Abstract
Liver cancer is one of the most common cancers worldwide. CDR3 sequencing-based immune repertoire can be closely associated with cancer prognosis and development. Identifying the specific interaction between the TCR and cellular antigens is important for developing novel immunotherapeutic approaches for the treatment of cancer. The rearranged TCRβ loci amplified using Vβ- and Jβ-specific primers by multi-PCR and sequenced using high-throughput sequencing (HTS) in liver cancers were compared with those of T cells from healthy adult peripheral blood and from adjacent liver tissue. The T-cell repertoires within each tumor show strong similarity to one another but are distinct from those of the circulating T-cell repertoire. In addition, our results demonstrate that there are significant differences in the T-cell repertoires of HCC (hepatocellular carcinoma), ICC (intrahepatic cholangiocarcinoma), and MHC (mixed hepatocellular and cholangiocellular carcinoma). Furthermore, we found that the highly expanded clone (HEC) ratio in blood samples from liver cancer patients differed significantly from those in the blood of healthy adults and hepatitis patients (p < 0.001). The above results suggest that comparison of the T-cell repertoires of tissue and blood could be used to distinguish liver cancer patients from healthy adults and from hepatitis patients. In the future, the diversity of CDR3 sequences in liver cancer may prove to be a useful and novel biomarker for detecting aggressive tumors with high invasive or metastatic capacity.
Keywords: hepatocellular carcinoma, high-throughput sequencing, highly expanded clone, T cell, TCRβ
Abbreviations
- HCC
hepatocellular carcinoma
- IR
immune repertoire
- HTS
high-throughput sequencing
- HBV
hepatitis B virus
- TCR
T-cell receptor
- TCRα
TCR α-chains
- TCRβ
TCR β-chains
- ICC
intrahepatic cholangiocarcinoma
- MHC
mixed hepatocellular and cholangiocellular carcinoma
- TILs
tumor-infiltrating lymphocytes.
Introduction
Primary liver cancer is the sixth most frequent cancer globally and the second leading cause of cancer deaths. In 2012, nearly 782,000 new cancer cases (50% in China alone) occurred worldwide and were responsible for 746,000 deaths.1 The most frequent liver cancer, accounting for approximately 75% of all primary liver cancers, is hepatic carcinoma (HCC) and originates in hepatocytes. Liver cancer can also originate in other structures of the liver, such as the bile duct, blood vessels, and immune cells. Cancers of the bile duct (cholangiocarcinoma and cholangiocellular cystadenocarcinoma) account for approximately 6% of primary liver cancers.2,3
HCC is the fifth most frequent cancer and the third most frequent cause of cancer mortality worldwide.4 Most cases of HCC are secondary to either viral hepatitis infection or cirrhosis.5-7 Approximately 5–10% of individuals infected with hepatitis B virus (HBV) become chronic carriers, and approximately 30% of these individuals acquire chronic liver disease that can lead to HCC.5,8 Cancerous cells often express aberrant peptides that are presented on the surface of cells and can be bound by T-cell receptors on the surface of T lymphocytes, the primary mediators of the cellular adaptive immune response. Healthy adults have approximately 2.5 × 108 distinct TCRs in the peripheral blood, allowing highly specific immune responses to a diverse range of foreign antigens.9 In the peripheral blood, more than 90% of T cells are αβ T cells.10 The TCRs expressed by αβ T cells are heterodimeric proteins that include two polypeptide chains, α and β. Each polypeptide has variable (V), joining (J) and constant (C) regions; β chains also have diversity (D) regions.11 In TCRs, the highly variable complementarity-determining region 3 (CDR3) is the primary source of antigen-specific recognition; its diversity is generated by rearrangement of the V, D, J, and C regions. The random insertion of non-germline–encoded nucleotides at the junctions of these rearranged segments provides additional diversity.12 Because the diversity of the TCR repertoire mirrors that of the human immune system, analysis of CDR3 diversity within the TCR is crucial for understanding the basic molecular mechanisms of adaptive immunity in health and disease.10,13 TCRβ sequencing may shed light on mechanisms of cancer immunity. The diversification and selection dynamics of TCR repertoires in healthy individuals and in those with infection, autoimmunity, immunodeficiency or cancer remain poorly understood but have important clinical implications. This method has been applied to a variety of cancers, including ovarian carcinoma, renal cell carcinoma, and cutaneous T-cell lymphoma.14-16 Researchers are currently attempting to identify biomarkers or prognostic factors in the T-cell receptor repertoire to facilitate the early detection, treatment and prognosis of tumors.
In this work, we aimed to analyze the T-cell receptor repertoire in peripheral blood samples and tissues from liver cancer and hepatitis B patients using HTS of the TCRβ CDR3 region. Our work focused on producing a comprehensive, unrestricted TCR immunogenetic characterization from the blood and tissue of healthy individuals, as well as those of hepatitis B and liver cancer patients.
Results
Analysis of the profile of TCRβ CDR3 in human cells using next-generation sequencing
To study the profile of the T-cell β receptor in human cells, primers were designed for multiplex PCR at the TRB V/D/J loci to amplify the CDR3 fragment at the RNA level (Fig. S1). To distinguish the sequences between samples, a 6-bp barcode sequence was added to the 5′ end of each primer. The total length of all PCR products (PCR product + barcode sequence) was approximately 250 bp (Fig. S2). The PCR products were purified using magnetic beads. The enriched products were used for library construction and then sequenced at a single-base resolution using the Illumina HiSeq2000 platform. To obtain sufficient coverage of the targeted regions for further analysis, 20 samples were pooled into a single lane. More detail information of the 160 samples was shown in Table S1. In each sequence run, we obtained approximately 300 M reads (average read length of 100 bp) per lane. Using the primer panel, we amplified all CDR3 sequences in 160 samples obtained from healthy adults and from patients with hepatitis, liver cancer, and colon cancer (CC). We obtained a total of 1.038 G raw reads representing 173,367 gigabase pairs from 160 samples.
