Abstract
Genetic linkage maps are critical for assembling draft genomes to a meaningful chromosome level and for deciphering the genomic underpinnings of biological traits. The estimates of recombination rates derived from genetic maps also play an important role in understanding multiple aspects of genomic evolution such as nucleotide substitution patterns and accumulation of deleterious mutations. In this study, we developed a high-throughput experimental approach that combines fluorescence-activated cell sorting, whole-genome amplification, and short-read sequencing to construct a genetic map using single-sperm cells. Furthermore, a computational algorithm was developed to analyze single-sperm whole-genome sequencing data for map construction. These methods allowed us to rapidly build a male-specific genetic map for the freshwater microcrustacean Daphnia pulex, which shows significant improvements compared to a previous map. With a total of mapped 1672 haplotype blocks and an average intermarker distance of 0.87 cM, this map spans a total genetic distance of 1451 Kosambi cM and comprises 90% of the resolved regions in the current Daphnia reference assembly. The map also reveals the mistaken mapping of seven scaffolds in the reference assembly onto chromosome II by a previous microsatellite map based on F2 crosses. Our approach can be easily applied to many other organisms and holds great promise for unveiling the intragenomic and intraspecific variation in the recombination rates.
Keywords: meiosis, fluorescence-activated cell sorting, single cell, whole-genome amplification
CONSTRUCTING a genetic linkage map that encompasses as much genomic sequence as possible is critical for current endeavors of de novo genomic assembly (e.g., Kawakami et al. 2014; International Cassava Genetic Map Consortium (ICGMC) 2015) and for deciphering the genomic underpinnings of the biological traits in the species of interest (Lynch and Walsh 1998). Genetic maps can be utilized in several ways to help achieve these goals. At the very least, a linkage map with an adequate number of genetic markers can serve as the backbone for orienting and assembling segments of DNA (i.e., scaffolds) into chromosomes. The developed genetic markers can also be used in QTL association mapping and genomic-scanning efforts to investigate genetic loci underlying ecological tolerance, adaptation, and disease. Furthermore, a linkage map provides genome-wide estimates of the meiotic recombination rate, which plays a significant role in the distribution of genetic diversity (Nachman 2001), rate of adaptation (Bachtrog and Charlesworth 2002), accumulation of deleterious mutations (Hussin et al. 2015), and nucleotide substitution (Duret and Arndt 2008).
Currently, the most common approach for constructing a genetic linkage map is based on genotyping a large number of molecular markers (e.g., SNPs, microsatellites) from a large number of offspring (usually on the order of hundreds) derived from various kinds of crossing schemes (e.g., backcrosses, F2s, recombinant inbred lines) or from family trios to estimate the frequencies of recombination between markers across the genome. Although most estimates of recombination rates in a diverse range of taxa come from studies based on this idea, crossing experiments are laborious, often inefficient (e.g., low hatching/survival rate of progeny), and unattainable in cases where manipulative crossing is impossible. Furthermore, this approach may introduce biases in estimating offspring genotype frequencies if certain classes of recombinant genotypes cause lethality and/or low viability.
With whole-genome sequencing becoming increasingly economical, interpretation of population-genomic data in a coalescence framework can generate estimates of historical recombination (i.e., crossover) rates needed for creating genetic maps (Stumpf and McVean 2003; McVean et al. 2004). This approach is gaining popularity and has been successfully applied to many organisms, including human (McVean et al. 2004; Myers et al. 2005), chimpanzee (Auton et al. 2012), dogs (Auton et al. 2013), and Arabidopsis (Choi et al. 2013), uncovering genetic factors that regulate crossover events such as PRDM9 in humans (Myers et al. 2010) and H2A.Z nucleosomes at promoters in Arabidopsis (Choi et al. 2013). Nonetheless, one prerequisite for applying this method is the availability of a genome assembly with reasonable quality for the species of interest or for a closely related species. This is because crossover events are inferred on the basis of patterns of linkage disequilibrium in the population of interest, which can be severely distorted if the mapping of short-sequence reads is performed on an assembly where physical ordering of genomic sites is erroneous. It should also be noted that the linkage-disequilibrium method can yield only estimates for historical population-level and sex-averaged crossover rates. Thus, this method is not suitable for examining the variation of recombination rate between individuals and sexes (e.g., Coop and Przeworski 2007; Kong et al. 2010; Brandvain and Coop 2012; Comeron et al. 2012; Bauer et al. 2013). Furthermore, it is well known that crossover, the reciprocal exchange of DNA between homologous chromosomes, only represents a small proportion (often <20%) of the total recombination events, with the rest generating nonreciprocal, gene-conversion events (Langley et al. 2000; Malkova et al. 2004; Morrell et al. 2006; Mancera et al. 2008; Yang et al. 2012; Lynch et al. 2014).
