Abstract
Heart disease is the leading cause of death worldwide. Therefore, assessing the risk of its occurrence is a crucial step in predicting serious cardiac events. Identifying heart disease risk factors and tracking their progression is a preliminary step in heart disease risk assessment. A large number of studies have reported the use of risk factor data collected prospectively. Electronic health record systems are a great resource of the required risk factor data. Unfortunately, most of the valuable information on risk factor data is buried in the form of unstructured clinical notes in electronic health records. In this study, we present an information extraction system to extract related information on heart disease risk factors from unstructured clinical notes using a hybrid approach. The hybrid approach employs both machine learning and rule-based clinical text mining techniques. The developed system achieved an overall microaveraged F-score of 0.8302.
1. Introduction
Heart disease is a collective term for conditions that affect the heart. Heart disease often leads to serious cardiovascular events such as heart attacks and stroke. It has been observed to be the leading cause of death worldwide in both men and women and has become a major burden on national healthcare expenditures around the world [1, 2]. Assessment of the risk of heart disease is very crucial in finding opportunities for prevention. Identifying and tracking the progression of heart disease risk factors are the basic steps in heart disease risk assessment. A few examples of heart disease risk factors are diabetes, coronary artery disease (CAD), hypertension, hyperlipidemia, obesity, medications, smoking history, and family history of premature CAD. Data for these risk factors are often specifically collected for the purpose of studies assessing the risk of heart disease.
The healthcare sector observed a rapid adoption of electronic health record (EHR) systems in the past decade. The primary purpose of EHR systems is to collect, store, and exchange patient data. EHRs are rich sources of valuable patient data such as comorbidities, medication history, social history, and family history. Data gathered from EHRs can be used as an alternative for data collected from studies specifically designed for heart disease risk assessment. However, most of these valuable patient data are buried in the form of unstructured format in EHRs [3, 4]. Manually extracting these unstructured data from EHRs can be very expensive and time consuming. Extracting unstructured data required for risk assessments can be automatically done using clinical text mining. This involves two major subtasks: identifying risk factors and tracking the progression of the disease. Automatic extraction of these heart disease risk factor data involves developing a highly specified system and may not be suitable for extracting risk factors for other diseases without necessary changes.
Recently, a great increase in information extraction (IE) systems catered for the clinical domain has been observed. There are various open source IE systems available to extract information from unstructured EHRs [5–12]. However, the types of heart disease risk factor information with temporality that can be extracted from these IE systems are limited. cTAKES is an open source IE system useful in extracting disease disorders, medications, symptoms, and anatomical locations [11]. HITEx is another clinical IE system based on the GATE framework capable of extracting disease disorders, medications, and smoking status [13]. MedEx is another IE system useful in extracting medication information [12]. TEMPTING, on the other hand, is an IE system capable of extracting temporal relations useful in tracking the progression of the disease from patient discharge summaries [5]. Byrd et al. developed a hybrid IE system to extract Framingham diagnostic criteria for heart failure with relevant disease progression information [14]. Another example is the rule-based FRSSystem capable of extracting Framingham risk factors used for predicting the risk of CAD [15]. Jonnagaddala et al. developed a machine learning-based IE system to identify disease disorder mentions [8]. The mentioned IE systems can be reused to identify heart disease risk factors but often require customization or addition of new modules. Savova et al. built a machine learning-based smoking classification module for cTAKES [16]. Goryachev et al. developed a module for HITEx to extract family history related information. None of these systems can identify a comprehensive number of heart disease risk factors that can be used for risk assessment.
In this study, we present an IE system capable of extracting unstructured data from EHRs. This is specifically developed for the purpose of identifying and tracking the progression of heart disease risk factors in diabetic patients. The system developed in this study is an extension to our baseline system which was developed as part of our participation in the 2014 i2b2/UTHealth shared tasks [17, 18]. The developed system performs risk factor concept recognition and assigns relevant time attributes to the recognized risk factors on longitudinal EHRs. The heart disease risk factors recognized by the system are diabetes, coronary artery disease (CAD), hypertension, hyperlipidemia, smoking status, obesity status, family history of premature CAD, and medications. The system extracts the above-mentioned heart disease risk factors and assigns an indicator attribute and a time attribute, if applicable. The system is a hybrid system with both rule-based and machine learning components. The evaluation of the system shows that it achieved an overall microaveraged F-score of 0.8302.
2. Materials and Methods
2.1. Dataset
The authors used the 2014 i2b2/UTHealth shared task 2 dataset in this study [18]. The dataset is a collection of unstructured longitudinal EHRs of diabetic patients provided by Partners Healthcare, USA. The EHRs are deidentified and annotated according to the guidelines. The annotations included heart disease risk factors and information of disease progression [19]. Gold standard annotations for this dataset were also available to evaluate the developed IE system. The dataset included 1304 unstructured EHRs (from here on referred to as records) from 297 patients divided into three sets: training set 1, training set 2, and test set. Training set 1 and training set 2 included 521 and 269 records, respectively, while the test set had 514 records. The dataset was also stratified into three different cohorts of diabetic patients: patients who had CAD, patients who develop CAD, and patients who did not develop CAD over a period of time [15]. Presence of heart risk factors and progression of the disease were defined in the form of risk factor, indicator attribute, and time attribute in the dataset. An overview of risk factors and their corresponding attributes is presented in Table 1. A sample (modified) EHR from the dataset is also illustrated in Figure 1. Each risk factor tag excluding family history and smoking history had time attribute that can take values, before document creation time (DCT), during DCT, and after DCT. The time attribute defines when a risk factor is known to have existed. The indicator attribute defines whether the identified risk factor is a mention, test, or lab value.
