Background: The formin INF2 can accelerate both actin polymerization and depolymerization.
Results: ATP hydrolysis continues even after apparent complete actin depolymerization by INF2, and profilin accelerates the process.
Conclusion: INF2 alone facilitates a cycle of polymerization and depolymerization, resulting in accelerated filament turnover and ATP hydrolysis.
Significance: In cells, the accelerated turnover induced by INF2 might result in assembly of short, transient filaments used for brief periods of time.
Keywords: actin, ATP, fluorescence, formin, profilin
Abstract
INF2 (inverted formin 2) is a formin protein with unique biochemical effects on actin. In addition to the common formin ability to accelerate actin nucleation and elongation, INF2 can also sever filaments and accelerate their depolymerization. Although we understand key attributes of INF2-mediated severing, we do not understand the mechanism by which INF2 accelerates depolymerization subsequent to severing. Here, we show that INF2 can create short filaments (<60 nm) that continuously turn over actin subunits through a combination of barbed end elongation, severing, and WH2 motif-mediated depolymerization. This pseudo-steady state condition occurs whether starting from actin filaments or monomers. The rate-limiting step of the cycle is nucleotide exchange of ADP for ATP on actin monomers after release from the INF2/actin complex. Profilin addition has two effects: 1) to accelerate filament turnover 6-fold by accelerating nucleotide exchange and 2) to shift the equilibrium toward polymerization, resulting in longer filaments. In sum, our findings show that the combination of multiple interactions of INF2 with actin can work in concert to increase the ATP turnover rate of actin. Depending on the ratio of INF2:actin, this increased flux can result in rapid filament depolymerization or maintenance of short filaments. We also show that high concentrations of cytochalasin D accelerate ATP turnover by actin but through a different mechanism from that of INF2.
Introduction
In eukaryotic cells, actin filaments are used for >20 different cellular processes, and these filaments vary greatly in length, lifetime, and higher order organization (1). For example, lamellipodial actin networks contain short, branched filaments that turn over on a time scale of seconds, whereas stress fibers contain longer filaments that turn over more slowly (2, 3). Recent findings reveal additional uses for actin-based structures around intracellular organelles such as Golgi and mitochondria (4–6), but detailed characterization has been challenging because of the transient nature of such structures.
At the foundation of all actin-based structures is the single actin filament, which can self-assemble and undergo a cycle of polymerization and depolymerization in the absence of other protein factors (7). Actin monomers alone can self-nucleate into filaments, and the filaments can elongate by further monomer addition at both the barbed and pointed ends, the barbed end being favored. Subsequent to filament incorporation, individual actin monomers (now called subunits) hydrolyze their bound ATP and release the phosphate (Pi) product while still in the filament. Both ATP hydrolysis (0.3 s−1) and phosphate release (0.002 s−1) are slow compared with filament incorporation (10 μm−1 s−1 at barbed end), resulting in a “cap” of ATP and ADP-Pi subunits toward the barbed end (8–11). As ADP-subunits accumulate at the pointed end, the equilibrium favors dissociation from the filament, resulting in net depolymerization from the pointed end. Thus, there is a net flow of ATP-actin onto the barbed end and ADP-actin off of the pointed end at steady state in the presence of excess ATP (12).
The variability in filament lengths and/or dynamics between cellular actin-based structures is caused by specific actin binding proteins. These proteins do not fundamentally change actin's polymerization/depolymerization cycle but alter the kinetics of specific steps. For example, the abundant actin monomer binding protein, profilin, has multiple effects on dynamics: 1) it inhibits pointed end addition of ATP-actin monomers (13–15), focusing elongation to the barbed end; 2) it inhibits spontaneous nucleation of actin monomers (13), thereby forcing de novo filament assembly to be triggered by nucleation factors such as Arp2/3 complex, formins, or COWs (compound WH2-containing proteins) (16, 17); and 3) it accelerates nucleotide exchange on actin monomers (18–20), allowing recently depolymerized ADP-actin monomers to recharge with ATP for subsequent rounds of polymerization.
Another important function of profilin is to work with formin proteins in controlling filament elongation. Formins can nucleate actin filaments de novo and then remain at the elongating barbed end because of their dimeric formin homology 2 (FH2)2 domain. Formins can also interact with profilin through their formin homology 1 (FH1) domain, which is directly N-terminal to the FH2 (21). The consequence of the formin/profilin interaction is an increase in barbed end elongation rate (8). There are 15 mammalian formins, and their abilities to nucleate and elongate actin vary considerably (8, 22–24), creating variation that cells can use for diverse actin-based structures.
INF2 (inverted formin 2) is a particularly unusual mammalian formin. In addition to accelerating nucleation and regulating elongation, INF2 has the unique capacity to accelerate filament depolymerization (23). A key component of depolymerization is the ability of INF2 to sever ADP-containing filament regions (25). In cells, INF2 acts in multiple processes, but the best understood is its role in mitochondrial fission, in which endoplasmic reticulum-bound INF2 assembles filaments that facilitate fission (26). INF2 is likely to have similar roles in other cytoplasmic actin-based processes (4).
This paper focuses on the biochemical mechanism behind the ability of INF2 to accelerate filament depolymerization. We show that INF2 works with the polymerization/depolymerization cycle of actin itself and by regulating key steps in the process accelerates the turnover of actin subunits considerably. In the presence of profilin, this process results in the assembly of highly dynamic filaments of relatively uniform filaments length, providing a potential mechanism for assembly of short, transient actin filaments.
Experimental Procedures
DNA Constructs
Human INF2 clone (CAAX variant, catalog no. SC313010) was obtained from OriGene Technologies, Inc. (Rockville, MD). INF2-FFC (amino acids 469–1249) and INF2-FC (amino acids 552–1249) were amplified using a GC-rich PCR system (Roche Applied Science) and subcloned into pGEX-KT vector (27) with a modified tobacco etch virus protease site for bacterial expression as a glutathione S-transferase fusion protein.
Buffers
The following buffers were used frequently: G buffer (2 mm Tris, pH 8, 0.5 mm DTT, 0.2 mm ATP, 0.1 mm CaCl2, and 0.01% NaN3), G-Mg buffer (same as G buffer but with 0.1 mm MgCl2 instead of CaCl2), 10× K50MEI (500 mm KCl, 10 mm MgCl2, 10 mm EGTA, and 100 mm imidazole, pH 7.0), 10xK50MEH (same as 10xK50MEI but with 100 mm HEPES, pH 7.4), 10× Na50MEI (same as 10× K50MEI but with 500 mm NaCl instead of KCl), and polymerization buffer (G-Mg buffer plus either 1× K50MEI, 1xK50MEH, or 1× Na50MEI and 0.5 mm thesit (the common name for the detergent nonaethylene glycol monododecyl ether (Sigma; P-9641), which was included to minimize protein adhesion to the tube/well). Polymerization buffer with 1× Na50MEI was used for pelleting assays because dodecyl sulfate precipitates as the potassium salt. Protein storage buffer (K50MEID) was composed of 1xK50MEI, 1 mm DTT. The TIRF buffer contained: 1× KMEI, 100 mm DTT, 0.2 mm ATP, 15 mm glucose, 0.5% methyl cellulose, 0.01 mg/ml catalase (Sigma; C3515), 0.05 mg/ml glucose oxidase (Sigma; G6125), and 0.1% BSA).
Protein Preparation and Purification
All formin constructs were expressed in Rosetta2 Escherichia coli (Stratagene Inc.) as GST fusion proteins, following procedures used previously (22, 24, 25, 28, 29). The constructs used were INF2-FFC (FH1-FH2-C construct, human CAAX variant, amino acids 469–1249), INF2-FC (FH2-C construct, human CAAX, 552–1249), FMNL3-FFC (mouse), mDia1-FFC (mouse), and mDia2-FFC (mouse). Briefly, expression was induced in log phase cultures with 0.5 mm isopropyl β-d-thiogalactopyranoside at 16 °C. After expression, extracts were passed over glutathione-Sepharose, cleaved with tobacco etch virus protease to elute the formin construct from GST, and further purified by ion exchange chromatography on SourceQ and/or gel filtration by Superdex 200 (GE Biosciences). INF2 and FMNL3 proteins were stored at −80 °C in K50MEIDT, whereas mDia1 and mDia2 proteins were stored as 50% glycerol stocks at −20 °C. Human profilin wild-type, profilin-Y6D, and profilin-R88E were expressed and purified as described previously (30). Rabbit skeletal muscle actin was purified from acetone powder (31) and labeled with pyrenyliodoacetamide (32) or TAMRA NHS ester (Invitrogen; C1171) (33). Both labeled and unlabeled actin were gel-filtered on Superdex 200 and stored in G buffer at 4 °C.
