Abstract
B-cell malignancies frequently colonizes the bone marrow (BM). The mechanisms responsible for this preferential homing are not entirely known. Using multiple myeloma (MM) as a model of a terminally differentiated B-cell malignancy that selectively colonizes the BM, we demonstrated that BM endothelial cells (BMECs), secrete cyclophilin A (eCyPA), which promotes migration, proliferation, and BM colonization of MM cells via binding to its receptor, CD147, on MM cells. The clinical and translational implications of this work are highlighted by the observation of significantly higher eCyPA levels in BM serum than in peripheral blood (PB) in MM persons, and that eCyPA-CD147 blockade supresses BM-homing and tumor growth in a mouse xenograft model of MM. eCyPA also promoted migration of CLL and LPL cells, two other B-cell malignancies that colonize the BM and express CD147. These findings offer a compelling rationale for exploring the eCyPA-CD147 axis as therapeutic target for these malignancies.
Keywords: multiple myeloma, lymphoplasmacytic lymphoma, chronic lymphocytic lymphoma, cyclophilin A, CD147, migration, invasion, proliferation bone marrow homing, BCL9, Wnt signaling pathway, tumor progression
Introduction
Like certain carcinomas that frequently metastasize to the BM1,2, B-cell malignancies such as MM, chronic lymphocytic leukemia (CLL), and lymphoplasmacytic lymphoma (LPL) preferentially colonize the BM3,4. Although the mechanisms driving this colonization are not completely understood, pathogenetic studies indicate that the BM-microenvironment (BM-ME), comprised of diverse cellular elements, plays a pivotal role3,5-7.
Here we used MM as a prototypical malignancy of terminally differentiated B-cell or plasma cells (PCs) to investigate the cellular and molecular drivers of BM colonization. MM is a post-germinal center malignancy that originate in the lymph nodes and accumulates in the BM during disease evolution. Interactions between MM cells and the BM-ME not only mediate their proliferation, but also protect them from apoptosis, resulting in lytic bone lesions and angiogenesis3. At end-stage disease, MM cells are able to survive and proliferate even in the absence of BM-ME. The number of MM cells circulating in the PB increases at this time, and growth outside the BM can occur7. As with the homing of hematopoietic stem cells (HSCs), factors implicated in BM homing of MM include among others CXCR4 and SDF-17-14.
Among the interactions between MM cells and the BM-ME, physical contact with BMECs is a major feature, and BM angiogenesis has been associated with MM progression, and patient survival8. Although several pro-angiogenic factors secreted by MM cells have been identified8, the molecules secreted by BMECs that promote BM homing and MM progression are not fully known8-11.
In the present report, we have investigated the role of BMECs in the colonization of MM cells to the BM niche. Having previously observed high BCL9 expression in BMECs, but not other BM cells, we chose to focus on the role of this transcriptional co-activator of the canonical Wnt-β-catenin pathway. We used an integrated approach combining in vitro assays with in vivo migration assays that simulate the human-human heterotypic interactions between MM and BM cells. Additionally, we performed proteomic analysis of signaling molecules secreted by BMECs, as well as shRNA-based loss-of-function assays, to identify and functionally validate eCyPA as a novel transcriptional target of the Wnt-β-catenin-BCL9 complex. eCyPA is secreted by BMECs and promotes signaling changes that enhance not only migration of MM cells toward the BM, but also proliferation mediated by binding to CD147 receptors on the MM cells. A comparison between BMECs and BM stromal cells (BMSCs) from the same person with MM demonstrated that these cells play different roles in the migration and BM colonization of MM cells. In contrast to primary BMECs, primary BMSCssecrete very little eCyPA but instead secrete SDF-1, thereby promoting migration and BM homing of MM cells, less efficiently than primary BMECs. Consistent with this finding, BMEC-induced migration of MM cells was inhibited by an anti-CD147 Ab, but not by an anti-CXCR4 Ab12. In addition, inhibition of the eCyPA-CD147 axis supressed migration, tumor growth, and BM-colonization in a mouse xenograt model of MM. Furthermore, we documented that eCyPA promotes migration of CLL and LPL cells, two other B-cell malignancies that colonize the BM and express CD147. Taken together our findings indicate that cells within the BM-ME play different roles in MM progression, and offer a potential link between chronic inflammation, immunomodulation, and the pathogenesis of MM, CLL and LPL. Moreover, our results provide a compelling rationale for exploring the role of eCyPA and CD147 as markers of disease progression and therapeutic targets.
Results
BCL9 promotes proliferation of BMECs
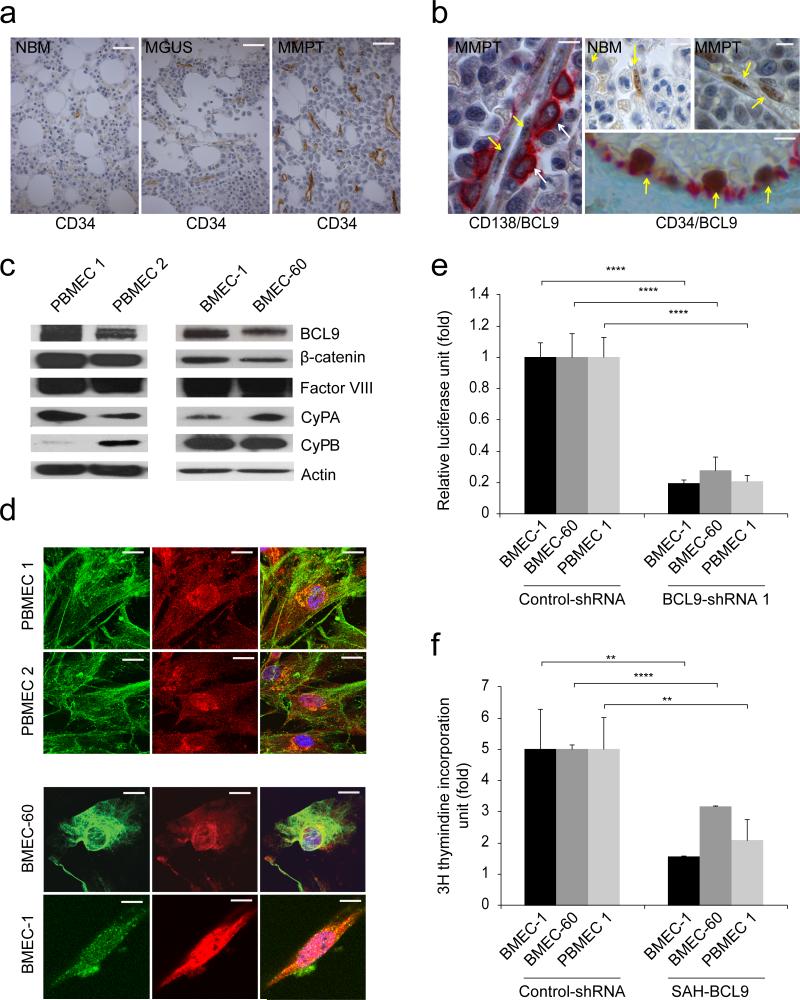
BM angiogenesis is a positive correlate of disease activity (Fig. 1a), suggesting that BMECs promote MM progression8-10. BCL9 is a transcriptional co-activator of β-catenin, and plays critical roles in the pathogenesis of various human cancers, including MM13,14-17. Since Stabilized Alpha-Helix peptides of BCL9 (SAH-BCL9) inactivate native β-catenin-BCL9 complexes, and ablate angiogenesis in a mouse xenograft model of MM17, we evaluated BCL9 expression in BMECs. High BCL9 nuclear stain was detected in cells in close physical contact with MM cells (Fig. 1b) from normal individuals (Figs. 1b and Supplementary Fig. 1a) and MM persons (Figs. 1b and Supplementary Fig. 1a). Double-immunostains, for BCL9 and CD34 confirmed BCL9 expression in BMECs (Fig. 1b). Nuclear co-localization of BCL9 and β-catenin in two primary BMECs from MM persons, and in BMEC-6018 and BMEC-119 cells, was confirmed by immunoblotts (Fig. 1c) and immunofluorescence (Fig. 1d). Lentiviral knockdown of BCL9 in BMEC-60, BMEC-1 and PBMEC 1 cells using BCL9-shRNAs13 (Supplementary Fig. 1b) was associated with decreased Wnt reporter activity (Fig. 1e) and cell proliferation (Supplementary Fig. 1c). Consistent with our previous studies17, BMCEs proliferation was likewise inhibited by SAH-BCL9 (Fig. 1f).
Figure 1. Analysis of BCL9 expression and canonical Wnt activity in BMECs.