Table 1.
Overview of risk factors, indicator attribute, and time attribute.
| Risk factor | Indicator attribute | Time attribute |
|---|---|---|
| CAD | Mention, event, test result, and symptom | Before DCT, during DCT, and after DCT |
|
| ||
| Diabetes | Mention, high A1c, and high glucose | Before DCT, during DCT, and after DCT |
|
| ||
| Family history | Present, not present | Not applicable |
|
| ||
| Hyperlipidemia | Mention, high cholesterol, and high LDL | Before DCT, during DCT, and after DCT |
|
| ||
| Hypertension | Mention, high blood pressure | Before DCT, during DCT, and after DCT |
|
| ||
| Medication | ACE inhibitors, ACE inhibitors ARBs, amylin, antidiabetes medications, aspirin, beta-blockers, calcium-channel blockers, DPP-4 inhibitors, ezetimibe, fibrates, GLP-1 agonists, insulin, meglitinides, metformin, niacin, nitrates, obesity, statins, sulfonylureas, thiazide diuretics, thiazolidinediones, and thienopyridines | Before DCT, during DCT, and after DCT |
|
| ||
| Obesity | Mention, BMI, and waist circumference | Before DCT, during DCT, and after DCT |
|
| ||
| Smoking history | Current, past, ever, never, and unknown | Not applicable |
Figure 1.
Sample EHR with annotations of heart disease risk factors.
2.2. System Description
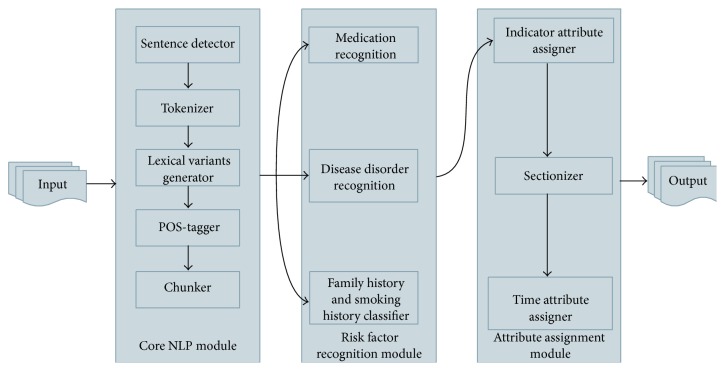
The heart disease risk factors system (HDRFSystem) in its current form includes three modules (i) core NLP module, (ii) risk factor recognition module, and (iii) attribute assignment module (Figure 2). The core NLP module identifies sentence boundaries (sentence detector), breaks sentences into tokens (tokenizer), assigns part of speech tags (POS-tagger), and identifies noun phrases (chunker). The core NLP module adopted components from the OpenNLP package (v1.5.3) available at https://opennlp.apache.org/. Processed information from the core NLP module is then passed to the risk factor recognition module where medications, disease disorder mentions, family history, and smoking history are identified. The risk factor recognition module is responsible for identifying all the heart disease risk factors. All the identified risk factors (except family history and smoking history) were then assigned indicator and time attributes by the components in the attribute assignment module. The components of the risk factor recognition module and the time attribute assignment module are explained in more detail in the following sections.
Figure 2.
Overview of heart disease risk factor information extraction system.
2.2.1. Medication Recognition
This component was used for identification of medications and is based on MetaMap [20, 21]. The noun phrase chunks identified by the chunker component in the core NLP module were passed to MetaMap. The component was configured to use MetaMap with UMLS2013AB as the knowledge source and USAbase as the data version and strict data model. For identifying medications, the component was restricted to use RxNorm terminology with a candidate score of 1000.
2.2.2. Disease Disorder Recognition
This component identifies the mentions of hypertension, hyperlipidemia, CAD, and obesity using MetaMap. To identify disease disorder MetaMap was configured to use SNOMEDCT_US terminology as source with a candidate score of 1000. Rules were developed for finding lab values such as blood pressure values, HDL count, and glucose level. For example, this component can identify BP value from text such as “BP: 158/72,” “blood pressure 149/96,” or “blood pressure elevated at 188/92.” Similarly, the authors developed rules to identify lipid levels (e.g., lipid levels: total cholesterol 164, TG 145, HDL 33, and LDL 102) and other blood tests (e.g., BUN is 27, creatinine is 4.7, and glucose is 79). Once values were identified, they were filtered out based on the levels mentioned in the annotation guidelines [19]. This component also filters out irrelevant disease disorders which are not considered as heart disease risk factors based on the rules using UMLS CUI. Furthermore, a custom-built dictionary was used to find abbreviation mentions. For example, DM2, DM Type II, and DMII refer to diabetes type 2. The rules in this component were implemented using Apache UIMA Ruta framework (https://uima.apache.org/ruta.html).