The most recent development in linkage map construction methodologies is the deployment of single-cell whole-genome sequencing on gametes such as sperm (Lu et al. 2012; Wang et al. 2012). Gametes are products of meiosis, bearing the genetic signature of meiotic recombination. Directly examining the haplotypes of gametes can offer an unbiased view of the recombination process and potentially yields insight into genetic factors regulating meiotic recombination events. The idea of utilizing single sperm to examine the recombination rate between a few markers on a small genomic scale coincided with the time when PCR (polymerase chain reaction) technology started revolutionizing the field of molecular biology in the 1980s (Li et al. 1988; Cui et al. 1989). However, this approach has not gained popularity until recently, mainly because of two technical hurdles, i.e., the rapid isolation of a large number of single gametes and the low amount of DNA present in each gamete. Problems inherent in sorting a large number of individual gametes made it implausible to gain enough power to estimate recombinant frequencies and disentangle individual gamete recombination patterns. Additionally, the extremely low amount of DNA present in each gamete (e.g., ∼3 pg of DNA in a single human sperm) prevented genotyping a sufficient number of genetic markers. Nonetheless, technological advances in microfluidics and flow cytometry have made it feasible to rapidly isolate single cells (Wang et al. 2012), and whole-genome amplification techniques now can propagate a single-copy genome to an amount of DNA that allows subsequent whole-genome sequencing/genotyping (Pan et al. 2008; Zong et al. 2012). Given the abundance of sperm cells, the combination of these techniques with whole-genome sequencing has led to the successful construction of a marker-dense male-specific genetic map for humans (Lu et al. 2012; Wang et al. 2012). Nonetheless, to our knowledge this single-sperm whole-genome sequencing approach has not been applied to other organisms.
In this study, we developed a high-throughput experimental workflow that isolates, whole-genome amplifies, and sequences single sperm from the North American microcrustacean Daphnia pulex (Crustacea, Anomopoda) to build a genetic map. Daphnia are keystone zooplankton species in global freshwater ecosystems (e.g., lakes and woodland ponds) and are model organisms for toxicological, evolutionary, and biomedical research (Colbourne et al. 2011). Daphnia typically reproduce by cyclical parthenogenesis. Under good environmental conditions, females produce directly developing and genetically identical daughters. However, with unfavorable conditions (e.g., food shortage), males are produced by environmental sex determination and engage in sexual reproduction to produce diapausing embryos. A major motivation for us to develop single-sperm sequencing for building a genetic map in D. pulex is the difficulty in efficiently hatching the diapausing embryos needed to form an offspring panel from crossing experiments (e.g., Cristescu et al. 2006).
Although there exists a microsatellite-based genetic map for the current Daphnia genome assembly, this map anchors only 73 scaffolds, which accounts for only 73.9 of the 200 Mbp Daphnia genome (Cristescu et al. 2006; Colbourne et al. 2011). Furthermore, there is a need for a genetic map specifically for D. pulex because the current Daphnia assembly and genetic map are both derived from Daphnia arenata, an endemic species to Oregon rather than D. pulex that occurs in most of the temperate regions in North America. Although D. arenata and D. pulex are nearly indistinguishable with respect to morphology, D. arenata shows significantly reduced heterozygosity relative to D. pulex, with a pairwise nucleotide diversity of 0.0013 vs. 0.0119 in D. pulex based on six protein-coding loci (Omilian and Lynch 2009). Although D. arenata is still paraphyletic with respect to D. pulex for the mitochondrial genome (Colbourne et al. 1998; Lynch et al. 2008), these two species display a nuclear genomic divergence of ∼2% (Tucker et al. 2013). Nonetheless, it remains largely an open question whether the nuclear genetic divergence between these two species involve any genetic map changes, i.e., chromosomal rearrangements such as inversions. Comparative analysis of the genetic maps of these two species will offer insight into the process of their genetic divergence and speciation.
The experimental procedure presented here employs fluorescence-activated cell sorting (FACS), the whole-genome amplification technique multiple annealing and looping-based amplification cycles (MALBAC) (Zong et al. 2012), and the Illumina Hi-Sequation 2500 sequencing platform. Because this procedure relies on equipment such as flow cytometers and standard PCR thermal cyclers that are readily available to the research community, it can be easily applied and/or adapted to many other organisms with or without existing reference genomic assemblies to rapidly generate genetic maps. Moreover, we provide a computer program for extraction of haplotype blocks that are free of recombination events (i.e., crossover and gene conversion) to build a male-specific genetic map in combination with the genetic map software MSTMap (Wu et al. 2008). This method constitutes a valuable approach for future studies that examine sex-specific genetic maps, fine-scale recombination patterns, and individual recombination variation.