(a) Representative CD34 immunostains in BM biopsies from normal individuals (NBM) (n=20) as well as MGUS (n=20) and MM persons (MMPT) (n=60). Bars: 50μm. (b) Representative BCL9 immunostains (brown color) in endothelial cells (arrows) in BM biopsies from MM persons (MMPT) or normal bone marrow (NBM) from otherwise healthy subjects. Selected representative cases are shown. Anti-CD138 staining (red color) is used as a marker of plasma cells on the left panel (arrows). Anti-CD34 staining (red color) is used as a marker of endothelial cells (right bottom panel). Bars: 10μm. Immunoblots (c) and immunofluorescence (d) analysis of BCL9 and β-catenin expression in primary endothelial cells derived from BM from two MM persons (PBMEC 1, PBMEC 1) and two BM endothelial cell lines (BMEC-1, BMEC-60). Note co-expression of BCL9 (Red color) and β-catenin (Greed color) by immunoblotting and by nuclear co-localization immunofluorescence. Factor VIII is used as marker of endothelial cells in immunoblots. Bars: 5μm. (e) Wnt reporter activity of BMEC-1, BMEC-60 and PBMEC 1 cells lentivirally transduced with BCL9-shRNA compared with cells lentivirally transduced with scrambled shRNAs (Control-shRNA). (f) Proliferation of BMEC-1, BMEC-60 and PBMEC 1 cells treated with medium alone (Vehicle) or in the presence of 10 uM SAH-BCL9. Proliferation and Wnt reporter data was normalized based on control or vehicle data. Results are means ± SD for assays performed in triplicate. Statistical significance of differences between groups was determined by applying the unpaired Student's t-test. (***P<0.001).
BMECs promote proliferation and survival of MM cells
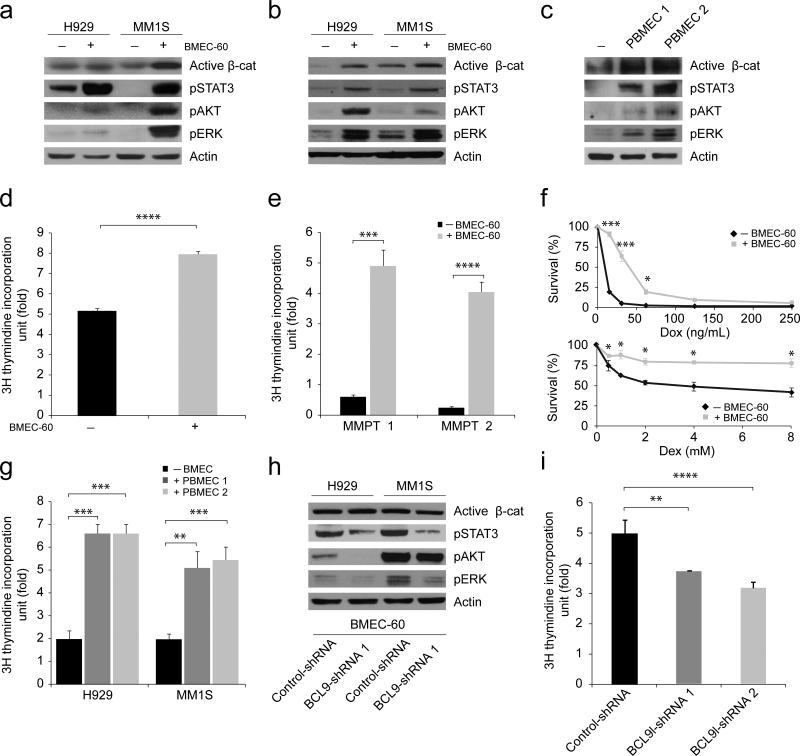
BMSCs were considered to be the only cell type with which MM cells interact functionally20. However, when BM angiogenesis was recognized as a hallmark of MM progression (Fig. 1a), it became clear that BMECs contribute to this process21. To understand the mechanisms by which BMECs promote MM progression, and to evaluate the possible role of BCL9 in this process, we performed biochemical and functional studies using co-cultured cells. Immunoblots revealed that incubation of MM cells with BMEC-60 cells activates several signaling pathways (Fig. 2a) known to promote survival, proliferation, and migration of MM cells22. Similar changes were observed when MM and BMEC-60 cells were co-cultured in separate chambers (transwell assays) (Fig. 2b), indicating that soluble factor(s) secreted by BMEC-60 cells promote(s) these signaling changes. Primary BMECs were as effective as BMEC-60 cells in secreting this factor(s) and promoting signaling changes (Fig. 2c). Co-culture with BMEC-60 cells likewise promoted proliferation of MM1S cells (Fig. 2d) and MM primary tumors (Fig. 2e), and elicited drug resistance (Fig. 2f). Primary BMECs were as effective as BMEC-60 cells in promoting proliferation of MM cells (Fig. 2g). BCL9 knockdown in BMEC-60 cells was associated with decreased pSTAT3, pAKT, and pERK activation (Fig. 2h), and slower proliferation of co-cultured MM1S cells (Fig. 2i).
Figure 2. Biochemical and functional analysis of MM cells upon interaction with BMECs.
Immunoblots of H929 and MM1S cells incubated in the absence (-) or presence (+) of BMEC-60 cells in the same (a) or separate (b) chambers. (c) Immunoblots of MM1S cells incubated in the absence (-) or presence (+) of endothelial cells derived from BM from two different MM persons (PBMEC 1, PBMEC 2) using transwell chambers. Proliferation of MM1S cells (d) and MM cells from two different MM persons (e, MMPT 1 MMPT 2), and incubated in the absence (-) or presence (+) of BMEC-60 cells using transwell chambers. (f) Cell viability using the tumor cell-specific in vitro bioluminescence imaging (CS-BLI) assay40 of MM1S-luc cells incubated in the presence or absence of increasing concentrations of doxorubicin (Dox) or dexamethasone (Dex), without (-) or with (+) BMEC-60 cells. (g) Proliferation of MM1S and H929 cells in the absence (-) or presence (+) of endothelial cells from BM of two different MM persons (PBMEC 1, PBMEC 2). (h). Immunoblots H929 and MM1S cells incubated in transwell chambers in the presence of γ-irradiated BMEC-60 cells lentivirally transduced with either Control-shRNAs or BCL9-shRNAs. (i) BCL9 knockdown in γ-irradiated BMEC-60 cells was associated with reduced proliferation of co-cultured MM1S cells. Proliferation data was normalized based on control data. Results are means ± SD for assays performed in triplicate. Statistical significance of differences between groups was determined by unpaired Student's t-test. (*P<0.05, **P<0.01, ***P<0.001).
BMECs promote migration, and BM colonization of MM cells
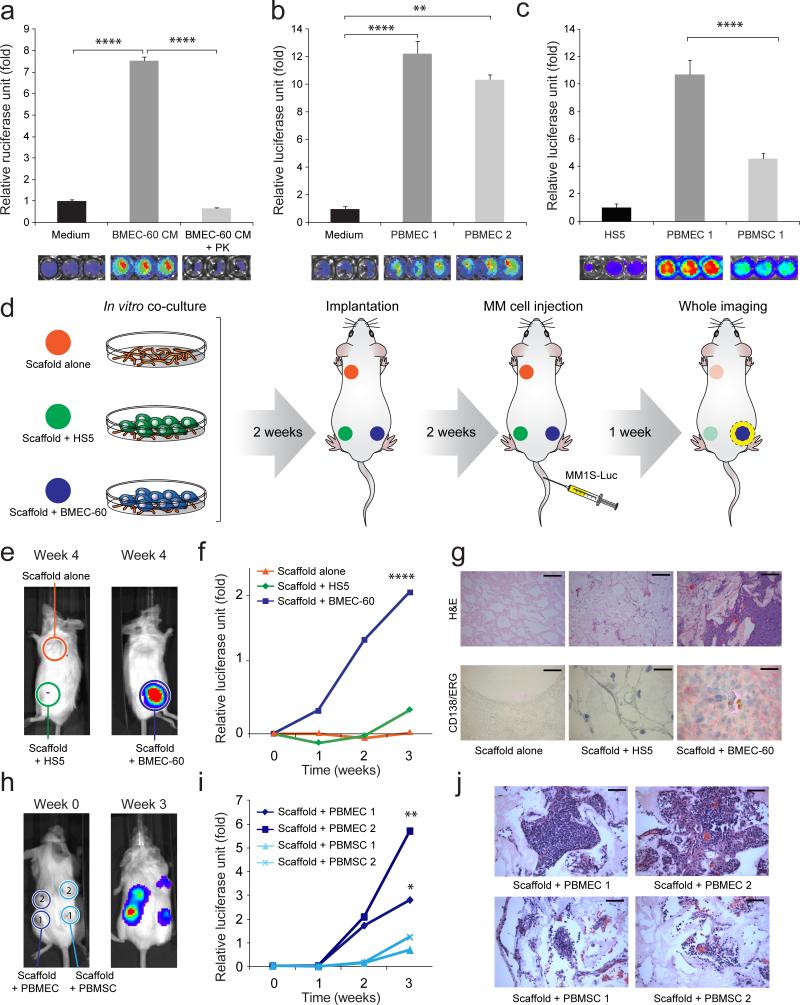
Since ERK activation has been implicated in cell proliferation23, our finding of enhanced pERK expression in MM cells co-cultured with BMEC-60 cells prompted us to ask whether this activation was also associated with increased migration. Conditioned medium (CM) from cultures of BMEC-60 cells (Fig. 3a) or primary BMECs (Fig. 3b), but not from BM derived stromal HS5 cells (Fig. 3c), promoted migration of MM1S-luc-cells (Fig. 3a). Protease treatment of BMEC-60-derived CM (Fig. 3a) reduced migration of MM1S-luc-cells. Migration of other MM cell lines was also enhanced by CM derived from cultures of primary BMECs but not primary BMSCs from the same person, or from HS5 cells (Supplementary Fig. 1d, e). Migration of MM1S-luc-cells was enhanced more strongly by primary BMECs than by primary BMSCs isolated from the same person (Fig. 3c).
