 1 h according to the supplier's instructions (New England BioLabs) and then analyzed by SDS-PAGE.
PG activity assay.
The PG activity was determined using the 3,5-dinitrosalicylic acid (DNS) method with d-(+)-galacturonic acid as the standard as described previously (7). The standard reaction mixture consisting of 900 μl of 0.33% (wt/vol) polygalacturonic acid in McIlvaine buffer (100 mM citric acid–200 mM Na2HPO4, pH 5.5) and 100 μl of appropriately diluted enzyme solution was incubated at 45°C for 10 min, followed by the addition of 1.5 ml of DNS reagent and 5 min of boiling. After the mixture was cooled to room temperature, its absorption at 540 nm was measured. One unit of PG activity was defined as the amount of enzyme that released reducing sugars equivalent to 1 μmol of d-(+)-galacturonic acid per minute under standard conditions (pH 5.5, 45°C, and 10 min).
pH activity profiles and effect of ionic strength.
The pH optima of wild-type PG8fn and its mutants were determined at 45°C for 10 min in McIlvaine buffer (100 mM citric acid–200 mM Na2HPO4, pH 3.0 to 8.0) containing 0.33% (wt/vol) polygalacturonic acid as the substrate. Considering the possible effect of residue substitution on surface charge-charge interaction, the enzyme activities in McIlvaine buffer and sodium acetate buffer of different concentrations (10 to 100 mM and 10 to 200 mM, respectively) were measured at pH 5.5 and 45°C to determine the effect of ionic strength.
Thermal properties of the wild type and its mutants.
The thermal properties of the wild type and three mutants were compared in terms of the temperature of maximum enzymatic activity (Tmax), the temperature at which 50% of the maximal activity of an enzyme is retained (T50), and the half-life of an enzyme at a particular temperature (t1/2). The Tmaxs of PG8fn and its three mutants were determined in the range of 20 to 80°C at pH 5.5 for 10 min. For the thermostability assay, wild-type PG8fn and its variants were incubated at a temperature range of 20 to 80°C for 30 min without a substrate, and the residual activities were then measured at pH 5.5 and the optimal temperature of each enzyme. The deglycosylated enzymes were subjected to determination of thermal properties as described above. The enzyme kinetic stabilities (an enzyme remains active for a certain duration before undergoing irreversible inactivation) were also determined at pH 5.5 and 50°C and 55°C without a substrate or at pH 5.5 and 60°C in the presence of polygalacturonic acid (0.1%, wt/vol). All enzymes were diluted to 50 μg/ml in McIlvaine buffer (pH 5.5) and incubated for different durations. The residual enzyme activities were measured under standard conditions described above. All reactions were performed in triplicate.
DSC.
The thermodynamic stabilities (Tms) of PG8fn and its mutants were analyzed by using differential scanning calorimetry (DSC). A Nano-DSC (TA Instruments) was run at a heating and scanning rate of 1°C/min over a temperature range of 25 to 80°C. The proteins, 0.25 mg of each sample, were dissolved in 1 ml of 10 mM phosphate-buffered saline (PBS) (pH 7.4). The test was repeated at least twice.
MD analysis.
Molecular dynamics (MD) simulation was conducted to analyze the thermal fluctuation of PG8fn and its three mutants at 323 K by using YASARA software (http://www.yasara.org) with Amber99 force field. The cutoff distance, 7.86 Å, of Van der Waals interactions and the long range electrostatics of particle mesh Ewald (PME) were employed. Trajectory analysis of data was performed with YASARA.
RESULTS
Selection of the mutagenesis site in PG8fn.
The total interaction energy between charged amino acids of points i and j (Eij) of wild-type PG8fn (see Fig. S1 in the supplemental material) was calculated using the ETSS, and 79 chargeable amino acids were identified in the PG8fn monomer, including 31 residues with a high positive value for Eij. Cysteine residues with a high isoelectric point were considered to be ionized. In combination with the four criteria, six residues were mutated to neutral A (D7A, D25A, E73A, E132A, D138A, and D244A) and three were mutated to positively charged K or R (E246K, E267K, and D299R) among the 31 candidates; these were all located on the protein surface and far from the catalytic center (Fig. 1).
