Significance
Proteins and enzymes are susceptible to oxidative damage and inactivation from reactive oxygen and nitrogen species formed as a consequence of oxidative stress. They also are at risk from oxidizing intermediates formed during enzymatic turnover. A survey of the protein structural database has revealed that one third of structurally characterized proteins contain chains of redox-active tyrosine and tryptophan residues. These chains can protect proteins from damage by transferring oxidizing equivalents away from critical active site regions and delivering them to surface sites for scavenging.
Keywords: iron oxygenases, electron transfer, reactive oxygen species, superoxide dismutase, cytochrome P450
Abstract
Living organisms have adapted to atmospheric dioxygen by exploiting its oxidizing power while protecting themselves against toxic side effects. Reactive oxygen and nitrogen species formed during oxidative stress, as well as high-potential reactive intermediates formed during enzymatic catalysis, could rapidly and irreversibly damage polypeptides were protective mechanisms not available. Chains of redox-active tyrosine and tryptophan residues can transport potentially damaging oxidizing equivalents (holes) away from fragile active sites and toward protein surfaces where they can be scavenged by cellular reductants. Precise positioning of these chains is required to provide effective protection without inhibiting normal function. A search of the structural database reveals that about one third of all proteins contain Tyr/Trp chains composed of three or more residues. Although these chains are distributed among all enzyme classes, they appear with greatest frequency in the oxidoreductases and hydrolases. Consistent with a redox-protective role, approximately half of the dioxygen-using oxidoreductases have Tyr/Trp chain lengths ≥3 residues. Among the hydrolases, long Tyr/Trp chains appear almost exclusively in the glycoside hydrolases. These chains likely are important for substrate binding and positioning, but a secondary redox role also is a possibility.
The dioxygen-rich atmosphere produced by the Great Oxidation Event that occurred some 2.3 billion years ago created tremendous opportunities and challenges for living organisms (1). Today, the oxidizing power of the O2/H2O redox pair is coupled to a vast array of essential biological transformations. Foremost among these processes is cellular respiration, consuming over 1013 mol of O2 per day in the terrestrial biosphere (2). Cellular organisms have evolved to exploit the oxidizing potential of O2 in myriad other processes. The power of O2-driven reactions is balanced against the risk from low-fidelity reactions in which substrate oxidation fails, leaving enzymes to cope with highly oxidizing intermediates. For extremely productive enzymes, even a modest descent below 100% fidelity can have extremely deleterious consequences. Dioxygen also is responsible for oxidative stress, one of the central challenges to biological survival in aerobic environments. Owing to their abundance in biological systems, proteins are primary targets of reactive oxygen (ROS) and nitrogen (RNS) species produced by oxidative stress (3). Protein responses to ROS and RNS attacks are managed by components of the redox proteome through a set of reversible and irreversible posttranslational modifications of amino acids (4). The sulfur-containing amino acids (Met and Cys) can undergo reversible changes in response to ROS and RNS, whereas the oxidative reactions of other susceptible residues (Trp and Tyr) and the protein backbone are generally considered to be irreversible (4). The redox proteome also may provide protection from high-potential reactive intermediates formed during unsuccessful turnover in O2-using enzymes (5, 6).
Electron transfer (ET) reactions are a critical component of the protein response to an oxidative environment. Several decades of experimental investigations (5, 7), guided by a transparent theoretical model (8), have produced a clear understanding of single-step ET in proteins. Biological electron transfers frequently are accompanied by proton transfers, introducing an additional complication (9–11). Although the details of protein structure and redox thermodynamics must be taken into account when considering individual cases (12), tunneling timetables indicate that 20–25 Å is the maximum distance that an electron (or hole, in the case of high-potential reactions) can transfer on submillisecond timescales (5, 7). When biological exigency requires electrons (holes) to move farther or faster, multistep tunneling (hopping) reactions come into play (13, 14).
Electron (or hole) hopping in proteins can involve a series of redox cofactors as in Complex I (NADH dehydrogenase) of the mitochondrial respiratory chain (15–17). The side chains of select amino acids also can serve as waystations for redox hopping reactions, with Cys, Met, Trp, and Tyr being the most likely candidates (18). Hole transfer reactions are more common in Tyr and Trp, whereas Cys and Met tend to undergo two-electron bond-making/breaking transformations (4, 19). Clusters of Tyr and Trp residues, therefore, could act as conduits for high-potential holes in proteins. Indeed, hole transfer chains composed of Tyr and Trp residues have been identified in several enzymes, including ribonucleotide reductase (20), DNA photolyase (21), cytochrome c peroxidase (22), and cytochrome c oxidase (23).
Aromatic amino acid clustering in protein primary sequences (24) and tertiary structures (25, 26) is usually attributed to polypeptide fold stability, protein–protein recognition, and ligand binding (27). In addition to these roles, we suggest that clusters and chains of Tyr and Trp residues also protect proteins from oxidative damage; this proposal is supported by findings from a search for Tyr/Trp chains in the protein structural database. More to the point, we have discovered correlations between the occurrence of these chains and the biological functions of the proteins in which they reside.
Results and Discussion
Using functions from the MATLAB Bioinformatics Toolbox, we searched for linear and branched redox chains in 89,761 of the 89,802 protein X-ray crystal structures available in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB; www.rcsb.org) (28) on April 2, 2015. We screened initially for linear chains comprised of redox-active amino acids (Tyr and Trp) and selected heteroatom residues (heme, Fe, Cu, NADH, and TPQ). As not all Tyr residues are redox active, owing to the acidity of the tyrosine radical cation (pKa ∼ −1) (29), only those that had suitable proton acceptors [carboxylate (Asp and Glu) O atoms, imidazole Nπ or Nτ atoms, and O atom from H2O] within 4 Å of the phenolic oxygen were included in the search. A matrix was generated describing distances among all redox-active residues of all polypeptide chains in a protein. Distances were defined as the shortest separation between aromatic side chain atoms of Trp and Tyr and any of the heteroatom residues. We refined the distance matrix by defining as infinite any separations greater than a specified ET cutoff (5, 7.5, and 10 Å). These cutoff distances correspond to estimated time constants for electron exchange (−ΔG° = 0; λ = 1 eV; T = 295 K) between residues of ∼20 ns, ∼250 ns, and 4 μs, respectively (5, 7, 8). The shortest paths between all possible pairs of redox-active residues were found using a MATLAB implementation of the Floyd–Warshall graph theory algorithm written by David Gleich (www.cs.purdue.edu/homes/dgleich/packages/matlab_bgl). This algorithm identified the shortest linear chains among all redox-active residues; these linear chains were subsequently analyzed for common residues to identify branched redox chains.