2.2.3. Family History and Smoking History Classifier
A rule-based classifier was employed to identify family history of premature CAD. This rule-based classifier identifies sentences containing CAD mentions that also has mention of familial relationships. More rules were applied to check whether the relative died prematurely (age < 55) due to CAD. If there are no such sentences in the document, then the document is simply classified as unknown for family history of premature CAD. In the dataset, smoking history is classified at document level using five classes: “current,” “past,” “never,” “ever,” and “unknown.” We developed Naïve Bayes algorithm-based supervised learning classifier to identify smoking history in conjunction with a few rules [22]. The Naïve Bayes classifier model was built using features illustrated in Table 2. Furthermore, we evaluated and selected features which were highly correlated with the classifier's predicative performance [23]. The smoking history classifier identifies smoking history by classifying each sentence. If multiple instances of smoking sentences were identified, rules were applied to select one. The training dataset was used to build a custom dictionary of smoking terms such as smoker, tobacco, and packs per year that can be used to identify sentences containing any mention of smoking history. Using this custom-built dictionary, sentences for smoking history mention were identified and were further classified into three classes, namely, “current,” “past,” and “never.” During the development of classifier, it was noticed that less than 1% of the records in the dataset belong to “ever” class. To improve classifier performance, “ever” class was completely ignored. If no mention of smoking terms was found in the document, then that document was simply classified as “unknown” for smoking history.
Table 2.
Features used by smoking history, sectionizer, and time attribute assigner classifiers.
| Component | Classification | Classifier | Classes | List of features |
|---|---|---|---|---|
| Smoking history | Sentence level | Naïve Bayes | Current, past, and never | Bag of words, POS tags |
|
| ||||
| Sectionizer | Sentence level | Conditional random fields | Section heading, section heading with text, and text | First word uppercased, all words uppercased, all words lowercased, dictionary match, first word, second word, previous sentence features, next sentence features, full stop, and containing colon |
|
| ||||
| Time attribute assigner | Phrase level | Naïve Bayes | Before DCT, during DCT, after DCT, and continuing | Identified risk factor spans, previous word, previous word POS tag, next word, next word POS tag, section information, and indicator attribute |
2.2.4. Indicator Attribute Assigner
The indicator attribute assigner takes input from the risk factor recognition module and assigns appropriate indicator attribute based on dataset annotation guidelines [19]. This component was developed by implementing various rules using Apache UIMA Ruta framework. The rules consider factors like how the risk factor was recognized and what the annotations made by the risk factor module were. For example, if a record contains text such as “type 1 diabetes,” the diabetes risk factor is recognized by the disease disorder component, and it is assigned with “mention” tag for the indicator attribute. Similarly, using the same rules, if the diabetes risk factor was recognized by A1c test values, it is assigned with “high A1c” indicator attribute. Medication type was assigned using a custom-built dictionary built from the training datasets using Wikipedia. The final dictionary file contained medications generic names and categories they belong to. In total, there were 474 medications in 21 categories (related to heart disease risk factors). Overall, we developed 26 rules to assign risk factor indicator attribute.
2.2.5. Sectionizer
Most of the EHRs in the dataset included section headings. The section headings information was useful in identifying family history and medication risk factors [17, 24]. At the same time, the same information was used as a feature for assigning time attribute to identified risk factors. For example, medications mentioned under section heading “medications” or “medications on admission” will always have a time attribute as before DCT, after DCT, and during DCT. Thus, we developed a conditional random field (CRF) based machine learning classifier to identify section information using features illustrated in Table 2 [25]. The classifier classified a sentence to either “section heading” or “section heading with text” or “text.” Section heading class was assigned if the sentence contains only a section heading (e.g., current medications). Section heading with text class was assigned if the sentence contains section heading with text (e.g., record date: 2073-12-14). Text class was assigned if the sentence contains text only and does not include any section headings (e.g., “s/p XRT to esophagus”).
2.2.6. Time Attribute Assigner
This component assigns time attribute for each of the risk factors identified by earlier components. Similar to the smoking history classifier, a supervised learning classifier based on Naïve Bayes algorithm was developed with addition of a few rules to complete the task [22]. We used risk factor phrases annotated by risk factor recognition module to train the model with features shown in Table 2. Each phrase was classified into either one of the four classes shown in Table 2. When a phrase is classified as continuing in the output, we assigned all three time attributes, before DCT, after DCT, and during DCT, as per annotation guidelines [19].
