Abstract
The 22q11.2 deletion syndrome (22q11DS; velocardiofacial/DiGeorge syndrome; VCFS/DGS) is the most common microdeletion syndrome and the phenotypic presentation is highly variable. Approximately 65% of individuals with 22q11DS have a congenital heart defect (CHD), mostly of the conotruncal type, and/or an aortic arch defect. The etiology of this phenotypic variability is not currently known. We hypothesized that copy-number variants (CNVs) outside the 22q11.2 deleted region might increase the risk of being born with a CHD in this sensitized population. Genotyping with Affymetrix SNP Array 6.0 was performed on two groups of subjects with 22q11DS separated by time of ascertainment and processing. CNV analysis was completed on a total of 949 subjects (cohort 1, n = 562; cohort 2, n = 387), 603 with CHDs (cohort 1, n = 363; cohort 2, n = 240) and 346 with normal cardiac anatomy (cohort 1, n = 199; cohort 2, n = 147). Our analysis revealed that a duplication of SLC2A3 was the most frequent CNV identified in the first cohort. It was present in 18 subjects with CHDs and 1 subject without (p = 3.12 × 10−3, two-tailed Fisher’s exact test). In the second cohort, the SLC2A3 duplication was also significantly enriched in subjects with CHDs (p = 3.30 × 10−2, two-tailed Fisher’s exact test). The SLC2A3 duplication was the most frequent CNV detected and the only significant finding in our combined analysis (p = 2.68 × 10−4, two-tailed Fisher’s exact test), indicating that the SLC2A3 duplication might serve as a genetic modifier of CHDs and/or aortic arch anomalies in individuals with 22q11DS.
Introduction
Congenital heart defects (CHDs) are the leading cause of birth defect-related deaths in newborns1 and are estimated to occur in 0.5% to 1% of live births.2 They can develop as an isolated abnormality or in conjunction with a syndromic condition. Approximately one third of CHDs result from malformations of the cardiac outflow tract and are collectively referred to as conotruncal heart defects (CTDs), examples of which include tetralogy of Fallot (TOF), pulmonary atresia with ventricular septal defect (VSD), truncus arteriosus, and interrupted aortic arch type B.3 Both genetic and environmental etiologies of CTDs have been described.4–6 With respect to genetic etiologies, CTDs have been identified in individuals with single gene disorders, gain or loss of entire chromosomes, and submicroscopic unbalanced structural rearrangements or copy-number variants (CNVs). One of the most common CNVs associated with CTDs is the 22q11.2 deletion.7,8
The 22q11DS (velocardiofacial syndrome; DiGeorge syndrome, VCFS/DGS [MIM: 192430, 188400]) is the most common microdeletion syndrome, affecting approximately 1 in 2,000–4,000 individuals.9,10 The vast majority of individuals with 22q11DS carry the typical 3 million base pair (3 Mb) deletion of one homolog of chromosome 22; nested, smaller interstitial 1.5–2 Mb 22q11.2 deletions are seen in <10% of individuals.11 Both the typical 3 Mb deletion and most nested interstitial deletions occur between low copy repeats that punctuate the 22q11.2 region.12 This deletion is usually de novo but can also be inherited.13 The 22q11DS phenotype is highly variable and includes CHDs, dysmorphic facial features, palatal anomalies, hypocalcemia, immunodeficiency, cognitive impairment, and various neuropsychiatric disorders. A variety of CHDs and/or aortic arch defects have been detected in approximately 65% of individuals with 22q11DS, the most prevalent of which are CTDs.14,15 The etiology of this cardiovascular phenotypic variability is not currently known, but it does not appear to correlate with sex, race, 22q11.2 deletion size, or parent of origin of the deletion.8,16,17
The variable expressivity and reduced penetrance of CHDs in 22q11DS (including aortic arch anomalies) is probably influenced by genetic factors because individuals with 22q11DS and a CHD are more likely to have an unaffected relative with an isolated CHD than individuals with 22q11DS that have normal intracardiac and aortic arch anatomy.8 These findings are not explained by the inheritance of the non-deleted chromosome 22, suggesting that the variants that influence the development of CHD in these families lie outside of the 22q11.2 region.8 More than 40 genes are in the typically deleted region in 22q11DS. One of the strongest candidate genes for CHD on 22q11DS is TBX1 (MIM: 602054), which encodes a T-box transcription factor.18–20 We previously sequenced coding exons of TBX1 in this cohort and did not find evidence for mutation on the remaining allele.21 Therefore, we hypothesized that individuals with 22q11DS and CHDs have structural variants that affect their risk of being born with intracardiac and/or aortic arch malformations, possibly through epistatic interactions with the dosage-sensitive gene(s) in the 22q11.2 deleted region.
Our study is an investigation in search of CNV genetic modifiers involved in the variable 22q11DS cardiac phenotype and represents the largest genomic study of a microdeletion syndrome performed to date. Genome-wide analysis of CNVs was performed on two separate cohorts of subjects with 22q11DS ascertained and processed in two different time periods: the first cohort consisted of 562 subjects (CHD, n = 363; no CHD, n = 199) and the second cohort comprised 387 subjects (CHD, n = 240; no CHD, n = 147) for a total of 949 subjects (CHD, n = 603; no CHD, n = 346). By analyzing 949 subjects with 22q11DS, we were able to identify a common CNV that was significantly enriched in 22q11DS-positive subjects with a CHD. This CNV and the gene it overlaps have not been previously reported in the CHD literature. The result supports the possibility that this is a genetic modifier of CHDs in individuals with 22q11DS.
Material and Methods
Subject Cohorts
Blood or saliva samples were obtained from subjects with 22q11DS, with their informed consent and in accordance with the ethical standards of the appropriate committees on human experimentation (Internal Review Board, 1999-201, Albert Einstein College of Medicine, NY; 07-005352_CR2 CHOP IRB). Two groups of subjects with 22q11DS were ascertained and processed at two distinct time points and were therefore treated as separate cohorts, referred to as cohort 1 and cohort 2. The recruitment goals for the two 22q11DS cohorts were a confirmed 22q11.2 deletion, self-reported as white of European descent, and (for familial cases) only one individual per family. Fluorescence in situ hybridization or multiplex ligation-dependent probe amplification (MLPA) testing was used to verify the 22q11.2 deletion in each subject and parents when available. Phenotypic information on intracardiac and aortic arch anatomy was obtained from echocardiograph and cardiology summary reports from the referring institutions; every subject enrolled in the study had an echocardiogram. The phenotypes of 227 of these subjects have been described in an earlier publication.21
A separate cohort of subjects with CTDs was recruited as part of a larger collaborative program (HD70454). These subjects tested negative for a 22q11.2 deletion and had no other recognizable genetic syndrome. A detailed description of the subject enrollment requirements, cardiac phenotypes, and array genotyping procedure for this non-deleted, CTD cohort has been published elsewhere.22
Genome-wide SNP Array, Quality Control, and CNV Detection
Genomic DNA samples from subjects with 22q11DS were analyzed with the Affymetrix SNP Array 6.0 platform according to the manufacturer’s instructions (Affymetrix) at the Genomics Core at Albert Einstein College of Medicine. Quality control values were calculated in Affymetrix Genotyping Console (Affymetrix) and any samples with contrast QC greater than 0.4 or mean absolute pairwise difference (MAPD) greater than 0.35 were excluded from further analysis. Only samples that had a typical 3 Mb 22q11.2 deletion or a proximal nested 22q11.2 deletion were included; all atypical deletions were excluded from the study. In addition, samples were removed if there was insufficient cardiac phenotype information about the subject or if the gender determined on the basis of X and Y chromosome SNP genotypes did not match their reported gender. SNP analysis was performed to exclude duplicate or related samples via estimation of identity by descent (IBD) with the PLINK software package.23 Figure S1A depicts this initial quality assessment and sample elimination.
