Abstract
Pectinase enzymes are one of the commercially important enzymes having great potential in various industries especially in food industry. Pectinases accounts for 25 % of global food enzymes produced and their market is increasing day by day. Therefore, the exploration of microorganism with novel characteristics has always been the focus of the research. Microorganism dwelling in unique habitat may possess unique characteristics. As such, a pectinase producing fungus Aspergillus niger strain MCAS2 was isolated from soil of Manaslu Conservation Area (MCA), Gorkha, Nepal. The optimum production of pectinase enzyme was observed at 48 h of fermentation. The pectinase enzyme was partially purified by cold acetone treatment followed by Sephadex G-75 gel filtration chromatography. The partially purified enzyme exhibited maximum activity 60 U/mg which was almost 8.5-fold higher than the crude pectinase. The approximate molecular weight of the enzyme was found to be 66 kDa as observed from SDS-PAGE. The pectinase enzyme was active at broad range of temperature (30–70 °C) and pH (6.2–9.2). Optimum temperature and pH of the pectinase enzyme were 50 °C and 8.2 respectively. The enzyme was stable up to 70 °C and about 82 % of pectinase activity was still observed at 100 °C. The thermostable and alkaline nature of this pectinase can meet the demand of various industrial processes like paper and pulp industry, in textile industry, fruit juice industry, plant tissue maceration and wastewater treatment. In addition, the effect of different metal ions on pectinase activity was also studied.
Keywords: Pectinase, Aspergillus niger strain MCAS2, Manaslu Conservation Area (MCA)
Background
Pectinase enzymes commonly represent a group of enzymes that involves in degradation of pectin. Pectins are the high molecular weight polysaccharides, primarily composed of α-1 → 4 linked D galacturonic acid residues with a small number of rhamnose residues in the main chain and arabinose, galactose and xylose on its side chain (Rangarajan et al. 2010). Pectin is found in middle lamella and primary cell wall of higher plants in the form of calcium pectate and magnesium pectate (Favela-Torress et al. 2003). It gives rigidity to plant. The amount of pectic substances present in plant materials varies (0.5–4.0 % of fresh weight plant material) depending upon the source of plant materials (Jayani et al. 2005; Sakai et al. 1993). During the developing stage of plants, the structure of these pectic substances is altered due to the activity of pectinases. These enzymes facilitate the cell wall extension and softening of some tissues during maturation of the part of plants such as fruits.
Enzymes are important biocatalyst for various industrial and biotechnological purposes and produced by microorganisms, animal and plant. They can work in many adverse condition compared to chemical catalyst. As such many microbial enzymes are being used for biotechnological and industrial purposes. Microorganisms are preferred as a source of enzyme because of their short life span, high productivity rate, cost effective, and also free of harmful chemicals that are found in enzyme from plant and animal source (Chaplin and Bucke 1990). Fifty percent of available enzymes are originated from fungi and yeast; 35 % from bacteria, while the remaining 15 % are either of plant or animal origin. Filamentous microorganisms are most widely used for pectinase production (Soares et al. 1999).
Pectinase enzymes are classified into polygalcturonase(PG), pectinesterase(PE), and pectin lyase(PL) based on their mode of action on the substrate (Jayani et al. 2005). Pectinase enzymes are extensively used in an industrial sector especially in food industry i.e. fruit juice extraction, coffee and tea fermentation, oil extraction, improvement of chromaticity and stability of red wine (Jayani et al. 2005). Besides food industry; pectinases are widely used in textile, paper and pulp industries, waste-water treatment (Solbak et al. 2005; Ahlawat et al. 2014). More recently, the enzyme has been used with cellulose enzyme for the production bioethanol from lignocellulosic biomass (Li et al. 2014).
Depending upon the pH requirement for optimum enzymatic activity, pectinase enzyme is also classified into acidic and alkaline pectinase. Acidic pectinases are useful in extraction, clarification and liquefaction of fruit juices (Kaur et al. 2004) and wines (Favela-Torres et al. 2005). Whereas, alkaline pectinases are widely used in the fabric industry, pulp and paper industry and in improving the quality of black tea (Sharma and Satyanarayan 2004; Favela-Torres et al. 2005).
Pectinase enzymes are produced by many microorganism such as bacteria (Takao et al. 2000; Kapoor et al. 2000; Hayashi et al. 1997), fungi (Patil and Dayanand 2006; Huang and Mahoney 1999), yeast (Blanco et al. 1999), and some actinomycete too (Bruhlman et al. 1994). Few pectinase enzymes also have been reported from agro-waste sources like mango peels (Mehrnoush et al. 2012) and Patiya peels (Zohdi and Mehrnoush 2013). Microbial production of pectinases has been extensively studied (Favela-Torres et al. 2006). In practice, the majority of commercially available microbial pectinase comes mainly from fungal source (Singh et al. 1999) of which Aspergillus species, predominate. The pectinase enzyme accounts for about 25 % of worldwide sales of industrial enzymes (Prathyusha and Suneetha 2011). Because of its potential application in biotechnology and industry, the demand for commercial pectinase with high stability and novel characteristics to overcome the limitation of existing commercial pectinase is increasing. As such, researchers have focused their attention towards the exploration of new microbial isolates with desirable biochemical and physicochemical characteristics and a low cost production (Silva et al. 2002; Malvessi and Silveira 2004; Phutela et al. 2005). There are many reports on pectinase enzyme from microorganism isolated from different geographical location and sources especially from agro/industrial waste or spoiled fruits. In the context of Nepal, very few studies have been carried out on the microbial pectinase (Gewali et al. 1994, 1997, 2007). In this regard, we attempted to isolate the fungal strain from highlands because the microorganism dwelling in unique habitat may possess unique characteristics. Considering the importance and application of pectinase enzyme the present study aims to characterize the pectinase enzyme produced by a newly isolated soil fungus, Aspergillus niger strain MCAS2 from the altitude of 3500 m, Manaslu Conservation Area (MCA), Gorkha district of Nepal.
