Abstract
Human height is a composite measurement, reflecting the sum of leg, spine, and head lengths. Many common variants influence total height, but the effects of these or other variants on the components of height (body proportion) remain largely unknown. We studied sitting height ratio (SHR), the ratio of sitting height to total height, to identify such effects in 3,545 African Americans and 21,590 individuals of European ancestry. We found that SHR is heritable: 26% and 39% of the total variance of SHR can be explained by common variants in European and African Americans, respectively, and global European admixture is negatively correlated with SHR in African Americans (r2 ≈ 0.03). Six regions reached genome-wide significance (p < 5 × 10−8) for association with SHR and overlapped biological candidate genes, including TBX2 and IGFBP3. We found that 130 of 670 height-associated variants are nominally associated (p < 0.05) with SHR, more than expected by chance (p = 5 × 10−40). At these 130 loci, the height-increasing alleles are associated with either a decrease (71 loci) or increase (59 loci) in SHR, suggesting that different height loci disproportionally affect either leg length or spine/head length. Pathway analyses via DEPICT revealed that height loci affecting SHR, and especially those affecting leg length, show enrichment of different biological pathways (e.g., bone/cartilage/growth plate pathways) than do loci with no effect on SHR (e.g., embryonic development). These results highlight the value of using a pair of related but orthogonal phenotypes, in this case SHR with height, as a prism to dissect the biology underlying genetic associations in polygenic traits and diseases.
Introduction
Human height has been used extensively as a model quantitative trait because it is highly heritable and polygenic, influenced by variants across many different loci.1 Recently, we and others identified 697 height-associated variants in a genome-wide association study (GWAS) of more than 300,000 individuals that together implicated many genes and pathways important for skeletal growth.2 However, human height is a composite phenotype, reflecting the sum of head, trunk, and leg lengths. Although the genetics of height has been extensively studied, little is known about the genetics of the relationship between these individual components with each other (body proportion). We hypothesize that studying the genetics of body proportion could provide complementary insights to those obtained by studying overall height.
Sitting height, the distance from the top of the head to the seated surface while a person is sitting upright, is a clinical measurement and sometimes measured in population-based studies.3 As such, it is an available measure that can provide insights into body proportion. Specifically, sitting height ratio (SHR), the ratio of sitting height to total height, can be used as a measure of body proportion: higher SHR corresponds to a higher spine/head to leg ratio, i.e., shorter leg length. Previous studies characterizing the SHR of individuals from childhood to adulthood show that SHR decreases rapidly from birth to teenage years (from 0.65 to 0.51 between ages 1 and 12 years) and increases slightly during puberty (from 0.51 to 0.52 between ages 12 and 18 years).3,4 Combined consideration of height and SHR can identify effects on height that act disproportionately on one body segment. For example, individuals with achondroplasia (MIM 100800) have short stature due to shortened limbs, reflected in an average SHR of 0.66 (for comparison, the 95% confidence interval of normal adult SHR of European-ancestry individuals is 0.51 to 0.55).5,6 On the other hand, individuals with spondyloepiphyseal (MIM 608361) and spondylometaepiphyseal (MIM 271665) dysplasias can have short stature but normal or decreased SHR values.7 SHR can also be indicative of future health outcomes. Previous studies indicate that SHR can be predictive of several traits and diseases including blood pressure, cholesterol levels, body mass index, intimal-medial thickness, type 2 diabetes, and insulin resistance.8–10
Although little is known about the heritability of SHR, this ratio is known to differ significantly across different ancestries. Adult individuals of African ancestry have lower SHR (SHR ∼ 0.51) than individuals of European ancestry (SHR ∼ 0.52) and individuals of Asian ancestry (SHR ∼ 0.53).11 The basis of these significant differences in SHR across populations is unclear.
This lack of knowledge about the genetics of SHR and the potential to gain new insights into the genetics of skeletal growth prompted us to investigate the heritability and the genetic architecture of this trait. We studied a total of 3,545 African Americans as well as 21,590 individuals of European ancestry that had both whole genome genotype data as well as the phenotypes needed to calculate SHR. We investigated the heritability and genetic architecture of SHR by performing several analyses including GCTA-REML12 to estimate heritability and admixture analysis and GWAS to identify common variants associated with SHR. Furthermore, given that we and others have recently identified many height-associated loci from GWASs,2 we used SHR like a prism to separate these loci into those that have an association with SHR and those that do not. The loci that have an association with SHR can be further separated into those that disproportionately affect the head/spine and those that disproportionately affect the long bones by testing whether the height-increasing allele increases or decreases SHR, respectively. We tested for enrichment of biological pathways for the various groups of loci and found that the loci associated with SHR, especially those that disproportionately affect the long bones, are significantly more enriched for genes in well-known pathways relevant to skeletal growth. On the other hand, the loci that are least associated with SHR tend to be more enriched for pathways involved in growth during embryogenesis. In general, our results highlight the capability of using an orthogonal phenotype like SHR to separate genetic loci known to be associated with height into their biological relevant components.