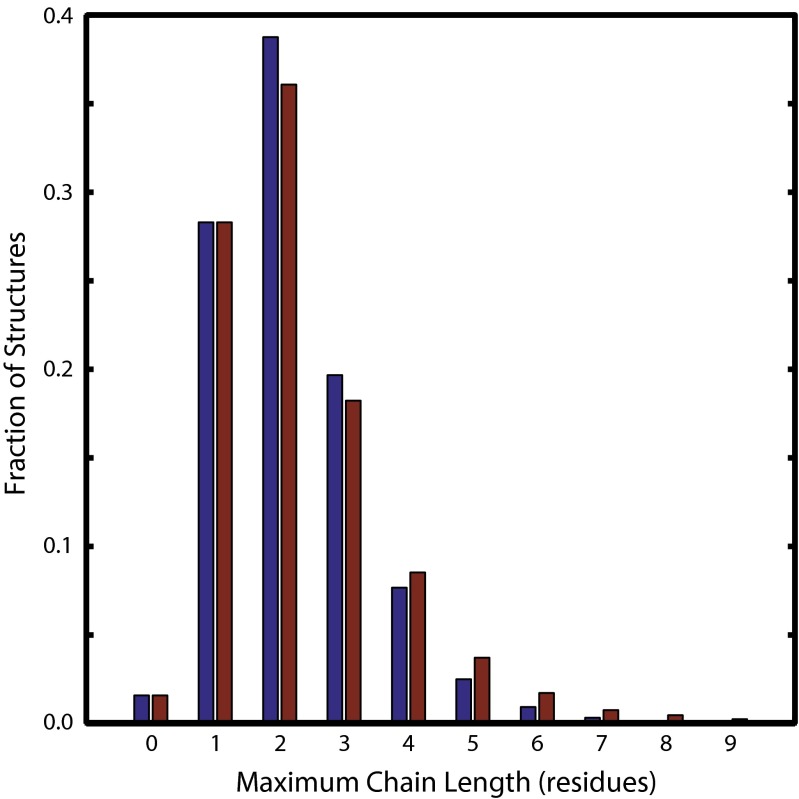
To avoid redundant structures, statistical analyses of the redox chains were restricted to protein structures defined by the PDB to have less than 90% sequence identity. This restriction reduced the size of the unique data set to 27,355 structures. For the purposes of the statistics discussion, we restrict consideration to chains found using the 5-Å ET cutoff and further require that at least one member of the chain be a solvent exposed residue. The distributions of linear and branched chain lengths in this dataset are shown in Fig. 1: the mean linear chain (MLC) length is 2.18 residues, and the mean branched chain (MBC) length is 2.34 residues. Results for the 7.5- and 10-Å cutoffs are given in SI Appendix, Figs. S1 and S2. As expected, the inclusion of redox chain branching produces a modest increase in mean chain length.
Fig. 1.
Distributions of maximum linear (blue) and branched (red) redox chain lengths (5-Å ET cutoff, at least one solvent-exposed residue) found in 27,355 unique protein X-ray crystal structures in the RCSB Protein Data Bank.
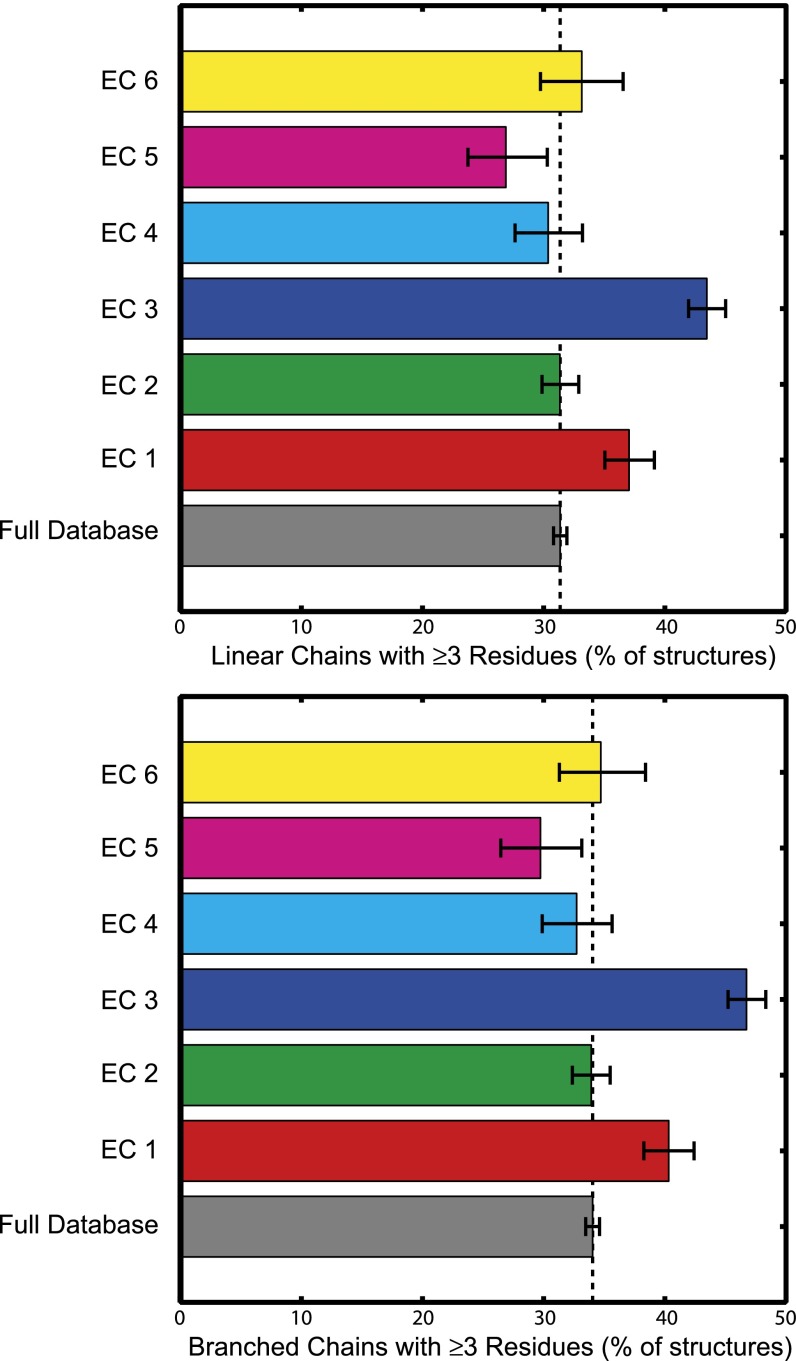
Approximately one third of the protein structures contain long redox chains composed of ≥3 residues. The distribution of these structures among the six enzyme classes (Fig. 2 and SI Appendix, Tables S1 and S2) reveals that long redox chains are most prevalent among oxidoreductases (EC 1, 2,200 structures, MLC 2.37) and hydrolases (EC 3, 3,999 structures, MLC 2.57). Long redox chains appear in transferases (EC 2, 3,590 structures, MLC 2.21), lyases (EC 4, 1,024 structures, MLC 2.12), and ligases (EC 6, 703 structures, MLC 2.22) with about the same frequency as in the full database, whereas they appear substantially less often in isomerases (EC 5, 703 structures, MLC 2.05). The higher frequency of long chains in oxidoreductases is consistent with a redox role; the large proportion among the hydrolases, where acid–base mechanisms tend to predominate, is somewhat surprising.