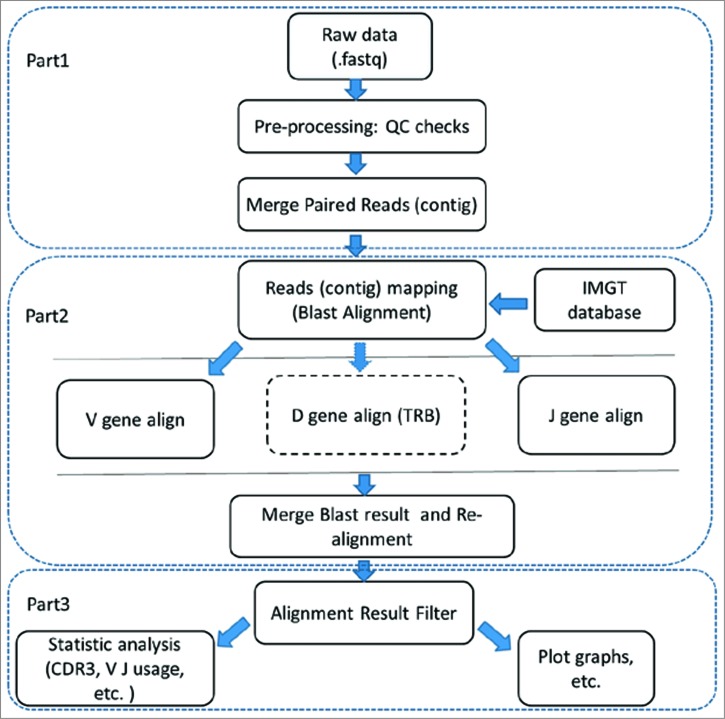
To retain the high-quality reads for further analysis, we filtered sequence reads (Fig. 1, Part 1) according to four strict criteria (described in detail in the Materials and Methods section). We then merged the high-quality paired reads using COPE and FqMerger (BGI) and designated the results as contigs.17 The contigs were aligned to the reference TCR Vβ/Dβ/Jβ gene sequences using BLAST (Fig. 1, Part 2). To ensure high accuracy of the results, the initial alignment results were realigned to the database to find precise V/D/J genes (Fig. 1, Part 3). A subset of these alignment results were then filtered (the criteria used in this step are described in the Materials and Methods section).
Figure 1.
Base process. The Base Process was performed using IMonitor (BGI, unpublished results). PART 1: Filtering of the reads; PART 2: Alignment with the IMGT database; PART 3: Filtering of the alignment result.
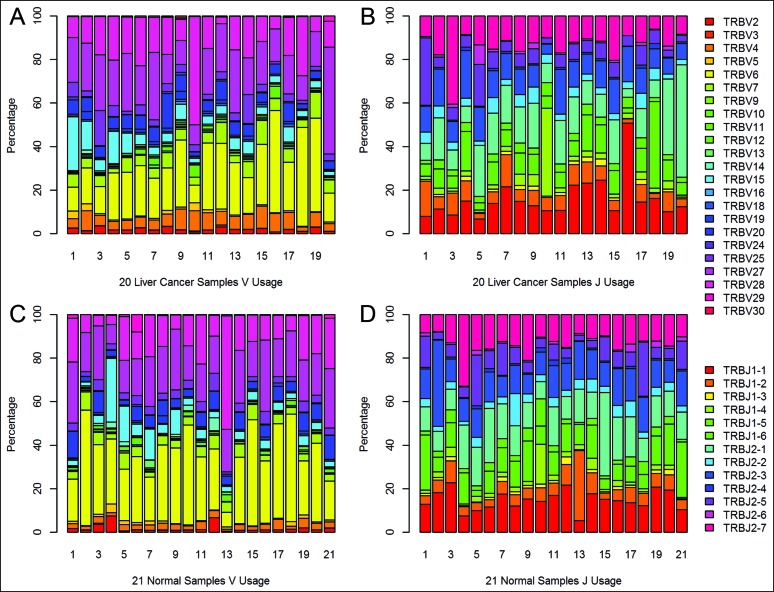
Finally, we obtained a total of 495,708,702 contigs of TCR CDR3 from 160 samples (Table S2). An average of 3,098,179 contigs was generated per sample. In the end, we identified 48 Vβ and 13 Jβ segments, of which 48 Vβ segments were merged into 23 Vβ segments. These results were used to analyze the Vβ and Jβ usage of CDR3 amino acid clonotypes in each sample (Fig. 2, Fig. S3), and the values of Vβ and Jβ segment usage was semi-quantitative.
Figure 2.
The semi-quantitative results of Vβ and Jβ segment usage in blood of liver cancer patients and healthy adults. (A) V segment usage in blood samples from 20 liver cancer patients. (B) J segment usage in blood samples from 20 liver cancer patients. (C) V segment usage in blood samples from 21 healthy adults. (D) J segment usage in blood samples from 21 healthy adults.
Differential expression profiling of TCRβ CDR3 in liver cancers
To identify the differential expression profile of CDR3 in various samples at the mRNA level, we selected all clones in each sample with a frequency of more than 0.1% for further analysis and defined these clones as HECs. The ratio of HECs to total clones was used to compare CDR3 differential expression across samples.
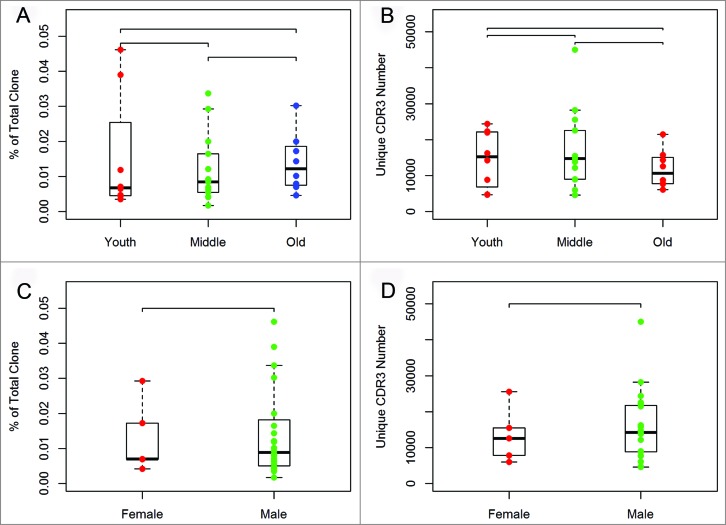
We first assessed the relationship of the HEC ratio for CDR3 or unique CDR3 with the age and gender in the samples from liver cancer patients. Our results indicated that there was no significant difference among these groups (Fig. 3).
