Abstract
Purpose: The aim of this study was to investigate the association between osteosarcoma (OS) and Fanconi anemia (FA) related pathways and the molecular mechanisms. Methods: siRNA for Fanconi anemia complementation group D2 (FANCD2) was constructed and transfected into the osteosarcoma cell line MG-63 cells. Expression of TP53INP1, p53, p21, caspase-9, and caspase-3 mRNA in MG-63 cells were examined by real-time fluorescence quantitative PCR, and the protein levels were also determined by western blot. Results: After silence of the FANCD2 gene in MG-63 cells, cell proliferation was inhibited, cell cycle was arrested and cell apoptosis was induced. The apoptosis was mediated by the p53 signaling pathway. After FANCD2 expression was inhibited, TP53INP1 gene expression was up-regulated, phosphorylation of p53 was promoted and the p21 protein was activated, leading to cell cycle arrested in G1, finally resulted in caspase-dependent cell apoptosis. Conclusions: Inhibition of FANCD2 gene expression can induce apoptosis of osteosarcoma cells, which indicated that FANCD2 played an important role in the development of osteosarcoma and it might be a potential target for treatment of osteosarcoma.
Keywords: Fanconi anemia complementation group D2, osteosarcoma, RNA interference, p53, cell apoptosis
Introduction
Osteosarcoma (OS) is the most common primary malignant tumor of the bone tissue, mainly invading the metaphysis of long bones. OS has high degree of malignancy and poor prognosis. New drugs and treatment strategies based on full understanding of the mechanism of OS are required [1]. Dysfunction of proto-oncogenes and tumor suppressor genes is one of the pathogenic factors for OS. Like most other malignancies, OS involves multiple oncogenes activations and tumor suppressor genes mutations, including proto-oncogene c-myc, ras, fos, etc., and tumor suppressor gene p16, p53, Rb, etc [2-5].
Fanconi anemia (FA) is a rare autosomal or X-linked recessive hereditary disorder. It is found that FA could result from missing or mutation of 15 kinds of FA-related genes. These related genes form a complex network named FA pathway, in which Fanconi anemia complementation group D2 (FANCD2) is a key member. After post-translational modifications like phosphorylation and ubiquitination, [6,7], FANCD2 protein plays important role in regulating expression of genes which are involved in tumorigenesis, apoptosis, and other important life processes [8-10]. A FA patient has considerably higher risk of cancer [11]. Studies have shown that 28% of FA patients would have non-hematopoietic tumors before the age of 40, indicating that there is a strong positive correlation between FA and malignant tumors [12]. FA patients often suffer from head and neck, skin or anogenital squamous cell carcinomas [13]. But few researches have investigated the association between osteosarcoma and the FA pathway.
OS is one of the complications of FA, and both FA and OS are likely to occur in adolescence, but the interactions between them remains unclear. It is thought that FANCD2 is an important protein related to OS, while the associations and mechanisms need further study.
In this study, siRNA of FANCD2 was constructed and transfected to the osteosarcoma MG-63 cells, gene expressions after silence of FANCD2 gene were detected by gene chip. Cell proliferation and apoptosis, and the apoptosis-related signaling pathway were investigated, in order to reveal the role of FANCD2 in OS development.
Materials and methods
Construction and transfection of the FANCD2 siRNA in MG-63 cells
siRNA-FANCD2 and a control siRNA plasmid were designed and synthesized by Santa Cruz Biotechnology, Inc. (Texas, USA). They were transfected into osteosarcoma MG-63 cells by liposome. Expression of FANCD2 proteins in MG-63 cells were detected by western blots 24 h and 48 h later in order to evaluate the efficacy of siRNA.
Cell proliferation
Cell Counting Kit8 (DOJINDO LABORATORIES, Kumamoto, JAPAN) was used to detect cell proliferations of MG-63 cells 4, 8, 12, 24, and 48 h after siRNA transfection.
Cell cycle and apoptosis detection
24 h and 48 h after siRNA transfection, PI staining combined with flow cytometry was used to detect the percentage of G0/G1, S-phase and G2/M cells and cell cycle arrest in MG-63 cells. Annexin V-FITC/PI staining was used to detect apoptosis in MG-63 cells. Four groups, namely the control group (Control), siRNA-control group, siRNA-FANCD2 24 h group and siRNA-FANCD2 48 h group were detected.
