The cellular prion protein (PrPc), a component of desmosomes, contributes to the intestinal barrier function. A nuclear pool of PrPc interacts with γ-catenin; up-regulates the transcriptional activity of the β-catenin/TCF7L2 complex, the main effector of the canonical Wnt pathway; and promotes intestinal cell proliferation.
Abstract
We reported previously that the cellular prion protein (PrPc) is a component of desmosomes and contributes to the intestinal barrier function. We demonstrated also the presence of PrPc in the nucleus of proliferating intestinal epithelial cells. Here we sought to decipher the function of this nuclear pool. In human intestinal cancer cells Caco-2/TC7 and SW480 and normal crypt-like HIEC-6 cells, PrPc interacts, in cytoplasm and nucleus, with γ-catenin, one of its desmosomal partners, and with β-catenin and TCF7L2, effectors of the canonical Wnt pathway. PrPc up-regulates the transcriptional activity of the β-catenin/TCF7L2 complex, whereas γ-catenin down-regulates it. Silencing of PrPc results in the modulation of several Wnt target gene expressions in human cells, with different effects depending on their Wnt signaling status, and in mouse intestinal crypt cells in vivo. PrPc also interacts with the Hippo pathway effector YAP, suggesting that it may contribute to the regulation of gene transcription beyond the β-catenin/TCF7L2 complex. Finally, we demonstrate that PrPc is required for proper formation of intestinal organoids, indicating that it contributes to proliferation and survival of intestinal progenitors. In conclusion, PrPc must be considered as a new modulator of the Wnt signaling pathway in proliferating intestinal epithelial cells.
INTRODUCTION
The cellular prion protein (PrPc) has been studied essentially for its ability to undergo structural conversion into a pathogenic conformer, which is a key process for the onset of transmissible spongiform encephalopathies (Prusiner, 1998). Besides this role in prion diseases, PrPc is expressed in a wide range of tissues and cell types, where it has been shown to contribute to the regulation of many basic biological processes, including cell proliferation, differentiation, survival, and adhesion (Westergard et al., 2007). It participates in several specific functions in specialized tissues, such as neuroprotection, synaptic activity, olfaction, immune response, and epithelial and endothelial barrier function (Linden et al., 2008; Petit et al., 2013). PrPc was found to be secreted mostly as a glycosylphosphatidyl inositol–anchored glycoprotein associated with lipid raft microdomains of the plasma membrane (Vey et al., 1996; Naslavsky et al., 1997), where it combines with several protein partners, including signaling molecules (Mouillet-Richard et al., 2000; Morel et al., 2004; Santuccione et al., 2005). The ability of PrPc to modulate cell signaling was proposed to mediate at least some of its biological effects (for reviews, see Westergard et al., 2007; Linden et al., 2008). However, the complete repertoire of PrPc biological functions has not been determined, and analyses of different subcellular localizations of the protein, as well as the identification of its site-specific partners, are important for this goal.
Among the various extraneuronal tissues that express PrPc, previous studies from our group focused on intestine, the first site for infectious agent entry into the organism. In enterocytes, which represent the most abundant population of intestinal epithelial cells, we demonstrated that PrPc is addressed to cell–cell junctions (Morel et al., 2004), where it interacts with several desmosomal proteins (desmoglein-2, desmoplakin, γ-catenin [plakoglobin], and plakophilin-2) and participates in the structure of desmosomes (Morel et al., 2008). We showed that PrPc is required for the proper organization of adherens and tight junctions as well, thereby contributing to the intestinal barrier function (Petit et al., 2012). We reported also a shortening of intestinal villi in PrP-knockout mice (Morel et al., 2008), suggesting involvement of PrPc in intestinal epithelium homeostasis.
The intestinal epithelium undergoes rapid renewal throughout life (Stappenbeck et al., 1998), which requires continuous coordination between proliferation, differentiation, and death programs. Stem cells and dividing transit-amplifying cells migrate from the crypt to the villus, where most of the differentiated cells are located. In proliferating intestinal epithelial cells, that is, in the intestinal crypts in vivo or in dividing human Caco-2/TC7 enterocytes in culture, we demonstrated that a PrPc pool was unexpectedly localized in the nucleus (Morel et al., 2008). A nuclear localization of the prion protein was reported also in neuronal cells for both the protease-resistant PrP form in prion-infected cells (Mange et al., 2004) and the normal PrPc in noninfected cells (Hosokawa et al., 2008). A nuclear localization signal (NLS) was described in the N-terminal domain of the mature PrP. However, this sequence was not efficient in targeting green fluorescent protein toward the nuclear compartment (Jaegly et al., 1998), and the neurotoxic truncated 23-230 PrP was shown to be addressed to the nucleus independently of its NLSs (Crozet et al., 2006). Although the mechanisms leading to the nuclear targeting of normal or pathological prion proteins are unknown, PrPc was found to be associated with the lectin CBP70 in the nucleus of the NB4 human promyelocytic leukemia cell line (Rybner et al., 2002) and with chromatin (Mange et al., 2004), histone H3, H1-0, and lamin B1 in neuronal and β cells of the endocrine pancreas (Strom et al., 2011). The latter findings suggested a role of PrPc in transcriptional regulation, but PrPc function in the nucleus needs to be deciphered.
To attain this objective, we first characterized the nuclear partners of PrPc through a proteomic approach and identified γ-catenin, a component of desmosomes in differentiated cells, as a nuclear PrPc partner in proliferating intestinal epithelial cells. γ-Catenin is a protein of the catenin family displaying a dual junctional and nuclear localization (Aktary and Pasdar, 2012). γ-Catenin is homologous to β-catenin, the core player of the canonical Wnt pathway, which drives cell proliferation in intestinal crypts and is involved in intestinal homeostasis (Pinto et al., 2003; Clevers and Nusse, 2012). β-Catenin is localized at adherens junctions, where it interacts with the E-cadherin cytodomain. In the absence of Wnt stimulation, cytoplasmic β-catenin is phosphorylated by a destruction complex, which involves casein kinase 1, glycogen synthase kinase 3β, adenomatous polyposis coli, and axin, and is addressed to the proteasome for degradation. In response to Wnt activation, the destruction complex is disrupted, leading to the stabilization of the cytoplasmic β-catenin and its targeting to the nucleus, where it acts as a coactivator of the transcription factors of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family to regulate the expression of many target genes involved in a variety of cellular processes (Clevers and Nusse, 2012). Although γ-catenin is known to interact with the TCF/LEF transcription factors, its exact role in the Wnt pathway is much less documented than that of β-catenin and seems to vary among different biological systems (Aktary and Pasdar, 2012).
The identification of PrPc as a new γ-catenin nuclear partner led us to investigate whether it could interfere with the canonical Wnt pathway. Therefore, we analyzed whether PrPc interacts with the Wnt signaling effectors β-catenin and transcription factor 7–like 2 (TCF7L2; also known previously as TCF4), the major member of the TCF family in intestine, and studied how PrPc affects β-catenin/TCF7L2 transcriptional activity on the expression of Wnt target genes or on cell proliferation.
RESULTS
The nuclear PrPc interacts with γ-catenin in proliferating Caco-2/TC7 enterocytes
As a first approach to determine the role of PrPc when it is addressed to the nucleus, we undertook a proteomic analysis after immunoprecipitation of PrPc from nuclear extracts of exponentially growing Caco-2/TC7 cells, in which we previously demonstrated the nuclear localization of a PrPc pool (Morel et al., 2008). This analysis revealed that the partners of PrPc in the nucleus included desmoplakin and γ-catenin (plakoglobin), identified previously as PrPc desmosomal partners in differentiated enterocytes (Petit et al., 2012), along with desmoglein 1 preproprotein, glyceraldehyde-3-phosphate dehydrogenase, squamous cell carcinoma antigen (SCCA2/SCCA1), and γ-actin (Table 1).
TABLE 1:
Proteomic analysis of nuclear PrPc partners.
| Identified protein | Accession number | Mr | Peptide matches | Coverage (%) |
|---|---|---|---|---|
| Desmoplakin I | gi 1147814 | 331,776 | 9 | 3.7 |
| γ-Catenin | gi 33875446 | 85,651 | 6 | 9.4 |
| Squamous cell carcinoma antigen | gi 239552 | 44,534 | 5 | 16.7 |
| Actin, γ1 propeptide | gi 4501887 | 41,792 | 5 | 19.5 |
| Desmoglein 1 preproprotein | gi 119703744 | 113,747 | 4 | 6.1 |
| Glyceraldehyde-3-phosphate dehydrogenase | gi 31645 | 36,054 | 4 | 16.4 |
| SCCA2/SCCA1 fusion protein isoform 1 | gi 33317676 | 44,648 | 4 | 12.6 |
Nuclear extracts from proliferative Caco-2/TC7 cells (2 d) were immunoprecipitated with anti-PrPc antibodies. The presence of PrPc in the resulting material was checked by Western blot before identification of the interacting proteins by liquid chromatography-MS/MS. The number of peptide matches obtained after trypsination for each protein, as well as the accession number (National Center for Biotechnology Information) and the molecular weight (Mr), are reported.
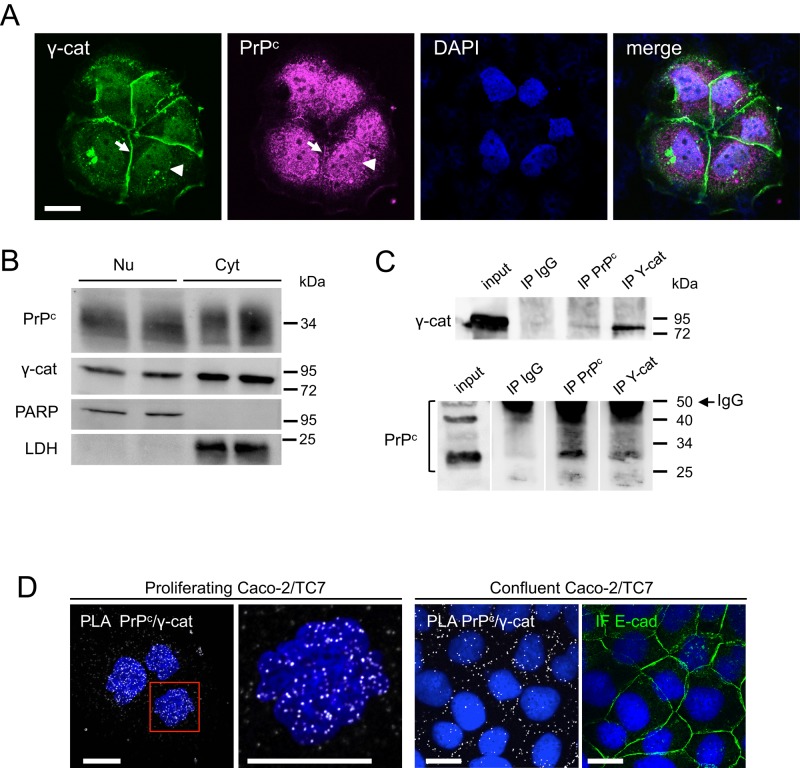
We further focused on the interaction between PrPc and γ-catenin. The nuclear colocalization of PrPc and γ-catenin was confirmed by confocal microscopy in exponentially growing Caco-2/TC7 cell clusters (Figure 1A). Both proteins were detected in the nucleus (arrowheads) and in the cytoplasm. Note that a portion of γ-catenin was also localized at cell–cell contacts, as was PrPc (arrows), but with a much lower intensity, suggesting that PrPc needs better maturation of cell–cell junctions than γ-catenin to be targeted to the membrane.
