Using an unbiased, microarray-based transcriptional profiling approach, this study identified a total of 121 gene transcripts in CD8 T cells that change significantly during intensification of antiretroviral therapy with Raltegravir.
Keywords: antiretroviral therapy, CD8 T cells, gene expression profiling, HIV-1, raltegravir
Abstract
Background. Intensification of antiretroviral therapy with raltegravir does not affect levels of residual human immunodeficiency virus (HIV)-1 viremia, but it has led to increased levels of episomal HIV-1 DNA in some patients, suggesting antiviral activity against otherwise unresponsive components of the viral reservoir. Effects of raltegravir on host cells remain less well understood.
Methods. We used comprehensive and unbiased microarray-based transcriptional profiling to analyze gene expression changes in CD8+ T cells from participants in a randomized clinical trial (AIDS Clinical Trials Group [ACTG] A5244) comparing raltegravir-intensified to nonintensified antiretroviral therapy.
Results. Although raltegravir intensification failed to induce statistically significant changes in HIV-1 DNA or residual plasma viremia, we observed significant increases in the expression intensity of 121 host gene transcripts. In functional annotations of these transcripts, we found that they were mainly involved in glucose and carbohydrate metabolism, immune regulation, control of cell proliferation, and tumor suppression. Two of the raltegravir-responsive gene transcripts were statistically correlated with levels of residual HIV-1 RNA, but none of the remaining 119 transcripts were associated with immunologic or virologic characteristics of the study patients.
Conclusions. Together, these findings demonstrate that raltegravir intensification can induce previously unrecognized, statistically significant gene expression changes in host CD8+ T lymphocytes.
Combination antiretroviral therapy (ART) can effectively suppress plasma levels of human immunodeficiency virus (HIV)-1 replication below the threshold of detection of commercial polymerase chain reaction (PCR) assays, but residual HIV-1 viremia and a reservoir of latently infected HIV-1 cells remain detectable in the majority of patients, and they contribute to viral persistence despite treatment [1]. Whether residual, low-level HIV-1 viremia in ART-treated patients reflects ongoing complete cycles of viral replication, cell-to-cell spread of HIV-1, or virus release from HIV-1 infected cells is an ongoing debate; however, antiretroviral intensification does not affect the levels of residual HIV-1 viremia [2–5], and most [6–8], but not all [9], studies do not find evidence of ongoing viral sequence evolution in residual plasma HIV-1 RNA. Nevertheless, ART intensified with raltegravir resulted in elevations of episomal 2-long terminal repeat (LTR) HIV-1 DNA in several patients [5, 10], suggesting that ongoing HIV-1 replication can occur in some ART-treated patients. Although effects of intensified ART on viral replication dynamics have been investigated in detail, only limited information is available regarding effects of raltegravir intensification on host cells. In previous studies, intensification of ART with raltegravir resulted in some degree of reduction of immune activation [2–4], which is inversely associated with reconstitution of CD4+ T cells [11, 12] and the frequency of HIV-1-associated comorbities [13, 14] in ART-treated individuals. However, effects of raltegravir on immune activation were of limited statistical significance, and not all studies showed an effect.
Transcriptional profiling allows for comprehensive high-throughput assessments of gene expression patterns, and it provides an opportunity to discover previously unrecognized changes in host gene expression signatures associated with distinct disease entities or treatment interventions. In this study, we used whole-genome transcriptional profiling to comprehensively analyze gene expression changes in T lymphocytes from patients participating in a randomized, placebo-controlled clinical study that evaluated the effects of raltegravir intensification in ART-treated patients [4] (AIDS Clinical Trials Group [ACTG] A5244 [NCT00515827]). This placebo-controlled clinical trial did not find evidence for an effect of raltegravir intensification on residual HIV-1 viremia, CD4+ T cell-associated total HIV-1 DNA, or 2-LTR HIV-1 DNA. In addition, there was no influence of raltegravir intensification on immune activation in CD8+ T cells or CD4+ T cells, defined by coexpression of CD38 and human leukocyte antigen (HLA)-DR, although a trend for a reduced percentage of CD38-positive cells in the CD8+ T cell compartment after raltegravir intensification was noted [4]. We hypothesized that despite the lack of a significant effect on persistent viremia or conventional measures of CD8+ T cell activation, raltegravir intensification may be associated with more subtle changes in gene expression and activation of CD8+ T cells that can only be detected using an unbiased, comprehensive analytic approach. We chose to focus on transcriptional changes in the CD8+ T compartment rather than the CD4+ T compartment, because abnormal immune activation in ART-treated HIV-1 patients is typically most pronounced in this cell subset.
