Abstract
Orchestration of cellular growth and development occurs during the life cycle of Aspergillus nidulans. A multi-copy genetic screen intended to unveil novel regulators of development identified the AN6578 locus predicted to encode a protein with the WOPR domain, which is a broadly present fungi-specific DNA-binding motif. Multi-copy of AN6578 disrupted the normal life cycle of the fungus leading to enhanced proliferation of vegetative cells, whereas the deletion resulted in hyper-active sexual fruiting with reduced asexual development (conidiation), thus named as osaA (Orchestrator of Sex and Asex). Further genetic studies indicate that OsaA balances development mainly by repressing sexual development downstream of the velvet regulator VeA. The absence of osaA is sufficient to suppress the veA1 allele leading to the sporulation levels comparable to veA + wild type (WT). Genome-wide transcriptomic analyses of WT, veA1, and ΔosaA veA1 strains by RNA-Seq further corroborate that OsaA functions in repressing sexual development downstream of VeA. However, OsaA also plays additional roles in controlling development, as the ΔosaA veA1 mutant exhibits precocious and enhanced formation of Hülle cells compared to WT. The OsaA orthologue of Aspergillus flavus is able to complement the osaA null phenotype in A. nidulans, suggesting a conserved role of this group of WOPR domain proteins. In summary, OsaA is an upstream orchestrator of morphological and chemical development in Aspergillus that functions downstream of VeA.
Introduction
Coordination of vegetative growth and reproduction in filamentous fungi requires finely regulated networks of diverse genetic elements, which integrate intrinsic signals with surrounding external cues [1–3]. In the ascomycete Aspergillus nidulans, the asexual life cycle starts with the germination of a conidium followed by the formation of an undifferentiated network of interconnected hyphal cells, collectively termed mycelium, which develops according to a functionally coherent plan. Asexual development (conidiation) begins by a series of morphogenetic differentiations results in the formation of conidia (asexual spores) bearing structure known as the conidiophore (reviewed in [4]).
Asexual reproduction is centrally controlled by BrlA, a highly conserved activator of conidiation in the genus Aspergillus [5, 6]. Sexual development in A. nidulans, a morphogenetic counterpart to conidiation, is achieved by the formation of a cleistothecium that bears ascospores (sexual spores). Each cleistothecium is surrounded by a nest-like structure made from thick-walled Hülle cells serving as nursing cells [7]. These two developmental processes are antagonistic, and initiation of one developmental process inhibits the other. Genetic and molecular investigations have revealed several activator of sexual reproduction (reviewed in [8]). To achieve the proper sporulation levels, the fungus utilizes a range of interacting genetic systems [3, 8].
The velvet regulators (veA, velB, velC and vosA) were found in many fungal species to assume a key role in regulating development; dictate and balance both types of reproductive spores (conidia and ascospores) [9–12]. VeA (interacting with VelB) is required to activate sexual reproduction and indirectly inhibits conidiation [10, 13–15]. In addition, it is involved in activating secondary metabolism (SM) through physically interacting with LaeA, the master activator of SM [10]. The veA1 knockdown mutation, which lacks nuclear localization signal (NLS), thereby defective in the translocation to the nucleus, causes significantly reduced activity, resulting in highly reduced sexual fruiting with enhanced conidiation, i.e., the velvet phenotype [14, 16].
The WOPR domain proteins are a newly defined family of regulators that started gaining attention for the past decade. WOPRs are a fungi-specific family of transcriptional factors that are involved in multiple biological processes in various fungi [17, 18]. The name WOPR is derived from the best-studied member Wor1, and its closely related members Pac2 and Ryp1 [18]. WOPR proteins regulate morphological transitions and pathogenesis in many fungi: e.g., Ryp1 in Histoplasma capsulatum [19] and Wor1 in Candida albicans [20] and Liv3 in Cryptococcus neoformans [21]. They were also found to play crucial roles in regulating sporulation in a number of fungi, including Fusarium spp. [22, 23]. The WOPR protein Ryp1 functions along with the velvet homologs (Ryp2 and Ryp3) in controlling hypha-to-yeast developmental switch in H. capsulatum [19, 24].
In an attempt to further understand the genetic networks underlying developmental regulation in A. nidulans, we carried out a gain-of-function multi-copy screen. OsaA with a predicted WOPR domain was identified as a potential regulator of development. Further genetic and genomic studies have revealed that OsaA plays a pivotal role in orchestrating asexual and sexual development primarily by down-regulating sexual fruiting downstream of VeA in A. nidulans. Our studies uncover a new genetic interaction between the WOPR protein OsaA and the key velvet regulator VeA, and they together function in coordinating developmental life cycle in A. nidulans. A new genetic model depicting the roles of VeA and OsaA is presented.
Material and Methods
Fungal strains and culture conditions
The Aspergillus strains used in this study are listed in Table 1. Standard culture and genetics techniques were used [25]. Strains were grown on minimal solid or liquid medium (simplified as MM) with appropriate supplements as previously described [26] at 37°C unless otherwise indicated. Induction of asexual development or sexual development was done as described previously [27, 28].
Table 1. Aspergillus strains used in this study.
| Strain | Genotype | Source/Reference |
|---|---|---|
| A. nidulans a | ||
| FGSC4 | veA + WT | FGSC b |
| FGSC26 | biA1 WT | FGSC |
| FGSC237 | pabaA1, yA2; trpC801 | FGSC |
| PW1 | biA1; argB2; methG1 | P. Weglenski |
| RRAW16 | pyrG89, yA2; veA + | R. A. Wilson and N. P. Keller |
| RYG1.9 | pabaA1, yA2; argB2, ΔfluG::trpC + | Guan and Yu, unpublished |
| TNI3.1 | argB2; pyroA4; ΔvosA::argB + | This study |
| RNIW5 | pyrG89; pyroA4 | This study |
| RNI11.2, 3, 4 C | biA1; ΔosaA::argB + | This study |
| RNI15.1, 2, 3C | biA1; ΔosaA::argB + ; veA + | This study |
| TNI18.1, 2, 3, 4, 5 C | pabaA1; yA2; gpdA(p)::osaA::trpC + | This study |
| TFA4.1, 2 C | biA1; ΔosaA::argB + ; biA + ; osaA + | This study |
| TFA5.1, 2 C | biA1; ΔosaA::argB + ; biA + ; AflwprA + | This study |
| A. flavus | ||
| NRRL 3375 | Wild type | [29] |
aAll A. nidulans strains carry the veA1 mutation, unless mentioned as veA +.
bFGSC: Fungal Genetics Stock Center
cMultiple isogenic strains.
Gain of function genetic screen
The recipient strain RNIW5 (pyrG89 pyroA4 veA +) was transformed with the pRG3-AMA1-NotI WT library [30], which confers multi-copy presence of a given gene. The transformants showing fluffy phenotypes were isolated. Genomic DNA was isolated from these fluffy transformants, and was transformed into Escherichia coli to recover the plasmids. The rescued plasmids were introduced back into the recipient strain to check whether they could still cause fluffy phenotype. By direct sequencing of the insert ends of the interested plasmids with the primer set OMN33 and OMN35 (all primers listed in S2 Table), and followed by genome search [31] identified several potential repressors of development. Four such developmentally altered transformants identified, including AN6578, which is renamed as osaA. To examine the positions of osaA’s introns, RT-PCR, followed by sequencing analyses, was carried out. Note that the gene structure of osaA is different from the predicted structure of AN6578.3 from the Broad Institute [31].
Construction of fungal strains
A. nidulans osaA deletion mutant (TNI3.1) was generated by transforming PW1 with the osaA deletion cassette containing argB + as the selective marker. The cassette was constructed by employing Double-Joint PCR [32]. RNI11.1 (ΔosaA veA1) was isolated from the cross between TNI2.1 and TNI3.1. RNI11.2, 3, 4 (ΔosaA veA1) were isolated from the cross between TNI3.1 and RYG1.9. RNI15.1, 2, 3 (ΔosaA veA +) were isolated from the cross between RNI11.1 and RRAW16. TFA4.1,2 (ΔosaA veA1 osaA(p)::osaA +) were generated by transforming RNI11.3 (biA1) with the osaA gene PCR fragment including 1.6 kb from each of 5’ and 3’ region. TFA5.1,2 (ΔosaA AflwprA(p)::AflwprA + veA1) were generated by transforming RNI11.3 (biA1) with the AflwprA gene PCR fragment including 1.6 kb from each of 5’ and 3’ region from A. flavus NRRL 3375 strain [29].
Nucleic acid isolation and manipulation
Genomic DNA and total RNA isolation and Northern blot analyses were carried out as previously described [27, 33]. The DNA probes were prepared by PCR-amplification of a coding region of individual genes with appropriate oligonucleotide pairs using FGSC4 genomic DNA as template (S2 Table).
Microscopy
The colony photographs were taken using a Sony DSC-F828 digital camera. Photomicrographs were taken using a Zeiss M2 BIO microscope equipped with AxioCam and AxioVision digital imaging software (Zeiss).
Sample preparation for mRNA sequencing
Three biological replicates were analyzed for each strain. All strains were cultured in agitating liquid-submerged medium (vegetative growth) for 18 h, and then shifted to an air-exposed medium to induce development. All samples were collected at time point 12 h following developmental induction, total RNA was extracted, and submitted to ProteinCT Biotechnologies (Madison, WI) for library preparation and sequencing.