3. Results
The HDFRSystem was evaluated using macro- and microaveraged precision, recall, and F-score [26]. An evaluation script provided with the dataset was used to calculate performance scores. The evaluation script is capable of reporting system performance at many levels including specific risk factors by indicator attribute and time attribute. The evaluation metrics are explained in more detail elsewhere [19]. The developed system achieved an overall microaveraged F-score of 0.8302 on the test set. Performance of the developed system on the test set categorized by indicator attribute is presented in Table 3. Every mention indicator attribute outperformed other indicator attributes, suggesting that our methods were effective in identifying risk factor mentions but not so effective in inferring risk factors from lab values or tests. A number of indicator attributes were not recognized, specifically CAD test result, high glucose, high cholesterol, obesity, medications, amylin, antidiabetes, and waist circumference. The smoking history classifier also underperformed when compared to other risk factors by achieving 0.5 and 0.7265 microaveraged F-score for “current” and “never” indicator attributes, respectively.
Table 3.
Performance on test set by indicator attributes.
| Risk factor | Macroaveraged | Microaveraged | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | |
| CAD | ||||||
| Mention | 0.3346 | 0.3405 | 0.3375 | 0.5029 | 1.0000 | 0.6693 |
| Event | 0.1148 | 0.1138 | 0.1143 | 0.8806 | 0.4245 | 0.5728 |
| Test result | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Symptom | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Diabetes | ||||||
| Mention | 0.6887 | 0.6907 | 0.6897 | 0.9219 | 0.9972 | 0.9581 |
| High A1c | 0.1109 | 0.106 | 0.1084 | 0.8906 | 0.6951 | 0.7808 |
| High glucose | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Family history | ||||||
| Present | 0.0097 | 0.0097 | 0.0097 | 1.0000 | 0.2632 | 0.4167 |
| Not present | 0.9630 | 0.9630 | 0.9630 | 0.9725 | 1.0000 | 0.9861 |
| Hyperlipidemia | ||||||
| Mention | 0.4436 | 0.4436 | 0.4436 | 0.8444 | 0.962 | 0.8994 |
| High cholesterol | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| High LDL | 0.0331 | 0.0331 | 0.0331 | 0.7391 | 0.5862 | 0.6538 |
| Hypertension | ||||||
| Mention | 0.7062 | 0.7101 | 0.7082 | 0.9553 | 0.9918 | 0.9732 |
| High blood pressure | 0.2996 | 0.2889 | 0.2942 | 0.4858 | 0.7897 | 0.6016 |
| Medication | ||||||
| ACE inhibitors | 0.3320 | 0.3482 | 0.3399 | 0.8797 | 0.8325 | 0.8555 |
| ACE inhibitors ARBs | 0.1096 | 0.1128 | 0.1112 | 0.8667 | 0.8756 | 0.8711 |
| Amylin | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Antidiabetes Medications | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Aspirin | 0.3709 | 0.3930 | 0.3817 | 0.9079 | 0.7168 | 0.8011 |
| Beta-blockers | 0.3891 | 0.4047 | 0.3967 | 0.9302 | 0.7186 | 0.8108 |
| Calcium-channel blockers | 0.2010 | 0.2160 | 0.2082 | 0.9064 | 0.8052 | 0.8528 |
| DPP-4 inhibitors | 0.0039 | 0.0039 | 0.0039 | 1.0000 | 1.0000 | 1.0000 |
| Ezetimibe | 0.0214 | 0.0253 | 0.0232 | 0.6471 | 0.9167 | 0.7586 |
| Fibrates | 0.0506 | 0.05447 | 0.0525 | 0.8966 | 0.8667 | 0.8814 |
| GLP-1 agonists | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Insulin | 0.1790 | 0.1887 | 0.1837 | 0.8598 | 0.6987 | 0.7709 |
| Meglitinides | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Metformin | 0.2069 | 0.2228 | 0.2145 | 0.8439 | 0.8598 | 0.8518 |
| Niacin | 0.0123 | 0.0175 | 0.0144 | 0.4524 | 0.7600 | 0.5672 |
| Nitrates | 0.1031 | 0.1148 | 0.1086 | 0.803 | 0.5867 | 0.6780 |
| Obesity | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Statins | 0.4617 | 0.4786 | 0.4700 | 0.9199 | 0.8715 | 0.8950 |
| Sulfonylureas | 0.1518 | 0.1595 | 0.1555 | 0.9286 | 0.8125 | 0.8667 |
| Thiazide Diuretics | 0.1226 | 0.1376 | 0.1297 | 0.3058 | 0.7441 | 0.4335 |
| Thiazolidinediones | 0.0396 | 0.04475 | 0.0420 | 0.8841 | 1.0000 | 0.9385 |
| Thienopyridines | 0.1543 | 0.1673 | 0.1606 | 0.8914 | 0.8380 | 0.8639 |
| Obesity | ||||||
| Mention | 0.1589 | 0.1693 | 0.1639 | 0.7632 | 1.0000 | 0.8657 |
| BMI | 0.0136 | 0.0123 | 0.0129 | 1.0000 | 0.4118 | 0.5833 |
| Waist circumference | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Smoking history | ||||||
| Current | 0.0234 | 0.0234 | 0.0234 | 0.8000 | 0.3636 | 0.5000 |
| Past | 0.1479 | 0.1479 | 0.1479 | 0.8636 | 0.6726 | 0.7562 |
| Ever | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Never | 0.1576 | 0.1576 | 0.1576 | 0.7864 | 0.6750 | 0.7265 |
| Unknown | 0.4728 | 0.4728 | 0.4728 | 0.7890 | 1.0000 | 0.8820 |
We also present the system's performance categorized by time attribute in Table 4. Similar to the trend noticed in Table 3, CAD and medication risk factors underperformed when compared to other risk factors. CAD and medication risk factors achieved a lower recall and F-score when compared to overall risk factors for all three time attributes. In other words, our time attribute classifier did not perform well on assigning time attributes for CAD and medication risk factors. However, our time attribute classifier performed well for diabetes and hypertension achieving microaveraged F-scores of 0.9203 and 0.9464, respectively.