PATIENT AND METHODS
Study Subjects
We performed whole-genome transcriptional profiling in isolated CD8+ T cells from patients participating in ACTG study A5244. In this study, 53 HIV-1-infected participants receiving suppressive ART with plasma HIV-1 RNA <50 copies/mL but detectable viremia by single-copy assay were randomized to either add raltegravir (group A) or placebo (group B) to their background ART regimen for 12 weeks; participants then crossed over to the other therapy (placebo in group A; raltegravir in group B) for 12 additional weeks while continuing prestudy ART. Clinical and demographic characteristics of the 32 patients included in this study did not significantly differ from the original study population (n = 53), and these characteristics are summarized in Table 1.
Table 1.
Clinical and Demographic Characteristics of Study Participants
| Study population | N = 32 subjects participating in ACTG 5244 |
| Age (median, range) | 52 (35–71) years |
| Sex (female/male ratio) | 3 female, 29 male |
| Median plasma HIV-1 RNA (25%, 75% percentile) (determined by single-copy assay) | 1.5 copies/mL (0.6–4 copies/mL) |
| Median CD4 T cell count (25%, 75% percentile) | 569 cells/µL (443–733 cells/µL) |
| Prestudy ART regimen | 11 Protease inhibitor-based |
| 21 NNRTI-based | |
| Race/ethnicity | Black non-Hispanic: 7 |
| Hispanic: 4 | |
| White non-Hispanic: 21 |
Abbreviations: ACTG, AIDS Clinical Trials Group; ART, antiretroviral therapy; HIV, human immunodeficiency virus; NNRTI, nonnucleoside reverse-transcriptase inhibitor.
Isolation of CD8 T Cells
Using an automated magnetic cell enrichment device that isolates target cell populations with >95% purity, CD3/CD8-positive T cells were isolated from peripheral blood mononuclear cell samples collected at baseline, week 12, and week 24 in 32 study subjects (n = 16 from Group A, n = 16 from group B).
Gene Microarrays
After mRNA extraction from the purified cells (mirVana RNA extraction kit, Ambion), whole-genome transcriptional profiling was performed using Illumina Human HT-12 V4 microarrays according to standard protocols [15]. Data retrieved from the Illumina platform were background corrected, and quantile normalized as previously described [16].
Statistical Analysis
A total of 5143 transcripts with detectable expression in at least 95% of all 96 study samples was used to calculate changes in gene expression intensities occurring between the beginning and the end of raltegravir-intensified (Δ1) or nonintensified treatment intervals (Δ2) for each of the study participants from group A and B. Subsequently, gene transcripts that changed significantly during Δ1 compared with Δ2 on a false discovery rate (FDR)-adjusted P value < .05 were identified using 2-tailed Student t tests and the Significant Analysis of Microarray program (SAM) implemented in samr [17]. Functional annotations of gene transcripts were analyzed using the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/). Correlations between gene expression intensity and biological or immunological parameters were performed by generalized estimated equations adjusted for repeated measures.