Library preparation and sequencing
Strand specific library was prepared from total RNA using Illumina TruSeq Strand specific RNA sample preparation system. Briefly, mRNA was extracted from total RNA using polyA selection, followed by RNA fragmentation. Strand specific library was constructed by first-strand cDNA synthesis using random primers, followed by sample cleanup and the second-strand synthesis using DNA Polymerase I and RNase H. A single 'A' base was added to the cDNA fragments followed by ligation of the adapters. Final cDNA library was obtained by further purification and enrichment with PCR, and the quality was checked using Bioanalyzer 2100. The library was sequenced (PE100bp) using the Illumina HiSeq2500, with final of over 10 million high quality reads per sample. RNA-Seq full workflow integrated service was provided by ProteinCT Biotechnologies (Madison, WI). The scattered plot in Supplementary S1 Fig shows a high correlation levels among triplicates, indicated by the correlation coefficient R (R-ΔosaA veA1 > 0.97, R-veA1 WT > 0.97 and R-veA + WT > 0.97), all with the p-value less than 0.01.
Data QC and analysis
The fastQC program was used to verify raw data quality of the Illumina reads. The A. nidulans FGSC4 genome and gene annotations (A_nidulans_FGSC_A4_version_s10-m03-r08_features.gff) were downloaded from AspGD [34, 35] and used for mapping. The raw sequence reads were mapped to the genome using Subjunc aligner from Subread [36], with majority of the reads (80–90% for all samples) aligned to the genome. The alignment bam files were compared against the gene annotation GFF file, and raw counts for each gene were generated using the featureCounts tool from Subread, with ~70–85% of reads assigned to genes overall. The raw counts data were normalized using voom method from the R Limma package [37], then used for differential expression analysis. For transcripts assembly, Cufflinks program [38–41] was used to assemble transcripts from RNA-Seq reads for each sample, and cuffmerge command from cufflinks was used to merge the transcripts from all samples into a single set of genes and transcripts. The gene annotation from AspGD was provided as reference in the process and included in the final assembly.
Results
Multi-copy screen identifies AN6578 as a regulator of development
To investigate the mechanisms governing fungal development, a gain-of-function (multi-copy) screen was carried out as described in (11). We envisioned that such regulators are unlikely to be defined via chemical mutagenesis, which leads to mostly loss of function mutations. As described (11), after screening more than 56,000 transformants, seven colonies exhibiting no or reduced developmental phenotypes were isolated, and four of them contained AN6578 in the multi-copy plasmid pRG3-AMA1 [30]. Re-introduction of the multi-copy plasmid containing the AN6578 locus alone resulted in a near complete loss of development with fluffy phenotype (Fig 1A). Further studies showed the AN6578 gene was required for balancing asexual and sexual development, thus named as osaA (orchestrator of sexual and asexual development). Constitutive expression of osaA by gpdA(p)::osaA delayed development about 24h (data not shown).
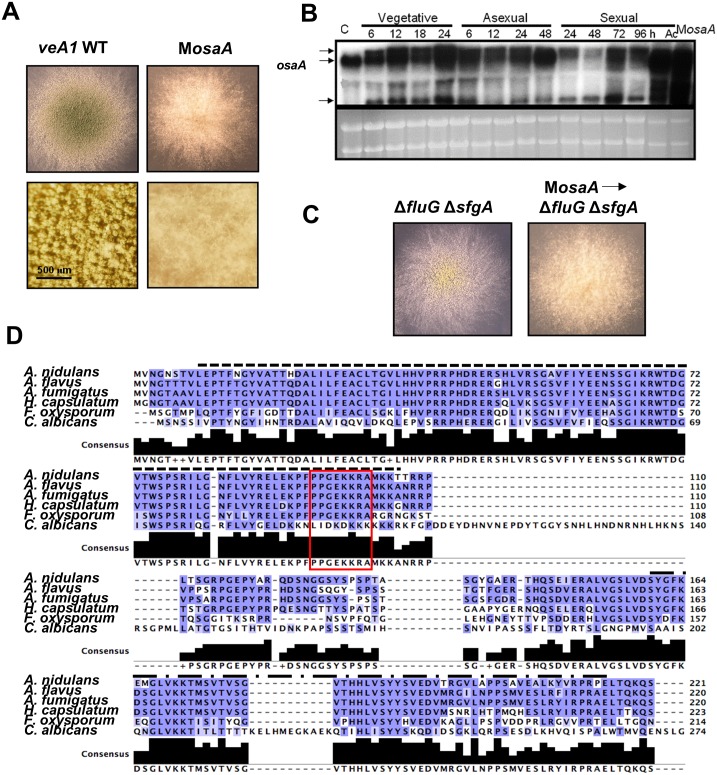
Fig 1. Summary of OsaA.
(A) Colony photographs of WT and the multi-copy osaA strains (upper panel) and close-up views (lower panel). (B) Northern blot showing the osaA transcripts [~3.2kb, ~2.8kb and ~1kb] levels throughout the life cycle. C: conidia, Ac: ascospores, MosaA: multi-copy osaA. (C) Colonies of ΔfluG ΔsfgA and multi-copy osaA (MosaA) in ΔfluG ΔsfgA strains grown on solid MM at 37°C for 3 d along with the close-up views (lower panel). (D) Alignment of the WOPR proteins in Aspergillus and other WOPR members. A. nidulans OsaA (AN6578), A. flavus (AFL2T_08419), A. fumigatus (Afu5g12960), (ABX74945.1), F. oxysporum Sge1 (AGA55574.1), F. graminearum (I1S5P3) and C. albicans Wor1 (Q5AP80). The dashed line indicates the conserved region 1, and the dash-dot line indicates the conserved region 2. The red box indicates the NLS sequence.
Northern blot and sequence analyses of the RT-PCR products suggested that the osaA gene consists of three overlapping transcripts (~3.2 kb, ~2.8 kb and ~1 kb; S4 Fig). Only the 2.8 kb transcript is present in conidia (C), while all three transcripts are detectable during vegetative growth, asexual and sexual development with the highest accumulation levels in sexual spores (ascospores; Ac in Fig 1B), suggesting OsaA might play a role during the life cycle of the fungus. The two long transcripts (3.2 kb and 2.8 kb) contain the identical 1,422 nt ORF, which is translated into a 474 amino acids (aa) polypeptide. The small transcript is derived from an alternative splicing of the 1,422 transcript, and the 43rd to 860th nt segment of the OsaA long ORF is removed in the final exon, leading to a 149 aa-length protein. All these findings imply that the activity of osaA might be regulated at the transcription level.
OsaA functions independently to the FluG mediated conidiation pathway
FluG is a key upstream activator of asexual development. The deletion of fluG results in complete blockage of conidiation [42]. FluG activates conidiation via removing repressive effects imposed by SfgA [43]. As the data indicate that OsaA may acts as a repressor of conidiation, we asked whether OsaA functions in the FluG—| SfgA governed conidiation pathway. We generated and examined the phenotypes of the ΔosaA ΔfluG mutant, which were similar to as those of ΔfluG (data not shown). Moreover, the ΔosaA alcA(p)::sfgA (overexpression of sfgA) double mutant showed phenotype similar to that of the alcA(p)::sfgA mutant (data not shown), indicating OsaA may function upstream of, or independent to, the FluG—| SfgA pathway. Importantly, multi-copy of osaA (MosaA) in the ΔfluG ΔsfgA background resulted in a developmental phenotype in-between the two mutants, (Fig 1C), indicating that OsaA does not function in upstream of FluG—|SfgA pathway.
OsaA encodes a WOPR domain protein
Through BLAST search [31] we found that the osaA gene is predicted to encode a WOPR domain protein at the N-terminus (Fig 1D) with a similarity in amino acid sequence to Wor1 N-terminus in C. albicans [18, 20, 44]. The predicted OsaA polypeptide (473 aa) is highly conserved in all Aspergillus species. The N-terminus, in particular, is conserved in almost all filamentous fungi and it harbors the WOPR domain (Fig 1D). WOPR was shown to have sequence-specific DNA binding property; therefore, it represents a novel DNA-binding protein superfamily [18, 24, 44]. The predicted OsaA protein contains the two-conserved WOPR subdomains, Conserved Region 1 (CR1) and Conserved Region 2 (CR2), separated by a characteristic less conserved linker [17, 18]. In addition, OsaA contains the nuclear localization signal (NLS: 95-PGEKKRA-102) within the WOPR domain as found in two other WOPR proteins, HcRyp1 [19, 24] and FoSge1 [23]. Like many fungi [17, 18], A. nidulans has two predicted WOPR proteins, OsaA and WprB.
OsaA represses sexual development downstream of VeA
To begin to understand the function of OsaA, through employing Double-Joint PCR [32], we first generated the osaA deletion, in a veA + strain mutant, with argB + as a selective marker (RRAW16, Table 1). VeA is required for sexual development, and the veA1 allele lacking the NLS is partially active thereby causing significantly suppressed sexual reproduction [14–16, 45]. Thus, to properly address the function of OsaA in developmental balancing, we further isolated ΔosaA veA1 (RNI11) and ΔosaA veA + (RNI15) recombinant strains through subsequent genetic crosses. As shown in Fig 2A, both ΔosaA mutant colonies displayed clearly altered developmental phenotypes with significantly higher levels of cleistothecia and much reduced levels of conidiation in comparison to their corresponding WTs. Quantitative analyses of conidiation and sexual fruiting revealed the absence of osaA resulted in 1.6 (veA +) and 2.1 (veA1) fold reduction in conidia formation (Fig 2B), while causing 2.5 (veA +) and ~ 7,000 (veA1) fold enhanced formation of cleistothecia (Fig 2C). It is important to note that the data showing the enhanced production of cleistothecia by ΔosaA are obtained from the solid air-exposed cultures grown for 6 days under the light. Light inhibits sexual fruiting in A. nidulans and almost completely abolishes cleistothecia formation in veA1 strains. These indicate that OsaA is a key repressor of sexual development and the absence of osaA can compensate the inhibitory effects on sexual development by light [1, 3].