Figure 3. In vitro and in vivo migration of MM cells toward BMECs.
Transwell migration of MM1S-luc cells incubated under different growth conditions: (a) in medium alone (Medium), conditioned medium from BMEC-60 cells (BMEC-60-CM) or conditioned medium derived from BMEC-60 cells and treated with proteinase K (BMEC-60-CM + PK); (b) in the absence or presence of endothelial cells derived from BM from two different MM persons (PBMEC 1, PBMEC 2); (c) in the presence of HS5 cells or PBMEC 1 and PBMSC 1 isolated from same person. Migration data was normalized based on data of Medium alone. Results are means ± SD for assays performed in triplicate. Statistical significance of differences between groups was determined by unpaired Student's t-test. (d) Diagram of the three-dimensional poly-ε-caprolactone scaffold xenograft mouse model. Xenogen data (e), time course (f), and histologic and immunohistochemical analysis (g) of MM1S-luc cell growth within non-coated scaffolds or within scaffolds coated with HS5 or BMEC-60 cells. ERG (Ets-related gene): Endothelial cell marker. Bars: Top 100μm, Bottom 20μm. Xenogen data (h), time course (i), and histologic analysis (j) of MM1S-luc cell growth within scaffolds coated with primary BM endothelial cells (PBMEC 1 and PBMEC 2) or primary BM stromal cells (PBMSC 1 and PBMEC 2) isolated from same person with MM. Bars: 50μm. Statistical analysis of tumor burden were done using factorial analysis in SPSS 13.0. The results of two representative experiments of three are shown. (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
To investigate the role of BMECs in the migration of MM cells in vivo, we used the scaffold mouse xenograft system24 (Fig. 3d). Only scaffolds coated with BMEC-60 cells could support growth of MM1S-luc-cells, whereas MM1S-luc-cells failed to propagate either in scaffolds without cells or in those coated with HS5 cells (Fig. 3e-g and Supplementary Fig. 1f). Migration and growth of H929-luc-cells were similarly observed in scaffolds coated with primary BMECs, but not in uncoated scaffolds (Supplementary Fig. 1g-i). Migration and proliferation of primary MM cells in scaffolds coated with BMEC-60 cells were observed (Supplementary Fig. 2a), albeit at lower frequency than that of MM1S-luc-cells. Primary BMECs were more efficient than primary BMSCs from the same person's BM in promoting migration and proliferation of MM1S-luc cells within scaffolds (Fig. 3h-j).
BCL9 knockdown in BMECs reduces migration and proliferation of MM cells
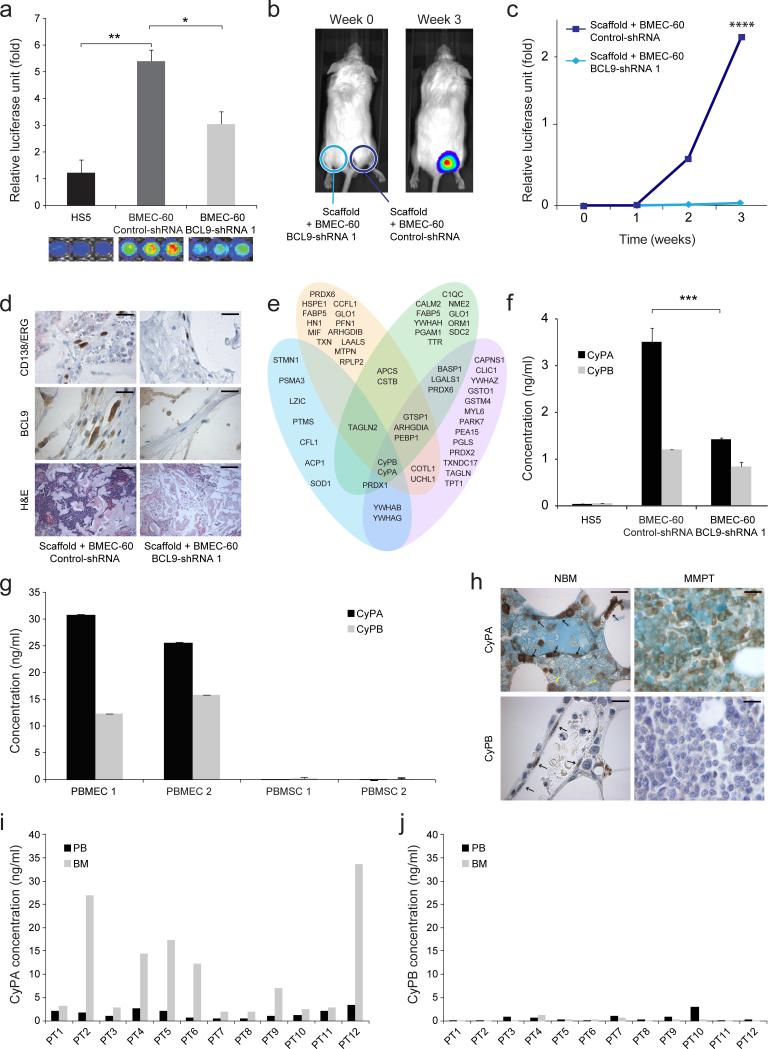
We knocked down BCL9 expression in BMECs (Supplementary Fig. 1b) to investigate whether it is needed to promote migration and proliferation of MM cells. In vitro migration of MM1S-luc (Fig. 4a) and H929 cells (Supplementary Fig. 2b) was decreased when co-cultured with BMEC-60 or BMEC-1 cells transduced with BCL9-shRNAs. In vivo migration and proliferation of MM1S-luc cells within scaffolds were also inhibited when the latter were transduced with BCL9-shRNA but not with Control-shRNA (Fig. 4b-d and Supplementary Fig. 2c,d). Histological analysis confirmed uniform scaffold coating by both BMEC-60-control-shRNA (Fig. 4d) and BMEC-60-BCL9-shRNA (Fig. 4d) cells, as well as the absence of MM1S-luc cells in scaffolds coated with BMEC-60-BCL9-shRNA cells. Similar results were observed with H929-luc-cells using scaffolds coated with BMEC-60 cells (Supplementary Fig. 2e-g).
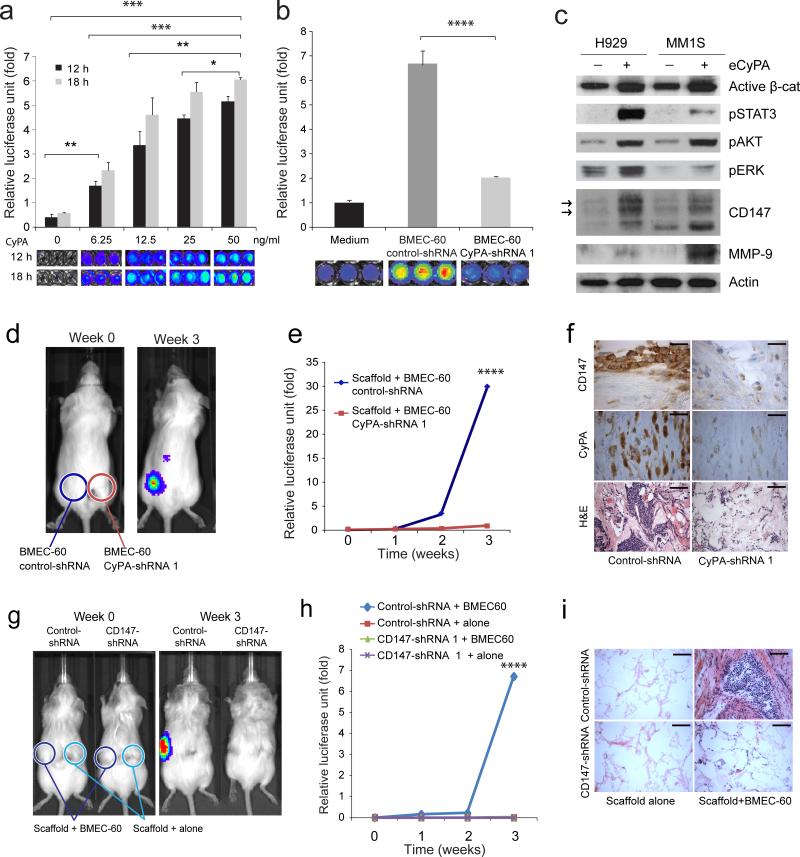
Figure 4. Secretion of eCyPA by BMEC and eCyPA levels in BM serum from MM persons.