FIGURE 1:
Nuclear localization of PrPc and γ-catenin and interaction between the two proteins in proliferating Caco-2/TC7 cells. (A) Confocal analysis of exponentially growing Caco-2/TC7 cells after immunofluorescence labeling of γ-catenin (green) and PrPc (magenta). Nuclei were stained by DAPI. Arrowheads, nuclear labeling; arrows, junctional labeling. The merge image shows that both proteins colocalize partially in the nucleus and that PrPc is also abundant in perinuclear compartments. Bar, 20 μm. (B) Western blot analysis of nuclear (Nu) and cytoplasmic (Cyt) extracts of exponentially growing Caco-2/TC7 cells. A 30-μg amount of protein was analyzed for each fraction in duplicate. PARP and LDH were used to check the purity of nuclear and cytoplasmic fractions. (C) Nuclear extracts were immunoprecipitated with anti-PrPc or anti–γ-catenin antibodies or with nonimmune IgG. Immunoprecipitated fractions (IP) and nuclear extracts (input) were analyzed by Western blot for γ-catenin (top) or PrPc (bottom). Molecular weight markers are shown on the right. In the bottom, IgG is detected for IP samples even though the membrane was cut at the level of the 50-kDa marker to avoid excessive trapping of the antibody. Note that the nonglycosylated, monoglycosylated, or diglycosylated forms of PrPc are clearly separated on this blot (whereas they may appear as a smear on other blots); a PrPc dimer (50 kDa) is also detected in the input line. (D) PLA showing in situ interaction between PrPc and γ-catenin (white spots) in the nucleus of proliferating Caco-2/TC7 cells (left and enlargement of the red square) and at cell–cell contacts in confluent cells. For confluent cells, immunofluorescence labeling of E-cadherin is shown in the same field. Nuclei were stained by DAPI. Bars, 10 μm.
PrPc and γ-catenin were detected in nuclear extracts by Western blot analysis (Figure 1B), and PrPc/γ-catenin interaction was shown by coimmunoprecipitation (Figure 1C). We further confirmed that these two proteins interact in the nucleus by using in situ proximity ligation assay (PLA) and confocal microscopy (Figure 1D). This approach showed that PrPc/γ-catenin interaction occurred mainly in the nucleus of proliferating Caco-2/TC7 cells. By contrast, this interaction was detected predominantly at cell–cell contacts in differentiated cells (Figure 1D), as observed also for the interaction between γ-catenin and another desmosomal protein, desmoplakin, as expected (unpublished data). The precision and reliability of this assay to localize interactions of endogenous proteins were assessed by the absence of PLA signal at cell–cell contacts between PrPc and the adherens junction–associated E-cadherin, which colocalize by immunofluorescence but do not interact, as we showed previously (Morel et al., 2004); in the same way, no PLA signal was detected in the nucleus between PrPc and the transcription factor HNF-4 α (Supplemental Figure S1).
Characterization of the expression and subcellular localization of PrPc, γ-catenin, and the Wnt effectors β-catenin and TCF7L2 (TCF4) in different intestinal cell lines
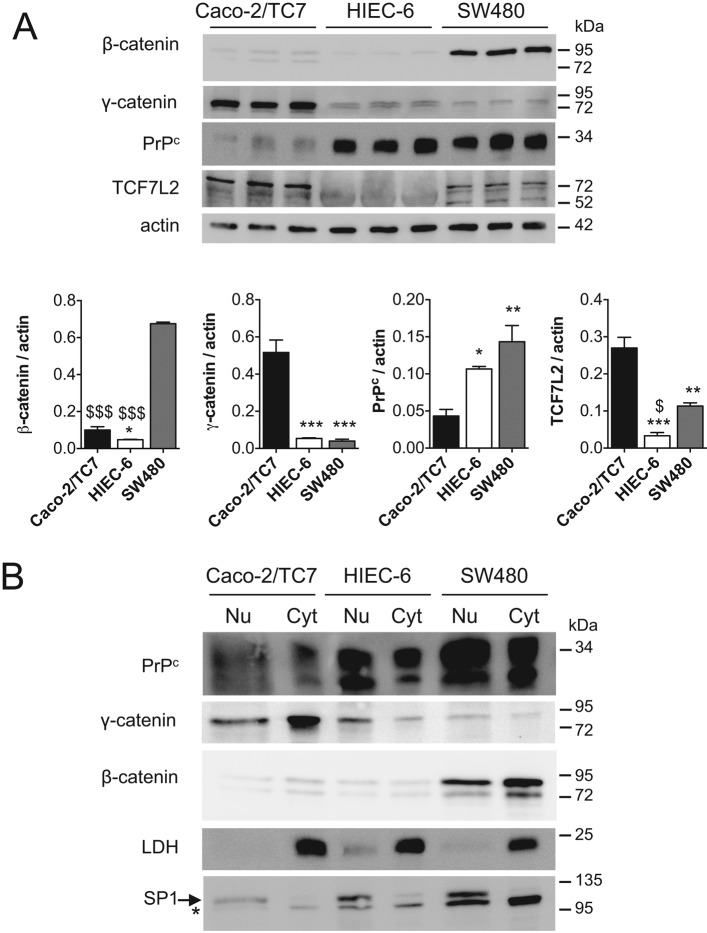
The identification of PrPc as a new γ-catenin nuclear partner led us to investigate the possible links between this complex and the canonical Wnt pathway. We therefore analyzed the expression of PrPc and γ-catenin, as well as that of β-catenin and TCF7L2, the major endpoint effectors of canonical Wnt signaling in intestine, during the exponential growth phase of several intestinal cell lines. With respect to their basal Wnt pathway activities, we selected three intestinal cell lines for study: the two cancer cell lines SW480, with a constitutively high activity of Wnt pathway, and Caco-2/TC7, in which Dickkopf-related protein 1 overexpression attenuates the constitutive Wnt signaling (Aguilera et al., 2007), and the nontumoral crypt-like human intestinal cells HIEC-6, with a weak basal activity of Wnt pathway (Guezguez et al., 2014). Consistent with Wnt pathway status, β-catenin protein level was much higher in SW480 cells than in the two other cell lines (Figure 2A). No correlation was observed between Wnt activity and PrPc protein levels, which were similar in HIEC-6 and SW480 cells and higher than in Caco-2/TC7 cells. Of interest, the highest γ-catenin level was detected in Caco-2/TC7 cells, which have the lowest β-catenin and PrPc levels (Figure 2A). TCF7L2 level was very low in HIEC-6 cells and twofold higher in Caco-2/TC7 than in SW480 cells. PrPc, γ-catenin, and β-catenin were present in the nuclear fraction of the three cell lines (Figure 2B). Confocal microscopy analyses confirmed the nuclear localization of the four proteins but highlighted differences in their subcellular partitioning between the cell lines. PrPc, γ-catenin, and β-catenin were visualized in the nucleus and the cytoplasm in both SW480 and HIEC-6 cells but with a much higher nuclear proportion in SW480 than in HIEC-6 cells (Figure 3). By contrast, even at this very low-density stage, Caco-2/TC7 cells exhibited partial junctional localization of PrPc, γ-catenin, and β-catenin, in accordance with the well-described ability of these cells to polarize, establish mature cell–cell junctions, and differentiate (Chantret et al., 1994), in contrast to the two other cell lines. The transcription factor TCF7L2 was concentrated mostly in the nucleus in SW480 and Caco-2/TC7 cells, as expected, but was much more diffusely distributed between the cytoplasm and the nucleus in HIEC-6 cells.
FIGURE 2:
Analysis of PrPc, γ-catenin, β-catenin, and TCF7L2 (TCF4) levels in proliferating Caco-2/TC7, HIEC-6 and SW480 cells. (A) Western blot analysis of total protein extracts from Caco-2/TC7, HIEC-6, and SW480 cells (2 d after plating). A 30-μg amount of protein was analyzed for PrPc, γ-catenin, β-catenin, and TCF7L2 levels in each cell line in triplicate, with actin as loading control. Bar graphs show the ratio of each protein to actin after densitometric analyses (mean ± SEM; *p < 0.05, **p < 0.01 ***p < 0.001 vs. Caco-2/TC7; $p < 0.05, $$$p < 0.001 vs. SW480). (B) Western blot analysis of nuclear (Nu) and cytoplasmic (Cyt) extracts of the three cell lines. A 30-μg amount of protein was analyzed for PrPc, γ-catenin, and β-catenin levels in each fraction and each cell line. Purity of nuclear and cytoplasmic fractions was checked by SP1 and LDH analyses; *nonspecific band revealed by the anti-SP1 antibody.
FIGURE 3:
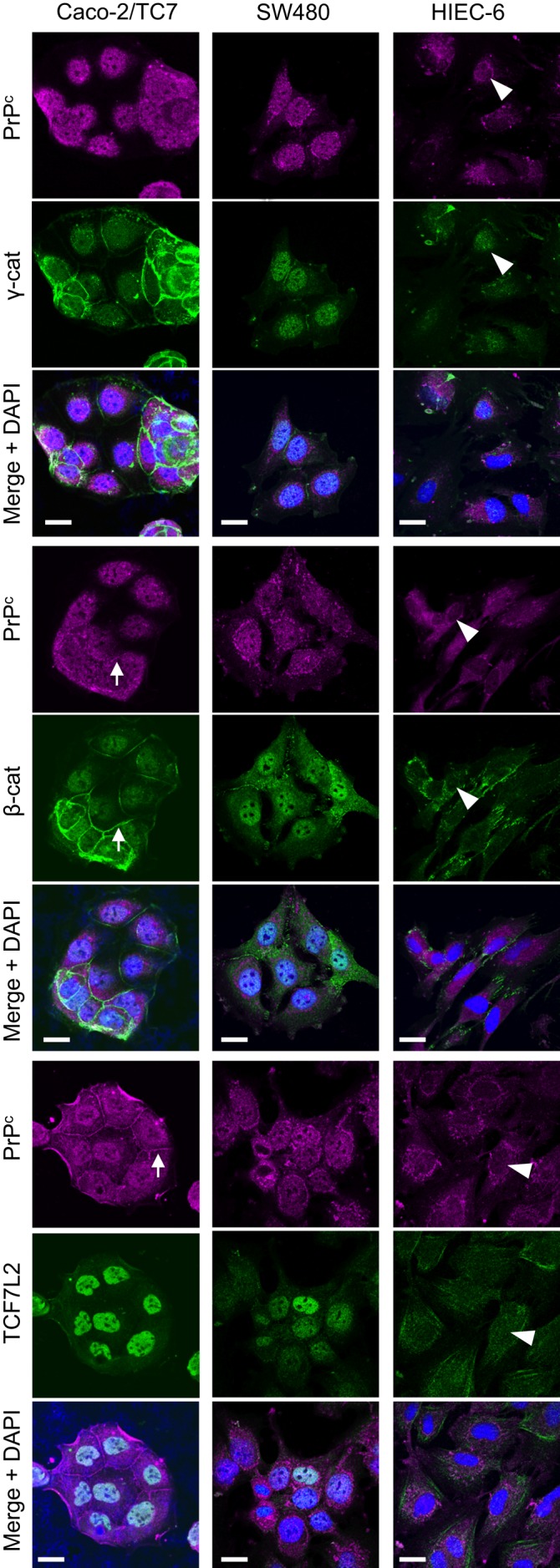
Subcellular distribution of PrPc, β-catenin, γ-catenin, and TCF7L2 in proliferating Caco-2/TC7, SW480, and HIEC-6 cells. Cells were immunolabeled for PrPc and γ-catenin (top three), PrPc and β-catenin (middle three), or PrPc and TCF7L2 (bottom three) and analyzed by confocal microscopy. Merge images with DAPI staining are shown for each labeling. Arrowheads show the faint nuclear labeling of all proteins in HIEC-6 cells, and arrows point out junctional labeling of PrPc, β-catenin, and γ-catenin in Caco-2/TC7 cells. Bars, 20 μm.