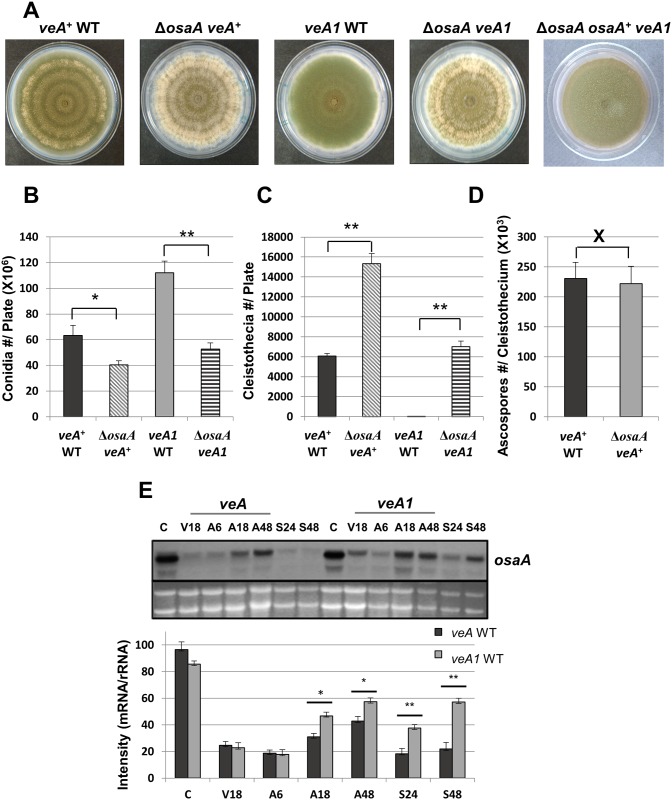
Fig 2. OsaA balances sporulation levels.
(A) Colony photographs of WT (veA + and veA1), osaA mutants (ΔosaA veA + and ΔosaA veA1), and osaA complementation (ΔosaA osaA veA1) strains. (B) A histogram depicting the number of conidia per plate. (C) A histogram depicting the number cleistothecia per plate. (D) A histogram depicting the number (103) ascospores per cleistothecium. In B, C and D error bars represent the standard deviation among three or more replicates; * represents p-value < 0.05; ** represents p-value < 0.01; X represents p-value > 0.5. (E) Northern blot showing osaA mRNA accumulation levels in veA + and veA1 WT strains (Upper). C: conidia; V18: vegetative 18 h; A6, 18, 48: asexual induction 6, 18, 24 h; S24, 48: sexual induction 24, 48 h. Band intensity estimation in reference to the rRNA levels using imageJ software [46] (Lower); * represents p-value < 0.05; ** represents p-value < 0.01.
On the other hand, the loss of osaA did not affect the spore viability or number. The number of ascospores per cleistothecium was similar between the ΔosaA and WT strains. However, factoring in the large number of cleistothecia produced by the ΔosaA mutant, the total number of ascospores per colony (each point inoculated and cultured for 6 days) is higher in ΔosaA compared to WT. In addition, germination and survival rates for both spore types were identical between ΔosaA and WT. Collectively, the primary role of OsaA is to repress the onset of sexual development.
Importantly, our data indicate that the loss of osaA is sufficient to suppress the veA1 mutation in development. As shown in Fig 2B and 2C, the ΔosaA veA1 mutant and veA + WT showed similar levels of conidiation and sexual fruiting, indicating that osaA is epistatic to veA1 and osaA likely functions downstream of veA in regulating sporulation. However, the absence of osaA was not sufficient to restore the mycotoxin sterigmatocystin production levels to that of veA + WT (data now shown), suggesting that OsaA is likely not involved in the VelB-VeA-LaeA controlled pathways for certain secondary metabolites [9, 10, 47]. Northern blot showed that levels of osaA mRNA are higher in veA1 WT in contrast to those in veA + WT, especially during sexual development initiation phase, 24 h and 48 h (Fig 2E). Taken together, we propose that OsaA is an upstream repressor of sexual development and VeA removes the repressive effects imposed by OsaA on sexual fruiting.
Distinct role of OsaA in repressing Hülle cell formation
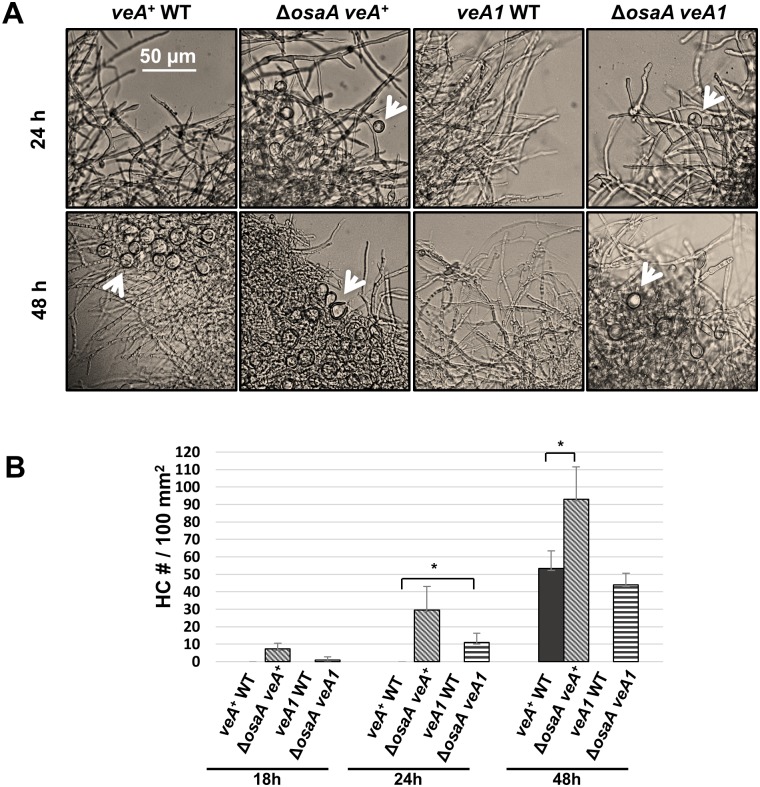
Hülle cell (HC) formation is coordinated with the cleistothecium developmental stages, yet, it is proposed to be under the regulation of a separate genetic pathway [7, 15, 48, 49]. Similar to the OsaA’s role in repressing sexual development on air-exposed solid medium, we found that OsaA represses the formation of HC in liquid submerged culture. The ΔosaA veA + mutant shows precocious production of HC (as early as 18 h) compared to veA + WT strain showing HC production at 48 h (Fig 3A and 3B). At 48 h ΔosaA veA + strain produces almost double the number of HC in comparison to veA + WT. Surprisingly, even the ΔosaA veA1 mutant started to produce HC at 24 h, earlier than veA + WT strain. These suggest that OsaA is a dual function upstream regulator of sexual development controlling both pathways governing cleistothecia and HC formation.
Fig 3. OsaA represses Hülle cell formation.
(A) Photomicrographs of WT (FGSC4 and FGSC26) and osaA mutant (ΔosaA veA + and ΔosaA veA1) strains grown in liquid-submerged culture up to 48 h. White arrows indicate Hülle cells. (B) Quantitative analysis of HCs within 100 mm2 area. Error bars represent the standard deviation among three replicates; * represents a p-value < 0.05.
Genome-wide suppression of veA1 by ΔosaA
Balancing asexual and sexual development involves the coordinated regulations of thousands of genes associated with cellular, morphological, and metabolic processes. The fact that the deletion of osaA was sufficient to suppress the developmental bias toward conidiation caused by the veA1 mutation suggests that genome-wide expression is likely restored to veA + WT level. To test this hypothesis, we sequenced total RNA populations in ΔosaA veA1, veA1 WT, and veA + WT strains at 12 h post asexual-developmental induction.
Fragments per kilobase of exon per million fragments mapped (FPKM) obtained for ΔosaA veA1, veA1 WT, and veA + WT strains were mapped for 10,536, 10,428 and 10,514 genes, respectively, representing 96.3%, 95.3% and 96% of the total of 10,943 genes predicted by the AspGD (S2 Fig) [34, 35]. FPKM mapped to the osaA locus in the three tested strains confirmed that no osaA transcript is detectable in ΔosaA veA1 strain and higher levels of osaA mRNA are present in veA1 compared to veA + WT (S3 Fig). From the data, any differentially expressed genes with a false discovery rate (FDR) greater than 0.05 was considered insignificant. Gene expression changes with slight fold change (FC), i.e., -1< log2FC <1, were not included in our analysis, unless stated otherwise.