FIG 1.
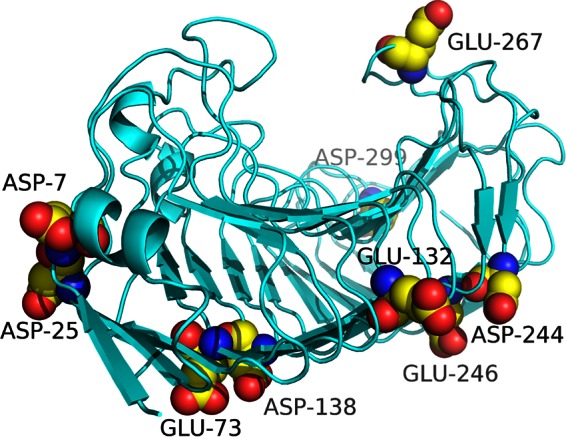
The modeled structure of PG8fn viewed from the N-terminal side. The mutated sites are indicated with balls.
Expression and purification of wild-type PG8fn and its mutants.
Wild-type PG8fn and its mutants were successfully generated and expressed in P. pastoris GS115 and secreted into the culture medium. The thermostability of wild-type PG8fn and its nine mutants were preliminarily determined by preincubating the enzymes (50 μg/ml in McIlvaine buffer, pH 5.5) at 55°C for 30 min without a substrate and measuring the residual activities. Among them, the D244A and D299R mutants showed significantly increased thermostability and retained activity comparable to that of the wild-type enzyme (data not shown). Thus, the single (D244A and D299R) and double (D244A/D299R) mutants were produced as described above and characterized.
Under standard conditions (pH 5.5, 45°C, and 10 min), the specific activities of wild-type PG8fn and the D244A, D299R, and D244A/D299R mutants toward polygalacturonic acid were 28,000 ± 293, 22,000 ± 126, 23,000 ± 167, and 23,000 ± 130 U/mg, respectively. SDS-PAGE revealed that wild-type PG8fn and its three mutants (D244A, D299R, and D244A/D299R) had apparent molecular masses of ∼38 to 42 kDa, which were higher than their theoretical molecular masses (35.4 kDa). After treatment with endo H (NEB), all enzymes showed a single band corresponding to the calculated mass (see Fig. S2 in the supplemental material).
pH optima and ionic-strength studies.
The pH activity profiles of wild-type PG8fn and its mutants were essentially quite similar (pH 5.5 to 6.0; see Fig. S3 in the supplemental material). The effect of ionic strength on enzyme activities with McIlvaine buffer and sodium acetate buffer of different ionic concentrations was also determined (Table 2). Wild-type PG8fn and its mutants showed the highest activity toward polygalacturonic acid in 50 mM McIlvaine buffer or 200 mM sodium acetate buffer, and the enzyme activity decreased with decreases in ionic strength. The results suggested that the substitution of surface charge residue has no effect on ionic strength for activity.
TABLE 2.
Effect of ionic strength at pH 5.5 on the activities of purified wild-type PG8fn and its mutants
| Enzyme | Relative activity (%) ina: |
||||||
|---|---|---|---|---|---|---|---|
| McIlvaine buffer (mM)b |
Sodium acetate buffer (mM)c |
||||||
| 100 | 50 | 10 | 200 | 100 | 50 | 10 | |
| PG8fn | 100.0 ± 2.5 | 133.1 ± 1.9 | 95.8 ± 0.7 | 100.0 ± 2.6 | 43.8 ± 1.9 | 41.7 ± 0.8 | 33.1 ± 1.2 |
| D244A mutant | 100.0 ± 2.4 | 109.8 ± 0.9 | 52.0 ± 1.9 | 100.0 ± 1.6 | 48.6 ± 3.1 | 35.5 ± 2.3 | 27.7 ± 2.2 |
| D299R mutant | 100.0 ± 2.3 | 106.6 ± 2.2 | 59.9 ± 1.3 | 100.0 ± 2.5 | 67.2 ± 1.0 | 56.1 ± 1.5 | 32.1 ± 2.2 |
| D244A/D299R mutant | 100.0 ± 2.4 | 126.2 ± 2.2 | 96.1 ± 1.0 | 100.0 ± 0.9 | 48.4 ± 0.6 | 43.7 ± 2.0 | 18.5 ± 0.3 |
Relative activity is defined as the percentage of enzymatic activity toward polygalacturonic acid against that in 100 mM McIlvaine buffer or 200 mM sodium acetate buffer (pH 5.5) at 45°C and is shown as the mean ± standard deviation (n = 3).