Material and Methods
Subjects
Individuals were recruited from various cohorts that have recording of sitting height, height, gender, age when measured, and body mass index (BMI). If there exists duplicated or closely related samples (PI_HAT > 0.05), one was randomly filtered out.
Atherosclerosis Risk in Communities
The participants of Atherosclerosis Risk in Communities (ARIC) study13 were recruited from four communities beginning in 1987: Forsyth County, NC; Jackson, MS; the northwest suburbs of Minneapolis, MN; and Washington County, MD (dbGaP phs000280.v3.p1). Whole genome genotyping performed with the Affymetrix 6.0 platform and genotype calling was performed with Birdseed calling algorithm.14 2,354 African Americans and 7,257 European Americans were used for the analyses.
Cardiovascular Health Study
The participants of Cardiovascular Health Study (CHS)15 were recruited from Forsyth County, NC; Sacramento County, CA; Washington County, MD; and Pittsburgh, PA (dbGaP phs000287.v4.p1). Whole genome genotyping was performed with the Illumina HumanCNV370v1 platform and genotype calling was performed with Illumina GenomeStudio software. 476 African Americans and 2,926 European Americans were used for the analyses.
Coronary Artery Risk Development in Young Adults
The participants of Coronary Artery Risk Development in Young Adults (CARDIA) study16 were recruited from four communities in the United States: Birmingham, AL; Chicago, IL; Minneapolis, MN; and Oakland, CA (dbGaP phs000285.v3.p2). Whole genome genotyping performed with the Affymetrix 6.0 platform and genotype calling was performed with Birdseed calling algorithm.14 715 African Americans and 1,047 European Americans were used for the analyses.
Framingham Heart Study
The participants of Framingham Heart Study (FHS)17 were recruited from town of Framingham, MA, starting from 1948 (dbGaP phs000007.v25.p9). Whole genome genotyping was performed by the Affymetrix 500K platform and genotype calling was performed with Birdseed calling algorithm.14 713 European Americans were used for the analyses.
The data for ARIC, CHS, CARDIA, and FHS cohorts were obtained from dbGaP (see Acknowledgments).
1958 British Birth cohort
The participants of 1958 British Birth Cohort (B58C)18 were born in England, Scotland, and Wales during a single week of 1958. Whole genome genotyping was performed with the Illumina HumanHap 550K/610K platform and genotype calling was performed with Illumina GenomeStudio software. Genome-wide typing and imputations were available for 6,296 cohort members with measurements of standing and sitting height at age 44–45 years.
Avon Longitudinal Study of Parents and Children
We used only the adult mothers from Avon Longitudinal Study of Parents and Children (ALSPAC) study19 for the analyses. Whole genome genotyping was performed with Illumina human660W-quad array platform and genotype calling was performed with Illumina GenomeStudio software. A total of 3,351 British adult females were used for analyses. A more complete description of ALSPAC can be found in the ALSPAC section below.
Ethical Approval
Ethical approval for the use of cohorts from dbGaP was obtained from the Institutional Review Board (IRB) of both Boston Children’s Hospital and the Broad Institute of Harvard and MIT. Ethical approval for the use of the 1958 British Birth Cohort was obtained from the South East Multicentre Research Ethics Committee (SE-MREC). Ethical approval for the use of ALSPAC was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees.
Quality Control
We filtered out samples that have ambiguous or incorrect gender, SNPs that have >5% missing rate, samples that have >2% missing SNPs, SNPs that have minor allele frequencies < 1%, and samples that have extreme heterozygosity (±4 SD). The SNP annotations for chromosome and base-pair positions were set to the coordinates of hg19 (GRCh37) via the liftOver tool.20 Pairwise IBD/IBS were calculated and individuals that have excessive matching with other individuals were removed. A filter of PI_HAT > 0.125 was used for ALSPAC. For all other cohorts, the threshold was set at PI_HAT > 0.05. Principal components analysis was performed with SMARTPCA21 and each study was projected onto HapMap v.322 space. Samples that did not cluster with the expected HapMap population were removed. SNPs that have excessive plate effects (p < 1 × 10−7) were dropped. For samples that are of European ancestry, SNPs that had excessive deviation from Hardy-Weinberg equilibrium (p < 1 × 10−7) were also dropped. Individuals who had no records of sitting height, height, age, or BMI or had very extreme sitting height ratios (SHR Z score > 4 SD or SHR Z score < −4 SD) were also removed. The QC protocol was performed with PLINK23 (v.1.07) and custom R and PERL scripts.