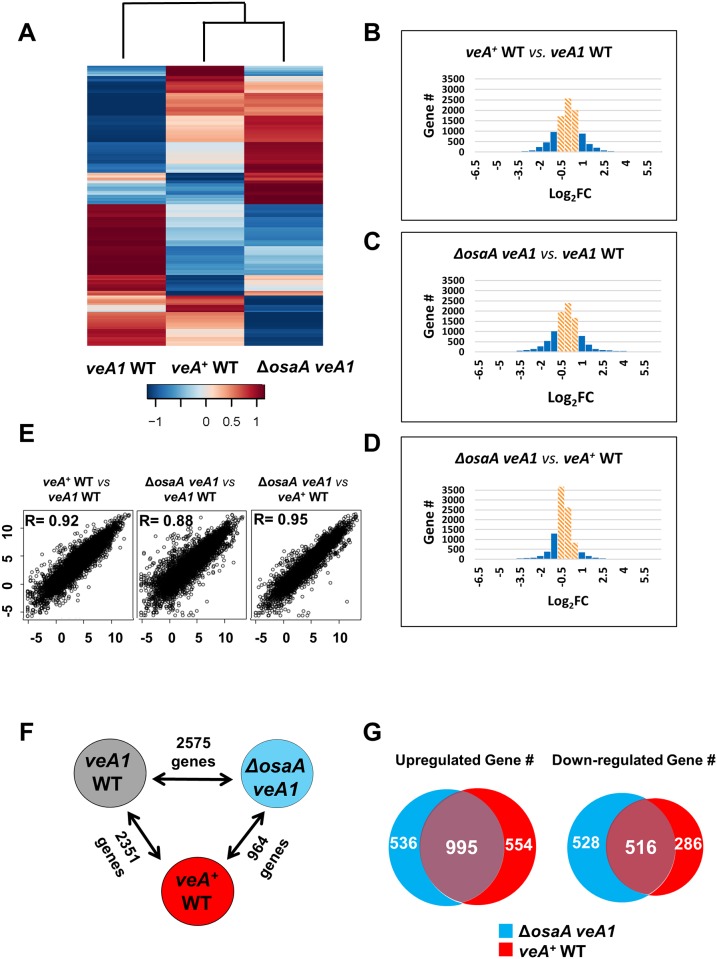
As shown in Fig 4A, the hierarchical clustering heat map based on genome-wide expression analysis shows that the transcriptome of ΔosaA veA1 and veA + WT strains are clustered in one group separated from that of the veA1 WT strain, suggesting that ΔosaA veA1 shows higher similarity of gene expression pattern to veA + WT strain than to veA1 WT. The degree of differential expression between ΔosaA veA1 and veA + WT is lower than those between veA1 WT and ΔosaA veA1, or veA1 WT and veA + WT strains (Fig 4B, 4C and 4D), further indicating that the distance between the gene expression profiles of ΔosaA veA1 and veA + WT strain is closer than those of the two strains and veA1 WT. Likewise, we examined the correlation among the overall transcriptomic profiles of the three strains via multidimensional scaling analysis (S5 Fig), and found that ΔosaA veA1 triplicates fall closer to veA + WT than to veA1 WT. Moreover, by using the gene expression profiles of ΔosaA veA1, veA + WT and veA1 WT, we calculated the correlation between differential expression values comparing veA + WT to veA1 WT, ΔosaA veA1 to veA1 WT, and ΔosaA veA1 to veA + WT. The obtained correlations have R-value equals to 0.92, 0.88, and 0.95, respectively (Fig 4E). These results further indicate that ΔosaA veA1 and veA + WT gene expression patterns are closer to each other than to veA1 WT, suggesting that suppression of veA1 by ΔosaA is likely due to restored genome-wide expression.
Fig 4. Genome-wide expression correlations among veA1 WT, veA + WT and ΔosaA veA1.
The included genes all have an FDR value less than 0.05 and fall in the -1<log2FC<1 fragment count range, unless indicated otherwise. (A) Heat map illustration of expression level changes among veA1 WT, veA + WT and ΔosaA veA1 strains. RPKM values were used for hierarchical clustering of genes. Scale bar shows the RPKM values (Z-score). In all analyses veA1 WT was used as a reference. (B, C, and D) Histograms showing general transcriptomic results of three analytical sets, veA + WT vs veA1 WT, ΔosaA veA1 vs veA1 WT and ΔosaA veA1 vs veA + WT, respectively. Columns in orange diagonal lines fall in the 1>log2FC>-1 fragment count range with low differential expression values. (E) Linear fitted models showing the correlation between three analytical sets, veA + WT vs veA1 WT, ΔosaA veA1 vs veA1 WT and ΔosaA veA1 vs veA + WT. The correlation coefficient R is indicated within each model. (F) The numbers of genes showing differential expression between two strains. (G) Venn diagrams showing up- and down-regulated genes in ΔosaA veA1 and veA + WT relative to veA1 WT. The numbers of common and unique genes are indicated.
To further analyze differentially expressed genes, pair-wise comparisons were carried out (Fig 4F and 4G). First, the veA1 and veA + data set comparison identified 2,351 differentially expressed genes, 1,549 up-regulated and 802 down-regulated genes. Second, the veA1 and ΔosaA veA1 data set comparison revealed 2,575 differentially expressed genes, 1,531 up-regulated and 1,044 down-regulated genes. The obtained high numbers of differentially expressed genes further indicate that both VeA and OsaA act upstream in regulating gene expression, affecting a wide range of downstream genes. Third, the veA + and ΔosaA veA1 data set comparison identified 964 differentially expressed genes, 480 up-regulated and 484 down-regulated genes (Fig 4F). Among the differentially regulated genes, 995 and 516 genes were up-regulated and down-regulated in both ΔosaA veA1 and veA + WT, respectively (Fig 4G). Many in the top 20 differentially expressed gene lists (Tables 2, 3, 4 and 5) are common between ΔosaA veA1 and veA + WT. In a simplified interpretation, common genes are regulated by the veA —| osaA pathway, whereas other genes are probably regulated by either VeA or OsaA independently.
Table 2. Top 20 up-regulated genes in ΔosaA veA1 relative to veA1 WT.
| ID | Log2FC | Notes |
|---|---|---|
| AN11049 | 11.566 | NmrA-like domain containing protein; predicted secondary metabolism gene cluster member |
| AN8105 | 10.679 | Putative NRPS-like enzyme; predicted secondary metabolism gene cluster member |
| AN1627 | 10.503 | Protein of unknown function |
| AN10044 | 8.893 | mdpK |
| AN3568 | 8.820 | Protein of unknown function |
| AN0146 | 8.631 | mdpC |
| AN5430 | 8.612 | Protein of unknown function |
| AN0223 | 8.491 | Has domain(s) with predicted sequence-specific DNA binding RNA polymerase II transcription factor activity, zinc ion binding activity, role in regulation of transcription, DNA-dependent and nucleus localization |
| AN11037 | 8.407 | Predicted transmembrane transporter; predicted secondary metabolism gene cluster member |
| AN0147 | 8.381 | mdpD |
| AN6784 | 8.319 | xptA |
| AN8106 | 8.058 | Predicted dioxygenase; role in secondary metabolite biosynthesis; predicted secondary metabolism gene cluster member |
| AN0149 | 7.688 | mdpF |
| AN1242 | 7.606 | Putative NRPS involved in nidulanin A biosynthesis; predicted backbone enzyme of the secondary metabolite gene cluster |
| AN10023 | 7.483 | mdpL |
| AN6314 | 7.399 | Has domain(s) with predicted nucleotide binding, oxidoreductase activity and role in metabolic process |
| AN2595 | 7.358 | Protein of unknown function |
| AN5539 | 7.254 | Has domain(s) with predicted methyltransferase activity and role in methylation |
| AN6446 | 7.080 | cicD |
| AN1309 | 7.077 | Has domain(s) with predicted GTP binding, GTPase activity |
*Genes that are up-regulated in both ΔosaA veA1 and veA + WT are in bold.
Table 3. Top 20 up-regulated genes in veA + WT relative to veA1 WT.
| ID | Log2FC | Notes |
|---|---|---|
| AN11049 | 8.119 | NmrA-like domain containing protein; predicted secondary metabolism gene cluster member |
| AN10650 | 6.821 | Ortholog of A. nidulans FGSC A4: AN11203, and A. fumigatus Af293: Afu4g00520 |
| AN10044 | 6.635 | mdpK |
| AN3568 | 6.536 | Protein of unknown function |
| AN5430 | 6.505 | Protein of unknown function |
| AN0146 | 6.435 | mdpC |
| AN3893 | 6.219 | Protein of unknown function |
| AN8105 | 6.165 | Putative NRPS-like enzyme; predicted secondary metabolism gene cluster member |
| AN6314 | 5.857 | Has domain(s) with predicted nucleotide binding, oxidoreductase activity and role in metabolic process |
| AN7116 | 5.835 | Protein of unknown function |
| AN6784 | 5.553 | xptA |
| AN8101 | 5.519 | Ortholog of A. fumigatus Af293: Afu5g02655, and A. oryzae RIB40: AO090102000386 |
| AN3983 | 5.448 | Ortholog of A. wentii: Aspwe1_0168245, and A. clavatus NRRL 1: ACLA_062140 |
| AN11203 | 5.438 | Ortholog of A. nidulans FGSC A4: AN10650, and A. fumigatus Af293: Afu4g00520 |
| AN9290 | 5.428 | Has domain(s) with predicted integral to membrane localization |
| AN0149 | 5.294 | mdpF |
| AN10023 | 5.291 | mdpL |
| AN11037 | 5.245 | Predicted transmembrane transporter; predicted secondary metabolism gene cluster member |
| AN1242 | 5.225 | Putative NRPS involved in nidulanin A biosynthesis; predicted backbone enzyme of the secondary metabolite gene cluster |
| AN8775 | 5.001 | Ortholog of Neosartorya fischeri NRRL 181: NFIA_004310 |
*Genes that are up-regulated in both ΔosaA veA1 and veA + WT are in bold.