RESULTS
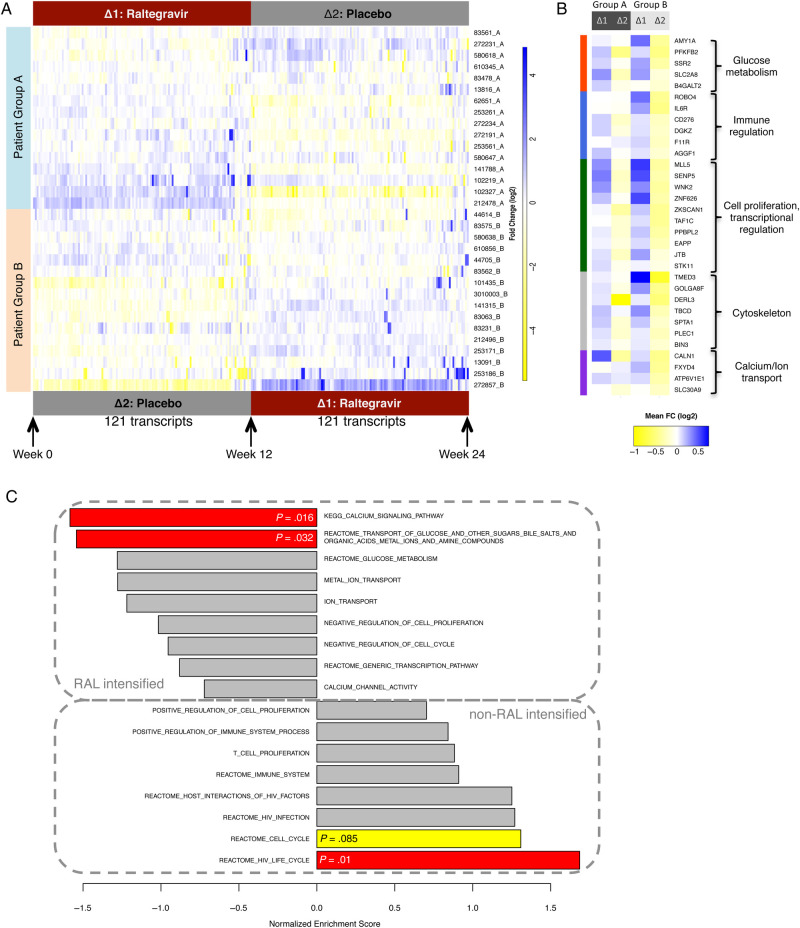
We observed a total of 121 gene transcripts with significant expression changes during raltegravir-intensified ART compared with nonintensified ART. Compared with when participants were receiving nonintensified ART, the expression intensity of all of these transcripts increased during raltegravir intensification; none decreased significantly (after FDR adjustment). Despite reaching statistical significance, changes in expression intensity for 79.3% of these transcripts were relatively small (<1.5 fold); however, expression levels of 20.7% of these transcripts demonstrated >1.5 fold changes during raltegravir intensification. Using a hierarchical clustering approach, we observed that gene expression patterns of all 121 differentially expressed genes allowed us to distinguish between intensified and the nonintensified treatment periods in both study groups (Figure 1A; Supplementary Table 1). Moreover, gene expression changes of these transcripts during raltegravir-intensified and nonintensified therapy in treatment group A closely resembled those observed in treatment group B (Figure 1A and B), indicating that they were independent of the relative timing of raltegravir intensification; however, differences appeared to be slightly more pronounced in study group B, which received raltegravir intensification in the second interval. These data demonstrate that raltegravir intensification can be associated with previously unrecognized, statistically significant changes in host gene expression profiles of CD8+ T lymphocytes.
Figure 1.
Differentially expressed gene transcripts in CD8 T cells during raltegravir-intensified antiretroviral therapy. A, Heatmap demonstrating the expression intensity of n = 121 gene transcripts with significantly (false discovery rate-corrected P < .05) different expression intensity during raltegravir intensified (Δ1) vs nonraltegravir-intensified (Δ2) antiretroviral therapy. B, Expression changes of selected gene transcripts with differential expression during raltegravir intensification. FC, fold change. C, Gene set enrichment scores of 17 predefined gene sets in raltegravir-intensified vs nonintensified treatment intervals.