Materials and Methods
Daphnia culture
The Daphnia isolate (PA42) used in this study was sampled from Portland Arch (latitude: 40°13, longitude: −87°20), Indiana, in May 2013. This isolate, which reproduces by cyclical parthenogenesis, was maintained under benign laboratory conditions at 20° and fed ad libitum with a suspension of Scenedesmus obliquus to enable essentially indefinite parthenogenetic reproduction.
Single-sperm isolation
We collected 15 mature, parthenogenetically produced males from the mass culture of the PA42 isolate. These males are genetically identical except for de novo mutations, which can be safely ignored given their low frequency (on the order of 10−9 events/base/generation). Sperm was collected from each male by squeezing the abdominal part of the individual in a drop of ultrapure distilled H2O under a cover slip. The presence of sperm was confirmed by examination under microscope. The pooled collection of sperm was transferred to 50 µl PBS buffer and stained using Hoechst 33528 (100 µg/µl, Sigma-Aldrich), a dye that binds to double-strand DNA. Then, we used a FACS Aria II SORP Flow Cytometer (BD Biosciences) to isolate single sperm. Lasers used were a 488 nm 100 mW for light scatter detection and a 355 nm 20 mW for Hoechst detection. A FSC–PMT was used for optimal small particle discrimination. A 70-µm nozzle was used at 45 psi. Sperm cells were dispensed into regular 96-well PCR plates. Each sperm was deposited into 5 µl cell lysis buffer (30 mM Tris, 2 mM EDTA, 20 mM KCl, 0.2% Triton-X100, 50 mM DTT, and 500 µm/ml protease) and lysed for 3 hr at 50°, 20 min at 75°, and 5 min at 80°.
Whole-genome amplification and sequencing
We whole-genome amplified 104 single-sperm cells following the MALBAC protocol (Zong et al. 2012). In brief, MALBAC consists of a preamplification stage and a second-stage PCR amplification. In the preamplification cycles, a pair of quasi-degenerate primers is used to initiate overlapped amplicons throughout the whole genome. The quasi-degenerate primers consist of a 27-bp fragment of 5ʹ-GTGAGTGATGGTTGAGGTAGTGTGGAG-3ʹ and eight variable nucleotides attached to the 3ʹ end. For each reaction, 3 µl ThermPol buffer (New England Biolabs), 1 µl dNTP (10 mM), 21 µl H2O, and 0.15 µl primers (50 µM) were added. Each sample is then denatured at 94° for 3 min and quenched on ice immediately. With samples staying on ice, 0.6 µl Bst large fragment (New England Biolabs) is added to each reaction. The following thermal regime is used to generate random amplicons across the genome: 10° for 45 sec, 15° for 45 sec, 20° for 45 sec, 30° for 45 sec, 40° for 45 sec, 50° for 45 sec, 65° for 2 min, 95° for 20 sec, followed by quenching on ice and adding 0.6 µl Bst large fragment to each sample. Subsequently, the samples are subject to five rounds of amplification, each of which consists of 10° for 45 sec, 15° for 45 sec, 20° for 45 sec, 30° for 45 sec, 40° for 45 sec, 50° for 45 sec, 65° for 2 min, 95° for 20 sec, 58° for at least 20 sec, followed by quenching on ice and adding 0.6 µl Bst large fragment to each sample. After the preamplification stage, a standard PCR amplification is performed with each sample to generate 1–2 µg DNA to be used for downstream applications. Each reaction consists of 3 µl ThermoPol Buffer (New England Biolabs), 1 µl dNTP (10 mM), 26 µl H2O, 0.15 µl primer 5ʹ-GTGAGTGATGGTTGAGGTAGTGTGGAG-3ʹ (100 mM), and 1 µl DeepVentR exo- (New England Biolabs). The PCR thermal regime consists of 22 rounds of 94° for 20 sec, 59° for 20 sec, 65° for 1 min, 72° for 2 min, followed by 72° for 5 min.
To eliminate the possibility of samples containing multiple sperm cells, 12 microsatellite markers, each from 1 of the 12 chromosomes in the Daphnia genome, were genotyped using an ABI 3730 genetic analyzer (Life Technologies). The allele sizes of the genotyped microsatellite loci were analyzed using Genemapper software 4.0 (Life Technologies). None of the samples presented evidence of multiple cells, i.e., more than one allele for the whole suite of loci.
Subsequently, the whole-genome-amplified DNA of each sperm was sheared to an average fragment size of 350 bp on a Covaris S2 shearing machine. Short-read sequencing library preparation followed the standard protocol of Illumina and was done by the Center for Genomics and Bioinformatics at Indiana University, Bloomington. Short-read sequencing was performed on an Illumina HiSeq2500 platform with 150-bp paired end reads.
