Abstract
Legionella organisms are environmental bacteria and accidental human pathogens that can cause severe pneumonia, termed Legionnaires' disease. These bacteria replicate within a pathogen-derived vacuole termed the Legionella-containing vacuole (LCV). Our understanding of the development and dynamics of this vacuole is based on extensive analysis of Legionella pneumophila. Here, we have characterized the Legionella longbeachae replicative vacuole (longbeachae-LCV) and demonstrated that, despite important genomic differences, key features of the replicative LCV are comparable to those of the LCV of L. pneumophila (pneumophila-LCV). We constructed a Dot/Icm-deficient strain by deleting dotB and demonstrated the inability of this mutant to replicate inside THP-1 cells. L. longbeachae does not enter THP-1 cells as efficiently as L. pneumophila, and this is reflected in the observation that translocation of BlaM-RalFLLO (where RalFLLO is the L. longbeachae homologue of RalF) into THP-1 cells by the L. longbeachae Dot/Icm system is less efficient than that by L. pneumophila. This difference is negated in A549 cells where L. longbeachae and L. pneumophila infect with similar entry dynamics. A β-lactamase assay was employed to demonstrate the translocation of a novel family of proteins, the Rab-like effector (Rle) proteins. Immunofluorescence analysis confirmed that these proteins enter the host cell during infection and display distinct subcellular localizations, with RleA and RleC present on the longbeachae-LCV. We observed that the host Rab GTPase, Rab1, and the v-SNARE Sec22b are also recruited to the longbeachae-LCV during the early stages of infection, coinciding with the LCV avoiding endocytic maturation. These studies further our understanding of the L. longbeachae replicative vacuole, highlighting phenotypic similarities to the vacuole of L. pneumophila as well as unique aspects of LCV biology.
INTRODUCTION
Legionella species are environmental bacteria that replicate within a unique vacuole in a range of protozoan hosts. For particular species of Legionella, this capacity for intracellular replication contributes to their pathogenic capacity. Upon human inhalation of contaminated aerosols, Legionella can enter the lungs and infect alveolar macrophages, leading to a severe pneumonia termed Legionnaires' disease. Legionella pneumophila and Legionella longbeachae are the primary causative agents of Legionnaires' disease. L. pneumophila is responsible for up to 90% of Legionnaires' disease in Europe and the United States, and L. longbeachae accounts for approximately 50% of cases in Australia and New Zealand (1, 2). Interestingly, in recent years there has been a global increase in the detection and incidence of infection caused by L. longbeachae (3, 4).
Despite causing clinically indistinguishable infections, L. longbeachae and L. pneumophila have distinct environmental niches. L. pneumophila is an aquatic organism, found in both natural and human-made aquatic environments, and L. longbeachae is predominantly found in soil environments, with most human infections associated with contaminated potting soil (5–7). These differences in environmental niches reflect important genetic and phenotypic differences between the two species. For example, L. longbeachae, but not L. pneumophila, has been observed to produce a capsule that may contribute to its environmental persistence (8).
L. pneumophila has a distinct biphasic life cycle in response to nutrient availability (9). Rapid replication and expression of few virulence traits occur under nutrient-rich conditions in what is termed the replicative phase. Once nutrients have been depleted, a complex regulatory system mediates a transition into the virulent, transmissive phase where the bacteria are highly motile and resistant to various stresses (10). In contrast, L. longbeachae is nonmotile, lacking flagellar biosynthesis genes. Furthermore, even though L. longbeachae possesses homologues of the regulatory genes that mediate the L. pneumophila biphasic life cycle, transcriptomic analysis demonstrated a significantly less pronounced life cycle switch between exponential and postexponential L. longbeachae bacteria (8). This is reflected in the observation that the ability of L. longbeachae to infect and replicate within host cells is independent of bacterial growth phase (11).
L. pneumophila replicates in free-living protozoa and mammalian macrophages within a unique vacuolar compartment termed the Legionella-containing vacuole (LCV). Upon entry into a host cell L. pneumophila rapidly remodels the LCV (forming the pneumophila-LCV) to evade endocytic maturation and subsequent degradation by the proteolytic lysosome. Instead, the bacterium directs the recruitment of secretory vesicles traveling between the endoplasmic reticulum (ER) and Golgi compartment to disguise the LCV as an ER-like organelle (reviewed in references 12 and 13). One hallmark of the LCV is the rapid recruitment of Rab1, the Rab GTPase that regulates ER-Golgi compartment vesicular trafficking (reviewed in reference 14). Rab1 plays an important role in the LCV, mediating the tethering and fusion of ER-derived vesicles by allowing the noncanonical pairing of the ER v-SNARE Sec22b and plasma membrane syntaxins (15). Characteristics of the LCV of L. longbeachae (longbeachae-LCV) have not been extensively studied. There is evidence to suggest that the longbeachae-LCV is distinct from the pneumophila-LCV (11). However, recent work has demonstrated that the replicating longbeachae-LCV is decorated with the ER protein calnexin (16). This indicates that these pathogens may indeed develop similar replicative niches inside eukaryotic cells, albeit through different mechanisms.
The Dot/Icm type IV secretion system (T4SS) is essential for intracellular replication of both L. pneumophila and L. longbeachae, as well as of the closely related Coxiella burnetii, and has been extensively examined in the context of L. pneumophila virulence (13, 17). This multiprotein apparatus delivers bacterial proteins, termed effectors, across both bacterial and vacuole membranes into the host cytosol. Collectively, these effectors act to remodel the LCV into a safe environment conducive to replication. Extensive research has identified over 300 L. pneumophila proteins as substrates of the Dot/Icm system. Genome sequencing has revealed that this effector repertoire is quite variable, with approximately 77% of these effectors present in all sequenced L. pneumophila genomes. This conservation is much lower between different Legionella species, and L. longbeachae possesses only 35% of the L. pneumophila effector cohort (8). Interestingly, this conservation is high compared to conservation between L. pneumophila and other non-pneumophila species. Indeed, a recent comprehensive comparative analysis demonstrated that, among a wide range of Legionella species, there is a core subset of only 24 Dot/Icm effectors (18).
Functional studies have revealed roles for some L. pneumophila Dot/Icm effectors although understanding the importance of individual effector functions is made particularly difficult by a high degree of functional redundancy. Many effectors work in concert to evade phagosome maturation, promote fusion of the LCV with the ER, and modulate host processes such as ubiquitination and transcription. Perhaps most well studied are the multiple effectors employed by L. pneumophila to manipulate all aspects of Rab1 dynamics (reviewed in reference 14). The effector DrrA (SidM) acts as a guanine nucleotide exchange factor (GEF) that facilitates recruitment of Rab1 to the pneumophila-LCV (19, 20). Conversely, L. pneumophila also encodes a GTPase-activating effector, LepB, that deactivates Rab1 (21). Furthermore, several L. pneumophila effectors, DrrA, AnkX, SidD, and Lem3, posttranslationally modify Rab1, impacting its localization and activity (22–27). Interestingly, all of these Rab1-modifying effectors, with the exception of LepB, are absent in L. longbeachae (8). This raises the question of whether L. longbeachae subverts Rab1 to mediate interaction with ER-derived vesicles or uses alternate means to establish an ER-like replicative vacuole.