To understand the possible function of the gene transcripts that are selectively upregulated during raltegravir intensification, we analyzed their functional annotations (Figure 1B). We observed that several of the raltegravir-responsive transcripts were involved in glucose and carbohydrate metabolism, which can be abnormally altered in persons infected with HIV-1 [18]. These transcripts included PFKFB2 (which encodes 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2); AMY1A (encoding for alpha-amylase 1); B4GALT2 (encoding for beta-1,4-galactosyltransferase 2), which is known to be involved in the biosynthesis of specific saccharide structures; SLC2A8, which encodes for GLUT8, an intracellular glucose transporter [19]; and SSR2, encoding for components of the somatostatin receptor. Another set of raltegravir-responsive gene transcripts plays important roles in immune regulation and immune activation, parameters that are independently associated with HIV-1 disease progression [20]. These genes included transcripts for the interleukin-6 receptor and for the costimulatory molecule B7-H3 (CD276), which can occur as a membrane-bound and soluble form and exerts context-dependent inhibitory or activating immune signals [21]. Moreover, we noted elevated expression of the gene transcripts encoding for DGK-zeta, a cell-intrinsic inhibitor of T-cell hyperactivation [22], and for the junctional cell adhesion molecule JAM-A, the lack of which has been associated with enhanced intestinal permeability, bacterial translocation, and immune activation [23]. It is interesting to note that AGGF1, which encodes a factor that inhibits vascular inflammation [24], was also upregulated during raltegravir intensification, as was ROBO4, a gene involved in stabilization of the vasculature and support of angiogenesis [25]. Another group of upregulated transcripts had known functions in regulating cell proliferation, gene transcription, and tumor suppression; these transcripts included MLL5, SENP5, WNK2, ZNF626, ZKSCAN1, TAF1C, PPBPL2, EAPP, JTB, and STK11. We also observed transcriptional changes in genes encoding for components of the cytoskeleton structure, the Golgi apparatus, and the endoplasmic reticulum, such as TMED3, GOLGA8J, DERL3, TBCD, SPTA1, PLEC1, and BIN3. Finally, transcripts encoding for proteins involved in calcium and ion transport and metabolism were elevated during raltegravir intensification; these included CALN1, FXYD4, ATP6V1E1, and SLC30A9.
We subsequently studied the relative over- or underexpression of specific genes during raltegravir-intensified ART by using gene set enrichment analysis (GSEA), a computational method that is independent of the detection of differentially expressed genes but can identify more modest but coordinated changes in the expression levels of functionally related genes between 2 distinct study populations [26]. For these investigations, we selected 17 different gene sets based on observations described in Figure 1A and B, and we calculated enrichment scores for the respective transcripts during treatment intervals with or without raltegravir intensification using the GSEA algorithm [27] (Figure 1C; Supplementary Table 2). Subsequently, these gene set enrichment scores were each tested for statistical significance by a permutation test with a test size of 1000, as described previously [27]. Overall, these data demonstrated a significant enrichment of genes involved in the HIV life cycle in nonraltegravir-intensified treatment periods; transcripts within this group included RANBP2 [28], NUP214 [29], FEN1 [30], and several additional transcripts that have recognized roles for supporting the efficacy of HIV-1 replication in human cells and were identified by high-throughput functional genomic screens [31, 32]. In addition, there was a trend for enrichment of transcripts involved in cell cycle regulation during nonraltegravir-intensified treatment periods, whereas genes encoding for components involved in transport and metabolism of glucose and calcium were enriched during raltegravir-intensified treatment periods (Figure 1C). Together, these findings support our initial observation that raltegravir intensification is associated with distinct transcriptional signatures of genes with presumed roles in HIV-1 disease pathogenesis.
To further investigate the role of the 121 raltegravir-responsive gene transcripts, we determined their association with immunologic or virologic characteristics of the study cohort. For this purpose, we used generalized estimating equations adjusted for repeated measures [33] implemented in geepack [34, 35] to correlate the gene expression intensity of all analyzed 5143 transcripts to the proportion of HLA-DR+/CD38+ CD4+ and CD8+ T cells, biomarkers of the viral reservoir size in ART-treated patients (total HIV-1 DNA, 2-LTR HIV-1 DNA) in CD4+ T cells, and residual HIV-1 plasma RNA measured by single copy assays; P values were adjusted for multiple comparisons using the Benjamini-Hochberg FDR. Overall, using a linear link function, we observed that 2 of the 121 raltegravir-responsive transcripts were statistically associated with residual HIV-1 plasma viremia (LOC728440 and LOC654103); the precise biological function of these transcripts is currently unknown. We identified an additional 339 transcripts that were statistically associated with 1 or more of the immunologic or virologic characteristics of the study patients (residual HIV-1 plasma RNA: n = 20 transcripts, proportion of HLA-DR+/CD38+ CD4+ T cells: n = 21 transcripts, proportion of HLA-DR+/CD38+ CD8+ T cells: n = 115 transcripts, CD4 T cell-associated total HIV-1 DNA: n = 4 transcripts, CD4 T cell-associated 2-LTR HIV-1 DNA (determined using a logit link function) n = 1 transcript); none of these transcripts were significantly affected by raltegravir intensification (Figure 2; Supplementary Table 3).