The concentration of McIlvaine buffer was based on the amount of citric acid.
The concentration of sodium acetate buffer was based on the amount of sodium acetate.
Temperature optima and thermostability studies.
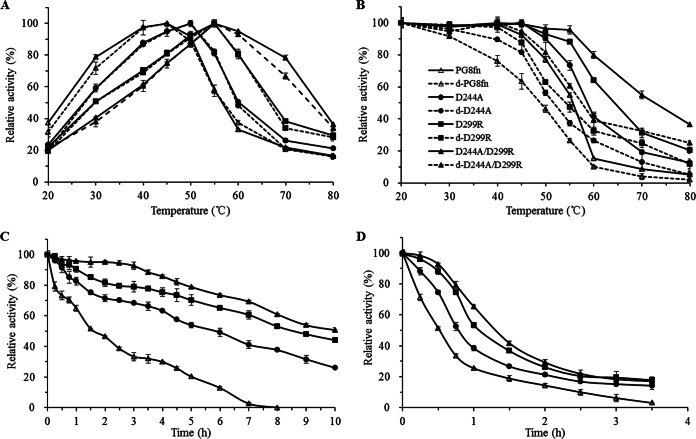
Three mutants showed improved activity at high temperatures (Fig. 2A). The Tmax values of the D244A, D299R, and D244A/D299R mutants were 50, 55, and 55°C, respectively, which were 5 to 10°C higher than that of wild-type PG8fn (45°C). Although the Tmax of the D299R mutant was the same as that of the D244A/D299R mutant, its activity beyond 55°C fell sharply. At 60°C, all mutants showed 50 to 95% of the maximal activity, which was much higher than the result for wild-type PG8fn (33% of the maximal activity). When the temperature was increased to 70°C, the D244A/D299R mutant maintained 78% of the activity, while the D299R mutant maintained only 38%. This indicates that the D244A/D299R mutant was more thermotolerant than the wild-type and other mutants.
FIG 2.
Enzymatic properties of wild-type PG8fn and its mutants (50 μg/ml). (A) Temperature-dependent activity profiles determined at pH 5.5 for 10 min; (B) enzyme inactivation assay at different temperatures for 30 min; (C and D) time courses of thermal inactivation at 50°C and 55°C, respectively. The enzymes with and without deglycosylation with endo H are marked with solid and dashed lines, respectively. Each data point represents an average from three independent experiments.
As shown in Fig. 2B, the T50 value of wild-type PG8fn and its mutants was determined at the temperature range of 20 to 80°C. The T50 value of wild-type PG8fn was determined to be 56°C, while the values of the D244A, D299R, and D244A/D299R mutants increased by 3°C, 8°C, and 17°C, respectively. After 30 min of incubation at 80°C (the feed pelleting process), PG8fn almost completely abolished the endo-PG activity (96% lost; 1,400 ± 57 U/mg), while the D244A/D299R mutant retained >35% (8,400 ± 83 U/mg) of the initial activity, which fully meets the requirements of feed processing.
N-Glycosylation is one of the most common posttranslational modifications in the methylotrophic yeast (32) and has an effect on enzyme secretion, activity, stability, and biosynthesis. The enzymatic properties of glycosylated and deglycosylated wild-type PG8fn and its variants were also compared (Fig. 2A and B). Although deglycosylated enzymes showed temperature profiles similar to those of the glycosylated counterparts, they showed lower enzyme activity and worse thermostability, as reported previously (7). This illustrates that residue substitutions D299R and D244A rather than N-glycosylation play the decisive role in the improvement of thermotolerance, and their combination has an additive effect.
