Abstract
Sterility testing as described in the pharmacopoeia compendia requires a 14-day incubation period to obtain an analytical result. Alternative methods that could be applied to evaluating product sterility are especially interesting due to the possibility of reducing this incubation period and thus the associated costs. The aims of this study were to evaluate the performance of the BacT/ALERTR 3D system in detecting microorganisms in large-volume parenteral solutions that were intentionally contaminated and to compare this system to pharmacopoeia sterility testing using the membrane filtration method. The results indicated that there were no significant differences between the methods regarding the ability to detect microbial contamination; however, detection with the BacT/ALERTR 3D system was faster compared to the pharmacopoeia method. Therefore, the BacT/ALERTR 3D system is a viable alternative for assessing the sterility of injectable products.
Keywords: sterility testing, BacT/ALERTR 3D, rapid microbiological method, large-volume parenteral solution
Introduction
Sterility is defined as the complete absence of viable microorganisms capable of developing and multiplying under favorable conditions. According to the pharmacopoeia compendia, the sterility of pharmaceutical products should be confirmed by subjecting a representative sample of the product to sterility testing; the product is considered to be non-sterile when microbial growth is detected during the test (Baird, 2004; Denyer and Baird, 2007; Pinto et al., 2010; Sandle, 2012).
Sterility testing is one of the oldest and most well-established microbiological tests. Sterility testing was first described in 1932 in the British Pharmacopeia and consisted of directly transferring the product to liquid culture media. It was only in the 1960s that membrane filtration methods were introduced in pharmacopoeia compendia (Bowman, 1969; Van Doorne et al., 1998). Since 1932, pharmacopoeia sterility testing has been used with only a few minor modifications, which primarily consisted of using culture medium to detect anaerobic microorganisms (in 1947, at USP XIII) (Bowman, 1969), using a single culture medium to detect both bacteria and fungi (in 1970, at USP XVIII) (Bowman, 1971; Marshall, 1998) and the introduction of the membrane filtration method (Bowman, 1969).
In membrane filtration sterility testing, microorganisms are collected on the surface of a 0.45-μm filter membrane by passing the product through the membrane, which is then divided into two parts; each part is then transferred to the appropriate culture medium. Fluid thioglycollate medium (FTM) is used because of its ability to support the growth of aerobic and anaerobic microorganisms, whereas tryptic soy broth (TSB) supports the growth of a broad range of aerobic microorganisms. The incubation time is 14 days at 30–35 °C for FTM and 20–25 °C for TSB, which allows for the growth of microorganisms that may be slow growing because they are in a dormant state or injured from the exposure to extreme environmental conditions during the manufacturing process. The results of the sterility testing are determined by a visual examination of the culture medium, which is generally clear and transparent (ANVISA, 2010).
The availability of a rapid sterility test could facilitate more rapid decision making for the implementation of corrective and preventive actions to ensure product quality and reduce the time and costs for the production of sterile products. The development of rapid microbiological methods for the isolation and detection of microorganisms has led to studies evaluating the suitability of these methods for use in sterility testing (Moldenhauer and Sutton, 2004; Kielpinski et al., 2005; PDA, 2013; USP, 2013). The BacT/ALERTR 3D system is one of these new technologies, in which microbial detection is based on measurements of biochemical or physiological parameters that reflect growth in liquid medium, such as the detection of CO2 production using colorimetric methods or by measuring changes in the headspace pressure. This is a fully automated system capable of incubating, shaking and continuously monitoring the culture media, in which readings are taken every 10 min throughout the incubation period, and in 2004, this system received FDA approval for use in evaluating the sterility of the short half-life products used in cell therapy, namely, Carticel (Thorpe et al., 1990; Kielpinski et al., 2005).
The aims of this study were to evaluate the microbial detection potential of the BacT/ALERTR 3D system compared to pharmacopoeia membrane filtration sterility testing and to determine the time required for detection.
Materials and Methods
Microbiological methods
The BacT/ALERTR 3D system (BioMerieux, France) was evaluated by comparing it to the membrane filtration sterility testing method, as recommended by the Brazilian Pharmacopoeia, 5th edition (ANVISA, 2010), regarding the detection of the microorganisms used to intentionally contaminate the following three matrices: 0.9% sodium chloride solution (P1), concentrated polyelectrolytes used in dialysis (P2) and a metronidazole solution (P3).