Figure 2.
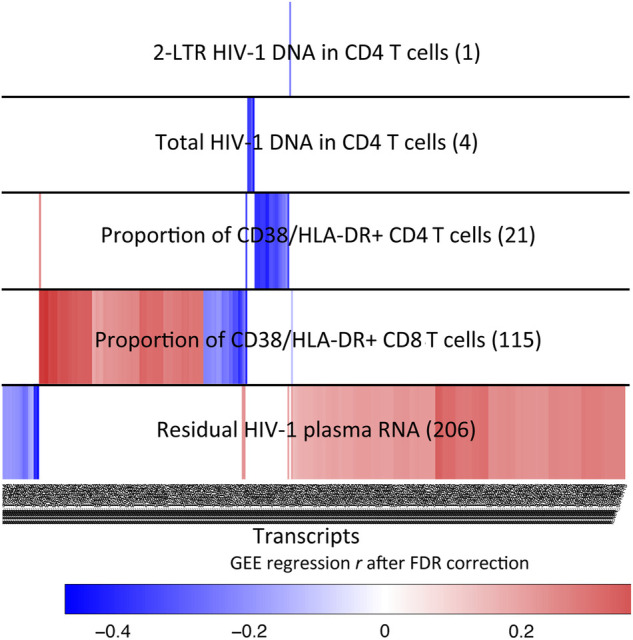
Associations between CD8+ T cell gene expression intensity and virological and immunological characteristics of study patients. Data reflect transcripts significantly (false discovery rate (FDR)-corrected P < .05) associated with indicated immunological or virological parameters. Numbers of significantly associated transcripts for each parameter are indicated in parentheses. A total of n = 206 transcripts was associated with residual human immunodeficiency virus (HIV)-1 plasma RNA; 2 of these transcripts were significantly altered during raltegravir intensification, whereas n = 204 were not. Colors reflect degree of association (generalized estimating equation (GEE) regression r).
DISCUSSION
In this study, we have identified a panel of gene transcripts in CD8+ T cells that are significantly increased by intensification of suppressive ART with raltegravir in a randomized clinical trial in which no significant effects of raltegravir intensification on biomarkers of the viral reservoir size or on the proportion of activated lymphocytes were observed. Thus, these data demonstrate more subtle, yet statistically significant effects of intensified ART on host lymphocytes. Whether such transcriptional changes in CD8+ T cells are clinically significant and relevant for determining disease outcome or disease progression will have to be investigated in future studies. Moreover, additional work will be required to determine whether the observed changes of transcriptional signatures indirectly result from the antiviral activities of raltegravir or from direct pharmaceutical effects of raltegravir on CD8+ T lymphocytes; HIV-1-negative individuals who receive raltegravir as part of postexposure prophylaxis or pharmacokinetic studies would be helpful to determine which of the observed effects are due to direct effects of the drug on host cells as opposed to changes related to the drug's antiviral activity.