Figure 3.
Comparison between groups of liver cancer patients separated by age and gender. The 29 liver cancer patients were divided into three age groups representing young (20 to 44 years of age), middle-aged (45 to 59 years of age), and elderly (greater than 60 years of age) patients. (A) Variance analysis of the HEC ratio among the three age groups. (B) Variance analysis of the unique CDR3 number among the three age groups. (C) Variance analysis of the HEC ratio between the two gender groups. (D) Variance analysis of the unique CDR3 number between the two gender groups.
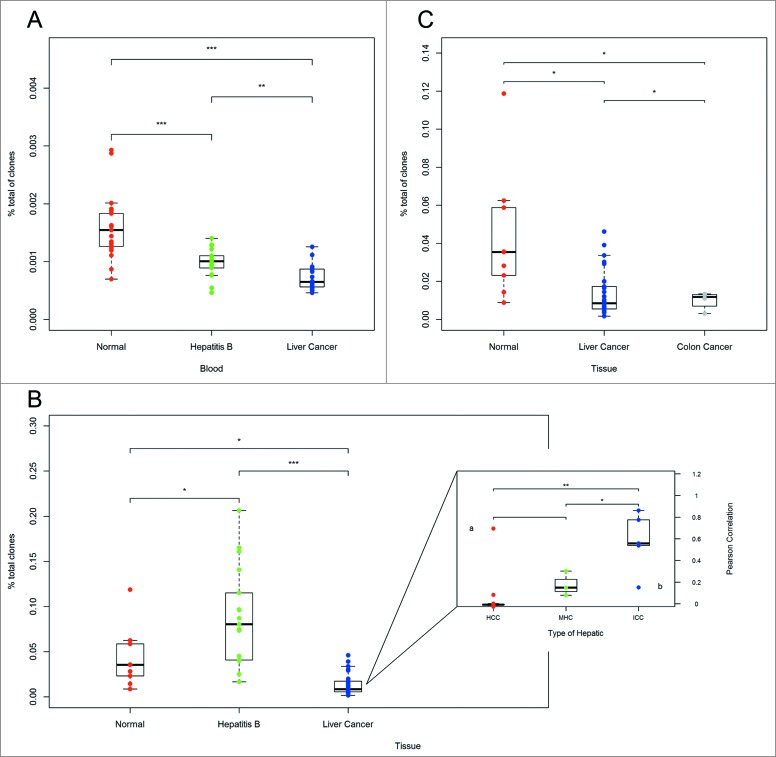
We then compared the HEC ratio of blood and tissue samples from healthy adults, hepatitis B patients and liver cancer patients. Basic information of healthy individuals and patients with hepatitis B or CC was summarized in Table S3. Interestingly, the results indicated that the HEC ratio in blood samples from liver cancer patients was significantly different from the HEC ratio in blood samples from healthy adults (p < 0.001) (Fig. 4A). We also observed significant differences between healthy adults and hepatitis B patients (p < 0.001) and between hepatitis B patients and liver cancer patients (p < 0.01) (Fig. 4A). In addition, we compared the HEC ratio in healthy adults, hepatitis B patients and liver cancer patients at the tissue level. We found significant differences between each two groups (Fig. 4B). The above results suggest that comparisons of HEC ratios in both tissue and blood can be used to distinguish liver cancer patients from healthy adults.
Figure 4.
Variance analysis. (A) Variance analysis of the HEC ratios in blood samples from healthy adults, hepatitis B patients and liver cancer patients. (B) Variance analysis of the HEC ratios in hepatic tissue from healthy adults, hepatitis B patients and liver cancer patients. The subgraph shows a comparison of the consistency between various types of hepatocellular carcinoma and adjacent tissues. The Pearson correlation is shown on the y2 axis, and the type of liver cancer is indicated on the x axis. (C) Variance analysis of the HEC ratio in tissues of healthy adults, liver cancer patients, and CC patients. *p < 0.05, **p < 0.01, ***p < 0.001 according to two-tailed t-tests.
We also analyzed the consistency of TCRβ CDR3 between tumor tissues and adjacent tissues of three types of liver cancers using the Pearson correlation coefficient method (Fig. 4B subgraph). Clinicopathologic informations of the 29 patients with liver cancer were summarized in Table S4.The two-sided t-test was then used to assess the consistency of these differences among three types of liver cancers: HCC, ICC, and MHC. For HCC, the tumor tissues and adjacent tissues displayed low consistency in the TCRβ CDR3 sequences, possibly indicating the low degree of malignancy of this tumor, whereas for ICC, the consistency was much higher (Fig. 4B subgraph), which may indicate that the cancer cells had already metastasized.
Differential expression in liver cancer patients and colon cancer (CC) patients
To determine whether we could distinguish CC patients from healthy adults in a similar fashion, we compared the HEC ratios of CC from healthy adults and liver cancer patients at the tissue level. There was a significant difference between CC patients and healthy adults (p < 0.05) (Fig. 4C). Interestingly, there was also a significant difference between CC patients and liver cancer patients.