PrPc and γ-catenin interact with the Wnt effectors β-catenin and TCF7L2 in proliferating intestinal cells
γ-Catenin interacts with the transcription factor TCF7L2 (Miravet et al., 2002). We hypothesized that PrPc could thus participate in molecular complexes involving γ-catenin, β-catenin, and TCF7L2. The PLA approach was chosen to visualize protein interactions and their subcellular distribution in situ. In exponentially growing SW480, Caco-2/TC7, and HIEC-6 cells, PrPc interacts with γ-catenin, β-catenin, and TCF7L2 (Figure 4). γ-Catenin/TCF7L2 and β-catenin/TCF7L2 interactions were detected also, as expected, and the presence of γ-catenin/β-catenin interactions confirmed the existence of multipartner complexes reported previously (Miravet et al., 2002). In SW480 and Caco-2/TC7 cells, the PLA fluorescence spots were concentrated in the nucleus for all interactions, although they could be observed also in the cytoplasm. By contrast, but in agreement with the distribution of all partners as observed by immunofluorescence (Figure 3), interactions between PrPc/γ-catenin, PrPc/β-catenin, and γ-catenin/β-catenin occurred in much higher proportions in the cytoplasm than in the nucleus of HIEC-6 cells (Figure 4). In this cell line, complexes involving TCF7L2 were very rare, in accordance with its low level (Figures 2 and 3).
FIGURE 4:
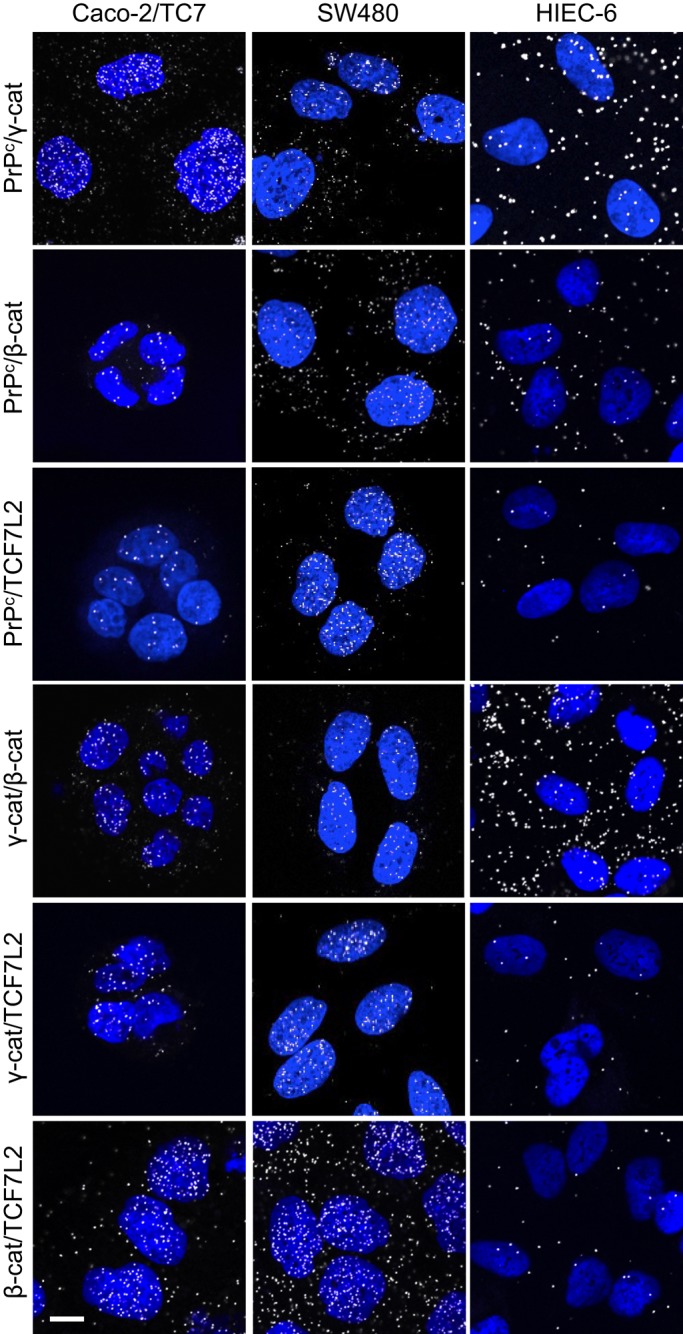
PrPc interacts with γ-catenin and the Wnt pathway effectors β-catenin and TCF7L2 in Caco-2/TC7, SW480, and HIEC-6 cells. PLA showing in situ interaction (white spots) between PrPc, γ-catenin, β-catenin, and TCF7L2 in proliferating cells for each cell line. PLA assays were performed between the pair of proteins indicated on the left. Nuclei were stained by DAPI. Note that interactions are detected mainly in the nucleus for Caco-2/TC7 and SW480 cells and mainly in the cytoplasm for HIEC-6 cells. Bar, 10 μm.
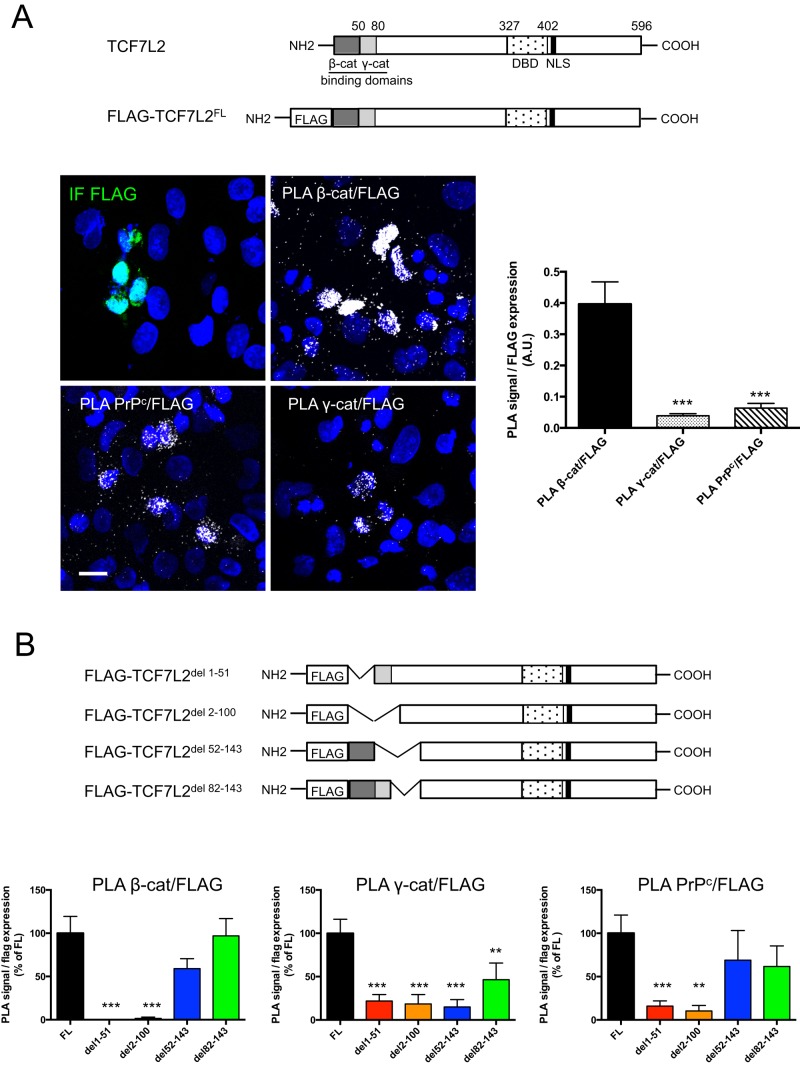
PrPc interaction with TCF7L2 depends mostly on its β-catenin binding domain
To determine whether PrPc interacts with TCF7L2 via β- or γ-catenin or independently of these proteins, we transfected SW480 cells with different fragments of a FLAG-TCF7L2 protein (Figure 5). Fragment expression was analyzed by immunofluorescence, and protein interactions were studied by PLA. Interactions of β-catenin, γ-catenin, and PrPc with the full-length FLAG-TCF7L2 were easily detected in transfected cells, although the PLA signal intensity (normalized to FLAG expression) was much higher for β-catenin/FLAG-TCF7L2 interaction than for γ-catenin/FLAG-TCF7L2 or PrPc/FLAG-TCF7L2 interactions (Figure 5A). These results are in accordance with the differences observed for interactions of each protein with the endogenous TCF7L2 (Figure 4). When FLAG-TCF7L2 constructs were deleted for the binding domains of β-catenin (del 1–51) or of both β- and γ-catenin (del 2–100), their interaction with β-catenin was completely lost (Figure 5B). This result is in agreement with the glutathione S-transferase pull-down approach, which was used previously to identify these binding sites (Miravet et al., 2002), thus demonstrating the accuracy of the PLA technique. The deletion of the catenin-binding domains decreased interactions of TCF7L2 with PrPc by 85 and 90% for del 1–51 and del 2–100 constructs, respectively, and with γ-catenin by 80% for both constructs. Interactions with β-catenin were restored partially for the construct deleted for the 52–143 fragment and totally for the construct deleted for the 82–143 fragment, as expected. Very similar results were obtained for interactions of PrPc with these constructs. By contrast, interactions with γ-catenin remained very low with the del 52–143 construct, as expected, and were only partially restored with the del 82–143 fragment, probably because this deletion affects the conformation of the γ-catenin– binding domain.