Here, we constructed a dotB deletion strain of L. longbeachae NSW150 to allow us to probe the Dot/Icm-dependent characteristics of the longbeachae-LCV, including the recruitment of Rab1 and Sec22b. In addition, we have investigated the activity of the L. longbeachae Dot/Icm system in comparison to that of L. pneumophila and demonstrated the translocation of a unique family of Rab GTPase-like effectors that may be important for longbeachae-LCV development.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. L. pneumophila 130b and L. longbeachae NSW150 strains were grown overnight in ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract (AYE) broth at 37°C or on buffered charcoal yeast extract (BCYE) agar plates for 2 or 3 days (28). Escherichia coli strains were grown in Luria-Bertani (LB) medium or on LB agar plates. Antibiotics were added to the media when appropriate at the following concentrations for Legionella spp. (or E. coli): ampicillin, 25 μg/ml (100 μg/ml); kanamycin, 25 μg/ml (100 μg/ml); and chloramphenicol, 6 μg/ml (25 μg/ml). To induce protein expression, the medium was supplemented with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source (restriction sites) |
|---|---|---|
| L. pneumophila strains | ||
| 130b | Clinical isolate | 40 |
| ΔdotA strain | 130b ΔdotA | 41 |
| 130b(pXDC61) | 130b expressing BlaM from pXDC61 | 41 |
| 130b(pXDC61-rleA) | 130b expressing BlaM-RleA fusion protein | This study |
| 130b(pXDC61-rleB) | 130b expressing BlaM-RleB fusion protein | This study |
| 130b(pXDC61-rleC) | 130b expressing BlaM-RleC fusion protein | This study |
| 130b(pXDC61-rleD) | 130b expressing BlaM-RleD fusion protein | This study |
| 130b(pXDC61-ralFLLO)a | 130b expressing BlaM-RalF from L. longbeachae | This study |
| ΔdotA(pXDC61) strain | ΔdotA strain expressing BlaM from pXDC61 | 41 |
| ΔdotA(pXDC61-rleA) strain | ΔdotA strain expressing BlaM-RleA fusion protein | This study |
| ΔdotA(pXDC61-rleB) strain | ΔdotA strain expressing BlaM-RleB fusion protein | This study |
| ΔdotA(pXDC61-rleC) strain | ΔdotA strain expressing BlaM-RleC fusion protein | This study |
| ΔdotA(pXDC61-rleD) strain | ΔdotA strain expressing BlaM-RleD fusion protein | This study |
| ΔdotA(pXDC61-ralFLLO) strain | ΔdotA strain expressing BlaM-RalF from L. longbeachae | This study |
| L. longbeachae strains | ||
| NSW150 | Clinical isolate | 8 |
| ΔdotB strain | NSW150 with the entire dotB gene deleted | This study |
| NSW150(pXDC61) | NSW150 expressing BlaM from pXDC61 | This study |
| NSW150(pXDC61-rleA) | NSW150 expressing BlaM-RleA fusion protein | This study |
| NSW150(pXDC61-rleB) | NSW150 expressing BlaM-RleB fusion protein | This study |
| NSW150(pXDC61-rleC) | NSW150 expressing BlaM-RleC fusion protein | This study |
| NSW150(pXDC61-rleD) | NSW150 expressing BlaM-RleD fusion protein | This study |
| NSW150(pXDC61-ralFLLO) | NSW150 expressing BlaM-RalF from L. longbeachae | This study |
| ΔdotB(pXDC61) strain | ΔdotB strain expressing BlaM from pXDC61 | This study |
| ΔdotB(pXDC61-rleA) strain | ΔdotB strain expressing BlaM-RleA fusion protein | This study |
| ΔdotB(pXDC61-rleB) strain | ΔdotB strain expressing BlaM-RleB fusion protein | This study |
| ΔdotB(pXDC61-rleC) strain | ΔdotB strain expressing BlaM-RleC fusion protein | This study |
| ΔdotB(pXDC61-rleD) strain | ΔdotB strain expressing BlaM-RleD fusion protein | This study |
| ΔdotB(pXDC61-ralFLLO) strain | ΔdotB strain expressing BlaM-RalF from L. longbeachae | This study |
| NSW150(pMMB207-4×HA-rleA) | NSW150 expressing HA-RleA | This study |
| NSW150(pMMB207-4×HA-rleB) | NSW150 expressing HA-RleB | This study |
| NSW150(pMMB207-4×HA-rleC) | NSW150 expressing HA-RleC | This study |
| NSW150(pMMB207-4×HA-rleD) | NSW150 expressing HA-RleD | This study |
| ΔdotB(pMMB207-4×HA-rleA) strain | ΔdotB strain expressing HA-RleA | This study |
| ΔdotB(pMMB207-4×HA-rleB) strain | ΔdotB strain expressing HA-RleB | This study |
| ΔdotB(pMMB207-4×HA-rleC) strain | ΔdotB strain expressing HA-RleC | This study |
| ΔdotB(pMMB207-4×HA-rleD) strain | ΔdotB strain expressing HA-RleD | This study |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB laclq ZΔM15 Tn10 (Tetr)] | Agilent Technologies |
| PIR2 | F− Δlac169 rpoS(Am) robA1 creC510 hsdR514 endA recA1 uidA(ΔMlul)::pir | Life Technologies |
| Plasmids | ||
| pGEM-T-Easy | Cloning vector; Ampr | Promega |
| pGEM-A | pGEM-T-Easy carrying 2-kb 5′ flanking segment of dotBLLOb | This study (BamHI, PstI) |
| pGEM-AB | pGEM-T-Easy carrying 2-kb 5′ and 3′ flanking segments of dotBLLO | This study (BamHI, SacI) |
| pSR47s | Cloning vector; Kanr | 42 |
| pSR47s-AB | pSR47s carrying 2-kb 5′ and 3′ flanking segments of dotBLLO | This study (BamHI, SacI) |
| pEGFP-Rab1 | Human Rab1 fused to GFP | Stow Laboratory |
| pEGFP-Sec22b | Human Sec22b fused to GFP | Stow Laboratory |
| pXDC61-rleA | Encodes IPTG-inducible expression of RleA with N-terminal BlaM fusion; Cmr | This study |
| pXDC61-rleB | Encodes IPTG-inducible expression of RleB with N-terminal BlaM fusion; Cmr | This study |
| pXDC61-rleC | Encodes IPTG-inducible expression of RleC with N-terminal BlaM fusion; Cmr | This study |
| pXDC61-rleD | Encodes IPTG-inducible expression of RleD with N-terminal BlaM fusion; Cmr | This study |
| pXDC61-ralFLLO | Encodes IPTG-inducible expression of L. longbeachae RalF with N-terminal BlaM fusion; Cmr | This study |
| pMMB207-4×HA-rleA | Encodes IPTG-inducible expression of RleA with N-terminal 4×HA tag; Cmr | This study |
| pMMB207-4×HA-rleB | Encodes IPTG-inducible expression of RleB with N-terminal 4×HA tag; Cmr | This study |
| pMMB207-4×HA-rleC | Encodes IPTG-inducible expression of RleC with N-terminal 4×HA tag; Cmr | This study |
| pMMB207-4×HA-rleD | Encodes IPTG-inducible expression of RleD with N-terminal 4×HA tag; Cmr | This study |
ralFLLO, ralF gene of L. longbeachae.
dotBLLO, dotB gene of L. longbeachae.
Molecular cloning.
Oligonucleotides used to amplify specific gene products and/or confirm mutations are listed in Table 2 with the restriction enzyme sites used for cloning. Plasmid DNA was purified using a QIAprep Spin Miniprep kit (Qiagen) or an Axyprep Plasmid MaxiPrep kit (Axygen). DNA-modifying enzymes were used according to the manufacturers' instructions (Promega and New England BioLabs).
TABLE 2.
Oligonucleotides used in this study
| Primera | Sequence (5′–3′)b | Description (restriction enzyme) |
|---|---|---|
| dotB-UPF | AAAGGATCCGACCAGTAAGGATGGTACGC | 5′ 2 kb upstream of dotBLLO (BamHI)d |
| dotB-UPR | AAACTGCAGATTCCCTCTTCTTTTCATTTT | 3′ 2 kb upstream of dotBLLO (PstI) |
| dotB-DOWNF | AAACTGCAGATGTTTTCATTAATGAATGT | 5′ 2 kb downstream of dotBLLO (PstI) |
| dotB-DOWNR | AAAGAGCTCGCACGATATTCGAGAAGCGC | 3′ 2 kb downstream of dotBLLO (SacI) |
| dotB-F | GGGAATCTATTGTTGGTATTCT | 5′ to amplify dotBLLO |
| dotB-R | CAATATTCATTTAATGCCTGATG | 3′ to amplify dotBLLO |
| dotB-UP50F | CACACCCTCAGATCCACAAA | 5′ 50 bp upstream of the 2-kb 5′ flanking segment of dotBLLO |
| dotB-DOWN50R | AGAAGGATTAAGACCCACGC | 3′ 50 bp downstream of the 2-kb 3′ flanking segment of dotBLLO |
| rleA-Fc | AAGGTACCATGGCAAAATTTAAAATAGTGCTT | 5′ to amplify rleA (KpnI) |
| rleA-Rc | AAGGATCCTTATAAACGGATTTCACGATTCC | 3′ to amplify rleA (BamHI) |
| rleB-Fc | AAGGTACCATGAAATTTTTTAAATTAGAAAATAAAG | 5′ to amplify rleB (KpnI) |
| rleB-Rc | AAGGATCCTTAATTTTTTGATGAAGTTTTTAAATT | 3′ to amplify rleB (BamHI) |
| rleC-Fc | AAGGTACCATGCTTATAAAATATTTAATTTCATC | 5′ to amplify rleC (KpnI) |
| rleC-Rc | AAGGATCCTTAATGATTTTTTGACCCGTTAC | 3′ to amplify rleC (BamHI) |
| rleD-Fc | AAGGTACCATGGAGCACAAAGAAGAATATG | 5′ to amplify rleD (KpnI) |
| rleD-Rc | AAGGATCCTTACCGTGTGGTTTTACAAAAAA | 5′ to amplify rleD (BamHI) |
| ralF-Fc | AAGGTACCATGATTAATCCAGAATTAGAAAAAG | 5′ to amplify ralFLLO (KpnI) |
| ralF-Rc | AAGGATCCCTATTTCAACCCATTAGATTTCA | 3′ to amplify ralFLLO (BamHI) |
F and R at the end of the primer designations indicate forward and reverse, respectively.
Restriction enzyme sites are underlined.
Oligonucleotides used for cloning into both pXDC61 and pMMB207.
dotBLLO, dotB gene of L. longbeachae.
Introduction of plasmids into Legionella.
L. longbeachae was grown on BCYE agar plates for 3 days at 37°C and then replated onto BCYE agar and grown at 37°C a further 2 days. Bacterial growth after 2 days was resuspended in ice-cold 20 mM MgCl with 2% sucrose. The cells were washed twice with ice-cold 20 mM MgCl with 2% sucrose and once with ice-cold 2% sucrose, and the final pellet was resuspended in ice-cold 2% sucrose. L. pneumophila was grown on BCYE agar plates for 3 days, and then stationary-phase bacteria were resuspended in ice-cold distilled H2O (dH2O) and adjusted to an optical density at 600 nm (OD600) of 1. The cells were washed twice with ice-cold dH2O and once with ice-cold 10% glycerol, and the final pellet was resuspended in ice-cold 10% glycerol.