Each of the samples was filtered onto a cellulose nitrate filter having a nominal pore size not greater than 0.45 μm and a diameter of approximately 50 mm, under aseptic conditions. After filtration of the samples, the membrane was washed three times by filtering 100 mL of Fluid A through it; then, the membrane was cut aseptically into two equal parts, and one half was transferred to conventional culture media.
Culture media and reagents
For the pharmacopoeia sterility testing, FTM with an indicator, TSB and Fluid A, supplied by BioMerieux (France), were used in 100-mL flasks.
For the BactTAlertR 3D, BacT/ALERTR SA (for aerobes) and BacT/ALERTR SN (for anaerobes) media, supplied by BioMerieux (France), were used in 40-mL flasks.
Incubation conditions
The flasks with the thioglycollate medium were incubated at 32.5 ± 2.5 °C, and the flasks with TSB were incubated at 22.5 ± 2.5 °C. Both types of culture media were evaluated daily for 14 days to detect the presence of microbial growth.
For the BactTAlertR 3D system, the flasks were incubated at 32.5 ± 2.5 °C for 14 days, and automatic readings were taken every 10 min.
Strains
The following strains of microorganisms were used as indicated in the pharmacopoeia compendia for evaluating the growth-promoting capacity of culture media used in sterility testing, all of which were in the form of bioballR SingleShots (BioMerieux, France) containing 30 cfu/unit: Staphylococcus aureus (ATCC 6538), Bacillus subtilis (ATCC 6633), Pseudomonas aeruginosa (ATCC 9027), Clostridium sporogenes (ATCC 19404), Candida albicans (ATCC 10231) and Aspergillus brasiliensis (ATCC 16404).
To confirm the microbial load, eight bioball RSingleShot units were tested; four units were used on each of two different days. Each bioball R SingleShot unit was dissolved with a 0.9% saline solution and inoculated onto an agar surface. The tryptic soy agar (TSA) plates for Staphylococcus aureus, Bacillus subtilis and Pseudomonas aeruginosa were incubated at 32.5 ± 2.5°C for 72 h. The TSA plates for Candida albicansand the dichloran rose bengal chloramphenicol agar (DRBC) plates for Aspergillus brasiliensis were incubated at 22.5 ± 2.5 °C for 5 days. The blood agar plates for Clostridium sporogeneswere incubated at 32.5 ± 2.5 °C for 72 h under anaerobic conditions.
After confirming the microbial load contained in each type of bioball R SingleShot, suspensions were prepared at three different concentrations (20 cfu/mL, 2 cfu/mL and 0.2 cfu/mL), which were used to intentionally contaminate the different matrices.
Experimental design
Phase 1: Proof of concept (PoC)
The validation of the performance of the BacT/ALERTR 3D system began with a proof-of-concept experiment designed to test the potential of the method to detect microbial contamination under the experimental conditions and to assess the possible differences between the alternative and pharmacopoeia methods.
For each microorganism, three units of each product were artificially contaminated with 1 mL of a microbial suspension containing a load of 20 cfu/mL and subjected to membrane filtration sterility testing. After 18 h of incubation, 10-mL aliquots of the conventional culture medium were transferred to the BacT/ALERTR 3D culture media.
Negative controls consisting of filtrations of each of the matrices that had not been intentionally contaminated were included in all of the assays. Negative controls of the culture media were also used to confirm the sterility of these reagents. Positive controls of the inocula were used to confirm their viability and ability to grow in the culture media.
All of the flasks containing culture media, both for the conventional and the BacT/ALERTR 3D methods, that had microbial growth were subjected to Gram staining and subcultured to identify the microorganism.
Phase 2: Performance evaluation
After defining the conditions for the use of the BacT/ALERTR 3D in Phase 1, units of each product were artificially contaminated with 1 mL of a suspension containing 2 cfu, and the same number of units were also contaminated with 1 mL of a suspension containing 0.2 cfu. All of the units underwent membrane filtration sterility testing, and after an 18-h incubation, 10-mL aliquots of the conventional culture media were transferred to the BacT/ALERTR 3D culture media.