FIGURE 5:
PrPc interaction with the transcription factor TCF7L2 occurs mainly via the β-catenin–binding domain of TCF7L2. (A) Schematic diagram of the human TCF7L2 protein and the full-length TCF7L2 tagged with the FLAG octapeptide (FLAG-TCF7L2FL) construct. SW480 cells were transfected with FLAG-TCF7L2 FL. Transfected cells were immunolabeled using an anti-FLAG antibody (IF FLAG), and PLA was performed using anti-FLAG and anti–β-catenin (PLA β-cat/FLAG), anti–γ-catenin (PLA γ-cat/FLAG), or anti-PrPc antibodies (PLA PrPc/FLAG). Nuclei were stained by DAPI (bar, 20 μm). Graphs present the quantifications of PLA signal for each interaction normalized to values of IF FLAG signal, measured in at least 10 random fields (∼1000 cells/experiment) in two independent experiments (A.U., arbitrary units; mean ± SEM; ***p < 0.001 vs. PLA β-cat/FLAG). NLS, nuclear localization signal; DBD, DNA-binding domain. (B) Schematic diagram of the different FLAG-TCF7L2 deletion constructs. TCF7L2 lacking the β-catenin interaction domain (TCF7L2del 1-51), lacking both β- and γ-catenin interaction domains (TCF7L2del 2-100), lacking the 52–82 γ-catenin interaction domain (TCF7L2del 52-143), or deleted for a sequence adjacent to the γ-catenin interaction domain (TCF7L2del 82-143). Experiments were conducted as in A. Graphs present the quantifications of PLA signal normalized to values of IF FLAG signal for each construct. Measurements were performed as described in A. For each interaction, results obtained with the different constructs are presented as percentage of the value obtained with the full-length (FLAG-TCF7L2FL) construct (mean ± SEM; **p < 0.01 and ***p < 0.001 vs. FL).
Interactions between β-catenin or PrPc and TCF7L2 can occur in the cytoplasm, as shown by analyses of TCF7L2 mutants obtained by serial deletions of ∼100 amino acids from the C-terminal part of the protein comprising or not the NLS (Supplemental Figure S2). Interactions of TCF7L2 with both proteins were detected mostly in the nucleus with fragments 1–500 and 1–420, in the cytoplasm, and around the nucleus with the 1–307 fragment, which is devoid of NLS, but were concentrated again in the nucleus with the 1–201 fragment, which is small enough to diffuse inside this compartment (Supplemental Figure S2).
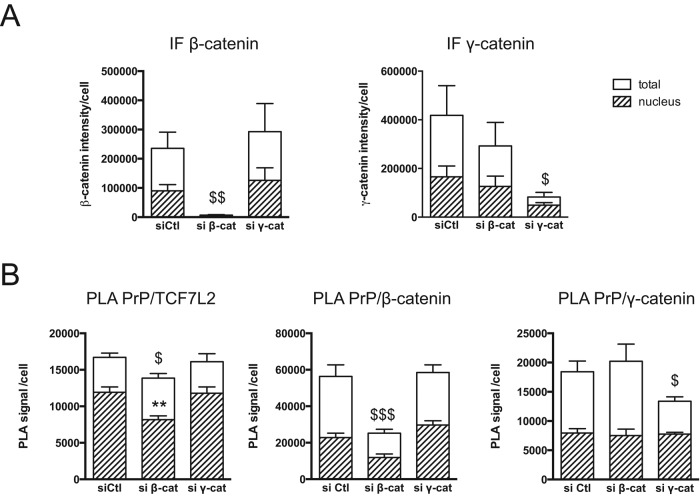
The foregoing results showed that PrPc interaction with TCF7L2 depends mostly on the β-catenin–binding domain. To examine further whether PrPc interacts with TCF7L2 through β-catenin, we studied the effect of β- or γ-catenin silencing by small interfering RNA (siRNA) on PrPc/TCF7L2 interactions in both nuclear and cytoplasmic compartments. On silencing of β-catenin, which was decreased by >90% in both cytoplasm and nucleus (Figure 6A), PrPc/TCF7L2 interactions were significantly decreased (Figure 6B). This loss of interactions affected the nucleus compartment (−25%), with no effect on cytoplasmic interactions (Figure 6B, compare hatched bars with white bars). However, this decrease of nuclear interactions was modest, most of them being maintained in the absence of β-catenin. γ-Catenin silencing (–80% for total level, –70% for nuclear level; Figure 6A) had no effect on PrPc/TCF7L2 interactions in the cytoplasm or the nucleus (Figure 6B). Moreover, the silencing of one catenin had no significant effect on the interaction of PrPc with the other catenin.
FIGURE 6:
Effect of β-catenin or γ-catenin silencing on PrPc/TCF7L2 interactions. SW480 cells were transfected with the indicated siRNAs and analyzed after 48 h. (A) Decrease of β-catenin or γ-catenin protein levels was evaluated by immunofluorescence labeling (IF). Graphs present the quantification of total (white bars) or nuclear (hatched bars) IF signal, measured in five random fields (∼300 cells; mean ± SEM; $p < 0.05 and $$p < 0.01 vs. siCtl for total signal). (B) Evaluation by PLA of the interactions of PrPc with TCF7L2, β-catenin, or γ-catenin. Graphs present the quantification of total (white bars) or nuclear (hatched bars) PLA signal per cell measured in at least 10 random fields (∼800 cells; mean ± SEM; $p < 0.05 and $$$p < 0.001 vs. siCtl for total PLA signal; **p < 0.01 vs. siCtl for nuclear signal).
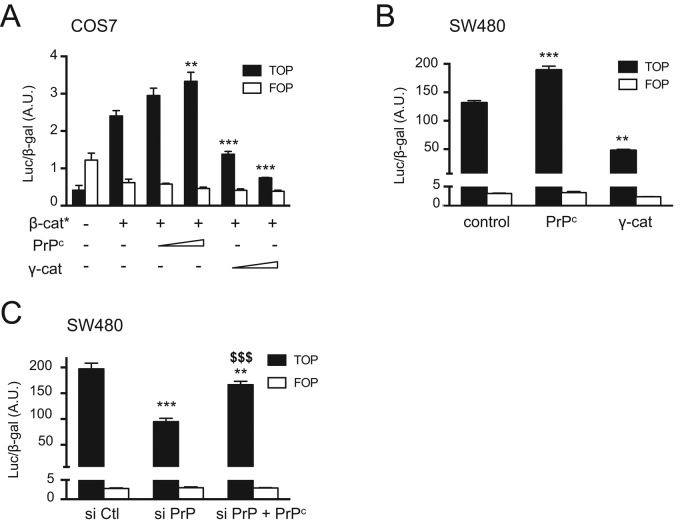
PrPc up-regulates the transcriptional activity of the β-catenin/TCF7L2 complex
We then addressed the functional role of PrPc in the Wnt signaling pathway, using a transcriptional reporter assay, and compared it to that of γ-catenin, which has been suggested to exert a negative effect on TCF7L2-mediated transcription (Miravet 2002). TOP or FOP reporter plasmids were cotransfected in COS7 cells together with a constitutively activated form of β-catenin and either PrPc or γ-catenin. PrPc increased the luciferase activity induced by β-catenin in a dose-dependent manner, whereas γ-catenin decreased it (Figure 7A). In SW480 cells, which have a constitutively high β-catenin level, we confirmed these opposite effects of PrPc and γ-catenin (Figure 7B). Positive regulation of the transcriptional activity of the β-catenin/TCF7L2 complex by PrPc was further demonstrated by decreased luciferase activity upon PrPc silencing by siRNA and its rescue upon cotransfection with mouse PrPc, which is not affected by the siRNA (Figure 7C; for siRNA efficiency, see Figure 8A and Supplemental Figure S3).
FIGURE 7:
The TCF7L2/β-catenin transcriptional activity is up-regulated by PrPc and down-regulated by γ-catenin. (A) COS7 cells were transfected with luciferase reporter plasmids containing TCF-binding sites (TOP) or mutated TCF-binding sites as negative control (FOP) and a LacZ expression plasmid as internal control. Expression vectors for constitutively active S33Y mutant of β-catenin (β-cat*), PrPc, or γ-catenin were cotransfected as indicated. Values represent mean ± SEM of luciferase activity normalized to corresponding β-Gal activity. **p < 0.01; ***p < 0.001 vs. β-cat* alone. (B) SW480 cells were transfected with the TOP/FOP reporters and LacZ plasmid and cotransfected with empty vector (control) or PrPc or γ-catenin expression plasmids as indicated. **p < 0.01, ***p < 0.001 vs. control. (C) SW480 cells were transfected with the TOP/FOP reporters and LacZ plasmid, together with a control siRNA (siCtl), PrPc siRNAs (siPrP), or PrPc siRNAs combined with a mouse PrPc expression vector (siPrP+PrPc). **p < 0.01, ***p < 0.001 vs. siCtl; $$$p < 0.001 vs. siPrP. Experiments were all performed in triplicate, and the graphs present one experiment representative of two or three independent experiments for each condition.
FIGURE 8:
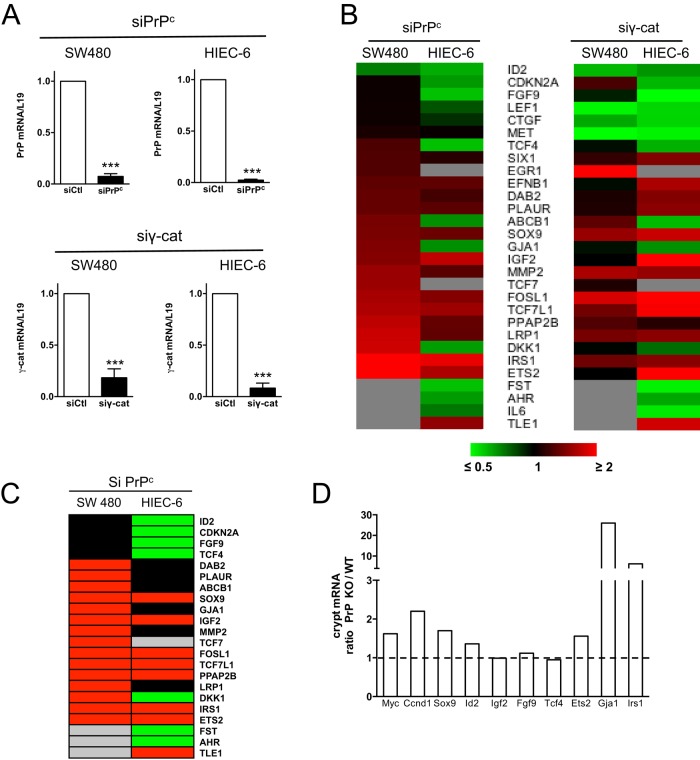
PrPc and γ-catenin silencing modulates the expression of several Wnt target genes with different effects in SW480 and HIEC-6 cells. (A) PrPc and γ-catenin knockdown in SW480 and HIEC-6 cells upon siRNA transfection. Decrease of PrPc or γ-catenin mRNA at 24 h after transfection with the corresponding pairs of siRNAs. mRNA levels were determined by RT-qPCR and normalized to L19 levels. For each cell line, results are normalized to mRNA levels in cells transfected with control siRNA, which were set at 1 (mean ± SEM from three independent experiments; ***p < 0.001 vs. cells transfected with control siRNA). (B) Heat maps illustrating the modifications of Wnt target gene expression 24 h after PrPc or γ-catenin siRNA transfection in each cell line (analysis of 84 Wnt target genes). Genes whose expression was modulated (≤0.7- or ≥1.4-fold) in at least one cell line by one siRNA were selected (mean of n = 3 independent experiments for each siRNA in each cell line; p < 0.05 for each gene in each cell line as compared with control siRNA). Genes were ranked according to the modulation factor of their expression in SW480 cells upon PrPc silencing. Gray indicates undetected expressions. Note that the TCF4-modulated gene appearing in the heat map (also known as ITF2 or SEF2) differs from TCF7L2 (widely known as TCF4). (C) Pseudo heat map after selection of the genes whose expression is increased ≥1.4-fold (red) or decreased ≥0.7-fold (green) after PrPc silencing in each cell line. Black, modulation factor between 0.7 and 1.4; gray, undetected expression. (D) Analysis of several Wnt target genes in the fraction of epithelial cells corresponding to the crypt bottom of wild-type (WT) and PrP-knockout (PrP KO) mice (pool of five mice for each genotype). mRNA levels were determined by RT-qPCR and normalized to cyclophilin levels. Results are expressed as the ratio of expression in PrPc-knockout vs. WT mice for each gene.