A custom copy number (CN) baseline reference was generated with 215 Affymetrix SNP 6.0 arrays from phenotypically normal, non-22q11DS control individuals that were ascertained concurrently with the 22q11DS cohorts. These reference arrays were run in the same facility, the Genomics Core at Albert Einstein College of Medicine, during the same time period and with an identical protocol as our experimental 22q11DS arrays in order to control for any batch variation. The reference arrays were subjected to and passed the same QC metrics, and equivalent ratio of male:female arrays were used to prevent gender bias (106 male, 109 female).
The CNV detection was performed on all three cohorts (both 22q11DS groups and the non-syndromic CTD group) via two methods: PennCNV24 and CNV workshop.25 The PennCNV-Affymetrix tool was used to extract the signal intensity data from the raw .cel files.24 The canonical genotype clustering file used for CNV calling with PennCNV was generated from our custom CN baseline reference set.24 The log2ratios generated by PennCNV were used in CNV Workshop to produce CNV calls via circular binary segmentation.25 The B allele frequency and log R ratio plots were visualized with the Affymetrix Chromosome Analysis Suite to support CNV calls. The final QC step occurred after CNV detection: any samples with elevated log2ratio SD (>0.44) or a large number of detected CNVs (>300) were excluded from further analysis, as shown in Figure S1B. A total of 949 22q11DS samples, 562 in the first cohort and 387 in the second cohort, passed all of the QC metrics and were included in our analysis.
CNV Analysis
A list of autosomal CNVs detected by greater than 10 contiguous probes for deletions and greater than 20 probes for duplications was generated with relevant annotations for analysis. Only CNVs detected by both PennCNV and CNV Workshop were included in the analysis, because these CNVs are less likely to be false positives due to variation in algorithms. CNV boundaries were determined by averaging the breakpoint locations predicted by CNV workshop and PennCNV. Any CNVs with a 50% or greater overlap with centromere, telomere, immunoglobulin regions, and/or segmental duplications were excluded. In addition, olfactory receptor genes were removed from further analysis. Finally, we merged CNVs separated by less than 10 kb to consider them as possible single contiguous events. CNV detection and analysis was performed with the GRCh36/hg18 build, and then CNV coordinates were converted to the GRCh37/hg19 build via the UCSC Genome Browser LiftOver tool. All genomic coordinates presented in figures and tables herein are based on the February 2009 Human Genome Build (GRCh37/hg19).
Common and rare CNVs were analyzed separately. We defined rare CNVs as those with a frequency of less than 0.1% of a previously published control population (dbVaR: nstd54);26 all remaining CNVs were categorized as common. Here we focus on common CNVs to determine the impact of common variants as genetic modifiers of the 22q11DS cardiac phenotype.
In Silico Analysis of Gene Function
CNVs in subjects with 22q11DS that passed all selection criteria were annotated with the RefSeq gene set downloaded from the UCSC Table browser of the hg18 build. Gene Ontology (GO) annotations for each RefSeq gene were obtained from the Ensembl database and their Mammalian Phenotype Ontology (MPO) term annotations were retrieved from the Mouse Genome Informatics (MGI) database (July 2014 available version). Previously published analytical methods were employed to expand the annotation of the Gene Ontology and Mammalian Phenotype Ontology terms.27 For each functional term (GO and MPO), we directly compared the frequency of occurrence between cases and controls via Fisher’s exact test. Duplication and deletion events were evaluated separately after excluding genes deleted in the 22q11.2 region.
Statistical Analysis
The Fisher exact test or the Wilcoxon rank sum test was used for gene and CNV enrichment analyses. The Benjamin-Hochberg false discovery rate procedure was adopted as our default multiple test correction method. Where appropriate, the permutation-based false discovery rate estimation was applied to correct multiple testing for functional analyses.28
CNV Validation by qPCR
CNVs selected for validation were screened by real-time PCR. SYBR Green detection on an ABI SDS-7500 Fast Real-Time PCR system was used to quantify copy number (Applied Biosystems). Primer Express 3.0 software (ABI) and Primer 3 were used to design primers to amplify the region of interest; the specificity of each primer pair was tested with the UCSC In-Silico PCR tool. The average length of each amplicon was 62 base pairs (range: 51–97 base pairs). For each CNV, primers were designed to amplify two regions within the deleted/duplicated region and at least one set of primers was designed to amplify a flanking region with normal copy number. Each qPCR run included amplification of an endogenous control with known copy number (RPPH1). The sequences for all of the qPCR primers used in the CNV validation are listed in Table S5. Two DNA samples with normal copy number (one CEPH subject and one 22q11DS-positive subject with normal copy number at the particular CNV) were used as controls in each run. SYBR Green qPCR was also used to amplify the DNA from the available parents of subjects with 22q11DS to determine whether CNVs were inherited or de novo. 10 μl reactions were performed with 12.5 ng of DNA according to the manufacturer’s recommended protocol.
Assessing SLC2A3 Duplication Frequency in Control Populations
Publically available databases were examined to determine the frequency of SLC2A3 duplications in phenotypically normal individuals. Many of the studies submitted to these databases did not provide robust phenotypic information for control samples or a sample ID associated with each CNV. It is not possible to know how many samples are redundant or uniquely presented in a database without adequate sample ID information, which prevents a reliable calculation of the frequency of the SLC2A3 duplication within the various databases. We were therefore restricted to previously published studies where sufficient sample ID information was provided and individuals were rigorously vetted to ensure a phenotypically “normal” control cohort (dbVaR: nstd2129 and nstd5426).
Mouse Embryo Dissections
Mouse embryos in the SW background were isolated in cold PBS at E9.5 and E10.5. Somite pairs were counted to define stages: 19–21 pairs of somites were defined as E9.5 and 30–32 pairs of somites were defined as E10.5.