For transformation, a 50-μl aliquot of electrocompetent cells was freshly mixed with 3 μg (L. longbeachae) or 0.2 μg (L. pneumophila) of plasmid DNA and electroporated at 2.3 kV, 200 Ω, and 0.25 μF (L. longbeachae) or at 2.4 kV, 200 Ω, and 0.25 μF (L. pneumophila). The cultures were recovered in AYE broth at 37°C with shaking for 5 h and then plated onto BCYE agar with the appropriate antibiotic.
Construction of L. longbeachae ΔdotB.
The 2-kb flanking regions of L. longbeachae dotB were amplified using the oligonucleotide pair dotB-UPF and dotB-UPR (upstream segment, A) and the pair dotB-DOWNF and dotB-DOWNR (downstream segment, B) using genomic DNA of L. longbeachae NSW150 as a template. Segment A was ligated into pGEM-T-Easy (Promega) to generate pGEM-A. Segment B was ligated into pGEM-A via PstI and SacI sites to generate pGEM-AB. This plasmid was digested with the enzymes BamHI and SacI, and the AB combined segment was ligated into pSR47s. The resulting plasmid, pSR47s-AB, was introduced into L. longbeachae NSW150 as described above, and transformants were plated onto BCYE agar with kanamycin. Resulting colonies were serially diluted in phosphate-buffered saline (PBS) and plated onto BCYE agar supplemented with 5% sucrose. Sucrose-sensitive clones were screened, and successful deletion was verified through PCR using the oligonucleotide pair dotB-F and dotB-R and the pair dotB-UP50F and dotB-DOWN50R.
Western blot analysis.
For protein expression analysis, overnight cultures were prepared to an OD600 of 1.5 and resuspended in 4× NuPage LDS Sample Buffer (Life Technologies) with dH2O. Dithiothreitol was added at 1:20 relative to the sample buffer. Samples were separated using NuPage Bis-Tris precast gels (Life Technologies), and then proteins were transferred to a polyvinylidene difluoride (PVDF) membrane using an iBLOT 2 Gel Transfer Device (Life Technologies). Mouse monoclonal anti-BlaM (QED Bioscience, Inc.), diluted 1:1,500, and mouse monoclonal anti-HA.11 (where HA is hemagglutinin) (Covance), diluted 1:1,000, were used as primary antibodies. Goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody (1:3,000) was used as the secondary antibody. Proteins were detected with Amersham ECL Western blotting detection reagent (GE Healthcare). The HRP signal was visualized using MF-ChemiBIS, version 3.2 (DNR Bio-Imaging Systems, Ltd.), and images were acquired using GelCapture, version 7.0.18 (DNR Bio-Imaging Systems, Ltd.).
Tissue culture conditions and intracellular replication assay.
The human monocytic cell line THP-1 was maintained in RPMI 1640 medium supplemented with GlutaMAX (Gibco) and 10% fetal calf serum (FCS) at 37°C with 5% CO2. Human alveolar epithelial A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with GlutaMAX (Gibco) and 10% FCS and incubated under the same conditions. THP-1 cells were seeded at a density of 5 × 105 cells/well in 24-well tissue culture trays (Corning) and differentiated into adherent macrophage-like cells with 10−8 M phorbol-12 myristate 13-acetate (PMA) for 48 to 72 h. A549 cells were seeded at a density of 2.5 × 105 cells/well and incubated for 24 h prior to infection. Stationary-phase Legionella bacteria were added at a multiplicity of infection (MOI) of 10 (THP-1) or 100 (A549) and incubated for 2 h. Cells were treated with 100 μg/ml gentamicin for 1 h to kill extracellular bacteria, washed three times with PBS, and then incubated with tissue culture maintenance medium. At specified time points postinfection, cells were lysed with 0.05% digitonin, and serial dilutions were plated onto BCYE agar with antibiotics where appropriate. To assess bacterial uptake and calculate the 3-h starting time point, THP-1 or A549 lysis and recovery of bacteria were performed directly following gentamicin treatment and PBS washes.
Translocation assay.
THP-1 cells were seeded at a density of 8 × 104 cells/well into clear-bottom 96-well trays (Corning) and differentiated as described above, and A549 cells were seeded at a density of 4 × 104 cells/well. Stationary-phase Legionella bacteria were added at an MOI as indicated, and the trays were centrifuged to synchronize the infection. At the specified time points, the wells were washed with Hanks' balanced salt solution (HBSS) with 5% HEPES and then incubated with the loading solution, containing CCF2-AM (Life Technologies) and 0.1 M probenecid, in the dark for 1 h and 45 min. Cells were then treated with HBSS–5% HEPES and 2.5 mM probenecid and read in a CLARIOstar Omega microplate reader. To prevent bacterial internalization, cells were treated with cytochalasin D (20 μM) for 30 min prior to infection.
Immunofluorescence microscopy.
HEK 293T cells expressing Fcγ receptor (HEK 293T FcγR cells) were maintained in DMEM (Gibco) supplemented with 10% FCS at 37°C and 5% CO2. HEK 293T FcγR cells were seeded at a density of 8 × 104 cells/well onto poly-l-lysine-treated coverslips in 24-well tissue culture trays 48 h prior to infection. Cells were transfected using FuGENE6 (Promega) 24 h later with pEGFP-Sec22b or pEGFP-Rab1 (carrying green fluorescent protein [GFP]), and AYE cultures were prepared 18 h prior to infection. On the day of the experiment, the bacteria were diluted to a concentration of 108 CFU/ml and coated with rabbit polyclonal anti-L. pneumophila (Meredian Life Science) or rabbit polyclonal anti-L. longbeachae (WEHI Antibody Services) at 37°C in 5% CO2 for 20 min prior to opsonization at an MOI of 1. For strains expressing the pMMB207-4×HA (carrying four copies of an HA tag) derivatives, 1 mM IPTG was added to the medium. Trays were centrifuged to synchronize the infection and incubated at 37°C and 5% CO2.
For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde for 20 min and then permeabilized with 0.5% saponin or with 0.1% Triton X-100, followed by treatment with 3% bovine serum albumin (BSA) in PBS. Cells were then treated with anti-L. pneumophila (1:1,000), anti-L. longbeachae (1:1,000), mouse monoclonal anti-lysosomal-associated membrane protein 1 (LAMP-1; 1:200 [Developmental Studies Hybridoma Bank]), or anti-HA.11 (1:50; Covance). Alexa Fluor-488-conjugated goat anti-mouse and Alexa Fluor-568-conjugated goat anti-rabbit (Life Technologies) secondary antibodies were used at a 1:2,000 dilution, followed by treatment with 4′,6′-diamidino-2-phenylindole (DAPI) at a 1:10,000 dilution. Images were acquired with a Zeiss LSM700 confocal laser scanning microscope and processed using ImageJ. Quantification of the association of specific host proteins with the LCV was performed using a Leica DMI4000b inverted microscope. At least 100 LCVs across two coverslips for each strain, host protein, and time point were blindly quantified.
RESULTS
Construction and characterization of a L. longbeachae dotB deletion strain.
In order to examine the Dot/Icm-dependent aspects of the longbeachae-LCV development, we constructed a deletion mutant of dotB, the gene encoding the ATPase that drives effector protein translocation (29). A chromosomal deletion of dotB was achieved using allelic exchange and confirmed via PCR. The ability of this mutant to replicate in the human monocytic cell line THP-1 was examined and compared to the abilities of the wild-type L. longbeachae NSW150, L. pneumophila 130b, and L. pneumophila 130b ΔdotA (Fig. 1A). As expected, similar to Dot/Icm-deficient L. pneumophila, L. longbeachae ΔdotB cannot replicate inside eukaryotic cells, confirming previous reports that the Dot/Icm secretion system is essential for intracellular replication of L. longbeachae (8). The wild-type L. longbeachae and L. pneumophila strains demonstrate comparable capacities for intracellular replication. However, we observed a significantly reduced ability of L. longbeachae to enter THP-1 cells (Fig. 1B). For intracellular growth curves, PMA-differentiated THP-1 cells were infected at an MOI of 10 for 2 h, followed by a 1-h gentamicin treatment to remove remaining extracellular bacteria. Bacterial numbers recovered at this 3-h postinfection point were significantly different (P = 0.0230) between L. pneumophila, with 0.295% ± 0.205% of the inoculum recovered, and L. longbeachae, with 0.070% ± 0.038% of the inoculum recovered (Fig. 1B). There was no difference in the proportions of the inoculum that were recovered when gentamicin was not used, indicating that the observed difference represents a diminished capacity for L. longbeachae to enter THP-1 cells.
FIG 1.