Pyrene Actin Assays
For depolymerization assays, actin (5.5 μm, 5% pyrene) was polymerized 16 h at 23 °C in polymerization buffer in the dark. This actin stock (54 μl) was diluted with 6 μl of formin protein with or without profilin (diluted in polymerization buffer) at the indicated final concentrations. For polymerization assays, 20 μl of 15 μm actin (5% pyrene) was mixed with 40 μl of formin with or without profilin in 1.5× polymerization buffer. Pyrene fluorescence (365/410 nm) was monitored in a 96-well fluorescence plate reader (Infinite M1000; Tecan, Mannedorf, Switzerland) within 1 min of mixing actin and formins. Starting ATP concentrations were 150–170 μm, with the variation caused by the variable concentrations of formins and/or profilin added. Concentration of depolymerized actin was calculated by normalizing to the actin alone curve, which represents 4.9 μm polymerized actin/0.1 μm depolymerized actin (7).
High Speed Sedimentation Assays
Actin monomers (5 μm) were polymerized either alone or in the presence of INF2 FFC (4 μm) with or without profilin (10 μm) for 20 min or 2 h at 23 °C in polymerization buffer (G-Mg buffer plus 1× NaMEI). For assays in the presence of cytochalasin D (CD), actin monomers (5 μm) were polymerized either alone or in the presence of 2 μm or 5 μm CD and 10 μm latrunculin B (LatB) for 20 min 23 °C in polymerization buffer. For determination of minimal length of actin filaments in pellets, 10 μm actin was incubated with indicated ratio of actin: capping protein and polymerized in polymerization buffer (G-Mg + KMEI) at 23 °C for 1 h. Samples were then diluted 10-fold with polymerization buffer containing 2.5 μm phalloidin and equilibrated at 23 °C for 2 h. After polymerization, all samples were transferred to polycarbonate 7 × 20 mm centrifuge tubes (Beckman 343775) with a final volume of 200 μl and centrifuged at 80,000 rpm for 20 min at 4 °C in a TLA-100.1 rotor (Beckman). 100 μl of supernatant was removed and mixed with SDS-PAGE sample buffer. The rest of the supernatant was removed and discarded. Pellets were washed briefly with 200 μl of 1× NaMEI and then resuspended in 100–400 μl of SDS-PAGE sample buffer (depending on experiment). Pellets were analyzed by Coomassie-stained SDS-PAGE and ImageJ software.
Velocity Analytical Ultracentrifugation
For INF2-FFC samples, 15 μm actin monomers +30 μm LatB, 7.5 μm INF2-FFC, or 15 μm actin monomers + 7.5 μm INF2-FFC was prepared in polymerization buffer and incubated for 1 h at 23 °C before starting the centrifugation. For actin sedimentation length calibration with capping protein, 20 μm actin monomers were polymerized in the presence of indicated ratios of capping protein in polymerization buffer for 1 h. After polymerization, samples were diluted to a final of 10 μm actin in polymerization buffer with 25 μm phalloidin and left to equilibrate for 2 h at 23 °C before centrifugation. Analytical ultracentrifugation was conducted using a Beckman Proteomelab XL-A and an AN-60 rotor. All samples were centrifuged at 25,000 rpm with monitoring at 290 nm. The data were analyzed by Sedfit to determine sedimentation coefficient, frictional ratio, and molecular weight. Sedimentation coefficient reported is that of the major peak (at least 80% of the total analyzed mass).
ATP Turnover Assays
ATP turnover was determined through two methods: 1H NMR, detecting ATP and ADP resonance peaks heights as previously described (34), and a colorimetric assay detecting the inorganic phosphate product of ATP hydrolysis (35). For colorimetric assays, reactions were started by mixing formins in polymerization buffer with actin monomers for a final concentration of 5 μm actin. Reactions were quenched at various time points by adding 20 μl of sample to 5 μl of 125 mm EDTA in clear flat-bottomed 96-well plate (Greiner). Five or six time points were acquired for each condition. Phosphate release was determined by addition of 150 μl of malachite green solution (1 mm Malachite Green (Sigma-Aldrich; 229105–100g), 10 mm ammonium molybdate (Sigma-Aldrich; A7302–100g) in 1 n HCl) to 25 μl quenched reactions and then measuring absorbance at 650 nm with a 96-well fluorescence plate reader (Infinite M1000; Tecan). ATP turnover rates were determined by plotting phosphate concentration as a function of time and then conducting linear regression analysis. Rates are given in units of s−1 and are understood to be μm ATP hydrolyzed per second per μm actin. We find similar rates of ATP turnover regardless of whether reactions were started from actin monomers or prepolymerized actin filaments. We calculated a theoretical ATP turnover rate for 5 μm actin under polymerizing conditions, with the following assumptions. First, the average filament length at steady state is 2000 subunits, based on filament length measurements we have made previously (23). Second, the rate-limiting step for ATP turnover at steady state is Pi release from actin subunits, which is 0.002 s−1 (11). Given these assumptions, we would expect an ATP turnover rate of 5 × 10−6 s−1.
Analysis of Nucleotides Bound to Actin Monomers
Reactions (1 ml) containing 5 μm ATP-actin monomer and 2.5 μm INF2-FFC were mixed for 20 min at 23 °C in K50MEH buffer containing 400 μm ATP, to establish the steady state condition. Phalloidin and latrunculin A (7.5 μm each) were added to “stop” the reaction (stabilizing any filaments present, sequestering any monomers present, and slowing nucleotide exchange) (36). Reactions were centrifuged in a TLA-120 rotor at 100,000 rpm for 20 min at 4 °C to sediment any long filaments present. To remove nucleotide that was not bound to actin, supernatant was gel-filtered on a Superdex 200 10 × 30 column (GE Biosciences) equilibrated in 1 mm HEPES, pH 7.4, 0.1 mm CaCl2. Protein-containing peak fractions were boiled 10 min and then centrifuged at 16,000 × g for 10 min to remove precipitated protein. Supernatant was analyzed by 1H NMR as described (34).
Negative Staining Electron Microscopy
The actin/INF2 or actin/capping protein mixtures were incubated for varying times (indicated in the figure legends) before applying to grids, to reach steady state. To make negatively stained grids, 30-μl aliquots of actin samples were adsorbed onto EM grids (Electron Microscopy Sciences; CF300-Cu) for 4 min and then blotted gently with filter paper. Grids were then stained by 30 μl of 1% uranyl acetate solution for 1 min. Prepared grids were imaged on a JEOL JEM 1010 transmission electron microscope operated at 100 keV acceleration. Images were taken with a XR-41B AMT digital camera and capture engine software (AMTV540; Advanced Microscopy Techniques). Filament lengths and densities were quantified with ImageJ analysis software.
