Abstract
P53-induced protein with a death domain (PIDD) has been described as primary p53 target gene, induced upon DNA damage. More than 10 years after its discovery, its physiological role in the DNA damage response remains enigmatic, as it seems to be able to execute life–death decisions in vitro, yet genetic ablation in mice failed to reveal an obvious phenotype. Nonetheless, evidence is accumulating that it contributes to the fine-tuning of the DNA-damage response by orchestrating critical processes such as caspase activation or nuclear factor κB translocation and can also exert additional nuclear functions, for example, the modulation of translesion synthesis. In this review, we aim to integrate these observations and propose possible unexplored functions of PIDD.
Keywords: PIDD, Caspase-2, RAIDD, p53, NFκB
INTRODUCTION
Genotoxic stress poses a dangerous threat to the integrity of the genome. It is therefore not surprising that cells have evolved multiple mechanisms to adequately process sustained DNA damage.1 One central mediator of the cellular signalling response after DNA damage is the tumor suppressor p53.2,3 In unstressed conditions, p53 is rapidly ubiquitinated by Mdm2 and subsequently degraded via the proteasomal pathway. However, in response to DNA damage p53 is posttranslationally modified at various residues, for example, by phosphorylation or acetylation that prevents or fine-tunes its ubiquitination and subsequent degradation.4 These events lead to stabilization and translocation of p53 into the nucleus, where it induces the transcription of numerous target genes containing p53-binding sites in their promoter. Those target genes can have such different outcomes as apoptosis (for example, Noxa, Bax and Puma), cell cycle arrest and repair (for example, p21 and GADD45), induction of autophagy (for example, DRAM and Sestrin2) or inhibition of glucose metabolism (for example, TIGAR). The exact mechanisms that regulate the differential expression of specific target genes are not definitely established until now. Transcription and subsequent translation of p53 target genes leads to arrest of the cell cycle and repair of the DNA damage and, if the damage is too severe and can not be repaired properly, cell death mediated mainly via the intrinsic Bcl-2-regulated apoptotic pathway.
One p53 target gene that can modulate the decision between cell cycle arrest and apoptosis in response to genotoxic stress downstream of p53 is the p53-induced protein with a death domain (PIDD). PIDD was first described in the year 20005,6 and contains a leucine-rich repeat domain at the N-terminus followed by two ZU5 domains and a death domain (DD) at the C-terminus. Over the last decade, there have been numerous publications reporting on possible functions for PIDD, none of them entirely convincing, summarized and discussed in this review.
PIDD DISCOVERY
PIDD was first described in the year 2000 by two research groups independently. Telliez et al.6 identified a protein dubbed LRDD (leucine-rich repeats and death domain containing protein) in a bioinformatics search for proteins with a DD, similar to the DD of receptor interacting protein (RIP-1). They already noted that LRDD/PIDD is processed within the cell (see below) and detected interactions with other (see Table 1), but not all, DD containing proteins. The second group5 used differential display of an erythroleukemia cell line transfected with a temperature sensitive p53 mutant (p53ts A135V). One gene upregulated after culturing the cell line at the permissive temperature was named PIDD, a splice variant of which being equivalent to LRDD. The promoter for the PIDD gene contained a bona fide p53-binding site, and the messenger RNA (mRNA) itself is strongly induced after overexpression of functional p53 or γ-irradiation (IR). No induction was observed in several cell types deficient for p53, which additionally had lower basal levels of PIDD mRNA. Overexpression of PIDD reduced cell proliferation and increased apoptosis comparable to p53 overexpression. Furthermore, antisense oligonucleotides targeting PIDD mRNA for degradation led to reduced apoptosis in response to γ-IR.5
Table 1.
PIDD interaction partners
| PIDD fragment | Interaction partner | Identified by | Effect | References |
|---|---|---|---|---|
| PIDD-FL | HSP90 | Co-IP and MS | Interaction enables PIDD cleavage and activation. | Tinel et al.7 |
| PIDD-FL | p23 | Co-IP and MS | ||
| PIDD-C | HSP70 | Co-IP and MS | Unknown | Tinel et al.7 |
| PIDD-C | RIP-1 | Co-IP | The RIP-1-PIDDosome leads to the sumoylation of NEMO and to subsequent activation of NFκB. | Janssens et al.8 |
| PIDD-C | Nemo | Co-IP | ||
| PIDD-C | PCNA | Co-IP and MS | PIDD displaces p21 from PCNA to enable translesion synthesis repair in response to UV irradiation. | Logette et al.9 |
| PIDD-CC | RAIDD | Co-IP | The RAIDD-PIDDosome can activate caspase-2. | Tinel and Tschopp10 |
| PIDD-CC | Caspase-2 | Co-IP | ||
| PIDD-C and PIDD-CC | Nucleolin | GST pulldown, Co-IP and Y-2H | Unknown | Pick et al.11 |
| PIDDa | MADD | Co-IP | Unknown | Telliez et al.6 |
| PIDDa | FADD | Co-IP | Unknown | Telliez et al.6 |
Abbreviations: FADD, Fas-Associated protein with Death Domain; GST, glutathione-S-transferase; IP, immunoprecipitation; MADD, MAP-kinase activating death domain; MS, mass spectrometry; PCNA, proliferating cell nuclear antigen; PIDD, P53-induced protein with a death domain; RIP-1, receptor interacting protein 1.