PrPc knockdown affects expression of Wnt and Hippo target genes
PCR array analyses were then performed to evaluate changes in the expression of a large panel of Wnt target genes in response to PrPc or γ-catenin silencing. Experiments were conducted on SW480 and HIEC-6 cells, which differ greatly in their Wnt pathway activity levels. Caco-2/TC7 cells were not analyzed because the presence of a desmosome-associated pool of PrPc and γ-catenin in these cells, even at very early stages of the culture, could render the results difficult to interpret. Efficiency of silencing by siRNA was shown by a net decrease of the corresponding mRNA levels (Figure 8A). Protein levels were reduced by 70–90% in both total cell lysates and nuclear extracts of SW480 or HIEC-6 cells from 24 h after transfection (Supplemental Figure S3). This processing time was then chosen for further analyses.
Among the 84 Wnt target genes that were analyzed, the expression of 29 was modulated in at least one cell line upon either PrPc or γ-catenin siRNA treatment (Figure 8B). Surprisingly, PrPc silencing in SW480 cells resulted in the increased expression of 15 genes (≥1.4-fold), with no decreased gene expression (≤0.7-fold; Figure 8C). The expression of half of these 15 genes was increased also upon PrPc silencing in HIEC-6 cells, in which basal Wnt activity is very low, but, in addition, the expression of seven genes was reduced in these cells (Figure 8C). The effect of γ-catenin silencing was similar to that of PrPc silencing for most of the genes whose expression was modulated in HIEC-6 cells but differed markedly from that of PrPc silencing in SW480 cells (Figure 8B). The classical transcriptional TCF7L2 targets, MYC, CCND1, and AXIN2, were not significantly modulated by PrPc or γ-catenin silencing, even though the expression of the three genes tended to increase when analyzed by quantitative PCR (qPCR) in SW480 cells (Supplemental Figure S4).
To determine whether PrPc could interfere with the expression of Wnt target genes in vivo, the mRNA level of eight genes whose expression was modulated in cell lines (Figure 8B), as well as that of Myc and Ccnd1, was analyzed in wild-type and PrP-knockout mice in the bottom of intestinal crypts, where high Wnt activity is observed in vivo (Figure 8D). The expression of seven genes was increased in crypt cells in the absence of PrPc. They include Myc and Ccnd1, whose expression was not significantly modulated in cell lines; Sox9, Ets2, Irs1, and Gja1, whose expression was increased in SW480 cells upon PrPc silencing; and also Id2, whose expression was decreased in HIEC-6 only. The expression of Igf2, Fgf9, and Tcf4 (new nomenclature) was not modified. None of the tested genes showed a decreased expression in crypt cells of PrP-knockout as compared with wild-type mice.
The apparently conflicting data in TOP/FOP reporter assays and PCR array analyses, as well as the effect of PrPc or γ-catenin silencing on the expression of Wnt target genes in HIEC-6 cells, in which nuclear β-catenin and TCF7L2 levels are very low, indicate that PrPc or γ-catenin could modulate Wnt target gene expression through other transcriptional effectors. Because some of these genes are also transcriptional targets of the Yes-associated protein (YAP)/TEAD complex, an effector of the Hippo pathway (Zhao et al., 2008), we analyzed whether interaction between PrPc and YAP could occur. Confirming this hypothesis, PLA analyses revealed the presence of PrPc/YAP complexes in both the cytoplasm and nucleus of proliferating HIEC-6, Caco-2/TC7, and SW480 cells (Figure 9).
FIGURE 9:
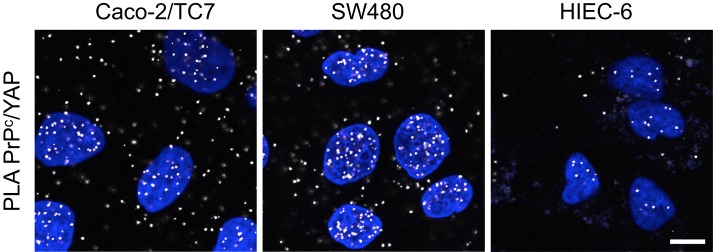
PrPc interacts with YAP, the effector of the Hippo pathway. PLA was performed in proliferating Caco-2/TC7, SW480, and HIEC-6 cells to reveal interactions between endogenous YAP and PrPc. Note that, although present also in the cytoplasm, interaction signals were concentrated in the nucleus, even for HIEC-6 cells. Bar, 10 μm.
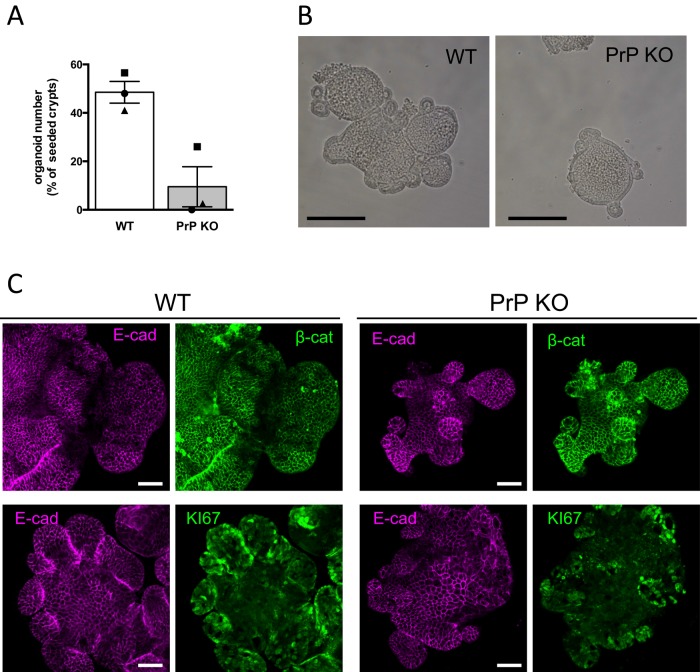
PrPc is required for proper intestinal organoid formation
The Wnt pathway is a driving force for cell proliferation in intestinal crypts (Clevers and Nusse 2012). The up-regulation of β-catenin/TCF7L2 transcriptional activity by PrPc prompted us to examine a possible effect of PrPc on cell proliferation. We observed previously a transient arrest of cell proliferation after PrPc knockdown by siRNA in Caco-2/TC7 cells (Morel et al., 2008). We confirmed this result in HIEC-6 cells but not in SW480 cells, in which, however, PrPc overexpression induced a slight increase in cell proliferation (unpublished data). We considered that the possible role of PrPc in cell proliferation could be studied more accurately in a model of normal intestinal progenitor cells. We isolated crypts from wild-type and PrP-knockout mice and performed intestinal organoid cultures (Sato et al., 2009). Figure 10A shows a net decrease of frequency in organoid formation from PrPc-knockout compared with wild-type mice. The few organoids that were obtained displayed the expected organization, with regular E-cadherin and β-catenin membrane staining (Figure 10C) and accumulation of apoptotic cells in the internal lumen (unpublished data) but were in general smaller (Figure 10B) and showed less-developed crypt domains (Figure 10, B and C) than the organoids developed from the wild-type crypts.
FIGURE 10:
Formation of intestinal organoids is impaired in the absence of PrPc. (A) Organoid initiation frequency from intestinal crypts of WT or PrP KO mice. The symbols represent the mean numbers of organoids per well 6 d after seeding (2–6 wells/experiment) expressed as percentage of plated crypts. Bars represent the mean ± SEM from the three independent experiments. (B) Phase contrast microscopy images 7 d after plating, showing the smaller size of organoids obtained from PrP KO mice. Bar, 200 μm. (C) E-cadherin, β-catenin, and KI-67 immunostaining of 7-d organoids. Although smaller and less abundant, PrP KO organoids have a normal epithelial organization, as shown by E-cadherin and β-catenin staining (top), but exhibit smaller, KI-67–positive crypt domains (bottom). Note that using the same focus for both genotypes, only part of the WT organoids may be visualized in the fields. Bar, 50 μm.
These results could be linked to weaker Wnt activity in intestinal epithelial crypt cells from PrPc-knockout mice. We compared β-catenin nuclear staining in the bottom of intestinal crypts from wild-type and PrP-knockout mice and observed a decreased frequency of cells exhibiting a clear nuclear localization of β-catenin in PrP-knockout crypts (Figure 11).
FIGURE 11:
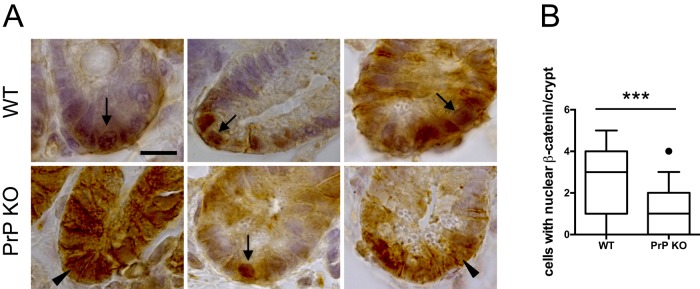
Altered nuclear β-catenin staining in the intestinal crypts of PrP-knockout mice. (A) Crypt sections of jejunum from WT and PrP-knockout mice were stained for β-catenin by immunohistochemistry. Arrows indicate examples of nuclear β-catenin, and arrowheads point out examples of diffuse and/or membranous staining. Bar, 10 μm. (B) Quantification of cells per crypt in which β-catenin was localized mainly in the nucleus. Thirty-four crypts from five WT mice and 55 crypts from five PrP-knockout mice were quantified. Whiskers mark the 10th and 90th percentiles, boxes mark the 25th and 75th percentiles, and the black circle represents an outlier. ***p < 0.001 (Student’s t test).
DISCUSSION
We identified the nuclear partners of PrPc in proliferating intestinal epithelial cells and showed a new role for this protein in the modulation of Wnt pathway. PrPc interacts not only with γ-catenin, one of its desmosomal partner (Morel et al., 2008), but also with β-catenin and TCF7L2, which are the main effectors of Wnt pathway in intestinal cells, the β-catenin–binding domain of TCF7L2 being crucial for its interaction with PrPc. Furthermore, we demonstrate that PrPc modulates the expression of several Wnt target genes, has a positive effect on the transcriptional activity of the β-catenin/TCF7L2 complex, and is required for establishing intestinal organoids ex vivo.