Table 4.
Performance on test set by time attributes.
| Risk factor | Macroaveraged | Microaveraged | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | |
| CAD | ||||||
| Before DCT | 0.3434 | 0.2628 | 0.2977 | 0.5599 | 0.5827 | 0.5711 |
| During DCT | 0.3405 | 0.3176 | 0.3286 | 0.5117 | 0.8102 | 0.6272 |
| After DCT | 0.3327 | 0.3288 | 0.3307 | 0.5000 | 0.9771 | 0.6615 |
| Diabetes | ||||||
| Before DCT | 0.6848 | 0.6683 | 0.6765 | 0.9152 | 0.9255 | 0.9203 |
| During DCT | 0.6907 | 0.6699 | 0.6801 | 0.9245 | 0.9293 | 0.9269 |
| After DCT | 0.6887 | 0.6887 | 0.6887 | 0.9219 | 0.9972 | 0.9581 |
| Hyperlipidemia | ||||||
| Before DCT | 0.4504 | 0.4429 | 0.4466 | 0.8419 | 0.9007 | 0.8703 |
| During DCT | 0.4426 | 0.4407 | 0.4416 | 0.8382 | 0.9421 | 0.8872 |
| After DCT | 0.4436 | 0.4436 | 0.4436 | 0.8444 | 0.962 | 0.8994 |
| Hypertension | ||||||
| Before DCT | 0.7043 | 0.6868 | 0.6954 | 0.9526 | 0.9403 | 0.9464 |
| During DCT | 0.644 | 0.7364 | 0.6871 | 0.7432 | 0.9557 | 0.8362 |
| After DCT | 0.7062 | 0.7062 | 0.7062 | 0.9553 | 0.9918 | 0.9732 |
| Medication | ||||||
| Before DCT | 0.6768 | 0.6600 | 0.6683 | 0.8332 | 0.7923 | 0.8122 |
| During DCT | 0.6613 | 0.6519 | 0.6565 | 0.8095 | 0.7858 | 0.7975 |
| After DCT | 0.6729 | 0.6648 | 0.6688 | 0.8200 | 0.7943 | 0.8069 |
| Obesity | ||||||
| Before DCT | 0.1537 | 0.1518 | 0.1527 | 0.7383 | 0.9753 | 0.8404 |
| During DCT | 0.1693 | 0.1634 | 0.1663 | 0.8246 | 0.9400 | 0.8785 |
| After DCT | 0.1537 | 0.1518 | 0.1527 | 0.7383 | 0.9753 | 0.8404 |
| All risk factors | ||||||
| Before DCT | 0.7727 | 0.7881 | 0.7803 | 0.8224 | 0.8146 | 0.8185 |
| During DCT | 0.7463 | 0.8187 | 0.7808 | 0.7835 | 0.8470 | 0.8140 |
| After DCT | 0.7706 | 0.8321 | 0.8002 | 0.8136 | 0.8688 | 0.8403 |
For comparison, we present the results of a cTAKES-based system (Table 5) versus the results we obtained (Table 6) using the test set [17]. Our system performed significantly better than the cTAKES-based system. The system developed in this study achieved a higher overall micro-F-score compared to the cTAKES-based system, 0.8302 versus 0.7151. Our methods outperformed the cTAKES-based system in all of the risk factors. Family history risk factor achieved the highest macro- and microaveraged F-score. All the risk factors achieved a microaveraged F-score above 0.80, except for CAD. Out of all the risk factors, CAD and medication risk factors achieved a lower macro- and microaveraged precision, recall, and F-score.
Table 5.