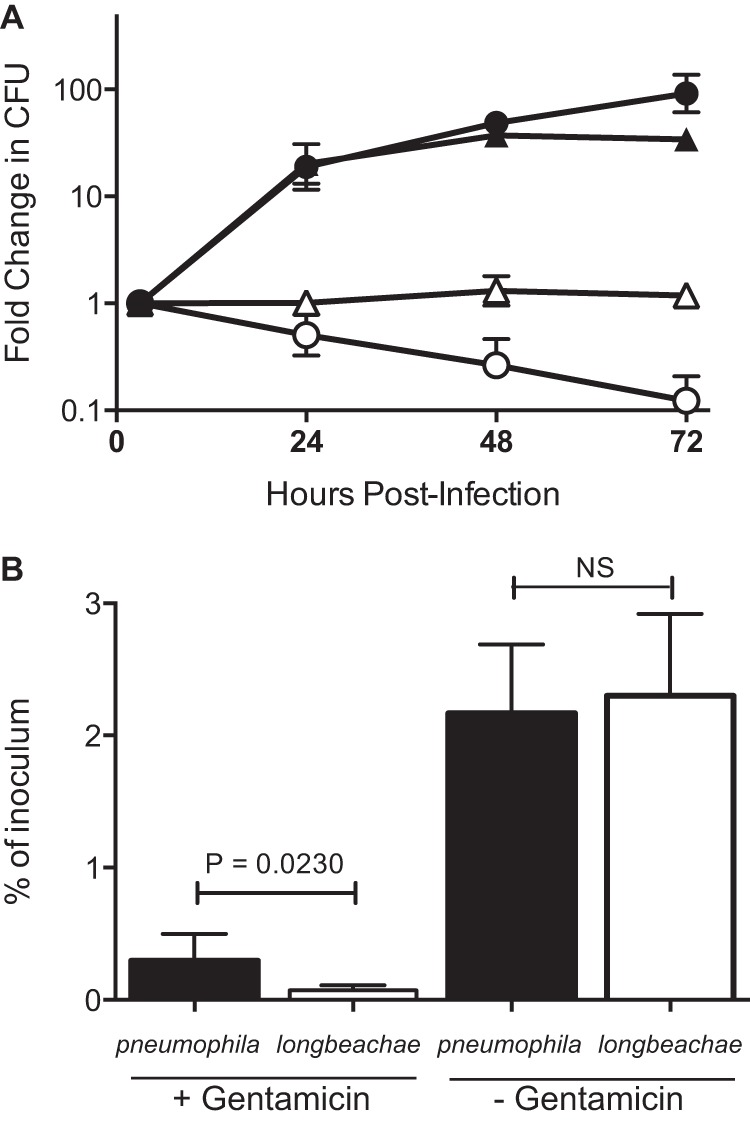
DotB is required for intracellular replication of L. longbeachae. (A) Replication of L. pneumophila 130b (filled triangles), L. pneumophila ΔdotA (open triangles), L. longbeachae NSW150 (filled circles) and L. longbeachae ΔdotB (open circles) in THP-1 macrophages. Following a 2-h infection and 1-h gentamicin treatment, the infection was allowed to proceed for 24, 48, or 72 h before cells were lysed and bacteria were enumerated. Results are the means ± standard deviations of the fold changes in CFU counts relative to the 3-h time point of at least three independent experiments. (B) Following gentamicin treatment, significantly more L. pneumophila bacteria (filled bars), as a percentage of the inoculum, were recovered than L. longbeachae bacteria (open bars). Without the gentamicin treatment, this difference was lost, and comparable proportions of the inoculum were recovered (P value calculated using an unpaired two-tailed t test; NS, not significant).
Dot/Icm-dependent effector translocation by L. longbeachae.
The Dot/Icm secretion system is essential for intracellular replication of L. longbeachae, presumably due to the necessary functions performed by effectors translocated into the host cell. We employed a β-lactamase reporter assay to examine Dot/Icm effector translocation by L. longbeachae. We investigated the translocation of the L. longbeachae homologue of RalF (RalFLLO; locus tag LLO1397). In L. pneumophila, RalF acts as a GEF to activate the small GTPase ADP-ribosylation factor (Arf) and recruit this host protein to the LCV (30). RalFLLO shares only 50% identity with L. pneumophila RalF; however, the glutamic finger E103 residue (E104 in RalFLLO) required for RalF function remains conserved (31). During infection of THP-1 cells BlaM-RalFLLO is translocated by the Dot/Icm systems of both L. longbeachae and L. pneumophila (Fig. 2).
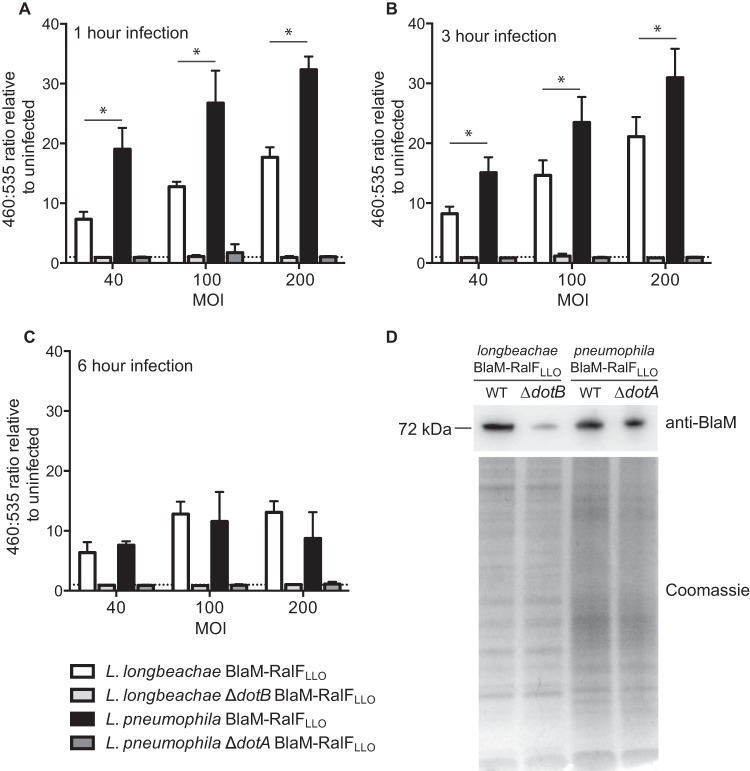
FIG 2.
Dot/Icm translocation into THP-1 cells by L. longbeachae is significantly less efficient than that by L. pneumophila. L. longbeachae NSW150 (white bars), L. longbeachae ΔdotB (light gray bars), L. pneumophila 130b (black bars), and L. pneumophila ΔdotA (dark gray bars) expressing BlaM-RalFLLO were used to infect THP-1 cells at an MOI of 40, 100, or 200. At 1 h (A), 3 h (B), and 6 h (C) postinfection, the BlaM substrate CCF2-AM was added. Translocation of the BlaM-RalFLLO fusion protein was determined by measuring the change in the 460 nm/535 nm fluorescence emission ratio when cells were excited at 415 nm. The mean ratio relative to the ratio in uninfected cells is presented ± standard deviation of at least three independent experiments. (D) Expression of the 72-kDa fusion protein BlaM-RalFLLO was confirmed by probing equivalent amounts of Legionella cell lysates, demonstrated by the visualization of protein loading on the Coomassie-stained gel, with anti-BlaM antibody. The dotted line represents a fluorescence ratio of 1, a value equivalent to that of an uninfected sample. *, P < 0.05, for the difference between L. longbeachae and L. pneumophila translocation. WT, wild type.
Interestingly, we observed that the translocation of BlaM-RalFLLO into THP-1 cells is consistently and significantly lower in an L. longbeachae system than the translocation efficiency in an L. pneumophila system. This is particularly evident at 1 h postinfection, the time point at which L. pneumophila Dot/Icm-dependent translocation appears to peak (Fig. 2A). The largest amount of L. longbeachae BlaM-RalFLLO translocation was observed at 3 h postinfection (Fig. 2B). At 6 h postinfection the amount of translocation of BlaM-RalFLLO that can be measured is reduced for both species (Fig. 2C), and the difference in translocation efficiencies is lost. This likely reflects THP-1 cell death that is observed, particularly at the higher MOI.
The reduced translocation of BlaM-RalFLLO into THP-1 cells by L. longbeachaecompared to that by L. pneumophila is not due to a difference in the expression of BlaM-RalFLLO as Fig. 2D demonstrates that the wild-type L. longbeachae and L. pneumophila strains express comparable levels of BlaM-RalFLLO. Instead, this difference could potentially arise from the reduced entry of L. longbeachae into THP-1 cells that was observed in the experiment shown in Fig. 1B. In order to address this, we examined whether Dot/Icm-dependent protein translocation by L. longbeachae required entry into host cells. Previously, it was reported that blocking L. pneumophila entry, by treating host cells with cytochalasin D, does not perturb the ability of the pathogen to translocate effectors (32). Translocation assays using BlaM-RalFLLO as a reporter demonstrated that Dot/Icm-dependent translocation by L. longbeachae also does not require bacteria to be internalized (Fig. 3). A similar trend for less translocation was observed for both Legionella species when the THP-1 cells were treated with 20 μM cytochalasin D. However, even in the absence of bacterial internalization, L. longbeachae maintains significantly less reporter translocation than L. pneumophila (P = 0.031).
FIG 3.