Functional annotations of raltegravir-responsive transcripts found that treatment intensification mainly influenced genes related to carbohydrate metabolism, immune homeostasis, and cell growth and proliferation, which are involved in immunological and metabolic abnormalities that are typically observed in HIV-1 patients, even when viral replication is suppressed by ART below the threshold of commercial PCR assays. As such, raltegravir intensification may induce subtle and previously unrecognized alterations of critical aspects of HIV-1 disease pathogenesis. However, our expression profiling experiments were limited to CD8+ T cells, and they did not involve other cell subsets such as endothelial cells, which may be affected by treatment with raltegravir [36]. Therefore, functional studies in additional cell subsets will be necessary to better define the precise consequences associated with raltegravir-associated gene changes. Moreover, our study was not designed to analyze the interplay between host gene expression profiles and the susceptibility or resistance of CD4+ T cells to HIV-1 replication steps; because the actual frequency of HIV-1-infected CD4+ T cells in ART-treated patients is very low, such studies are better conducted in ex vivo-infected CD4+ T cells [37].
CONCLUSIONS
Our data suggest that raltegravir intensification seems to have previously unrecognized, significant effects on host genes that may be functionally involved in specific aspects of HIV-1 disease pathogenesis. In this way, this study highlights advantages of a comprehensive, unbiased analysis approach for evaluation of biological effects of treatment interventions in clinical antiretroviral treatment studies, and it may have implications for the future evaluation and monitoring of antiretroviral treatment effects that are independent from traditional outcome parameters.
Supplementary Material
Supplementary material is available online at Open Forum Infectious Diseases (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
We acknowledge all the members of the A5244 study team and particularly thank Dr. David Margolis for helpful comments on this manuscript. We also thank all of the Clinical Research Sites as well as all study participants of the A5244 study.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Financial support. This study was supported by a research grant for an investigator-initiated study from Merck & Co, Inc. (grant 40262). M. L. is supported by the American Foundation for AIDS Research (grant 108302-51-RGRL), the Doris Duke Charitable Foundation (grant 2009034), and National Institutes of Health (NIH) grants AI098487 and AI106468. X. G. Y. is supported by NIH grants AI098484 and HL126554. R. T. G. receives support from grants to the AIDS Clinical Trials Group (NIH U01 AI694722) and the Harvard University Center for AIDS Research (NIH 2P30 AI060354-06).
The trial described in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH under Award Number UM1 AI068636 and supported in part by a grant(s) funded by National Center for Advancing Translational Sciences or subsequently designated NIH centers, including the National Institute of Mental Health, the National Heart, Lung and Blood Institute, the National Cancer Institute for OHARA and Malignancy Studies, the National Institute on Drug Abuse, and the National Institute of Dental and Craniofacial Research. The project was also supported by a grant from NIAID to the Pittsburgh Virology Support Laboratory (UM1 AI106701) and the Statistical and Data Analysis Center (UM1 AI 068634).
Potential conflicts of interest. R. T. G.'s institution has received educational grant support from Janssen, Abbott, and Viiv for projects in which he is involved.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Katlama C, Deeks SG, Autran B et al. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 2013; 381:2109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llibre JM, Buzon MJ, Massanella M et al. Treatment intensification with raltegravir in subjects with sustained HIV-1 viraemia suppression: a randomized 48-week study. Antivir Ther 2012; 17:355–64. [DOI] [PubMed] [Google Scholar]
- 3.Hatano H, Hayes TL, Dahl V et al. A randomized, controlled trial of raltegravir intensification in antiretroviral-treated, HIV-infected patients with a suboptimal CD4+ T cell response. J Infect Dis 2011; 203:960–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi RT, Zheng L, Bosch RJ et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med 2010; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buzon MJ, Massanella M, Llibre JM et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med 2010; 16:460–5. [DOI] [PubMed] [Google Scholar]
- 6.Bailey JR, Sedaghat AR, Kieffer T et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 2006; 80:6441–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kieffer TL, Finucane MM, Nettles RE et al. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis 2004; 189:1452–65. [DOI] [PubMed] [Google Scholar]