TIRF Microscopy
Glass flow chambers were prepared from VWR micro cover glasses (22 × 22 mm and 18 × 18 mm no. 1.5) to hold 10 μl volume. Cover glasses were silanized using the procedure adapted from Gell et al. (37) and explained previously (25). Briefly, cover glasses were washed in acetone (50 min), ethanol (10 min), and water (1 min) and then incubated in Pirhana Solution (1:2 ratio of 30% H2O2 and H2SO4) for 1 h. Glasses were then rinsed with water, 0.1 m KOH, and then water again and dried with inert gas before silanization. Glasses were silanized overnight in a 0.0025% solution of dichlorodimethyl silane (Sigma 85126) in chloroform, washed with methanol, dried with inert gas, and stored in clean sealed containers. For silanized cover glasses, chambers were incubated with 1% Pluronic F127 (Sigma P2443) in BRB80 buffer (80 mm PIPES/KOH, pH 6.9, 1 mm EGTA, 1 mm MgCl2) for 1 min and then equilibrated in TIRF buffer. Nucleation assays were conducted as previously mentioned (25). Briefly, unlabeled actin monomers were mixed with 20% TAMRA-labeled actin monomers (2 μm) in G buffer, diluted with 2× TIRF buffer without or with 1 nm GFP-INF2 construct and profilin or profilin mutant (3 μm), and flowed into the flow chamber. The filaments were visualized as quickly as possible (∼60 s) on a Nikon Eclipse Ti-E inverted microscope with perfect focus and 488- and 561-nm lasers. Simultaneous two-color images were acquired every 1 s at 100-ms exposure with TIRF objective (60 × 1.49 N.A.) and two iXON Ultra 897 cameras.
Results
INF2 Causes Continuous ATP Turnover on Actin after Apparent Depolymerization
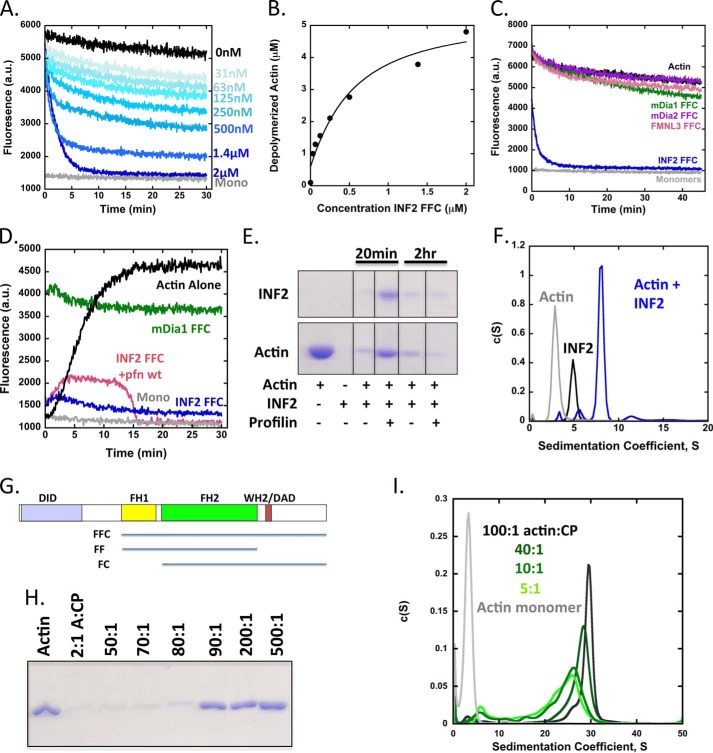
Previously, we showed that an INF2 construct containing the FH1, FH2, and C-terminal domains (FFC; Fig. 1) potently severed and depolymerized filaments (23, 25). However, these experiments used an actin concentration (1 μm) close to the pointed end critical concentration (0.6 μm) (38), which complicated interpretation of depolymerization mechanism. To analyze INF2-FFC depolymerization activity in more detail, we repeated these pyrene-actin assays using 5 μm prepolymerized actin filaments. Under these conditions, INF2-FFC depolymerizes actin filaments in a concentration-dependent manner in the absence of any additional factors (Fig. 1A). This effect is not merely a 1:1 sequestration of actin monomers by INF2-FFC, because 2 μm formin causes apparent complete depolymerization, and lower concentrations cause depolymerization well in excess of their concentration (Fig. 1B). The effect is specific to INF2, because FFC constructs of three other formins do not induce depolymerization (Fig. 1C). We note that similar apparent steady state pyrene fluorescence is obtained when starting with 5 μm actin monomers instead of filaments (Fig. 1D), similar to our earlier findings (23).
FIGURE 1.
INF2 depolymerizes actin to a limiting 1:4 INF2:actin complex. A, actin depolymerization assay using pyrene-labeled actin filaments (5 μm actin, 5% pyrene, prepolymerized overnight) and the indicated concentrations of INF2-FFC. Dead times between INF2 addition and data acquisition were ∼30–60 s. monomers (Mono) are 5 μm actin incubated overnight in the presence of 50 μm LatB. B, quantification of depolymerized actin based on plateau heights at 30 min from pyrene-actin assays in A. C, actin depolymerization assay similar to A, using 2 μm of the FFC constructs for mDia1, mDia2, FMNL3, and INF2. D, pyrene actin assays starting from actin monomers. Reactions contain 5 μm actin monomers (5% pyrene-labeled) alone, with 2 μm INF2-FFC, with 2 μm INF2-FFC and 10 μm profilin, or with 2 μm mDia1 FFC. E, high speed sedimentation assay with 5 μm actin (prepolymerized overnight) and 2 μm INF2-FFC ± 10 μm profilin, incubated for indicated times at 23 °C then ultracentrifuged for 20min at 4 °C. All lanes are from the same gel, with intervening lanes cropped. F, velocity analytical ultracentrifugation of 15 μm actin monomers (Actin), 7.5 μm INF2-FFC (INF2), or 15 μm actin mixed with 7.5 μm INF2-FFC (actin + INF2) in polymerizing conditions. Samples were preincubated for 60 min at 23 °C before starting the centrifugation. The actin sample also contained 30 μm LatB to maintain monomers. G, bar diagram of INF2 (1249 amino acids) showing domain structure (to scale with their lengths) and constructs used in this study. H, high speed sedimentation assay with indicated ratio of actin:CP and a final concentration of 1 μm actin. Samples were polymerized at 10× concentration for 1 h, then diluted 10×, and equilibrated 2 h before ultracentrifugation for 20 min at 4 °C. I, velocity analytical ultracentrifugation of 10 μm actin monomers mixed with varying ratios of capping protein in polymerizing conditions. Actin and capping protein were polymerized at 2× final concentration for 1 h at 23 °C, then diluted 2×, and equilibrated for 2 h at 20 °C before starting the centrifugation. The actin alone sample also contained 20 μm LatB to maintain monomers.
High speed sedimentation assays confirm the apparent depolymerization effect of INF2, showing that addition of INF2-FFC causes a dramatic reduction of sedimentable actin in less than 20 min (Fig. 1E). By sedimentation velocity analytical ultracentrifugation, a 2:1 ratio of actin:INF2-FFC results in a predominant 8S particle (Fig. 1F), similar to the 8S complex, we previously observed when INF2-FFC is mixed with an excess of latrunculin A (LatA)-stabilized actin monomers (23). Calculation of apparent mass of this complex from the S value and optimal frictional coefficient (2.061) gives 347.7 kDa. Our interpretation is that this particle represents a complex of one INF2 dimer (170 kDa) with four actin monomers (43 kDa each). A significantly less abundant peak at 11.5 S is also present. No higher S peaks are detectable above background. Our previous conclusion was that the 8S INF2/LatA-actin peak represented two actins and one INF2 dimer, but we revise this assessment here based on our greater confidence in the fitting algorithm and the fact that the residual actin monomer peak is minimal under these conditions, whereas a more substantial peak would be present if 1:1 binding were occurring.
To provide calibration for these results, we polymerized actin in varying ratios with capping protein (CP) to create differential filament lengths. Ratios of actin:CP lower than 90:1 do not pellet significantly in high speed sedimentation assays (Fig. 1G). Sedimentation velocity analytical ultracentrifugation experiments of CP/actin filaments show that the predominant sedimentation coefficient varies from 24S at 5:1 to 30S at 100:1 actin:CP (Fig. 1H). Thus, even mini-filaments of 5:1 actin:CP sediment much more readily than the predominant 8S complex formed by 4:1 actin:INF2-FFC dimer, suggesting that this is indeed a complex of INF2 and a small number of actin molecules. We note, however, that there is some uncertainty as to the exact stoichiometry, because of the assumptions that go into the mass calculations.