Construction of genetic maps
Our approach can be applied to organisms with or without an existing reference assembly. This is because the whole-genome sequences of all sperm samples can be used for creating a de novo assembly, although the completeness of the assembly depends on the whole-genome amplification coverage across the entire genome. To demonstrate this, we built a de novo sperm reference assembly using the assembler Platanus (Kajitani et al. 2014) based on the pooled whole-genome sequences of all sperm. The pooled sequences for de novo assembly were normalized using the software BBNorm (http://sourceforge.net/projects/bbmap/) to reduce the redundancy and remove errors of raw reads. We used the default settings in Platanus for building contigs and scaffolds. All scaffolds that were possibly from contaminant DNA (e.g., bacteria, algae, and human) or shorter than 1000 bp were removed from the final sperm reference assembly.
The 27-bp primer sequences for the whole-genome amplification reactions were computationally removed from the ends of raw reads when present using the software CLC Genomics Workbench (v. 7, CLC Bio). The processed raw reads for each sample were mapped to the Daphnia reference assembly (Colbourne et al. 2011) and the sperm reference assembly using the short-read mapping function implemented in CLC Genomics Workbench with default settings. However, reads mapped to multiple locations were removed from further analysis. The haplotype for each position of a single sperm was determined using a consensus approach, where a base call is made with the support of >80% of the reads. To avoid sequencing errors, PCR artifacts, and potential mapping errors, we also require at least two forward and two reverse reads to validate the consensus call. Because only heterozygous loci are informative for analyzing recombination events, only sites where two nucleotides were found across the entire set of sperm samples were kept.
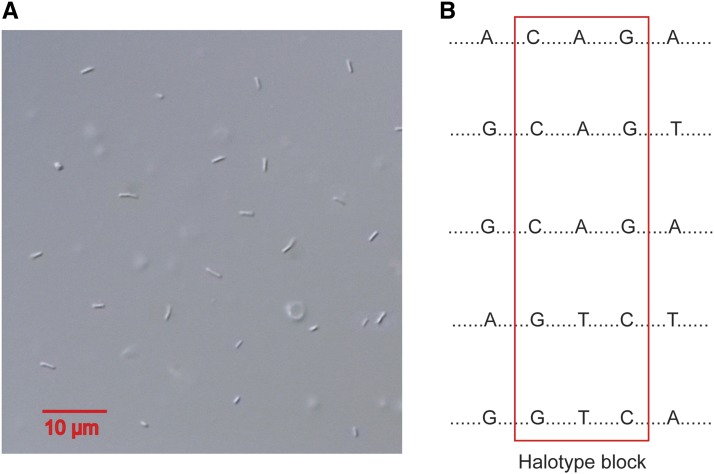
We developed an algorithm implemented in Python (Supporting Information, File S5, phasingHaplotype.py) to detect haplotype blocks that are free of recombination events to be used as markers for genetic map construction. Because this set of sperm is derived from the recombination of two parental haplotypes, we randomly assign either 0 or 1 (designating the two parental haplotypes) to the same two-locus haplotype for the first pair of sites on each scaffold (Figure 1B). Every pair of sites across the samples has a maximum of four haplotypes when a crossover or gene conversion event happens, three haplotypes for biased gene conversion events, and two haplotypes for no recombination. We then consecutively examine each pair of sites and extend the haplotype assignment. A switching of phase occurs only when three or four haplotypes evidently exists.
Figure 1.
(A) Image of sperm extracted from males of the D. pulex PA42 isolate from Portland Arch, Indiana. The sperm is rod shaped, with a length ∼2 µm. (B) A hypothetical example of haplotype block indicated by the red box. Each haplotype is represented by five nucleotide sites. A haplotype block is identified where only two haplotypes occur across the whole set of samples.
Once the haplotype-phase assignment was done, we selected haplotype blocks (a minimum of two SNP sites) free of evident recombination events in the set of sequenced sperm samples as genetic markers for genetic map construction. The selected haplotype blocks were coded as either 0 or 1. We used the software MSTMap (Wu et al. 2008) to construct the linkage map with its default settings. MSTMap implements an efficient algorithm to determine the correct order of a large number of genetic markers (10,000–100,000) by computing the minimum spanning tree. MSTMap is significantly better at recovering the correct order of markers from noisy data compared to most other software, which is helpful for dealing with the missing data in our data set.
MareyMap database for estimating recombination rate
Integrating the genetic map data into a physical map allows the estimation of recombination rate for genomic regions along a chromosome using different curve fitting methods (Rezvoy et al. 2007). This is the so-called Marey map method (Chakravarti 1991). We combined the sperm genetic map data with the current Daphnia reference assembly (Colbourne et al. 2011) and created a database (File S3) using the R package MareyMap (Rezvoy et al. 2007), which allows users to estimate recombination rates for most of the regions on major scaffolds in the current assembly.