Performance of baseline system on test set.
| Risk factor | Macroaveraged | Microaveraged | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | |
| CAD | 0.2135 | 0.2311 | 0.2220 | 0.6652 | 0.5599 | 0.6080 |
| Diabetes | 0.6576 | 0.6745 | 0.6660 | 0.8692 | 0.9517 | 0.9086 |
| Family history | 0.9689 | 0.9689 | 0.9689 | 0.9689 | 0.9689 | 0.9689 |
| Hyperlipidemia | 0.4465 | 0.4412 | 0.4439 | 0.8434 | 0.9254 | 0.8825 |
| Hypertension | 0.3429 | 0.4833 | 0.4012 | 0.5579 | 0.6148 | 0.5850 |
| Medication | 0.5486 | 0.6534 | 0.5964 | 0.6227 | 0.7409 | 0.6767 |
| Obesity | 0.1402 | 0.1419 | 0.141 | 0.8447 | 0.8511 | 0.8479 |
| Smoking history | 0.6284 | 0.6284 | 0.6284 | 0.6284 | 0.6309 | 0.6296 |
|
| ||||||
| Overall | 0.6954 | 0.7634 | 0.7278 | 0.6779 | 0.7566 | 0.7151 |
Table 6.
Performance of HDRFSystem on test set.
| Risk factor | Macroaveraged | Microaveraged | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | |
| CAD | 0.3455 | 0.2985 | 0.3203 | 0.5261 | 0.7334 | 0.6127 |
| Diabetes | 0.6876 | 0.6724 | 0.6799 | 0.9202 | 0.9483 | 0.9341 |
| Family history | 0.9728 | 0.9728 | 0.9728 | 0.9728 | 0.9728 | 0.9728 |
| Hyperlipidemia | 0.4504 | 0.4451 | 0.4477 | 0.8415 | 0.9334 | 0.8851 |
| Hypertension | 0.6970 | 0.7375 | 0.7166 | 0.8531 | 0.9613 | 0.9040 |
| Medication | 0.6703 | 0.6731 | 0.6717 | 0.8209 | 0.7908 | 0.8056 |
| Obesity | 0.1589 | 0.1652 | 0.1620 | 0.7683 | 0.9618 | 0.8542 |
| Smoking history | 0.8113 | 0.8113 | 0.8113 | 0.8113 | 0.8145 | 0.8129 |
|
| ||||||
| Overall | 0.8053 | 0.8515 | 0.8277 | 0.8138 | 0.8472 | 0.8302 |
4. Discussion
We performed an extensive error analysis to understand our results in depth. Several interesting findings about the system and the dataset, in general, were observed. A few errors in the gold set annotations were also noticed. For example, the smoking history did not have annotations for all documents in the test set. Thus, being a document level classification problem, the evaluation metrics precision, recall, and F-score are not the same for smoking risk factor. Previously, we believed that our methods did not recognize amylin and antidiabetes as shown in Table 5. However, upon further inspection, the test set did not have any entities with amylin or antidiabetes. The training sets also had very few negated mentions of risk factors. Moreover, we found that there were very few instances (<1%) for “ever” smoking history class in the training set. This created an imbalance classification problem where the performance of classifier may not represent the full capabilities of the classifier [27]. So we simply removed “ever” class from our classification problem, to make it a balanced classification problem.
During the development of the system, we noticed that by employing simple rules we can drastically improve the performance of the system. As a result, we implemented rules in our components. Table 7 summarizes the number of rules in each component with examples. From the results, it can be interpreted that for most of the risk factors MetaMap outperformed rule-based lab value extractor component. For example, diabetes mention had an F-score value of 0.8897 while diabetes A1c had 0.7808. This means that the rule-based lab value extractor was not as effective as we expected. The lab value extractor failed to recognize values represented as ranges. For example, the blood pressure value “120–130/88–92” was extracted as “130/88,” and as a result it was not detected as high blood pressure by our rules. One simple rule applied for time attribute assignment turned to be effective for diabetes, hypertension, hyperlipidemia, and obesity but not for CAD and medication. We believe that the poor performance of the time attribute assigner on the CAD and medication risk factors is due to usage of limited features.
Table 7.
Examples of rules used in HDRFSystem components.
| Component | Number of rules | Examples |
|---|---|---|
| Medication recognition | 12 | If the medication identified by MetaMap is from RxNorm terminology, assign risk factor with identified medication name. |
| If the medications identified by MetaMap include abbreviations from custom abbreviations dictionary, assign medication risk factor with full medication name. | ||
|
| ||
| Disease disorder recognition | 22 | If the disease identified by MetaMap is from SNOMED CT terminology and is either CAD or obesity or diabetes or hypertension or hyperlipidemia, assign risk factor with identified disease name. |
| If annotated text is identified by blood pressure lab value extractor and diastolic >90 or systolic >140, assign risk factor = “hypertension.” | ||
|
| ||
| Family history | 05 | If a sentence contains “cad” or “coronary artery disease” and contains “father,” “mother,” or brother, assign sentence as family history sentence. |
| If family history sentence contains age of death and age <45, assign family history = “present” or else “unknown.” | ||
|
| ||
| Smoking history | 07 | If a sentence contains terms from custom smoking terms dictionary, assign sentence as smoking history sentence. |
| If document does not contain smoking terms, assign smoking history = “unknown.” | ||
|
| ||
| Sectionizer | 04 | If a sentence is classified as “text” but contains terms from custom section headings dictionary, assign label “section heading.” |
| If a sentence is classified as “section heading with text” and contains “:”, extract text before “:” to obtain section information. | ||
|
| ||
| Indicator attribute assigner | 26 | If annotated text is identified by MetaMap, assign attribute = “mention.” |
| If annotated text is identified by blood pressure lab value extractor and diastolic >140 or systolic >90, assign indicator attribute = “high BP.” | ||
|
| ||
| Time attribute assigner | 01 | If time attribute assigner assigned class is “continuing,” assign time attributes = “before DCT,” “after DCT,” and “during DCT.” |
We formulated the time attribute assignment problem as a classification problem and assigned one of the three time attributes to each of the risk factors identified by the system. Even though very few features like indicator type, identified token, and section information were used in building a Naïve Bayes model, the classifier performed well, achieving overall microaveraged F-scores of 0.8185, 0.814, and 0.8403 for before DCT, during DCT, and after DCT time attributes, respectively. At an individual risk factor level, the time attribute assigner component performed very well for hypertension. However, the performance for CAD risk factor was observed to be low. We believe this is due to the disease disorder component, which failed to recognize CAD risk factor effectively. The performance of the time attribute assigner component can be further improved by adding context and negation based features [28].