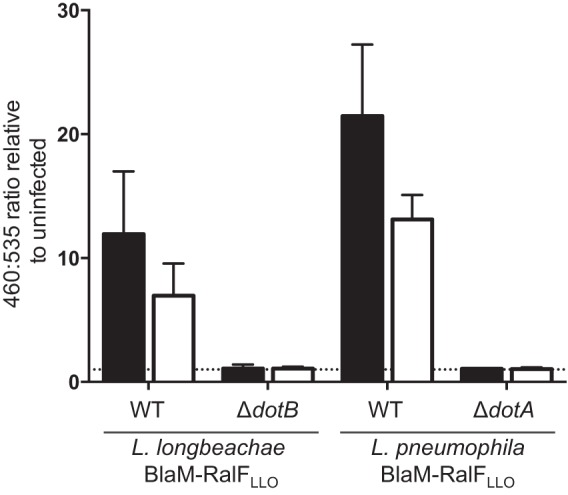
L. longbeachae Dot/Icm-dependent protein translocation occurs in the absence of host cell uptake. L. longbeachae and L. pneumophila strains expressing BlaM-RalFLLO were used to infect THP-1 cells that were treated with 20 μM cytochalasin D (white bars) or the equivalent volume of the carrier dimethyl sulfoxide (black bars) at an MOI of 40 for 1 h before the BlaM substrate CCF2-AM was added and translocation was measured using a fluorescence plate reader. Cells were excited at 415 nm, and the 460 nm/535 nm emission ratio was calculated relative to the ratio in uninfected cells. The mean ratio ± standard deviation is presented from at least three independent experiments. There is a statistically significant difference between the 460 nm/535 nm ratio of cells infected with L. longbeachae wild type (WT) and that of L. pneumophila wild type with cytochalasin D treatment (unpaired, two-tailed t test; P = 0.031). The dotted line represents a fluorescence ratio of 1, a value equivalent to that of an uninfected sample.
Entry and Dot/Icm effector translocation in alveolar epithelial cells.
In order to determine whether L. longbeachae is less efficient at translocating BlaM-RalFLLO or whether these results reflect the reduced capacity for L. longbeachae to enter host cells, we examined the efficiency of infection in the A549 alveolar epithelial cell line. A549 cells were incubated with L. longbeachae NSW150 or L. pneumophila 130b at an MOI of 100 for 2 h, followed by a further 1 h of incubation, either with or without gentamicin treatment, before cells were lysed and bacteria were enumerated through serial dilution plating on BCYE agar (Fig. 4A). This approach demonstrated that equivalent proportions of L. longbeachae and L. pneumophila are internalized by these nonphagocytic cells (P = 0.499) (Fig. 4A, gentamicin treatment) and, interestingly, that significantly more L. longbeachae bacteria are attached to A549 cells than L. pneumophila bacteria after a 3-h infection (P = 0.017) (Fig. 4A). The finding that L. longbeachae and L. pneumophila comparably infect A549 cells provided an opportunity to determine whether the reduced L. longbeachae effector translocation is due to decreased entry into host cells or is a universal trait of L. longbeachae. The translocation of BlaM-RalFLLO was compared during infection of A549 cells by L. longbeachae and L. pneumophila at 1 h, 3 h, and 6 h postinfection (Fig. 4B). Strikingly, no difference was observed between the translocation efficiencies of L. pneumophila and L. longbeachae at 1 h (P = 0.331) or 3 h (P = 0.238). At 6 h postinfection the 460/535-nm ratio was higher for L. longbeachae-infected cells (P = 0.044); however, this reflects the significant host cell death that occurs during L. pneumophila infection. These A549 infections highlight that the L. longbeachae Dot/Icm secretion system is as efficient as the L. pneumophila Dot/Icm secretion system when the bacteria are internalized by a host cell.
FIG 4.
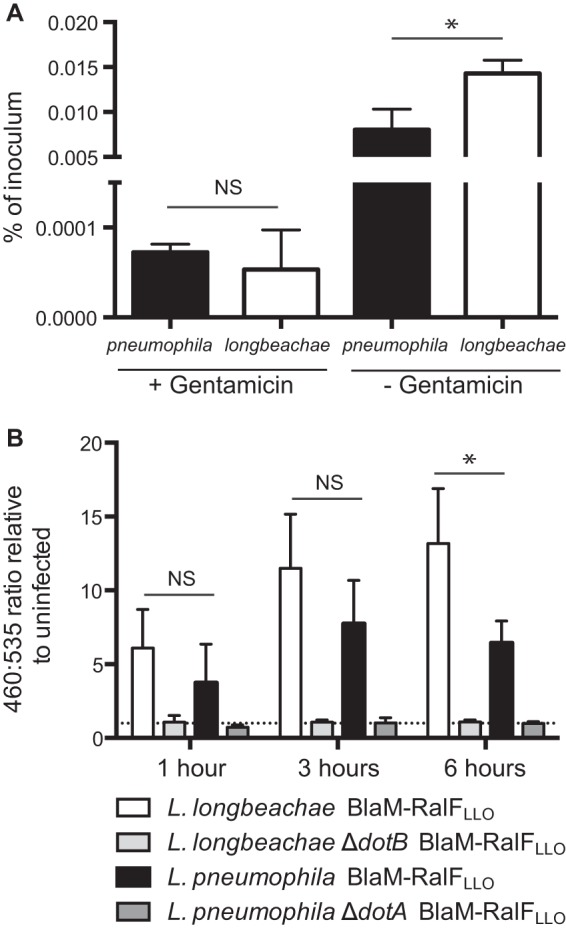
L. longbeachae and L. pneumophila behave similarly in nonphagocytic cells. (A) A549 cells were infected with L. pneumophila (black bars) and L. longbeachae (white bars) at an MOI of 100 for 2 h, followed by a further 1 h either with or without gentamicin before cells were lysed and bacteria were enumerated. Following gentamicin treatment, equivalent numbers of bacteria, as a percentage of the inoculum, were recovered (NS, not significant; P = 0.499, calculated using an unpaired two-tailed t test). Without the gentamicin treatment significantly more L. longbeachae bacteria were recovered than L. pneumophila bacteria (*, P = 0.016), indicating that more L. longbeachae bacteria attach to the A549 cells. (B) Dot/Icm effector translocation during A549 infection was examined at 1 h, 3 h, and 6 h postinfection. L. longbeachae NSW150 (white bars), L. longbeachae ΔdotB (light gray bars), L. pneumophila 130b (black bars), and L. pneumophila ΔdotA (dark gray bars) expressing BlaM-RalFLLO were used to infect A549 cells at an MOI of 200. At appropriate times postinfection, the BlaM substrate CCF2-AM was added. Translocation of the BlaM-RalFLLO fusion protein was determined by measuring the change in the 460 nm/535 nm fluorescence emission ratio when cells were excited at 415 nm. The mean ratio relative to the ratio in uninfected cells is presented ± standard deviation of at least three independent experiments. At 1 h and 3 h postinfection there is no significant difference between the wild-type L. longbeachae and L. pneumophila infections (NS); however, at 6 h postinfection more translocation by L. longbeachae is observed (*, P = 0.044).
Rab-like proteins are translocated by the Dot/Icm T4SS.
We employed our newly developed ability to examine Dot/Icm effector translocation by L. longbeachae to investigate whether four proteins with homology to eukaryotic Rab GTPases are Dot/Icm effectors. Both wild-type L. longbeachae and the ΔdotB derivative strain were transformed with plasmids to express each of the Rab-like proteins with an N-terminal BlaM fusion (Fig. 5A). These strains were used to infect THP-1 cells and examine translocation (Fig. 5C). As the highest translocation for BlaM-RalFLLO into THP-1 cells was observed at 3 h postinfection with an MOI of 200, these conditions were used to examine translocation of the Rab-like proteins. Under these assay conditions, even though considerable translocation of BlaM-RalFLLO was observed, only BlaM-2249 and BlaM-3288 demonstrated detectable translocation in the wild-type strain in comparison to that in the L. longbeachae ΔdotB mutant (P = 0.0482 and P = 0.0016, respectively). This result may reflect that BlaM-1716 and BlaM-2329 were less efficiently expressed in L. longbeachae.
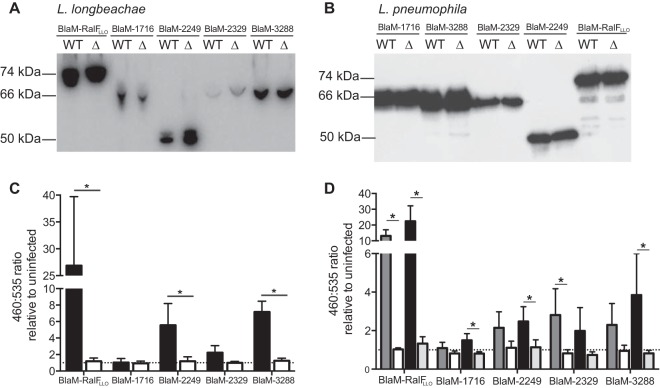
FIG 5.