Negative controls consisting of filtrations of each of the matrices that had not been intentionally contaminated were included in all of the assays. Negative controls of the culture media were also used to confirm their sterility. Positive controls of the inocula were used to confirm their viability and ability to grow in the culture media.
All of the flasks containing culture medium, both for the conventional and the BacT/ALERTR 3D methods, that had microbial growth were subjected to Gram staining and subcultured to identify the microorganism.
Results and Discussion
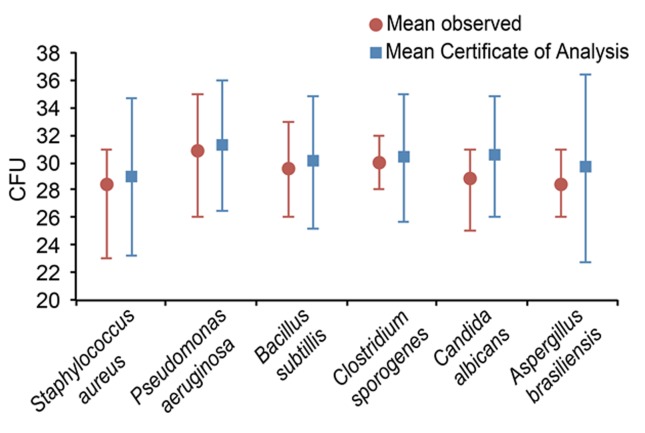
Figure 1 depicts the data from the tests used to confirm the microbial load contained in each type of bioball R SingleShot. The bioball R SingleShots of Candida albicans and Aspergillus brasiliensis had mean microbial loads that were 5.58% and 4.14% lower, respectively, than those stated on the Certificates of Analysis (CoAs), whereas the differences in the microbial loads for the other microorganisms were less than 2%.
Figure 1. Microbial loads in the assays confirming the microbial load of the bioball R SingleShots.
Despite the differences in the microbial loads compared to the means reported on the CoAs, all the different types of bioball R SingleShots were suitable for use in these experiments, especially considering the variability allowed in estimating microbial populations (± 0.5 logs for fungi and ± 0.3 logs for bacteria) (PDA, 2013). In the case of Candida albicans and Aspergillus brasiliensis, the variabilities observed were 0.2 logs and 0.1 logs, respectively.
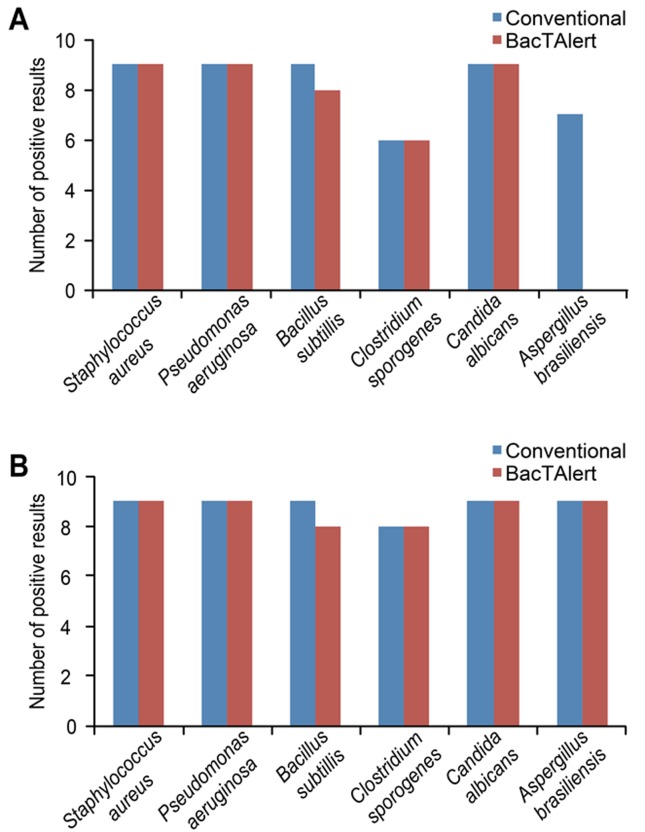
In the first phase of the experiment (PoC), the level of agreement between the methodologies applied to detect microbial growth was 75.9% (41 positive results in the 54 trials performed). As shown in Figure 2A, we observed conflicting results in all the trials where Aspergillus brasiliensis was used, with positive results using the conventional method and negative results using the BacT/ALERTR 3D. These differences were likely caused by problems in recovering the microorganism due to the dispersion of the hyphae in the conventional culture medium (100 mL), which interfered with our ability to collect a homogeneous aliquot for transfer to the BacT/ALERTR 3D medium. Therefore, for