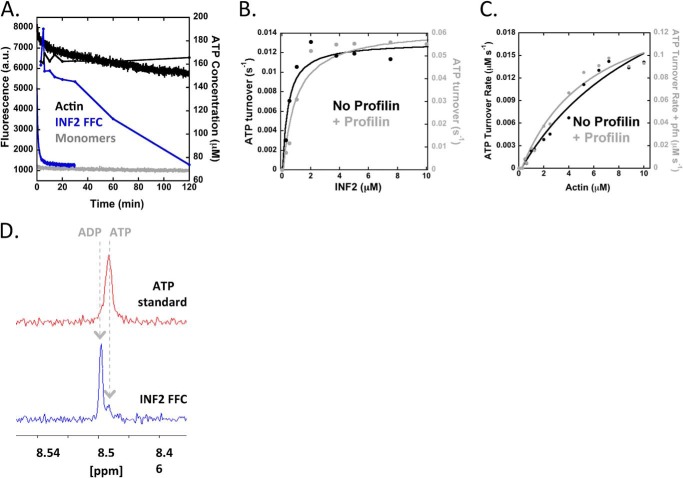
One interpretation of these results is that each INF2-FFC monomer sequesters at least two actin monomers after severing, preventing those monomers from participating in any further polymerization after the initial depolymerization reaction. To test this hypothesis, we measured ATP turnover kinetics during the depolymerization reaction, because actin ATP hydrolysis accelerates ∼10,000-fold upon polymerization (9, 39). We used two assays to monitor ATP turnover: 1) 1H NMR, detecting ATP and ADP resonance peaks heights that are proportional to nucleotide concentration (34), and 2) a colorimetric assay detecting the inorganic phosphate product of ATP hydrolysis (35). ATP turnover of 5 μm actin alone under polymerizing conditions is below the level of detection of these assays over 24 h (Fig. 2A and not shown), as would be predicted based on the kinetic cycle of actin at steady state (calculated at 5 × 10−6 s−1, see “Experimental Procedures”). Surprisingly, INF2 induces an approximate 3000-fold increase in ATP turnover (1.45 × 10−2 ± 3.08 × 10−3 s−1) over that of actin alone, and this turnover continues with linear kinetics long after actin depolymerization reaction reaches plateau (Fig. 2A and Table 1).
FIGURE 2.
INF2-FFC accelerates ATP turnover by actin after depolymerization. A, pyrene actin depolymerization assay as in Fig. 1A (left axis), overlaid with plot of ATP concentration (measured by 1H NMR) as a function of time (right axis; 2 μm INF2-FFC, 5 μm actin). B, double-Y plot of concentration dependence of INF2-FFC for ATP turnover of 5 μm actin monomers in the absence (black, left axis) or presence (gray, right axis) of 10 μm profilin. ATP turnover measured by phosphate assay. Rates represent μm ATP hydrolyzed per second per μm actin. C, double-Y plot of concentration dependence of actin monomers for ATP turnover in the presence of 2 μm INF2 FFC alone (black, left axis) or with 10 μm profilin (gray, right axis). ATP turnover measured by phosphate assay. D, 1H NMR of nucleotide bound to actin monomers in the steady state actin/INF2-FFC reaction. Reactions containing 5 μm ATP-actin monomers and 2.5 μm INF2-FFC were incubated for 20 min at 23 °C to establish steady state, then processed as described under “Experimental Procedures” to isolate monomer-bound nucleotide, and analyzed by 1H NMR.
TABLE 1.
ATP turnover rates with and without profilin (wild-type or mutants)
The conditions were 5 μm actin, 2 μm formin or CD, and 10 μm profilin. ND, not determined.
| ATP turnover rate |
||||
|---|---|---|---|---|
| Alone | With profilin WT | With profilin Y6D | With profilin R88E | |
| s−1 | ||||
| INF2 FFC | 0.014 ± 0.003 | 0.083 ± 0.03 | 0.037 ± 0.0083 | 0.017 ± 0.003 |
| INF2 FC | 0.0068 ± 2.3 × 10−4 | 0.026 ± 0.007 | ND | ND |
| CD | 0.0038 ± 5.8 × 10−4 | 0.0044 ± 6.6 × 10−4 | ND | ND |
This result is unexpected because the actin filament starting material should contain ADP-bound actin subunits exclusively, and the combination of severing and depolymerization by INF2 should not result in any additional ATP hydrolysis. In fact, similar ATP turnover is observed whether the starting material is ADP-actin filaments or ATP-actin monomers. INF2-FFC alone has no appreciable ATPase activity (not shown). The saturating concentration of INF2-FFC for ATP turnover (∼2 μm) is similar to the concentration that affects complete apparent actin depolymerization in pyrene-actin assays (Fig. 2B, compare with Fig. 1B). ATP turnover scales with actin concentration, with the x intercept at 0.07 μm actin (Fig. 2C), which is similar to actin's barbed end critical concentration 0.1 μm (40) and suggests that ATP turnover requires some form of polymerization to occur.
In view of these results, we hypothesize that INF2-FFC rapidly establishes a pseudo-steady state condition, rather than simply depolymerizing the starting filaments. The system reaches this pseudo-steady state condition rapidly, whether starting from ADP-actin filaments or from ATP-actin monomers. Because of experimental ease, most subsequent experiments start from actin monomers. We hereafter refer to the “steady state,” assuming an adequate supply of ATP under most conditions.
Profilin Accelerates ATP Turnover and Shifts the Polymerization Equilibrium of INF2-FFC/Actin
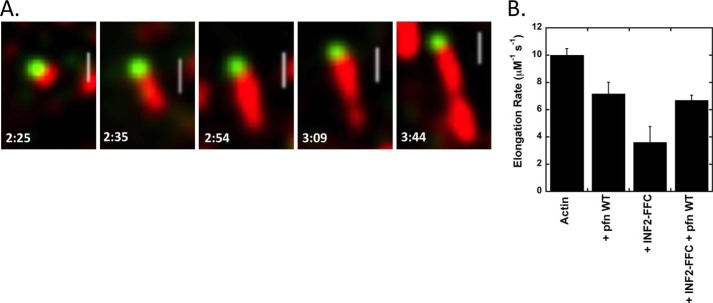
The rate constant of ATP turnover in this cycle is similar to both those of phosphate (Pi) release from actin filaments (0.0022 s−1) (11) and of ADP release from actin monomers (0.009 s−1) (19, 20). By 1H NMR, the actin monomers present in the steady state condition are >90% ADP-bound (Fig. 2D). To test the rate-limiting step in this cycle, we perturbed the system by the addition of profilin. The two major effects of profilin in this system are 1) to accelerate nucleotide exchange on actin monomers (9, 18, 20), and 2) to accelerate formin-mediated barbed end elongation to a variable degree, depending on the formin (8). Because the effect of profilin on INF2-FFC mediated barbed end elongation is not known, we measured this parameter by TIRF microscopy and found that profilin increases barbed end elongation 2-fold, from 3.6 to 7 μm−1 s−1 (Fig. 3).
FIGURE 3.
TIRF elongation rates of INF2 FFC in the absence and presence of profilin. A, TIRF microscopy time lapse montage of elongating actin filament (red) with bound GFP-INF2-FFC (green) at barbed end. Reaction was initiated by mixing 1 μm actin (20% TAMRA-labeled) with 0.2 nm GFP-INF2-FFC. Under these conditions, INF2 is confined almost exclusively to the barbed end. Numbers indicate time (min:s). Scale bar, 2 μm. B, bar graph of elongation rates for each condition tested. For these measurements, the INF2-FFC is not coupled to GFP. Rates determined from at least six filaments, and error bars denote standard deviation. pfn, profilin.