Principal component analysis for differential Vβ and Jβ genes in HCC patients and in healthy adults
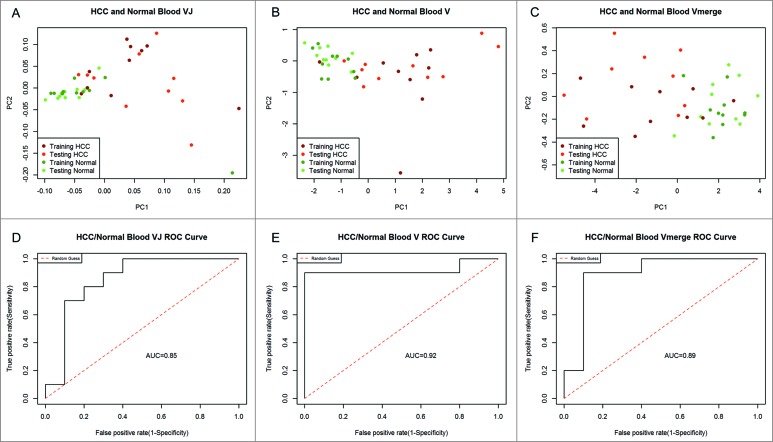
To find the best factor for distinguishing HCC patients and healthy adults, we compared V–J usage, V usage and V merged usage in HCC blood and healthy blood. The 48 Vβ segments could be merged into 23 Vβ subclasses (Table S5). Of which 23 Vβ subclasses were used for V merged subclasses usage, while the 48 Vβ segments were used for V and J recombination usage and V usage. 10 out of 20 healthy cases and 10 out of 20 HCCs were random selected as training set, and other 10 healthy samples and 10 HCCs were used as testing set. The results revealed that blood samples from healthy adults and HCC patients can be clearly separated using three types of indexes (Fig. 5A–C). Compared with VJ and Vmerge, the V usage displayed a higher AUC (the area under the ROC curve) value of 0.92 (Fig. 5D–F) in testing set by ROC curve analysis and was superior at distinguishing HCC patients and healthy adults. To validate the conclusion, we also used linear discriminant analysis (LDA) for classification analysis (Fig. S4). The resulting data strongly support V usage may be a potential classifier for HCC.
Figure 5.
Principal component analysis. To distinguish HCC patients and healthy adults, we used three indexes for PCA: V and J recombination usage (A), V subclasses usage (B), and V merged subclasses usage (C). The PCA1 values of testing set were used for further ROC analysis. The AUC values for classifier of VJ, V, and V merge were 0.85 (D), 0.92 (E), and 0.89 (F), respectively. PC1 refers to the first principal component, whereas PC2 indicates the second principal component.
Discussion
TCR high-throughput sequencing technology show obvious advantages
In this work, we coupled HTS with semi-quantitative multiplex PCR amplification of TCRβ CDR3 sequences in mRNA to characterize the basic properties of T cells in hepatitis B and liver cancer patients and to compare those properties with the properties of T cells from adjacent liver tissue or from the peripheral blood of healthy adults.
TCR CDR3 diversity has been estimated previously via spectratyping.18,19 This technique is inexpensive and rapid but ignores the actual sequence content of the CDR3 regions. Traditional Sanger sequencing can be used to determine CDR3 sequence identity,20 but the cost of this sequencing and the constraints on the number of cells that can reasonably be analyzed makes the approach impracticable for assessing CDR3 diversity.21 However, HTS technologies have enabled millions of TCR clonotypes to be identified.22-24 The use of ultra-deep TCR-sequencing technology reveals the clonal composition of T-cell populations and has proven helpful in efforts to better understand the immune response in patients with liver cancer.
HEC of T-cell repertoire may be the potential biomarkers for HCCs
In this work, we determined the contribution of HECs with frequencies of over 0.1% to the total T-cell repertoire, whereas Klarenbeek et al. determined the contribution of HECs occurring at frequencies that exceeded 0.5%. These authors observed that 84% of the clones were of low frequency (<0.1% of total TCR analyzed); above this value, the distribution decreased quickly, which it approaches at a clonal frequency of 0.4–0.5%.25 Through data analysis and comparison, we selected 0.1% as the standard of HECs in our current workflow, which proven to be an effective choice. Comparison of the T-cell repertoires of both tissue and blood could distinguish liver cancer patients from healthy adults or hepatitis patients based on the HEC ratio.
Cancer is a disease for which an early diagnostic would be immediately beneficial. Early cancer detection using cancer biomarkers is an exciting field. However, the complex response of the body to disease makes it difficult to characterize this response using only a few biomarkers. A recent study indicated that some complex heterogeneous diseases could be distinguished from other cancers and from conditions of health using immune markers, a finding that demonstrates the potential power of the immunosignature approach in the accurate, simultaneous classification of disease.26 In our research, we characterized the basic properties of T cells across multiple dimensions, including Vβ–Jβ combination usage. We compared peripheral blood samples from HCC patients with samples from healthy adults according to the total Vβ–Jβ usage analysis and found no significant differences. However, the TRBV18, TRBV4-1, TRBV4-2, and TRBV6-9 displayed higher AUC value, suggesting that these V usages may be potential classifier to distinguish HCCs from healthy adults. The conclusion was confirmed by LDA analysis.
When tumor tissues and adjacent tissues were compared, significant differences were found in TRBV6-4TRBJ1-1, TRBV6-4TRBJ2-2, TRBV6-4TRBJ2-3, and TRBV6-5TRBJ1-6. The observed usage bias of Vβ and Jβ is likely due to a combination of proximity effects such as recombination signal sequence compatibilities that influence initial TCR development, thymic selection and immune challenges that modify the representation of selected clones in the extant repertoires.27 Cancers are themselves heterogeneous, and individual response to disease, at a molecular level, can vary considerably. Here, we identified some specific TRBV and TRBJ combinations that distinguish the TCR repertoires of liver cancer patients and healthy adults. These specific TRBV and TRBJ combinations may offer new biomarkers.