Unclear at present which fragment constitutes the minimal unit for binding, that is full length, CC- , C- or N-terminal fragment.
PIDD ISOFORMS
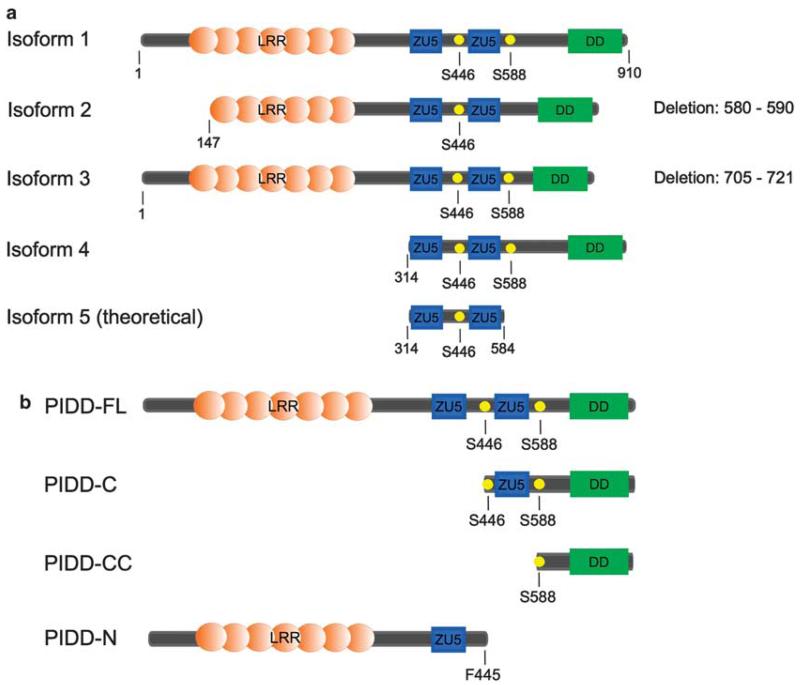
The human PIDD gene is located on chromosome 11 and contains 16 exons. The full length PIDD protein (isoform 1) is made up by 910 amino acids and contains seven N-terminal leucine-rich repeat domains, followed by two ZU5 domains and a C-terminal DD. Several other isoforms have been described:12,11,13 isoform 2 contains a deletion between amino acids 705 and 721; isoform 3, also known as LRDD, lacks the first 147 amino acids and harbors a deletion between amino acids 580 and 590; isoform 4 lacks the first 314 amino acids, and therefore the leucine-rich repeat domain; isoform 5, which has so far only been identified with bioinformatic methods12 and was not further investigated biologically, contains only the two ZU5 domains and is alternatively spliced giving rise to a frameshift and therefore a different C-terminus (Figure 1a).
Figure 1.
Generation of PIDD isoforms by splicing or proteolysis. (a) Currently known PIDD isoforms. Isoform 1 contains seven leucine-rich repeat (LRR) domains at the C-terminus followed by two ZU5 domains and an N-terminal DD. In isoform 2, the first 146 amino acids as well as amino acids 580–590 are deleted. Isoform 3 harbors a deletion of amino acids 705–721. Isoform 4 is composed of the two ZU5 and the DD, whereas isoform 5 has not yet been experimentally validated. (b) PIDD processing products. Full length PIDD-FL is processed at S446 to give rise to PIDD-N and PIDD-C. PIDD-C can be further autoproteolytically cleaved at S588, generating PIDD-CC.
Whereas isoforms 1 and 3 are expressed in all tissues examined, with the exception of skeletal muscle, isoform 2 mRNA is only detectable in liver, pancreas and leukocytes. Isoform 4 is expressed in all tissues including skeletal muscle, but not in the heart.12,13 The lack of reliable antibodies recognizing and discriminating endogenously expressed mouse PIDD isoforms, however, make validation of these findings at the protein level difficult.