We showed the presence of a nuclear pool of PrPc in two adenocarcinoma cell lines, SW480 and Caco-2/TC7 cells, as well as in normal crypt-like cells, HIEC-6 (Figure 2), in accordance with our previous results in human intestinal crypts (Morel et al., 2008). However, it should be noted that PrPc was less concentrated in the nucleus of HIEC-6 cells, which exhibit a very low Wnt activity, than in the two other cell lines, in which the effectors of Wnt pathway are present in the nucleus (Figure 3). Because PrPc lacks a functional NLS (Jaegly et al., 1998), it likely needs a partner to be imported into the nucleus. TCF7L2 could be such a partner, as was suggested for β-catenin (Shitashige et al., 2008). By the use of PLA, which allows visualization of the subcellular localization of protein interactions, we showed that PrPc can interact with β-catenin, γ-catenin, and TCF7L2 outside the nucleus. This was observed for the interactions of endogenous proteins (Figure 4) and for PrPc or β-catenin interactions with TCF7L2 constructs lacking their NLS (Supplemental Figure S2). PrPc interactions with β-catenin, γ-catenin, and TCF7L2 were concentrated in nucleus only in the context of high Wnt activity and TCF7L2 levels, that is, in SW480 and Caco-2/TC7 cells and not in HIEC-6 cells (Figure 4). These results suggest that multipartner complexes are formed in the cytoplasm before their nuclear import via the classical NLS of TCF7L2.
Combining the use of TCFL2 mutants and β-catenin silencing (Figures 5 and 6), we showed that β-catenin mediates a portion of PrPc/TCF7L2 interactions through the β-catenin–binding domain of TCF7L2 but that other intermediate proteins and/or a direct interaction between PrPc and TCF7L2 may also exist. By contrast, the presence of the γ-catenin–binding domain of TCF7L2 has no influence on its interaction with PrPc, and, accordingly, γ-catenin silencing does not modulate these interactions (Figures 5 and 6), even though PrPc/γ-catenin and γ-catenin/TCF7L2 complexes are observed (Figures 1 and 4). Finally, our results in HIEC-6 cells suggest that PrPc and β-catenin may interact also independently of TCF7L2 (Figure 4). Involvement of PrPc in these multiple combinations of protein complexes, whose composition most probably differs between cytoplasm and nucleus, could modify either the nuclear import of the Wnt effectors or their activity in the nucleus.
The transcription factor complex β-catenin/TCF7L2 is responsible for the transcriptional modulation of Wnt target gene expression in intestinal epithelial cells (Hatzis et al., 2008). The γ-catenin/TCF7L2 complex was shown to be inefficient in binding to DNA (Zhurinsky et al., 2000; Miravet et al., 2002). It was suggested previously that γ-catenin exerts by itself a negative regulation of TCF7L2 transcriptional activity (Miravet et al., 2002) but, through its ability to displace β-catenin from adherens junctions or from the destruction complex, can also enhance the β-catenin/TCF7L2 pool (Salomon et al., 1997; Aktary and Pasdar, 2012). In the present study, we observed a negative modulation of TCF7L2 transcriptional activity by γ-catenin. By contrast, we establish for the first time a positive regulation of this activity by PrPc (Figure 7). Because activation of the Wnt pathway is necessary for proliferation of intestinal epithelial cells, this result is in agreement with 1) the impairment of growth and survival of intestinal organoids from PrP-knockout mice (Figure 10), and 2) the alteration of nuclear β-catenin localization in intestinal crypts of PrP-knockout mice (Figure 11). This could explain the shortening of the villi that we described previously in PrPc-knockout mice (Morel et al., 2008) and is in accordance with the positive role of PrPc in gastric cancer cell proliferation mediated by phosphoinositide 3-kinase/Akt (Liang et al., 2007), a pathway that may interfere with several steps of Wnt signaling (Yan and Lackner, 2012).
In this context, the increased expression of several Wnt target genes upon PrPc silencing seems contradictory—in particular, in crypts of PrP-knockout mice, the up-regulation of Myc and Ccnd1, which are well known to mediate effects of Wnt signaling on cell proliferation (van de Wetering et al., 2002). Compensatory effects in knockout mice cannot explain these discrepancies, since the same tendency was observed in SW480 cells soon after PrPc silencing. In HIEC-6 cells, in which nuclear β-catenin and TCF7L2 levels are very low, PrPc and γ-catenin are able to modulate the expression of genes identified as Wnt targets, suggesting that both proteins interfere with the activity of other transcriptional effectors as well. Multiple cross-talks between the Wnt and Hippo pathways have been shown (Azzolin et al., 2012; Rosenbluh et al., 2012). In this study, we showed an interaction of PrPc with the transcriptional coactivator YAP, which is one effector of Hippo pathway (Figure 9). Thus PrPc could be also a partner of this pathway, as shown recently for desmosomal γ-catenin in arrhythmogenic cardiomyopathy (Chen et al., 2014). PrPc interacts with YAP in both cytoplasm and nucleus, two compartments where YAP may exert opposite effects on Wnt effectors and cell proliferation (Moroishi et al., 2015). Whether and how PrPc modulates the transcriptional activity of YAP/ or TAZ/TEAD complexes remain to be explored. Nevertheless, our results argue strongly for a role of PrPc in the regulation of gene transcription beyond the β-catenin/TCF7L2 complex and Wnt classical targets associated with cell proliferation.
During the past decade, several studies have established a positive correlation between PrPc expression and tumor aggressiveness or adenoma-to-carcinoma progression (for review, see Antony et al., 2012). Our results, which establish an interaction of PrPc with effectors of Wnt and Hippo pathways, both clearly involved in cancer (Clevers and Nusse, 2012; Moroishi et al., 2015), opens a new field of research on the mechanisms and signaling pathways that link PrPc—not only its expression, but also its subcellular localization and the complexes in which it is involved—to colorectal cancers.
In conclusion, PrPc is targeted toward desmosomes or nucleus in intestinal epithelial cells and shares roles with the armadillo family proteins β- and γ-catenin in cell–cell adhesion and cell signaling leading to the regulation of the Wnt pathway. We propose that nuclear PrPc acts as a coregulator able to finely tune the final steps of Wnt signaling and potentially other related pathways involved in the regulation of intestinal epithelium homeostasis.
MATERIALS AND METHODS
Cell culture
All culture media were purchased from Life Technologies/Invitrogen (Cergy-Pontoise, France). Caco-2/TC7 cells (Chantret et al., 1994) were cultured as previously described (Morel et al., 2008). SW480 and COS7 cells were cultured in high-glucose DMEM GlutaMAX I supplemented with 10% heat-inactivated fetal bovine serum (Eurobio/Abcys, Les Ulis, France) and with (SW480) or without (COS7) 1% nonessential amino acids. Nontumoral crypt-like human intestinal cells HIEC-6, kindly provided by J.-F. Beaulieu (Department of Anatomy and Cell Biology, University of Sherbrooke, Sherbrooke, Canada), were cultured in OPTIMEM-GlutaMAX supplemented with 5% fetal bovine serum, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Life Technologies/Invitrogen), and 5 ng/ml EGF (BD Biosciences, Le Pont de Claix, France). Depending on experiments, cells were plated on 3-μm–pore size microporous polyethylene terephthalate filters (Transwell; Corning, Fisher, Illkirch, France), glass coverslips (Polylabo, Strasbourg, France), or plastic (Corning, Fisher).
Mass spectrometry analysis
Proliferating Caco-2/TC7 cells were washed twice in 10 mM Tris-HCl, pH 7.5, containing 20 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 0.2 mM spermidine (TKCM buffer) and scraped in TKCM containing 1% Triton X-100, 1 mM phenylmethylsulfonylfluoride, antiproteases, and antiphosphatases. Nuclei were pelleted by centrifugation at 1000 × g for 10 min at 4°C and washed in TKCM buffer, and nuclear proteins were extracted with 2 M NaCl in TKCM buffer for 1 h at 4°C. Excess NaCl was removed by overnight dialysis against phosphate-buffered saline (PBS) at 4°C. Nuclear proteins were immunoprecipitated with anti-PrPc antibodies (Ab703; rabbit polyclonal antibody; Abcam, Cambridge, United Kingdom) and separated on 4–12% SDS/polyacrylamide gels. After staining with colloidal Coomassie blue (G250; Bio-Rad, Hercules, CA), the visualized bands were cut into 1-mm slices. Gel slices were then reduced, alkylated, and subjected to digestion with trypsin (Roche Diagnostics, Meylan, France). Extracted peptides were dried and solubilized in solvent A (95/5 water/acetonitrile in 0.1% [wt/vol] formic acid). The total digestion product of a gel slice was used per liquid chromatography–tandem mass spectrometry (MS/MS) analysis. The extracted peptides were concentrated and separated on an LC-Packing system (Dionex; Thermo Fisher Scientific, Illkirch, France) coupled to the nano-electrospray II ionization interface of a QSTAR Pulsar i (Applied Biosystems, Life Technologies, Saint Aubin, France) using a PicoTip (10-mm inner diameter; New Objectives, Woburn, MA). The MS/MS data were searched twice by using MASCOT (Matrix Science, London, United Kingdom) and PHENYX (Geneva Bioinformatics, Geneva, Switzerland) software on internal servers, first without taxonomic restriction to reveal the presence of proteins of interest and mammalian contaminants, then in the National Center for Biotechnology Information Human database (National Library of Medicine, Bethesda, MD). All data were manually verified in order to minimize errors in protein identification and/or characterization.
Immunofluorescence analyses and proximity ligation assay
Cells were seeded on glass coverslips (SW480 and HIEC-6 cells) or on Transwell filters (Caco-2/TC7 cells). After 2 or 8 d, cells were fixed with paraformaldehyde (4%, 30 min, room temperature) and permeabilized with Triton X-100 (0.1%, 30 min). Alternatively, for some antibodies, cells were fixed and permeabilized with methanol (5 min, –20°C). The following antibodies were used: anti-PrPc (12F10, mouse monoclonal antibody, S.P.I. BIO, Montigny le Bretonneux, France; or Ab703, rabbit polyclonal antibody, Abcam), γ-catenin and β-catenin (mouse monoclonal antibodies; BD Biosciences, Erembodegem, Belgium), γ-catenin (rabbit polyclonal antibody; Abcam), E-cadherin (ECCD2, rat monoclonal antibody; TaKaRa Bio Europe, Saint-Germain-en-Laye, France), β-catenin and TCF7L2 (rabbit polyclonal antibodies; Cell Signaling, St Quentin en Yvelines, France), FLAG (mouse monoclonal antibody; Sigma-Aldrich, St Quentin-Fallavier, France), YAP1 (rabbit monoclonal antibody; WuXi AppTec, San Diego, CA), and KI-67 (rabbit polyclonal antibody; Abcam). Secondary antibodies were Alexa 488 and Alexa 546–conjugated anti–immunoglobulin G (IgG; Molecular Probes, St Aubin, France).