(a) Migration of MM1S-luc cells incubated in the presence of HS5 cells or BMEC-60 cells transduced with Control-shRNA or BCL9-shRNA. Xenogen data (b), time course (c), and immunohistochemical and histologic analysis (d) of MM1S-luc cell growth within scaffolds coated with BMEC-60 cells transduced with Control-shRNA or BCL9-shRNA. Bars: Top and midle 20μm, Bottom 100μm. Statistical analyses of tumor burden were done using factorial analysis in SPSS 13.0. (e) Histogram of proteins identified by mass spectrometry of excised bands (blue) and whole protein supernatants from BMEC-60 transduced with Control-shRNA (pink), as well as PBMEC 1 (yellow) and PBMEC 2 (green) cells. At the intersection are eCyPA and eCyPB identified by both procedures. (f) ELISA levels of eCyPA and eCyPB in CM from HS5 and BMEC-60 cells transduced with Control-shRNAs or BCL9-shRNA. (g) ELISA of eCyPA and eCyPB in CM from PBMEC 1 and PBMEC 2 or PBMSC 1 and PBMSC 2 cells isolated from same person with MM. Results are means ± SD for assays performed in triplicate. Statistical significance of differences between groups was determined by unpaired Student's t-test. (h) Representative immunostains of CyPA and CyPB expression in BM from healthy subjects (NBM) (n=20) and MM persons (n=60) (MMPT). Black and yellow arrows indicate expression of CyPA or CyPB in BMECS and myeloid cells, respectively, in a NBM. Bars: 30μm. ELISA quantification of eCyPA (i) and eCyPB (j) levels in serum from BM and PB isolated from same MM persons (n= 12). (**P<0.01, ***P<0.001, ****P<0.0001).
Proteomic analysis identified eCyPA as a signaling factor secreted by BMEC
The foregoing results prompted us to perform proteomic analysis to identify signaling molecules secreted by BMECs, whose expression is regulated by BCL9, and could promote migration, and proliferation of MM cells. Silver-stained agarose gels revealed differences, among lower-molecular weight proteins, in CM from BMEC-60 cells transduced with Control-shRNAs or BCL9-shRNAs. The same was observed in HS5 cells, indicating secretion of a discrete protein by the control BMEC-60 cells, but not the others (Supplementary Fig. 2h). To identify the low-molecular weight proteins secreted by BMEC-60 cells, we performed proteomic analysis of the six excized major bands of <28 kD from silver-stained gels, and also whole CM from BMEC-60, and two primary BMECs (Fig. 4e). This procedure provided a large number of potential candidates that are known to be secreted into media, have known signaling functions, and were detected by both procedures in BMEC-60 and in primary BMECs. Among these, the most likely were eCyPA and eCyPB25,26 (Fig. 4e and Supplementary Table 1 and 2). ELISA assays confirmed the presence of eCyPA and eCyPB in CM from BMEC-60-control-shRNA cells, and their reduced levels in CM from BEMC-60-BCL9-shRNA and HS5 cells (Fig. 4f), and showed that primary BMECs secrete much more eCyPA than primary BMSCs from the same person (Fig. 4g). BMEC-60 cells and primary BMECs secreted much more eCyPA than eCyPB when compared with HS5 and primary BMSCs (Fig. 4f, g). Immunoblots of transduced BMEC-60 cells confirmed stable BCL9 knockdown and decreased cellular CyPA and CyPB expression in BMEC-60-BCL9-shRNA cells (Supplementary Fig. 2h), indicating that CyPA and CyPB, which have overlapping signaling functions27 are both regulated by BCL9. Immunoblots in other BM-derived cells including primary BMECs, and primary BMSCs from the same person (Fig. 1c and Supplementary Fig. 1d), and immunostains of BM biopsies from healthy subjects (Fig. 4h and Supplementary Fig. 3a) confirmed that most BMECs express CyPA, and that CyPB expression is decreased compared with CyPA. Immunostains also revealed that BM myeloid cells express remarkable amounts of CyPA, but not CyPB (Fig. 4h, left and Supplementary Fig. 3b). Although, immunostains revealed that MM cells also expressed CyPA (Fig. 4h, right and Supplementary Fig. 3c), they secreted it at very low levels in comparison with BMECs (Supplementary Fig. 3d). In addition, immunostains revealed much lower CyPA expression in endothelial cells from other tissues than from BM (Supplementary Fig. 4a).
We next measured serum levels of eCyPA and eCyPB in MM persons whose BM and PB was collected at time of diagnosis. As documented by ELISA, eCyPA was more abundant in BM serum than in PB serum (BM: 10.54 ng per ml ± 10.72; PB: 1.59 ng per ml ± 0.94) (Fig. 4i). In the same samples, serum levels of eCyPB were much lower than those of eCyPA, and no major differences were detected between BM and PB (Fig. 4j). eCyPA levels in PB serum were higher in MM than MGUS persons (Supplementary Fig. 4b), indicating that this parameter could be a marker of disease progression.
eCyPA promotes migration, proliferation, and BM colonization of MM cells
Because of the documented role of eCyPA in neutrophil migration27, it was of interest to evaluate the role of eCyPA in MM cell migration in vitro and in vivo. eCyPA promoted MM migration in vitro in a time- and concentration-dependent manner (Fig. 5a). Although maximal responses were observed at 50 ng per ml, migration was induced at eCyPA concentrations typically found in the BM serum of MM persons (Fig. 4i). eCyPA also induced migration of various MM cell lines (Supplementary Fig. 4c) and primary MM cells (Supplementary Fig. 4d), with no major differences between primary MM cells incubated with BMEC-60 cells or with 50 ng per ml eCyPA. BM serum from MM persons also promoted migration of MM cells to an extent that varied with eCyPA levels (Supplementary Fig. 4e), and major differences were likewise not observed in the migration of cells treated with 50 ng per ml CyPA or CyPB, respectively (Supplementary Fig. 5a). Migration of MM cells was decreased when BMEC-60 (Fig. 5b) and BMEC-1 cells (Supplementary Fig. 5b-d) were transduced with CyPA-shRNAs (Supplementary Fig. 5b). As would be expected from an on-target effect of eCyPA-shRNAs, addition of eCyPA restored migration of MM cells incubated with BMEC-1 transduced with CyPA-sh-RNAs (Supplementary Fig. 5d). eCyPA enhanced several components of signaling pathways in MM cells (Fig. 5c), including the higher-glycosylated form of CD147 that induces matrix metalloprotease production and metastasis26,27 (Fig. 5c). Matrix metalloprotease 9 (MMP-9) expression increased in cells treated with eCyPA (Fig. 5c). eCyPA increased CD147 expression in a concentration- and time-dependent manner (Supplementary Fig. 5e). The signaling changes induced by eCyPA were reversed when MM cells were incubated with BMEC-60 cells with CyPA knockdown (Supplementary Fig. 5f). eCyPA also enhanced proliferation of MM cells in a concentration-dependent manner (Supplementary Fig. 5g).
Figure 5. eCyPA promotes signaling changes, migration, and proliferation of MM cells through the CD147 receptor.
Transwell migration assays of MM1S-luc cells incubated under different conditions: (a) increased concentrations of recombinant eCyPA.; (b) medium alone or BMEC-60 cells lentivirally transduced with Control-shRNA or shRNAs against CyPA (CyPA-shRNA). Migration data was normalized based on data of medium alone. Results are means ± SD for assays performed in triplicate. Statistical significance of differences between groups was determined by unpaired Student's t-test. (c) Immunoblot of total protein extracts from H929 and MM1S cells incubated in the absence (-) or presence (+) of eCyPA at 50ng per ml. Xenogen data (d), time course (e), and histologic analysis (f) of MM1S-luc cell growth within scaffolds coated with BMEC-60 lentivirally transduced with ControlshRNAs or CyPA-shRNA. Bars: Top and midle 20μm, Bottom 100μm. The results of one representative of three independent experiment is shown Xenogen data (g), time course (h), and histologic analysis (i) of cell growth of MM1S-luc transduced with Control-shRNA or CD147-shRNA within empty scaffolds or scaffolds coated with BMEC-60 cells. Bar: 100μm. Statistical analyses of tumor burden were done using factorial analysis in SPSS 13.0. (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Migration and proliferation of MM1S-luc-cells within scaffolds was inhibited when the scaffolds were coated with BMEC-60 cells transduced with CyPA-shRNA (Fig. 5d-f). Immunostains confirmed uniform scaffold coating with both BMEC-60-control-shRNA and BMEC-60-CyPA-shRNA cells (Fig. 5f). These results were replicated in two other experiments (Supplementary Fig. 6a), and with H929-luc cells and scaffolds coated with BMEC-60 cells (Supplementary Fig. 6b-d).
CyPA promotes migration and proliferation of MM through the CD147 receptor
Because CD147 is the principal receptor mediating chemotaxis by eCyPA27,28, we evaluated CD147 expression in MM cells. Flow cytometry and immunostains revealed that most MM cell lines, including MM1S-luc-cells (Supplementary Fig. 7a) and primary tumors (Supplementary Fig. 7b), expressed CD147. At the mRNA level, CD147 was present in most MM samples, with higher levels detected in MM samples than in normal PCs (Supplementary Fig. 7c), indicating that CD147 could be a marker of disease progression.