Thermostabilities of wild-type PG8fn and its mutants were assessed by their half-lives (t1/2s) at pH 5.5 and 50°C and 55°C without a substrate for various periods (Fig. 2C and D, respectively). The t1/2 values of PG8fn and the D244A, D299R, and D244A/D299R mutants at 50°C and at 55°C were 1.6, 6.0, 8.6, and 10.0 h and 33, 48, 66, and 78 min, respectively. The D244A and D299R mutants were much more thermostable than the wild type, with t1/2s increased up to ∼2.8- and 4.4-fold at 50°C and ∼0.5- and 1.0-fold at 55°C, respectively. The D244A/D299R mutant showed the greatest improvement in thermostability, with the t1/2 value increased 5.3-fold at 50°C and 1.4-fold at 55°C.
In the presence of 0.1% polygalacturonic acid, wild-type PG8fn and its mutants all showed improved stability at 60°C in comparison with that without a substrate (Fig. 3). The presence of a substrate may stabilize the enzyme structure at high temperatures. In summary, the double mutant D244A/D299R thus demonstrated the best thermostability, as observed for Tmax, T50, t1/2, and thermostability in the presence of a substrate.
FIG 3.
Thermostability of PG8fn and its mutants after preincubation without (dotted line) or with (solid line) 0.1% polygalacturonic acid at 60°C for various periods. Each value represents the mean ± SD (n = 3).
DSC analysis.
DSC was performed to determine the Tm values of wild-type PG8fn and its mutants over the temperature range of 25°C to 80°C. In comparison with the Tm of PG8fn (43.8°C), the Tms of the D244A, D299R, and D244A/D299R mutants showed increases of ∼2.2°C (46.0°C), 7.6°C (51.4°C), and 10.2°C (54.0°C), respectively. The most dramatic increase in stability of the D244A/D299R mutant further confirmed the above-described results showing that the combination of double residue substitutions, D244A and D299R, makes the most remarkable contribution to the improvement of thermostability.
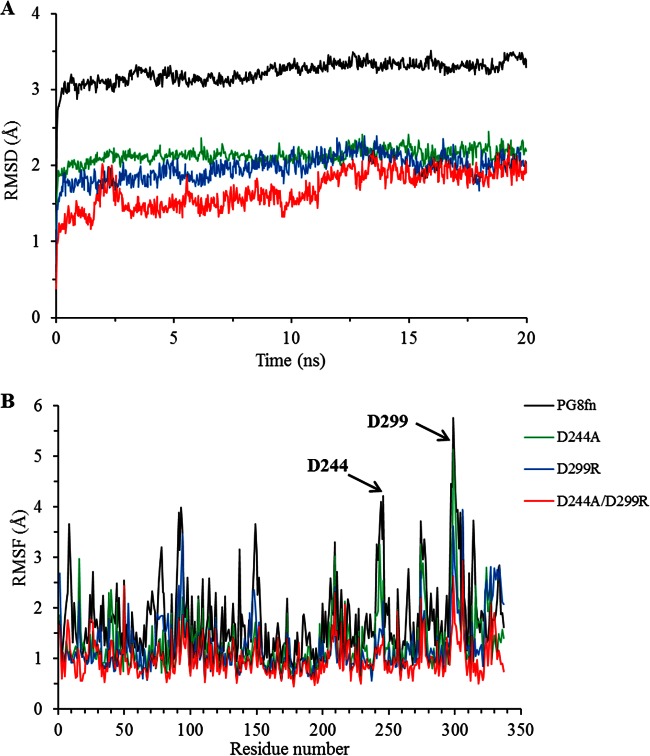
MD studies.