Data availability
The binary alignment files for sperm samples were deposited at NCBI Sequence Read Archive under study no. SRP058678.
Results
Single-cell genome sequencing, mapping, and SNP density
We sequenced the whole genomes of 104 single sperm (Figure 1A) from the D. pulex isolate PA42 from Portland Arch, Indiana, with 150-bp paired end reads. After trimming the PCR and Illumina adapter sequences from the raw reads and removing reads that mapped to multiple locations in the Daphnia reference assembly (Colbourne et al. 2011), we found that the aligned reads covered on average 77.3 Mbp ± 14.7 (SD) of the 200-Mbp Daphnia genome, which is equivalent to 52.8% of the resolved regions in the reference assembly. The average coverage per site per sperm sample is 12.5 ± 4.5 (SD).
We identified a total of 1,537,288 heterozygous sites from the parental diploid genotype PA42 following a set of stringent criteria (see Materials and Methods). With a 2% estimated heterozygosity based on whole-genome sequencing of the PA42 genotype, these recovered sites account for 38.4% of the total heterozygous sites expected in the genome.
Furthermore, we built a de novo assembly using the pooled sperm genomic sequences. The sperm assembly spans 109 Mbp with 1.7% gap regions and consists of 32,549 scaffolds. The largest scaffold size is 90,435 bp, whereas the smallest scaffold is 1001 bp. After mapping the processed raw reads for each sperm to the sperm assembly, the aligned reads covered on average 49.4 Mbp ± 11.9 of the assembly. The observation that the average coverage is lower than the total assembly length is mainly because the sperm assembly is based on the pooled sequence of all samples and each whole-genome amplified sperm sample does not contain all sequences in the assembly.
Genetic linkage map
For the analyses based on the Daphnia reference genomic assembly (Colbourne et al. 2011), we selected a total of 1672 marker regions where at least two consecutive SNP loci show the same haplotype in >90 sperm samples; as they contained zero recombination events, such spans serve as single markers in the map construction. The lengths of the marker regions range from 50 to 269,519 bp, with a mean of 22,771 ± 21,710 (SD). The total length of the marker regions is ∼38.1 Mbp, comprising 19.1% of the ∼200-Mbp Daphnia genome. Based on these markers, we constructed a male-specific Daphnia linkage map using the software MSTmap (Wu et al. 2008). This genetic map consists of 12 linkage groups, corresponding to the 12 chromosomes in Daphnia genome (Zaffagnini and Sabelli 1972), and spans a total genetic distance of 1451 Kosambi cM (Figure S1), with an average intermarker distance of 0.87 cM. The number of haplotype blocks on each linkage group ranges between 67 and 202, with a mean of 139. The map distance for each linkage group varies between 81 and 149 Kosambi cM, with a mean of 121 ± 22 (SD) cM. This map anchors to chromosomes a total of 187 of 5191 scaffolds from the Daphnia genome reference assembly (Colbourne et al. 2011). These scaffolds encompass 131.9 Mbp of DNA sequence, which is equivalent to 90.0% of the resolved portion of current assembly.
To demonstrate that our approach can be used for organisms without preexisting reference assembly, we built a genetic map based on sperm de novo assembly. We recovered 12 linkage groups using 350 haplotype blocks (File S4). The total length of the 350 haplotype blocks is 2.36 Mbp, corresponding to 2.2% of the sperm assembly. The lengths of the haplotype blocks range from 50 to 47,702 bp, with a mean of 6742 bp. The total genetic map distance is 823 Kosambi cM. The map distance for each lineage group ranges between 23 and 137 cM, with a mean of 67 ± 34 (SD) cM. The average intermarker distance is 2.35 cM. Furthermore, this map anchors in total 343 scaffolds into the 12 linkage groups.
Differences of scaffold assignment between maps of D. pulex and D. arenata
We compared the male-specific genetic map of D. pulex with the prior microsatellite map of D. arenata to detect possible chromosomal rearrangements. Because all of the microsatellite markers in D. arenata have known physical locations in the reference assembly, we were able to compare the differences in anchored locations of scaffolds. Most notably, a large chunk of chromosome II spanning from 0 to 29.9 cM in D. arenata maps to chromosome XII in D. pulex; this map difference involves seven scaffolds (5, 27, 58, 63, 70, 84, and 86). Furthermore, we detected numerous cases in which segments of one scaffold in the current reference assembly do not form a continuous tract on the genetic map but are split by segments from other scaffolds (Figure S1), which indicates problematic genomic assembly or possible rearrangements. An example of this category of observations is that one portion of scaffold 260,935–1,006,942 bp maps to chromosome XI, while the rest of scaffold 8 is anchored to chromosome IV (File S1). In addition to these cases of split scaffolds, in many of the largest scaffolds (e.g., scaffolds 1, 2, 3) in the current Daphnia assembly, physical orders of haplotype blocks do not agree with the genetic map (File S1). For example, scaffold 1: 260,935–1,006,942 bp is mapped on chromosome II with a map distance from 42.68 to 44.35 cM, whereas the physically downstream segment scaffold 1 3806402–3934086 is mapped from 34.34 to 42.11 cM on the same chromosome.