Gene expression
24 h and 48 h after siRNA transfection, total RNA was extracted using TRIzol reagent (Invitrogen, NY, USA) and the changes in gene expression in MG-63 cells after FANCD2 gene silence were analyzed by Affymetrix GeneChip microarray system (Affymetrix, CA, USA). Differential gene expression was detected by real-time fluorescence quantitative PCR (Q-PCR).
The clustering graph and the differentially expressed genes were obtained, and a fold change of 2.0 was set as threshold for the screening of differentially expressed genes.
Apoptosis-related proteins
Tumor protein 53-induced nuclear protein 1 (TP53INP1), p53, p21 caspase-9 and caspase-3 mRNA expression in MG-63 cells were examined by Q-PCR method. And western blot assay was performed to detect the production of TP53INP1, phos-p53, p21 protein, activation of caspase-9 and caspase-3. As mentioned above, four groups, namely the control group (Control), siRNA-control group, siRNA-FANCD2 24 h group and siRNA-FANCD2 48 h group were detected.
Statistic analysis
The experimental data were expressed as mean±standard deviation (SD), and the two groups were compared using independent sample t-test after test for the homogeneity of variance. More than two groups were compared using ANOVA. P<0.05 indicates statistical significance.
Results
FANCD2 siRNA interfere FANCD2 expression in MG-63 cells
FANCD2 protein was detected both in the control group and the siRNA-control group, and no significant difference was observed between the two groups, indicating that the negative control did not prevent FANCD2 expression in MG-63 cells. The production of FANCD2 protein was significantly inhibited in the siRNA-FANCD2 group at 24 h (siRNA-FANCD2 24 h) and 48 h (siRNA-FANCD2 48 h), and the interfere effect was more pronounced at 48 h (Figure 1).
Figure 1.
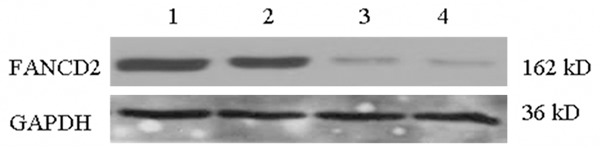
FANCD2 protein expression in MG-63 cells after RNAi. Lane 1: Control; Lane 2: siRNA-Control; Lane 3: siRNA-FANCD2 24 h; Lane 4: siRNA-FANCD2 48 h.
Cell proliferation
In the control group and the siRNA-control group, MG-63 cells showed rapid cell proliferation. There was no significant difference between the proliferation rates of the two groups. While the proliferation rates of the siRNA-FANCD2 group were significantly lower than the control group at 8, 12, 24 and 48 h (Table 1).
Table 1.
Absorbance of MG-63 cells after siRNA-FANCD2 interfere by CCK-8 assay (x̅±s, n = 6)
| Groups | Changes of absorbance value (A 450) | ||||
|---|---|---|---|---|---|
|
| |||||
| 4 h | 8 h | 12 h | 24 h | 48 h | |
| Control | 0.268±0.012 | 0.297±0.006 | 0.289±0.007 | 0.397±0.016 | 0.463±0.02 |
| siRNA-control | 0.275±0.007 | 0.288±0.009 | 0.281±0.013 | 0.382±0.014 | 0.483±0.018 |
| siRNA-FANCD2 | 0.268±0.006 | 0.263±0.004*** | 0.263±0.004*** | 0.369±0.017* | 0.384±0.009*** |
P<0.05 vs. Control;
P<0.001 vs. Control.
Cell cycle
The siRNA-control group had similar percentage of cells in G0/G1, S and G2/M phase as the control group. While after siRNA-FANCD2 transfection, cells in the G2/M and G0/G1 phase increased significantly, and the percentage of S phase was significantly lower than the control group (P<0.001); and the percentage of cells in G0/G1 phase at 24 h after siRNA interference was significantly higher than that of the 48 h group (P<0.01) (Table 2). These results indicated that the RNAi resulted in G0/G1 and G2/M phase arrest.