MD simulations of wild-type PG8fn and its three mutants at 323 K were carried out for 20 ns. The four enzymes showed an equilibrium state within the last 15 ns, as shown by the root mean square deviation (RMSD) calculated for the backbone atoms (Fig. 4A). Generally, lowering the overall RMSD is one way to modify the enzyme structure to make it more rigid and consequently enhance its thermostability (33). All mutant enzymes had lower RMSD values than that of wild-type PG8fn, suggesting that their structures are more stable. Taking the root mean square fluctuation (RMSF) value as the index (Fig. 4B), residues D244 and D299 were more flexible in the wild type than in the mutants. Thus, the crucial role of aspartic acid substitutions in thermostability was also supported by the MD results. We can conclude that the substitution of charged amino acids (aspartic acid) is one key factor responsible for the improved thermal properties of the PG8fn mutants.
FIG 4.
MD analysis of PG8fn and its mutants using YASARA at 323 K. (A) RMSD values during a 20-ns MDS; (B) RMSF values calculated over the last 10 ns.
DISCUSSION
PG is one of the most widely used commercial enzymes. It is a common practice to use PG to supplement food and animal feed to increase nutrient utilization. For example, endo-PG from T. arenaria XZ7 had a significant effect on disaggregation of soybean meal (7). However, most PGs are mesophilic, with an optimal temperature of 30 to 50°C, and completely inactivated during feed pelleting. Thus, improvement of PG thermostability is of great importance to broaden its industrial applications. In this study, we first employed the combination of open-source software ETSS (28), four residue selection criteria, and site-directed mutagenesis to identify two key residues, D244 and D299, to be crucial contributors to thermostability of PG8fn from Achaetomium sp. strain Xz8. Most PGs have negatively charged D or E at position 244 and no oppositely charged residues at this position; thus, residue D244 was mutated to neutral A. Similarly, most PGs have at position 299 negatively charged D and E, neutral N, or positively charged R in endo-PG I from Stereum purpureum (1K5C), which has a remarkable stability at 70°C (34, 35); we then mutated D299 to R. Thermophilic enzymes are more rigid and more stable than mesophilic enzymes and show lower RMSD values (33). Substitution of weak residues may cause a significant alteration of surface potential and lower the overall RMSD values.
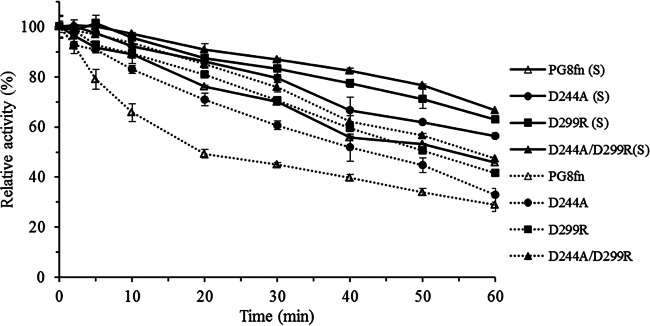
In enzyme engineering, it is important to retain the enzyme's activity when enhancing enzyme stability. Earlier attempts to improve enzyme thermostability have resulted in the reduction of enzyme activity. For example, Wang et al. (36) improved the Tm of Streptomyces sp. strain S9 GH10 xylanase by 7.0°C but lowered the catalytic activity by 75%. In contrast, our main concern in this study was to maintain the enzyme activity while enhancing thermal stability. The catalytic activities of the mutants generated in this study were comparable to that of wild-type PG8fn (22,000 to 23,000 versus 28,000 U/mg). The great care in selecting mutation sites far from the catalytic center bore fruit. After 60 min of incubation at 50 or 55°C, the mutants retained higher residual activities than that of PG8fn (Fig. 5).
FIG 5.
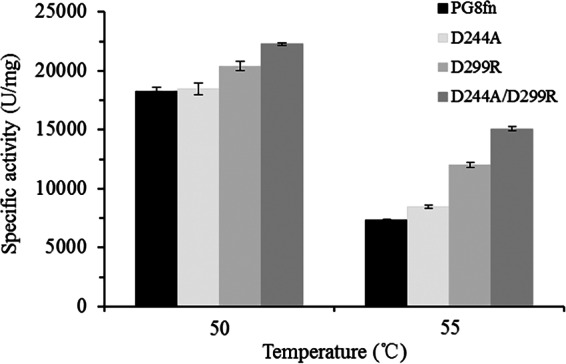
Activity profiles of PG8fn and its mutants at 50°C and 55°C.