Additionally, we determined the contribution of HECs occurring with a frequency of over 0.1% to the total T-cell repertoire. Whether the comparison among liver cancer, hepatitis and healthy adults was performed using blood or liver tissue, liver cancer and hepatitis B patients showed clear differences from healthy adults at the HEC level. And comparison of the HEC ratios in blood could distinguish liver cancer or hepatitis B patients from healthy controls. This provides a possible basis for non-invasive detection of liver cancer. Furthermore, the differences in observations in peripheral blood and tissues between hepatitis and liver cancer patients deserve our attention. We observed significant differences between patients with hepatitis B and liver cancer in blood (p < 0.01) and in tissues (p < 0.001), and these differences indicate that comparison of TCRB CDR3 in tissues is superior to comparison in the blood for the identification of hepatitis B. However, comparing the HEC ratio in blood was able to distinguish liver cancer and hepatitis B patients, thus providing an additional possible basis for non-invasive detection of liver cancer.
Difference of TCR repertoires characteristic in disease and health
We used pairwise comparison to analyze samples of the same type and calculated the proportion of CDR3 amino acid sequences they shared (Fig. S5–S6). We compared the proportion of shared clones in blood samples of healthy adults, hepatitis B and liver cancer patients (Fig. S5). The TCRβ repertoire of healthy adults is more diverse than that of patients with diseases. Comparison of blood and tissues of hepatitis B patients suggests that fewer immune cell species types were found in the tissue than in blood with the same initial amount of RNA (200 ng).
The shared unique clones of all blood samples of HCC and CC patients were also analyzed. Out of 82119, 51.36% (42178/82119) derived from healthy adults. While there was only 48.64% (39941/82119) shared clones between HCCs and CCs (data not shown). Pairwise analysis displayed the clone number ranged from 617 to 3247 among HCCs (Fig. S6A), 187 to 2473 among CCs (Fig. S6B), 556 to 2200 among health adults (Fig. S6C), suggesting that CDR3 clone has a strong heterogeneity between different individuals.
T cell recognition of a particular antigen needs the presentation of an HLA molecule and differences in HLA types may influence TCR repertoires. Warren et al. inferred that HLA-matched individuals may display increased TCRβ CDR3 repertoire overlap, which suggesting an influence of HLA type on T-cell repertoire features.28 But other deep profiling studies of unrelated subjects or monozygotic twins suggest that repertoire overlap between individuals is generally independent of HLA type.29-30 In the current results, we didn't detect the HLA type for individuals who participated in this study. In the future, we will simultaneously detect HLA type and TCR sequencing and study the relationship of HLA and TCR repertoires.
The ability to mount a protective immune response depends on the diversity of T cells, and the aging process threatens this diversity.31-33 A recent study indicates that TCRβ CDR3 diversity declines throughout life.34 The authors of that study directly quantified and compared T-cell repertoire diversity in samples obtained from 39 healthy donors 6–90 years of age using an advanced deep TCRβ sequencing approach. In our study, we observed no significant change in TCR diversity with age. One likely reason for this discrepancy in the findings is intrinsic differences in the samples used for comparison; in our case, the samples were obtained from liver cancer patients, in whom TCRβ diversity had already decreased. Thus, the increase in the proportion of expanded clones that occurs with age was hidden by the presence of disease. The infiltration of human tumors by T cells is a common phenomenon, and the nature of these intratumoral T-cell populations can predict the course of disease.35
Tumor infiltrating lymphocyte in cancer
In this research, we analyzed the consistency of the TCR repertoires of tumor tissues and adjacent tissues in HCC, ICC and MHC patients. In HCC and ICC patients, the consistency of tumor tissues and adjacent tissues was significantly different (p < 0.01), a finding that may be associated with the range of malignancy observed in these tumors and that emphasizes the reliability of our detection method and demonstrates its potential power in the classification of liver cancer. Two patients, referred to as ‘a’ and ‘b’ here, for whom the consistency values were far from the average values of their respective groups, caught our attention. Patient ‘a’ presented a poorly differentiated HCC that was classified as grade III according to Edmondson–Steiner's classification,36 whereas ICC patient ‘b’ presented a highly differentiated mucinous adenocarcinoma according to histopathological examination. These observations suggest that our results are consistent with diagnoses made by histopathological examination.
Recent evidence suggests that in colorectal and ovarian carcinoma patients, the presence or absence of tumor-infiltrating lymphocytes (TILs) provides a strong prognostic marker for survival independent of current staging methods.37,38 The importance of TILs in the prognosis of melanoma patients has also been known for many years.39 It has been postulated that biomarkers could be developed to capture the TIL response for both cancer prognosis and the prediction of therapeutic response. Many groups are testing various types of TIL measurements as predictive biomarkers for immunotherapeutic responses.40,41 T cells directed toward cancer cells can significantly impact clinical outcome, as shown by a number of studies that have found a correlation between increased numbers of TILs and improved survival in a variety of tumor types.42
In conclusion, high-throughput analyses of the TCR repertoire of a tissue can be performed using a deep sequencing platform, and these analyses help provide a better understanding of the immune response in liver cancer patients in whom the multi-faceted T-cell response is comparable to that found in healthy volunteers and HBV-infected patients. Further studies are needed to clarify the functional basis of TCRβ CDR3 clonalities underlying the persistence and/or eradication of cancer cells. The diversity of CDR3 sequences in liver cancer tissue may be a novel biomarker for detecting aggressive tumors with high invasive or metastatic capacity.
Materials and Methods
Patients and sample collection
Peripheral blood of healthy adults and patients was collected in PAXgene blood RNA collection tubes. All HCC tissue specimens were obtained from patients who underwent surgical resection for their tumors and who provided informed consent prior to liver surgery. The primary tumor specimens were immediately frozen at −80°C until RNA extraction. Specimens (approximately 1 cm3) of the tumor and adjacent liver tissue were collected from each patient, and the diagnosis of HCC was confirmed through pathological examination. This project and protocols involving human and animal tissues were approved by the ethics committee of the Chinese National Human Genome Center.