specifically assaying Aspergillus brasiliensis, a pre-incubation step was performed in 20 mL of TSB. We also noted a lack of recovery of Clostridium sporogenes in both methods when this microbe was inoculated into the metronidazole solution, indicating that the experimental conditions were not sufficient to neutralize the bacteriostatic effect of the metronidazole on the anaerobic microorganism. Therefore, the analytical procedure required a modification that increased the number of membrane filter washes to eliminate residual metronidazole prior to introducing the inoculum. When the tests were repeated under these new conditions, the level of agreement was 96.3% for the trials performed (Figure 2B).
Figure 2. Distribution of the positive results obtained for the 20-cfu contamination level according to the methodology employed, before (A) and after (B) modification of the analytical procedure.
A chi-square test was used to evaluate the sensitivity of the BacT/ALERTR3D system; the number of positive cultures detected with the BacT/ALERTR3D system was compared to the number detected using the conventional method at this level of intentional contamination (20 cfu). The difference between the abilities of these two methods to detect microbial contamination was not significant (p > 0.05), similar to results reported in other studies that compared the performance of the BacT/ALERTR 3D system in detecting microbial contamination (Thorpe et al., 1990; Khuu et al., 2004; Akcam et al., 2006; Parveen et al., 2011; Yonetani et al., 2012).
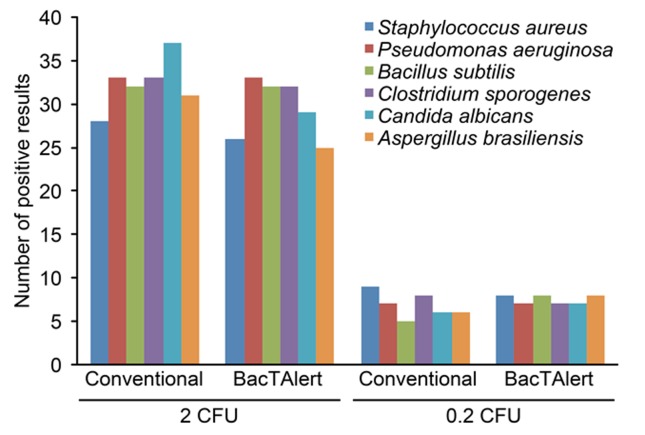
In the second phase of the experiment, the additional levels of intentional contamination, 2 cfu and 0.2 cfu, were considered. Figure 3 shows that the absolute number of contaminated samples detected using these two methods decreased as the microbial load decreased.
Figure 3. Distribution of the positive results for each level of contamination according to the methodology employed.
For these levels of contamination, the chi-square test was used to evaluate whether the BacT/ALERTR 3D system was equivalent to the conventional method. There were no significant differences between the two methodologies at a significance level of 5% (p > 0.05), thus indicating that these two methods have equivalent sensitivities.