Due to the increasing interest and potential markets for feed enzyme, thermophilic and thermostable PGs have been obtained from various sources, cloned, and characterized (11–13); however, few PGs are economically efficient for feed applications, owing to their low activity or poor stability under the required processing conditions (70 to 80°C for 5 min). Therefore, PGs with higher activity and stability under the feed processing conditions are in great demand. Currently, several microbial pectinases have been made commercially available for various industrial applications. For example, pectinases from A. niger (≥5 U/mg), Aspergillus aculeatus (3,800 U/ml), and Rhizopus sp. (400 to 800 U/g) are available from Sigma-Aldrich, and that from A. niger, widely used in the guava juice production, has a specific activity of 54 U/mg and retains less than half of its activity after incubation at 45°C for 30 min (37). In comparison with these commercial counterparts, the D244A/D299R double mutant exhibits higher specific activity and better thermostability, which would be more useful for industrial applications, especially in the feed industry.
The experimental results and bioinformatics analysis in combination showed that the D299R and D244A/D299R mutants had the best thermostability among the wild type and mutants. Based on the ETSS analysis, when D299 was mutated to R299, the Eij value decreased from 25.82 kJ/mol to −26.51 kJ/mol in the D299R mutant and −28.23 kJ/mol in the D244A/D299R mutant. This indicates a shift of the whole energy contribution from an unfavorable to a favorable state, that is, a significant potential improvement of thermostability. It also illustrates that the introduction of mutations on the chargeable residues to those of opposite electric charges would be better than to the unprotonable ones.
In conclusion, the thermostability of a highly active endopolygalacturonase (PG8fn) from Achaetomium sp. strain Xz8 was remarkably improved via the optimization of charge-charge interactions. Another distinguishing characteristic of the mutants was that their activity was comparable to that the wild type, which is rare in engineered proteins. This study revealed the importance of charge-charge interactions in protein conformation and provided a viable strategy to enhance protein stability.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National High Technology Research and Development Program of China (2012AA022208), the National Science and Technology for the Rural Development in China (2013BAD10B01-2), the National Science Fund for Distinguished Young Scholars (31225026), and the China Modern Agriculture Research System (CARS-42).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01363-15.
REFERENCES
- 1.Gupta S, Kapoor M, Sharma KK, Nair LM, Kuhad RC. 2008. Production and recovery of an alkaline exo-polygalacturonase from Bacillus subtilis RCK under solid-state fermentation using statistical approach. Bioresour Technol 99:937–945. doi: 10.1016/j.biortech.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Mohnen D. 2008. Pectin structure and biosynthesis. Curr Opin Plant Biol 11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Jayani RS, Saxena S, Gupta R. 2005. Microbial pectinolytic enzymes: a review. Process Biochem 40:2931–2944. doi: 10.1016/j.procbio.2005.03.026. [DOI] [Google Scholar]
- 4.Sharma N, Rathore M, Sharma M. 2013. Microbial pectinase: sources, characterization and applications. Rev Environ Sci Biol 12:45–60. doi: 10.1007/s11157-012-9276-9. [DOI] [Google Scholar]
- 5.Henrissat B, Bairoch A. 1993. New families in the classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem J 293:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kashyap DR, Vohra PK, Chopra S, Tewari R. 2001. Applications of pectinases in the commercial sector: a review. Bioresour Technol 77:215–227. doi: 10.1016/S0960-8524(00)00118-8. [DOI] [PubMed] [Google Scholar]