RNA extraction
The PAXgene Blood RNA System* was used as a blood collection tube (Becton, Dickinson and Company, USA). Total RNA was extracted from blood samples using a nucleic acid purification kit (PAXgene Blood RNA Kit) according to the manufacturer's instructions. To reduce the risk of genomic DNA contamination, 1–2 μg RNA was incubated with 2 U DNase I (Invitrogen, Carlsbad, CA, USA), 1 μl DNase buffer and 0.4 μl RNaseOut for 15 min at room temperature. The RNA concentration of the sample was determined using spectrophotometry, and the total RNA integrity was examined by visualization of the 28S and 18S RNA transcripts on a 1.2% agarose gel. The quality of RNA was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA).
Library construction
In this study, we used the HTBI primers and Arm-PCR from iRepertoire to construct the libraries including PCR1 and PCR2, inclusively and semi-quantitatively. During the first round of PCR1, only 15 cycles were used to amplify CDR3 fragments using the specific primers against each V and J genes. And in the second round, PCR was performed using universal primers.
PCR1: RNA reverse transcription and amplification of the T-cell receptor β CDR3 using the HTBI primers (Huntsville, Alabama, America) was carried out using Qiagen OneStep RT-PCR. The first round of PCR was performed using 200 ng of total RNA mixed with 4 μl random iRepertoire primers, 5 μl 5 × buffer, 1 μl dNTP mix, 0.25 μl RNasin (40 U/μl), and 1 μl enzyme mix, with nuclease-free water added to reach a total volume of 25 μl. After mixing and centrifugation, the reactions were transferred to a thermal cycler that carried out the following program: one cycle of 50°C for 40 min; one cycle of 95°C for 15 min; 15 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 40 min, and extension for 30 s at 72°C; 10 cycles of denaturation at 94°C for 30 s, annealing and extension at 72°C for 2 min; and a final extension at 72°C for 10 min. The samples were then held at 4°C.
PCR2: A 2 μl sample of the PCR1 product was used as template for a second step of amplification following the addition of 5 μl communal primers, 25 μl Multiplex MM prepared using the Multiplex PCR Kit (Hilden, Nordrhein-Westfalen, Germany), and 18 µl nuclease-free water to reach a total volume of 50 μl. The reactions were then transferred to a thermal cycler that carried out the following program: one cycle of 95°C for 15 min; 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s; and final extension at 72°C for 5 min. The samples were then held at 4°C. Size selection was used to purify 250-bp PCR products on magnetic beads (Agencourt No. A63882, Beckman, Beverly, MA, USA). After gel purification, the PCR product was subjected to HTS using the Illumina Hiseq2000 platform.
Sequencing using the Illumina Hiseq2000
We used the same workflow described elsewhere to perform cluster generation,43 template hybridization, isothermal amplification, linearization, blocking and denaturation and hybridization of the sequencing primers. Paired-end sequencing of samples was carried out with a read length of 100 bp using the Illumina Hiseq2000 platform.
Analysis of Illumina sequence data
We amplified all CDR3 sequences present in 160 samples obtained from healthy adults and patients with hepatitis, liver cancer and CC. To reduce the impact of sequencing errors, we first filtered the sequence reads according to four strict criteria, removing the following: (1) reads contaminated by the adapter sequence; (2) reads with more than 5% uncalled bases (N); (3) reads with an average Phred-type Q-score <15; and (4) PE reads with low-quality base readings(Q-score <10)at the ends of reads or short reads (Reads1 length <60 bp; Reads2 length <50 bp). We then merged the high-quality paired reads using COPE and FqMerger (BGI, S henzhen, China), designating the results as contigs. The contigs were subsequently aligned to reference TCR Vβ/Dβ/Jβ gene sequences (http://www.imgt.org/download/GENE-DB/) using BLAST. We referenced directory sets of sequences containing the human TRB V-REGION, D-REGION, and J-REGION alleles (http://www.imgt.org/). The TCRβ CDR3 regions were identified within the sequencing reads according to the definition established by the International ImMunoGeneTics (IMGT) collaboration.44 Finally, we filtered the alignment results to remove the following: (1) low frequency contigs for which the number of the supported reads was less than 2; (2) contigs that failed to match V or J reference sequences; (3) contigs merged onto the Vβ and Jβ in the opposite direction; (4) contigs containing stop codons; and (5) contigs for which the length of the CDR3 sequence was not a multiple of 3. In the end, we identified 48 Vβ and 13 Jβ segments, of which 48 Vβ segments were merged into 23 Vβ segments. These results were used to analyze the Vβ and Jβ usage of the CDR3 amino acid clonotypes in each sample.
Statistical analysis
Clones with a frequency that exceeded 0.1% were considered to be HECs. The Pearson correlation coefficient (r) was used to measure the linear correlation between pairs of variables. Comparisons between groups were performed using two-tailed t-tests. Two-sided p values <0.05 were considered statistically significant.
Principal component analysis
We performed principal component analysis by using the fast.prcomp function in the package ‘gmodels’ in R. 10 out of 20 healthy cases and 10 out of 20 HCCs were random selected as training set, and other 10 healthy samples and 10 HCCs were used as testing set. We first calculate the relative abundance of VJ (V and V merge) of each sample, and selected differential genes with p values less than 0.01 (t-test) in training set for PCA to determine the coefficient of linear combination of the first principal component and the second principal component. Then we calculate the value of linear combination of the PCA1 and PCA2 of the samples from testing set according to the determine coefficient of linear combination of training set. The corresponding values of PCA1 of testing set were used for further ROC curve estimation, assessing the generalization of the classifier.
Linear discriminant analysis
LDA was used to evaluate separability of the two subject cohorts (HCC and control) of testing set using V usage. 10 out of 20 healthy cases and 10 out of 20 HCCs were random selected as training set, and other 10 healthy samples and 10 HCCs were used as testing set. We first calculate the relative abundance of V of each sample and selected differential V genes (p < 0.01) of training set to construct model by using LDA module in R. Then, we used predict function to calculate the corresponding LD1 value of testing set for validation.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
JH and XY conceived and designed the study. YXH, YQW, XLW, YFG, YXZ, HML, and JH performed immune repertoire sequencing and analyzed the sequence data set. XL, YO, QY, WQH, YW, RHW, MZ, DXZ, HMZ, and LY performed molecular experiments. BPZ, XCC contributed the blood and tissue samples. JH and YXH integrated, analyzed and interpreted all the data. JH, YXH and CH contributed to the supervision of the work. JH and YXH wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
We gratefully acknowledge support from the National High Technology Research and Development Program of China (863 Program, 2012AA02A205), the National Natural Science Foundation of China (81272306 and 81472639), the Shanghai Commission for Science and Technology (11JC1408800), the Program of Shanghai Subject Chief Scientist (12XD1421400), and the Program of Shenzhen Science Technology and Innovation Committee (JCYJ20130329171031740, CXZZ20130515163643 and JCYJ20120831144704366).