Discussion
Combining single-cell isolation based on flow cytometry, whole-genome amplification, and short-read sequencing, we established a rapid experimental workflow for constructing a genetic map in the microcrustacean D. pulex using single sperm. Compared to the extensive time (e.g., weeks or months) required by conventional crossing experiments, this procedure can gather the data needed for a map in a time frame of a few days. Unlike the population-genomic sequencing approach that requires a reference genomic assembly (e.g., Auton et al. 2012), the sperm-based method does not necessarily rely on a preexisting reference assembly. Although we did use a Daphnia reference assembly to guide the identification of SNP sites in this study, we were also able to use spermwhole-genome sequences to create a de novo assembly as an alternative basis for downstream genetic linkage map construction. Our results show that although this map is less marker dense than the one generated with the existing Daphnia reference assembly, the correct number of linkage groups is recovered. More importantly, it should be noted that this represents an extreme situation in which there are no previous genomic resources at all. As a result, the quality of the genetic map can be significantly improved by incorporating a few more paired-end and mate-pair sequencing libraries into the de novo assembly process. The discussion below focuses only on the genetic map generated using the existing Daphnia genomic reference assembly.
Our experimental workflow heavily relies on FACS for isolating single sperm. There are a few other alternative approaches for isolating single cells such as manual micromanipulation and laser dissection (Macaulay and Voet 2014). However, to achieve accurate isolation of single cells in a high-throughput manner, FACS provides a highly reliable platform with advantages that alternative approaches cannot offer. Considering that the small size of Daphnia sperm (a length of ∼2–3 µm, Figure 1) makes them indistinguishable from dust particles if they were sorted only by size on the flow cytometer, we stained sperm cells with a Hoechst dye, which binds to double-stranded DNA. Combining sorting by size and wavelength of fluorescence emission on the flow cytometer ensures highly accurate and rapid isolation of single sperm. For example, it takes only ∼1 hr to sort ∼1000 single sperm and the accuracy for single-cell isolation is 100% in our data set (see Materials and Methods).
A major concern with whole-genome amplifying an extremely small amount of DNA in a single cell is the uneven amplification on different regions of the genome, which can lead to biased genome-wide coverage (for a recent review see Macaulay and Voet 2014). A major reason that we chose MALBAC in our workflow is that this procedure limits the further amplification of the genomic regions that are amplified early on in the reaction, resulting in much improved coverage across the genome (Zong et al. 2012). Although the average breadth of coverage across the sperm samples is only 52.8% of the resolved region in the reference assembly, the mapped scaffolds cover 90.0% of the resolved regions. Therefore, with this efficient amplification procedure, our genetic map achieves the goal of encompassing as much actual genome sequence as possible.
These state-of-the-art technologies greatly aided our generation of an ultradense male-specific genetic map for D. pulex based on short-read sequencing, yielding a substantial improvement over the previous microsatellite-based Daphnia genetic map (Table 1), which actually required substantially more work. The total genetic map distance of the new map is 1451 Kosambi cM, slightly greater than that of the D. arenata map (1206 cM) and similar to D. magna (1483 cM; Routtu et al. 2014). This map anchors to chromosomes 187 scaffolds (131.9 Mbp), in comparison to 73 anchored scaffolds (73.9 Mb) from the D. arenata map. Furthermore, the new map provides much refined estimates of the recombination rate between markers (0.87 vs.7 cM), allowing the mapping of 90% of the D. pulex genome to on average <1 cM to the nearest genetic marker. This intermarker distance is also smaller than that in the SNP-based D. magna genetic map (1.13 cM; Routtu et al. 2014). Therefore, this map provides a framework to examine the role of recombination rate in shaping the various aspects of the genomic architecture of Daphnia genome such as patterns of nucleotide substitution and codon usage, which to date have not been explored in detail.
Table 1. Comparison between the sperm-based genetic map for D. pulex and the microsatellite genetic map for D. arenata (Cristescu et al. 2006).
| D. pulex map | D. arenata map | |
|---|---|---|
| Total map distance (Kosambi cM) | 1451 | 1206 |
| No. of markers | 1672 | 185 |
| No. of scaffolds mapped | 187 | 73 |
| Basepairs of genome mapped (Mbp) | 131.9 | 73.9 |
| Average intermarker distance (cM) | 0.87 | 7 |
The statistics for the D. arenata map were compiled from Cristescu et al. (2006) and Colbourne et al. (2011).