Methods
Isolation of fungal species
The fungi were isolated from soil collected from the altitude of 3500, Manaslu Conservation area, Gorkha by soil dilution plate and soil plate method (Phutela et al. 2005). Morphologically and microscopically different colonies were picked up and subcultured. The spores of pure culture were maintained in potato dextrose agar (PDA) medium in test tubes sealed with parafilm and stored at 4 °C for further use.
Screening of crude pectinase enzymes
The isolated fungal spores were aseptically inoculated into separate flasks containing Potato Dextrose (PD) broth and incubated at 30 °C and 200 rpm for 14 days. The mycelial mass and cell debris were removed by centrifugation at 4000 rpm for 20 min (FISONS centrifuge, Centaur-2, England). The supernatant thus obtained was used as the crude pectinase enzyme. Cup plate method was used for the screening of pectinase enzyme. The supernatant was added on a PDA media supplemented with 0.5 % pectin substrates and incubated at 37 °C f or 48 h. The plates were stained with 0.1 % iodine solution and clear zone formed around the hole was observed.
Production and purification of pectinase enzyme
The fungus was grown on defined liquid media as described by Bhardwaj (2010) containing 1 % pure Pectin, 0.1 % (NH4)2SO4, 0.6 % K2HPO4, 0.2 % KH2PO4, MgSO4·7H2O, pH 6.0 and incubated at 30 °C. After 48 h, the cell debris was removed by using Whatman No. 1 filter paper. The crude enzyme (filtrate) was mixed with three volumes of ice cold acetone and allowed to stand for 15 min in ice cold condition (Rajendran et al. 2011).The entire content was centrifuged at 4000 rpm for 20 min. The supernatant was discarded. The precipitate was dissolved in minimum volume of sodium acetate buffer (0.1 M, pH 4.2) and further subjected to Sephadex G-75 (35 × 1.5 cm, bed volume 12–15 ml/g, Sigma company) as per the standard method (Keller et al. 2006) with certain modifications. The molecular weight and the degree of purity were determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDSPAGE) (Sigma, USA) using the wide range protein molecular weight marker (GeNei, India) according to Laemmli (1970). The protein marker contains myosin (205 kDa), Phosphorylase B (97.4 kDa), Bovine Serum Albumin (66 kDa), Ovalbumin (43 kDa), Carbonic Anhydrase (29 kDa), Soybean Trypsin Inhibitor (20.1 kDa), and Lysozyme (14.3 kDa). Protein bands are visualized using coomassie brilliant blue dye.
Pectinase assay
The Pectinase activity of partially purified enzyme was assayed by using 3, 5-dinitrosalicylic acid (DNS) as described by (Miller 1959). The reaction mixture contained 1 ml of 0.5 % pectin, 0.5 ml sodium acetate buffer (0.1 M, pH 4.2) and 0.5 ml enzyme. The reaction mixture was incubated at 30 °C in water bath (JulaboLabortechnik GMBH, Germany, and Model-TW12) for 10 min. After 10 min, 2 ml of DNS reagent (Sigma, USA) were added and boiled for 15 min. After cooling, the absorbance was measured in spectrophotometer at 575 nm (6715UV/vis. Spectrophotometer, JENWAY, UK). The reaction mixture containing 1 ml of 0.5 % pectin, 1 ml of sodium acetate buffer (0.1 M, pH 4.2) and 2 ml of DNS reagent was used as a control. The standard curve was simultaneously prepared for reducing sugars with galacturonic acid. One unit of pectinase activity was defined as the amount of enzyme needed to catalyze the reaction 1 mg of galacturonic acid per hour under analysis condition.
Determination of protein
The concentration of soluble protein was estimated by Biuret method (Gornall et al. 1949) using bovine serum albumin (BSA) as the standard. Specific activity of an enzyme was defined as enzyme units per mg protein.