References
- 1.World Cancer Report 2014 World Health Organization. 2014. pp. Chapter 1.1. ISBN 9283204298 [Google Scholar]
- 2.Irfan A, Dileep NL. Malignant tumours of the liver. Surgery (Oxford) 2009; 27(1):30-7; http://dx.doi.org/ 10.1016/j.mpsur.2008.12.005 [DOI] [Google Scholar]
- 3.Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD, Thillainayagam AV et al.. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 2012; 61(12):1657-69; PMID:22895392; http://dx.doi.org/ 10.1136/gutjnl-2011-301748 [DOI] [PubMed] [Google Scholar]
- 4.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007; 132(7):2557-76; PMID:17570226; http://dx.doi.org/ 10.1053/j.gastro.2007.04.061 [DOI] [PubMed] [Google Scholar]
- 5.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 2013; 13(2):123-35; PMID:23344543; http://dx.doi.org/ 10.1038/nrc3449 [DOI] [PubMed] [Google Scholar]
- 6.Rosen HR. Clinical practice. Chronic hepatitis C infection. N Engl J Med 2011; 364(25):2429-38; PMID:21696309; http://dx.doi.org/ 10.1056/NEJMcp1006613 [DOI] [PubMed] [Google Scholar]
- 7.Bolondi L, Gramantieri L. From liver cirrhosis to HCC. Intern Emerg Med 2011; Suppl 1:93-8; PMID:22009618; http://dx.doi.org/24172704 10.1007/s11739-011-0682-8 [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Canc Epidemiol Biomarkers Prev 2010; 19(8):1893-907; PMID:20647400; http://dx.doi.org/24172704 10.1158/1055-9965.EPI-10-0437 [DOI] [PubMed] [Google Scholar]
- 9.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood 2009; 114(19):4099-107; PMID:19706884; http://dx.doi.org/ 10.1182/blood-2009-04-217604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodsworth DJ, Castellarin M, Holt RA. Sequence analysis of T-cell repertoires in health and disease. Genome Med 2013; 5(10):98; PMID:24172704; http://dx.doi.org/ 10.1186/gm502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature 1988; 334(6181):395-402; PMID:3043226; http://dx.doi.org/ 10.1038/334395a0 [DOI] [PubMed] [Google Scholar]
- 12.Rubtsova K, Scott-Browne JP, Crawford F, Dai S, Marrack P, Kappler JW. Many different Vbeta CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci U S A 2009; 106(19):7951-6; PMID:19416894; http://dx.doi.org/ 10.1073/pnas.0902728106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolotin DA, Mamedov IZ, Britanova OV, Zvyagin IV, Shagin D, Ustyugova SV, Turchaninova MA, Lukyanov S, Lebedev YB, Chudakov DM. Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. Eur J Immunol 2012; 42(11):3073-83; PMID:22806588; http://dx.doi.org/ 10.1002/eji.201242517 [DOI] [PubMed] [Google Scholar]
- 14.Emerson RO, Sherwood AM, Rieder MJ, Guenthoer J, Williamson DW, Carlson CS, Drescher CW, Tewari M, Bielas JH, Robins HS. High-throughput sequencing of T cell receptors reveals a homogeneous repertoire of tumor-infiltrating lymphocytes in ovarian cancer. J Pathol 2013; 231(4):430-40; PMID:24027095; http://dx.doi.org/24122851 10.1002/path.4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlinger M, Quezada SA, Peggs KS, Furness AJ, Fisher R, Marafioti T, Shende VH, McGranahan N, Rowan AJ, Hazell S et al.. Ultra-deep T-cell receptor sequencing reveals the complexity and intratumour heterogeneity of T-cell clones in renal cell carcinomas. J Pathol 2013; 231(4):424-32; PMID:24122851; http://dx.doi.org/ 10.1002/path.4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng WK, Armstrong R, Arai S, Desmarais C, Hoppe R, Kim YH. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci Transl Med 2013; 5(214):214ra171; PMID:24307695; http://dx.doi.org/ 10.1126/scitranslmed.3007420 [DOI] [PubMed] [Google Scholar]
- 17.Liu B, Yuan J, Yiu SM, Li Z, Xie Y, Chen Y, Shi Y, Zhang H, Li Y, Lam TW et al.. COPE: an accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics 2012; 28(22):2870-4; PMID:23044551; http://dx.doi.org/ 10.1093/bioinformatics/bts563 [DOI] [PubMed] [Google Scholar]
- 18.Gorski J, Yassai M, Zhu X, Kissela B, Kissella B, [corrected to Kissela B], Keever C, Flomenberg N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. Correlation with immune status. J Immunol 1994; 152(10):5109-19; PMID:8176227 [PubMed] [Google Scholar]
- 19.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci U S A 1993; 90(9):4319-23; PMID:8483950; http://dx.doi.org/ 10.1073/pnas.90.9.4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K et al.. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 2004; 21(6):793-803; PMID:15589168; http://dx.doi.org/ 10.1016/j.immuni.2004.10.010 [DOI] [PubMed] [Google Scholar]
- 21.Kircher M, Kelso J. High-throughput DNA sequencing-concepts and limitations. BioEssays 2010; 32(6):524-36; PMID:20486139; http://dx.doi.org/ 10.1002/bies.200900181 [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Sanders CM, Yang Q, Schroeder HW Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR Jr, Davis RW et al.. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A 2010; 107(4):1518-23; PMID:20080641; http://dx.doi.org/ 10.1073/pnas.0913939107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM et al.. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A 2009; 106(48):20216-21; PMID:19875695; http://dx.doi.org/ 10.1073/pnas.0909775106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren RL, Freeman JD, Zeng T, Choe G, Munro S, Moore R, Webb JR, Holt RA. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res 2011; 21(5):790-7; PMID:21349924; http://dx.doi.org/ 10.1101/gr.115428.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klarenbeek PL, de Hair MJ, Doorenspleet ME, van Schaik BD, Esveldt RE, van de Sande MG, Cantaert T, Gerlag DM, Baeten D, van Kampen AH et al.. Inflamed target tissue provides a specific niche for highly expanded T-cell clones in early human autoimmune disease. Ann Rheum Dis 2012; 71(6):1088-93; PMID:22294635; http://dx.doi.org/ 10.1136/annrheumdis-2011-200612 [DOI] [PubMed] [Google Scholar]