Fig. 2.
Distribution among six enzyme classes (EC 1, oxidoreductases; EC 2, transferases; EC 3, hydrolases; EC 4, lyases; EC 5, isomerases; EC 6, ligases) of protein structures with (Upper) linear or (Lower) branched redox chains ≥3 residues in length (5-Å ET cutoff, at least one solvent exposed residue). Error bars indicate 95% confidence interval from bootstrap sampling statistics.
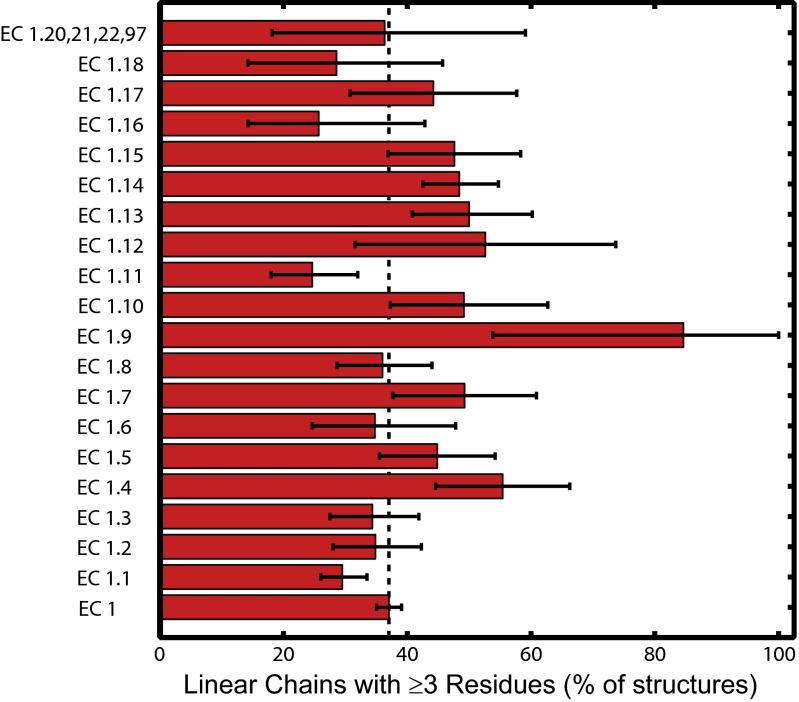
The breakdown of long redox chains among subclasses of the oxidoreductases (Fig. 3 and SI Appendix, Table S3) reveals that about half (EC 1.4, 1.5, 1.7, 1.9, 1.10, 1.12, 1.13, 1.14, 1.15, and 1.17) have above-average (>37%) long-chain frequencies. Among the oxidoreductases that react with O2, with or without incorporation of O atoms into substrate (542 structures), only 12% have no chains, and ∼50% have linear chains composed of three or more residues.
Fig. 3.
Distribution among oxidoreductase subclasses of protein structures with linear redox chains ≥3 residues in length (5-Å ET cutoff, at least one solvent exposed residue). Error bars indicate 95% confidence interval from bootstrap sampling statistics.
The 13 structures in the EC 1.9 subclass are composed largely of cytochrome c oxidases. EPR investigations have identified a potential radical transfer pathway from Tyr244 through Trp236 to Tyr129 (bovine numbering) during catalysis (23). Our search of the bovine enzyme structure [PDB ID 3AG4 (30)] identified a six-member branched pathway (7.5-Å ET cutoff, no solvated residues) that included these residues along with the CuB center, Trp126, and Trp288. With the 10-Å ET cutoff criterion, we identified several branched chains of >30 residues spanning multiple polypeptide chains.