Despite initially being described as a gene upregulated in response to DNA damage in a p53-dependent manner, not all genotoxic stressors induce PIDD. Surprisingly, PIDD mRNA levels of isoforms 1 and 2 remain unchanged in response to ultraviolet (UV) irradiation or treatment with etoposide or doxorubicin, whereas isoform 3 was induced after UV and etoposide but downregulated after doxorubicin.12 Of note, basal levels of PIDD protein, presumably isoform 1 or its autoprocessing products (see below), can also be detected in cell lines lacking functional p53, such as in p53−/− HCT116 colon cancer cells, indicating that basal protein expression does not rely on this transcription factor.12
PIDD PROCESSING
PIDD is involved in both pro- and anti-apoptotic pathways (see below), accordingly there should be a means to regulate participation of PIDD in one or the other. Surprisingly, the decision which path to take is imprinted (at least in part) within PIDD itself. PIDD contains two conserved ‘HFS motifs’, also present in the nuclear pore protein Nup98 and ZUD/Unc5cl, a positive regulator of nuclear factor (NF)κB signalling, related to PIDD.14 Similar to Nup98,15 PIDD uses an intramolecular reaction to cleave the peptide bond between the phenylalanine F445 and the serine S446, leading to the generation of the fragments PIDD-N and PIDD-C.7 Cleavage at a second HFS site at serine S588 within PIDD-C leads to the generation of the PIDD-CC fragment (see Figure 1b). PIDD-N contains the leucine-rich repeat domain and one ZU-5 domain, the biological function of this polypeptide remains unknown. The PIDD-C fragment is composed of one ZU-5 domain and the DD, whereas the PIDD-CC fragment only harbors the DD. As the full length PIDD protein is hardly detectable in most cell types, it is believed that it is rapidly processed to PIDD-C. The second cleavage occurs in response to high or prolonged genotoxic stress, therefore leading to accumulation of the PIDD-CC fragment. However, in thymocytes or primary keratinocytes, the CC fragment appears to be the dominant form detectable, implicating that processing of PIDD can be the default program, even in the absence of genotoxic stress.9,16 Another report shows additional processing products upon PIDD or LRDD overexpression; however, the exact nature of those fragments has not been investigated in more detail,11 but N-terminal protein sequencing may help to identify the nature of these PIDD-derived polypeptides.
So how is cleavage regulated? PIDD can be part of a complex containing various molecular chaperones like HSP70, HSP90 or p23, where HSP90 and p23 only interact with full length PIDD in the cytoplasm.17 Accordingly, HSP90 is required for PIDD processing into the PIDD-N and PIDD-C fragments, presumably by facilitating a conformation of PIDD that is favorable for the cleavage reaction. After processing, HSP90 is released from PIDD and the latter can participate in complex formation with various interaction partners (see below). HSP90 seems to be required for both processing to PIDD-C as well as PIDD-CC, although it is at present unclear how exactly processing to PIDD-CC is regulated by HSP90, as both PIDD-CC and HSP90 have been shown not to interact with each other. One possibility would be the involvement of HSP70, which is able to bind both PIDD-C and -CC, for the second processing step, however this has not yet been tested experimentally.
A further mode of regulation of PIDD processing occurs posttranscriptionally via generation of the aforementioned isoforms by differential splicing. Isoform 2 carries a small deletion, which includes the second HFS motif, and is therefore only able to be processed to PIDD-N and PIDD-C, but not to the PIDD-CC fragment.12
PIDD is mainly localized in the cytoplasm, but can also be present in the nucleus.8 In response to DNA-damaging agents or heat shock, PIDD can translocate to the nucleus where it localizes to nucleoli11 and is able to interact with the nucleolar protein nucleolin or chromatin, either directly or via interaction with DNA repair proteins, which remains to be determined.9
THE ROLE OF PIDD IN CASPASE-2 ACTIVATION AND APOPTOSIS
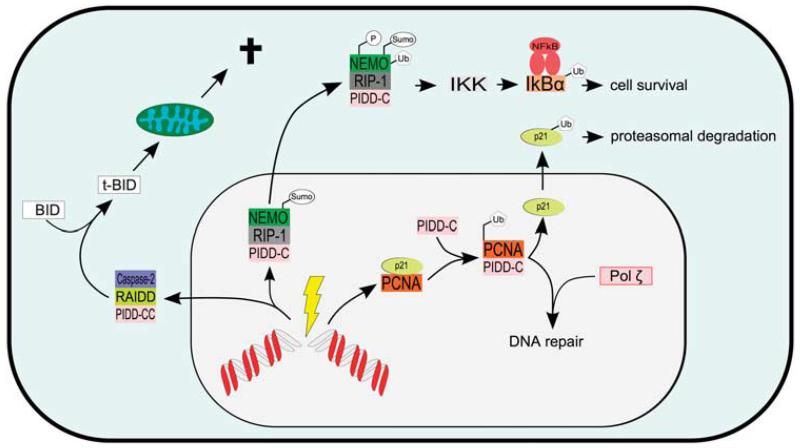
Various binding partners of PIDD have been described (see Table 1), implicating PIDD in different cellular signalling pathways (see Figure 2). The most studied interaction of PIDD is with caspase-2 and RIP-associated ICH1/CED3-homologous protein with death domain (RAIDD) in a complex termed the PIDDosome.10 Caspases are cysteine-driven aspartases involved in several cellular pathways, including cell death, inflammation, migration or differentiation.18 Apoptotic caspases in mammals can be grouped into initiator caspases (8, 9 and 10) and effector caspases (3, 6 and 7). Initiator caspases become activated upon hetero- or homodimerization in high molecular weight complexes formed in response to apoptotic stimuli like the death-inducing signalling complex (DISC) for caspases-8 and -10 or the apoptosome for capsase-9. Once activated and processed within these complexes, the initiator caspases cleave and thereby activate the effector caspases. Of note here, processing/cleavage of initiator caspases can also occur as a secondary event by effector caspase-mediated proteolysis, and is therefore not equivalent to activation. Hence, detection of initiator caspase fragments by western blotting is not indicative for their activation that requires proximity-induced dimerization.19,20
Figure 2.