Table 2.
Cell cycle distribution of MG-63 cells after siRNA-FANCD2 interfere (x̅±s, n = 4)
| Groups | Positive cell percentage (%) | ||
|---|---|---|---|
|
| |||
| G0/G1 | S | G2/M | |
| Control | 61.11±1.56 | 33.22±1.60 | 5.67±0.32 |
| siRNA-control | 62.14±0.87 | 31.81±0.35 | 6.08±0.58 |
| siRNA-FANCD2 24 h | 72.16±0.31*** | 16.19±0.32*** | 11.64±0.54*** |
| siRNA-FANCD2 48 h | 77.25±1.52***,## | 12.66±2.71*** | 10.09±1.55** |
*P<0.05 vs. Control;
P<0.01 vs. Control;
P<0.001 vs. Control;
P<0.01 vs. siRNA-FANCD2 24 h.
siRNA-FANCD2 induced apoptosis of MG-63
Apoptotic percentages of MG-63 cells after FANCD2 siRNA transfection were detected by Flow cytometry (Table 3). Compared to the control group, siRNA-Control group had a higher percentage of apoptosis, which may be caused by the affects of transfection reagents. The percentage of normal cells were significantly decreased, while the percentage of apoptotic cells was significantly increased in the siRNA-FANCD2 24 h and siRNA-FANCD2 48 h group (P<0.01, P<0.001, Table 3).
Table 3.
Apoptotic percentages of MG-63 cells after FANCD2 siRNA interfere by Flow Cytometry (x̅±s, n = 4)
| Groups | Positive percentage, % | ||
|---|---|---|---|
|
| |||
| Normal | Necrosis | Apoptosis | |
| Control | 86.68±0.35 | 0.31±0.12 | 13.01±0.36 |
| siRNA-control | 85.56±0.45* | 0.29±0.01 | 14.15±0.46* |
| siRNA-FANCD2 24 h | 77.45±0.69*** | 0.36±0.23 | 22.17±0.81*** |
| siRNA-FANCD2 48 h | 64.00±1.71**,### | 0.53±0.30 | 35.47±1.42***,### |
P<0.05 vs Control;
P<0.01 vs Control;
P<0.01 vs Control;
P<0.01 vs. FANCD2 siRNA 24 h.
Gene expression profiles of MG-63 cells after siRNA interference by gene microarray
Compared with the control group, 35 genes were up-regulated and 53 were down-regulated in the FANCD2 siRNA 24 h group, involving 37 pathways according to the KEGG database. And 103 genes were up-regulated and 30 were down-regulated in the FANCD2 siRNA 48 h group, involving 54 pathways according to the KEGG database. These differentially expressed genes were involved in many aspects, including cell proliferation, apoptosis, cell cycle control, DNA damage repair and signal transduction.
mRNA expression related to the p53 pathway
Compared to the Control group, TP53INP1 mRNA expression in the siRNA-control group was similar, while the p53, p21, caspase-9 and caspase-3 mRNA expression increased. For the siRNA-FANCD2 24 h and siRNA-FANCD2 48 h group, TP53INP1, p53, p21, caspase-9 and caspase-3 mRNA expression were significantly increased (Table 4), up to 25 times, and this effect increased with time.
Table 4.
Relative expression of mRNAs in MG-63 cells after FANCD2 siRNA interfere (x̅±s, n = 3)
| Groups | mRNA relative expression | ||||
|---|---|---|---|---|---|
|
| |||||
| TP53INP1 | p53 | p21 | caspase-9 | caspase-3 | |
| Control | 1 | 1 | 1 | 1 | 1 |
| siRNA-control | 1.10±0.02 | 1.34±0.04* | 1.63±0.02* | 1.24±0.04* | 1.29±0.03* |
| siRNA-FANCD2 24 h | 5.41±0.29** | 6.64±0.32** | 4.23±0.11** | 12.42±0.54*** | 14.88±0.60*** |
| siRNA-FANCD2 48 h | 13.00±0.21***,# | 13.40±1.60***,# | 11.14±0.21***,# | 21.19±0.23***,## | 25.05±1.52***,## |
P<0.05 vs. Control;
P<0.01 vs. Control;
P<0.001 vs. Control;
P<0.05 vs. FANCD2 siRNA 24 h;
P<0.01 vs. FANCD2 siRNA 24 h.