Whole-Mount In Situ Hybridization
Digoxigenin-labeled RNA probe for Slc2a3 was amplified by PCR from cDNA, with the primers 5′-TCCCCTCAGCTGCAGCCTACTT-3′ and 5′-TTGTTCAATCCCCCAGGGCCCT-3′, forward and reverse, respectively. The forward primer contained the T3 polymerase priming sequence and the reverse primer contained the T7 polymerase priming sequence. The PCR products were purified with the PCR Purification Kit (QIAGEN), and antisense RNA was in vitro transcribed and labeled with T7 RNA polymerase (Roche) and the DIG RNA Labeling Mix (Roche), via the Digoxigenin Labeling Method. A sense RNA was generated with T3 RNA polymerase following the same procedure as the antisense RNA. The sense RNA served as control for the specificity of the expression pattern observed with the antisense RNA. Digoxigenin-labeled RNA probes were purified with mini Quick Spin RNA Columns (Roche). Whole-mount in situ hybridization was performed as previously described.30
Results
CNV Analysis in Two Consecutively Ascertained Cohorts of Subjects with 22q11DS
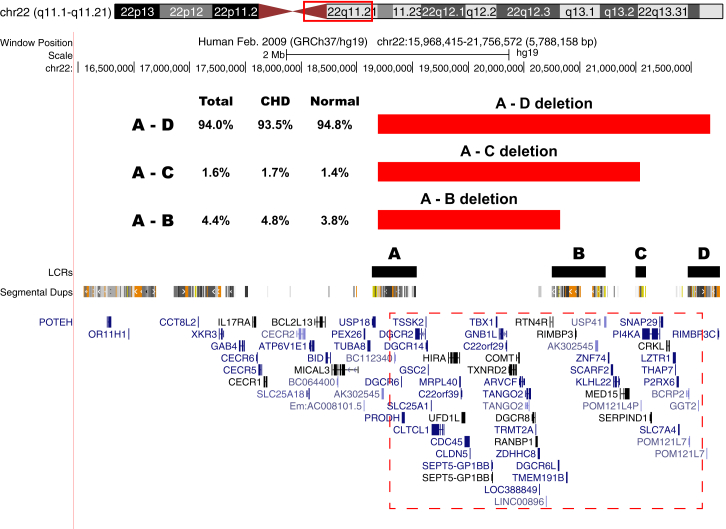
CNV analysis was performed with PennCNV and CNV workshop on 949 Affymetrix SNP6.0 arrays from two cohorts of subjects with 22q11DS, cohort 1 and cohort 2. The two cohorts were ascertained over different but consecutive time periods. When combined there were 603 individuals (cohort 1, n = 363; cohort 2, n = 240) with intracardiac defects and/or aortic arch defects and 346 (cohort 1, n = 199; cohort 2, n = 147) that had normal heart and aortic arch anatomy. The specific defects observed in these subjects are listed in Table 1. Each subject had a 22q11.2 deletion that included TBX1 (Figure 1), a strong candidate gene for the physical defects of the syndrome including CHDs.18–20 Although parental DNA was not available to test from all subjects, the majority of the 22q11.2 deletions in the study cohort were de novo events. Analysis of the 22q11.2 deletion sizes (Table S1) revealed that 93.5% of subjects (n = 564) with a CHD carried the typical 3 Mb (LCR-A to LCR-D) 22q11.2 deletion12,31 compared to 94.8% of subjects (n = 328) with a normal heart (p = 0.48, two-tailed Fisher’s exact test). Figure 1 illustrates the various 22q11.2 deletions and Table S1 contains the deletion size data broken down by cohort in addition to the overall frequencies. These findings indicate that the size of the 22q11.2 deletion does not play a role in the development of congenital cardiac defects.
Table 1.
Frequency of CHDs in Subjects with 22q11DS
| Type of CHD |
First Cohort |
Second Cohort |
||
|---|---|---|---|---|
| Totala | No. of Isolatedb | Totala | No. of Isolatedb | |
| ASD | 56 | 12 | 59 | 18 |
| IAAB | 41 | 3 | 25 | 0 |
| PS | 105 | 3 | 21 | 2 |
| PTA | 35 | 0 | 19 | 0 |
| RAA | 114 | 30 | 41 | 14 |
| TOF | 149 | 40 | 59 | 47 |
| VSDc | 118 | 15 | 81 | 47 |
Abbreviations are as follows: ASD, atrial septal defect; IAAB, interrupted aortic arch type B; PS, pulmonary atresia stenosis; PTA, persistent truncus arteriosus; RAA, right aortic arch; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Total number of subjects with each type of CHD. Subjects with >1 heart defects are included in multiple categories.
Number of subjects with isolated CHDs.
This category does not include VSDs found in association with TOF and PTA.
Figure 1.
Chromosome 22 Deletion Sizes
UCSC Genome Browser view of the 22q11.2 deletion sizes in the 949 subjects with 22q11DS from both cohorts. The typical 22q11.2 deletions are mediated by low copy repeats (LCRs); the deletions start at LCR A and end within LCR B, C, or D. The LCRs are shown in black directly above the segmental duplication track. Table S1 contains the exact frequency and distribution for each of the typical deletions. Individuals with atypical 22q11.2 deletions were excluded from analysis. The red hatched box contains the genes that are typically deleted.
A total of 13,518 CNVs outside of the 22q11.2 region were detected by both algorithms in the 949 subjects (Table 2). These CNVs are unlikely to be false positives because they were identified by two methods and passed the probe cutoffs that were chosen based on extensive validation of such CNVs in the past. There was no significant difference in the number of CNVs detected (14.29 ± 5.34 versus 13.88 ± 5.01, p = 0.37) or the average size of CNVs (607.72 ± 484.05 kb versus 575.48 ± 437.78 kb, p = 0.22) between subjects with CHDs including aortic arch anomalies and those with normal cardiovascular anatomy in either cohort or when the cohorts were combined (Table 2). CNVs affecting genes implicated in cardiac development and/or overrepresented in case or control subjects were chosen for qPCR validation (number of unique CNVs = 23; Table S2). The validation rate for CNVs that were identified by both algorithms (CNV Workshop and PennCNV, n = 70) was 100%, but was lower for CNVs predicted by only one algorithm (validation rate for CNVs identified only by CNV workshop = 35.5%; validation rate for CNVs identified only by PennCNV = 71.4%).
Table 2.
Total Number and Length of All Autosomal CNVsa in 22q11DS-Positive Subjects with and without CHDs
|
First Cohort |
Second Cohort |
Cohorts Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
CHD |
No CHD |
p Value |
CHD |
No CHD |
p Value |
CHD |
No CHD |
p Value | |
| (n = 363) | (n = 199) | (n = 240) | (n = 147) | (n = 603) | (n = 346) | ||||
| No. of male subjects (Percent male) | 174 (47.8%) | 107 (53.8%) | 0.22b | 119 (49.6%) | 68 (46.3%) | 0.53b | 293 (48.5%) | 175 (50.6%) | 0.59b |
| Mean no. of CNVs per subject ±SD | 14.34 ± 5.49 | 13.90 ± 5.39 | 0.31c | 14.09 ± 4.57 | 13.86 ± 4.45 | 0.83c | 14.29 ± 5.34 | 19.25 ± 5.01 | 0.37c |
| Mean CNV length (kb) per subject ±SD (kb) | 627.67 ± 519.78 | 568.15 ± 397.87 | 0.14c | 577.55 ± 423.61 | 585.39 ± 487.87 | 0.87c | 607.72 ± 484.05 | 575.48 ± 437.78 | 0.22c |
| No. with ≥1 CNV ≥500 kb | 47 | 24 | 0.79b | 25 | 19 | 0.51b | 72 | 43 | 0.84b |
Excluding the 22q11.2 deletion.
Fisher’s exact test.
Wilcoxon rank-sum test.