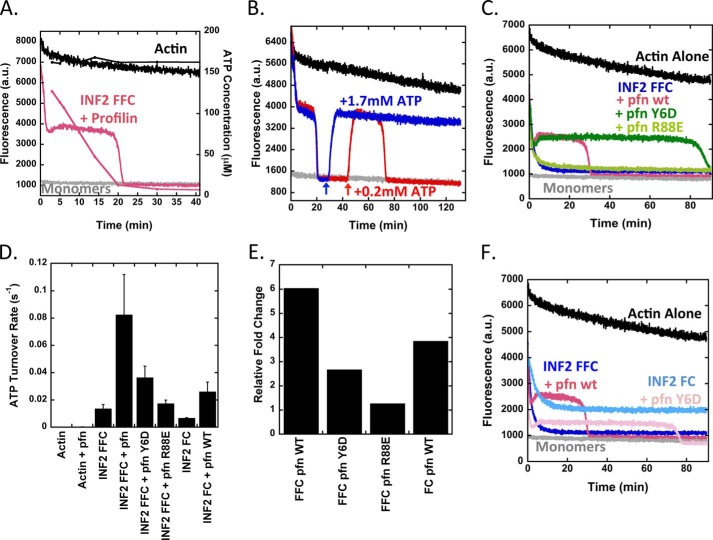
Addition of profilin to our depolymerization reaction increases INF2-FFC-induced ATP turnover ∼6-fold over that of INF2-FFC alone (to 8.25 × 10−2 ± 2.9 × 10−2 s−1) (Fig. 4, A, D, and E, and Table 1). Furthermore, the equilibrium shifts toward polymerization in the presence of profilin, as shown by both elevated pyrene-actin fluorescence (Fig. 4A) and increased sedimented actin at high speed (Fig. 1E). This equilibrium shift also occurs when starting from actin monomers (Fig. 1D). Upon complete depletion of ATP, the pyrene-actin curve drops to baseline fluorescence of actin monomers (Fig. 4A), suggesting that the polymerization plateau is dependent on recharging monomers with ATP. To test this hypothesis further, we added additional ATP after initial ATP depletion, which causes re-establishment of the previous plateau level (Fig. 4B). The duration of the plateau, but not its height, is dependent on the amount of ATP added.
FIGURE 4.
Profilin accelerates ATP turnover and shifts the equilibrium state for INF2/actin. A, pyrene-actin depolymerization curves overlaid with ATP concentration curve as in Fig. 1E, but with 10 μm profilin added. Actin, 5 μm (5% pyrene) prepolymerized overnight. INF2-FFC, 2 μm. B, effect of ATP readdition to polymerization equilibrium established by INF2-FFC and profilin. Reactions contain 5 μm prepolymerized actin (5% pyrene), 2 μm INF2-FFC, and 10 μm profilin. Additional ATP added at times indicated by arrows. In the red curve, 0.2 mm ATP was added. In the blue curve, 1.7 mm ATP was added. C, pyrene actin depolymerization assays containing 5 μm prepolymerized actin (5% pyrene), 2 μm INF2-FFC, and 10 μm of profilin or profilin mutants Y6D (poly proline-binding mutant) or R88E (actin-binding mutant). D, ATP turnover for reactions starting from 5 μm actin monomers alone or with combinations of 2 μm INF2-FFC, 2 μm INF2-FC, and 10 μm profilin or profilin mutants as indicated. Turnover determined based on four to six time points for each condition, bars represent standard deviation for three to eight separate experiments. E, fold change in ATP turnover for INF2-FFC or INF2-FC with or without profilin or profilin mutants. Determined from ATP turnover in Fig. 3D. F, pyrene-actin depolymerization assay containing 5 μm prepolymerized actin (5% pyrene) with 2 μm INF2-FFC or INF2-FC with or without 10 μm of profilin. pfn, profilin.
We tested the role of profilin further by using two profilin mutants: profilin-Y6D, which displays reduced FH1 domain binding and has diminished ability to accelerate formin-mediated barbed end elongation (8, 41), and profilin-R88E, which displays reduced actin monomer binding and is diminished in both nucleotide exchange and formin-mediated barbed end elongation (41). Pyrene-actin depolymerization assays show that profilin-R88E has no apparent effect on INF2-FFC mediated depolymerization (Fig. 4C). In contrast, profilin-Y6D behaves similarly to profilin in that it causes a similar polymerization plateau, but this plateau is prolonged ∼3-fold (Fig. 4C). In terms of ATP turnover rate, profilin-R88E does not increase the effect of INF2-FFC, whereas profilin-Y6D has a 3.4-fold reduced effect when compared with profilin at 10 μm (Fig. 4D).
One interpretation of these results is that, in the absence of profilin, nucleotide exchange is the rate-limiting step of this ATP turnover cycle. Profilin addition shifts the rate-limiting step to barbed end elongation, with profilin being more effective at elongation than profilin-Y6D because it can interact with the FH1 domain of INF2. To test this hypothesis, we examined the effect of the INF2-FH2-C construct (INF2-FC) on ATP turnover. Lacking the FH1 domain, INF2-FC cannot bindprofilin or allow profilin-mediated elongation acceleration. INF2-FC causes a 2-fold lower rate of ATP turnover than INF2-FFC (Fig. 4D). Profilin addition, however, increases turnover rate 4-fold for INF2-FC, similar to the 6-fold increase of profilin for INF2-FFC (Fig. 4E). This result suggests that FH1 binding is not necessary for profilin acceleration of ATP turnover. In depolymerization assays, INF2-FC causes less of a reduction in filament concentration at steady state than does INF2-FFC (Fig. 4F). Profilin addition still causes the steady state condition to terminate for INF2-FC, but after a 3-fold longer time period than for INF2-FFC, consistent with slower ATP turnover. These results suggest that the major effect of profilin in this system is to accelerate nucleotide exchange, the rate-limiting step.
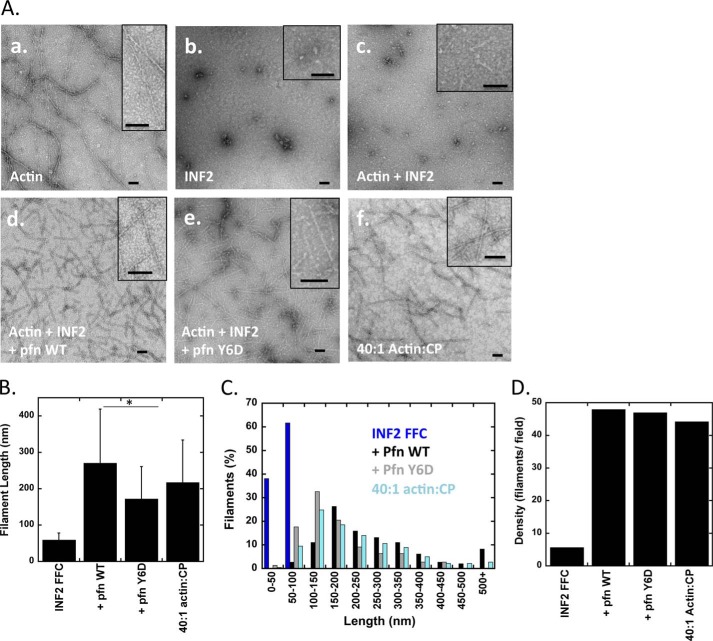
We used negative stain electron microscopy to examine visually the actin-based structures assembled by INF2-FFC in the absence and presence of profilin, allowing the mixtures to reach steady state before processing. Actin alone assembles into long filaments (Fig. 5A), such that they generally span the viewing field at 15,000× magnification (1.9 × 1.9-μm field). A 40:1 ratio of actin:CP displays a large number of shorter filaments, averaging 217 ± 117 nm (Fig. 5, A and B), which is larger than expected theoretically (108 nm). However, the most abundant filament population is in the 100–150-nm range, with a broad distribution of longer filaments that skews the average (Fig. 5C). INF2-FFC alone displays no filament-like structures. With actin:INF2-FFC at a 2:1 ratio, linear structures averaging 59.6 ± 18.7 nm in length are apparent (Fig. 5A). However, these structures are much less abundant than the filaments observed with 40:1 actin:CP or with INF2-FFC/profilin mixes (Fig. 5D). In our velocity analytical ultracentrifugation experiments, we did not detect a peak representing these filaments (Fig. 1F), but their concentration relative to the predominant 8 S particle is probably too low for detection.