Another goal of this study was to examine whether chromosomal rearrangement is involved in the divergence of D. pulex and D. arenata. It should be noted that genomic assemblies are not perfect and could contain misassembled regions (e.g., physically distant regions assembled adjacent to each other). Misassembled regions can disrupt the map orders of nearby markers, leading to false conclusions about true genomic rearrangements. In the current map, we observed many cases of split scaffolds and discrepant intrascaffold orderings of markers between genetic and physical maps. Unfortunately, we are not able to distinguish between these assembly errors and true rearrangements for these problematic genomic areas (Figure S1). Nonetheless, given the numerous occurrences of these observations in many different parts of the reference assembly, which was built with only 8.7× coverage of Sanger sequencing reads and with little data from long insert mate-pair libraries (Colbourne et al. 2011), our observations of aberrant mappings are more likely to reflect assembly errors than true rearrangements.
The most notable difference between the D. arenata and D. pulex map involves changes between chromosome II and chromosome XII. The scaffolds mapped to between 0 and 29.9 cM on chromosome II in D. arenata are mapped onto chromosome XII in D. pulex. Although this may potentially represent an interesting case of chromosomal translocation, a few observations collectively suggest that this is an error from the previous mapping study. First, this map interval in D. arenata was affected by segregation distortion (Cristescu et al. 2006), showing severe homozygote deficiency for the markers in this region. The map distance between this region and the closest genetic marker in the D. arenata map is 40.6 cM, which indicates its weak genetic linkage with the rest of the chromosome (because 50 cM map distance means free recombination). In fact, while the entire scaffold 5 in the reference assembly is unambiguously mapped to chromosome XII in the current map, the D. arenata genetic map shows that this scaffold is split between chromosome II and XII, indicating potential problems in assigning genetic markers to chromosomes.
Because of the high heterozygosity in the Daphnia genome and the great number of heterozygous sites recovered from single-sperm whole-genome sequencing, our data set offers a great opportunity to locate genomic intervals containing the breakpoints for crossover events and gene-conversion tracts. However, accomplishing such a task requires a reference assembly that embodies the correct physical order of all nucleotide sites in the genome. Because of the great number of potentially problematic assembled regions in the current Daphnia assembly, we are working on a de novo assembly for the PA42 D. pulex isolate using the sperm sequencing strategy in combination with a range of sequencing libraries with different insert sizes. In fact, there is a growing interest in using genome sequences of mapping panels to facilitate de novo genome assembly and alleviate problems of falsely assembling the two divergent alleles of the same locus into paralogous loci (Hahn et al. 2014). With the new genome assembly, we will hopefully gain sufficient power to reveal the genomic location of crossover and gene-conversion events and characterize the possible genetic elements that control the occurrence of recombination events.
Supplementary Material
Acknowledgments
We thank C. Hassel at the Indiana University Flow Cytometry Facility for technical assistance, K. Young for maintaining Daphnia culture, and W. Sung for bioinformatics assistance. The computational analyses were supported in part by National Science Foundation (NSF) grants CNS-0723054 and CNS-0521433, which support computational facilities at Indiana University. This work is supported by NSF grant DBI-1229361 to W.K.T. and National Institutes of Health grant R01GM101672 to M.L.
Footnotes
Communicating editor: J. Shendure
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179028/-/DC1.