- 8.Palmer S, Maldarelli F, Wiegand A et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2008; 105:3879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiu C, Cunningham CK, Greenough T et al. Identification of ongoing human immunodeficiency virus type 1 (HIV-1) replication in residual viremia during recombinant HIV-1 poxvirus immunizations in patients with clinically undetectable viral loads on durable suppressive highly active antiretroviral therapy. J Virol 2009; 83:9731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatano H, Strain MC, Scherzer R et al. Increase in 2-LTR circles and decrease in D-dimer after raltegravir intensification in treated HIV-infected patients: a randomized, placebo-controlled trial. J Infect Dis 2013; 208:1436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunt PW, Martin JN, Sinclair E et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis 2003; 187:1534–43. [DOI] [PubMed] [Google Scholar]
- 12.Anthony KB, Yoder C, Metcalf JA et al. Incomplete CD4 T cell recovery in HIV-1 infection after 12 months of highly active antiretroviral therapy is associated with ongoing increased CD4 T cell activation and turnover. J Acquir Immune Defic Syndr 2003; 33:125–33. [DOI] [PubMed] [Google Scholar]
- 13.Sandler NG, Wand H, Roque A et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 2011; 203:780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lok JJ, Hunt PW, Collier AC et al. The impact of age on the prognostic capacity of CD8+ T-cell activation during suppressive antiretroviral therapy. AIDS 2013; 27:2101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigneault F, Woods M, Buzon MJ et al. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. J Virol 2011; 85:3015–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolstad BM, Irizarry RA, Astrand M et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19:185–93. [DOI] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98:5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez AD, Balasubramanyam A. Dysregulation of glucose metabolism in HIV patients: epidemiology, mechanisms, and management. Endocrine 2012; 41:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt S, Joost HG, Schurmann A. GLUT8, the enigmatic intracellular hexose transporter. Am J Physiol Endocrinol Metab 2009; 296:E614–8. [DOI] [PubMed] [Google Scholar]
- 20.Deeks SG, Kitchen CM, Liu L et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood 2004; 104:942–7. [DOI] [PubMed] [Google Scholar]
- 21.Chapoval AI, Ni J, Lau JS et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol 2001; 2:269–74. [DOI] [PubMed] [Google Scholar]
- 22.Zhong XP, Guo R, Zhou H et al. Diacylglycerol kinases in immune cell function and self-tolerance. Immunol Rev 2008; 224:249–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khounlotham M, Kim W, Peatman E et al. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity 2012; 37:563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu FY, Wu C, Li Y et al. AGGF1 is a novel anti-inflammatory factor associated with TNF-alpha-induced endothelial activation. Cell Signal 2013; 25:1645–53. [DOI] [PubMed] [Google Scholar]
- 25.London NR, Li DY. Robo4-dependent Slit signaling stabilizes the vasculature during pathologic angiogenesis and cytokine storm. Curr Opin Hematol 2011; 18:186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, Backes C, Gerasch A et al. A novel algorithm for detecting differentially regulated paths based on gene set enrichment analysis. Bioinformatics 2009; 25:2787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, Mehla R, Chauhan A. Perturbation of host nuclear membrane component RanBP2 impairs the nuclear import of human immunodeficiency virus-1 preintegration complex (DNA). PloS One 2010; 5:e15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Nunzio F, Danckaert A, Fricke T et al. Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration. PloS One 2012; 7:e46037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rumbaugh JA, Fuentes GM, Bambara RA. Processing of an HIV replication intermediate by the human DNA replication enzyme FEN1. J Biol Chem 1998; 273:28740–5. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H, Xu M, Huang Q et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 2008; 4:495–504. [DOI] [PubMed] [Google Scholar]
- 32.Brass AL, Dykxhoorn DM, Benita Y et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008; 319:921–6. [DOI] [PubMed] [Google Scholar]
- 33.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 1986; 42:121–30. [PubMed] [Google Scholar]
- 34.Halekoh U, Hojsgaard S, Yan J. The R package geepack for Generalized Estimating Equations. J Stat Softw 2006; 15:1–11. [Google Scholar]
- 35.R Development Core Team, R: A Language and Environment for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing; Available at: http://www.R-project.org. Accessed 26 April 2015. [Google Scholar]
- 36.Masia M, Martinez E, Padilla S et al. Endothelial function in HIV-infected patients switching from a boosted protease inhibitor-based regimen to raltegravir: a substudy of the SPIRAL study. J Antimicrob Chemother 2013; 68:409–13. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi P, Desfarges S, Bartha I et al. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog 2013; 9:e1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.