We also evaluated whether CD147 inactivation on MM cells could block the action of eCyPA in vitro and in vivo. MM cells transduced with CD147-shRNAs (Supplementary Fig. 7d) showed decreased migration in the presence of BMEC-60 cells or CyPA (Supplementary Fig. 7e,f). The signaling changes induced by CyPA in MM cells (Fig. 5C) were reversed when the cells were transduced with CD147-shRNAs (Supplementary Fig. 7g). Using the scaffold xenograft mouse model we observed that knockdown of CD147 in MM1S-luc-cells inhibited migration and proliferation within scaffolds coated with BMEC-60 cells (Fig. 5g-i).
Targeting the eCyPA-CD147 complex for MM therapy
To test the targeting of the eCyPA-CD147 complex as a potential strategy for clinical MM therapy, we first evaluated whether an anti-CD147 Ab known to inhibit neutrophil migration in a mouse model of inflammation29 also inhibits MM cell migration. Treatment with the anti-CD147 Ab reduced eCyPA-induced MM1S-luc-cell migration (Supplementary Fig. 7h) and pERK, pSTAT3, and pAKT activation as well as activate PARP in a concentration-dependent manner (Supplementary Fig. 8a,b). An anti-CXCR4 Ab12 did not inhibit CyPA-induced migration of MM1S-luc-cells (Supplementary Fig. 8c). Migration of MM1S-luc-cells was also reduced by the anti-CD147 Ab when cells were incubated with primary BMECs, but not primary BMSCs, both from the same person (Supplementary Fig. 8d). Treatment with anti-CXCR4 Ab did not inhibit migration of MM1S-luc-cells induced by primary BMECs, but inhibited migration induced by primary BMSCs (Supplementary Fig. 8d). We then evaluated secretion of eCyPA and SDF-1 in the same cells by ELISA. As shown in Supplementary Fig. 8e, eCyPA was detected in CM from primary BMECs, but not primary BMSCs, and the converse was observed with SDF-1.
Our observation of decreased pSTAT in MM cells transduced with CD147-shRNA or treated with anti-CD147 Ab prompted us to next use the scaffold system to evaluate whether anti-CD147 Ab could be therapeutic for MM in vivo. Local injection of anti-CD147 Ab but not isotype Ab reduced tumor burden in mice implanted with scaffolds pre-coated with BMEC-60 cells and MM1S-luc-cells (Fig. 6a,b and Supplementary Fig. 9a). More immunostaining for caspase 3 was observed in MM1S-luc cells within scaffolds of mice treated with anti-CD147 Ab (Supplementary Fig. 9a) than in mice treated with isotype Ab. Analysis of immune-mediated activity revealed that complement-dependent cytotoxicity was not induced by anti-CD147 Ab (Supplementary Fig. 9b). Although these studies indicate that targeting the CyPA-CD147 complex could be therapeutic for MM, additional preclinical studies including other mouse models of MM will be needed to further validate this hypothesis. In addtion, due to the broad tissue pattern of expression of CD147, unintended side effects should be carefully and systematically evaluated.
Figure 6. Targeting eCyPA-CD147 complex is associated with anti MM activity. Decreased CD147 expression in circulating MM cells.
(a) Time course of Xenogen imaging of MM1S-luc cell growth in scaffolds implanted inCB17.Cg-PrkdcscidLystbg-J-Crl mice and treated with local injections of isotype control or anti-CD147 Abs. Xenogen data (b) of MM1S-luc cell growth within scaffolds coated with BMEC-60 cells and implanted subcutaneously in CB17.Cg-PrkdcscidLystbg-J-Crl mice. Groups of 4 mice were subsequently treated with either isotype Ab or anti-CD147 Ab, and tumor growth within the scaffolds was evaluated by Xenogen imaging every five days. (c) Left panel, Immunofluorescence analysis of CD147 expression in MM plasma cells from BM (top) and PB (bottom) from one person with MM (Case 1). Right panel, Immunofluorescence analysis of CD147 expression in normal plasma cells from BM (top) and lymph node (LN) (bottom) in two different normal donors (Case 3 and 4). Bars: 5μm. (d) Proposed model of BM homing of MM cells based on eCyPA secreted by BMECs and on CD147 expression by MM cells. Statistical analysis of tumor burden were done using factorial analysis in SPSS 13.0. (*P<0.05, **P<0.01).
Decreased CD147 expression in circulating MM cells
Since CD147 is the known receptor of eCyPA27,28, we evaluated its expression in plasma cells (PCs) from the lymph nodes and BM of healthy subjects, as well as in PCs from BM and PB of MM persons. Due to the paucity of PCs in the PB of MM persons32, CD147 expression was determined by immunofluorescence on cytospin preparations using CD138 as PCs marker30. BM PCs in 10 in10 MM persons expressed high CD147 levels while 9 in10 samples of PB PCs from the same person with MM did not (Fig. 6c and Supplementary Fig. 9c). In contrast, 9 in10 normal PCs from BM and 8 in10 cells from lymph nodes (Fig. 6c and Supplementary Fig. 9d) expressed CD147 only at low or undetectable levels.
CLL and LPL cells express CD147 and migrate in response to eCyPA
Since CLL and LPL, like MM, preferentially colonize the BM4, it was of interest to examine whether tumor cells express CD147, and whether eCyPA is chemotactic to them. Immunostains and flow cytometry (Supplementary Fig. 10a,b) revealed that most CLL and LPL primary cells expressed CD147. The CLL cell line HG3 and a primary tumor CLLPT 1(Supplementary Fig. 10c) as well as the LPL cell line BCWM.1 and a primary tumor LPLPT 1 (Supplementary Fig. 10d) used in subsequent in vitro migration studies expressed high CD147 levels. Both BMEC-60-derived CM and eCyPA enhanced migration of CLL (Supplementary Fig. 10e) and LPL (Supplementary Fig. 10f) cells.
Discussion
The mechanisms responsible for the preferential colonization of MM and certain other B-cell malignancies to the BM are not entirely known5-8. In a manner similar to HSC homing to the BM12,31-33, it was also shown that the CXCR4-SDF-1 axis plays an important role in the migration and BM homing of MM cells13. However, the specific cell that secretes SDF-1 within the BM is not known13. Here we offer evidence of a novel molecular basis for the migration and BM colonization of MM cells. We document that eCyPA is secreted by BMECs, but not endothelial cells from other vascular beds, or by other stromal cells in the BM, and that eCyPA acts as a signaling factor to promote migration, proliferation, and BM colonization of MM cells via binding to its cognate receptor, CD147, on MM cells. Through these studies we have identified eCyPA as a novel target of the β-catenin-BCL9 complex, and have provided evidence for a functional role of the Wnt pathway and of BMECs in MM pathogenesis. Furthermore, we have identified BMECs as the main source of eCyPA, but not SDF-1, which is secreteted by BMSCs. This is the first suggestion that BMECs and BMSCs play different roles in MM pathogenesis, and do so via different molecular mechanisms.
Although it was initially recognized as the intracellular target for the immunosuppressive drug Cyclosporine A, recent studies have revealed that cells can secrete eCyPA in response to inflammatory stimuli34. Extracellualr CyPA can initiate a signaling response in target cells and is a potent chemoattractant25-28. Studies aimed at establishing the mechanism whereby eCyPA exerts chemotaxis have identified CD147 as its principal receptor31.
CD147, also known as Extracellular Matrix Metalloproteinase Inducer (EMPRIN), plays critical roles in intercellular communication involved in chronic inflammation, tumor metastasis and angiogenesis34,29. CD147 induces expression of the matrix metallopeptidases required for tumor invasion and metastasis via cell-cell and cell-matrix interactions28. Recently, expression of CD147 has been broadly correlated with progression in various carcinomas, as well as hematological malignancies such as MM28,35,36. In regard to MM, it was documented that CyPB induces proliferation of MM cells40, although tested at much higher concentrations than those in BM serum of MM persons. Of relevance to our studies, however, is the observation that CD147 is not expressed in normal PCs, yet its expression correlates with disease progression36.
On the basis of the present findings and the generally recognized trafficking model of normal lymphocytes and BM recruitment of HSCs6,7, we propose a mechanism that could explain both preferential BM colonization of MM cells during early-stage disease and loss of preferential homing during the late-stage progression (Fig. 6d). According to this model, normal PCs do not accumulate in the BM because they don't express CD14735, whereas MM cells can accumulate in BM because they do express CD14735, the receptor for eCyPA27 which acts as chemokine to promote migration of MM cells along a concentration gradient between the PB and BM. The gradient is generated because endothelial cells from the BM, but not those from other organ sites, produce and secrete eCyPA in remarkable amounts. In the BM, the effect of eCyPA from endothelial cells can be enhanced by local production and secretion by myeloid cells. Although MM cells also express CyPA, they secrete it at very low rates relative to BMECs, indicating that MM are less efficient at Rho-kinase-mediated CyPA secretion37, and that MM-derived eCyPA does not play a significant role in MM recruitment to the BM, at least during early-stage disease, when tumor burden is low. As MM cells circulate in the PB and ecounter progressively higher concentrations of eCyPA, binding of eCyPA to CD147 further increases CD147 expression by MM cells, therby enhancing their migration. Additionally, extravasation of MM cells, required for penetration through the sub-endothelial basement membrane, is mediated by proteolytic enzymes such as MMP-9, whose level rise when eCyPA binds to CD14728. Once MM cells enter the BM, colonization is further enhanced by direct exocrine action of eCyPA and by local production and secretion of other growth factors and chemoattractants (e.g. laminin-1, SDF-1, VEGF)6,21.