FIGURE 5.
Evaluation of INF2 and profilin effects on actin filament length. A, negative-stain electron microscopy with 15,000× magnification. Actin monomers were incubated with INF2-FFC with or without profilin or profilin-Y6D in polymerization buffer for the indicated amounts of time (to attain steady state) before spotting on EM grid. Insets represent close-up of actin filaments. Scale bars, 100 nm. Panel a, 1 μm actin filaments polymerized for 10 min. Panel b, 1 μm INF2-FFC alone. Panel c, 2 μm actin filaments and 1 μm INF2-FFC incubated for 30 min. Panel d, 2 μm actin filaments, 1 μm INF2-FFC, and 4 μm profilin incubated for 15 min. Panel e, 2 μm actin filaments, 1 μm INF2-FFC, and 4 μm profilin-Y6D incubated for 15 min. Panel f, 2 μm actin polymerized in the presence of 50 nm capping protein. B, comparison of average filament lengths quantified from negative-stain EM images such as in A. Bars represent standard deviations. 34, 139, 139, and 170 filaments quantified for INF2-FFC alone, profilin, profilin-Y6D, and 40:1 actin:CP, respectively. The asterisk indicates Student's t test values of p < 1 × 10−5. C, histogram of filament length distributions from negative stain EM in A. D, density of actin filaments per visualization field from conditions in A.
When actin, INF2-FFC, and profilin are mixed in a 2:1:4 molar ratio, the predominant structure is a much more dense field of filaments, with an average length of 270.8 ± 148.2 nm (Fig. 5). Profilin-Y6D gives a similar result, but with shorter average length (172.3 ± 88.7 nm; Fig. 5C). These results show that profilin addition allows filaments to elongate to comparatively uniform lengths at steady state, despite continuous actin turnover in the filament. This uniformity is in contrast to the wide range of lengths of actin polymerized alone (23).
Cytochalasin D Accelerates ATP Turnover by Actin through a Different Mechanism
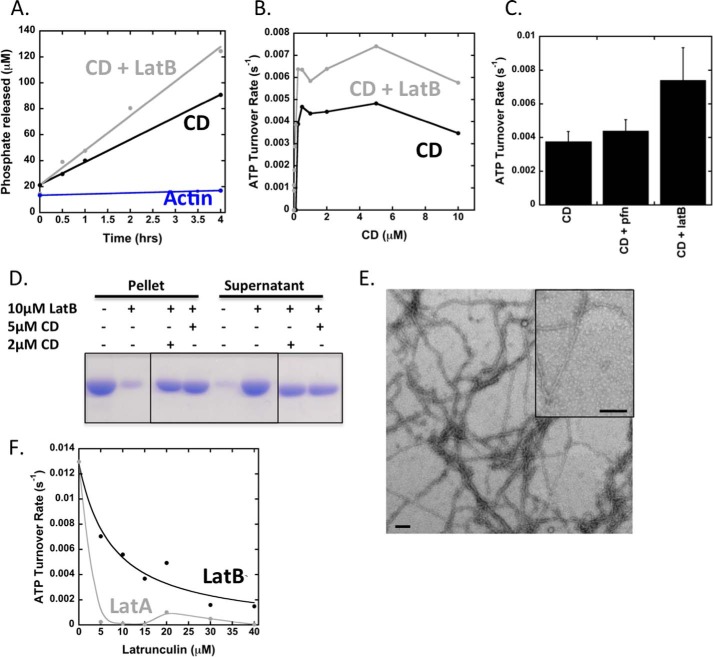
Previous studies have shown that CD at a high concentration can accelerate ATP turnover by actin, through a mechanism involving actin dimerization (42). We wished to test whether the ATP turnover effect of CD was similar to that of INF2. CD causes ATP turnover to a level ∼4-fold lower than INF2-FFC (Fig. 6, A and B). Interestingly, profilin has no effect on this turnover (Fig. 6C), suggesting that the rate-limiting step is not nucleotide exchange on ADP-actin monomers and that profilin does not compete with CD in this process. The addition of LatB does not inhibit the effect of CD on actin turnover but actually stimulates it 1.5–2-fold (Fig. 6C). By high speed pelleting and electron microscopy, substantial actin filaments still exist even in the presence of both CD and LatB (Fig. 6, D and E). In contrast, both LatA and LatB alone inhibit ATP turnover by INF2-FFC/actin substantially, with LatA being considerably more potent and causing complete inhibition at 1:1 ratio to actin (Fig. 6F). Thus, the mechanism of CD-mediated ATP turnover on actin appears to be significantly different from that of INF2-FFC.
FIGURE 6.
Effect of cytochalasin D on ATP turnover by actin. A, time course of ATP turnover for 5 μm actin alone or 5 μm actin + 5 μm CD in the presence or absence of 10 μm LatB. B, concentration curve of ATP turnover for reactions starting with 5 μm actin monomers in the presence or absence of 10 μm LatB and varying concentrations of CD. C, ATP turnover quantified from reactions starting with 5 μm actin monomers and 2 μm CD in the presence or absence of 10 μm LatB or 10 μm profilin. Error bars represent standard deviation for three or four separate experiments. D, high speed sedimentation assay starting with 5 μm actin monomers with indicated concentrations of CD and LatB. Samples were incubated for 20 min at 23 °C before centrifugation. Pellets and supernatants are shown. E, negative-stain EM of sample with 2 μm actin, 1 μm CD, and 4 μm LatB and incubated for 1 h. Insets represent close-up of actin filaments. Scale bars, 100 nm. F, ATP turnover for reactions starting with 5 μm actin monomers and 2 μm INF2 FFC either alone or with indicated concentrations of LatB or LatA.
Discussion
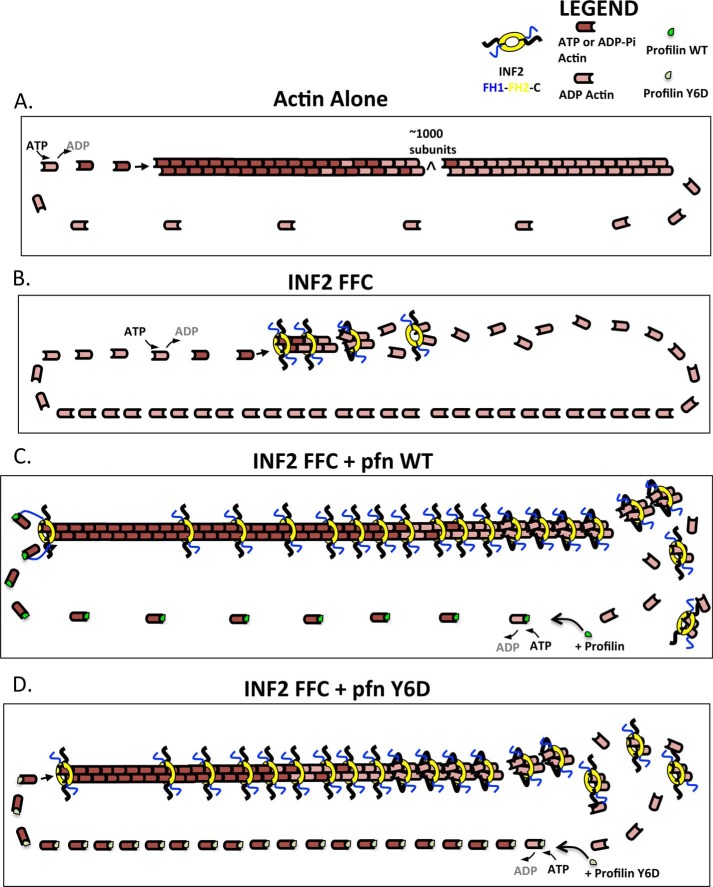
We show that INF2 creates short actin filaments with accelerated turnover through the ensemble of its multiple activities, including nucleation acceleration, barbed end elongation regulation, severing, and actin monomer binding (through the WH2 motif). These activities can also result in net depolymerization, depending on the ratios of actin, INF2, and profilin. In this manner, the activities of INF2 are similar to those of the Cordon Bleu protein, which has been described as a “dynamizer” of actin (43). Below, we describe our model to explain the INF2 cycle on actin filaments in the absence and presence of profilin (Fig. 7).