- 7.Tu T, Meng K, Huang H, Luo H, Bai Y, Ma R, Su X, Shi P, Yang P, Wang Y, Yao B. 2014. Molecular characterization of a thermophilic endo-polygalacturonase from Thielavia arenaria XZ7 with high catalytic efficiency and application potential in the food and feed industries. J Agric Food Chem 62:12686–12694. doi: 10.1021/jf504239h. [DOI] [PubMed] [Google Scholar]
- 8.Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan MC, Henrissat B, Coutinho PM, Lombard V, Natvig DO, Lindquist E, Schmutz J, Lucas S, Harris P, Powlowski J, Bellemare A, Taylor D, Butler G, de Vries RP, Allijn IE, van den Brink J, Ushinsky S, Storms R, Powell AJ, Paulsen IT, Elbourne LDH, Baker SE, Magnuson J, LaBoissiere S, Clutterbuck AJ, Martinez D, Wogulis M, de Leon AL, Rey MW, Tsang A. 2011. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol 29:922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- 9.Chan L, Cross HF, She JK, Cavalli G, Martins HFP, Neylon C. 2007. Covalent attachment of proteins to solid supports and surfaces via sortase-mediated ligation. PLoS One 2:e1164. doi: 10.1371/journal.pone.0001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park HJ, Park K, Kim YH, Yoo YJ. 2014. Computational approach for designing thermostable Candida antarctica lipase B by molecular dynamics simulation. J Biotechnol 192:66–70. doi: 10.1016/j.jbiotec.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Martins ES, Silva D, Leite RS, Gomes E. 2007. Purification and characterization of polygalacturonase produced by thermophilic Thermoascus aurantiacus CBMAI-756 in submerged fermentation. Antonie Van Leeuwenhoek 91:291–299. doi: 10.1007/s10482-006-9114-6. [DOI] [PubMed] [Google Scholar]
- 12.Mathew A, Eldo AN, Molly AG. 2008. Optimization of culture conditions for the production of thermostable polygalacturonase by Penicillium SPC-F 20. J Ind Microbiol Biotechnol 35:1001–1005. doi: 10.1007/s10295-008-0375-0. [DOI] [PubMed] [Google Scholar]
- 13.Schnitzhofer W, Weber HJ, Vršanská M, Biely P, Cavaco-Paulo A, Guebitz G. 2007. Purification and mechanistic characterisation of two polygalacturonases from Sclerotium rolfsii. Enzyme Microb Technol 40:1739–1747. doi: 10.1016/j.enzmictec.2006.11.005. [DOI] [Google Scholar]
- 14.Dumon C, Varvak A, Wall MA, Flint JE, Lewis RJ, Lakey JH, Morland C, Luginbuhl P, Healey S, Todaro T, DeSantis G, Sun M, Parra-Gessert L, Tan X, Weiner DP, Gilbert HJ. 2008. Engineering hyperthermostability into a GH11 xylanase is mediated by subtle changes to protein structure. J Biol Chem 283:22557–22564. doi: 10.1074/jbc.M800936200. [DOI] [PubMed] [Google Scholar]
- 15.Xiao S, Patsalo V, Shan B, Bi Y, Green DF, Raleigh DP. 2013. Rational modification of protein stability by targeting surface sites leads to complicated results. Proc Natl Acad Sci U S A 110:11337–11342. doi: 10.1073/pnas.1222245110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Tan Z, Wu M, Li J, Wu J. 2014. Improving the thermostability of a mesophilic family 10 xylanase, AuXyn10A, from Aspergillus usamii by in silico design. J Ind Microbiol Biotechnol 41:217–225. [DOI] [PubMed] [Google Scholar]
- 17.Pace CN, Fu H, Fryar KL, Landua J, Trevino SR, Shirley BA, Hendricks MM, Iimura S, Gajiwala K, Scholtz JM, Grimsley GR. 2011. Contribution of hydrophobic interactions to protein stability. J Mol Biol 408:514–528. doi: 10.1016/j.jmb.2011.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. 2001. Crystal structure of Bacillus stearothermophilus α-amylase: possible factors determining the thermostability. J Biochem 129:461–468. doi: 10.1093/oxfordjournals.jbchem.a002878. [DOI] [PubMed] [Google Scholar]
- 19.Szilágyi A, Zavodszky P. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8:493–504. doi: 10.1016/S0969-2126(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 20.Chakravarty S, Varadarajan R. 2002. Elucidation of factors responsible for enhanced thermal stability of proteins: a structural genomics based study. Biochemistry 41:8152–8161. doi: 10.1021/bi025523t. [DOI] [PubMed] [Google Scholar]