Table 4. Top 20 down-regulated genes in ΔosaA veA1 relative to veA1 WT.
| ID | Log2FC | Notes |
|---|---|---|
| AN7836 | -9.872 | Ortholog of A. fumigatus Af293: Afu7g01060, and A. oryzae RIB40: AO090003000833 |
| AN7839 | -9.739 | Has domain(s) with predicted ATP binding, ATPase activity, coupled to transmembrane movement of substances activity, role in transmembrane transport and integral to membrane localization |
| AN11574 | -9.614 | Protein of unknown function |
| AN12330 | -9.021 | Protein of unknown function |
| AN7834 | -8.786 | Protein of unknown function |
| AN12331 | -7.868 | Putative PKS-like enzyme |
| AN6476 | -7.423 | Protein of unknown function |
| AN6464 | -7.305 | Has domain(s) with predicted hydrolase activity, acting on ester bonds activity and role in lipid metabolic process |
| AN5032 | -6.489 | Has domain(s) with predicted role in transmembrane transport and integral to membrane localization |
| AN8609 | -6.396 | Predicted glycosylphosphatidylinositol-anchored protein |
| AN11027 | -6.061 | Has domain(s) with predicted acyl-CoA hydrolase activity and role in acyl-CoA metabolic process |
| AN5558 | -6.035 | prtA |
| AN8969 | -5.893 | Has domain(s) with predicted cation binding, lysozyme activity and role in carbohydrate metabolic process, cell wall macromolecule catabolic process, peptidoglycan catabolic process |
| AN7402 | -5.613 | glaB |
| AN3996 | -5.602 | Has domain(s) with predicted methyltransferase activity and role in metabolic process |
| AN2380 | -5.569 | Protein of unknown function |
| AN5267 | -5.402 | faeC |
| AN7396 | -5.382 | bglM |
| AN7962 | -5.247 | pepJ |
| AN12183 | -5.091 | Protein of unknown function |
*Genes that are down-regulated in both ΔosaA veA1 and veA + WT are in bold.
Table 5. Top 20 down-regulated genes in veA + WT relative to veA1 WT.
| ID | Log2FC | Notes |
|---|---|---|
| AN2380 | -6.560 | Protein of unknown function |
| AN6798 | -6.255 | Has domain(s) with predicted catalytic activity and role in metabolic process |
| AN1567 | -5.711 | Ortholog of A. fumigatus Af293: Afu5g00910, and Neosartorya fischeri NRRL 181 |
| AN6476 | -4.876 | Protein of unknown function |
| AN8476 | -4.583 | Has domain(s) with predicted aminopeptidase activity, dipeptidyl-peptidase activity and role in proteolysis |
| AN0011 | -4.445 | Possible pseudogene |
| AN2538 | -4.311 | Ortholog of A. versicolor: Aspve1_0026536 and A. sydowii: Aspsy1_0028980 |
| AN6464 | -4.245 | Has domain(s) with predicted hydrolase activity, acting on ester bonds activity and role in lipid metabolic process |
| AN7402 | -4.058 | glaB |
| AN8609 | -4.050 | Predicted glycosylphosphatidylinositol-anchored protein |
| AN4817 | -3.997 | Has domain(s) with predicted role in transmembrane transport and integral to membrane localization |
| AN5315 | -3.976 | Ortholog of A. nidulans FGSC A4: AN7859, and A. niger CBS 513.88 |
| AN2465 | -3.785 | Has domain(s) with predicted substrate-specific transmembrane transporter activity, role in transmembrane transport and integral to membrane localization |
| AN7870 | -3.767 | Ortholog of A. fumigatus Af293: Afu3g01650, Afu6g11970, and Neosartorya fischeri NRRL 181: NFIA_002730 |
| AN6820 | -3.726 | hk-8-3 |
| AN3205 | -3.605 | Putative aldehyde dehydrogenase; ortholog of A. fumigatus Afu4g02830 |
| AN6470 | -3.604 | Protein with lysozyme activity, involved in carbohydrate catabolism |
| AN3218 | -3.597 | Ortholog of A. oryzae RIB40: AO090701000492 and A. flavus NRRL 3357: AFL2T_06113 |
| AN4816 | -3.554 | Ortholog of A. oryzae RIB40: AO090020000221, and A. versicolor: Aspve1_0128410 |
| AN2395 | -3.505 | Putative beta-glucuronidase with a predicted role in polysaccharide degradation |
*Genes that are down-regulated in both ΔosaA veA1 and veA + WT are in bold.
The majority of the top differentially expressed genes obtained for both VeA and OsaA are related to metabolism (Tables 2, 3, 4 and 5); including polyketide synthases (PKS), ATPases, non-ribosomal peptide synthases (NRPS), and a plethora of various enzymes. In Aspergillus several metabolites are involved in regulating sporulation levels [1, 8, 50, 51]. For instance, the veA —| osaA pathway is necessary for the proper expression of the ivo and wA clusters that play different roles in A. nidulans sporulation [52, 53]. Interestingly, OsaA down-regulates expression of the monodictyphenone gene cluster [54] through the veA —| osaA pathway. Genes within the monodictyphenone cluster were in the top 20 up-regulated genes in ΔosaA veA1 (Table 2). Taken together, biosynthesis of several metabolites are subject to the OsaA-mediated orchestration.
Expression analyses of developmental genes
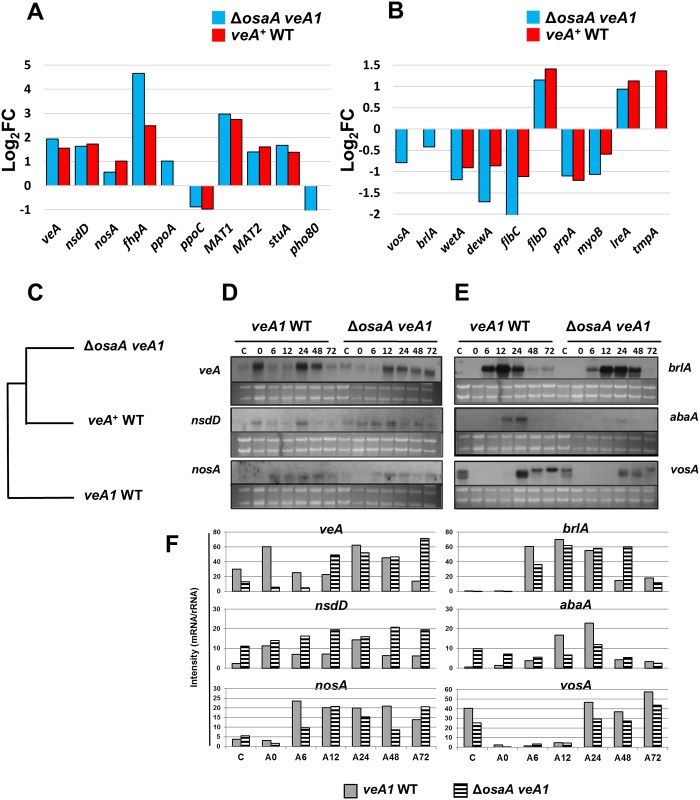
Relative to veA1 WT, in both veA + WT and ΔosaA veA1 strains the majority of sexual activators were up-regulated (Fig 5A and S1 Table) [8]. In contrast, many downstream asexual regulators (Fig 5B and S1 Table) were down-regulated in both veA + WT and ΔosaA veA1 strains compared to veA1 WT [3, 55, 56]. These findings correspond to the distinct developmental phenotypes of veA + WT and ΔosaA veA1, which show significantly higher levels of cleistothecia and much reduced levels of conidiation in comparison to veA1 WT. The hierarchical clustering-based of all developmental gene expression available in AspGD [34, 35] shows that ΔosaA veA1 and veA + WT cluster in one group separated from the veA1 WT clade. Hence, loss of osaA has competency in suppressing the veA1 mutation’s effect on developmental gene expression (Fig 5C). S1 Table shows the list of differentially expressed development-related genes.
Fig 5. Influence of OsaA on expression of developmental genes.
(A) A histogram showing log2FC values of select sexual regulators in veA + WT and ΔosaA veA1 relative to veA1 WT. (B) A histogram showing log2FC values of select asexual regulators in veA + WT and ΔosaA veA1 relative to veA1 WT. (C) Phylogenetic analysis of select developmental gene expression changes among veA1 WT, veA + WT and ΔosaA veA1 strains. Only those genes with significant changes in pair-wise comparisons (p-value < = 5%) were analyzed using their RPKM values for hierarchical clustering. (D) Northern blot showing mRNA accumulation levels of three sexual activators, veA, nsdD, and nosA, in veA1 WT and ΔosaA veA1. (E) Northern blot showing mRNA accumulation levels of three asexual regulators, brlA, abaA and vosA, in veA1 WT and ΔosaA veA1. (F) Densitometry analysis of the northern blot data shown in (Fig 5D and 5E). Band intensity was estimated in reference to the rRNA levels using imageJ software [46].