Identification and Analysis of the SLC2A3 Duplication
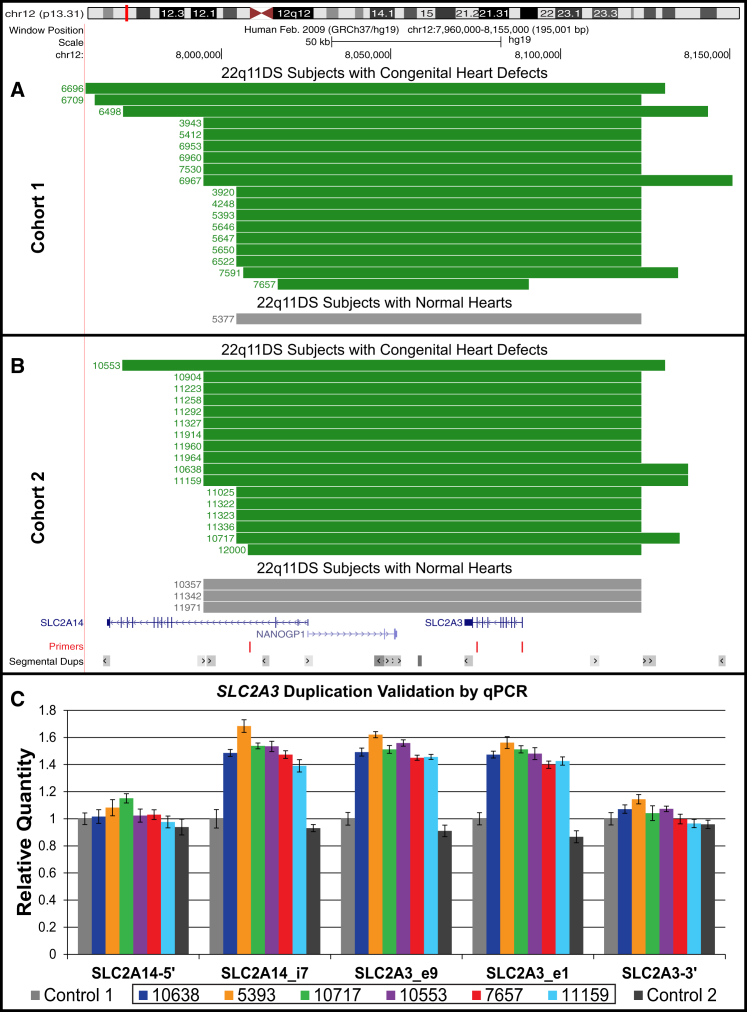
In the analysis of the first cohort, only a single CNV showed a statistically significant difference in frequency between 22q11DS-positive subjects with and without CHDs. A common duplication of chromosome 12p13.31 was detected in 18 (5.0%) 22q11DS-positive subjects with a CHD and in 1 (0.5%) 22q11DS-positive subject with a normal heart (p = 3.12 × 10−3, two-tailed Fisher’s exact test; Figure 2A). This duplication encompasses the entirety of SLC2A3 (solute carrier family 2, member 3 [MIM: 138170]), the pseudogene NANOGP1, and part of SLC2A14 (solute carrier family 2, member 14 [MIM: 611039]). SLC2A14 is expressed only in the testes32 and the pseudogene NANOGP1 is transcribed but not translated and therefore neither is relevant. Thus, this CNV at 12p13.31 is referred to as a duplication of SLC2A3 hereafter.
Figure 2.
SLC2A3 Duplication
(A and B) UCSC Genome Browser view depicting the duplication of the entire SLC2A3 gene, found in 39 subjects with 22q11DS. The duplications, which range from 74 to 172 kb in size, span the pseudogene NANOGP1 and part of SLC2A14, neither of which are relevant because the pseudogene is not translated and SLC2A14 expression occurs only in the testis.46 Each track corresponds to a unique subject with 22q11DS; the duplication is shown in green for individuals with CHDs and gray for those with a normal heart.
(A) The 19 individuals from the first cohort, 18 with a CHD and 1 without.
(B) The 20 individuals from the second cohort, 17 with a CHD and 3 without.
(C) SYBR green qPCR validation of the SLC2A3 duplication in 6 representative samples from both cohorts (10638, blue; 5393, orange; 10717, green; 10553, purple; 7657, red; 11159, aqua) compared to two controls (shown in gray). qPCR was performed in triplicate with primers designed to amplify regions proximal and distal to the SLC2A3 duplication as well as three regions within the predicted duplication as shown in (A) and (B). These graphics were constructed with the UCSC Genome Browser (GRCh37/hg19). The qPCR primer sequences are listed in Table S5.
The second cohort was then examined and an additional 20 SLC2A3 duplications were identified: 17 (7.1%) subjects had a CHD and 3 (2.0%) did not. The SLC2A3 duplication was also significantly enriched in individuals with CHDs within the second cohort (p = 3.30 × 10−2, two-tailed Fisher’s exact test; Figure 2B). In total, 39 individuals with a duplication of SLC2A3 were identified: 35 in 22q11DS-positive subjects with CHD (5.8%) and in 4 (1.1%) of the 22q11DS-positive subjects with normal hearts (p = 2.68 × 10−4, two-tailed Fisher’s exact test; Figure 2). A duplication of the entire SLC2A3 gene has been previously observed in 45/2,026 (2.2%) of healthy individuals.29 Thus, it appears that the overall frequency of this duplication in our entire cohort with 22q11DS (4.1%) is substantially greater than that seen in healthy controls.
The SLC2A3 duplication was validated with qPCR in all 37 individuals for whom DNA samples were still available (Figure 2C shows representative qPCR data). We investigated whether the SLC2A3 duplication was de novo or inherited in the 13 probands with parental DNA available for qPCR analysis (11 case subjects with CHDs and 2 controls without). As expected for a common variant, 100% of the individuals tested had inherited the SLC2A3 CNV from an unaffected parent that did not have a 22q11.2 deletion or a CHD (9 maternal; 3 paternal; 1 case where mother and father both carried the SLC2A3 duplication). A deletion of the SLC2A3 CNV was identified in 9 subjects with a CHD (1.4%) and 4 subjects with a normal heart (1.1%) (p = 0.78, two tailed Fisher’s exact test). This indicates that only a duplication of SLC2A3 and not the hemizygous deletion is associated with CHDs in subjects with 22q11DS.
The CNV analysis of the non-deleted CHD cohort (627 subjects with CHDs and 2,980 normal controls of European descent) was performed as previously described.27 The detected CNVs were examined to determine whether SLC2A3 duplications were associated with non-syndromic CHDs. A duplication of SLC2A3 was identified in 19 of the 627 individuals with a CHD (3.0%) and in 75 of the 2,980 control subjects with no reported CHD (2.5%). The SLC2A3 duplication was not enriched among individuals with CHDs in this non-syndromic cohort (p = 0.49, two tailed Fisher’s exact test). Deletions of SLC2A3 were identified in 3 individuals with a non-syndromic CHD and 22 normal controls (p = 0.60, two tailed Fisher’s exact test), which is a similar distribution as was seen in the 22q11DS cohorts.