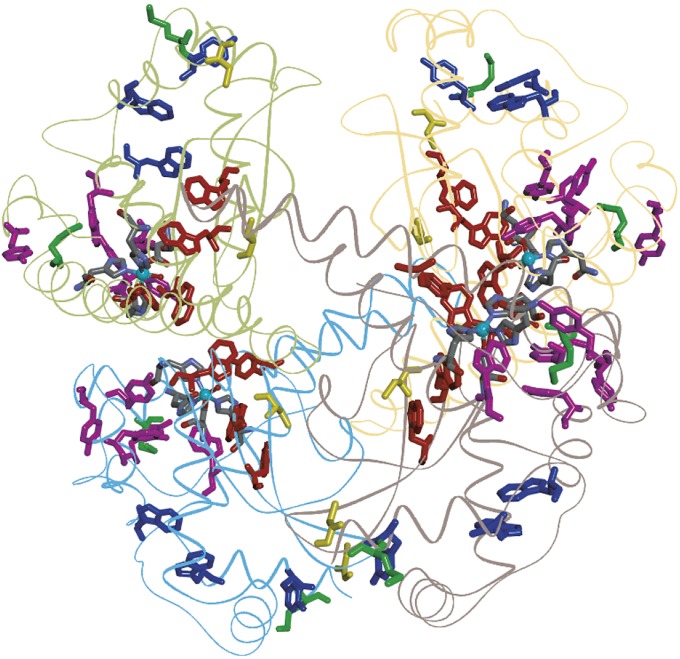
Enzymes that react with superoxide as an acceptor (EC 1.15, 84 structures) exhibit above-average chain lengths. Superoxide dismutases (SOD) feature prominently in this subclass. Three SOD enzymes have been identified in humans: a cytoplasmic CuZn protein (SOD1), a mitochondrial Mn enzyme (SOD2), and an extracellular CuZn enzyme (SOD3). Human SOD1 [PDB ID 1HL5 (31)] contains a single Trp residue and no Tyr residues; SOD3 [PDB ID 2JLP (32)] has just 2 Trp and 2 Tyr in each 222-residue chain. The longest chain in SOD3 (Trp139, Trp200, and Tyr141) is ∼20 Å from the CuZn active site. Human SOD2 [PDB ID 1N0J (33)] provides a striking contrast to SOD1 and SOD3, exhibiting above-average numbers of Tyr/Trp residues (9 Tyr, 6 Trp in each of four 199-residue polypeptides) assembled into long branched chains (5-Å ET cutoff, 4 residues; 7.5 Å, 9 residues; 10 Å, 12 residues; Fig. 4). Tyr34, a residue previously implicated in enzymatic turnover, lies ∼5.5 Å from the Mn center and is a member of each chain (34). Residues in the 5-Å cutoff chain extend from Tyr34 and reach the solvent-filled central tunnel at Trp125. The thiolate S atom of Cys140 is just 4 Å from the Trp125 indole. Indeed, S atoms of the two Cys and two Met residues in each peptide are within 8 Å of members of the Tyr/Trp chains (Fig. 4). It is striking that long Tyr/Trp chains are so much more prevalent in the mitochondrial enzyme than in the cytoplasmic or extracellular proteins. Mitochondrial enzymes as a group (402 structures) exhibit below-average occurrence of long chains (31% of structures, MLC 2.16). The role of SOD2 in managing high levels of mitochondrial ROS (35) might necessitate additional endogenous antioxidant protection afforded by the Tyr/Trp chains (36). The Tyr/Trp chains could deliver oxidizing equivalents to Cys or Met residues during conditions of oxidative stress. Alternatively, ROS-generated Tyr or Trp radicals at the protein surface could be reduced by the pool of glutathione (GSH) available in the mitochondrion (37).
Fig. 4.
Structural model of the human SOD2 homotetramer [PDB ID 1N0J (33)]. Each polypeptide contains one five-coordinate Mn center (cyan ball and five ligands). Members of Tyr/Trp chains: red, 5-Å ET cutoff (Tyr34, Trp125, Trp123, and Trp161); magenta, 7.5-Å cutoff (Tyr11, Trp78, Tyr169, Tyr165, and Tyr176); blue, 10-Å cutoff (Trp181, Trp186, and Tyr193). Met residues are shown in green, and Cys residues are shown in yellow.
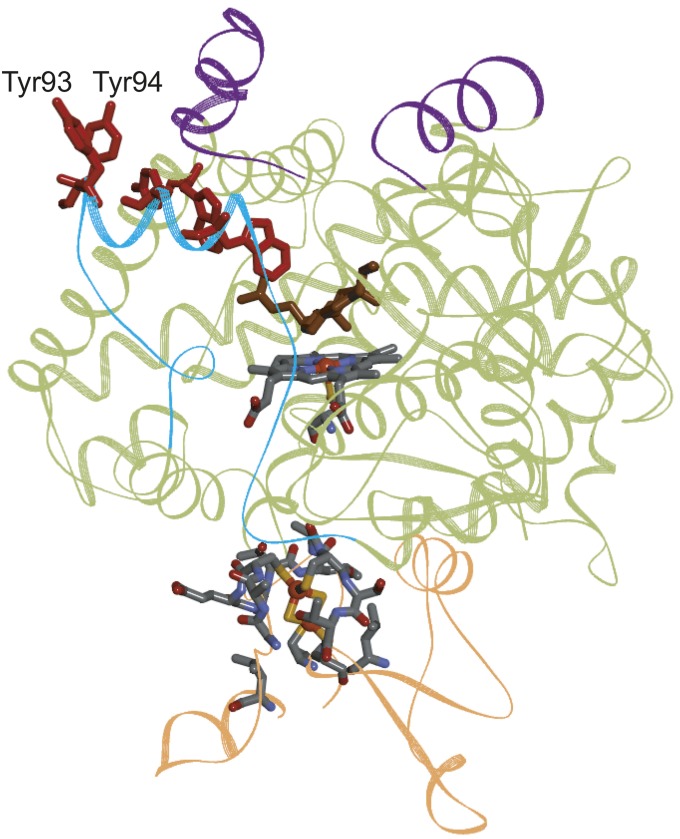
The EC 1.14 oxidoreductase subclass of enzymes acts on paired donors, with incorporation or reduction of molecular oxygen. These enzymes include the 2-oxoglutarate dependent nonheme iron oxygenases and the cytochromes P450. The five-residue linear hole-hopping chain (5-Å ET cutoff) identified in a mitochondrial cholesterol metabolizing cytochrome P450 [CYP11A1, PDB ID 3N9Y (38)] is a notable example. CYP11A1 is expressed in steroidogenic tissues and is responsible for the first step in the biosynthesis of steroid hormones, catalyzing the conversion of cholesterol to pregnenolone in three turnovers, requiring one O2 and one NADH molecule per step (39). A high-potential Fe-oxo (Cpd I) species has been implicated in each of the two hydroxylation and one carbon–carbon bond cleavage reactions required to produce pregnenolone (38, 40). If substrate is not available and properly positioned for reaction, the powerfully