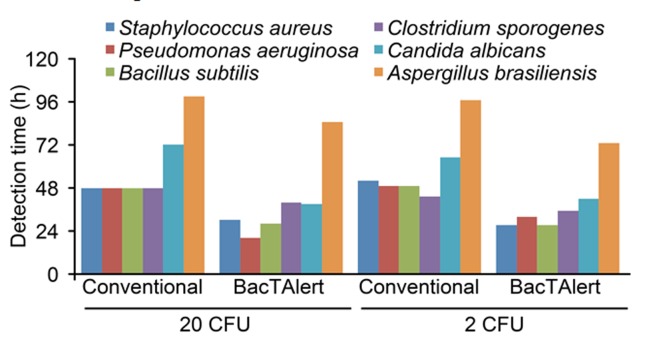
Figure 4 displays the mean time required to detect microbial growth using the conventional method and the BacT/ALERTR3D system at contamination levels of 20 cfu and 2 cfu, wherein for the BacT/ALERTR 3D system, the time required to detect growth included the 18-h incubation before an aliquot was transferred to its culture medium. The data obtained for the 0.2-cfu contamination level were not used to evaluate the time of detection because approximately 84.4% of the assays had no microbial growth at this level of contamination.
Figure 4. Mean time required to detect microbial growth for each of the methodologies employed.
We performed an analysis of variance (ANOVA) to examine the differences in the time required to detect microbial growth and found that the BacT/ALERTR 3D system required significantly shorter mean times compared to the conventional sterility testing for both contamination levels (p < 0.05).
The results of this study indicate that there are no significant differences between the abilities of these methods to detect bacterial contamination. However, the BacT/ALERTR 3D system detected microbial growth more rapidly than the pharmacopoeia method, which indicates superior performance, especially considering that the BacT/ALERTR 3D system uses an incubation temperature of 32.5 °C to detect all of the microorganisms evaluated, whereas the conventional method requires incubation temperatures of between 20 °C and 25 °C to detect molds and yeasts. In addition, the BacT/ALERTR 3D system detected fungi in a significantly shorter time than the conventional method, consistent with other studies (Thorpe et al., 1990; Weinstein et al., 1995; Bourbeau et al., 1998; Vigano et al., 2002; Khuu et al., 2004; Kielpinski et al., 2005; Parveen et al., 2011; Ericson et al., 2012).
Despite the limitations of sterility testing in demonstrating the absolute sterility of a product, the conventional method has adequately ensured the sterility of products. However, a method that can generate results more quickly with the same degree of sterility assurance as the conventional method is highly desirable. The results obtained in this study indicate that the BacT/ALERTR 3D system can be an alternative method for assessing the sterility of injectable products, similar to the procedures already performed with other products (Khuu et al., 2004; Kielpinski et al., 2005; Parveen et al., 2011).
Nevertheless, collaborative studies between industry, academia and regulatory agencies should be conducted to ensure that replacements for the conventional methodologies are adequate. These studies should not be limited to only comparing the performance of the methods but should also take into consideration the volume of samples that can be evaluated, the systems ease of operation, the risk factors for accidental contamination during execution, the necessary technical qualifications, the cost of reagents, the availability of technical support.
Acknowledgments
The authors would like to thank FAPESP for funding the project and BioMérieux for providing the BacT/ALERTR 3D system used in this study and for providing the reagents necessary for its execution.
References
- Agência Nacional de Vigilância Sanitária (ANVISA) Farmacopéia Brasileira. 5th ed. 2010. [Accessed 28 November 2013]. Available at: http://www.anvisa.gov.br/hotsite/farmacopeiabrasileira.