- 26.Stafford P, Cichacz Z, Woodbury NW, Johnston SA. Immunosignature system for diagnosis of cancer. Proc Natl Acad Sci U S A 2014; 111(30):E3072-80; PMID:25024171; http://dx.doi.org/ 10.1073/pnas.1409432111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat Immunol 2003; 4(7):624-30; PMID:12830137; http://dx.doi.org/ 10.1038/ni0703-624 [DOI] [PubMed] [Google Scholar]
- 28.Warren RL, Freeman JD, Zeng T, Choe G, Munro S, Moore R, Webb JR, Holt RA.. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res 2011; 21:790-97; PMID:21349924; http://dx.doi.org/ 10.1101/gr.115428.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med 2010; 2:1-9; PMID:20811043; http://dx.doi.org/24711416 10.1126/scitranslmed.3001442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zvyagin IV, Pogorelyy MV, Ivanova ME, Komech EA, Shugay M, Bolotin DA, Shelenkov AA, Kurnosov AA, Staroverov DB, Chudakov DM et al.. Distinctive properties of identical twins' TCR repertoires revealed by high-throughput sequencing. PNAS 2014; 111:5980-5; PMID:24711416; http://dx.doi.org/ 10.1073/pnas.1319389111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larbi A, Pawelec G, Wong SC, Goldeck D, Tai JJ, Fulop T. Impact of age on T cell signaling: a general defect or specific alterations? Ageing Res. Rev 2011; 10(3):370-8; PMID:20933612; http://dx.doi.org/ 10.1016/j.arr.2010.09.08 [DOI] [PubMed] [Google Scholar]
- 32.Li G, Yu M, Lee WW, Tsang M, Krishnan E, Weyand CM, Goronzy JJ. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med 2012; 18(10):1518-24; PMID:23023500; http://dx.doi.org/ 10.1038/nm.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naylor K, Li G, Vallejo AN, Lee WW, Koetz K, Bryl E, Witkowski J, Fulbright J, Weyand CM, Goronzy JJ. The influence of age on T cell generation and TCR diversity. J Immunol 2005; 174(11):7446-52; PMID:15905594; http://dx.doi.org/ 10.4049/jimmunol.174.11.7446 [DOI] [PubMed] [Google Scholar]
- 34.Britanova OV, Putintseva EV, Shugay M, Merzlyak EM, Turchaninova MA, Staroverov DB, Bolotin DA, Lukyanov S, Bogdanova EA, Mamedov IZ et al.. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol 2014; 192(6):2689-98; PMID:24510963; http://dx.doi.org/ 10.4049/jimmunol.1302064 [DOI] [PubMed] [Google Scholar]
- 35.Linnemann C, Mezzadra R, Schumacher TN. TCR repertoires of intratumoral T-cell subsets. Immunol Rev 2014; 257(1):72-82; PMID:24329790; http://dx.doi.org/ 10.1111/imr.12140 [DOI] [PubMed] [Google Scholar]
- 36.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48 900 necropsies. Cancer 1954; 7(3):462-503; PMID:13160935; http://dx.doi.org/ 10.1002/1097-0142(195405)7:3%3c462::AID-CNCR2820070308%3e3.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- 37.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: Ameta-analysis. Gynecol Oncol 2012; 124(2):192198; PMID:22040834; http://dx.doi.org/ 10.1016/j.ygyno.2011.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galon J, Pagès F, Marincola FM, Thurin M, Trinchieri G, Fox BA, Gajewski TF, Ascierto PA. The immune score as a new possible approach for the classification of cancer. J Transl Med 2012; 10:1; PMID:22214470; http://dx.doi.org/ 10.1186/1479-5876-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, Saw RP, Thompson JF. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 2012; 30(21):2678-83; PMID:22711850; http://dx.doi.org/ 10.1200/JCO.2011.37.8539 [DOI] [PubMed] [Google Scholar]
- 40.Ascierto PA, Kalos M, Schaer DA, Callahan MK, Wolchok JD. Biomarkers for immunostimulatory monoclonal antibodies in combination strategies for melanoma and other tumor types. Clin Cancer Res 2013; 19(5):1009-20; PMID:23460532; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-2982 [DOI] [PubMed] [Google Scholar]
- 41.Liu C, Peng W, Xu C, Lou Y, Zhang M, Wargo JA, Chen JQ, Li HS, Watowich SS, Yang Y et al.. BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin Cancer Res 2013; 19(2):393-403; PMID:23204132; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour‐infiltrating lymphocytes in cancer: a systematic review with meta‐analysis. Br J Cancer 2011; 105(1):93-103; PMID:21629244; http://dx.doi.org/ 10.1038/bjc.2011.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y et al.. The diploid genome sequence of an Asian individual. Nature 2008; 456(7218):60-5; PMID:18987735; http://dx.doi.org/ 10.1038/nature07484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giudicelli V, Lefranc MP. IMGT/junction analysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR). Cold Spring Harb Protoc 2011; 2011(6):716-25; PMID:21632777; http://dx.doi.org/ 10.1101/pdb.prot5634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.