Unique Rab-like L. longbeachae proteins are novel substrates of the Dot/Icm system. (A) Wild-type (WT) L. longbeachae and the ΔdotB derivative strain (Δ) were transformed with pXDC61 constructs to express BlaM fusion proteins. Expression of the BlaM fusion proteins was confirmed by an anti-BlaM Western blotting of equivalent amounts of bacterial cell lysate. (B) Similarly, wild-type (WT) L. pneumophila and the ΔdotA derivative strains (Δ) were transformed with the same plasmids, and expression was confirmed by Western blotting. For both L. longbeachae and L. pneumophila strains the fusion proteins were detected at the appropriate sizes: BlaM-RalFLLO at 74 kDa, BlaM-1716 at 66 kDa, BlaM-2249 at 49.8 kDa, BlaM-2329 at 67.9 kDa, and BlaM-3288 at 65.2 kDa. (C) Translocation assays were performed with L. longbeachae strains in THP-1 cells at an MOI of 200 for 3 h before the CCF2-AM was added, and translocation was measured by exciting cells at 415 nm and detecting the emission signal at 460 nm and 535 nm. Wild-type L. longbeachae (black bars) demonstrated Dot/Icm-dependent translocation of BlaM-RalFLLO, BlaM-2249, and BlaM-3288. Results are expressed as the mean 460 nm/535 nm ratio relative to the ratio in uninfected cells ± standard deviation of at least three independent experiments. (D) Similarly, translocation by L. pneumophila was examined in THP-1 cells infected at an MOI of 40 for 1 h with wild-type L. pneumophila (dark gray bars) or L. pneumophila ΔdotA (white bars) or for 3 h with wild-type L. pneumophila (black bars) or L. pneumophila ΔdotA (light gray bars). Results are expressed as the mean 460 nm/535 nm ratio relative to the ratio in uninfected cells ± standard deviation of at least three independent experiments. The dotted lines in panels C and D represent a fluorescence ratio of 1, a value equivalent to that of an uninfected sample. *, P < 0.05, for the 460 nm/535 nm ratios of the wild-type strains compared to those of the Dot/Icm-deficient equivalents.
Given that we had previously observed more robust translocation in THP-1 cells by L. pneumophila, we examined whether the Rab-like proteins could be translocated by the Dot/Icm system of L. pneumophila. All plasmids encoding the BlaM fusion proteins were introduced into L. pneumophila 130b, and these strains were used to infect THP-1 cells at an MOI of 40 for 1 h or 3 h to measure translocation (Fig. 5B and D). Under these conditions, we could demonstrate that all four of the Rab-like proteins can be translocated by the Dot/Icm type IV secretion system. Therefore, we propose to name this novel family of effectors Rab-like effectors A to D (RleA is LLO1716, RleB is LLO2249, RleC is LLO2329, and RleD is LLO3288).
RleA, -B, -C, and -D show distinct patterns of localization during infection.
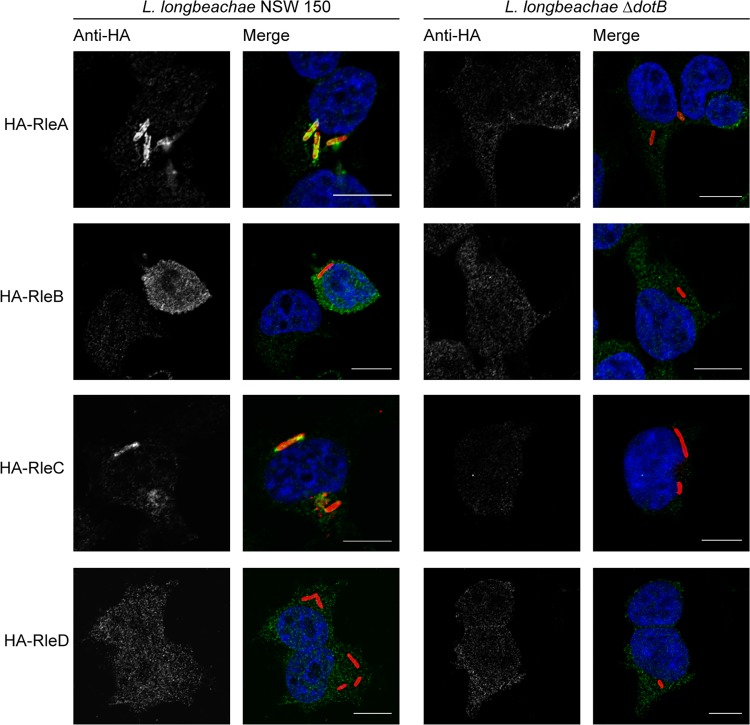
Given that our β-lactamase translocation assay showed low levels of translocation of RleA to RelD and in order to further characterize these novel effectors, we undertook immunofluorescence examination of the localizations of these proteins during infection. RleA to RelD were expressed with an N-terminal 4×HA tag in both wild-type L. longbeachae and L. longbeachae ΔdotB. These strains were used to infect HEK 293T FcγR cells for 3 h, and the localizations of the Rle proteins were determined using an anti-HA antibody (Fig. 6). Interestingly, we observed distinct patterns of localization for each of the Rle proteins, dependent on a functional Dot/Icm system. RleA specifically decorated the LCV in defined puncta, RleB was observed throughout the cytoplasm, and RleC showed some association with the LCV. The localization of RleD could not be definitively determined as specific staining could not be consistently detected above the background HA signal.
FIG 6.
The Rle proteins have distinct subcellular localizations during infection. L. longbeachae NSW150 and the ΔdotB derivative strain were transformed with pMMB207-HA constructs to express Rle proteins with an N-terminal 4×HA tag. These strains were used to infect HEK 293T FcγR cells at an MOI of 1, and the infected cells were fixed and stained at 3 h postinfection with anti-L. longbeachae (red), anti-HA (green), and DAPI (blue). Representative images demonstrate that RleA and RleC associate with the longbeachae-LCV and that RleB is found throughout the cytoplasm in a Dot/Icm-dependent manner. RleD could not be reliably detected above the background anti-HA signal. Scale bar, 10 nm.
Bioinformatic analysis of these effectors indicates that they have different levels of conservation within the Rab GTPase functional domains and that their capacity to act as GTPases remains to be investigated (Fig. 7). The G1 box, with a consensus sequence of GXXXXGK(S/T), acts as a purine nucleotide binding signature, with the serine (S) or threonine (T) residue important for coordinating an essential Mg2+ ion (33). All Rle proteins, except RleB, harbor a functional G1 box. The switch I and II regions (G2 box and G3 box) are the regions of Rab GTPases that change confirmation upon nucleotide binding (34). The switch I region, with a consensus sequence of YDPTIEDSY, provides major components of the effector binding surface, with only the threonine highly conserved, and the switch II region, with a DXXGL (where L is a hydrophobic residue) consensus, is required to bind a nucleotide-associated Mg2+ ion via the aspartic acid residue (D) (34). RleA shows conservation of these regions, with the exception of a serine residue instead of a hydrophobic residue within the switch II region. This alteration is, however, also present in the functional host Rab GTPase Rab24, leading us to predict that RleA could act as a functional Rab GTPase within the host cell. Similarly, both RleC and RleD possess the switch I threonine residue and show appropriate conservation within the G1 box and switch II region. RleB demonstrates the least conservation in these regions. This suggests that RleB does not possess Rab GTPase activity. However, as a likely inactive Rab GTPase, RleB may still impact host vesicular trafficking by binding and sequestering downstream Rab effectors. Interestingly, none of the Rle proteins has a C-terminal prenylation motif carried by the eukaryotic Rab GTPases. Lipidation of this moiety allows membrane tethering of active Rab GTPases. Therefore, RleA and RleC must use other means to localize to the LCV. RleA does harbor two predicted transmembrane domains toward the C terminus that may assist with membrane localization.
FIG 7.
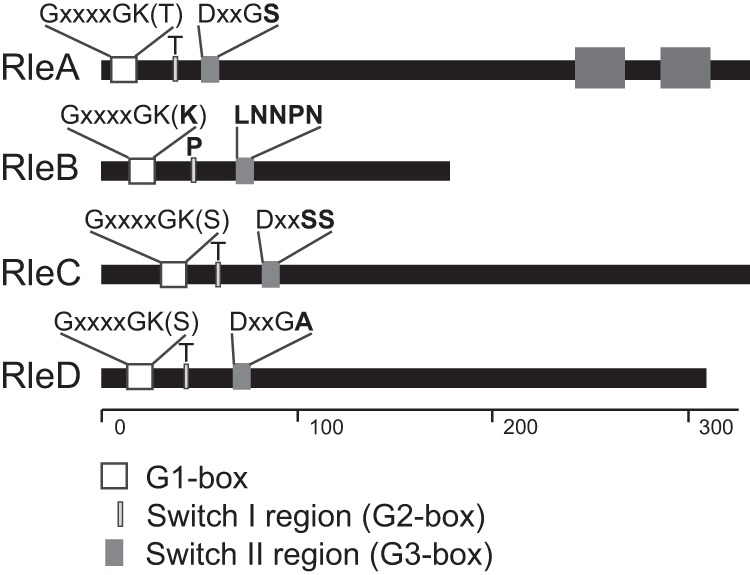
Rle proteins of L. longbeachae show various degrees of catalytic residue conservation. The figure shows a schematic representation of the L. longbeachae Rab-like effectors, with the catalytic residues highlighted. The scale along the bottom signifies the number of amino acids of each effector, highlighting that RleB is significantly smaller than the other effectors. RleA is predicted to carry two C-terminal transmembrane domains, represented by the dark gray boxes. The N-terminal white box represents the highly conserved G1 box with a consensus sequence of GXXXXGK(S/T), where X is any amino acid. The light gray box represents the switch I region which requires a conserved threonine (T) for function. The dark gray box represents the switch II region with a consensus sequence of DXXGL, where L is a hydrophobic residue. Residues that do not match the consensus sequence are shown in bold, demonstrating that RleB is unlikely to have GTPase activity.
The replicative longbeachae-LCV has characteristics similar to those of the pneumophila-LCV.