Characterization of partially purified pectinase
For the characterization, partially purified pectinase enzyme was used. To determine the optimum temperature of pectinase enzyme, the purified enzyme (5 units) was incubated in sodium acetate buffer pH 4.2 in presence of 0.5 % pectin at different temperature ranging from 30 to 100 °C. To study the effect of pH on enzyme activity, the reaction was carried out using 5 units enzyme and 0.5 % pectin in different buffer system: Sodium acetate buffer (3.2–6.2), sodium phosphate buffer (7.2–8.2), and 0.1 M sodium borate (9.2–10.2) at 30 °C for 10 min. In the same way, the effect of substrate concentration on pectinase activity was carried out by using 0.5–3.0 % pectin in the reaction mixture. The influence of incubation time was measured by incubating the reaction mixture containing 0.5 % pectin in sodium acetate buffer pH 4.2, 5 units enzyme at fixed 30 °C over the period of 1 h at the interval of 10 min. The effects of metal ions on the pectinase activity were determined in the presence of 5 mM Na+, Mg+2, Pb+2, Ba+2, Zn+2, Ca+2, Cd+2 and Fe+3 in the reaction mixture.
Result
Screening of isolates
Ten different fungi were isolated from soil samples collected from Manaslu Conservation Area, Gorkha, by using spread plate and soil dilution method. Out of ten isolate, only eight isolates were found pectinase producer. Among them, strain MCAS2 showed the highest pectinase activity and thus selected for the production and characterization of pectinase enzyme. Two isolates, MCAS1 and MCAS6 did not show any pectinase activity (Fig. 1). Based on the colony morphology, microscopic observation and ITS nucleotide sequence homology, the isolate MCAS2 was identified as Aspergillus niger strain MCAS2. The Gene Bank accession no for ITS sequence of Aspergillus niger strain MCAS2 is KM103363.
Fig. 1.
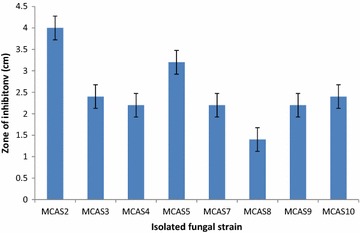
Pectinase activity of different isolated fungal strains
Production and purification of enzyme
The production of pectinase was carried out in defined liquid medium. The crude enzyme was harvested from 48 h old culture and acetone precipitated which was followed by gel filtration on Sephadex G-75 column chromatography. Table 1 shows purification summary for pectinase enzyme. It showed that the enzyme was purified 1.2-fold with specific activity of 8.33 U/mg by using cold acetone purification method and 8.5-folds with specific activity of 60 U/mg by using Sephadex method. The apparent molecular weight of pectinase enzyme was found to be 66 kDa as revealed form SDS-PGE analysis (Fig. 2).
Table 1.
Overall scheme of purification and activity of pectinase from Aspergillus niger strain MCAS2
| Phase | Volume (ml) | Total activity (U) | Protein (mg/ml) | Protein total (mg) | Specific activity (U/mg) | Yield (%) | Purification fold (X) |
|---|---|---|---|---|---|---|---|
| Crude extract | 100 | 370 | 0.525 | 52.5 | 7.05 | 100 | 1 |
| Acetone ppt. | 7 | 70 | 1.2 | 8.4 | 8.33 | 19 | 1.2 |
| Sephadex G-75 | 3 | 9 | 0.05 | 0.15 | 60 | 16 | 8.5 |
Fig. 2.
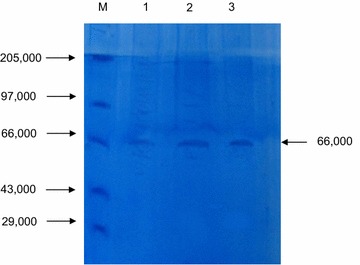
SDS-PAGE of Pectinase enzyme from isolated Aspergillus niger strain MCAS2. Lane M protein marker in Dalton, Lane 1 crude pectinase, Lane 2 partially purified pectinase and Lane 3 Sephadex G-75 purified pectinase
Characterization of pectinase
Effect of temperature and pH
The enzyme activity was highly influenced by the factors like temperature and pH. In this study, pectinase was found to be highly active at the temperature range of 50–70 °C with the temperature optimum at 50 °C. A slight decrease in the pectinase activity was observed with the increase in temperature. About 82 % of pectinase activity was still retained at 100 °C (Fig. 3). This shows that the pectinase enzyme from Aspergillus niger strain MCAS2 was thermostable. In the same way, pectinase enzyme was found active in the pH range 6.2–9.2 with the optimum pH 8.2. This result indicates that the pectinase enzyme is alkaline in nature (Fig. 3).
Fig. 3.
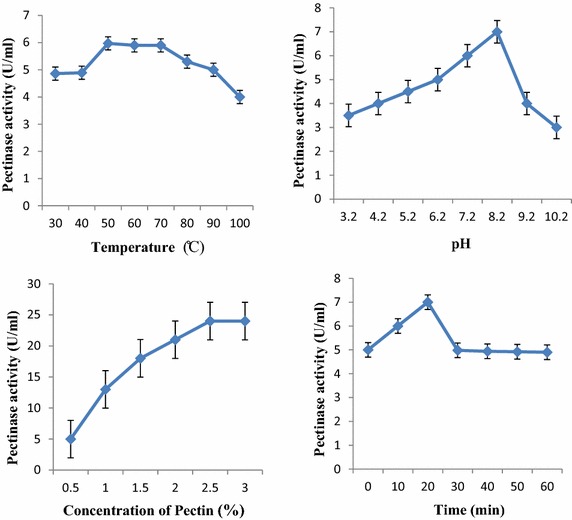
Effect of temperature, pH, pectin concentration, and incubation time on pectinase activity
Effect of substrate concentration and incubation time
The pectinase enzyme was found to be optimally active when the assay was carried out in presence of 2.5 % pectin. Further increase in substrate did not show any significant effect on pectinase activity. Similarly, the optimum pectinase activity was observed when the reaction mixture was incubated at 30 °C for 20 min. Although the pectinase activity decreased with increase in incubation time, 74.4 % of pectinase activity was still observed after incubating for 60 min (Fig. 3).