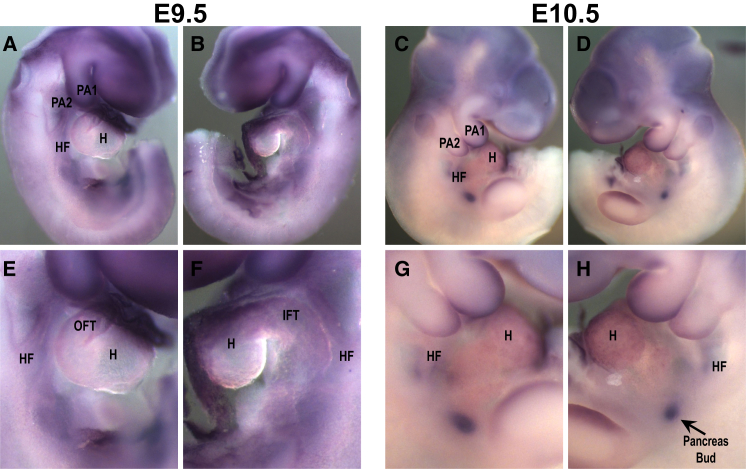
Slc2a3 RNA In Situ Hybridization of Mouse Embryos
Whole-mount in situ hybridization was performed to determine whether Slc2a3 is expressed in the murine pharyngeal apparatus and/or heart during development. Cells from the pharyngeal apparatus migrate into the cardiac outflow tract to form the conotruncal region. Slc2a3 was expressed in the brain, pharyngeal arches, and outflow tract but less so in the heart and placenta at embryonic days 9.5 and 10.5 (Figure 3). It was also expressed in the pancreatic bud at embryonic day 10.5.
Figure 3.
Slc2a3 RNA In Situ Hybridization
(A–D) Whole-mount in situ hybridization on wild-type mouse embryos showing the expression pattern of Slc2a3 (Glut3) at embryonic stages E9.5 (A and B) and E10.5 (C and D). Panels (A) and (C) show the right side of the embryos, and panels (B) and (D) show the left side. Slc2a3 is expressed in the brain, pharyngeal arches 1 and 2 (PA1 and PA2), in the heart field (HF), and in the heart (H) at both embryonic stages.
(E–H) Higher magnification of the central part of the embryos shown in (A)–(D). Slc2a3 is also expressed in the placenta (not shown) and in the pancreas bud at E10.5 (H, white arrow).
Abbreviations are as follows: OFT, outflow tract; IFT, inflow tract.
In Silico Analysis of CNV Function
To determine relevance of the common CNVs observed in subjects with 22q11DS to cardiac development, we used phenotype data from Gene Ontology and Mouse Genome Informatics Resource as previously described.28 Gene Ontology (GO) analysis was performed in order to examine the annotated biological processes, cellular components, and/or molecular functions of genes impacted by CNVs in CHD case versus control subjects. Mammalian Phenotype (MP) analysis was performed to investigate the various phenotypes associated with genes impacted by CNVs in CHD case versus control subjects. The common and rare CNVs were evaluated separately for each analysis. The rare CNV analysis was less informative and will be reported elsewhere (E.E.M. and B.S.E., unpublished data). The results from the GO analysis are listed in Table S3. The only GO terms that were statistically significant after B-H/FDR adjustment were all related to SLC2A3 ontologies and function. The results from the MP analyses are listed in Table S4. Several of the statistically significant MP terms enriched in 22q11DS-positive subjects with CHDs involved physiological defects and lethality such as “abnormal extraembryonic tissue physiology” and “complete embryonic lethality” (Table S4).
Discussion
The goal of our study was to determine whether structural genetic variants outside the 22q11.2 commonly deleted region explain the incomplete penetrance of CHDs in 22q11DS. Our analysis of subjects with 22q11DS, divided into two cohorts based upon time period of ascertainment and processing, revealed that the number of autosomal CNVs in those with CHDs compared to subjects with a normal heart was not significantly different in either cohort (Table 2). Furthermore, there was no correlation between the size of the 22q11.2 deletion and cardiac phenotype (Figure 1 and Table S1). However, one common CNV was found to be significantly associated with subjects with CHDs, suggesting that it might have an influence on cardiac development in the presence of a 22q11.2 deletion.
Duplication of SLC2A3
One common CNV, the duplication of SLC2A3, was significantly enriched in 22q11DS-positive subjects with a CHD. In total, the SLC2A3 duplication was detected in 35 22q11DS-positive subjects with CHD (5.8%) and in 4 22q11DS-positive subjects with normal hearts (1.1%) (p = 2.68 × 10−4, two-tailed Fisher’s exact test; Figure 2). This is the first study to report an association between a gain of SLC2A3 and CHD. The SLC2A3 duplication does not correlate with a specific type of heart defect in our 22q11DS-positive population, as shown in Table 3. This CNV is present in 2.2% of healthy individuals (dbVaR: nstd21)29 and was inherited from a parent in all 13 of the subjects with 22q11DS with available parental DNA. Cooper et al. identified a SLC2A3 duplication in 1/575 subjects with cardiovascular disease (0.17%) and in 143/8,329 controls (1.7%) (dbVaR: nstd54).26 Collectively, these data suggest that the SLC2A3 duplication is benign unless it is inherited in combination with the 22q11.2 deletion. This intriguing finding seems to be even more compelling because it was initially found in excess in the first cohort and then it was “replicated” in a separate, consecutively ascertained group of subjects with 22q11DS (i.e., in cohort 2).
Table 3.
Cardiac Phenotype for 22q11DS-Positive Subjects with the SLC2A3 Duplication
| ID | Gender | Inherited | Intracardiac Phenotype | Aortic Arch Phenotype |
|---|---|---|---|---|
| Cohort 1 | ||||
| 6960 | F | paternal | TOF, PDA | RAA, PS |
| 5412 | M | ND | TOF | PS |
| 5393 | F | ND | TOF | RAA |
| 3920 | M | ND | TOF | aberrant RSCA |
| 3943 | F | ND | TOF | RAA, aberrant LSCA |
| 7657 | F | ND | TOF | normal |
| 5647 | F | ND | TOF | normal |
| 6522 | M | ND | VSD, ASD, bicuspid aortic valve, PDA | IAAB |
| 6953 | M | ND | VSD, PFO | aberrant RSCA, IAAB |
| 6498 | F | maternal | VSD, ASD | aberrant RSCA, TGA, PS |
| 7591 | M | ND | VSD, ASD | aberrant RSCA |
| 5646 | F | ND | VSD, bicuspid aortic valve | RAA, PS |
| 6709 | M | maternal | VSD | RAA, aberrant LSCA |
| 5650 | F | ND | ASD | normal |
| 6696 | F | ND | PTA | aberrant RSCA |
| 4248 | F | maternal | normal | aberrant RSCA |
| 6967 | F | ND | normal | aberrant LSCA |
| 7530 | F | maternal | normal | aberrant RSCA |
| 5377 | M | ND | normal | normal |
| Cohort 2 | ||||
| 10638 | M | ND | VSD, ASD, PDA | coarctation |
| 11025 | F | ND | VSD, ASD | normal |
| 11336 | M | ND | VSD, ASD | normal |
| 10553 | M | paternal | VSD | IAAB |
| 11964 | F | botha | VSD | IAAB |
| 10717 | F | maternal | VSD | normal |
| 11223 | F | ND | VSD | normal |
| 11914 | M | ND | VSD | normal |
| 11292 | F | ND | VSD | normal |
| 11322 | M | ND | VSD | PS |
| 10904 | M | ND | ASD, PDA | normal |
| 11327 | F | ND | ASD | normal |
| 11258 | M | maternal | PDA | RAA, aberrant LSCA |
| 11960 | F | paternal | bicuspid aortic valve | aberrant RSCA |
| 11323 | F | ND | unspecified congenital heart defectb | normal |
| 11159 | F | maternal | normal | RAA, aberrant LSCA, vascular ring |
| 12000 | F | ND | normal | aberrant LSCA |
| 10357 | F | ND | normal | normal |
| 11342 | M | maternal | normal | normal |
| 11971 | F | maternal | normal | normal |
Abbreviations are as follows: LSCA, left subclavian artery; RSCA, right subclavian artery; PDA, patent ductus arteriosus; PS, pulmonary atresia stenosis; PTA, persistent truncus arteriosus; RAA, right aortic arch; TGA, transposition of the great arteries; ND, DNA not available for testing.