Protein production related to the p53 pathway
As shown in Figure 2, compared to the control group, the production of p53, phos-p53, p21, TP53INP1, cleaved caspase-9 and cleaved caspase-3 were similar in the siRNA-control group. Production of p53 in the siRNA-FANCD2 24 h and siRNA-FANCD2 48 h group did not change significantly, but phos-p53, p21 protein and TP53INP1 increased significantly. Caspasae-9 was cleaved into 37 and 35 kD, caspase-3 was cleaved into 19 and 17 kD, and the expression was significantly increased, especially the cleaved caspase-3.
Figure 2.
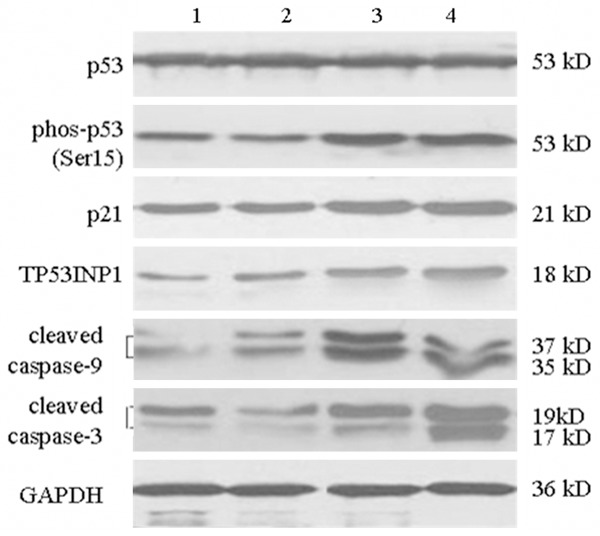
Western blotting picture of p53, phos-p53, p21, TP53INP1, cleaved caspase-9 and-3 protein expression after RNAi. Lane 1: Control; Lane 2: siRNA-Control; Lane 3: siRNA-FANCD2 24 h; Lane 4: siRNA-FANCD2 48 h.
Discussion
In previous studies, it has been found that FANCD2 plays an important role in the FA/BRCA pathway. Other studies have shown that FANCD2 is involved in DNA damage repair, cell cycle regulation, chromatin remodeling, DNA methylation and apoptosis, and it is essential to cell growth, differentiation and maintaining normal function of the body [14-16]. However, its associations with osteosarcoma remain unclear. In this study, we successfully constructed an efficient FANCD2 targeting siRNA, almost no expression of FANCD2 was observed after transfection into the MG-63 cells. And it caused suppressed cell proliferation, cell cycle arrest and apoptosis. Down-regulated or absent of the FANCD2 gene seriously altered the biological processes of tumor cell proliferation. Numerous studies have showed that tumor cells have unlimited proliferation, migration, invasion [17,18]. Thus, induction of tumor cells apoptosis is the usual method to control tumor. After interference of FANCD2 in MG-63 cells, apoptosis was observed.
In order to illuminate the involved signal pathways, the gene expression changes in MG-63 cells were further detected using gene chip method. There are three major apoptotic pathway, namely the mitochondrial pathway, endoplasmic reticulum pathway and death receptor pathway. Mitochondrial pathway is activated when the Bcl-2 family members (including Bcl-2, Bax, Bak and other molecules) received the death signal of the cell, then the mitochondrial membrane permeability is changed and the transmembrane potential is lost, finally leads to apoptosis. p53 is a transcription factor located on the mitochondrial, which can interact directly with Bax and induce change of mitochondrial permeability, then cause the release of various pro-apoptotic factors into the cytoplasm, finally leads to apoptosis [19-21]. Endoplasmic reticulum path way is mainly mediated by caspase family members. Caspase 12 is located on the endoplasmic reticulum, whose over-expression results in transfer of caspase 7 from cytoplasm to the endoplasmic reticulum surface, and then further cleave caspase 3 and induce apoptosis [22]. Death receptor pathway is mediated by the death receptors, which are a class of transmembrane proteins belonging to the tumor necrosis factor receptor super family [23-25]. All these three pathways in general depend on the final caspase-mediated apoptosis.