To confirm the RNA-Seq data and to further examine the expression pattern of developmental genes, we examined mRNA accumulation levels, via Northern blot, in ΔosaA veA1 comparing to veA1 WT (Fig 5D and 5E). Similar to the RNA-Seq data, mRNA levels of upstream sexual activators (veA [14], nsdD [57]) were overall higher in ΔosaA veA1 in comparison to veA1 WT (Fig 5D). The veA transcript in ΔosaA veA1 accumulated 12 h earlier and it was detectable up to 72 h, whereas in veA1 WT the transcript signal disappeared at 72 h. However, the downstream positive sexual regulator nosA [58] exhibited varying mRNA levels through out the tested time points (Fig 5D). One the other hand, the asexual regulators (brlA [5], abaA [59] and vosA [11]) showed an overall reduced and altered levels of mRNA accumulation in ΔosaA veA1 comparing to veA1 WT (Fig 5E). The brlA transcript in ΔosaA veA1 showed lower accumulation levels at 6 h and its signal stays high at 48 h (Fig 5E), whereas in veA1 WT the brlA transcript started accumulating from 6 h and disappeared at 48 h.
VeA and NsdD are proposed to down-regulate brlA [13, 60]. Moreover, the early accumulation of the veA transcript (Fig 5E) is suggested to inhibit the conidiophore formation through interfering with the competence time required to initiate conidiation [45, 61]. Accordingly, the lower brlA mRNA accumulation levels and the reduced conidiation levels in the ΔosaA mutants might be caused by the higher and early accumulation levels of nsdD and veA transcripts, respectively.
WOPR might be functionally conserved in Aspergillus
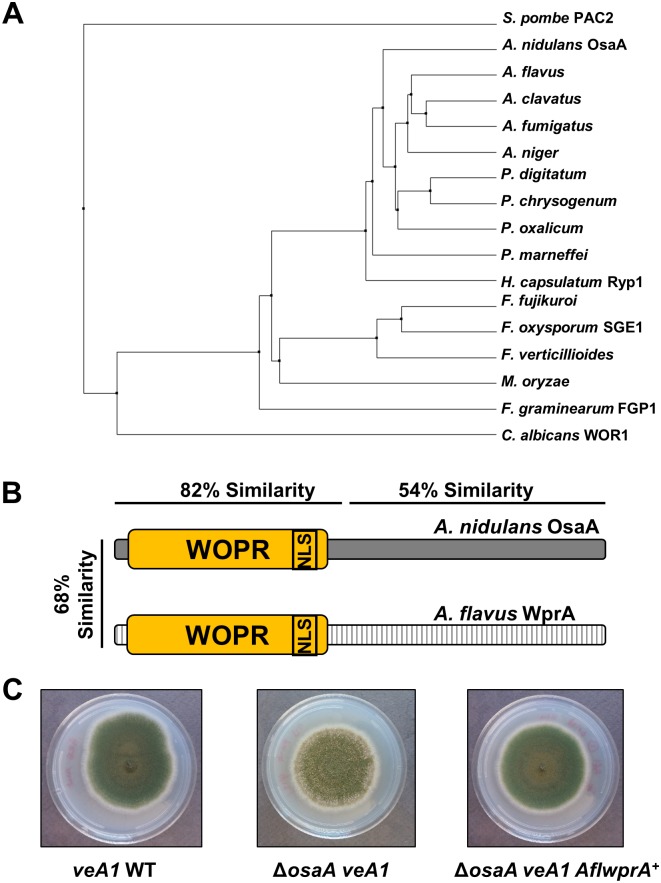
Pair-wise alignment of the Aspergillus WOPR proteins (using only the ones that showed the highest similarity score to A. nidulans OsaA) revealed a high homology amongst all Aspergillus WOPR proteins even at the less conserved C-terminus (data not shown). Aspergillus WOPRs showed higher than 80% aa similarity, except, A. nidulans OsaA that showed the lowest similarity index comparing to all the examined Aspergillus WOPRs (~68%). The C-terminal region showed above 50% homology among all aspergilli WOPRs. We constructed a phylogenetic tree using only the WOPR domain in various fungal species (Fig 6A). Aspergillus WOPRs cluster with the C. albicans Wor1 clade indicating that Aspergillus OsaA homologs are more related to Wor1 than to the closely related S. pombe Pac2 protein [20, 62].
Fig 6. A. flavus wprA can complement ΔosaA in A. nidulans.
(A) A phylogenetic tree of the WOPR proteins homologous to OsaA in several fungi. A. nidulans OsaA (AN6578), A. flavus (AFL2T_08419), A. fumigatus (Afu5g12960), A. clavatus (XP_001270095.1), A. niger (XP_001396650.1), H. capsulatum Ryp1 (ABX74945.1), F. oxysporum Sge1 (AGA55574.1), F. graminearum Fgp1 (I1S5P3), P. digitatum (EKV12249.1), P. chrysogenum (XP_002563706.1), P. oxalicum (EPS28657.1), P. marneffei (XP_002153295.1), F. fujikuroi (S0E3H0), F. verticillioides (W7MPI5.1), M. oryzae (XP_003713871.1), C. albicans Wor1 (Q5AP80) and S. pombe Pac2 (BAC54908.1). (B) AflWprA and OsaA polypeptides showing the amino acid similarity. Yellow box indicates the WOPR domain, NLS: nuclear localization sequence. (C) Colony photographs showing veA1 WT, osaA mutant (ΔosaA veA1) and cross-complementation (ΔosaA veA1 AflwprA) strains.
Through BLAST search (http://www.ncbi.nlm.nih.gov) we were able to identify OsaA’s homolog in A. flavus (AflWprA). OsaA and AflWprA proteins share 68% aa similarity. The homology percentage of the N-terminal region containing the WOPR domain is 82%, whereas the C-terminal region shows 54% similarity (Fig 6B). To test whether OsaA and AflWprA are functionally conserved, we carried out a cross-complementation experiment by introducing an AflwprA genomic DNA fragment to a ΔosaA strain, yielding the ΔosaA veA1 AflwprA + strains. As shown in Fig 6C, an example of such strains exhibited restored balance of sexual and asexual development to veA1 WT. These results indicate that WOPRs in Aspergillus are likely functionally conserved. The downstream gene targets for each WOPR protein, however, might be different in each species.
Discussion
The WOPR-domain family of global regulators (WOPRs) play critical roles in almost all fungal species. Despite the critical biological roles of WOPRs, no clear bioinformatics predictions of known-functional domains could have been made. For the past decade, extensive studies have been carried out to reveal the molecular mechanisms of WOPRs function. The WOPR domain consists of two regions (80 and 50 aa) conserved across many fungal species, separated by a non-conserved linker region with variable length (25 ~ 100 aa) [17, 18, 44]. Importantly, recent studies have solved the crystal structures of two WOPRs, S. cerevisiae WOPR YHR177w [44] and the C. albicans Wor1 segments [20], in complex with dsDNA containing a consensus binding sequence. Both studies show that the two conserved regions are tightly bound to each other through a β-sheet, with β-strands from one conserved region interdigitated with those from the other, with the linker looped out away from the DNA [17]. However, Lohse et al found that the most important contacts with DNA are made by an adjacent loop (contributed by Arg1), which is inserted into an especially narrow minor groove where it makes base-specific and backbone contacts. These studies unveil that the WOPR domain represents a new family of fungi-specific DNA-binding proteins, one with key roles for fungal morphogenesis and pathogenesis.
The protein-DNA interactions are essential for WOR1 transcriptional regulation and white-to-opaque switching. The best studied Wor1 (white-opaque switching regulator 1) protein, a master regulator of the white-opaque switching in Candida albicans, binds to a consensus 9-nucleotide core motif (TTAAAGTTT) in three different fungal species, C. albicans Wor1, H. capsulatum Ryp1, and Saccharomyces ceriviseae Mit1, indicating conservation of the WOPR domain–DNA sequence interactions over a period of 600 million to 1.2 billion years of divergence [18, 24, 44]. These WOPRs exhibit transcriptional regulation of downstream gene targets by directly interacting with the cis-regulatory elements.
In this study, we show that the A. nidulans WOPR regulator OsaA is an orchestrator of development. OsaA functions as an upstream repressor of sexual development. Sexual fruiting is subject to a tight regulation due to its high metabolic and structural costs [1, 2, 4, 8]. In addition to OsaA, a number of sexual fruiting repressors have previously been characterized in A. nidulans. RosA is an upstream transcriptional factor that represses sexual development under low glucose conditions [63]. Light inhibits sexual development mainly through the activity of the red light-sensing phytochrome FphA [64]. G-protein signaling components, RasA (GTPase) [65] and GprD (G protein coupled receptor) [66], also play critical roles in repressing sexual development. PpoC, an oxylipin biosynthetic oxygenase, represses sexual development through the biosynthesis of the 10’ oxylipins that signal the deactivation of sexual sporulation [67]. Taken together, regulating sexual fruiting requires a dynamic network that is able to integrate intrinsic signals with surrounding extrinsic cues.