Mother and father both carry the SLC2A3 duplication.
Required surgical intervention 3 months after birth.
SLC2A3, formerly known as GLUT3, encodes a facilitated glucose transporter. SLC2A3 was first isolated from neurons and was originally considered to be a neuronal-specific glucose transporter, but studies have shown that SLC2A3 is expressed in a variety of human tissues.33 SLC2A3/GLUT3 is important in tissues with heightened energy demands and high metabolic rates because GLUT3 has the highest glucose affinity and greatest transport capacity in the GLUT protein family; expression is most abundant in the brain, high in adult cardiac myocytes, liver, placenta, and at a barely detectable level in kidney.33,34 Here we have shown that murine Slc2a3 is expressed in the pharyngeal apparatus and cardiac outflow tract at embryonic days 9.5 and 10.5 during murine cardiac morphogenesis (Figure 3). Previous work done in rats showed that GLUT3 is the prevailing glucose transporter in cardiomyoblasts and therefore it has been suggested that GLUT3 has a predominant role during cardiac development.35 Grover-McKay et al. determined that SLC2A3 is expressed during human heart development because SLC2A3 protein was present in the fetal myocardium at 10 weeks, increased protein levels were detected at 15 weeks, and the levels then decreased at 20 weeks of gestation.36 Recent work has shown that there are dynamic expression changes in fetal myocardium during development: SLC2A3 transcripts were detected at 15.9-fold higher levels than in newborn infants.37 Together these observations indicate that SLC2A3 might be involved in cardiac development, but the specific function of SLC2A3 in the heart during embryogenesis has not been delineated.
The importance of SLC2A3/GLUT3 during development has been well documented. SLC2A3 is the main glucose transporter responsible for transplacental transport of maternal glucose, thereby controlling the rate at which glucose is delivered to the fetus.38 SLC2A3 expression adaptively responds to glucose demands of fetal growth during normal development. Aberrant levels of SLC2A3 have been linked to intrauterine growth retardation and pregnancy loss.39–41 Animal models have shown that SLC2A3 alterations can cause extremely deleterious developmental defects. The homozygous null deletion of Slc2a3 is embryonic lethal in mice and the heterozygous deletion is not lethal but results in intrauterine growth retardation.39,42 Knockdown of the SLC2A3 zebrafish ortholog, slc2a3a, increased apoptosis and was embryonic lethal.43 Thus, although animal models have clearly demonstrated that loss of SLC2A3 can cause significant defects, currently there are no reports in the literature about the developmental effect of a SLC2A3 duplication or overexpression in animal models. However, it has been shown in a variety of human cell types that a duplication of SLC2A3 results in significantly increased expression and protein levels.44,45
There are two recent reports of SLC2A3 CNVs in association with human disease phenotypes.45,46 Both duplications and deletions of SLC2A3 were identified in a genetic study of rheumatoid arthritis (MIM: 180300). The deletions were deemed protective against rheumatoid arthritis whereas duplications of SLC2A3 had no effect.46 SLC2A3 duplications and deletions were also detected in subjects with Huntington disease (MIM: 143100); the duplication correlated with delayed age of onset.45 It is important to note that the individuals in the Huntington disease and rheumatoid arthritis studies do not carry the 22q11.2 deletion. In the absence of the 22q11.2 deletion, individuals with a duplication of SLC2A3 do not present with congenital heart defects, indicating that both mutations might be required for the manifestation of a CHD.
Gains overlapping SLC2A3 have not been reported in studies of non-syndromic CHDs although they have been detected at the same frequency as the general population.47 In our analysis of a different non-syndromic cohort, the SLC2A3 duplication showed no enrichment (p = 0.49, two-tailed Fisher’s exact test) (unpublished data). These results are consistent with the fact that, in the 22q11.2-deleted cohorts, the SLC2A3 duplication is inherited from unaffected parents. Thus, it appears likely that there might be an epistatic interaction between the SLC2A3 duplication and dosage-sensitive gene(s) in the 22q11.2-deleted region that increase the likelihood of a structural cardiac defect. Therefore, SLC2A3 is probably a modifier of the 22q11DS cardiac phenotype that exemplifies a “two-hit” model.
Genetic modifiers and the two-hit model are not unique to 22q11DS, but thus far the two-hit model has been used to explain only the variable expressivity of CNVs associated with neurodevelopmental phenotypes.48 The SLC2A3 duplication might be a genetic modifier of the 22q11DS cardiac phenotype. However, it does not completely explain the etiology of heart defects in the 22q11DS-positive population because the CNV was seen in only 5.8% of subjects with a CHD and also in 4 individuals with a purportedly “normal” heart. We classified these 4 subjects as controls because intracardiac defects were not noted in their echocardiogram report. Although these individuals have a normal left arch, it is quite possible they have an aberrant right subclavian that was not reported because very few look for this type of abnormality or because the imaging was insufficient to detect it. Alternatively, the combined effect of the SLC2A3 duplication with the 22q11.2 deletion increases the risk of having a CHD but might not be sufficient to cause a structural defect, and perhaps something additional in the genetic background and/or possibly exposure to epigenetic, environmental, or maternal factors in utero might be necessary for the manifestation of a CHD.
Previously Identified CNVs Associated with Non-syndromic CHDs
A number of recent studies have examined the prevalence of CNVs in non-syndromic individuals with CHDs. These studies have illustrated that rare CNVs can play a role in the pathogenesis of non-syndromic CHDs.47,49–53 Both 22q11DS cohorts were assessed for the CNVs reported in these non-syndromic studies. With the exception of the 22q11.2 deletion reported by Greenway et al.,52 none of the previously identified CNVs were detected or significantly associated with CHDs in our 22q11DS cohorts. This is not surprising because the non-syndromic CHD studies focused predominantly on rare CNVs, which are mechanistically distinct from the goal of this study. Rather than looking for causal primary lesions in the form of rare CNVs, we started with the 22q11.2 deletion and investigated the genomes of deleted individuals for CNVs that act as possible genetic modifiers.
Conclusion
This study sheds new insight onto the idea that copy-number changes at two different loci might cause or affect penetrance of a structural birth defect during development.48 Individuals with 22q11DS carry an initial genetic lesion that significantly elevates their risk of developing CHDs with variable expressivity. The 22q11.2 deletion was the only CNV present in every 22q11DS-positive individual with a CHD; however, one common CNV was significantly enriched and associated with CHDs in our two cohorts of subjects with 22q11DS. It suggests that variability of CHD phenotype in 22q11DS might be in part due to a duplication of SLC2A3. This finding supports a possible “two-hit” model where CNVs outside of the deleted region might explain the incomplete penetrance of the 22q11.2 deletion as a facilitator of congenital heart defects in a subset of subjects with 22q11DS. In the future, functional validation studies with a model system will help elucidate the role of SLC2A3 duplication as a cardiac modifier.