Effect of metal ions
The effects of various metal ions (5 mM) on the pectinase activity were investigated at pH 4.2 and 30 °C. The enzyme activity without metal ion was taken as 100 % and relative activity was determined in the presence of metal ions. Metal ions have found to influence the activity of pectinase from Aspergillus niger strain MCAS2. Metal ions viz. Cd+2, Mg+2, Ba+2 and Fe+3 were found to stimulate the pectinase activity while Na+, Pb+2, Zn+2, and Ca+2 inhibited the pectinase activity. The maximum inhibition was observed by calcium ion followed by sodium ion as shown in Table 2.
Table 2.
Effect of different metal ions on pectinase activity
| Metal ions | Activity (%) |
|---|---|
| None | 100 |
| Cd+2 | 147 |
| Mg+2 | 134 |
| Ba+2 | 124 |
| Fe+3 | 113 |
| Pb+2 | 89 |
| Zn+2 | 86 |
| Na+2 | 84 |
| Ca+2 | 59 |
Discussion
Pectinase enzyme has been the focus of research for many years due to the potential and wide applications in various industrial processes. An increasing demand of pectinase has rendered the need of exploring microbial strains capable of producing novel pectinases with improved activities. In this study, an attempt has been made to isolate the fungal strains producing pectinase enzyme and to characterize the pectinase enzyme. Out of ten isolated fungal strain, only eight of them were found to be pectinase producer. Among them, strain MCAS2, being the highest producer of pectinase enzyme, was selected for the characterization of pectinase enzyme.
Cultivation of Aspergillus niger strain MCAS2 in a liquid medium containing 1 % pectin as a carbon source resulted into maximum production of pectinase at 48 h. The pectinase production time is less than pectinase from other reported fungal strains. Oyeleke et al. (2012) has reported that the production of pectinase after 96 h from Aspergillus niger. The pectinase from Aspergillus flavus was best produced after 120 h of incubation (Gewali et al. 2007). However, the result was in agreement with the pectinase enzyme produced from Bacillus subtilis isolated from soil. It produced the pectinase enzyme after 48 h of incubation (Tripathi et al. 2014). Optimum production of pectinase depends upon the microbial sources as well as the composition of culture media. For the industrial process, shorter fermentation cycle is beneficial from the commercial point of view. Hence, this isolate could have potential biotechnological as well as other industrial application.
Crude pectinase enzyme obtained by the cell culture of Aspergillus niger MCAS2 was partially purified by the cold acetone treatment and followed by Sephadex G-75 column chromatography. The activity of partially purified enzyme was increased by 8.5-fold. The molecular mass of isolated pectinase enzyme was 66 kDa as revealed from sodium dodecyl sulphate(SDS)-PAGE. The result is in agreement with the other fungal pectinases such as 63 kDa from Penicillium frequentans (Barense et al. 2001), 68 kDa from Sclerotinia sclerotiorum (Riou et al. 1992), 60 kDa from Cochliobolus carbonum (Scott-Craig et al. 1998), 78 kDa from Aspergillus tubingensis (Kester et al. 1996), and 74 kDa from Fusarium oxysporum sps. (Di Pietro and Roncero 1996). De Vries and Visser (2001) have reported the molecular mass of pectinase in a range of 35–80 kDa. This range is in agreement with the apparent molecular mass of the pectinase enzyme produced by Aspergillus niger strain MCAS2.
The partially purified pectinase from Aspergillus niger strain MCAS2 is significantly active over the broad range of temperature range 50–70 °C and pH 6.2–9.2 with optimum temperature and pH 50 °C and 8.2 respectively. These results indicate that the pectinase enzyme was thermostable and alkaline in nature. These properties make the enzyme advantageous in various industrial processes that employ high temperature and pH. The fungal pectinase enzymes, especially from Aspergillus species, are active in the range of 30–50 °C. Pectinase from Aspergillus species have been reported to inactive due to denaturation at temperature above 50 °C (Galiotou-Panayotou et al. 1997). However, the result obtained is in agreement with the pectinase enzyme produced by Penicillium chrysogenum, and a thermophilic fungus Aspergillus fumigatus which showed that the optimum pectinase activity at 50 °C (Banu et al. 2010; Phutela et al. 2005). Similarly pectinase enzyme from Aspergillus niger URM4645 is active at 50–80 °C and Thermomucorindicae_seudaticae N31 at 60 °C (Maciel et al. 2011; Martin et al. 2010).