After FANCD2 siRNA interference for 48 h, 103 genes were up-regulated, 30 were down-regulated, involving 54 pathways in KEGG database. In these differentially expressed genes, up-regulation of XAF1 and TP53NP1 attracted our attention, because these two genes are closely related to p53 pathway. It seemed that the siRNA-FANCD2 induced apoptosis is mediated by the p53 signaling pathway, which was confirmed by the results of Q-PCR and western blot.
Mutant p53 gene was found in a variety of cancers, such as breast cancer, colorectal cancer, lung cancer, liver cancer, osteosarcoma, the mutation rate of up to 50% [26]. Wild-type p53 plays a role in monitoring cell growth, cell cycle regulation, cell cycle, DNA damage repair and induction of apoptosis [27,28]. TP53 protein is encoded by the p53 gene, which can regulate the cell cycle and prevent transformation of cancer cells. TP53INP1 is a target gene of p53, whose expression is regulated by the transcription factor p53, p73 and E2F1, and its activation can induce cell cycle arrest and p53-mediated apoptosis [29]. Studies have shown that over expression of TP53INP1 can cause inhibition of cell proliferation and apoptosis, and cells arrest in the G1 and G2 [30,31]. XAF1 is a negative regulatory protein of the apoptosis inhibitor, which widely present in normal human tissues, but low or absent [32] in most cancer cells, which is located on chromosome 17p13.2 locus, adjacent to p53. Studies have shown that, XAF1 gene was down-regulated in a variety of malignancies, such as gastric cancer, non-small cell lung cancer, liver cancer and other tumors [33-35]. In recent years, researchers have found that XAF1 is a p53 target gene, wild-type p53 gene can down-regulate the expression of XAF1 mRNA, and accumulation of protein p53 in the nucleus leads to apoptosis, indicating that there is a feedback loop between the p53 and XAF1 genes [36]. TP53INP1, XAF1 and p53 are closely related, and therefore we believe that the apoptosis of MG-63 by siRNA-FANCD2 is mediated by the p53 signaling pathway.
TP53INP1 (upstream protein of p53), phosphorylation of p53 (phos-p53), p21 protein (downstream protein which is involved in regulation of G1 phase), activation of caspase-9 and caspase-3 (apoptosis related proteins) were detected by western blot methods. After siRNA-FANCD2 was transfected into MG-63 cells for 48 hours, TP53INP1, p53, p21, caspase-9 and caspase-3 mRNA expression were significantly elevated; and production of phos-p53, p21 and TP53INP1 proteins were also increased. The experimental results showed clearly the MG-63 cell apoptosis was mediated by p53 signaling pathway. After transfection of siRNA-FANCD2 into MG-63 cells, FANCD2 gene expression was suppressed, and TP53INP1 was activated in the transcription level. Over-expression of TP53INP1 promoted phosphorylation of the Ser15 site of p53 protein; the phosphorylated p53 activated p21, and then started the p53 apoptotic signaling pathway. p21 gene is at the downstream of the p53 gene, which is a cyclin dependent kinase inhibitor, whose interaction with p53 leads to cell cycle arrest. Interaction of activated p53 with p21 inhibits tumor cell proliferation, and keeps the tumor cells arrest in the G1 phase. In addition, phosphorylation of p53 mediates the mitochondrial pathway of apoptosis. It induces change of the mitochondrial permeability, and then cause release of a variety of pro-apoptotic factors into the cytoplasm, finally induces apoptosis in MG-63 cells.
p53 also can activate the caspase pathway, and the activation of caspase associated proteins would promote apoptosis. Caspase belongs to cysteine protease family, and plays a key role in the process of apoptosis [37]. In the caspase pathway, caspase 7 is firstly activated, then caspase 12, and the activated caspase 12 further cleaves caspase 9 and caspase 3, finally triggering apoptosis [38-40]. Caspase 3 is in the downstream of this process, which is the key enzyme of caspase family. It is widely expressed in various tumor tissues [41,42]. This study showed that caspase 3 was cleaved and activated after RNAi. The activated caspase 3 finally induced the apoptosis.