PLA was performed on cells processed as described for immunofluorescence, using two primary antibodies from mouse and rabbit and according to manufacturer’s instructions (Olink Bioscience, Sigma-Aldrich). PLA PLUS and MINUS probes for mouse and rabbit and the Duolink Orange detection kit were used.
Nuclei were stained by 4′-6-diamidino-2-phenylindole (DAPI), and cells were examined by confocal microscopy (LSM 710 microscope; Carl Zeiss, Jena, Germany) using ZEN 2011 software. Quantifications were performed using a macro of ImageJ software (2.0.0; National Institutes of Health, Bethesda, MD).
Immunoprecipitation and Western blots
For total protein extraction, the cell layer was washed in cold PBS and scraped in TNE buffer (10 mM Tris, pH 8, 150 mM NaCl, 1 mM EDTA)/Nonidet P-40 (1%) supplemented with antiprotease and antiphosphatase cocktails (Sigma-Aldrich). Alternatively, nuclear/cytoplasmic protein fractions were purified using the Pierce NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific, Illkirch, France) according to the manufacturer’s instructions.
Protein concentrations were determined using the BC Assay (Uptima/Interchim, Montluçon, France).
Immunoprecipitations were performed on nuclear extracts with rabbit anti-PrPc and anti–γ-catenin antibodies (Abcam) or nonimmune rabbit IgG (Sigma-Aldrich) as control coupled to protein A–Sepharose CL 4B (Amersham Biosciences, GE Healthcare Europe, Orsay, France). For Western blots, samples were boiled for 10 min in Laemmli buffer and fractionated through 10 or 12% polyacrylamide gels under reducing conditions. The following antibodies were used: anti-PrPc (mouse monoclonal SAF32; S.P.I. BIO), anti–γ-catenin and β-catenin (mouse monoclonal antibodies; BD Biosciences), and anti-TCF7L2 (rabbit polyclonal antibody; Cell Signaling). Anti-actin (Millipore), poly[ADP-ribose] polymerase 1 (PARP-1), specificity protein 1 (SP1), and lactate dehydrogenase (LDH; Santa Cruz Biotechnology, Santa Cruz CA) antibodies were used for cell fraction purity control. Bound antibodies were detected by chemiluminescence (ECL, Amersham Biosciences, GE Healthcare Europe; or ECL2 Pierce, Thermo Fisher Scientific) on a Luminescence Image Analyzer (LAS-4000; Fujifilm, Courbevoie, France). Densitometric semiquantitative analyses were performed using ImageJ software (2.0.0).
Site-directed mutagenesis, transfection experiments, and PLA quantifications
Mutagenesis was achieved by PCR using 50 ng of pFLAG-TCF7L2 (Idogawa et al., 2005), the high-fidelity thermostable Phusion DNA polymerase (New England BioLabs, Evry, France) and the complementary mutagenic oligonucleotides listed in Supplemental Table S1. After PCR, the starting template was eliminated by DpnI digestion, and the final products were used to transform competent bacteria from New England BioLabs. All selected mutants were sequenced before use.
SW480 cells were seeded on glass coverslips in 24-well plates (40,000 cells/well) and transfected 24 h later with the different Flag-TCF7L2 constructs (500 ng) using X-treme GENE HP DNA (Roche Diagnostics, Meylan, France) according to the manufacturer’s instructions. PLA experiments were performed 2 d after transfection as described using mouse monoclonal anti-FLAG combined with rabbit anti-PrPc, anti–β-catenin, or anti–γ-catenin antibodies. Expression of the different constructs was analyzed by immunofluorescence with the anti-FLAG antibody in parallel wells because PLA and immunofluorescent detection of FLAG tag could not be performed simultaneously owing to competition of PLA probes and secondary antibodies for fixation on primary anti-FLAG antibodies.
For confocal microscopy analysis, acquisition settings were chosen so that PLA signals for β-catenin/FLAG interactions, which gave the highest intensities, were close to the saturation level. The same settings were then kept constant for all interactions and all constructs, maximizing the dynamic range of quantification. Quantification of PLA signals for the different Flag-TCF7L2 constructs and for the different interactions was performed using a macro of ImageJ software (2.0.0). Integrated intensity of PLA signal normalized to cell number was measured in at least 10 random fields for each condition (total of ∼1000 cells). After subtraction of background (mean intensity obtained for PLA assays with FLAG-empty vector), values were normalized to mean values of FLAG immunofluorescence signal/cell, to correct for variations in transfection efficiency.
Small interfering RNA transfection
siRNAs were purchased from Qiagen SA Biosciences (Courtaboeuf, France). Two different siRNA sequences were combined for each gene silencing (Supplemental Table S2). Cells were seeded at 20,000 cells/cm2 on plastic or on glass coverslips and transfected 48 h after plating using X-tremeGENE siRNA Transfection Reagent (Roche Diagnostics). The total concentration of siRNA in the media was 200 nM.
TCF/β-catenin reporter assays
The TCF-responsive TOP-FLASH, expressing luciferase driven by multiple TCF-responsive elements, or FOP-FLASH with mutated TCF-responsive elements, were purchased from Millipore. The pRSV-β-gal encoding β-galactosidase was used as internal control. The vectors encoding human γ-catenin and mouse PrPc were obtained from Eric Fearon (Addgene plasmid #16827; Caca et al., 1999) and Susan Lindquist (Addgene plasmid #22109; Jackson et al., 2009), respectively. The vector encoding a constitutively active mutant S33Y of β-catenin was obtained from Corinne Quittau-Prevostel (U1194, Institut de Recherche en Cancérologie, Montpellier, France).
COS7 or SW480 cells were plated into 12-well plates (120,000 and 80,000 cells/well, respectively). Transfection was performed 24 h after plating using X-tremeGENE HP DNA transfection reagent (Roche Diagnostics). Cells were transfected with 200 ng of TOP or FOP vector, together with 100 ng of pRSV-β-gal. COS7 received in addition 50 ng of S33Y β-catenin vector and either γ-catenin or PrPc vector (100 or 300 ng). SW480 received only γ-catenin or PrPc vector because β-catenin is constitutively stabilized in this cell line. The PTZ18R plasmid was used to adjust the quantity of plasmid DNA to 1 μg for all conditions. Cells were harvested 48 h after transfection, and β-galactosidase and luciferase activities were analyzed using a multifunctional microplate reader (FLUOSTAR Omega; BMG Labtech, Ortenberg, Germany).
TOP/FOP assays were also performed in SW480 after PrPc silencing by siRNA. In this case, reporter plasmids and the mouse PrPc expression vector were transfected 24 h after plating, and siRNAs were transfected the next day, as described. We showed previously that mouse PrPc was not targeted by siRNA directed against the human mRNA (Petit et al., 2012).
Purification of epithelial cells from mouse intestinal crypts
PrPc-knockout mice, backcrossed on C57BL/6, and their wild-type counterparts were housed in pathogen-free conditions (Petit et al., 2012). Ileum of 3-mo-old wild-type (five animals) or PrPc-knockout mice (five animals) were collected, flushed with PBS containing 1 mM CaCl2 and 0.5 mM MgCl2, minced in 1-mm pieces, and transferred, as a pool, in a chelation buffer (27 mM trisodium citrate, 5 mM Na2HPO4, 96 mM NaCl, 8 mM KH2PO4, 1.5 mM KCl, 0.5 mM dithiothreitol, 55 mM d-sorbitol, 44 mM sucrose) at 4°C for shaking. According to the strength and number of agitations, five fractions of epithelial cells were obtained, corresponding to the villus tip (fraction 1) to the crypt bottom (fraction 5). Each cell fraction was centrifuged (1500 rpm, 5 min, 4°C); the cell pellet was resuspended in 800 μl of PBS+ and stored at −80°C until RNA extraction. Analysis of the differential expression of a set of genes between the top and bottom fractions of the crypts and comparison with the results reported in Mariadason et al. (2005) confirmed the purity of the bottom crypt fraction.
RNA extraction and gene expression analyses
Total RNA was isolated from mouse epithelial crypt cells or cultured cell lines with Tri-Reagent (Molecular Research Center, Cincinnati, OH) according to manufacturer’s instructions. After rDNase digestion (Macherey-Nagel, Hoerdt, France), RNA integrity was checked by gel electrophoresis. Reverse transcription (RT) was performed with 1 μg of RNA using Roche reagents (Roche Diagnostics). Real-time PCR was conducted with cDNA and both the sense and antisense oligonucleotides in a volume of 10 μl of SYBR green PCR mix (Agilent, Massy, France) and monitored and assessed in a Stratagene Mx3000P (Thermo Fisher Scientific) system. Values were normalized to L19 or 18S for human cells or to cyclophilin expression for mouse cells. Primer sequences are given in Supplemental Table S3.
PCR array analysis
SW480 and HIEC-6 cells were transfected with siRNA 48 h after plating and collected 24 h after transfection for RNA extraction as described. After reverse transcription with the RT2 First Strand Kit (Qiagen SA Biosciences) using 1 μg of RNA, silencing efficiency was checked by semiquantitative real-time PCR. A minimum of 80% extinction of the siRNA targets was a prerequisite to proceed further for PCR array analysis. Human WNT Signaling Targets RT2 Profiler PCR Array Plate (Qiagen SA Biosciences), which profiles the expression of 84 target genes and eight control genes, was used with the RT2 SYBR Green qPCR Mastermix and the Mx3000P Stratagene system. Genes were selected for heat map profiles when modulations of their expression, after normalization by manufacturer’s controls, were ≥1.4× or ≤0.7× upon either PrPc or γ-catenin silencing in at least one cell line, with similar values in three independent experiments and p < 0.05.
Immunohistochemistry on mouse intestinal crypts
For β-catenin staining in mouse crypts, wild-type and PrP-knockout mice were killed, and their intestines were removed, flushed gently with PBS, and fixed overnight at 4°C in alcohol/Formalin/acetic acid before being embedded in paraffin. Immunostaining with anti–β-catenin (rabbit polyclonal; Cell Signaling) was performed on 5-μm paraffin sections after antigen retrieval in boiling 10 mM citrate buffer (pH 6) for 10 min and permeabilization with 0.1% Triton X-100 (20 min). A horseradish peroxidase–labeled anti-rabbit antibody (Amersham Biosciences) and 3,3′-diaminobenzidine were used for revelation. β-Catenin staining was examined on a Zeiss Imager-M2 microscope using ZEN 2011 software.