The pectinase enzyme retained 60–90 % activity over the range 6.2–9.2. The similar pectinase enzyme with similar pH profiling have been reported from Aspergillus niger URM4645 over the wide pH range (3.5–11.0) (Maciel et al. 2011). Contrary to the result obtained in this study, the pectinase enzyme from Aspergillus flavus, Aspergillus niger, and Botrytis cinerea were active at pH 4.2 (Gewali et al. 1994, 1997, 2007). Fahmy et al. (2008) reported that the pectinase from Aspergillus niger has optimum activity at pH 5.0. Similarly, Pectinase from Penicillium viridicatum was optimally active at pH ranging from 5.0 to 8.5 (Silva et al. 2002). The fungal pectinase especially from Aspergillus species are active in pH range of 4.0–6.0 i.e. the pectinase enzyme is acidic in nature (Tutobello and Mill 1961). The previous reports have shown that alkaline pectinase are mostly produced by bacteria especially Bacillus species (Kumar and Sharma 2012; Sonnotel and Nigam 2002; Sonia Ahlawat et al. 2008). The pectinase enzyme from Bacillus pumilus dcsr1, Bacillus stearothermophilus, Paenibacillus xylanolyticusm, Bacillus halondurans M29 were shown to be active at high temperature and pH (Sharma and Satyanarayan 2006; Karbassi and Vaughn 1980; Giacobbe et al. 2014; Mei et al. 2013). This thermostability may be attributed to the cystein residue present in the amino acid sequence as observed in pectinase from Bacillus licheniformis (Singh et al. 2012). The presence of cystein residue in not only provides the thermostability due to the formation of disulfide bond but also due to strong hydrophobic effect (You et al. 2010). Such residue may be present in the pectinase enzyme isolated from Aspergillus niger strain MCAS2 that confers the thermostability to the enzyme. Temperature and pH stability are the most important characteristics of a biocatalyst for their use in industrial applications. So, this fungal pectinase could be the best alternative to bacterial thermostable alkaliphilic pectinase and may find its use in degumming and retting of fiber crops, pulp and paper industries, and pretreatment of pectic waste water from fruit juice industries.
Incubation of pectinase with different concentration of pectin indicated that the enzyme showed maximum activity of 24 U/ml at 2.5 % pectin concentration. Further increase in pectin concentration had no effect in pectinase activity. At high substrate concentration all the enzymes are occupied with substrate so no more substrate can be catalyzed. The maximum enzyme activity was obtained at incubation time of 20 min which can be utilized in the industrial sector for quick product recovery. The metal ions play an important role on activity and stability of enzyme. The pectinase enzyme was strongly inhibited by Ca+2 and fairly by Na+, Pb+2, and Zn+2. This correlates with the finding form Yadav et al. that Ca+2, Zn+2 and Mg+2 inhibited the pectinase activity of Aspergillus flavus MTCC7589 at 1.0 mmol/l concentration (Yadav et al. 2008). However, the result is in contrary to pectinase enzyme from Bacillus sp. KSMp576, Penicillium chrysogenum, and mango peel that Ca+2 ions found to enhance the pectinase activity (Sonnotel and Nigam 2002; Yadav et al. 2008; Mehrnoush et al. 2012). The isolated pectinase activity was highly activated by Cd+2 (more than 50 %) followed by Mg+2, Ba+2 and Fe+3. Whereas, the pectinase activity from Penicillium chrysogenum was inhibited by 5 mM Mg+2 and Ba+2 ions (Banu et al. 2010). This suggests that the requirement of metal ions for the pectinase activity vary depending upon their sources.
Conclusion
Thermostable and alkaline pectinase from Aspergillus niger strain MCAS2 was isolated from Manaslu Conservation Area, Gorkha district, Nepal. This pectinase enzyme with high stability at temperature and pH can be used for various industrial applications including extraction and clarification of fruit juices, processing of cotton fabrics in textile industries, waste water treatment and maceration of tea leaves.
Authors’ contributions
BPK performed the laboratory work. JM and SS conceptualized this study. BPK and JM drafted the manuscript. JM, TBB, and SS provided the guidance and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Nepal Academy of Science and Technology (NAST) for providing all necessary funds and laboratory facilities to carry out this research.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- ITS
internal transcribed spacer
- MCA
Manaslu Conservation Area
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- kDA
kilo dalton
Contributor Information
Bhim Prakash Khatri, Email: bhimbiotech@gmail.com.
Tribikram Bhattarai, Email: drtbb@mail.com.np.
Sangita Shrestha, Email: sangita7@hotmail.com.
Jyoti Maharjan, Phone: 977-1-5547714, FAX: 977-1-5547714, Email: jyotimaharjan@yahoo.com.