Cellular responses reported to engage PIDD. PIDD is induced in response to DNA damage in a p53-dependent manner, and depending on the type and severity of the damage is recruited to different complexes. A low amount of DNA damage leads to autoproteolytical cleavage of full length PIDD to the PIDD-C form. This fragment associates with NEMO and receptor interacting protein (RIP-1), thereby facilitating postranslational modifications on NEMO leading to activation of NFκB. After more severe genotoxic stress, PIDD-C is further autoproteolytically cleaved to the PIDD-CC fragment, which can bind to RAIDD and caspase-2, which is subsequently processed and activated. In response to UV-IR, PIDD-C can displace p21 from proliferating cell nuclear antigen (PCNA), thereby facilitating recruitment of the translesion synthesis repair machinery.
Caspase-2 possesses some unique features.21 As it contains a caspase recruitment domain (CARD) in its long prodomain, it can structurally be classified as initiator caspase. However, in contrast to the other initiator caspases, caspase-2 is not able to cleave effector caspases under physiological conditions. In addition, caspase-2 is the only caspase that can be detected within the nucleus, either constitutively, or upon induction of DNA damage, although its precise function there is still under discussion.
As mentioned above, initiator caspases need high molecular weight complexes for activation, and for caspase-2 this complex was found to involve the proteins RAIDD and PIDD.10 Within this complex, RAIDD acts as an adaptor between PIDD and caspase-2, making use of its CARD domain to bind the CARD domain of caspase-2 and binding the DD of PIDD with its own DD. This complex is formed spontaneously upon incubation of cell extracts at 37 °C and leads to processing and activation of caspase-2. Furthermore, RAIDD seems to be the only CARD domain-containing protein capable of binding caspase-2, and RAIDD only interacts with caspase-2 among all CARD domain-containing caspases. However, RAIDD may not interact exclusively with caspase-2 and PIDD, as it was initially found as an interaction partner of RIP-1.22
Several reports describe PIDDosome formation in response to different death-inducing conditions. Transient global cerebral ischemia in hippocampal CA1 pyramidal neurons in rats leads to increased levels of the PIDD-CC fragment, PIDDosome formation and processing of caspase-2.23 Cell death accompanied with ischemia/reperfusion injury in the brain seems to be dependent on PIDD, as in vivo knockdown of PIDD decreases apoptosis as well as caspase-2 processing. Of note, recent data suggest that PIDD, in contrast to caspase-2 and RAIDD, is not required for cell death induced by β-amyloid, a neurotoxic protein involved in Alzheimers disease (C. Troy et al., submitted).
Treatment with the histone deacetylase inhibitor LBH589 was recently shown to trigger caspase-2 processing in adult T cell leukemia/lymphoma cell lines and patient-derived cell samples.24 In this study, knockdown of PIDD did not cause differences in tumor cell survival upon LBH589 treatment. However, both RAIDD and caspase-2 seem to be essential for cell death induction and able to act upstream of caspase-9. Additionally, whereas caspase-2 cleavage was still observed after knockdown of PIDD, it was impaired after knockdown of RAIDD, yet formal evidence that caspase-2 is indeed the most apical caspase activated in this setting was not provided.
Another chemotherapeutic drug shown to induce caspase-2 processing is fluorouracil, a pyrimidine analog. Fluorouracil-induced apoptosis is dependent on p53 and caspase-2, which is in this setting reportedly activated before caspase-3.25 However, the observed caspase-2 processing again seems to be independent of the PIDDosome, as knockdown of PIDD or RAIDD had no influence on apoptosis. Although the exact mechanism is not clear, fluorouracil was shown to upregulate CD95 expression and to induce the formation of a complex involving caspase-2, caspase-8, FADD and CD95 that may orchestrate its activation.26
An alternative cell death pathway engaging caspase-2 has been described based on experiments with zebrafish embryos.27 In this model, caspase-2 is selectively activated in response to DNA damage in p53-mutated zebrafish embryos if checkpoint kinase 1 is inhibited simultaneously. This treatment apparently leads to apoptosis of p53-deficient cells, frequently resisting DNA damage-induced killing, in a caspase-2-dependent manner. It will be interesting to investigate whether PIDD and/or RAIDD are involved in this specific pathway of caspase-2 activation and, if so, how they communicate with the DNA-damage response machinery.