Acknowledgments
Thanks to Colleen Franconi, Meghan McNamara, April Hacker, Tracy Busse, Mark Bowser, Anne Marie Higgins, Chad Haldeman-Englert, Petra Warner, Jenna Walck, and Danica Kuncio for technical assistance. Special thanks go to the subjects and their families for their willing participation. We also thank the Molecular Cytogenetics and Genomics Cores at Einstein for preparing DNA and genotyping with Affymetrix 6.0 arrays. This work was funded by NIH (MH87636, HL84410, HD070454, and HD026979) and in part by NIH/NCATS (National Center for Advancing Translational Sciences), Grant UL1TR000003.
Published: April 16, 2015
Footnotes
Supplemental Data include one figure and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.03.007.
Web Resources
The URLs for data presented herein are as follows:
Gene Ontology Consortium, http://geneontology.org/
Mouse Genome Informatics, http://www.informatics.jax.org/
OMIM, http://www.omim.org/
Primer3, http://bioinfo.ut.ee/primer3
UCSC Genome Browser, http://genome.ucsc.edu
UCSC In-Silico PCR, http://genome.ucsc.edu/cgi-bin/hgPcr?command=start
Supplemental Data
References
- 1.Rosano A., Botto L.D., Botting B., Mastroiacovo P. Infant mortality and congenital anomalies from 1950 to 1994: an international perspective. J. Epidemiol. Community Health. 2000;54:660–666. doi: 10.1136/jech.54.9.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Bom T., Zomer A.C., Zwinderman A.H., Meijboom F.J., Bouma B.J., Mulder B.J. The changing epidemiology of congenital heart disease. Nat. Rev. Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- 3.Goldmuntz E., Clark B.J., Mitchell L.E., Jawad A.F., Cuneo B.F., Reed L., McDonald-McGinn D., Chien P., Feuer J., Zackai E.H. Frequency of 22q11 deletions in patients with conotruncal defects. J. Am. Coll. Cardiol. 1998;32:492–498. doi: 10.1016/s0735-1097(98)00259-9. [DOI] [PubMed] [Google Scholar]
- 4.Pierpont M.E., Basson C.T., Benson D.W., Jr., Gelb B.D., Giglia T.M., Goldmuntz E., McGee G., Sable C.A., Srivastava D., Webb C.L., American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H., Kartiko S., Finnell R.H. Importance of gene-environment interactions in the etiology of selected birth defects. Clin. Genet. 2009;75:409–423. doi: 10.1111/j.1399-0004.2009.01174.x. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins K.J., Correa A., Feinstein J.A., Botto L., Britt A.E., Daniels S.R., Elixson M., Warnes C.A., Webb C.L., American Heart Association Council on Cardiovascular Disease in the Young Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 7.Lammer E.J., Chak J.S., Iovannisci D.M., Schultz K., Osoegawa K., Yang W., Carmichael S.L., Shaw G.M. Chromosomal abnormalities among children born with conotruncal cardiac defects. Birth Defects Res. A Clin. Mol. Teratol. 2009;85:30–35. doi: 10.1002/bdra.20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaby J.A., Silversides C.K., Bekeschus S.C., Piran S., Oechslin E.N., Chow E.W., Bassett A.S. Complex congenital heart disease in unaffected relatives of adults with 22q11.2 deletion syndrome. Am. J. Cardiol. 2011;107:466–471. doi: 10.1016/j.amjcard.2010.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burn J., Goodship J. Developmental genetics of the heart. Curr. Opin. Genet. Dev. 1996;6:322–325. doi: 10.1016/s0959-437x(96)80009-8. [DOI] [PubMed] [Google Scholar]
- 10.Robin N.H., Shprintzen R.J. Defining the clinical spectrum of deletion 22q11.2. J. Pediatr. 2005;147:90–96. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Emanuel B.S. Molecular mechanisms and diagnosis of chromosome 22q11.2 rearrangements. Dev. Disabil. Res. Rev. 2008;14:11–18. doi: 10.1002/ddrr.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaikh T.H., Kurahashi H., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Emanuel B.S. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 13.McDonald-McGinn D.M., Tonnesen M.K., Laufer-Cahana A., Finucane B., Driscoll D.A., Emanuel B.S., Zackai E.H. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net! Genet. Med. 2001;3:23–29. doi: 10.1097/00125817-200101000-00006. [DOI] [PubMed] [Google Scholar]
- 14.McDonald-McGinn D.M., LaRossa D., Goldmuntz E., Sullivan K., Eicher P., Gerdes M., Moss E., Wang P., Solot C., Schultz P. The 22q11.2 deletion: screening, diagnostic workup, and outcome of results; report on 181 patients. Genet. Test. 1997;1:99–108. doi: 10.1089/gte.1997.1.99. [DOI] [PubMed] [Google Scholar]
- 15.Ryan A.K., Goodship J.A., Wilson D.I., Philip N., Levy A., Seidel H., Schuffenhauer S., Oechsler H., Belohradsky B., Prieur M. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J. Med. Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandrin-Garcia P., Abramides D.V., Martelli L.R., Ramos E.S., Richieri-Costa A., Passos G.A. Typical phenotypic spectrum of velocardiofacial syndrome occurs independently of deletion size in chromosome 22q11.2. Mol. Cell. Biochem. 2007;303:9–17. doi: 10.1007/s11010-007-9450-5. [DOI] [PubMed] [Google Scholar]
- 17.Goldmuntz E., Driscoll D.A., Emanuel B.S., McDonald-McGinn D., Mei M., Zackai E., Mitchell L.E. Evaluation of potential modifiers of the cardiac phenotype in the 22q11.2 deletion syndrome. Birth Defects Res. A Clin. Mol. Teratol. 2009;85:125–129. doi: 10.1002/bdra.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merscher S., Funke B., Epstein J.A., Heyer J., Puech A., Lu M.M., Xavier R.J., Demay M.B., Russell R.G., Factor S. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 19.Jerome L.A., Papaioannou V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 20.Lindsay E.A., Vitelli F., Su H., Morishima M., Huynh T., Pramparo T., Jurecic V., Ogunrinu G., Sutherland H.F., Scambler P.J. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 21.Guo T., McDonald-McGinn D., Blonska A., Shanske A., Bassett A.S., Chow E., Bowser M., Sheridan M., Beemer F., Devriendt K., International Chromosome 22q11.2 Consortium Genotype and cardiovascular phenotype correlations with TBX1 in 1,022 velo-cardio-facial/DiGeorge/22q11.2 deletion syndrome patients. Hum. Mutat. 2011;32:1278–1289. doi: 10.1002/humu.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agopian A.J., Mitchell L.E., Glessner J., Bhalla A.D., Sewda A., Hakonarson H., Goldmuntz E. Genome-wide association study of maternal and inherited loci for conotruncal heart defects. PLoS ONE. 2014;9:e96057. doi: 10.1371/journal.pone.0096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gai X., Perin J.C., Murphy K., O’Hara R., D’arcy M., Wenocur A., Xie H.M., Rappaport E.F., Shaikh T.H., White P.S. CNV Workshop: an integrated platform for high-throughput copy number variation discovery and clinical diagnostics. BMC Bioinformatics. 2010;11:74. doi: 10.1186/1471-2105-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper G.M., Coe B.P., Girirajan S., Rosenfeld J.A., Vu T.H., Baker C., Williams C., Stalker H., Hamid R., Hannig V. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White P.S., Xie H.M., Werner P., Glessner J., Latney B., Hakonarson H., Goldmuntz E. Analysis of chromosomal structural variation in patients with congenital left-sided cardiac lesions. Birth Defects Res. A Clin. Mol. Teratol. 2014;100:951–964. doi: 10.1002/bdra.23279. [DOI] [PubMed] [Google Scholar]