We also noticed that the dataset included numerous abbreviated disease and medication mentions. ASA (acetyl salicylic acid), NTG (nitroglycerin), TNG (trinitroglycerin), DM (diabetes mellitus), and HTN (hypertension) are few examples to mention. Even though a custom abbreviations dictionary was prepared by the authors using the training sets, the test set included several other abbreviations which were not included in the custom dictionary list. Employing a more sophisticated abbreviation handling technique which is not specific to a particular dataset will improve the performance of system overall [29, 30]. Unified medical language system (UMLS) should be used to disambiguate abbreviations. We also found that there were a few misspelled mentions like pravastatin which was misspelled as “pravastain” and obese as “obeise.” Similar to abbreviations custom dictionary, another custom list was developed for misspellings from training sets. However, this approach is not generic and very specific to a given dataset; employing a generic spelling correction is necessary [31].
5. Conclusion
In summary, we described an approach to extract heart disease risk factors in diabetic patients from longitudinal unstructured EHRs. The approach was based on both rules and machine learning techniques. We also described an IE system developed using this approach followed by a comprehensive evaluation of the system. The system was developed using one dataset and might not perform well on other datasets, especially with the rules that were developed. The limitations of the system include issues in lab value extractor and absence of negation and context aware components. In the future, we would like to improve the performance of sectionizer component and also build negation components into the system. We also would like to explore a more sophisticated method to disambiguate abbreviations and handle misspellings effectively. The developed system is available for free at https://github.com/TCRNBioinformatics/HDRFSystem.
Acknowledgments
The authors would like to thank the organizers of 2014 i2b2/UTHealth Shared-Tasks. Deidentified health records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by Grants 2U54LM008748 and 1R13LM01141101 from National Institute of Health (NIH). This study was conducted as part of the Electronic Practice Based Research Network (ePBRN) and Translational Cancer Research Network (TCRN) research programs. ePBRN is funded in part by the School of Public Health & Community Medicine, Ingham Institute for Applied Medical Research, UNSW Medicine, and South West Sydney Local Health District. TCRN is funded by Cancer Institute of New South Wales and Prince of Wales Clinical School, UNSW Medicine.
Disclaimer
The content of this paper is solely the responsibility of the authors and does not necessarily reflect the official views of the funding bodies.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Yach D., Hawkes C., Gould C. L., Hofman K. J. The global burden of chronic diseases: overcoming impediments to prevention and control. The Journal of the American Medical Association. 2004;291(21):2616–2622. doi: 10.1001/jama.291.21.2616. [DOI] [PubMed] [Google Scholar]
- 2.Mackay J., Mensah G. A. The Atlas of Heart Disease and Stroke. WHO; 2004. [Google Scholar]
- 3.Ohno-Machado L., Nadkarni P., Johnson K. Natural language processing: algorithms and tools to extract computable information from EHRs and from the biomedical literature. Journal of the American Medical Informatics Association. 2013;20(5):p. 805. doi: 10.1136/amiajnl-2013-002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonnagaddala J., Dai H.-J., Ray P., Liaw S.-T. Mining electronic health records to guide and support good clinical decision support systems. In: Moon J., Galea M. P., editors. Improving Health Management through Clinical Decision Support Systems. IGI-Global; 2015. [Google Scholar]
- 5.Chang Y.-C., Dai H.-J., Wu J. C.-Y., Chen J.-M., Tsai R. T.-H., Hsu W.-L. TEMPTING system: a hybrid method of rule and machine learning for temporal relation extraction in patient discharge summaries. Journal of Biomedical Informatics. 2013;46(supplement):S54–S62. doi: 10.1016/j.jbi.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Fiszman M., Rosemblat G., Ahlers C. B., Rindflesch T. C. Identifying risk factors for metabolic syndrome in biomedical text. AMIA Annual Symposium Proceedings. 2007;2007:249–253. [PMC free article] [PubMed] [Google Scholar]
- 7.Goryachev S., Kim H., Zeng-Treitler Q. Identification and extraction of family history information from clinical reports. AMIA Annual Symposium Proceedings. 2008;2008:247–251. [PMC free article] [PubMed] [Google Scholar]
- 8.Jonnagaddala J., Liaw S., Rayb P., Kumarc M., Dai H. TMUNSW: identification of disorders and normalization to SNOMED-CT terminology in unstructured clinical notes. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval '15); 2015; Association for Computational Linguistics; [Google Scholar]