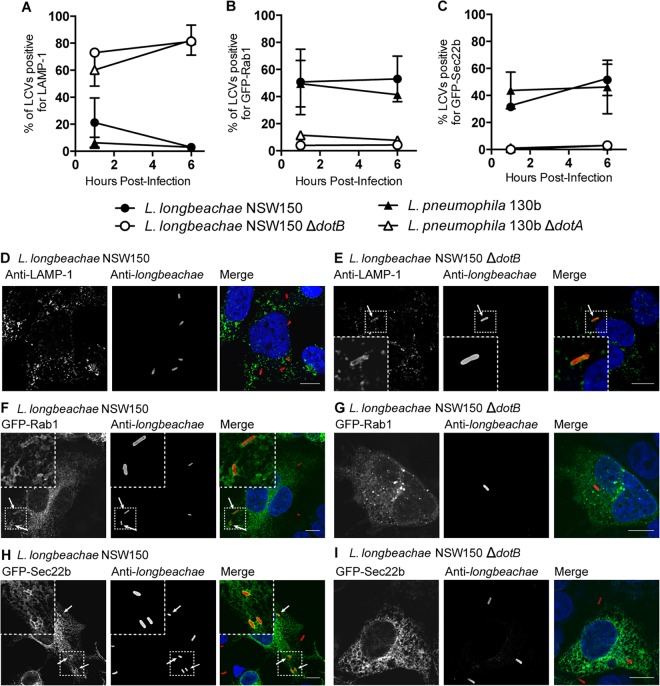
Previous reports have suggested that the longbeachae-LCV is distinct from the pneumophila-LCV and that it associates with both early endosomal antigen 1 (EEA1) and lysosomal-associated membrane protein 2 (LAMP-2) (11). However, more recently it has been confirmed that the longbeachae-LCV formed in the amoebal host Dictyostelium discoideum is decorated with ER markers (16). We have clarified that the longbeachae-LCV is characteristically similar to the pneumophila-LCV by demonstrating that the longbeachae-LCV avoids endocytic maturation and acquisition of the late endosome and lysosome marker LAMP-1 in HEK 293T FcγR cells (Fig. 8A, D, and E). Interestingly, at 1 h postinfection a small but variable proportion of longbeachae-LCVs, 21.15% ± 18.21%, are positive for LAMP-1. This contrasts with just 6.25% ± 4.03% LAMP-1-positive pneumophila-LCVs. By 6 h postinfection the vast majority of longbeachae-LCVs show no association with LAMP-1 (only 3.00% ± 2.00% LAMP-1 positive), with a level equivalent to the LAMP-1 avoidance seen for pneumophila-LCVs (3.00% ± 1.73% LAMP-1 positive).
FIG 8.
The developing longbeachae-LCV evades fusion with lysosomes and recruits Rab1 and Sec22b. HEK 293T FcγR cells expressing GFP-Rab1 or GFP-Sec22b or not transfected were infected with Legionella strains at an MOI of 1 for 1 h or 6 h before being fixed and stained for examination of the association between LCVs and the host proteins LAMP-1, Rab1, and Sec22b. LAMP-1 was detected using anti-LAMP-1, and the bacteria were stained red using either anti-L. longbeachae or anti-L. pneumophila. The percentages of LCVs positive for LAMP-1 (A), Rab1 (B), and Sec22b (C) were enumerated at 1 h and 6 h postinfection. Results are the mean percentages of positive LCVs ± the standard deviations from at least three independent infections where 100 LCVs were blindly quantified for each infection. L. longbeachae NSW150 and L. pneumophila 130b evaded association with LAMP-1 and recruited both GFP-Rab1 and GFP-Sec22b to the LCV. Conversely, L. longbeachae ΔdotB and L. pneumophila ΔdotA associated with LAMP-1 but showed no recruitment of GFP-Rab1 and GFP-Sec22b to the LCV. Representative pictures demonstrate that LCVs containing L. longbeachae NSW150 show no association with LAMP-1 (D) and are positive for GFP-Rab1 (F) and for GFP-Sec22b (H) at 1 h postinfection. LCVs formed by L. longbeachae ΔdotB were observed to be LAMP-1 positive (E) and negative for GFP-Rab1 (G) and GFP-Sec22b (I). Nucleic acid is stained blue with DAPI. Arrows highlight LCVs positive for the host protein. Scale bar, 10 nm.
Rab1 and Sec22b are recruited to the pneumophila-LCV during the early stages of infection. We were interested to determine whether Rab1 and Sec22b associate with the longbeachae-LCV as the genome sequence of L. longbeachae revealed the absence of homologues of the L. pneumophila Dot/Icm effectors responsible for recruiting these host proteins. Furthermore, our demonstration that L. longbeachae translocates novel Rab-like effectors and that both RleA and RleC localize to the longbeachae-LCV suggests that it may be unnecessary for L. longbeachae to commandeer host Rab GTPases to remodel the LCV. Using HEK 293T FcγR cells transfected with either GFP-Rab1 or GFP-Sec22b, we observed that L. longbeachae does, however, recruit both Rab1 and Sec22b to the longbeachae-LCV and that this trait is dependent on the Dot/Icm system (Fig. 8).
The dynamics of the association of Rab1 and Sec22b are similar for the longbeachae-LCV and pneumophila-LCV. However, as has previously been reported, fewer pneumophila-LCVs are positive for these proteins as infection progresses. Strikingly, at between 1 and 6 h postinfection there is no decline in the percentage of longbeachae-LCVs positive for Rab1 (50.75% ± 24.17% at 1 h and 53.00% ± 16.87% at 6 h) (Fig. 8B), and the percentage of longbeachae-LCVs positive for Sec22b actually increases in this time (31.50% ± 1.73% at 1 h and 51.50% ± 11.56% at 6 h; P = 0.014) (Fig. 8C).
DISCUSSION
L. longbeachae is an under-studied human pathogen that causes Legionnaires' disease that is clinically indistinguishable from that caused by L. pneumophila. Despite the clinical similarities of the diseases, L. longbeachae occupies a different environmental niche, and comparative genomics has revealed that only 65.2% of L. longbeachae genes are orthologous to L. pneumophila genes and that the L. longbeachae genome, at 4,149,158 bp for the NSW150 strain, is 500 kb larger than the L. pneumophila genome, at 3,503,610 bp for the Paris strain (8). Such differences would be expected to phenotypically manifest in the interactions between these pathogens and eukaryotic host cells. Our study has begun to characterize both similarities and unique aspects of the longbeachae-LCV compared to those of the pneumophila-LCV. Importantly, we have demonstrated that, similar to the pneumophila-LCV, the developing longbeachae-LCV evades endocytic maturation and recruits the host proteins Rab1 and Sec22b in a Dot/Icm-dependent manner. Yet the dynamics of LCV development and the effectors that mediate this process are distinct.
We confirmed that the Dot/Icm type IV secretion system is essential for intracellular replication of L. longbeachae by creating and characterizing a deletion mutant of the gene encoding the Dot/Icm ATPase DotB. In conducting intracellular growth curves in THP-1 cells, comparing wild-type and Dot/Icm-deficient L. longbeachae and L. pneumophila, we observed a diminished capacity for L. longbeachae to enter the host cell compared to that of L. pneumophila. This reduction in intracellular bacteria was not dependent on the Dot/Icm system and may reflect a different mode of entry for these two pathogens in THP-1 cells. Interestingly, the infection kinetics of L. longbeachae and L. pneumophila in nonphagocytic A549 cells are equivalent, with similar rates of entry. These results suggest that L. longbeachae may not be phagocytosed as efficiently as L. pneumophila, perhaps due to the expression of a capsule, but that bacterially mediated host cell entry is not perturbed.
Our L. longbeachae ΔdotB strain also provided the capacity to investigate the translocation of Dot/Icm effectors by this pathogen. Controlling when Dot/Icm effectors are introduced into the host cell is very important for L. pneumophila as effectors are immediately required upon phagocytosis to evade LCV endocytic maturation and subsequent degradation of the bacteria. Transmissive-phase L. pneumophila bacteria show upregulated expression of components of the Dot/Icm secretion apparatus and effectors (10). As such, it has been demonstrated that L. pneumophila Dot/Icm effector translocation occurs immediately upon contact with the host cell (32). This is in stark contrast to observations in Coxiella burnetii, another pathogen entirely reliant on a Dot/Icm system, which does not activate effector translocation until it reaches an acidified vacuole (35). We applied a β-lactamase reporter assay to examine the dynamics of Dot/Icm-dependent protein translocation by L. longbeachae (36). This approach has confirmed that the L. longbeachae homologue of the L. pneumophila effector RalF, RalFLLO, is translocated by L. longbeachae in a Dot/Icm-dependent manner. As with L. pneumophila, translocation can occur even in the absence of bacterial internalization. In the THP-1 model, we consistently observed that L. longbeachae translocated less BlaM-RalFLLO than L. pneumophila when it translocated the same BlaM-RalFLLO. This reduced capacity for Dot/Icm effector translocation was not observed during infection of nonphagocytic A549 cells. This indicates that the reduced translocation of BlaM-RalFLLO likely reflects the reduced phagocytosis of L. longbeachae by THP-1 cells.