- 21.Jeong MY, Kim S, Yun CW, Choi YJ, Cho SG. 2007. Engineering a de novo internal disulfide bridge to improve the thermal stability of xylanase from Bacillus stearothermophilus No. 236. J Biotechnol 127:300–309. doi: 10.1016/j.jbiotec.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Strickler SS, Gribenko AV, Gribenko AV, Keiffer TR, Tomlinson J, Reihle T, Loladze VV, Makhatadze GI. 2006. Protein stability and surface electrostatics: a charged relationship. Biochemistry 45:2761–2766. doi: 10.1021/bi0600143. [DOI] [PubMed] [Google Scholar]
- 23.Schweiker KL, Zarrine-Afsar A, Davidson AR, Makhatadze GI. 2007. Computational design of the Fyn SH3 domain with increased stability through optimization of surface charge-charge interactions. Protein Sci 16:2694–2702. doi: 10.1110/ps.073091607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanford C, Kirkwood JG. 1957. Theory of protein titration curves. I. General equations for impenetrable spheres. J Am Chem Soc 79:5333–5339. [Google Scholar]
- 25.Richmond TJ. 1984. Solvent accessible surface area and excluded volume in proteins: analytical equations for overlapping spheres and implications for the hydrophobic effect. J Mol Biol 178:63–89. doi: 10.1016/0022-2836(84)90231-6. [DOI] [PubMed] [Google Scholar]
- 26.Bashford D, Karplus M. 1991. Multiple-site titration curves of proteins: an analysis of exact and approximate methods for their calculation. J Phys Chem 95:9556–9561. doi: 10.1021/j100176a093. [DOI] [Google Scholar]
- 27.Havranek JJ, Harbury PB. 1999. Tanford-Kirkwood electrostatics for protein modeling. Proc Natl Acad Sci U S A 96:11145–11150. doi: 10.1073/pnas.96.20.11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Tang X, Cui D, Yao Z, Gao B, Jiang S, Yin B, Yuan Y, Wei D. 2014. A method to rationally increase protein stability based on the charge-charge interaction, with application to lipase LipK107. Protein Sci 23:110–116. doi: 10.1002/pro.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu T, Meng K, Bai Y, Shi P, Luo H, Wang Y, Yang P, Zhang Y, Zhang W, Yao B. 2013. High-yield production of a low-temperature-active polygalacturonase for papaya juice clarification. Food Chem 141:2974–2981. doi: 10.1016/j.foodchem.2013.05.132. [DOI] [PubMed] [Google Scholar]
- 30.van Santen Y, Benen JAE, Schröter KH, Kalk KH, Armand S, Visser J, Dijkstra BW. 1999. 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J Biol Chem 274:30474–30480. doi: 10.1074/jbc.274.43.30474. [DOI] [PubMed] [Google Scholar]
- 31.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 32.Shental-Bechor D, Levy Y. 2008. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci U S A 105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Yu H, Liu C, Liu J, Shen Z. 2013. Improving stability of nitrile hydratase by bridging the salt-bridges in specific thermal-sensitive regions. J Biotechnol 164:354–362. doi: 10.1016/j.jbiotec.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Miyairi K, Senda M, Watanabe M, Hasui Y, Okuno T. 1997. Cloning and sequence analysis of cDNA encoding endopolygalacturonase I from Stereum purpureum. Biosci Biotechnol Biochem 61:655–659. doi: 10.1271/bbb.61.655. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu T, Nakatsu T, Miyairi K, Okuno T, Kato H. 2002. Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution. Biochemistry 41:6651–6659. doi: 10.1021/bi025541a. [DOI] [PubMed] [Google Scholar]
- 36.Wang K, Luo H, Tian J, Turunen O, Huang H, Shi P, Hua H, Wang C, Wang S, Yao B. 2014. Thermostability improvement of a Streptomyces xylanase by introducing proline and glutamic acid residues. Appl Environ Microbiol 80:2158–2165. doi: 10.1128/AEM.03458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kant S, Vohra A, Gupta R. 2013. Purification and physicochemical properties of polygalacturonase from Aspergillus niger MTCC 3323. Protein Expr Purif 87:11–16. doi: 10.1016/j.pep.2012.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.