Surprisingly, despite the compelling evidence for a participation of the PIDDosome in cell death induced by various apoptotic substances, both caspase-2, RAIDD as well as PIDD knockout mouse models do not support a critical role of the PIDDosome in developmental or stress-induced apoptosis.16,28,29 Furthermore, activation of caspase-2 is proficient in cells lacking PIDD, indicating that other pathways for caspase-2 activation are likely to exist. In addition, both PIDD- and RAIDD-deficient cells were able to induce the formation of high molecular weight complexes containing caspase-2.16 In contrast, the caspase-2 processing observed was dependent on mitochondrial outer membrane permeabilization and effector caspase activation, placing caspase-2 downstream in the apoptotic cascade. Conflicting data regarding a rate-limiting role for the PIDDosome in cell death triggered by agents disrupting microtubule dynamics, such as vincristine or paclitaxel, has been reported. One study reported cell death in primary low passage mouse embryonic fibroblasts (MEF) that was dependent on all three PIDDosome components,30 yet our own observations in primary as well as SV40-immortalized MEF lacking caspase-2 or PIDD do not support these findings.16 A similar discrepancy appears to exist in MEF transformed with different oncogenes and exposed to γ-IR.16,31 Although the mode of immortalization (E1a/Ras vs SV40 large T antigen) may well have an impact on the DNA damage response, the different findings reported for primary MEF exposed to microtubule poisons remain currently unexplained.
One phenotype that has been reported for caspase-2-deficient mice was an increased sensitivity of oocytes after genotoxic stress.29 However, this sensitivity was not observed in PIDD-deficient oocytes, again indicating a PIDD-independent function for caspase-2,28 whereas the role of RAIDD in oocytes cell death has not been investigated so far.
Overall, these studies suggest that caspase-2 can be a secondary target of other executioner caspases where it might act as part of an amplification loop rather than an essential initiator caspase. However, in certain tumor cell types, frequently lacking p53, caspase-2 can be activated upstream of mitochondria, but this activation does not strictly depend on PIDD. Consistently, the pro-apoptotic function of PIDD may engage other effectors than caspase-2 for cell killing.32
PIDD IN NFκB ACTIVATION AND CELL SURVIVAL
DNA damage not necessarily leads to programmed cell death. Several mechanisms have evolved to allow cells to repair the sustained damage in order to avoid excessive cell death or the elimination of cells that are not irreparably damaged or paramount to maintain organ function, such as stem cells. Next to the DNA repair machinery, an important prosurvival pathway induced by genotoxic stress is the one leading to activation of NFκB.33,34 Although NFκB appears to exert its most prominent role in the immune system,35 where it participates in the host defence by inducing the expression of various mediators of inflammation, NFκB has also been demonstrated to be activated in response to DNA damage, and PIDD seems to be an essential component of this event. The aforementioned PIDD-C fragment can form a complex with NEMO (NFκB essential modulator), a component of the IKK (inhibitor of NFκB kinase) complex and the serine/threonine kinase RIP-1. This protein complex shuttles from the cytoplasm to the nucleus, where NEMO is sumoylated.36 This sumoylation is facilitated by PIDD,8 although at the moment it is not clear whether PIDD directly influences this reaction, recruits ligases for sumoylation or prevents desumoylation that has recently been shown to rely on SENP2.37 Sumoylated NEMO is subsequently bound by ATM (ataxia telangiectasia mutated) and phosphorylated at Serine-85.38 This phosphorylation is required for mono-ubiquitination by cIAP1 and export back to the cytoplasm, as cells harboring a phosphorylation mutant of NEMO are deficient in both ubiquitination and nuclear export, but not sumoylation.39
Recently, several publications show further evidence of a highly complex mode of regulation leading to activation of IKK in response to DNA damage. Immediately after the damage, Poly (ADP-ribose) polymerase-1 (PARP1) becomes activated and poly-ADP-ribosylated, which serves as platform for the recruitment of ATM and protein inhibitor of activated STAT-4 (PIASy).40 In this signalosome, PIASy acts as a sumo ligase for NEMO, leading to the retention of sumoylated NEMO in the nucleus.41 Phosphorylation by ATM bound to NEMO in turn facilitates NEMO export back into the cytoplasm.38 Additionally, ATM is also exported to the cytoplasm in a Ca2+dependent manner, where it binds TRAF6.42 This in turn leads to TRAF6 and Ubc13-dependent generation of K63-linked ubiquitin chains on TRAF6 and ELKS. Ubiquitinated TRAF and ELKS serve as a platform for the recruitment of TAK1 by TAB2/3, which then phosphorylates and activates IKKβ.42-44 At the same time, sumoylated NEMO is mono-ubiquitinated and forms part of a complex with IKKα, IKKβ and ELKS that is essential for IKK activation. This complex formation is dependent on binding of NEMO to a yet to be identified protein. If any of these molecular events also involves PIDD remains to be established.