Therefore, induction of selective colonization of MM cells by eCyPA results from preferential migration of MM cells through the blood to the BM, followed by selective survival and proliferation within the BM. It is also possible that eCyPA in the BM serves as a “retention signal” for MM cells, which needs to be further investigated. During disease progression, the number of circulating MM cells may increase because of genetic and epigenetic changes in the tumor that decrease CD147 expression. Consequently MM cells, now unable to respond to eCyPA, can leave the BM to colonize extramedullary sites.
The role we have uncovered for the eCyPA-CD147 complex in MM identifies this interaction as an attractive target for therapeutic intervention. Agents potentially disruptive of this interaction can be grouped into those that can directly target CD147, eCyAP, or their mutual interaction27, or those which act indirectly via inhibition of the Wnt-β-catenin-BCL9 complex17. Using anti-CD147 Ab to block eCyPA- and BMECs-induced migration of MM cells, we provide evidence that inhibition of the eCyPA-CD147 complex is associated with anti-MM activity. This approach may also be useful to treat CLL and LPL, which likewise express CD147 and migrate in respond to eCyPA, indicating that in addition to other mechanims implicated in homing of lymphoid cells38, the CyPA-CD147 axis may also play an important role. Interestingly, CD147 inhibition using an anti-CD147 Ab is of therapeutic benefit in hepatocellular carcinoma39.
In summary, our studies uncovered a pivotal role of the eCyPA-CD147 axis in the functional interaction between MM cells and BMECs, and have established a functional link between the Wnt-β-catenin-BCL9 pathway and the eCyPA-CD147 system, suggesting a possible role for chronic inflammation in MM pathogenesis. Logical translational extension of these studies into the clinical setting could include: identifying biomarkers of disease progression within the downstream signaling cascade triggered by eCyPA-CD147 binding, and developing therapies targeting eCyPA-CD147 interactions in MM and other B-cell malignancies that preferentially colonize within the BM.
Materials and Methods
Humansamples and cell lines
BM specimens were obtained from persons with MM or from normal donors in accordance with Dana-Farber Cancer Institute Review Board protocols, and with informed consent in compliance with the Helsinki Declaration. BM mononuclear cells were isolated with the aid of a Ficoll gradient, and BMECs as well as MM primary cells were isolated using CD34 or CD138 magnetic beads (Miltenyi Biotec, Auburn, CA), respectively, as described41. Primary BMECs and primary BMSCs from the same person with MM were prepared as follows: After Ficoll gradient, BM mononuclear cells were treated with collagenase, and single-cell suspensions were placed on plastic dishes. When only attached cells remained in the cultures, they were divided into two fractions and one was stored without further processing for subsequent use as BMSCs. From the other fraction, we purified BMECs using CD34 immunomagnetic beads. Purified BMECs cells were further expanded in culture ex-vivo and their identity confirmed by FC and immunoblot analysis using several endothelial cell markers (CD31, VEGF R1, CD138, and Factor VIII). Primary CLL and LPL cells were purified using CD19 magnetic beads (Miltenyi Biotec, Auburn, CA). All primary cells were >90% pure and fresh for functional studies orstored in liquid nitrogen for other subsequent studies. Established MM cell lines (MM1S, ABNL6, U266, H929, OPM2, and RPMI) were all from the Carrasco laboratory41,13. All cells were grown at 37°C under a 5% CO2 humidified atmopsphere in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). All cell lines were routinely tested using Human Cell Line Genotyping System (Promega), and were periodically surveyed for mycoplasma contamination with the help of a commercial detection kit (LT07-218, Lonza). Serum from BM and PB of MM persons were obtained from the Jerome Lipper Multiple Myeloma Center, Dana-Farber Cancer Institute. Studies with human subjects were approved by the Dana-Farber Cancer Institute Review Board Committee (IRB 01-206), and informed consent was obtained from all subjects.
Scaffold mouse xenograft model
2 X 106 BMEC-60 cells, HS5 cells,PBMEC or PBMSC were trypsinized and seeded into poly-ε-caprolactone polymeric scaffolds generously provided by Dr. Tassone24. Scaffolds were first coated with 50 μg per ml fibronectin (Santa Cruz). The seeded scaffolds were cultured in vitro for 2 weeks and implanted s.c. in the right or left flank of five-week old non-γ-irradiated CB17.Cg-PrkdcscidLystbg-J-Crl male mice (Charles River). For animal studies using the xenograft scaffold system, experiments were done with a single mouse and confirmed in at least three independent occasions. Two weeks later, 5 X106 luciferase-labeled MM1S or H929 cells were injected via the tail-vein into scaffold-implanted mice. Tumor burden was analyzed on the basis of luciferase bioluminescence using an LAS-4000 Luminescent Imager Analyzer (Fujifilm). Mice were euthanized 4 weeks later and the scaffolds removed for histological and IHC examination as described13. Since transplanted human MM cell lines can grow within the BM of γ-irradiated mice13, non-irradiated mice were used to reduce the background signal and delay the spread of MM1S-luc cells to the spine Experiments were repeated independently three times. Animal experiments were approved by the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee (IRB10-039) and were in compliance with ethical practice. All the experiments were done blindly, asthe investigator who implanted the mouse didn't know the content of the scaffolds. In animal studies using the Xenograft scaffold system, one experiment was excluded because MM1S-luc or H929-luc cells grew within both the BMEC-60 coated scaffold and the spine and skull of the mouse, precluding optimal evaluation using the Xenogen system.
For in vivo treatment with anti-CD147 Ab, the mouse scaffold system was modified as follows: scaffolds were pre-coated with BMEC-60 cells, incubated with MM1S-luc cells, and then implanted into the flank of non-γ-irradiated CB17.Cg-PrkdcscidLystbg-J-Crl mice. One week post-implantation the mice were subjected to whole-body imaging, and those with a comparable tumor burden were selected for Ab therapy. Mice were injected with 100 μl of a solution containing either 100 ng of isotype Ab or anti-CD147 Ab s.c every other day in a region adjoing the scaffold. Tumor growth within each scaffold was evaluated by Xenogen imaging every five days. After 15 days, the scaffolds were removed and processed for histological and immunohistochemical analysis.
Complement-dependent cytotoxicity (CDC)
MM.1S-Luc cells (1 × 106) were labeled with Calcein-AM (1 μg per ml, Technologies, Grand Island, NY) for 30 min in a CO2 incubator at 37 °C, washed twice with fresh mouse medium, and then cultured for 1 h with anti-CD147 Ab or isotype control Ab (10 μg per ml), in the absence or presence of mouse serum (20% final concentration). Cells were spun down and fluorescence in the supernatant was measured using a 490 nm excitation filter and a 520 nm emission filter. Percent specific lysis was calculated from the formula % Lysis = (Sample RFU - Background RFU over Total RFU - Background RFU) × 100.
Immunoblot analysis
Total protein samples were prepared from non-attached H929 and MM1S cells grown in CM from cultures of BMEC-60 or BMEC-1 cells attached for 72 hr. In some experiments, the cells were co-cultured with BMEC-60 cells using transwell chambers (Corning Costar, 0.45 μm pore diameter). Immunoblotting was performed as described17. When MM1S and H929 cells were treated with eCyPA or CD147 Ab, they were first incubated with eCyPA or CD147 Ab for 5 hrs in serum-free medium. Then 0.5% serum was added, and incubation was resumed for another 72 hrs before performing protein preparation. Anti-human primary antibodies included: BCL9 (2D4, Abnova, 1:1000), CyPA (ERPR7511, Abcam, 1:1000), CD147 (13287, Cell Signaling, 1:1000), pSTAT3 (9131, Cell Signaling, 1:1000), pERK (4370, Cell Signaling, 1:1000), PARP (9542, Cell Signaling, 1:1000), pAKT (9271, Cell Signaling, 1:1000), Active-β-catenin (05-665, Millipore, 1:1000), Factor VIII (A0082, DAKO, 1:1000), MMP-9 (MAB911, R&D, 1:1000), CyPB (MAB5410, R&D, 1:500), and horseradish peroxidase(HRP)-conjugated actin (C-11, Santa Cruz, 1:200). Actin Ab was used as a loading control. Secondary antibodies included: anti-rabbit IgG (HRP)-conjugated (W402b, Promega, 1:5000), and anti-mouse IgG (HRP)-conjugated (W401b, Promega, 1:5000). Protein concentrations were measured in triplicate by Bradford assay (BioRad). Optimal antibody concentrations were used according to the manufacturer's recomendations.