oxidizing Cpd I intermediate could attack surrounding residues or the porphyrin itself (41). The configuration of the five-residue Tyr/Trp chain in CYP11A1 can protect the enzyme from such damaging side reactions. The chain begins at Trp87, located 9.6 Å from the heme Fe atom. The position of this gateway residue is critical for effective enzyme protection: too close to the heme and it outcompetes substrate for reaction with Cpd I; too far away and it fails to prevent active-site damage. Once a hole reaches Trp87, it would rapidly transit through Trp231 and Tyr90 and arrive at a pair of surface-exposed Tyr residues (Tyr93 and Tyr94; Fig. 5 and SI Appendix, Fig. S3). The two Tyr residues lie near the water channel that faces the mitochondrial matrix (38). ET kinetics simulations (SI Appendix) indicate that a high-potential (E° ∼ 1 V vs. NHE) hole localized on the heme would have a ∼3-μs (SI Appendix, Fig. S4) survival time before it transferred to the protein surface along this pathway. Hence, substrate reaction with Cpd I must occur on a submicrosecond timescale to prevent hole migration to the surface Tyr93/94 residues. Although pregnenolone production is reported to be reasonably well coupled to NADH consumption in CYP11A1 (42), the pathway illustrated in Fig. 5 and SI Appendix, Fig. S3, would protect the enzyme active site in the event of unsuccessful substrate reaction with Cpd I during catalysis. Alignment of the amino acid sequences of CYP11A1 from the 13 species identified in the UniProt/Swiss-Prot database (www.uniprot.org) indicates that the five residues in the Tyr/Trp chain are almost fully conserved (SI Appendix, Table S4). The presence of analogous hole transfer pathways in cytochromes P450 may explain the difficulty in isolating Cpd I intermediates in these enzymes (43, 44). In this regard, it is notable that long hole-hopping chains are relatively uncommon among enzymes that act on peroxide (Fig. 3, EC 1.11 and 150 structures) and that Cpd I intermediates have been identified in several peroxidases.
Fig. 5.
Structural model of human CYP11A1 (green ribbon) [PDB ID 3N9Y (38)] in complex with its redox partner adrenodoxin (orange ribbon). The purple ribbon highlights the inner mitochondrial membrane interaction zone, the cyan ribbon shows the hydrophilic water channel, and the cholesterol substrate is brown (38). The linear five-member Tyr/Trp chain is shown in red.
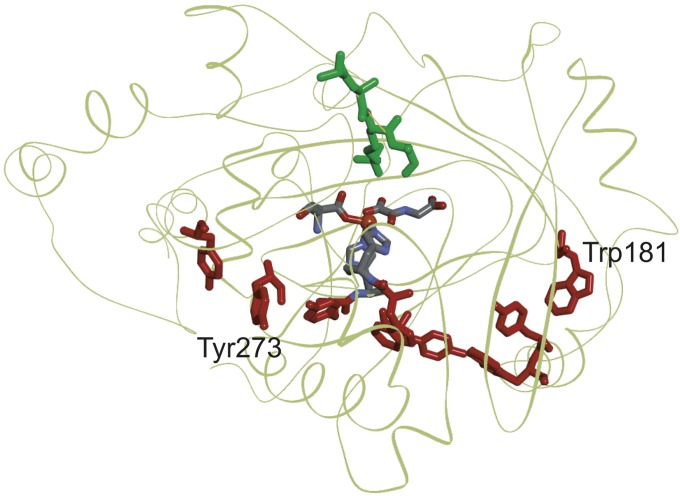
Long Tyr/Trp chains are also present in nonheme iron oxygenases. The structure of the catalytic domain of the human 2-oxo-glutarate dependent nonheme iron oxygenase Nε-methyllysine demethylase (KDM4A) is characterized by an eight-residue linear Tyr/Trp chain [5-Å ET cutoff, PDB ID 4W2V (45)]. The enzyme is believed to catalyze lysine demethylation in a hydroxylation/demthylation sequence involving an FeIV(O) intermediate (45). The central portion of the Tyr/Trp chain (Tyr275 and Trp187) passes ∼9 Å from the active site Fe center and terminates near surface exposed residues (Tyr273 and Trp181; Fig. 6 and SI Appendix, Fig. S5). ET kinetics simulations (SI Appendix) suggest that the survival time of an FeIV(O) center at the active site is ∼0.4 μs (SI Appendix, Fig. S6).
Fig. 6.
Structural model of human KDM4A (green ribbon) [PDB ID 4V2W (45)]. The linear eight-member Tyr/Trp chain is shown in red. A model peptide substrate is shown in green, and the Fe center is represented as an orange ball.