References
- Ahlawat S, Mandhan RP, Dhiman SS, Kumar R, Sharma J. Potential application of alkaline pectinase from Bacilllus subtilis SS in pulp and paper industry. Appl Biochem Biotechnol. 2008;149(3):287–293. doi: 10.1007/s12010-007-8096-9. [DOI] [PubMed] [Google Scholar]
- Ahlawat S, Battan B, Dhiman SS, Sharma J, Mandhan RP. Production of thermostable pectinase and xylanase for their potential application in bleaching of kraft pulp. J Ind Microbiol Biotechnol. 2014;34(12):763–770. doi: 10.1007/s10295-007-0251-3. [DOI] [PubMed] [Google Scholar]
- Banu AR, Devi MK, Ganaprabhal GR, Pradep BV, Palaniswamy M. Production and characterization of pectinase from Penicillium chrysogenum. Indian J Sci Technol. 2010;3(4):377–381. [Google Scholar]
- Barense RI, Chellegatti MASC, Fonseca MJV, Said S. Partial purification and characterization of exopolygalacturonase II and III of Penicillium frequentans. Braz J Microbiol. 2001;32:327–330. doi: 10.1590/S1517-83822001000400014. [DOI] [Google Scholar]
- Bhardwaj V (2010) Exploitation of micro-organisms for isolation and screening of pectinase from environment. In: Globelics 2010 8th International Conference, University of Malaya, Kuala Lumpur, Malaysia, 1–3 November 2010
- Blanco P, Sieiro C, Villa TG. Production of pectic enzymes in yeast. FEMS Microbiol Lett. 1999;175:1–9. doi: 10.1111/j.1574-6968.1999.tb13595.x. [DOI] [PubMed] [Google Scholar]
- Bruhlman F, Kim KS, Zimmerman W, Fiechter A. Pectinolytic enzymes from actinomycetes for the degumming of ramie bast fibers. Appl Environ Microbiol. 1994;60:2107–2112. doi: 10.1128/aem.60.6.2107-2112.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin MF, Bucke C. Enzyme technology. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- De Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 2001;65(4):497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A, Roncero MI. Purification and characterization of an exo-polygalacturonase from the tomato vascular wilt pathogen Fusarium oxysporum f.sp. lycopersici. FEMS Microbiol Lett. 1996;145(2):295–299. doi: 10.1111/j.1574-6968.1996.tb08592.x. [DOI] [PubMed] [Google Scholar]
- Fahmy AS, El-beih FM, Mohamed SA, Abdel-Gany SS, Abd-Elbaky EA. Characterization of an exopolygalacturonase from Aspergillus niger. Appl Biochem Biotechnol. 2008;149:205–217. doi: 10.1007/s12010-007-8107-x. [DOI] [PubMed] [Google Scholar]
- Favela-Torress E, Aguilar C, Esquivel-Contreras CJ, Gustavo GV. Enzyme technology. Delhi: Asiatech Publisher Inc.; 2003. Pectinase; pp. 273–296. [Google Scholar]
- Favela-Torres E, Aguiler CN,Contrara-Equivel JC, Viniegra-Gonzalez G (2005) Pectinase. In: Pandey A, Webb C, Soccol CR, Larroche C (eds) Enzyme technology. Asiatech Publishers Inc., New Delhi, India, pp 265–267
- Favela-Torres E, Sepulveda TV, Gonzalez GV. Production of hydrolytic depolymerising pectinases. Food Technol Biotechnol. 2006;44:221–227. [Google Scholar]
- Galiotou-Panayotou MPR, Kapantai M, Kalantzi O. Growth conditions of Aspergillus sp. ATHUM-3482 for polygalacturonase production. Appl Microbiol Biotechnol. 1997;47:425–429. doi: 10.1007/s002530050951. [DOI] [PubMed] [Google Scholar]
- Gewali MB, Thapa S, Baral A, Bhattarai S, Shrestha S (1994) Studies on polygalacturonase from Botrytis cinerea. In: Proceeding of Second National Conference on Science and Technology, Kathmandu
- Gewali MB, Manandhar S, Thapa S, Banskota A, Shrestha JK. Study of natural and microbial inhibitors of pectinolytic enzymes from Botrytis Cinerea and Aspergillus niger. Sci World. 1997;16:23–26. [Google Scholar]
- Gewali MB, Maharjan J, Thapa S, Shrestha JK. Studies on polygalacturonase from Aspergillus flavus. Sci World. 2007;5(5):19–22. [Google Scholar]
- Giacobbe S, Pepe O, Ventorino V, Birolo L, Viciguerra R, Faraco V. Identification and characterization of a pectinolytic enzyme form Paenibacillus xylanolyticus. Bioresources. 2014;9(3):4873–4887. doi: 10.15376/biores.9.3.4873-4887. [DOI] [Google Scholar]
- Gornall AG, Bardwill CJ, David MM. Determination of serum proteins by means of biuret reagent. J Biol Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- Hayashi K, Inoue Y, Shiga M, Sato S, Takano R, Hirayae K, HibiT Hara S. Pectinolytic enzymes from Pseudomonas marginalis MAFF 03-01173. Phytochemistry. 1997;45(7):1359–1363. doi: 10.1016/S0031-9422(97)00191-X. [DOI] [PubMed] [Google Scholar]