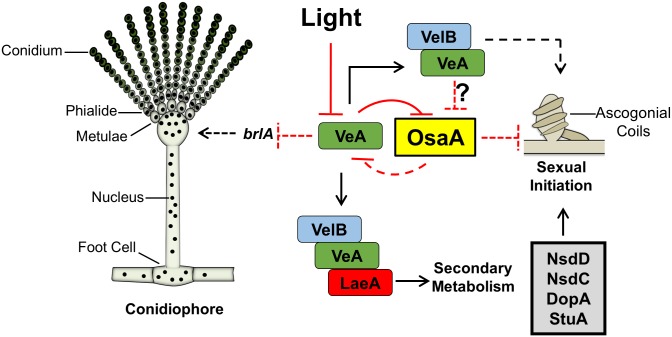
Through genetic and transcriptomic approaches we found that osaA functions downstream of the velvet regulator veA; i.e., the loss of osaA is sufficient to suppress the veA1 mutation. It is proposed that VeA is required to remove OsaA’s repressive effects on sexual development. Both velvets and WOPRs are fungi-specific regulators that are found in a large number of fungal genomes [9, 10, 17, 18, 68]. The veA1 mutation interferes with VeA nuclear translocation due to the lack of NLS [16]. The osaA gene exhibited higher mRNA accumulation levels in veA1 WT, suggesting a possibility of VeA’s role in down-regulating transcription of osaA. As a consequence, sexual development is significantly reduced in veA1 WT (Fig 2). The VeA regulatory pathways for development and SM are separated. The VelB-VeA dimer is involved in regulating sexual development, while the VelB-VeA-LaeA complex is required for SM [10, 47]. The loss of osaA did not affect the levels of the mycotoxin sterigmatocystin production in comparison to WT. Thus, OsaA is possibly regulated through the VelB-VeA dimer, or by an alternative VeA-regulated genetic pathway (Fig 7).
Fig 7. Genetic model for balanced development in A. nidulans.
Blunt ended lines indicate negative regulation, arrowheads indicate positive regulation, and dashed lines indicate potential/indirect relationship. See the main text.
Our data show that virtually all critical asexual activators were down-regulated, while most known activators of sexual development were relatively up-regulated in both ΔosaA veA1 and veA + WT (Fig 5A)[8]. This shift in gene expression relative to veA1 WT might explain the basis for the higher levels of sexual sporulation in veA + WT and ΔosaA veA1 strains. Up-regulation of many sexual activators, which in turn have a global suppressive effect on conidiation, explains the observed ΔosaA phenotype [1, 3]. OsaA also appears to down-regulate veA mRNA levels; the veA transcript exhibits earlier and higher accumulation levels in ΔosaA veA1 comparing to WT. Early accumulation of veA transcript might interfere with the competence time required to initiate asexual development [45, 61]. This could explain: 1) the suppressed conidiation levels in ΔosaA strains comparing to WT, and 2) reduced brlA mRNA levels at early asexual development in ΔosaA veA1 comparing to WT. These findings propose that osaA and veA may have a cross-repression genetic relationship that controls development and maintains a tight balance of asexual-to-sexual spore ratio (Fig 7).
The sporulation block represented by the fluffy phenotype obtained from the multi-copy expression of osaA is likely triggered by mis-regulation of the finely-tuned spatial and temporal genetic control of downstream morphologic and metabolic pathways. This speculation is based on the genome-wide expression changes of more than 2,500 genes by the deletion of osaA. Furthermore, our data show that osaA is not involved in the FluG—|SfgA main conidiation activating pathway. Taken together, instead of directly affecting asexual development, OsaA influences conidiation by repressing sexual development. On the other hand, the transcriptomic analysis shows that the veA —| osaA pathway might not be specific to morphological development, and might also regulate genes related to metabolism. This promiscuous functional behavior is due to the fact that development is a type of fungal morphogenesis, which requires both a significant epigenetic and biochemical shifts. Therefore, the genes related to processes other than development are possibly necessary to cater to the need of differentiated reproductive structures.
Each WOPR protein plays a distinct role in the fungal species it belongs to [18–20, 22, 23, 69]. We show that AflwprA was sufficient to repress enhanced sexual development in ΔosaA veA1 strain and restore the sporulation ratios back to that of veA1 WT (Fig 6C). This cross-complementation experiment suggests that WOPRs in Aspergillus are functionally conserved and might recognize the same cis-regulatory elements, despite the 450 million years’ of divergence between A. nidulans and A. flavus [70]. This further proposes an idea that the differential roles of WOPR controlling species-specific biology might be due to the modifications in each fungal genome rather than changes in the WOPR domain.
In summary, through genetic and transcriptomic studies we found that OsaA is a key upstream regulator of development primarily acting as a repressor of sexual fruiting in A. nidulans. OsaA acts downstream of the velvet regulator veA and orchestrates the developmental balance and progression. A major part of VeA role in activating sexual fruiting is likely removing OsaA’s repressive effects. In Aspergillus, the function of each WOPR protein is species dependent, and variations in WOPR roles are due to variations in the fungal genomes rather than variations in the conserved WOPR domain. Further studies are in progress to understand OsaA’s role in regulating development and SM in other Aspergillus species, and to determine direct targets of OsaA.
Supporting Information
Scattered plot showing the correlation levels among triplicates of each sample. The correlation coefficient R for ΔosaA veA1 was > 0.97, veA1 WT was > 0.97, and veA + WT was > 0.97, all with p-value less than 0.01, indicating the high quality of the RNA-Seq data sets.
(TIF)
FPKM obtained for ΔosaA veA1, veA1 WT, and veA + WT strains are mapped to 10,536, 10,428 and 10,514 genes, respectively, representing 96.3%, 95.3% and 96% coverage of a total of 10,943 genes predicted by AspGD.
(TIF)
A snapshot from Integrative Genomics Viewer (IGV) software showing the osaA locus (AN6578) in ΔosaA veA1, veA1 WT, and veA + WT strains.
(TIF)
Open box represents osaA locus; grey box represents ORF; arrow line represents transcript; open triangle in transcript ~1kb indicates alternative splicing.
(TIF)
The overall transcriptomic profiles of ΔosaA veA1, veA1 WT, and veA + WT strains examined by a two-dimensional plot. Black circles indicate replicates.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
The authors express special thanks to Dr. Heesoo Park and ProteinCT Biotechnologies for technical assistance. This work was supported by Kuwait University Graduated Fellowship from the Ministry of Higher Education to FA, and the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology (2011–0031955) to JHY and SCK.
Data Availability
RNA Seq data has been deposited in NCBI GEO database. Series no. GSE72316 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72316.
Funding Statement
This work was supported by the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology, 2011-0031955, SCK and JHY; and Kuwait University Graduated Fellowship from the Ministry of Higher Education, kuniv.edu, FA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ugalde U, Rodriguez-Urra AB. The Mycelium Blueprint: insights into the cues that shape the filamentous fungal colony. Applied microbiology and biotechnology. 2014;98(21):8809–19. 10.1007/s00253-014-6019-6 . [DOI] [PubMed] [Google Scholar]
- 2. Yu JH. Regulation of Development in Aspergillus nidulans and Aspergillus fumigatus . Mycobiology. 2010;38(4):229–37. Epub 2010/12/01. 10.4489/MYCO.2010.38.4.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park HS, Yu JH. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol. 2012;15(6):669–77. Epub 2012/10/25. 10.1016/j.mib.2012.09.006 . [DOI] [PubMed] [Google Scholar]
- 4. Adams TH, Wieser JK, Yu JH. Asexual sporulation in Aspergillus nidulans . Microbiol Mol Biol Rev. 1998;62(1):35–54. Epub 1998/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adams TH, Boylan MT, Timberlake WE. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans . Cell. 1988;54(3):353–62. Epub 1988/07/29. . [DOI] [PubMed] [Google Scholar]
- 6. Chang YC, Timberlake WE. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics. 1993;133(1):29–38. Epub 1993/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sohn KT, Yoon KS. Ultrastructure Study of the Cleistothecium Dvevelopment in Aspergillus nidulans . microbiology. 2002;30(3):117–27. [Google Scholar]
- 8. Dyer PS, O'Gorman CM. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev. 2012;36(1):165–92. Epub 2011/11/19. 10.1111/j.1574-6976.2011.00308.x . [DOI] [PubMed] [Google Scholar]
- 9. Bayram O, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2012;36(1):1–24. Epub 2011/06/11. 10.1111/j.1574-6976.2011.00285.x . [DOI] [PubMed] [Google Scholar]
- 10. Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320(5882):1504–6. Epub 2008/06/17. 10.1126/science.1155888 . [DOI] [PubMed] [Google Scholar]
- 11. Ni M, Yu JH. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans . PLoS One. 2007;2(10):e970 Epub 2007/10/04. 10.1371/journal.pone.0000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park HS, Nam TY, Han KH, Kim SC, Yu JH. VelC positively controls sexual development in Aspergillus nidulans . PLoS One. 2014;9(2):e89883 10.1371/journal.pone.0089883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato N, Brooks W, Calvo AM. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryotic cell. 2003;2(6):1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim H, Han K, Kim K, Han D, Jahng K, Chae K. The veA gene activates sexual development in Aspergillus nidulans . Fungal Genet Biol. 2002;37(1):72–80. Epub 2002/09/12. . [DOI] [PubMed] [Google Scholar]
- 15. Yager LN. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans . Biotechnology. 1992;23:19–41. . [PubMed] [Google Scholar]
- 16. Kafer E. Origins of translocations in Aspergillus nidulans . Genetics. 1965;52(1):217–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lohse MB, Rosenberg OS, Cox JS, Stroud RM, Finer-Moore JS, Johnson AD. Structure of a new DNA-binding domain which regulates pathogenesis in a wide variety of fungi. Proc Natl Acad Sci U S A. 2014;111(29):10404–10. Epub 2014/07/06. 10.1073/pnas.1410110111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lohse MB, Zordan RE, Cain CW, Johnson AD. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A. 2010;107(32):14105–10. Epub 2010/07/28. 10.1073/pnas.1005911107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A. 2008;105(12):4880–5. Epub 2008/03/15. 10.1073/pnas.0710448105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A. 2006;103(34):12813–8. Epub 2006/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans . Cell. 2008;135(1):174–88. 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jonkers W, Dong Y, Broz K, Kistler HC. The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog. 2012;8(5):e1002724 Epub 2012/06/14. 10.1371/journal.ppat.1002724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michielse CB, van Wijk R, Reijnen L, Manders EM, Boas S, Olivain C, et al. The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathog. 2009;5(10):e1000637 Epub 2009/10/24. 10.1371/journal.ppat.1000637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beyhan S, Gutierrez M, Voorhies M, Sil A. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 2013;11(7):e1001614 Epub 2013/08/13. 10.1371/journal.pbio.1001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans . Adv Genet. 1953;5:141–238. . [DOI] [PubMed] [Google Scholar]
- 26. Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. . [DOI] [PubMed] [Google Scholar]
- 27. Seo JA, Guan Y, Yu JH. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans . Genetics. 2003;165(3):1083–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ni M, Rierson S, Seo JA, Yu JH. The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans . Eukaryotic cell. 2005;4(8):1465–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown SH, Zarnowski R, Sharpee W, Keller N. Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus . Applied and environmental microbiology. 2008;74(18):5674–85. 10.1128/AEM.00565-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osherov N, Mathew J, May GS. Polarity-defective mutants of Aspergillus nidulans . Fungal Genet Biol. 2000;31(3):181–8. Epub 2001/03/29. 10.1006/fgbi.2000.1236 . [DOI] [PubMed] [Google Scholar]
- 31.Broad Institute of Harvard and MIT (http://www.broadinstitute.org/).