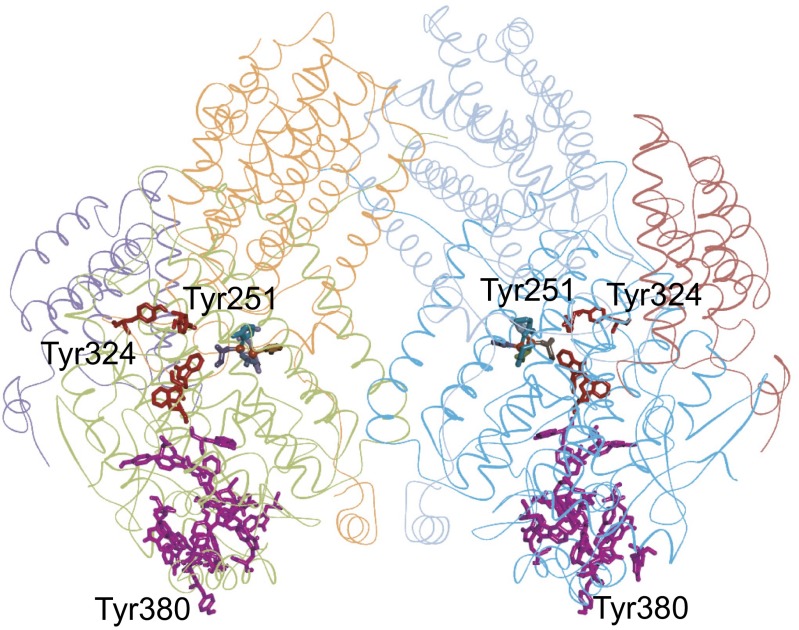
The structure of the methane monooxygenase from Methylococcus capsulatus (Bath) [PDB ID 1MTY (46)] exhibits a remarkably long branched Tyr/Trp chain (19 residues, 5-Å ET cutoff; SI Appendix) adjacent to (8.5-Å separation) a four-residue linear chain (Tyr251, Tyr324, Trp313, and Trp317; Fig. 7 and SI Appendix, Fig. S7). Both the linear and branched chains are coupled to surface Tyr residues (Tyr-324, Tyr251, and Tyr380). The di-iron active site is closest to the linear chain (11.5 Å to Trp317 and 13.3 Å to Trp317); the distance to the closest branched chain residue (Trp308) is 14.5 Å. The estimated survival time for a high-potential FeIV center in the enzyme active site is ∼9 μs (SI Appendix, Fig. S8). As with CUP11A1 and KDM4A, the positions of the residues closest to the active site are critical for determining survival times of the reactive intermediates.
Fig. 7.
Structural model of methane monooxygenase from Methylococcus capsulatus (Bath) [PDB ID 1MTY (46)]. The linear 4-residue Tyr/Trp chain is shown in red; the 19-residue branched chain is shown in magenta. The Fe centers are represented as orange balls.
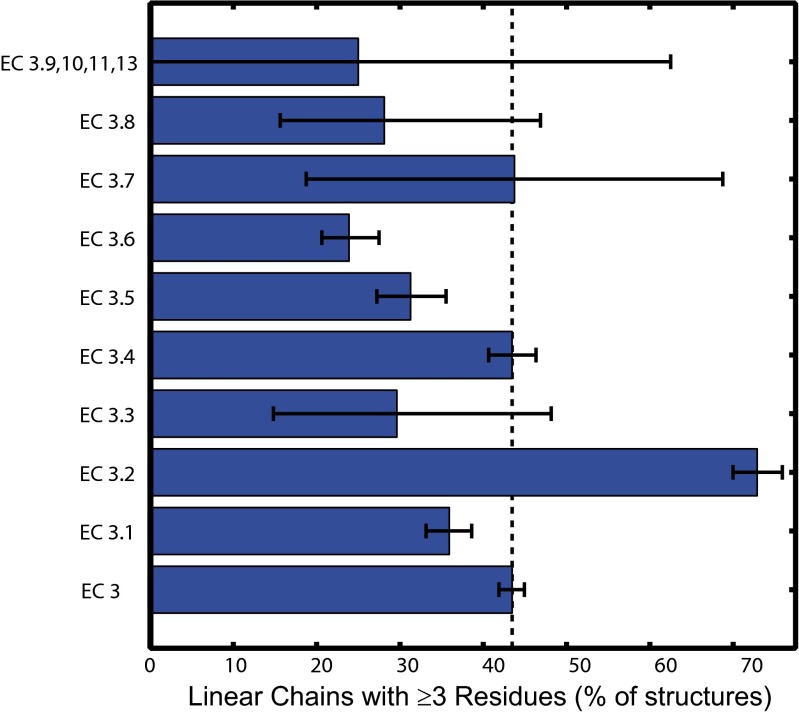
A breakdown among subclasses of the hydrolase data reveals that long chains occur with greatest frequency in glycosylases (EC 3.2; Fig. 8 and SI Appendix, Table S5). Over 70% of the 863 glycosylase structures have redox chains of three or more residues (MLC 3.54), compared with a value of 43% for all hydrolase structures. Just two of the hydrolase subclasses have long redox chain frequencies comparable with the EC 3 average (EC 3.4, peptide hydrolases, 1,144 structures, MLC 2.50; EC 3.7, C−C bond hydrolases, 16 structures, MLC 2.38); the remaining subclasses have frequencies far below average. Clearly, it is the glycosylases alone that are responsible for the high frequencies of long chains among hydrolases. The occurrence frequencies of Tyr and Trp residues among classified (UniProt/Swiss-Prot) EC 3.2 protein sequences are 39% and 100% higher, respectively, than database averages, whereas the occurrence frequencies of the other two aromatic amino acids are just slightly above averages (Phe, 5%; His, 8%). The two members of the glycosylase subclass hydrolyze O- and S-glycosyl compounds (glycoside hydrolases, 3.2.1, 747 structures, MLC 3.75, 79% ≥3 residues) and N-glycosyl compounds (3.2.2, 114 structures, MLC 2.23, 35% ≥3 residues). Clearly, the glycoside hydrolases have Tyr/Trp chains far in excess of PDB averages. The consensus mechanism for glycoside hydrolases involves one amino acid residue to serve as a proton donor and another to act as a nucleophile (47). The preponderance of aromatic residues in glycoside hydrolases has been attributed to a central role in substrate recognition and binding (48–50). No redox function has been identified in most of these enzymes, and it is probable that long Tyr/Trp chains are involved simply in substrate recognition and binding as well as fold stability. Given the susceptibility of Tyr/Trp chains to long-range hole migration, a protective redox function during times of oxidative stress is an additional possibility. The discovery that some members of the glycoside hydrolase family are lytic polysaccharide mono-oxygenases (51), however, raises the possibility that more members of the glycoside hydrolase subclass might use high-potential redox reactions in catalysis.