Intestinal organoids
Isolation and culture of intestinal crypts were performed as previously described (Andersson-Rolf et al., 2014), with some modifications. Briefly, the small intestines of 4- to 5-mo-old wild-type or PrP-knockout mice were isolated, cut into 5-cm-long pieces, and washed in cleaning solution (PBS-C, phosphate-buffered saline, calcium/magnesium-free; 2% penicillin-streptomycin, 1% gentamicin; Life Technologies). Intestinal pieces were opened longitudinally, and villi were scraped off by using a coverslip. The tissue was washed by vigorous shaking in precooled PBS-C and transferred into a 1 mM EDTA solution in PBS-C for 30 min at 4°C on a rotating wheel. Villi were then removed by vigorous shaking (∼20 times), and the tissue was incubated at 4°C for 1 h on a rotating wheel in Leibovitz medium (Sigma-Aldrich) supplemented with 5 mM EDTA, 2% penicillin-streptomycin, 1× GlutaMAX, and 25 mM HEPES (Life Technologies). To isolate the crypts, the tissue fragments were transferred into a precooled Leibovitz EDTA-free solution and vigorously shaken (∼40 times). The presence of crypts was confirmed under the microscope and their number counted in a 30-μl drop of crypt solution. A volume containing 300 crypts was spun down at 300 × g for 5 min at 4°C, supernatant was discarded, and the pellet was resuspended in 150 μl of Matrigel (Corning) half-diluted in DMEM/F12 (Life Technologies). Crypts were then seeded into 48-well flat-bottom plates and incubated for 15 min at 37°C. Then, Matrigel was overlaid by 300 μl of ENR medium (DMEM/F12, 20 ng/ml EGF [Peprotech, Neuilly sur Seine, France], 10 ng/ml FGF [Peprotech], 100 ng/ml Noggin [Peprotech], 2.5% [vol/vol] GlutaMAX [Life Technologies], 500 ng/ml R-Spondin [R&D Systems, Lille, France], 1× B27 [Life Technologies], 1× N2 [Life Technologies]). The crypt number was evaluated after 6 d of culture by manual counting. Immunofluorescence staining was performed and analyzed as described after organoid fixation in 4% paraformaldehyde and permeabilization with Triton X-100 (0.3%, 10 min).
Statistical analysis
Values are expressed as mean ± SEM. Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). The statistical significance of the differences between groups was determined by Student’s t test, one-way or two-way analysis of variance (ANOVA), or nonparametric Mann–Whitney or Kruskal–Wallis test as appropriate. ANOVA and Kruskal–Wallis analyses were followed by Sidak or Tukey and Dunn multiple-comparison posttests, respectively. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
HIEC-6 cells were a generous gift from J. F. Beaulieu (University of Sherbrooke, Sherbrooke, Canada). We thank Magashi Idogawa (Sapporo Medical University, Sapporo, Japan) and Tesshi Yamada (National Cancer Center Research Institute, Tokyo, Japan) for the plasmid pFLAG-TCF4 (TCF7L2) and Corinne Quittau-Prevostel (Institut de Recherche en Cancérologie, Montpellier, France) for the plasmid encoding the S33Y mutant of β-catenin. We thank Ryad Boukherrouf for his participation in the study. Confocal microscopy was performed at the Centre d’Imagerie Cellulaire et de Cytométrie (Centre de Recherche des Cordeliers, UMRS 1138, Paris, France). Mice were housed in the SPF facility of the Centre de Recherche des Cordeliers. This work was supported by institutional funding from the Institut National de la Santé et de la Recherche Médicale, Université Pierre et Marie Curie and the Ecole Pratique des Hautes Etudes and a grant from the Agence Nationale pour la Recherche (ANR-09-JCJC-0052-01). L.S.B. was the recipient of a fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche.
Abbreviations used:
- PLA
proximity ligation assay
- PrPc
cellular prion protein
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- TCF7L2
transcription factor 7–like 2
- YAP
Yes-associated protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.3313) on July 29, 2015.
REFERENCES
- Aguilera O, Pena C, Garcia JM, Larriba MJ, Ordonez-Moran P, Navarro D, Barbachano A, Lopez de Silanes I, Ballestar E, Fraga MF, et al. The Wnt antagonist DICKKOPF-1 gene is induced by 1alpha,25-dihydroxyvitamin D3 associated to the differentiation of human colon cancer cells. Carcinogenesis. 2007;28:1877–1884. doi: 10.1093/carcin/bgm094. [DOI] [PubMed] [Google Scholar]
- Aktary Z, Pasdar M. Plakoglobin: role in tumorigenesis and metastasis. Int J Cell Biol. 2012;2012:189521. doi: 10.1155/2012/189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Rolf A, Fink J, Mustata RC, Koo BK. A video protocol of retroviral infection in primary intestinal organoid culture. J Vis Exp. 2014:e51765. doi: 10.3791/51765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony H, Wiegmans AP, Wei MQ, Chernoff YO, Khanna KK, Munn AL. Potential roles for prions and protein-only inheritance in cancer. Cancer Metastasis Rev. 2012;31:1–19. doi: 10.1007/s10555-011-9325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Caca K, Kolligs FT, Ji X, Hayes M, Qian J, Yahanda A, Rimm DL, Costa J, Fearon ER. Beta- and gamma-catenin mutations, but not E-cadherin inactivation, underlie T-cell factor/lymphoid enhancer factor transcriptional deregulation in gastric and pancreatic cancer. Cell Growth Differ. 1999;10:369–376. [PubMed] [Google Scholar]
- Chantret I, Rodolosse A, Barbat A, Dussaulx E, Brot-Laroche E, Zweibaum A, Rousset M. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: evidence for glucose-dependent negative regulation. J Cell Sci. 1994;107:213–225. doi: 10.1242/jcs.107.1.213. [DOI] [PubMed] [Google Scholar]
- Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res. 2014;114:454–468. doi: 10.1161/CIRCRESAHA.114.302810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Crozet C, Vezilier J, Delfieu V, Nishimura T, Onodera T, Casanova D, Lehmann S, Beranger F. The truncated 23–230 form of the prion protein localizes to the nuclei of inducible cell lines independently of its nuclear localization signals and is not cytotoxic. Mol Cell Neurosci. 2006;32:315–323. doi: 10.1016/j.mcn.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Guezguez A, Pare F, Benoit YD, Basora N, Beaulieu JF. Modulation of stemness in a human normal intestinal epithelial crypt cell line by activation of the WNT signaling pathway. Exp Cell Res. 2014;322:355–364. doi: 10.1016/j.yexcr.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Tsuchiya K, Sato I, Takeyama N, Ueda S, Tagawa Y, Kimura KM, Nakamura I, Wu G, Sakudo A, et al. A monoclonal antibody (1D12) defines novel distribution patterns of prion protein (PrP) as granules in nucleus. Biochem Biophys Res Commun. 2008;366:657–663. doi: 10.1016/j.bbrc.2007.11.163. [DOI] [PubMed] [Google Scholar]
- Idogawa M, Yamada T, Honda K, Sato S, Imai K, Hirohashi S. Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/beta-catenin complex. Gastroenterology. 2005;128:1919–1936. doi: 10.1053/j.gastro.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Jackson WS, Borkowski AW, Faas H, Steele AD, King OD, Watson N, Jasanoff A, Lindquist S. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63:438–450. doi: 10.1016/j.neuron.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaegly A, Mouthon F, Peyrin JM, Camugli B, Deslys JP, Dormont D. Search for a nuclear localization signal in the prion protein. Mol Cell Neurosci. 1998;11:127–133. doi: 10.1006/mcne.1998.0675. [DOI] [PubMed] [Google Scholar]
- Liang J, Pan Y, Zhang D, Guo C, Shi Y, Wang J, Chen Y, Wang X, Liu J, Guo X, et al. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21:2247–2256. doi: 10.1096/fj.06-7799com. [DOI] [PubMed] [Google Scholar]
- Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- Mange A, Crozet C, Lehmann S, Beranger F. Scrapie-like prion protein is translocated to the nuclei of infected cells independently of proteasome inhibition and interacts with chromatin. J Cell Sci. 2004;117:2411–2416. doi: 10.1242/jcs.01094. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Nicholas C, L’Italien KE, Zhuang M, Smartt HJ, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Miravet S, Piedra J, Miro F, Itarte E, Garcia de Herreros A, Dunach M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. J Biol Chem. 2002;277:1884–1891. doi: 10.1074/jbc.M110248200. [DOI] [PubMed] [Google Scholar]
- Morel E, Fouquet S, Chateau D, Yvernault L, Frobert Y, Pincon-Raymond M, Chambaz J, Pillot T, Rousset M. The cellular prion protein PrPc is expressed in human enterocytes in cell-cell junctional domains. J Biol Chem. 2004;279:1499–1505. doi: 10.1074/jbc.M308578200. [DOI] [PubMed] [Google Scholar]
- Morel E, Fouquet S, Strup-Perrot C, Thievend CP, Petit C, Loew D, Faussat AM, Yvernault L, Pincon-Raymond M, Chambaz J, et al. The cellular prion protein PrP(c) is involved in the proliferation of epithelial cells and in the distribution of junction-associated proteins. PLoS One. 2008;3:e3000. doi: 10.1371/journal.pone.0003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, Kellermann O. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem. 1997;272:6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- Petit CS, Barreau F, Besnier L, Gandille P, Riveau B, Chateau D, Roy M, Berrebi D, Svrcek M, Cardot P, et al. Requirement of cellular prion protein for intestinal barrier function and mislocalization in patients with inflammatory bowel disease. Gastroenterology. 2012;143:122–132. doi: 10.1053/j.gastro.2012.03.029. e115. [DOI] [PubMed] [Google Scholar]
- Petit CS, Besnier L, Morel E, Rousset M, Thenet S. Roles of the cellular prion protein in the regulation of cell adhesion, cell-cell junctions and barrier function. Tissue Barriers. 2013;1 doi: 10.4161/tisb.24377. e24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybner C, Finel-Szermanski S, Felin M, Sahraoui T, Rousseau C, Fournier JG, Seve AP, Botti J. The cellular prion protein: a new partner of the lectin CBP70 in the nucleus of NB4 human promyelocytic leukemia cells. J Cell Biochem. 2002;84:408–419. doi: 10.1002/jcb.10017. [DOI] [PubMed] [Google Scholar]
- Salomon D, Sacco PA, Roy SG, Simcha I, Johnson KR, Wheelock MJ, Ben-Ze’ev A. Regulation of beta-catenin levels and localization by overexpression of plakoglobin and inhibition of the ubiquitin-proteasome system. J Cell Biol. 1997;139:1325–1335. doi: 10.1083/jcb.139.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuccione A, Sytnyk V, Leshchyns’ka I, Schachner M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol. 2005;169:341–354. doi: 10.1083/jcb.200409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Shitashige M, Satow R, Honda K, Ono M, Hirohashi S, Yamada T. Regulation of Wnt signaling by the nuclear pore complex. Gastroenterology. 2008;134:1961–1971. doi: 10.1053/j.gastro.2008.03.010. 1971.e1–e4. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Wong MH, Saam JR, Mysorekar IU, Gordon JI. Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Curr Opin Cell Biol. 1998;10:702–709. doi: 10.1016/s0955-0674(98)80110-5. [DOI] [PubMed] [Google Scholar]
- Strom A, Wang GS, Picketts DJ, Reimer R, Stuke AW, Scott FW. Cellular prion protein localizes to the nucleus of endocrine and neuronal cells and interacts with structural chromatin components. Eur J Cell Biol. 2011;90:414–419. doi: 10.1016/j.ejcb.2010.11.015. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard L, Christensen HM, Harris DA. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Lackner MR. FOXO3a and beta-catenin co-localization: double trouble in colon cancer? Nat Med. 2012;18:854–856. doi: 10.1038/nm.2799. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze’ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol. 2000;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.