Lentiviral infection
Validated expression plasmids for pLKO-shCyPA: 1,CCGGGTTCCTGCTTTC ACAGAATTACTCGAGTAATTCTGTGAAAGCAGGAACTTTTTG; 2,CCGGCT GACTGTGGACAACTCGAATCTCGAGATTCGAGTTGTCCACAGTCAGTTTTTG and pLKO-shCD147: 1,CCGGCCAGAATGACAAAGGCAA GAACTCGAGT TCTTGCCTTTGTCATTCTGGTTTTT; 2, CCGGACAGTCTTCACTACCGTAG AACTCGAGTTCTACGGTAGTGAAGACTGTTTTTTG were purchased from Sigma. The pLKO-Control-shRNA and pLKO-shBCL9 expression vectors have been described 13. Lentiviral packaging and infection of MM1S-luc and BMEC-60 cells was done according to manufacturer's protocol (Sigma). To minimize off-target effects of shRNAs, we implemented a series of assay conditions: i) shRNAs were designed to target the 3’UTR region, ii) the effect of the target shRNAs had to differ from that of a control shRNA containing scrambled nucleotide sequences, iii) the phenotypic response had to be reproducible using two distinct target shRNAs (1 and 2), and iv) identical results had to be obtained with more than one targeted cell. Only cells with >70% knockdown of BCL9 expression were selected for in vitro and in vitro assays.
Cell migration assays
For these cell migration assays, the top chamber of transwell plates (8 μM pore diameter, Corning Costar) was seeded with 2 X 105 MM1S-luc cells, and the bottom chamber was seeded with either medium (0.1% FBS) alone, BMEC-60 cells, BMEC-60 cell-conditioned medium, or plain medium containing CyPA and(or) CD147. After 12 hrs of incubation, MM1S-luc cells that had migrated to the bottom chamber were collected and quantified by Xenogen Imaging. Migration of RPMI, U266, CLL, LPL, or primary tumor cells was evaluated by counting the number of unlabeled cells in the lower chamber. Each experiment was done in triplicate and repeated twice. Recombinant HPLC-purified eCyPA was purchased from BioMart Inc. and eCyPB (PPIB) human recombinant protein was purchased from Abnova. In most instances, 50 ng per ml of eCyPA or eCyPB diluted in serum-free RPMI medium was used. CM was collected from confluent cultures of BMEC-60 cells incubated for 48hr. When needed, the CM was treated with 100 μg per ml proteinase K (19133, Qiagen) for 2 hrs and then deactivated for 15 min at 70 °C. Anti-CD147(UM-8D6) antibody and monoclonal mouse IgG1 (MOPC31C) were obtained from Ancell;anti-CXCR4 (MAB171) was from R&D systems. Migration data were normalized using the data obtained with medium alone. Results are means ± SD for triplicate assays.
Reporter assays
Luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega), as described13. To measure Wnt reporter activity, BMEC-60 cells were transfected with TOP-FLASH, FOP-FLASH plasmid (Millipore), along with an internal Renilla control plasmid (hRL-null). Transfection was accomplished using FuGENE (Roche) according to the manufacturer's protocol. The results were normalized to control for Renilla activity. The reported data represent the average of three independent transfection experiments in triplicate.
Cell proliferation and viability assays
Cell proliferation was evaluated by 3H thymidine incorporation as described13. When proliferation of MM cells was determined in the presence of HS5 or BMEC-60 cells, the latter were previously gamma-irradiated (10,000 rads) and dispensed into 96-well plates. Then, MM1S and H929 cells were co-cultured with either BMEC-60 cells, CM alone, CyPA, or CyPB for 72 hrs. Viability of MM1s-Luc cells were co-cultured with BMEC-60 cells and then treated with drugs as described was assessed, using the tumor cell-specific in vitro bioluminescence imaging (SS-BLI)40.
Histopathological and immunohistochemica analysis
Tissue sections were processed as described13. Human tissue samples were obtained from the Department of Pathology, Brigham and Women's Hospital. Sections were incubated with primary antibodies (5 μg per ml) or the corresponding IgG fraction of pre-immune serum overnight at 4 °C in blocking solution (3% BSA in PBS). Anti-human primary specific antibodies included: BCL9 (ab37305, Abcam, 1:200), CD138 (PN IM 2757, Beckman Coulter, 1:1000), CD34 (M71165, DAKO, 1:400), ERG (5115-1, Epitomics, 1:3000), Caspase-3 (9664, Cell Signaling, 1:150), CD147 (MEM-M6-1, LifeSpan BioSciences, 1:300), CyPA (ERPR7511, Abcam, 1:1500) and CyPB (AV44365, Sigma, 1:100); and were visualized with the aid of the corresponding biotinylated antibody coupled to streptavidin-peroxidase complex (Vector Labs). For CD147 immunohistochemistry, non-decalcified BM clots were used. For negative controls, tissue sections were incubated in the absence of primary antibodies or pre-immune serum from the species of origin of the primary antibody. Optimal antibody concentrations were used according to manufacturer recomendations.
Immunofluorescence
Single-cell suspensions were spun onto glass slides using a cytocentrifuge (Shandon) or were grown on polylysine-coated slides (p8920, Sigma), as described13. Cells were fixed at room temperature in 2% paraformaldehyde for 20 min, permeabilized in TBS-Tween 20 for 20 min, washed three times in PBS, and then blocked with 5% bovine serum albumin in PBS for 2 h before addition of primary antibodies against BCL9 (ab37305, Abcam, 1:200), β-catenin (CAT5-H10, Zymed, 1:100), Myeloperoxidase (A0398, DAKO, 1:100), CyPA (ERPR7511, Abcam, 1:200), Alexa Fluor 647-conjugated CD147 (HIM6, Biolegend, 1:200), or FITC-conjugated CD138 (MI15, Becton Dickinson, 1:200). Cells were incubated overnight with primary antibodies at 4 °C, and then washed three times in PBS before staining with secondary antibodies conjugated to Alexa Fluor 488 (A11034, Molecular Probes, 1:200) or Alexa Fluor 546 (A11035, Molecular Probes, 1:200). Images were acquired with the aid of a Bio-Rad Radiance 2000 laser scanning confocal or Nikon Eclipse E800 phase-contrast microscope. Optimal antibody concentrations were used according to manufacturer's recomendations.
Flow cytometry (FC)
Harvested cells in aliquots of up to 1 X 106 cells per 100 μL were dispensed into FACS tubes and stained for FC, as described 13. Anti-human antibodies included: CD147-Alexa Fluor 647 (HIM6, Biolegend, 1:100), CD147-PE (8D12, eBioscience, 1:100), CD19-FITC (SJ25C1, Becton Dickinson, 1:100), CD19-APC (HIB19, Becton Dickinson, 1:100), CD5-FITC (53-7.3, Becton Dickinson, 1:100), CD138-APC (MI15, Becton Dickinson, 1:100), and isotype control mouse IgG1 ƙ (P3.6.2.8.1, Bioscience, 1:100). Optimal antibody concentrations were used according to manufacturer's recomendations.
ELISA
Serum levels of eCyPA and eCyPB in BM and peripheral blood samples from MM persons were measured by ELISA according to the manufacturer's protocols for eCyPA (KA1176, Abnova), eCyPB (ABIN414776, USCN Life Science), and SDF-1 (DSA00, R&D), respectively. For the measurement of eCyPA, eCyPB, and SDF-1 from cell supernatants, 1 X 106 cells were dispensed in triplicate into 12-well plates and cultured for different times. The spent medium was then replaced with an equal volume of fresh medium, the incubation was resumed for another 12 h, and finally triplicate samples of supernatant containing equal amounts of total protein were used for ELISA analysis. Standard curves were linear, and 100 ul of each sample was used for analysis in triplicate.
Mass spectrometry (MS)
Bands excised from silver-stained gels were cut into approximately 1 mm3 pieces, and the latter were analyzed by mass spectrometry, as described42,43. BMEC secretomes were analyzed directly, according to a previously described protocol18. Approximately 1 X 106 BMEC-60 cells were lentivirally transduced with control shRNA or shBCL9. These, along with HS5 cells, PBMEC 1,and PBMEC 2, were dispensed into 12-well plates, which were kept for 6 h at 37 °C under 5% CO2, and rinsed twice with 2 ml of PBS. Then, 200 μl of fresh PBS was added and the cells were incubated at 37 °C under 5% CO2 for another 24 h. Supernatants were collected and centrifuged for 5 min at 300 rpm to remove floating cells, and the amount of total protein in each sample was measured. Total protein was loaded onto agarose gels in amonts commensurate with the decreased levels resulting from BCL9 knockdown;after electrophoresis, the gels were silver-stained according to the manufacturer's protocol (Bio-Rad 161-0449). The remaining aliquots were processed for LC-MS and MS analysis. Proteins were reduced with 10 mM DTT at 56 °C for 1 h, alkylated with 22.5 mM IAA for 30 min at room temperature in the dark, and digested with 2.5 μg of trypsin (Promega) at 37 °C overnight. Peptides were desalted using POROS10R2 (Applied Biosystems) and reconstituted with 0.1% TFA44. Peptides were then analyzed by LC-MS and MS on an Orbitrap-XL mass spectrometer (Thermo Scientific), as described45. MS and MS spectra were searched against a forward-reversed human NCBI Refseq database using Mascot (Matrix Science, version 2.2.1), and were filtered to a 1% false-discovery rate. Five different criteria were used to select proteins from MS raw data: 1) molecular weights had to be <28 kDa; 2) more than two unique target peptides had to be identified for each protein; 3) peptides giving an overly weak signal were discarded; 4) the protein was cytoplasmic or secreted; 5) nucleoprotein or cytoskeleton proteins were excluded.