Fig. 8.
Distribution among hydrolase subclasses of protein structures with linear redox chains ≥3 residues in length (5-Å ET cutoff, at least one solvent exposed residue). Error bars indicate 95% confidence interval from bootstrap sampling statistics.
Concluding Remarks
The susceptibility of Tyr and Trp residues to oxidation at potentials near 1 V vs. NHE creates opportunities for their involvement in a wide assortment of biological redox processes. Incorporation of these residues into extended chains can enable long-distance transfer of oxidizing equivalents across proteins. The mitochondrial SOD2 and CYP11A1 enzymes operate in the O2-processing compartment of the cell where the risks of ROS and RNS are high and maximum protection is required. The Tyr/Trp chains identified in these proteins could provide protection by transporting potentially destructive high-potential holes away from the enzyme active sites. Our analysis of the protein structural database has revealed that extended Tyr/Trp chains are quite common, even among proteins that have no well-defined redox function. The mere presence of such chains does not imply a redox function, but current understanding of long-range ET reactions highlights their potential to serve as hole conduits in polypeptide structures.
Materials and Methods
Structure Searches.
The RCSB Protein Data Bank coordinate files were accessed using functions from the MATLAB (The MathWorks, Inc.) Bioinformatics Toolbox. Redox chain results for individual proteins are available from the authors upon request.
Confidence Intervals.
Confidence intervals from bootstrap statistics were determined using 104 bootstrap samples with the bootci function from the Statistics Toolbox in MATLAB.
Kinetics Simulations.
The rate laws for intraprotein electron hopping reactions are a set of coupled first-order linear differential equations. Numerical solutions of these equations were obtained using the eig function in MATLAB. Electron transfer rate constants for individual reaction steps (kET) were calculated according to Eq. 1 (where ΔG° is the standard free energy change for the electron transfer reaction and R is the gas constant) (5–8). Reorganization energies (λ) for all reaction steps were set equal to 0.8 eV, the exponential distance decay parameter (β) was taken to be 1.1 Å–1, the temperature (T) was taken to be 295 K, and the contact distance (ro) was taken to be 3 Å. Distances (r) were defined as described in the main text, except for hemes where distances were defined to be to the Fe center. The driving force for the first electron transfer step to the chain was defined as –ΔG1°, and the driving force for electron transfer from the originating residue to the terminal residue(s) was defined as –ΔG2°. All intermediate members of the redox chain were assumed to have the same reduction potential, hence as ΔG° = 0 for transfers among nonoriginating and nonterminal members of the chain. Hopping times (τhop) were taken from the time integral of the normalized originating-residue concentration profile. Estimated active-intermediate survival times correspond to –ΔG1° = –0.1 eV and –ΔG2° = 0.1 eV. Contour maps of hopping times as functions of –ΔG1° and –ΔG2° appear in SI Appendix, Figs. S4, S6, and S8.
| [1] |
Supplementary Material
Acknowledgments
For many years we enjoyed collaborative work with the late John H. (Jack) Richards; discussions with him influenced our current view of the roles of hole hopping in biology. Research reported in this publication was supported by The National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award R01DK019038 (to H.B.G. and J.R.W.). Additional support was provided by the Arnold and Mabel Beckman Foundation.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 10821.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512704112/-/DCSupplemental.
References
- 1.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506(7488):307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 2.Watson RT, et al., editors. Land Use, Land-Use Change, and Forestry—A Special Report of the Intergovernmental Panel on Climate Change. Cambridge Univ Press; Cambridge, UK: 2000. p. 375. [Google Scholar]
- 3.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703(2):93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Go Y-M, Jones DP. The redox proteome. J Biol Chem. 2013;288(37):26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler JR, Gray HB. Long-range electron tunneling. J Am Chem Soc. 2014;136(8):2930–2939. doi: 10.1021/ja500215j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler JR, Gray HB. 2015. Could tyrosine and tryptophan serve multiple roles in biological redox processes? Philos Trans R Soc A 373(2037):20140178.
- 7.Winkler JR, Gray HB. Electron flow through metalloproteins. Chem Rev. 2014;114(7):3369–3380. doi: 10.1021/cr4004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim Biophys Acta. 1985;811(3):265–322. [Google Scholar]
- 9.Dempsey JL, Winkler JR, Gray HB. Proton-coupled electron flow in protein redox machines. Chem Rev. 2010;110(12):7024–7039. doi: 10.1021/cr100182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migliore A, Polizzi NF, Therien MJ, Beratan DN. Biochemistry and theory of proton-coupled electron transfer. Chem Rev. 2014;114(7):3381–3465. doi: 10.1021/cr4006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammes-Schiffer S, Stuchebrukhov AA. Theory of coupled electron and proton transfer reactions. Chem Rev. 2010;110(12):6939–6960. doi: 10.1021/cr1001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beratan DN, Onuchic JN, Winkler JR, Gray HB. Electron-tunneling pathways in proteins. Science. 1992;258(5089):1740–1741. doi: 10.1126/science.1334572. [DOI] [PubMed] [Google Scholar]
- 13.Shih C, et al. Tryptophan-accelerated electron flow through proteins. Science. 2008;320(5884):1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 14.Warren JJ, Ener ME, Vlček A, Jr, Winkler JR, Gray HB. Electron hopping through proteins. Coord Chem Rev. 2012;256(21-22):2478–2487. doi: 10.1016/j.ccr.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verkhovskaya ML, Belevich N, Euro L, Wikström M, Verkhovsky MI. Real-time electron transfer in respiratory complex I. Proc Natl Acad Sci USA. 2008;105(10):3763–3767. doi: 10.1073/pnas.0711249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roessler MM, et al. Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance. Proc Natl Acad Sci USA. 2010;107(5):1930–1935. doi: 10.1073/pnas.0908050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkhovskaya M, Bloch DA. Energy-converting respiratory Complex I: On the way to the molecular mechanism of the proton pump. Int J Biochem Cell Biol. 2013;45(2):491–511. doi: 10.1016/j.biocel.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Close DM. Calculated vertical ionization energies of the common α-amino acids in the gas phase and in solution. J Phys Chem A. 2011;115(13):2900–2912. doi: 10.1021/jp200503z. [DOI] [PubMed] [Google Scholar]