In summary, after silence of the FANCD2 by siRNA-FANCD2 in MG-63 cells, inhibited cell proliferation, cell cycle arrest and induction of apoptosis were detected. The apoptosis was likely mediated by p53 signaling pathway. After FANCD2 expression was inhibited, TP53INP1 gene expression was promoted and then further enhanced phosphorylation of p53, thereby activated the p21 protein, leading to cell cycle arrest in G1, finally induced caspase-dependent cell apoptosis. It was suggested that FANCD2 plays an important role in the development of osteosarcoma, and inhibition of FANCD2 gene expression can effectively promote apoptosis of osteosarcoma cells, which providing new target for the treatment of osteosarcoma.
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. s. 81350013, 81250016).
Disclosure of conflict of interest
None.
References
- 1.Hu X, Liu Y, Qin C, Pan Z, Luo J, Yu A, Cheng Z. Up-regulated isocitrate dehydrogenase 1 suppresses proliferation, migration and invasion in osteosarcoma: in vitro and in vivo. Cancer Lett. 2014;346:114–121. doi: 10.1016/j.canlet.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeda J, Yurkon CR, Fujisawa H, Kaneko M, Genet SC, Roybal EJ, Rota GW, Saffer ER, Rose BJ, Hanneman WH, Thamm DH, Kato TA. Genomic instability and telomere fusion of canine osteosarcoma cells. PLos One. 2012;7:e43355. doi: 10.1371/journal.pone.0043355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan ML, Choong PFM, Dass CR. Osteosarcoma: Conventional treatment vs. gene therapy. Cancer Biol Ther. 2009;8:106–117. doi: 10.4161/cbt.8.2.7385. [DOI] [PubMed] [Google Scholar]
- 4.Smida J, Baumhoer D, Rosemann M, Walch A, Bielack S, Poremba C, Remberger K, Korsching E, Scheurlen W, Dierkes C, Burdach S, Jundt G, Atkinson MJ, Nathrath M. Genomic alterations and allelic imbalances are strong prognostic predictors in osteosarcoma. Clin Cancer Res. 2010;16:4256–4267. doi: 10.1158/1078-0432.CCR-10-0284. [DOI] [PubMed] [Google Scholar]
- 5.Ta H, Dass C, Choong P, Dunstan D. Osteosarcoma treatment: state of the art. Cancer Metasis Rev. 2009;28:247–263. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury I, Sareen A, Raghunandan M, Sobeck A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013;41:6444–6459. doi: 10.1093/nar/gkt348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rego MA, Kolling FW, Vuono EA, Mauro M, Howlett NG. Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood. 2012;120:2109–2117. doi: 10.1182/blood-2012-02-410472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koptyra M, Stoklosa T, Hoser G, Glodkowska-Mrowka E, Seferynska I, Klejman A, Blasiak J, Skorski T. Monoubiquitinated Fanconi anemia D2 (FANCD2-Ub) is required for BCR-ABL1 kinase-induced leukemogenesis. Leukemia. 2011;25:1259–1267. doi: 10.1038/leu.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo P, Bogliolo M, Surralles J. Coordinated action of the Fanconi anemia and ataxia telangiectasia pathways in response to oxidative damage. DNA Repair. 2011;10:518–525. doi: 10.1016/j.dnarep.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Marietta C, Thompson LH, Lamerdin JE, Brooks PJ. Acetaldehyde stimulates FANCD2 monoubiquitination, H2AX phosphorylation, and BRCA1 phosphorylation in human cells in vitro: Implications for alcohol-related carcinogenesis. Mutat Res. 2009;664:77–83. doi: 10.1016/j.mrfmmm.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]