- Huang LK, Mahoney RR. Purification and characterization of an endo-polygalacturonase from Verticillum alboatrum. J Appl Microbiol. 1999;86:145–146. doi: 10.1046/j.1365-2672.1999.00645.x. [DOI] [Google Scholar]
- Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: a review. Process Biochem. 2005;40:2931–2944. doi: 10.1016/j.procbio.2005.03.026. [DOI] [Google Scholar]
- Kapoor M, Beg QK, Bhushan B, Dadhich KS, Hoondal GS. Production and partial purification and characterization of a thermo-alkali stable polygalacturonase from Bacillus sp. MG-cp-2. Process Biochem. 2000;36:467–473. doi: 10.1016/S0032-9592(00)00238-7. [DOI] [Google Scholar]
- Karbassi A, Vaughn RH. Purification and properties of polygalacturonic acid trans-eliminase from Bacillus stearothermophilus. Can J Microbiol. 1980;26(2):377–384. doi: 10.1139/m80-061. [DOI] [PubMed] [Google Scholar]
- Kaur G, Sarkar BC, Sharma HK. Production, characterization and application of a thermostable polygalcturonase of a thermophilic mould Sporotricumthermophile. Bioresour Technol. 2004;94:239–243. doi: 10.1016/j.biortech.2003.05.003. [DOI] [PubMed] [Google Scholar]
- Keller V, Jen JJ, Brunner RJ. Purification of commercial pectinase by hydrophobic chromatography. J Food Sci. 2006;47:2076–2077. doi: 10.1111/j.1365-2621.1982.tb12954.x. [DOI] [Google Scholar]
- Kester HCM, Kusters-Van-Someren MA, Müller Y, Visser J. Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. Eur J Biochem. 1996;240:738–746. doi: 10.1111/j.1432-1033.1996.0738h.x. [DOI] [PubMed] [Google Scholar]
- Kumar A, Sharma R. Production of alkaline pectinase by bacteria (Cocci sps) isolated from decomposting fruit materials. J Phytol. 2012;4(1):1–5. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li J, Zhou P, Liu H, Lin J, Gong Y, Xiao W, Liu Z. Monosaccharides and ethanol production from superfine ground sugarcane bagasse using enzyme cocktail. Bioresources. 2014;9(2):2529–2540. [Google Scholar]
- Maciel MDC, Herculano PN, Porto TS, Teixeira MFS, Moreira KA, Souza-Motta CM. Production and partial characterization of pectinases from forage palm by Aspergillus niger URM4645. Afr J Biotechnol. 2011;10(13):2469–2475. [Google Scholar]
- Malvessi E, Silveira MM. Influence of medium composition and pH on the production of polygalacturonases by Aspergillus oryzae. Braz Arch Biol Technol. 2004;47:693–702. doi: 10.1590/S1516-89132004000500004. [DOI] [Google Scholar]
- Martin N, Guez MAU, Sette LD, Da Silva R, Gomes E. Pectinase production by a Brazilian thermophilic fungus Thermomucorindicae_seudaticae N13 in solid-state and submerged fermentation. Mikrobiologiia. 2010;79(3):321–328. [PubMed] [Google Scholar]
- Mehrnoush A, Mustafa S, Yazid AMM. Characterization of pectinase from mango (Mangiferaindica cv. Chokanan) peel. J Food Agric Environ. 2012;10(1):85–88. [Google Scholar]
- Mei Y, Chen Y, Zhai R, Liu Y. Cloning, purification and biochemical properties of a thermostable pectinase from Bacillus halodurans M29. J Mol Catal B Enzym. 2013;94:77–81. doi: 10.1016/j.molcatb.2013.05.004. [DOI] [Google Scholar]
- Miller LG. Use of dinitrosalicylic acid reagent for the determination of reducing sugar. Anal Chem. 1959;31:426–473. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Oyeleke SB, Oyewole OA, Egwim EC, Dauda BEN, Ibeh EN. Cellulase and pectinase production potentials of Aspergillus niger isolated from corn cob. Bayero J Pure Appl Sci. 2012;5(1):78–83. [Google Scholar]
- Patil R, Dayanand A. Exploration of regional agrowastes for the production of pectinase by Aspergillus niger. Food Technol Biotechnol. 2006;44:289–292. [Google Scholar]
- Phutela U, Dhuna V, Sandhu S, Chadha BS. Pectinase and polygalacturonase production by a thermophilic Aspergillus fumigatus isolated from decomposting orange peels. Braz J Microbiol. 2005;36:63–69. doi: 10.1590/S1517-83822005000100013. [DOI] [Google Scholar]
- Prathyusha K, Suneetha V. Bacterial pectinases and their potent biotechnological application in fruit processing/juice production industry: a review. J Phytol. 2011;3(6):6–19. [Google Scholar]
- Rajendran R, Sundaram KS, Radhai R, Rajapriya P, Balakumar C. Production and optimization of fungal pectinase from Fusarium sp. Int J Curren Res. 2011;3(4):254–258. [Google Scholar]
- Rangarajan V, Rajasekharan M, Ravichandran R, Sriganesh K, Vaitheeswaran V. Pectinase production from orange peel extract and dried orange peel solid as substrate using Aspergillus niger. Int J Biotechnol Biochem. 2010;6(3):445–453. [Google Scholar]