Literature cited
- Auton A., Fledel-Alon A., Pfeifer S., Venn O., Ségurel L., et al. , 2012. A fine-scale chimpanzee genetic map from population sequencing. Science 336: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A., Li Y. R., Kidd J., Oliveira K., Nadel J., et al. , 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9: e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Charlesworth B., 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416: 323–326. [DOI] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y., Coop G., 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics 190: 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., 1991. A graphical representation of genetic and physical maps: the Marey Map. Genomics 11: 219–222. [DOI] [PubMed] [Google Scholar]
- Choi K. H., Zhao X. H., Kelly K. A., Venn O., Higgins J. D., et al. , 2013. Arabidopsis meiotic crossover hot spots overlap with H2A. Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne J. K., Crease T. J., Weider L. J., Hebert P. D. N., Dufresne F., et al. , 1998. Phylogenetics and evolution of a circumarctic species complex (Cladocera: Daphnia pulex). Biol. J. Linn. Soc. Lond. 65: 347–365. [Google Scholar]
- Colbourne J. K., Pfrender M. E., Gilbert D., Thomas W. K., Tucker A., et al. , 2011. The ecoresponsive genome of Daphnia pulex. Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Przeworski M., 2007. An evolutionary view of human recombination. Nat. Rev. Genet. 8: 23–34. [DOI] [PubMed] [Google Scholar]
- Cristescu M. E., Colbourne J. K., Radivojac J., Lynch M., 2006. A microsatellite-based genetic linkage map of the waterflea, Daphnia pulex: on the prospect of crustacean genomics. Genomics 88: 415–430. [DOI] [PubMed] [Google Scholar]
- Cui X. F., Li H. H., Goradia T. M., Lange K., Kazazian H. H., et al. , 1989. Single-sperm typing: determination of genetic distance between the G-gamma globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc. Natl. Acad. Sci. USA 86: 9389–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Arndt P. F., 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4: e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Zhang S. V., Moyle L. C., 2014. Sequencing, assembling, and correcting draft genomes using recombinant populations. G3 Genes Genomes Genetics 4: 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin J. G., Hodgkinson A., Idaghdour Y., Grenier J.-C., Goulet J.-P., et al. , 2015. Recombination affects accumulation of damaging and disease-associated mutations in human populations. Nat. Genet. 47: 400–404. [DOI] [PubMed] [Google Scholar]
- International Cassava Genetic Map Consortium (ICGMC) , 2015. High-resolution linkage map and chromosome-scale genome assembly for Cassava (Manihot esculenta Crantz) from 10 populations. G3 Genes Genomes Genetics 5: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., et al. , 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Smeds L., Backstrom N., Husby A., Qvarnstrom A., et al. , 2014. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol. Ecol. 23: 4035–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D. F., Masson G., Sigurdsson A., et al. , 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Langley C. H., Lazzaro B. P., Phillips W., Heikkinen E., Braverman J. M., 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(w(a)) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. H., Gyllensten U. B., Cui X. F., Saiki R. K., Erlich H. A., et al. , 1988. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335: 414–417. [DOI] [PubMed] [Google Scholar]
- Lu S., Zong C., Fan W., Yang M., Li J., et al. , 2012. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338: 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA. [Google Scholar]
- Lynch M., Seyfert A., Eads B., Williams E., 2008. Localization of the genetic determinants of meiosis suppression in Daphnia pulex. Genetics 180: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Xu S., Maruki T., Jiang X. Q., Pfaffelhuber P., et al. , 2014. Genome-wide linkage-disequilibrium profiles from single individuals. Genetics 198: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay I. C., Voet T., 2014. Single cell genomics: advances and future perspectives. PLoS Genet. 10: e1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Swanson J., German M., McCusker J. H., Housworth E. A., et al. , 2004. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Myers S. R., Hunt S., Deloukas P., Bentley D. R., et al. , 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Morrell P. L., Toleno D. M., Lundy K. E., Clegg M. T., 2006. Estimating the contribution of mutation, recombination and gene conversion in the generation of haplotypic diversity. Genetics 173: 1705–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C., et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman M. W., 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17: 481–485. [DOI] [PubMed] [Google Scholar]
- Omilian A. R., Lynch M., 2009. Patterns of intraspecific DNA variation in the Daphnia nuclear genome. Genetics 182: 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X. H., Urban A. E., Palejev D., Schulz V., Grubert F., et al. , 2008. A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc. Natl. Acad. Sci. USA 105: 15499–15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvoy C., Charif D., Gueguen L., Marais G. A. B., 2007. MareyMap: an R-based tool with graphical interface for estimating recombination rates. Bioinformatics 23: 2188–2189. [DOI] [PubMed] [Google Scholar]
- Routtu J., Hall M. D., Albere B., Beisel C., Bergeron R. D., et al. , 2014. An SNP-based second-generation genetic map of Daphnia magna and its application to QTL analysis of phenotypic traits. BMC Genomics 15: 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M. P. H., McVean G. A. T., 2003. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 4: 959–968. [DOI] [PubMed] [Google Scholar]
- Tucker A. E., Ackerman M. S., Eads B. D., Xu S., Lynch M., 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl. Acad. Sci. USA 110: 15740–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. B., Fan H. C., Behr B., Quake S. R., 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150: 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. H., Bhat P. R., Close T. J., Lonardi S., 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4: e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. H., Yuan Y., Wang L., Li J., Wang W., et al. , 2012. Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. USA 109: 20992–20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini F., Sabelli B., 1972. Karyologic observations on the maturation of the summer and winter eggs of Daphnia pulex and Daphnia middendorffiana. Chromosoma 36: 193–203. [DOI] [PubMed] [Google Scholar]
- Zong C., Lu S., Chapman A. R., Xie X. S., 2012. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338: 1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The binary alignment files for sperm samples were deposited at NCBI Sequence Read Archive under study no. SRP058678.