- 28.Gai X., Xie H.M., Perin J.C., Takahashi N., Murphy K., Wenocur A.S., D’arcy M., O’Hara R.J., Goldmuntz E., Grice D.E. Rare structural variation of synapse and neurotransmission genes in autism. Mol. Psychiatry. 2012;17:402–411. doi: 10.1038/mp.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaikh T.H., Gai X., Perin J.C., Glessner J.T., Xie H., Murphy K., O’Hara R., Casalunovo T., Conlin L.K., D’Arcy M. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowotschin S., Liao J., Gage P.J., Epstein J.A., Campione M., Morrow B.E. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 31.Edelmann L., Pandita R.K., Morrow B.E. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X., Freeze H.H. GLUT14, a duplicon of GLUT3, is specifically expressed in testis as alternative splice forms. Genomics. 2002;80:553–557. doi: 10.1006/geno.2002.7010. [DOI] [PubMed] [Google Scholar]
- 33.Simpson I.A., Dwyer D., Malide D., Moley K.H., Travis A., Vannucci S.J. The facilitative glucose transporter GLUT3: 20 years of distinction. Am. J. Physiol. Endocrinol. Metab. 2008;295:E242–E253. doi: 10.1152/ajpendo.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shepherd P.R., Gould G.W., Colville C.A., McCoid S.C., Gibbs E.M., Kahn B.B. Distribution of GLUT3 glucose transporter protein in human tissues. Biochem. Biophys. Res. Commun. 1992;188:149–154. doi: 10.1016/0006-291x(92)92362-2. [DOI] [PubMed] [Google Scholar]
- 35.Abel E.D. Glucose transport in the heart. Front. Biosci. 2004;9:201–215. doi: 10.2741/1216. [DOI] [PubMed] [Google Scholar]
- 36.Grover-McKay M., Walsh S.A., Thompson S.A. Glucose transporter 3 (GLUT3) protein is present in human myocardium. Biochim. Biophys. Acta. 1999;1416:145–154. doi: 10.1016/s0005-2736(98)00216-8. [DOI] [PubMed] [Google Scholar]
- 37.Kong B., Liu Y.L., Lü X.D. Microarray-bioinformatics analysis of altered genomic expression profiles between human fetal and infant myocardium. Chin. Med. J. (Engl.) 2008;121:1257–1264. [PubMed] [Google Scholar]
- 38.Fowden A.L., Forhead A.J., Coan P.M., Burton G.J. The placenta and intrauterine programming. J. Neuroendocrinol. 2008;20:439–450. doi: 10.1111/j.1365-2826.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- 39.Ganguly A., McKnight R.A., Raychaudhuri S., Shin B.C., Ma Z., Moley K., Devaskar S.U. Glucose transporter isoform-3 mutations cause early pregnancy loss and fetal growth restriction. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1241–E1255. doi: 10.1152/ajpendo.00344.2006. [DOI] [PubMed] [Google Scholar]
- 40.Janzen C., Lei M.Y., Cho J., Sullivan P., Shin B.C., Devaskar S.U. Placental glucose transporter 3 (GLUT3) is up-regulated in human pregnancies complicated by late-onset intrauterine growth restriction. Placenta. 2013;34:1072–1078. doi: 10.1016/j.placenta.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesage J., Hahn D., Léonhardt M., Blondeau B., Bréant B., Dupouy J.P. Maternal undernutrition during late gestation-induced intrauterine growth restriction in the rat is associated with impaired placental GLUT3 expression, but does not correlate with endogenous corticosterone levels. J. Endocrinol. 2002;174:37–43. doi: 10.1677/joe.0.1740037. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt S., Hommel A., Gawlik V., Augustin R., Junicke N., Florian S., Richter M., Walther D.J., Montag D., Joost H.G., Schürmann A. Essential role of glucose transporter GLUT3 for post-implantation embryonic development. J. Endocrinol. 2009;200:23–33. doi: 10.1677/JOE-08-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carayannopoulos M.O., Xiong F., Jensen P., Rios-Galdamez Y., Huang H., Lin S., Devaskar S.U. GLUT3 gene expression is critical for embryonic growth, brain development and survival. Mol. Genet. Metab. 2014;111:477–483. doi: 10.1016/j.ymgme.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurent L.C., Ulitsky I., Slavin I., Tran H., Schork A., Morey R., Lynch C., Harness J.V., Lee S., Barrero M.J. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vittori A., Breda C., Repici M., Orth M., Roos R.A., Outeiro T.F., Giorgini F., Hollox E.J., REGISTRY investigators of the European Huntington’s Disease Network Copy-number variation of the neuronal glucose transporter gene SLC2A3 and age of onset in Huntington’s disease. Hum. Mol. Genet. 2014;23:3129–3137. doi: 10.1093/hmg/ddu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veal C.D., Reekie K.E., Lorentzen J.C., Gregersen P.K., Padyukov L., Brookes A.J. A 129-kb deletion on chromosome 12 confers substantial protection against rheumatoid arthritis, implicating the gene SLC2A3. Hum. Mutat. 2014;35:248–256. doi: 10.1002/humu.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soemedi R., Wilson I.J., Bentham J., Darlay R., Töpf A., Zelenika D., Cosgrove C., Setchfield K., Thornborough C., Granados-Riveron J. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am. J. Hum. Genet. 2012;91:489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girirajan S., Rosenfeld J.A., Cooper G.M., Antonacci F., Siswara P., Itsara A., Vives L., Walsh T., McCarthy S.E., Baker C. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erdogan F., Larsen L.A., Zhang L., Tümer Z., Tommerup N., Chen W., Jacobsen J.R., Schubert M., Jurkatis J., Tzschach A. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J. Med. Genet. 2008;45:704–709. doi: 10.1136/jmg.2008.058776. [DOI] [PubMed] [Google Scholar]
- 50.Lalani S.R., Shaw C., Wang X., Patel A., Patterson L.W., Kolodziejska K., Szafranski P., Ou Z., Tian Q., Kang S.H. Rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities. Eur. J. Hum. Genet. 2013;21:173–181. doi: 10.1038/ejhg.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomita-Mitchell A., Mahnke D.K., Struble C.A., Tuffnell M.E., Stamm K.D., Hidestrand M., Harris S.E., Goetsch M.A., Simpson P.M., Bick D.P. Human gene copy number spectra analysis in congenital heart malformations. Physiol. Genomics. 2012;44:518–541. doi: 10.1152/physiolgenomics.00013.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenway S.C., Pereira A.C., Lin J.C., DePalma S.R., Israel S.J., Mesquita S.M., Ergul E., Conta J.H., Korn J.M., McCarroll S.A. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat. Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silversides C.K., Lionel A.C., Costain G., Merico D., Migita O., Liu B., Yuen T., Rickaby J., Thiruvahindrapuram B., Marshall C.R. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet. 2012;8:e1002843. doi: 10.1371/journal.pgen.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.