Studies mentioned above using RNA interference and overexpression as well as our own studies using PIDD-deficient cells show significant defects in NFκB activation in response to genotoxic stress, but not TNF, at least in tissue culture (Bock et al., submitted). This seemingly makes PIDD one of the few genes that are exclusively required for NFκB activation in response to DNA damage, but the analysis of cytokine release upon systemic challenge of mice with IR suggests that PIDD may still have a role in this process (Bock et al., submitted). Surprisingly, as mentioned before, PIDD-deficient cells and mice show no difference in cell survival in response to DNA damage.16,28 This is in stark contrast to other mouse models lacking genes required for NFκB activation, which generally show decreased survival and frequently die in utero.45 Furthermore, different models of DNA damage-induced cancer formation failed to show any signs of PIDD-dependency for onset and severity of disease (Bock et al., submitted, Manzl et al., submitted). Interestingly, also absence of PARP1, limiting DNA damage-dependent activation of NFκB, fails to modulate IR-driven tumorigenesis, but, as loss of PIDD, affected cytokine release in response to acute IR challenge in vivo (Bock et al., submitted). These results suggest that NFκB activation in response to DNA damage is not sufficient to protect cells from apoptosis or increase repair efficacy in premalignant cells and in the end has no significant influence on cancer initiation. We propose it mainly orchestrates cytokine production, as during an immune response, but do not rule out that it may still be important for tumor cell survival, once disease is established. In fact, it is well known that different inflammation-induced cancers are highly dependent on NFκB, mainly by constant secretion of prosurvival and proliferative cytokines as well as production of prosurvival molecules.46 However, direct evidence that DNA damage triggered tumor formation is dependent on NFκB is lacking, because all known components of DNA damage-induced NFκB activation have additional roles, either in the DNA repair response or in DNA damage-independent NFκB activation making a clean genetic dissection of individual functions difficult.
PIDD IN DNA REPAIR, CANCER AND DRUG RESISTANCE
Yet another complex containing PIDD was identified by large-scale IP and mass spectrometry analysis.9 Within this complex, PIDD interacts in the nucleus with components of the replication factor complex 5 (RFC5) and proliferating cell nuclear antigen (PCNA) in response to UV-IR. This leads to detachment of the CDK-inhibitor p21, bound to PCNA, and subsequent degradation of p21 considered as a prerequisite for DNA repair in response to UV-IR. Additionally, PIDD positively influences mono-ubiquitination of PCNA, thereby facilitating recruitment of the low fidelity repair DNA polymerase η to chromatin. How exactly PIDD promotes ubiquitination of PCNA remains unknown, as well as whether the mechanism involves similar posttranslational modifications facilitated by PIDD on NEMO. Of note, PIDD-deficiency sensitized the human keratinocyte cell line HaCaT to UV-C-mediated cell death as well as primary mouse keratinocytes to the lethal effects of the more physiological UV-B irradiation in vivo. It will be interesting to see whether this effect culminates in reduced rates of mutational load and/or altered latency of UV-driven skin cancer.
The PIDD interaction partner caspase-2 is implicated to act as a tumor suppressor in a model of B-cell lymphomas.31 However, until now there are no reports whether PIDD has a similar role in tumorigenesis as caspase-2 or whether the tumor suppressor function of caspase-2 depends on PIDD. Our own studies confirm tumor suppression by caspase-2 in response to aberrant c-Myc expression but this effect was independent of PIDD, at least in this model system (Manzl et al., submitted). The latter observation would be in line with a role in low fidelity DNA repair responses, such as non-homologous end-joining (NHEJ) also triggered in response to oncogenic stress.47 Of note here, PIDD was also found to interact with the catalytic subunit of DNA-activated protein kinase (DNA-PK),9 but the physiological relevance of this observation remains to be reinvestigated.
Expression analysis of various oral squamous cell carcinomas revealed an association between the apoptotic index and PIDD expression, in line with its proposed role in apoptosis.48 Surprisingly, no correlation between p53 status and PIDD expression was found, arguing for p53-independent PIDD expression changes in these tumors. Another, although indirect, cue on the role of PIDD in cancer was shown in cell lines or primary cells derived from patients with high-risk myelodysplastic syndrome or acute myeloid leukemia.49 Those cells are characterized by constitutive NFκB activation that is essential for their survival, as blocking NFκB activation by ATM inhibition leads to apoptosis. In the model proposed by the authors, NEMO is activated by nuclear PIDD and ATM, thereby triggering NFκB signalling.
Furthermore, expression of PIDD was upregulated in the testicular germ cell carcinoma cell line NT2/D1 after cisplatin treatment, suggesting a role for PIDD in the cell death response triggered by this drug, as these tumors are frequently cured by cisplatin-based chemotherapy.50 Consistently, in a mouse model of lung cancer, PIDD was shown to be highly upregulated in tumors in response to in vivo cisplatin treatment.51 Surprisingly, however, PIDD overexpression in turn facilitated resistance to cisplatin treatment. Hence, it will be interesting to see whether high levels of PIDD associate with treatment failure in patients suffering from lung cancer or testicular germ cell carcinoma.