FIGURE 7.
Models of steady state actin turnover in the presence of INF2 and profilin. Actin subunits are colored dark red for ATP- or ADP-Pi state, and light red for ADP state. INF2-FFC in blue, yellow, and black (for FH1, FH2, and C terminus, respectively). Profilin is shown in green (wild type) or gray (Y6D mutant). A, actin alone. 1) ATP-bound actin adds to the barbed end (10 μm−1 s−1). 2) The incorporated actin hydrolyzes its ATP and releases the phosphate product in a random manner (0.002 s−1). 3) Net dissociation of ADP-actin at the pointed end (0.06 s−1). 4) Nucleotide exchange: the dissociated ADP-actin monomer releases ADP slowly (0.009 s−1) and then binds ATP, enabling another round of barbed end addition. B, actin with INF2-FFC (2:1 ratio). 1) ATP-bound actin adds to the INF2-bound barbed end (3.6 μm−1 s−1). 2) INF2 side binding to filament (calculated rate 0.0014 μm−1 s−1). 3) ATP hydrolysis and phosphate release (unknown in presence of bound INF2). 4) INF2-mediated filament severing (rate unknown). 5) Release of ADP-actins from WH2 of INF2 after severing (rate unknown). 6) nucleotide exchange (0.003 s−1). C, actin with INF2-FFC and profilin (2:1:4 ratio). 1) Profilin-bound ATP-actin adds to the INF2-bound barbed end (6.9 μm−1 s−1). 2) INF2 side binding to filament (calculated rate, 0.0014 μm−1 s−1). 3) ATP hydrolysis and phosphate release (unknown in presence of bound INF2). 4) INF2-mediated filament severing (rate unknown). 5) release of ADP-actins from WH2 of INF2 after severing (rate unknown). 6) Profilin-enhanced nucleotide exchange (1.4 s−1). D, actin with INF2-FFC and profilin-Y6D (2:1:4 ratio). 1) Profilin-bound ATP-actin adds to the INF2-bound barbed end but is not facilitated by the profilin-FH1 interaction (3.6 μm−1 s−1). 2) INF2 side binding to filament (calculated rate 0.0014 μm−1 s−1). 3) ATP hydrolysis and phosphate release (unknown in presence of bound INF2). 4) INF2-mediated filament severing (rate unknown). 5) release of ADP-actins from WH2 of INF2 after severing (rate unknown). 6) Profilin-enhanced nucleotide exchange (1.4 s−1).
In the absence of profilin, INF2-FFC nucleates actin filaments and then stays at the barbed end and allows elongation at a rate ∼40% that of actin alone (Fig. 7B). Additional INF2 binds to the filament side with an affinity independent of the nucleotide state of actin (25). Actin subunits in the filament hydrolyze ATP and release Pi. The length of the ATP and ADP-Pi caps are relatively short, because elongation is slow. Upon Pi release, side-bound INF2-FFC severs the filament, with the C-terminal WH2 motifs binding actin subunits at the barbed end side of the FH2, as suggested by our previous results (25). After severing, the two WH2-bound subunits release from the filament, thereby affecting a net depolymerization. The fact that we observe a predominant complex of four actins and one INF2-FFC dimer at steady state suggests that two additional actins remain bound to the FH2 domain in this complex, in addition to the actins bound to each WH2. The depolymerized actin subunits release from the WH2, exchange ADP for ATP, and participate in another round of filament addition. Nucleotide exchange is the rate-limiting step of the process, resulting in a low ATP:ADP ratio on recycling actin monomers.
Addition of profilin to this system has two effects (Fig. 7C). First, profilin accelerates nucleotide exchange, which raises the ATP:ADP ratio in the pool of recycling actin monomers and shifts the rate-limiting step to elongation. For profilin to bind and accelerate nucleotide exchange on ADP-actin monomers, the depolymerized monomer must first release from the INF2 WH2 motif, because profilin and WH2 compete for a similar binding site (24). Second, profilin increases the barbed end elongation rate through its interaction with the FH1 domain of INF2. This elongation effect is mediated by barbed end-bound INF2, which is distinct from the side-bound INF2 molecules mediating severing. Inhibiting the profilin/INF2 interaction either by the profilin-Y6D mutant (Fig. 7D) or by removal of the FH1 domain slows elongation while still allowing rapid nucleotide exchange, resulting in even higher ATP:ADP ratio in the pool of recycling actin monomers. We are puzzled as to why the polymerization plateau for profilin Y6D is similar to that of wild-type profilin, given the difference in barbed end elongation rates. Further examination of this feature, including kinetic modeling, will be useful.
The following results suggest that nucleotide exchange is the rate-limiting step in the absence of profilin. First, profilin addition increases ATP turnover 6-fold, as opposed to its 2-fold effect on barbed end elongation from INF2-FFC bound ends. Second, turnover increases with added profilin even when INF2/profilin interaction is compromised (by profilin mutation or FH1 domain deletion).
One puzzling feature of this cycle is that Pi release from ADP-Pi subunits does not appear to be the rate-limiting step of the cycle, even though its measured rate (0.002 s−1) (9, 11) is slower than the cycle even in the absence of profilin. Two possible explanations are that 1) INF2-FFC binding to filaments increases Pi release rate or 2) INF2-FFC is able to sever ADP-Pi filaments at a rate sufficient to drive the cycle. Because of technical challenges in measuring Pi release during the rapid polymerization/depolymerization cycle induced by INF2-FFC, we have been unable to test the first possibility. As to the second possibility, our previous results show that addition of Pi to the buffer potently inhibits severing (25). However, this high concentration of added Pi is different from the situation of random ATP hydrolysis and Pi release from an elongating filament (44, 45). Thus, it is possible that severing does not rely on complete Pi release from a filament segment.
Our results with CD confirm those reported by Goddette and Frieden (42) indicating that high concentrations of CD accelerate ATP turnover by actin. However, our results are inconsistent with the model by which CD causes actin dimer assembly that results in ATP hydrolysis, followed by ADP-actin dissociation and nucleotide exchange (42). Our results suggest, rather, that CD causes assembly of an actin filament-like structure that turns over ATP without cycling through the monomer state. The following results support this model: high concentrations of LatB do not inhibit ATP turnover, as they should if actin monomers exist at any point in the cycle; profilin does not alter ATP, as it should if nucleotide exchange is the rate-limiting step; and filament-like structures assemble in the presence of both high CD and LatB. One possible explanation for the difference between our results and those of Goddette and Frieden (42) is the buffer conditions; 50 mm KCl and 1 mm MgCl2 are present in our assays, as opposed to low ionic strength G buffer used in the previous study.
The result of fundamental interest from this work is that the combination of INF2 and profilin is capable of generating short actin filaments that are also highly dynamic. These filaments can be of relatively uniform size, and their length (200–300 nm) can be on the order of those postulated to participate in mitochondrial fission through the ER-bound INF2-CAAX isoform (46). Given that a cytosolic isoform, INF2-non-CAAX, participates in Golgi dynamics (4), it is possible that INF2-generated short, transient actin filaments might be used in other organellar membrane fission processes. Indeed, small actin “puncta” are present at the trans-Golgi network (4, 6), and we have found that the puncta number decreases upon INF2 suppression (4). Other proteins, such as Cordon Bleu, might play similar roles for other actin-based processes.
The length and stability of these filaments depends on INF2 concentration and the presence of profilin. Profilin is at 50–100 μm in mammalian cytosol (38), well in excess of the concentration required to produce this effect. The cytosolic concentration of INF2 is much lower, ranging from 10 to 100 nm in the cytosol from quantitative immune-blotting experiments.3 In addition, the two distinct splice variants of INF2 have very different localization patterns (4, 47). However, local enrichment of INF2, such as on the ER membrane, might drive short filament assembly. Testing this cellular hypothesis will be important in the future.