Establishing the β-lactamase reporter assay in L. longbeachae has allowed us to characterize a family of four unique Rab-like proteins as novel Dot/Icm effectors. The only other L. longbeachae effector that has been described to date is SidC, a phosphatidylinositol 4-phosphate [PtdIns(4)P]-binding protein. SidC is a Dot/Icm effector conserved in both L. pneumophila and L. longbeachae. As previously described in L. pneumophila, SidC localizes to the longbeachae-LCV in a Dot/Icm-dependent manner, and, in both organisms, SidC acts to promote ER recruitment to the LCV (16, 37). Translocation of SidC was demonstrated using immunofluorescence microscopy and an antibody specific to SidC (16). The proteins BlaM-RleA to -RelD were translocated at low levels by both L. longbeachae and L. pneumophila. Despite very low levels of translocation using our reporter system, we were able to confirm Dot/Icm-dependent translocation of these effectors, with the exception of RleD, using immunofluorescence microscopy to visualize 4×HA-tagged effectors expressed by L. longbeachae. This technique revealed that, during early stages of infection, the Rle proteins have distinct subcellular localizations when translocated into the eukaryotic host cell and therefore likely serve distinct functions in the host cell. RleA and, to a lesser extent, RleC are localized to the longbeachae-LCV. RleB is distributed throughout the cytoplasm. Our inability to detect HA-RleD indicates that the protein was not present at a high enough concentration for detection. This may be due to low expression, poor translocation, diffuse localization within the host cell, and/or proteolytic processing of the N terminus of the protein upon translocation.
It is exciting to hypothesize that the presence of bacterially derived, putative Rab GTPases on the longbeachae-LCV could provide a novel mechanism for L. longbeachae to remodel the LCV. Given the presence of these effectors and the absence of homologues of the L. pneumophila effectors that modulate Rab1, we hypothesized that the Rle proteins may provide a Rab1-independent means to mediate the recruitment and fusion of ER-derived vesicles with the LCV. However, we surprisingly observed that Rab1 and Sec22b are still recruited to the longbeachae-LCV. Our finding that the longbeachae-LCV recruits both Rab1 and Sec22b in a Dot/Icm-dependent manner provides an example of L. longbeachae remodeling the phagosome in a manner similar to that of L. pneumophila. However, the mechanism used by L. longbeachae to recruit these host proteins is novel as the L. pneumophila Dot/Icm effectors required for Rab1 recruitment to the LCV are not encoded by L. longbeachae. Of all the known L. pneumophila effectors that modulate Rab1, only the Rab1 GTPase-activating protein LepB has an L. longbeachae homologue. Strikingly, even the LepB homologue, LLO0796, is not well conserved, retaining only 42% identity with L. pneumophila LepB. Interestingly, L. longbeachae also possesses a homologue of the L. pneumophila effector PieE, LLO3131 with 60% identity, which was recently shown to interact with a subset of Rab GTPases, including Rab1 (38).
Many intracellular bacterial pathogens manipulate the function of host Rab GTPases to modify vesicular trafficking (reviewed in reference 39). Understanding the mechanisms used to manipulate these fundamental host processes provides great insight into both bacterial pathogenesis and aspects of the eukaryotic biology. Further research will likely allow us to elucidate the novel L. longbeachae Dot/Icm effectors that lead to the LCV recruitment of Rab1 and Sec22b. In addition, L. longbeachae is the only bacterial pathogen known to encode homologues of Rab GTPases. The functional characterization of the Rle L. longbeachae effector family identified here may inform our understanding of the unique aspects of the longbeachae-LCV development.
ACKNOWLEDGMENTS
This work was supported by project grants awarded to H.J.N. from the Australian National Health and Medical Research Council (grant numbers 1062383 and 1063646). R.E.W. and E.A.L. are supported by Australian Postgraduate Awards.
We are indebted to Craig Roy, Yale University, for supplying HEK 293T cells stably expressing FcγR. Jenny Stow, The University of Queensland, kindly provided Rab1- and Sec22b-expressing constructs. Confocal imaging was performed at the Biological Optical Microscopy Platform, The University of Melbourne (www.microscopy.unimelb.edu.au).
REFERENCES
- 1.NNDSS Annual Report Writing Group. 2013. Australia's notifiable disease status, 2011: annual report of the National Notifiable Diseases Surveillance System. Commun Dis Intell Q Rep 37:E313–E393. [DOI] [PubMed] [Google Scholar]
- 2.NNDSS Annual Report Writing Group, Milton A, Stirzaker S, Trungove M, Knuckey D, Martin N, Hastie C, Pennington K, Sloan-Gardner T, Fitzsimmons G, Knope K, Martinek S, Mills L, Barry C, Wright P, Power M. 2012. Australia's notifiable disease status, 2010: annual report of the National Notifiable Diseases Surveillance System. Commun Dis Intell Q Rep 36:1–69. [DOI] [PubMed] [Google Scholar]
- 3.Whiley H, Bentham R. 2011. Legionella longbeachae and legionellosis. Emerg Infect Dis 17:579–583. doi: 10.3201/eid1704.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potts A, Donaghy M, Marley M, Othieno R, Stevenson J, Hyland J, Pollock KG, Lindsay D, Edwards G, Hanson MF, Helgason KO. 2013. Cluster of Legionnaires disease cases caused by Legionella longbeachae serogroup 1, Scotland, August to September 2013. Euro Surveill 18(50):pii=20656 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20656. [DOI] [PubMed] [Google Scholar]
- 5.Fliermans CB, Cherry WB, Orrison LH, Smith SJ, Tison DL, Pope DH. 1981. Ecological distribution of Legionella pneumophila. Appl Environ Microbiol 41:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steele TW, Lanser J, Sangster N. 1990. Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl Environ Microbiol 56:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steele TW, Moore CV, Sangster N. 1990. Distribution of Legionella longbeachae serogroup 1 and other legionellae in potting soils in Australia. Appl Environ Microbiol 56:2984–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, Sansom FM, Jarraud S, Zidane N, Ma L, Bouchier C, Etienne J, Hartland EL, Buchrieser C. 2010. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet 6:e1000851. doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66:3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol 8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 11.Asare R, Abu Kwaik Y. 2007. Early trafficking and intracellular replication of Legionella longbeachaea [sic] within an ER-derived late endosome-like phagosome. Cell Microbiol 9:1571–1587. doi: 10.1111/j.1462-5822.2007.00894.x. [DOI] [PubMed] [Google Scholar]
- 12.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 14.Hardiman CA, McDonough JA, Newton HJ, Roy CR. 2012. The role of Rab GTPases in the transport of vacuoles containing Legionella pneumophila and Coxiella burnetii. Biochem Soc Trans 40:1353–1359. doi: 10.1042/BST20120167. [DOI] [PubMed] [Google Scholar]
- 15.Arasaki K, Toomre DK, Roy CR. 2012. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe 11:46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolinsky S, Haneburger I, Cichy A, Hannemann M, Itzen A, Hilbi H. 2014. The Legionella longbeachae Icm/Dot substrate SidC selectively binds PtdIns(4)P with nanomolar affinity and promotes pathogen vacuole-endoplasmic reticulum interactions. Infect Immun 82:4021–4033. doi: 10.1128/IAI.01685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton HJ, Roy CR. 2011. The Coxiella burnetii Dot/Icm system creates a comfortable home through lysosomal renovation. mBio 2(5):e00226-11. doi: 10.1128/mBio.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez-Valero L, Rusniok C, Rolando M, Neou M, Dervins-Ravault D, Demirtas J, Rouy Z, Moore RJ, Chen H, Petty NK, Jarraud S, Etienne J, Steinert M, Heuner K, Gribaldo S, Medigue C, Glockner G, Hartland EL, Buchrieser C. 2014. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires' disease. Genome Biol 15:505. doi: 10.1186/s13059-014-0505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 20.Machner MP, Isberg RR. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 22.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr, Yergey A, Machner MP. 2011. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y, Arnold RJ, Luo ZQ. 2011. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A 108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y, Luo ZQ. 2011. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardiman CA, Roy CR. 2014. AMPylation is critical for Rab1 localization to vacuoles containing Legionella pneumophila. mBio 5(1):e01035-13. doi: 10.1128/mBio.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelstein PH. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol 14:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol 186:1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 31.Amor JC, Swails J, Zhu X, Roy CR, Nagai H, Ingmundson A, Cheng X, Kahn RA. 2005. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem 280:1392–1400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 32.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A 102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colicelli J. 2004. Human RAS superfamily proteins and related GTPases. Sci STKE 2004:RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol 186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. 2008. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 10:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 38.Mousnier A, Schroeder GN, Stoneham CA, So EC, Garnett JA, Yu L, Matthews SJ, Choudhary JS, Hartland EL, Frankel G. 2014. A new method to determine in vivo interactomes reveals binding of the Legionella pneumophila effector PieE to multiple Rab GTPases. mBio 5(4):e01148-14. doi: 10.1128/mBio.01148-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherwood RK, Roy CR. 2013. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14:256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engleberg NC, Pearlman E, Eisenstein BI. 1984. Legionella pneumophila surface antigens cloned and expressed in Escherichia coli are translocated to the host cell surface and interact with specific anti-Legionella antibodies. J Bacteriol 160:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedmaier P, Sansom FM, Sofian T, Beddoe T, Schuelein R, Newton HJ, Hartland EL. 2014. Multiple ecto-nucleoside triphosphate diphosphohydrolases facilitate intracellular replication of Legionella pneumophila. Biochem J 462:279–289. doi: 10.1042/BJ20130923. [DOI] [PubMed] [Google Scholar]
- 42.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun 65:2497–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]