In a subsequent study it was proposed that PIDD-driven drug resistance to cisplatin might be caused by sustained p53 activation and increased p21 levels, noted upon PIDD overexpression, because of induction of a positive feedback loop.52 This loop seems to be initiated by PIDDosome-dependent Mdm2 cleavage at D367, removing the C-terminal RING domain required for its E3-ligase function and ΔC-Mdm2/p53 interaction-dependent stabilization of p53. Although intriguing and in line with a discussed role of caspase-2 in cell cycle arrest,31 a number of inconsistencies remain. For example, why and how does small interfering RNA against PIDD or caspase-2, that shortens the time to p53 degradation because of lack of Mdm2 cleavage, sensitize cells to DNA damage-mediated apoptosis after doxorubicin treatment? Presumably, the argument would be that less effective induction of p21 should sensitize to cell death induction, yet impaired p21 induction was not formally demonstrated in PIDDosome defective cells. A recent study also suggests that knockdown of caspase-2 can negatively affect p21 levels after DNA damage, but this effect did not require the proteolytic activity of caspase-2, nor PIDD or RAIDD, suggesting translational control.53 Nonetheless, sensitization of PIDD-knockdown cells to cell death was also reported after exposure to UV, but there it correlated with impaired p21 degradation.9 Furthermore, doxorubicin-driven cell death in cells wild type for p53 usually depends on p53-driven expression of PUMA and/or Noxa, pro-apoptotic proteins of the Bcl-2 family that should then be activated less effectively as well, culminating in reduced death rates in the absence of the PIDDosome, but the opposite was observed in U2OS cells lacking PIDD.52 Analysis of primary lymphocytes and MEF lacking PIDD or caspase-2, however, did not provide evidence for differences in cell survival in response to several DNA-damaging agents.16,28
FINAL REMARKS
Despite a growing number of research articles, the exact physiological role of PIDD is still poorly defined. Although PIDD was shown to be a part of several different protein complexes mediating various signalling pathways when overexpressed, the absence of a phenotype of PIDD-deficient cells and mice so far excludes an essential role of physiological importance within those pathways. It is therefore likely that PIDD is either highly redundant with other proteins, is only required for fine-tuning of certain pathways, which could not be clearly defined in past work, or, alternatively, may rather be involved in cellular responses secondary to or different from the DNA-damage response. Production of inflammatory cytokines downstream of NFκB activation or participation in the sensing of other types of DNA or damage, for example, in the innate immune response, may be considered as well. Clearly more and refined research is required to define the biological significance of PIDD.
ACKNOWLEDGEMENTS
This review is dedicated to the late Jürg Tschopp, who spearheaded research on PIDD, a great scientist, colleague and mentor. Research on PIDD in our laboratory is supported by grants from the Austrian Science Fund (FWF; Y212-B12 and SFB021), EU-FP6 RTN ‘Apoptrain’, the Tyrolean Science Fund (TWF) and the ‘Krebshilfe-Tirol’.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 3.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 4.Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- 6.Telliez JB, Bean KM, Lin LL. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta. 2000;1478:280–288. doi: 10.1016/s0167-4838(00)00029-7. [DOI] [PubMed] [Google Scholar]
- 7.Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. Embo J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 9.Logette E, Schuepbach-Mallepell S, Eckert MJ, Leo XH, Jaccard B, Manzl C, et al. PIDD orchestrates translesion DNA synthesis in response to UV irradiation. Cell Death Differ. 2011;18:1036–1045. doi: 10.1038/cdd.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 11.Pick R, Badura S, Bosser S, Zornig M. Upon intracellular processing, the C-terminal death domain-containing fragment of the p53-inducible PIDD/LRDD protein translocates to the nucleoli and interacts with nucleolin. Biochem Biophys Res Commun. 2006;349:1329–1338. doi: 10.1016/j.bbrc.2006.08.176. [DOI] [PubMed] [Google Scholar]
- 12.Cuenin S, Tinel A, Janssens S, Tschopp J. p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappaB and caspase-2 in response to genotoxic stress. Oncogene. 2008;27:387–396. doi: 10.1038/sj.onc.1210635. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, Han D, Yang X, Qin B, Ji G, Yu L. PIDD4, a novel PIDD isoform without the LRR domain, can independently induce cell apoptosis in cytoplasm. Biochem Biophys Res Commun. 2011;407:86–91. doi: 10.1016/j.bbrc.2011.02.114. [DOI] [PubMed] [Google Scholar]