- 19.Rinalducci S, Murgiano L, Zolla L. Redox proteomics: Basic principles and future perspectives for the detection of protein oxidation in plants. J Exp Bot. 2008;59(14):3781–3801. doi: 10.1093/jxb/ern252. [DOI] [PubMed] [Google Scholar]
- 20.Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc Chem Res. 2013;46(11):2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brettel K, Byrdin M. Reaction mechanisms of DNA photolyase. Curr Opin Struct Biol. 2010;20(6):693–701. doi: 10.1016/j.sbi.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Jiang N, et al. Distance-independent charge recombination kinetics in cytochrome c-cytochrome c peroxidase complexes: Compensating changes in the electronic coupling and reorganization energies. J Phys Chem B. 2013;117(31):9129–9141. doi: 10.1021/jp401551t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu MA, et al. Two tyrosyl radicals stabilize high oxidation states in cytochrome C oxidase for efficient energy conservation and proton translocation. J Am Chem Soc. 2012;134(10):4753–4761. doi: 10.1021/ja210535w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlin S, Bucher P. Correlation analysis of amino acid usage in protein classes. Proc Natl Acad Sci USA. 1992;89(24):12165–12169. doi: 10.1073/pnas.89.24.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlin S, Zuker M, Brocchieri L. Measuring residue associations in protein structures. Possible implications for protein folding. J Mol Biol. 1994;239(2):227–248. doi: 10.1006/jmbi.1994.1365. [DOI] [PubMed] [Google Scholar]
- 26.Burley SK, Petsko GA. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science. 1985;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 27.Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG. Aromatic-aromatic interactions in proteins: Beyond the dimer. J Chem Inf Model. 2011;51(7):1623–1633. doi: 10.1021/ci200062e. [DOI] [PubMed] [Google Scholar]
- 28.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costentin C, Louault C, Robert M, Savéant JM. The electrochemical approach to concerted proton--electron transfers in the oxidation of phenols in water. Proc Natl Acad Sci USA. 2009;106(43):18143–18148. doi: 10.1073/pnas.0910065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muramoto K, et al. Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate. Proc Natl Acad Sci USA. 2010;107(17):7740–7745. doi: 10.1073/pnas.0910410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strange RW, et al. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J Mol Biol. 2003;328(4):877–891. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 32.Antonyuk SV, Strange RW, Marklund SL, Hasnain SS. The structure of human extracellular copper-zinc superoxide dismutase at 1.7 A resolution: Insights into heparin and collagen binding. J Mol Biol. 2009;388(2):310–326. doi: 10.1016/j.jmb.2009.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Borgstahl GEO, et al. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71(1):107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- 34.Perry JJP, et al. Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis. Biochemistry. 2009;48(15):3417–3424. doi: 10.1021/bi8023288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93(26):15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marí M, Morales A, Colell A, García-Ruiz C, Fernández-Checa JC. Mitochondrial glutathione, a key survival antioxidant. Antioxid Redox Signal. 2009;11(11):2685–2700. doi: 10.1089/ars.2009.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strushkevich N, et al. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc Natl Acad Sci USA. 2011;108(25):10139–10143. doi: 10.1073/pnas.1019441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mast N, et al. Structural basis for three-step sequential catalysis by the cholesterol side chain cleavage enzyme CYP11A1. J Biol Chem. 2011;286(7):5607–5613. doi: 10.1074/jbc.M110.188433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davydov R, Gilep AA, Strushkevich NV, Usanov SA, Hoffman BM. Compound I is the reactive intermediate in the first monooxygenation step during conversion of cholesterol to pregnenolone by cytochrome P450scc: EPR/ENDOR/cryoreduction/annealing studies. J Am Chem Soc. 2012;134(41):17149–17156. doi: 10.1021/ja3067226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Bosch I, Sharma SK, Karlin KD. A selective stepwise heme oxygenase model system: An iron(IV)-oxo porphyrin π-cation radical leads to a verdoheme-type compound via an isoporphyrin intermediate. J Am Chem Soc. 2013;135(44):16248–16251. doi: 10.1021/ja405739m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanukoglu I, Rapoport R, Weiner L, Sklan D. Electron leakage from the mitochondrial NADPH-adrenodoxin reductase-adrenodoxin-P450scc (cholesterol side chain cleavage) system. Arch Biochem Biophys. 1993;305(2):489–498. doi: 10.1006/abbi.1993.1452. [DOI] [PubMed] [Google Scholar]
- 43.Rittle J, Green MT. Cytochrome P450 compound I: Capture, characterization, and C-H bond activation kinetics. Science. 2010;330(6006):933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 44.Yosca TH, et al. Iron(IV)hydroxide pK(a) and the role of thiolate ligation in C-H bond activation by cytochrome P450. Science. 2013;342(6160):825–829. doi: 10.1126/science.1244373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams ST, et al. Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics. 2014;9(12):1596–1603. doi: 10.4161/15592294.2014.983381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenzweig AC, et al. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): Implications for substrate gating and component interactions. Proteins. 1997;29(2):141–152. [PubMed] [Google Scholar]
- 47.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 48.Vyas NK. Atomic features of protein-carbohydrate interactions. Curr Opin Struct Biol. 1991;1(5):732–740. [Google Scholar]
- 49.Payne CM, et al. Multiple functions of aromatic-carbohydrate interactions in a processive cellulase examined with molecular simulation. J Biol Chem. 2011;286(47):41028–41035. doi: 10.1074/jbc.M111.297713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asensio JL, Ardá A, Cañada FJ, Jiménez-Barbero J. Carbohydrate-aromatic interactions. Acc Chem Res. 2013;46(4):946–954. doi: 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
- 51.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.