- Riou C, Freyssinet G, Fevre M. Purification and characterization of extracellular pectinolytic enzymes produced by Sclerotinia sclerotiorum. Appl Environ Microbiol. 1992;58(2):578–583. doi: 10.1128/aem.58.2.578-583.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Sakamoto T, Hallaert J, Vandamme J. Pectin, pectinase and protopectinase: production, properties and applications. Adv Appl Microbiol. 1993;39:213–294. doi: 10.1016/S0065-2164(08)70597-5. [DOI] [PubMed] [Google Scholar]
- Scott-Craig JS, Cheng YQ, Cervone F, de Lorenzo G, Pitkin JW, Walton JD. Targeted mutants of Cochliobolus carbonum lacking the two major extracellular polygalacturonases. Appl Environ Microbiol. 1998;64:1497–1503. doi: 10.1128/aem.64.4.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DC, Satyanarayan T. Production and application of pectinolytic enzymes of Sporotrichum thermopohile and Bacillus pumilus. In: Reddy MS, Khanna S, editors. Biotechnolgical approaches for sustainable development. India: Allied Publishers; 2004. pp. 164–169. [Google Scholar]
- Sharma DC, Satyanarayan T. A marked enhancement in the production of a highly alkaline and thermostable pectinase by Bacillus pumilus dcsr1 in submerged fermentation by using statistical methods. Bioresour Technol. 2006;9(5):727–733. doi: 10.1016/j.biortech.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Silva D, Martins ESD, Silva RD, Gomes E. Pectinase production by Penicillium viridicatum RFC3 by solid state fermentation using agricultural wastes and agro-industrial by-products. Braz J Microbiol. 2002;33:318–324. [Google Scholar]
- Singh SA, Ramakrishna M, Rao AGA. Optimization of downstream processing parameters for the recovery of pectinase from the fermented broth of Aspergillus carbonarious. Process Biochem. 1999;35:411–417. doi: 10.1016/S0032-9592(99)00089-8. [DOI] [Google Scholar]
- Singh R, Dhawan S, Singh K, Kaur J. Cloning, expression and characterization of a metagenome derived thermoactive/thermostable pectinase. Mol Biol Rep. 2012;39:8353–8361. doi: 10.1007/s11033-012-1685-x. [DOI] [PubMed] [Google Scholar]
- Soares MNC, Silva Da, Gomes R. Screening of bacterial strains for pectinolytic activity. Characterization of polygalacturonase produced by Bacillus sp. Rev Microbiol. 1999;30:299–303. [Google Scholar]
- Solbak AI, Richardson TH, McCann RT, Kline KA, Bartnek F, Tomlinson G, Tan X, Parra-Gessert L, Frey GJ, Podar M, Luginbuhl P, Gray KA, Mathur EJ, Robertson DE, Burk MJ, Hazlewood GP, Short JM, Kerovuo J. Discovery of pectin degrading enzymes and directed evolution of a novel pectatelyase for processing cotton fabric. J Biol Chem. 2005;280:9431–9438. doi: 10.1074/jbc.M411838200. [DOI] [PubMed] [Google Scholar]
- Sonnotel O, Nigam P. Pectinolytic activity of bacteria isolated form soil and two fungal strains during submerged fermentation. World J Microbiol Biotechnol. 2002;18:835–839. doi: 10.1023/A:1021209123641. [DOI] [Google Scholar]
- Takao M, Nakaniwa T, Yoshikawa K, Terashita T, Sakai T. Purification and characterization of thermostable pectatelyase with protopectinase activity from thermophilic Bacillus sp. TS 47. Biosci Biotechnol Biochem. 2000;64:2360–2367. doi: 10.1271/bbb.64.2360. [DOI] [PubMed] [Google Scholar]
- Tripathi GD, Javed Z, Sushma, Singh A (2014) Pectinase production and purification from Bacillus subtilis isolated from soil. Adv Appl Sci Res 5(1):103–105
- Tutobello R, Mill PJ. Thepectic enzymes of Aspergillus niger. Part II. Endopolygalacturonase. Biochem J. 1961;79:57–64. doi: 10.1042/bj0790051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Yadav PK, Yadav D, Yadav KDS. Purification and characterization of an alkaline pectin lyase from Aspergillus flavus. Process Biochem. 2008;43:547–552. doi: 10.1016/j.procbio.2008.01.015. [DOI] [Google Scholar]
- You C, Huang Q, Xue H, Xu Y, Lu H. Potential hydrophobic interaction between two cysteines in interior hydrophobic region improve thermostability of a family 11 xylanase from Neocallimastix patriciarum. Biotechnol Bioeng. 2010;105:861–870. doi: 10.1002/bit.22623. [DOI] [PubMed] [Google Scholar]
- Zohdi NK, Mehrnoush A. Optimization of extraction of novel pectinase enzyme discovered in red Pitaya (Hylocereuspolyrhizus) peel. Molecule. 2013;18(11):14366–14380. doi: 10.3390/molecules181114366. [DOI] [PMC free article] [PubMed] [Google Scholar]