- 12.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 13.Kachnic LA, Li L, Fournier L, Willers H. Fanconi Anemia pathway heterogeneity revealed by cisplatin and oxaliplatin treatments. Cancer Lett. 2010;292:73–79. doi: 10.1016/j.canlet.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Barroso E, Pita G, Arias JI, Menendez P, Zamora P, Blanco M, Benitez J, Ribas G. The Fanconi anemia family of genes and its correlation with breast cancer susceptibility and breast cancer features. Breast Cancer Res Treat. 2009;118:655–660. doi: 10.1007/s10549-009-0439-5. [DOI] [PubMed] [Google Scholar]
- 15.Garcia MJ, Fernandez V, Osorio A, Barroso A, Llort G, Lazaro C, Blanco I, Caldes T, de la Hoya M, Cajal TRY, Alonso C, Tejada MI, Roman CS, Robles-Diaz L, Urioste M, Benitez J. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res Treat. 2009;113:545–551. doi: 10.1007/s10549-008-9945-0. [DOI] [PubMed] [Google Scholar]
- 16.Garcia MJ, Fernandez V, Osorio A, Barroso A, Fernandez F, Urioste M, Benitez J. Mutational analysis of FANCL, FANCM and the recently identified FANCI suggests that among the 13 known Fanconi Anemia genes, only FANCD1/BRCA2 plays a major role in high-risk breast cancer predisposition. Carcinogenesis. 2009;30:1898–1902. doi: 10.1093/carcin/bgp218. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y, Lee JS, Xie N, Li E, Hurtado-Coll A, Fazli L, Cox M, Plymate S, Gleave M, Dong XS. Prostate stromal cells express the progesterone receptor to control cancer cell mobility. PLos One. 2014;9:e92714. doi: 10.1371/journal.pone.0092714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennon FE, Mirzapoiazova T, Mambetsariev B, Poroyko VA, Salgia R, Moss J, Singleton PA. The Mu opioid receptor promotes opioid and growth factor-induced proliferation, migration and Epithelial Mesenchymal Transition (EMT) in human lung cancer. PLos One. 2014;9:e91577. doi: 10.1371/journal.pone.0091577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin JZ, Lin GF, Huang H, Xu D, Yu H, Ma X, Zhu LS, Ma DY, Jiang HL. Capsaicin mediates cell cycle arrest and apoptosis in human colon cancer cells via stabilizing and activating p53. Int J Biol Sci. 2014;10:285–295. doi: 10.7150/ijbs.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rengarajan T, Nandakumar N, Rajendran P, Haribabu L, Nishigaki I, Balasubramanian MP. D-pinitol promotes apoptosis in MCF-7 cells via induction of p53 and Bax and inhibition of Bcl-2 and NF-kB. Asian Pac J Cancer Prev. 2014;15:1757–1762. doi: 10.7314/apjcp.2014.15.4.1757. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Qian RQ, Xiao J, Xu D, Fu HZ, Chen YY. Kirenol, a compound from Herba Siegesbeckiae, induces apoptosis in human chronic myeloid leukemia K562 cells. Pharmazie. 2014;69:148–153. [PubMed] [Google Scholar]
- 22.Mishra R, Karande AA. Endoplasmic reticulum stress-mediated activation of p38 MAPK, Caspase-2 and Caspase-8 Leads to abrin-Induced apoptosis. PLoS One. 2014;9:e92586. doi: 10.1371/journal.pone.0092586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman MA, Sundaram K, Mitra S, Gavrilin MA, Wewers MD. Receptor interacting protein-2 plays a critical role in human lung epithelial cells survival in response to Fas-induced cell-death. PLoS One. 2014;9:e92731. doi: 10.1371/journal.pone.0092731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Li J, Zhou T, Wang C, Zhang H, Wang H. Colistin-induced apoptosis in PC12 cells: involvement of the mitochondrial apoptotic and death receptor pathways. Int J Mol Med. 2014;33:1298–1304. doi: 10.3892/ijmm.2014.1684. [DOI] [PubMed] [Google Scholar]
- 25.Alshatwi AA, Shafi G, Hasan TN, Syed NA, Khoja KK. Fenugreek induced apoptosis in breast cancer MCF-7 cells mediated independently by fas receptor change. Asian Pac J Cancer Prev. 2013;14:5783–5788. doi: 10.7314/apjcp.2013.14.10.5783. [DOI] [PubMed] [Google Scholar]