Statistical analysis
Statistical differences between groups were estimated by means of the unpaired Student's t-test, with p≤0.05 being considered significant. Analysis of tumor burden was done using factorial analysis in SPSS 13.0. mRNA expression of CD147 (GSE6477) was measured in BM plasma cells from a normal subject or a personwith MM. All experiments were done blindly, without the investigator's knowing the identity of the samples, which were labelled only with code numbers.
Supplementary Material
Acknowledgments
DRC is supported by a senior award from the Multiple myeloma Research Foundation, Doctor's Cancer Foundation, as well as 1R01CA151391 and 1P01CA15525801 from the National Institutes of Health.. Human BM-derived endothelial cell lines BMEC-6018 and HBME-119 were kindly provided by Drs. van der Schoot and Giuliani, respectively.
Footnotes
Authorship contributions
D.Z. performed most of the experiments, analyzed the data, and prepared the manuscript; W.Z. performed ELISA studies and helped with immunoblots; J.A.M. planned and coordinated proteomic experiments; Z.J. generated reagents; C.M. helped with scaffold experiments; S.B.F. performed total proteomic analysis; D.M., and M.C., studied cell viability using tumor cell-specific in vitro bioluminescence imaging (CS-BLI); D.D. selected person cases and analyzed CD147 by flow cytometry; T. H. performed CDC assay; H.T. performed animal imaging; Y.K performed protemocis analysis; G.P. performed IHC studies; Y.T., C.W., S.T., Z.H., F.M., N.M., provided clinical samples and critically reviewed the manuscript; KA provided clinical samples and edited the manuscript; J.A.M. supervised the proteomic studies; P.T. provided scaffolds along with technical and scientific assistance with scaffold experiments; R.D.C. designed experiments, analyzed data, and wrote the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. The Journal of clinical investigation. 2012;122:3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller-Hermelink HK ME, Catovsky D, Campo E, Harris NL, Stein H. Chronic lymphocytic leukemia/small lymphocytic lymphoma. World Health Organization Classification of Tumor, Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2008:180–182. [Google Scholar]
- 5.Anderson KC. Oncogenomics to target myeloma in the bone marrow microenvironment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:1225–1233. doi: 10.1158/1078-0432.CCR-10-3366. [DOI] [PubMed] [Google Scholar]
- 6.Vande Broek I, Vanderkerken K, Van Camp B, Van Riet I. Extravasation and homing mechanisms in multiple myeloma. Clin Exp Metastasis. 2008;25:325–334. doi: 10.1007/s10585-007-9108-4. [DOI] [PubMed] [Google Scholar]
- 7.Ghobrial IM. Myeloma as a model for the process of metastasis: implications for therapy. Blood. 2012;120:20–30. doi: 10.1182/blood-2012-01-379024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakob C, et al. Angiogenesis in multiple myeloma. Eur J Cancer. 2006;42:1581–1590. doi: 10.1016/j.ejca.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Ribatti D, Vacca A. The role of microenvironment in tumor angiogenesis. Genes & nutrition. 2008;3:29–34. doi: 10.1007/s12263-008-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribatti D, Nico B, Vacca A. Importance of the bone marrow microenvironment in inducing the angiogenic response in multiple myeloma. Oncogene. 2006;25:4257–4266. doi: 10.1038/sj.onc.1209456. [DOI] [PubMed] [Google Scholar]
- 11.Ria R, et al. Gene expression profiling of bone marrow endothelial cells in patients with multiple myeloma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:5369–5378. doi: 10.1158/1078-0432.CCR-09-0040. [DOI] [PubMed] [Google Scholar]
- 12.Alsayed Y, et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109:2708–2717. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani M, et al. BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 2009;69:7577–7586. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012;31:2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 16.de la Roche M, et al. An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid. Nat Commun. 2012;3:680. doi: 10.1038/ncomms1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takada K, et al. Targeted disruption of the BCL9/beta-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med. 2012;4:148ra117. doi: 10.1126/scitranslmed.3003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rood PM, Calafat J, von dem Borne AE, Gerritsen WR, van der Schoot CE. Immortalisation of human bone marrow endothelial cells: characterisation of new cell lines. Eur J Clin Invest. 2000;30:618–629. doi: 10.1046/j.1365-2362.2000.00672.x. [DOI] [PubMed] [Google Scholar]
- 19.Giuliani N, Colla S, Rizzoli V. Angiogenic switch in multiple myeloma. Hematology. 2004;9:377–381. doi: 10.1080/10245330400018524. [DOI] [PubMed] [Google Scholar]
- 20.Mitsiades CS, Mitsiades NS, Munshi NC, Richardson PG, Anderson KC. The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: interplay of growth factors, their receptors and stromal interactions. Eur J Cancer. 2006;42:1564–1573. doi: 10.1016/j.ejca.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 21.De Bruyne EM, Eline, Van Valckenborgh Els, De Raeve Hendrik, Van Camp Ben, Van Riet Ivan, Vanderkerken Karin. Myeloma Cells and Their Interactions With the Bone Marrow Endothelial Cells. Current Immunology Reviews. 2007;3(15):41–55. [Google Scholar]
- 22.Hideshima T, Anderson KC. Molecular mechanisms of novel therapeutic approaches for multiple myeloma. Nat Rev Cancer. 2002;2:927–937. doi: 10.1038/nrc952. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, et al. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009;69:9228–9235. doi: 10.1158/0008-5472.CAN-09-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calimeri T, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia. 2011;25:707–711. doi: 10.1038/leu.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song F, et al. Cyclophilin A (CyPA) induces chemotaxis independent of its peptidylprolyl cis-trans isomerase activity: direct binding between CyPA and the ectodomain of CD147. J Biol Chem. 2011;286:8197–8203. doi: 10.1074/jbc.C110.181347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yurchenko V, Constant S, Eisenmesser E, Bukrinsky M. Cyclophilin-CD147 interactions: a new target for anti-inflammatory therapeutics. Clin Exp Immunol. 2010;160:305–317. doi: 10.1111/j.1365-2249.2010.04115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidle UH, Scheuer W, Eggle D, Klostermann S, Stockinger H. Cancer-related issues of CD147. Cancer Genomics Proteomics. 2010;7:157–169. [PubMed] [Google Scholar]
- 29.Damsker JM, et al. Targeting the chemotactic function of CD147 reduces collagen-induced arthritis. Immunology. 2009;126:55–62. doi: 10.1111/j.1365-2567.2008.02877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanderson RD, Borset M. Syndecan-1 in B lymphoid malignancies. Annals of hematology. 2002;81:125–135. doi: 10.1007/s00277-002-0437-8. [DOI] [PubMed] [Google Scholar]
- 31.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 32.Avecilla ST, et al. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nature medicine. 2004;10:64–71. doi: 10.1038/nm973. [DOI] [PubMed] [Google Scholar]
- 33.Kollet O, et al. HGF, SDF-1, and MMP-9 are involved in stress-induced human CD34+ stem cell recruitment to the liver. The Journal of clinical investigation. 2003;112:160–169. doi: 10.1172/JCI17902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agrawal SM, Yong VW. The many faces of EMMPRIN - roles in neuroinflammation. Biochim Biophys Acta. 2011;1812:213–219. doi: 10.1016/j.bbadis.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Arendt BK, et al. Increased expression of extracellular matrix metalloproteinase inducer (CD147) in multiple myeloma: role in regulation of myeloma cell proliferation. Leukemia. 2012;26:2286–2296. doi: 10.1038/leu.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacono KT, Brown AL, Greene MI, Saouaf SJ. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol. 2007;83:283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki J, Jin ZG, Meoli DF, Matoba T, Berk BC. Cyclophilin A is secreted by a vesicular pathway in vascular smooth muscle cells. Circulation research. 2006;98:811–817. doi: 10.1161/01.RES.0000216405.85080.a6. [DOI] [PubMed] [Google Scholar]
- 38.Comerford I, Kara EE, McKenzie DR, McColl SR. Advances in understanding the pathogenesis of autoimmune disorders: focus on chemokines and lymphocyte trafficking. British journal of haematology. 2014;164:329–341. doi: 10.1111/bjh.12616. [DOI] [PubMed] [Google Scholar]
- 39.Niu H, Wang R, Cheng J, Gao S, Liu B. Treatment of (131)I-labeled anti-CD147 monoclonal antibody in VX2 carcinoma-induced liver tumors. Oncology reports. 2013;30:246–252. doi: 10.3892/or.2013.2418. [DOI] [PubMed] [Google Scholar]
- 40.McMillin DW, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nature medicine. 2010;16:483–489. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sukhdeo K, et al. Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci U S A. 2007;104:7516–7521. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng J, Gygi SP. Proteomics: the move to mixtures. Journal of mass spectrometry : JMS. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 43.Levanon K, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29:1103–1113. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adelmant GO CJ, Ficarro SB, Sikorski TW, Zhang Y, Marto JA. Affinity and chemical enrichment for mass spectrometry-based proteomics analyses. Sample preparation in biological mass spectrometry. 2011:437–486. [Google Scholar]
- 45.Ficarro SB, et al. Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Analytical chemistry. 2009;81:3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.