- 14.Heinz LX, Rebsamen M, Rossi DC, Staehli F, Schroder K, Quadroni M, et al. The death domain-containing protein Unc5CL is a novel MyD88-independent activator of the pro-inflammatory IRAK signaling cascade. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.147. e-pub ahead of print 9 December 2011; doi:10.1038/cdd.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenblum JS, Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci U S A. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, et al. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinel A, Eckert MJ, Logette E, Lippens S, Janssens S, Jaccard B, et al. Regulation of PIDD auto-proteolysis and activity by the molecular chaperone Hsp90. Cell Death Differ. 2011;18:506–515. doi: 10.1038/cdd.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 19.Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol. 2002;159:739–745. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen’s view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 23.Niizuma K, Endo H, Nito C, Myer DJ, Kim GS, Chan PH. The PIDDosome mediates delayed death of hippocampal CA1 neurons after transient global cerebral ischemia in rats. Proc Natl Acad Sci U S A. 2008;105:16368–16373. doi: 10.1073/pnas.0806222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasegawa H, Yamada Y, Tsukasaki K, Mori N, Tsuruda K, Sasaki D, et al. LBH589, a deacetylase inhibitor, induces apoptosis in adult T-cell leukemia/lymphoma cells via activation of a novel RAIDD-caspase-2 pathway. Leukemia. 2011;25:575–587. doi: 10.1038/leu.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene. 2006;25:5683–5692. doi: 10.1038/sj.onc.1209569. [DOI] [PubMed] [Google Scholar]
- 26.Olsson M, Vakifahmetoglu H, Abruzzo PM, Hogstrand K, Grandien A, Zhivotovsky B. DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene. 2009;28:1949–1959. doi: 10.1038/onc.2009.36. [DOI] [PubMed] [Google Scholar]
- 27.Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim IR, Murakami K, Chen NJ, Saibil SD, Matysiak-Zablocki E, Elford AR, et al. DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis. 2009;14:1039–1049. doi: 10.1007/s10495-009-0375-1. [DOI] [PubMed] [Google Scholar]
- 29.Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho LH, Read SH, Dorstyn L, Lambrusco L, Kumar S. Caspase-2 is required for cell death induced by cytoskeletal disruption. Oncogene. 2008;27:3393–3404. doi: 10.1038/sj.onc.1211005. [DOI] [PubMed] [Google Scholar]
- 31.Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci U S A. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, et al. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci U S A. 2005;102:14314–14320. doi: 10.1073/pnas.0506475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 2006;13:773–784. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto S. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 2011;21:116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 36.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 37.Lee MH, Mabb AM, Gill GB, Yeh ET, Miyamoto S. NF-kappaB induction of the SUMO Protease SENP2: a negative feedback loop to attenuate cell survival response to genotoxic stress. Mol Cell. 2011;43:180–191. doi: 10.1016/j.molcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 39.Jin HS, Lee DH, Kim DH, Chung JH, Lee SJ, Lee TH. cIAP1, cIAP2, and XIAP act cooperatively via non-redundant pathways to regulate genotoxic stress-induced nuclear factor-kappaB activation. Cancer Res. 2009;69:1782–1791. doi: 10.1158/0008-5472.CAN-08-2256. [DOI] [PubMed] [Google Scholar]
- 40.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 41.Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 42.Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, et al. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-kappaB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol. 2011;31:2774–2786. doi: 10.1128/MCB.01139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, et al. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell. 2010;40:75–86. doi: 10.1016/j.molcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 46.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 47.Heinen CD, Schmutte C, Fishel R. DNA repair and tumorigenesis: lessons from hereditary cancer syndromes. Cancer Biol Ther. 2002;1:477–485. doi: 10.4161/cbt.1.5.160. [DOI] [PubMed] [Google Scholar]
- 48.Bradley G, Tremblay S, Irish J, MacMillan C, Baker G, Gullane P, et al. The expression of p53-induced protein with death domain (Pidd) and apoptosis in oral squamous cell carcinoma. Br J Cancer. 2007;96:1425–1432. doi: 10.1038/sj.bjc.6603745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grosjean-Raillard J, Tailler M, Ades L, Perfettini JL, Fabre C, Braun T, et al. ATM mediates constitutive NF-kappaB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2009;28:1099–1109. doi: 10.1038/onc.2008.457. [DOI] [PubMed] [Google Scholar]
- 50.Kerley-Hamilton JS, Pike AM, Li N, DiRenzo J, Spinella MJ. A p53-dominant transcriptional response to cisplatin in testicular germ cell tumor-derived human embryonal carcinoma. Oncogene. 2005;24:6090–6100. doi: 10.1038/sj.onc.1208755. [DOI] [PubMed] [Google Scholar]
- 51.Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, et al. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell. 2011;43:57–71. doi: 10.1016/j.molcel.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sohn D, Budach W, Janicke RU. Caspase-2 is required for DNA damage-induced expression of the CDK inhibitor p21(WAF1/CIP1) Cell Death Differ. 2011;18:1664–1674. doi: 10.1038/cdd.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]