- 26.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12:3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jochemsen AG. Reactivation of p53 as therapeutic intervention for malignant melanoma. Curr Opin Oncol. 2014;26:114–119. doi: 10.1097/CCO.0000000000000033. [DOI] [PubMed] [Google Scholar]
- 28.Huang YY, Dai L, Gaines D, Droz-Rosario R, Lu HM, Liu JM, Shen ZY. BCCIP suppresses tumor initiation but Is required for tumor progression. Cancer Res. 2013;73:7122–7133. doi: 10.1158/0008-5472.CAN-13-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seillier M, Peuget S, Gayet O, Gauthier C, N’Guessan P, Monte M, Carrier A, Iovanna JL, Dusetti NJ. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ. 2012;19:1525–1535. doi: 10.1038/cdd.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Cheng C, Yuan X, He JT, Pan QH, Sun FY. microRNA-155 acts as an oncogene by targeting the tumor protein 53-induced nuclear protein 1 in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:602–610. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79. doi: 10.1186/1423-0127-20-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fong WG, Liston P, Rajcan-Separovic E, St Jean M, Craig C, Korneluk RG. Expression and genetic analysis of XIAP-associated factor 1 (XAF1) in cancer cell lines. Genomics. 2000;70:113–122. doi: 10.1006/geno.2000.6364. [DOI] [PubMed] [Google Scholar]
- 33.Kim SK, Park HJ, Seok H, Jeon HS, Kim JW, Chung JH, Kwon KH, Woo SH, Lee BW, Baik HH. Missense polymorphisms in XIAP-associated factor-1 (XAF1) and risk of papillary thyroid cancer: correlation with clinicopathological features. Anticancer Res. 2013;33:2205–2210. [PubMed] [Google Scholar]
- 34.Lin Y, Li W. Assessment of XAF1 as A Biomarker to Differentiate Hepatocellular Carcinoma from Nonneoplastic Liver Tissues. Chin J Cancer Res. 2012;24:201–206. doi: 10.1007/s11670-012-0201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tse MK, Cho CK, Wong WF, Zou B, Hui SK, Wong BCY, Sze KH. Domain organization of XAF1 and the identification and characterization of XIAPRING-binding domain of XAF1. Protein Sci. 2012;21:1418–1428. doi: 10.1002/pro.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou B, Chim CS, Pang R, Zeng H, Dai Y, Zhang RX, Lam CSC, Tan VPY, Hung IFN, Lan HY, Wong BCY. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol Carcinog. 2012;51:422–432. doi: 10.1002/mc.20807. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Park HS, Kim JA, Hong GE, Nagappan A, Park KI, Kim GS. Flavonoids identified from Korean Scutellaria baicalensis induce apoptosis by ROS generation and caspase activation on human fibrosarcoma cells. Am J Chin Med. 2014;42:465–483. doi: 10.1142/S0192415X14500311. [DOI] [PubMed] [Google Scholar]
- 38.Walsh JG, Cullen SP, Sheridan C, Luthi AU, Gerner C, Martin SJ. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci U S A. 2008;105:12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci U S A. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao SP, Zeng Z, Wang XB, Bin JP, Xu DL, Liao YL. Pravastatin slows the progression of heart failure by inhibiting the c-JunN-terminal kinase-mediated intrinsic apoptotic signaling pathway. Mol Med Rep. 2013;8:1163–1168. doi: 10.3892/mmr.2013.1622. [DOI] [PubMed] [Google Scholar]
- 41.Mazumder S, Plesca D, Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol Biol. 2008;414:13–21. doi: 10.1007/978-1-59745-339-4_2. [DOI] [PubMed] [Google Scholar]
- 42.Shen T, Yang C, Ding L, Zhu Y, Ruan Y, Cheng H, Qin W, Huang X, Zhang H, Man Y, Liu D, Wang S, Bian Y, Xiao C, Zhao Y, Li J. Tbx20 functions as an important regulator of estrogen-mediated cardiomyocyte protection during oxidative stress. Int J Cardiol. 2013;168:3704–3714. doi: 10.1016/j.ijcard.2013.06.018. [DOI] [PubMed] [Google Scholar]