- 32. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41(11):973–81. Epub 2004/10/07. 10.1016/j.fgb.2004.08.001 . [DOI] [PubMed] [Google Scholar]
- 33. Han KH, Seo JA, Yu JH. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Galpha) signalling. Molecular microbiology. 2004;53(2):529–40. . [DOI] [PubMed] [Google Scholar]
- 34. Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40(Database issue):D653–9. 10.1093/nar/gkr875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42(Database issue):D705–10. 10.1093/nar/gkt1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47 10.1093/nar/gkv007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–9. 10.1093/bioinformatics/btr355 . [DOI] [PubMed] [Google Scholar]
- 42. Lee BN, Adams TH. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 1994;8(6):641–51. . [DOI] [PubMed] [Google Scholar]
- 43. Seo JA, Guan Y, Yu JH. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics. 2006;172(3):1535–44. Epub 2006/01/03. 10.1534/genetics.105.052258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics. 2012;190(2):511–21. Epub 2011/11/19. 10.1534/genetics.111.134080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Champe SP, Kurtz MB, Yager LN, Butnick NJ, Axelrod DE. Spore formation in Aspergillus nidulans: competence and other developmental processes. The fungal spore: morphogenetic controls. 1981:255–76. [Google Scholar]
- 46. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park HS, Ni M, Jeong KC, Kim YH, Yu JH. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans . PLoS One. 2012;7(9):e45935 Epub 2012/10/11. 10.1371/journal.pone.0045935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barron G. Emericella nidulans—cleistothecium with hulle cells. University of Guelph; 2013. [Google Scholar]
- 49. Geiser DM, Timberlake WE, Arnold ML. Loss of meiosis in Aspergillus . Mol Biol Evol. 1996;13(6):809–17. Epub 1996/07/01. . [DOI] [PubMed] [Google Scholar]
- 50. Gerke J, Bayram O, Feussner K, Landesfeind M, Shelest E, Feussner I, et al. Breaking the silence: protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans . Applied and environmental microbiology. 2012;78(23):8234–44. Epub 2012/09/25. 10.1128/AEM.01808-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rodriguez-Urra AB, Jimenez C, Nieto MI, Rodriguez J, Hayashi H, Ugalde U. Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans . ACS chemical biology. 2012;7(3):599–606. Epub 2012/01/12. 10.1021/cb200455u . [DOI] [PubMed] [Google Scholar]
- 52. Clutterbuck AJ. The genetics of conidiophore pigmentation in Aspergillus nidulans . Journal of general microbiology. 1990;136(9):1731–8. . [DOI] [PubMed] [Google Scholar]
- 53. Mayorga ME, Timberlake WE. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol Gen Genet. 1992;235(2–3):205–12. . [DOI] [PubMed] [Google Scholar]
- 54. Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5(7):462–4. 10.1038/nchembio.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kwon NJ, Garzia A, Espeso EA, Ugalde U, Yu JH. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans . Molecular microbiology. 2010;77(5):1203–19. Epub 2010/07/14. 10.1111/j.1365-2958.2010.07282.x . [DOI] [PubMed] [Google Scholar]
- 56. Krijgsheld P, Bleichrodt R, Van Veluw G, Wang F, Müller W, Dijksterhuis J, et al. Development in Aspergillus . Studies in mycology. 2013;74:1–29. 10.3114/sim0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han KH, Han KY, Yu JH, Chae KS, Jahng KY, Han DM. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans . Molecular microbiology. 2001;41(2):299–309. Epub 2001/08/08. . [DOI] [PubMed] [Google Scholar]
- 58. Vienken K, Fischer R. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans . Molecular microbiology. 2006;61(2):544–54. Epub 2006/06/20. . [DOI] [PubMed] [Google Scholar]
- 59. Andrianopoulos A, Timberlake WE. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol. 1994;14(4):2503–15. Epub 1994/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee MK, Kwon NJ, Choi JM, Lee IS, Jung S, Yu JH. NsdD is a key repressor of asexual development in Aspergillus nidulans . Genetics. 2014;197(1):159–73. 10.1534/genetics.114.161430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Axelrod DE. Kinetics of differentiation of conidiophores and conidia by colonies of Aspergillus nidulans . Journal of general microbiology. 1972;73(1):181–4. . [DOI] [PubMed] [Google Scholar]
- 62. Kunitomo H, Sugimoto A, Wilkinson CR, Yamamoto M. Schizosaccharomyces pombe pac2+ controls the onset of sexual development via a pathway independent of the cAMP cascade. Curr Genet. 1995;28(1):32–8. Epub 1995/06/01. . [DOI] [PubMed] [Google Scholar]
- 63. Vienken K, Scherer M, Fischer R. The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics. 2005;169(2):619–30. Epub 2004/11/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenberg-Dinkel N, et al. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Current biology: CB. 2005;15(20):1833–8. . [DOI] [PubMed] [Google Scholar]
- 65. Hoffmann B, Wanke C, Lapaglia SK, Braus GH. c-Jun and RACK1 homologues regulate a control point for sexual development in Aspergillus nidulans . Molecular microbiology. 2000;37(1):28–41. . [DOI] [PubMed] [Google Scholar]
- 66. Han KH, Seo JA, Yu JH. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans . Molecular microbiology. 2004;51(5):1333–45. 10.1111/j.1365-2958.2003.03940.x . [DOI] [PubMed] [Google Scholar]
- 67. Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP. Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans . Microbiology. 2005;151(Pt 6):1809–21. Epub 2005/06/09. . [DOI] [PubMed] [Google Scholar]
- 68. Ahmed YL, Gerke J, Park HS, Bayram O, Neumann P, Ni M, et al. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-kappaB. PLoS Biol. 2013;11(12):e1001750 Epub 2014/01/07. 10.1371/journal.pbio.1001750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mirzadi Gohari A, Mehrabi R, Robert O, Ince IA, Boeren S, Schuster M, et al. Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici . Mol Plant Pathol. 2014;15(4):394–405. Epub 2013/12/18. 10.1111/mpp.12102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae . Nature. 2005;438(7071):1105–15. Epub 2005/12/24. 10.1038/nature04341 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scattered plot showing the correlation levels among triplicates of each sample. The correlation coefficient R for ΔosaA veA1 was > 0.97, veA1 WT was > 0.97, and veA + WT was > 0.97, all with p-value less than 0.01, indicating the high quality of the RNA-Seq data sets.
(TIF)
FPKM obtained for ΔosaA veA1, veA1 WT, and veA + WT strains are mapped to 10,536, 10,428 and 10,514 genes, respectively, representing 96.3%, 95.3% and 96% coverage of a total of 10,943 genes predicted by AspGD.
(TIF)
A snapshot from Integrative Genomics Viewer (IGV) software showing the osaA locus (AN6578) in ΔosaA veA1, veA1 WT, and veA + WT strains.
(TIF)
Open box represents osaA locus; grey box represents ORF; arrow line represents transcript; open triangle in transcript ~1kb indicates alternative splicing.
(TIF)
The overall transcriptomic profiles of ΔosaA veA1, veA1 WT, and veA + WT strains examined by a two-dimensional plot. Black circles indicate replicates.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
RNA Seq data has been deposited in NCBI GEO database. Series no. GSE72316 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72316.