- Akcam FZ, Yayli G, Uskun E, et al. Evaluation of the Bactec microbial detection system for culturing miscellaneous sterile body fluids. Res Microbiol. 2006;157:433–436. doi: 10.1016/j.resmic.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Baird RM. Sterility assurance: concepts, methods and problems. In: Fraise AP, Lambert PA, Maillard JY, editors. Principle and Practice of Disinfection, Preservation and Sterilization. Blackwell Scientific Publications; Oxford: 2004. pp. 526–539. [Google Scholar]
- Bourbeau P, Riley J, Heiter BJ, et al. Use of BacT/Alert blood cultures system for culture of sterile body fluids other than blood. J Clin Microbiol. 1998;36:3273–3277. doi: 10.1128/jcm.36.11.3273-3277.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman FW. The sterility testing of pharmaceuticals. J Pharm Sci. 1969;58:1301–1307. doi: 10.1002/jps.2600581102. [DOI] [PubMed] [Google Scholar]
- Bowman FW, White M, Calhoum MP. Collaborative study of aerobic media for sterility testing by membrane filtration. J Pharm Sci. 1971;60:1087–1088. doi: 10.1002/jps.2600600721. [DOI] [PubMed] [Google Scholar]
- Denyer SP, Baird RM. Guide to Microbiology Control in Phamaceuticals and Medical Devices. Taylor & Francis Group; New York: 2007. [Google Scholar]
- Ericson EL, Klingspor L, Ulbberg M, et al. Clinical comparison of the Bactec Mycosis IC/F, BacT/Alert FA, and BacT/Alert FN blood cultures vials for the detection of candidemia. Diag Microbiol Infect Disease. 2012;73:153–156. doi: 10.1016/j.diagmicrobio.2012.02.020. [DOI] [PubMed] [Google Scholar]
- Khuu HM, Stock F, McGann M, et al. Comparison of automated culture systems with a CFR/USP-compliance method for sterility testing of cell-therapy products. Cytotherapy. 2004;6:183–195. doi: 10.1080/14653240410005997. [DOI] [PubMed] [Google Scholar]
- Kielpinski G, Prinzzi J, Duguid J, et al. Roadmap to approval: use of an automated sterility test method as a lot release test for CarticelR, autologous cultured chondrocytes. Chemotherapy. 2005;7:531–541. doi: 10.1080/14653240500361079. [DOI] [PubMed] [Google Scholar]
- Moldenhauer J, Sutton S. Towards an improved sterility test. PDA J Pharm Sci Technol. 2004;58:284–286. [PubMed] [Google Scholar]
- Parenteral Drug Association (PDA) Evaluation, Validation and Implementation of Alternative and Rapid Microbiological Methods. Parenteral Drug Association, Inc.; Bethesda, MD: 2013. Technical Report No. 33 (Revised 2013) [Google Scholar]
- Parveen S, Kaur S, David SAW, et al. Evaluation of growth based rapid microbiological methods for sterility testing of vaccines and other biological products. Vaccine. 2011;29:8012–8023. doi: 10.1016/j.vaccine.2011.08.055. [DOI] [PubMed] [Google Scholar]
- Pinto TJA, Kaneko TM, Pinto AA. Controle Biológico de Qualidade de Produtos Farmacêuticos, Correlatos e Cosméticos. Atheneu Editora; São Paulo: 2010. [Google Scholar]
- Sandle T. Sterility test failure investigations. J GXP Compliance. 2012;16:66–73. [Google Scholar]
- Thorpe TC, Wilson ML, Turner JE, et al. BacT/Alert: an automated colorimetric microbial detection system. J Clin Microbiol. 1990;28:1608–1612. doi: 10.1128/jcm.28.7.1608-1612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Pharmacopeia (USP) USP36/NF 31. The United States Pharmacopeial Convention; Rockville: 2013. [Google Scholar]
- Van Doorne H, Van Kampen BJ, Van der Lee RW, et al. Industrial manufacture of parenteral products in the Netherlands. A survey of eight years of media fills and sterility testing PDA. J Pharm Sci Technol. 1998;52:159–164. [PubMed] [Google Scholar]
- Vigano EF, Vasconi E, Agrappi C, et al. Use of simulated blood cultures for time to detection comparison between BacT/ALERTTM and BACTECTM 9240 blood culture systems. Diagn Microbiol Infect Disease. 2002;44:235–240. doi: 10.1016/s0732-8893(02)00451-0. [DOI] [PubMed] [Google Scholar]
- Weinstein MP, Mirret S, Reimer LG, et al. Controlled evaluation of BacT/Alert standard aerobic and FAN aerobic blood culture bottles for detection of bacteremia and fungemia. J Clin Microbiol. 1995;33:978–981. doi: 10.1128/jcm.33.4.978-981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani S, Okazaki M, Araki K, et al. Direct inoculation method using BacT/ALERT 3D and BD Phoenix System allows rapid and accurate identification and susceptibility testing for both Gram-positive cocci and Gram-negative rods in aerobic blood cultures. Diagn Microbiol Infect Disease. 2012;73:129–134. doi: 10.1016/j.diagmicrobio.2012.03.004. [DOI] [PubMed] [Google Scholar]