- 9.Jonnagaddala J., Liaw S.-T., Ray P., Kumar M., Dai H.-J. HTNSystem: hypertension information extraction system for unstructured clinical notes. In: Cheng S.-M., Day M.-Y., editors. Technologies and Applications of Artificial Intelligence. Vol. 8916. Springer; 2014. pp. 219–227. (Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 10.Kraus S., Blake C., West S. L. Medinfo 2007: Proceedings of the 12th World Congress on Health (Medical) Informatics. Building Sustainable Health Systems. IOS Press; 2007. Information extraction from medical notes. [Google Scholar]
- 11.Savova G. K., Masanz J. J., Ogren P. V., et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H., Stenner S. P., Doan S., Johnson K. B., Waitman L. R., Denny J. C. MedEx: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association. 2010;17(1):19–24. doi: 10.1197/jamia.m3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng Q. T., Goryachev S., Weiss S., Sordo M., Murphy S. N., Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Medical Informatics and Decision Making. 2006;6, article 30 doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrd R. J., Steinhubl S. R., Sun J., Ebadollahi S., Stewart W. F. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. International Journal of Medical Informatics. 2014;83(12):983–992. doi: 10.1016/j.ijmedinf.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonnagaddala J., Liaw S. T., Ray P., Kumar M., Chang N. W., Dai H. J. Coronary artery disease risk assessment from unstructured electronic health records using text mining. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2015.08.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savova G. K., Ogren P. V., Duffy P. H., Buntrock J. D., Chute C. G. Mayo clinic NLP system for patient smoking status identification. Journal of the American Medical Informatics Association. 2008;15(1):25–28. doi: 10.1197/jamia.m2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang N.-W., Dai H.-J., Jonnagaddala J., Chen C.-W., Hsu W.-L. A context-aware approach for progression tracking of medical concepts in electronic medical records. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2015.09.013. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stubbs A., Kotfila C., Xu H., Uzuner O. Practical applications for NLP in clinical research: the 2014 i2b2/UTHealth shared tasks. Proceedings of the i2b2 Shared Task and Workshop Challenges in Natural Language Processing for Clinical Data; 2014. [Google Scholar]
- 19.Stubbs A., Uzuner O. Annotating risk factors for heart disease in clinical narratives for diabetic patients. Journal of Biomedical Informatics. 2015 doi: 10.1016/j.jbi.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aronson A. R., Lang F.-M. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aronson A. R. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the AMIA Symposium. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 22.John G. H., Langley P. Estimating continuous distributions in Bayesian classifiers. Proceedings of the 11th Conference on Uncertainty in Artificial Intelligence; 1995; Morgan Kaufmann Publishers; [Google Scholar]
- 23.Hall M. A. Correlation-Based Feature Selection for Machine Learning. The University of Waikato; 1999. [Google Scholar]
- 24.Dai H.-J., Syed-Abdul S., Chen C.-W., Wu C.-C. Recognition and evaluation of clinical section headings in clinical documents using token-based formulation with conditional random fields. doi: 10.1155/2015/873012. BioMed Research International. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutton C., McCallum A. An introduction to conditional random fields for relational learning. In: Getoor L., Taskar B., editors. Introduction to Statistical Relational Learning. MIT Press; 2006. pp. 93–128. [Google Scholar]
- 26.Yang Y., Liu X. A re-examination of text categorization methods. Proceedings of the 22nd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR '99); August 1999; Berkeley, Calif, USA. ACM; pp. 42–49. [DOI] [Google Scholar]
- 27.Song L., Li D., Zeng X., Wu Y., Guo L., Zou Q. nDNA-prot: identification of DNA-binding proteins based on unbalanced classification. BMC Bioinformatics. 2014;15(1, article 298) doi: 10.1186/1471-2105-15-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harkema H., Dowling J. N., Thornblade T., Chapman W. W. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of Biomedical Informatics. 2009;42(5):839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y., Denny J. C., Rosenbloom S. T., Miller R. A., Giuse D. A., Xu H. A comparative study of current clinical natural language processing systems on handling abbreviations in discharge summaries. AMIA Annual Symposium Proceedings. 2012;2012:997–1003. [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H., Stetson P. D., Friedman C. A study of abbreviations in clinical notes. AMIA Annual Symposium Proceedings. 2007;2007:821–825. [PMC free article] [PubMed] [Google Scholar]
- 31.Patrick J., Sabbagh M., Jain S., Zheng H. Spelling correction in clinical notes with emphasis on first suggestion accuracy. Proceedings of the 2nd Workshop on Building and Evaluating Resources for Biomedical Text Mining; March 2010. [Google Scholar]