Author Contributions
P. S. G. designed concepts and experiments, analyzed and interpreted data, co-wrote the paper, and contributed to all figures. M. A. designed, performed, and analyzed the experiments in Fig. 3. B. G. performed experiments related to NMR in Fig. 2. R. S. performed the experiments in Fig. 1. D. F. M. coordinated the NMR experiments. H. N. H. conceived and coordinated the study, analyzed and interpreted data, and co-wrote the paper. All authors approved the final version of the manuscript.
Acknowledgments
We thank Enrique de la Cruz and Tom Pollard for key insights on this project that put us on the right path; Laurent Blanchoin, Dave Sept, and Dave Kovar for additional helpful advice; and Ron Vutre for being able to adapt quickly.
Note Added in Proof
Figs. 1E and 6D were not formatted correctly in the version of this article that was published on June 29, 2015 as a Paper in Press. Figs. 1E and 6D have been revised to show that they were assembled by cutting out the intervening lanes from the original gel.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM069818 (to H. N. H.) and T32 GM008704 (to P. S. G.). The authors declare that they have no conflicts of interest with the contents of this article.
H. N. Higgs, unpublished data.
- FH
- formin homology
- CD
- cytochalasin D
- CP
- capping protein
- FC
- construct containing the FH2 and C-terminal domains
- FFC
- construct containing the FH1 FH2 and C-terminal domains
- LatA
- latrunculin A
- LatB
- latrunculin B.
References
- 1. Chhabra E. S., Higgs H. N. (2007) The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 2. Blanchoin L., Pollard T. D. (1999) Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 [DOI] [PubMed] [Google Scholar]
- 3. Theriot J. A., Mitchison T. J. (1991) Actin microfilament dynamics in locomoting cells. Nature 352, 126–131 [DOI] [PubMed] [Google Scholar]
- 4. Ramabhadran V., Korobova F., Rahme G. J., Higgs H. N. (2011) Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell 22, 4822–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gurel P. S., Hatch A. L., Higgs H. N. (2014) Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr. Biol. 24, R660–R672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miserey-Lenkei S., Chalancon G., Bardin S., Formstecher E., Goud B., Echard A. (2010) Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat. Cell Biol. 12, 645–654 [DOI] [PubMed] [Google Scholar]
- 7. Pollard T. D., Cooper J. A. (1986) Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55, 987–1035 [DOI] [PubMed] [Google Scholar]
- 8. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 9. Blanchoin L., Pollard T. D. (2002) Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry 41, 597–602 [DOI] [PubMed] [Google Scholar]
- 10. Carlier M. F., Pantaloni D., Evans J. A., Lambooy P. K., Korn E. D., Webb M. R. (1988) The hydrolysis of ATP that accompanies actin polymerization is essentially irreversible. FEBS Lett. 235, 211–214 [DOI] [PubMed] [Google Scholar]
- 11. Melki R., Fievez S., Carlier M. F. (1996) Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry 35, 12038–12045 [DOI] [PubMed] [Google Scholar]
- 12. Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 13. Pollard T. D., Cooper J. A. (1984) Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry 23, 6631–6641 [DOI] [PubMed] [Google Scholar]
- 14. Pantaloni D., Carlier M. F. (1993) How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell 75, 1007–1014 [DOI] [PubMed] [Google Scholar]
- 15. Pring M., Weber A., Bubb M. R. (1992) Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry 31, 1827–1836 [DOI] [PubMed] [Google Scholar]
- 16. Campellone K. G., Welch M. D. (2010) A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chesarone M. A., Goode B. L. (2009) Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mockrin S. C., Korn E. D. (1980) Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry 19, 5359–5362 [DOI] [PubMed] [Google Scholar]
- 19. Vinson V. K., De La Cruz E. M., Higgs H. N., Pollard T. D. (1998) Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37, 10871–10880 [DOI] [PubMed] [Google Scholar]
- 20. Selden L. A., Kinosian H. J., Estes J. E., Gershman L. C. (1999) Impact of profilin on actin-bound nucleotide exchange and actin polymerization dynamics. Biochemistry 38, 2769–2778 [DOI] [PubMed] [Google Scholar]
- 21. Paul A. S., Pollard T. D. (2009) Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li F., Higgs H. N. (2003) The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 23. Chhabra E. S., Higgs H. N. (2006) INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281, 26754–26767 [DOI] [PubMed] [Google Scholar]
- 24. Heimsath E. G. Jr., Higgs H. N. (2012) The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J. Biol. Chem. 287, 3087–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gurel P. S., Ge P., Grintsevich E. E., Shu R., Blanchoin L., Zhou Z. H., Reisler E., Higgs H. N. (2014) INF2-mediated severing through actin filament encirclement and disruption. Curr. Biol. 24, 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korobova F., Ramabhadran V., Higgs H. N. (2013) An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hakes D. J., Dixon J. E. (1992) New vectors for high level expression of recombinant proteins in bacteria. Anal. Biochem. 202, 293–298 [DOI] [PubMed] [Google Scholar]
- 28. Ramabhadran V., Gurel P. S., Higgs H. N. (2012) Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J. Biol. Chem. 287, 34234–34245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li F., Higgs H. N. (2005) Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 280, 6986–6992 [DOI] [PubMed] [Google Scholar]
- 30. Harris E. S., Li F., Higgs H. N. (2004) The mouse formin, FRLa, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 31. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 32. Cooper J. A., Walker S. B., Pollard T. D. (1983) Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4, 253–262 [DOI] [PubMed] [Google Scholar]
- 33. Isambert H., Venier P., Maggs A. C., Fattoum A., Kassab R., Pantaloni D., Carlier M. F. (1995) Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270, 11437–11444 [DOI] [PubMed] [Google Scholar]
- 34. Guo B., Gurel P. S., Shu R., Higgs H. N., Pellegrini M., Mierke D. F. (2014) Monitoring ATP hydrolysis and ATPase inhibitor screening using 1H NMR. Chem. Commun. (Camb.) 50, 12037–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leonard M., Song B. D., Ramachandran R., Schmid S. L. (2005) Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol. 404, 490–503 [DOI] [PubMed] [Google Scholar]
- 36. Bubb M. R., Lewis M. S., Korn E. D. (1991) The interaction of monomeric actin with two binding sites on Acanthamoeba actobindin. J. Biol. Chem. 266, 3820–3826 [PubMed] [Google Scholar]
- 37. Gell C., Bormuth V., Brouhard G. J., Cohen D. N., Diez S., Friel C. T., Helenius J., Nitzsche B., Petzold H., Ribbe J., Schäffer E., Stear J. H., Trushko A., Varga V., Widlund P. O., Zanic M., Howard J. (2010) Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 95, 221–245 [DOI] [PubMed] [Google Scholar]
- 38. Pollard T. D., Blanchoin L., Mullins R. D. (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 [DOI] [PubMed] [Google Scholar]
- 39. McCullagh M., Saunders M. G., Voth G. A. (2014) Unraveling the mystery of ATP hydrolysis in actin filaments. J. Am. Chem. Soc. 136, 13053–13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pollard T. D. (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu J., Pollard T. D. (2001) Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell 12, 1161–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goddette D. W., Frieden C. (1986) Actin polymerization. The mechanism of action of cytochalasin D. J. Biol. Chem. 261, 15974–15980 [PubMed] [Google Scholar]
- 43. Husson C., Renault L., Didry D., Pantaloni D., Carlier M. F. (2011) Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell 43, 464–477 [DOI] [PubMed] [Google Scholar]
- 44. Ohm T., Wegner A. (1994) Mechanism of ATP hydrolysis by polymeric actin. Biochim. Biophys. Acta 1208, 8–14 [DOI] [PubMed] [Google Scholar]
- 45. Jégou A., Niedermayer T., Orbán J., Didry D., Lipowsky R., Carlier M. F., Romet-Lemonne G. (2011) Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 9, e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hatch A. L., Gurel P. S., Higgs H. N. (2014) Novel roles for actin in mitochondrial fission. J. Cell Sci. 127, 4549–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chhabra E. S., Ramabhadran V., Gerber S. A., Higgs H. N. (2009) INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 122, 1430–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]