Genotype Phasing
For ARIC, CHS, CARDIA, FHS, and ALSPAC, the genotypes were phased with SHAPEIT2 (v.2.644)24 and imputed with IMPUTE2 (v.2.3).25 For B58C, the genotypes were phased with MACH24 and imputed with Minimac.25
Genotype Imputation
The imputation for all cohorts was performed with a 1000 Genomes version 1 phase 3 reference panel26 containing 379 Europeans, 246 Africans and African Americans, 286 Asians, and 181 Latin Americans. The imputation panel consists of approximately 22 million variants (SNPs and indels). For the X chromosome, only the non-pseudo autosomal region was imputed. The phasing and imputation were done separately for males and females for the X chromosome.
ALSPAC
Description of Study Numbers
ALSPAC recruited 14,541 pregnant women resident in Avon, UK with expected dates of delivery 1st April 1991 to 31st December 1992. 14,541 is the initial number of pregnancies for which the mother enrolled in the ALSPAC study and had either returned at least one questionnaire or attended a “Children in Focus” clinic by 19/07/99. Of these initial pregnancies, there were a total of 14,676 fetuses, resulting in 14,062 live births and 13,988 children who were alive at 1 year of age. We used only the enrolled adult mothers that had data on sitting height, height, and body mass index. After filtering away individuals that were not genotyped or were removed after going through the quality-control pipeline, there was a final tally of 3,351 individuals.
Quality Control and Preparation
Centre National de Génotypage (CNG) carried out DNA genotyping on the Illumina human660W-quad array and genotypes were called with Illumina GenomeStudio. PLINK (v.1.07) was used to carry out quality-control measures on an initial set of 10,015 subjects and 557,124 directly genotyped SNPs. SNPs were removed if they displayed more than 5% missingness or a Hardy-Weinberg equilibrium p value of less than 10−6. Additionally, SNPs with a minor allele frequency of less than 1% were removed. Samples were excluded if they displayed more than 5% missingness or had indeterminate X chromosome heterozygosity or extreme autosomal heterozygosity. Samples showing evidence of population stratification were identified by multidimensional scaling of genome-wide identity by state pairwise distances with the four HapMap populations as a reference, and then excluded. Cryptic relatedness was assessed with a PI_HAT of more than 0.125, which is expected to correspond to roughly 12.5% alleles shared IBD or a relatedness at the first-cousin level. Related subjects that passed all other quality-control thresholds were retained during subsequent phasing and imputation. 9,048 subjects and 526,688 SNPs passed these quality-control filters. We combined 477,482 SNP genotypes of these subjects with genotype data of a sample of 9,115 children, removed SNPs with a missingness above 1% due to poor quality (n = 11,396), and removed a further 321 subjects due to potential ID mismatches. This resulted in a dataset of 17,842 subjects containing 6,305 duos and 465,740 SNPs (112 were removed during liftover and 234 were out of HWE after combination). We estimated haplotypes with ShapeIT (v.2.r644), which utilizes relatedness during phasing. We imputed SNPs against the 1000 Genomes phase I version 3 integrated set via IMPUTE2 (v.2.3). Analysis was carried out in SNPtest (v.2.5) including the ten principal components of population ancestry from EIGENSTRAT.
Data Dictionary
The ALSPAC study website contains details of all the data that are available through a fully searchable data dictionary (see Web Resources).
Overall European Ancestry in African-American Individuals
Overall European admixture (ARIC, CARDIA, and CHS) was calculated by SMARTPCA from the CEU and YRI samples of HapMap v.3.22 CEU individuals were used as proxies of European ancestry and YRI individuals were used as proxies of African ancestry. The principal components were calculated using only the CEU and YRI individuals while projecting them onto the ARIC, CARDIA, and CHS African Americans. The first principal component (PC1) was used to calculate the degree of overall European admixture for each individual. The percentage overall European admixture was calculated by (IndividualPC1 – YRIPC1)/(CEUPC1 – YRIPC1) × 100%, where IndividualPC1 is PC1 for each individual and CEUPC1 and YRIPC1 are the mean PC1 values for the CEU and YRI individuals, respectively.
SHR Phenotype for Comparison between European and African Americans
Both European-American and African-American individuals from ARIC, CARDIA, and CHS were pooled together and the SHR was calculated by taking the sitting height divided by overall height. SHR residuals were obtained by performing linear regression of SHR against overall height, age, gender, BMI, and cohort as covariates. The SHR Z scores were then obtained by taking the resulting residuals divided by the estimated standard deviation of the residuals. This was performed with custom R scripts.
SHR Phenotype for GWAS and GCTA-REML Analysis
SHR was calculated by taking the sitting height divided by overall height. The SHR residuals were obtained by performing linear regression of SHR against overall height, age, gender, and BMI as covariates separately for each cohort. For the ARIC, CARDIA, and CHS cohorts, this was performed separately for the African Americans and European Americans. For the African Americans, SHR was also adjusted for overall European ancestry by including it as a covariate. For ALSPAC, the top ten principal components were included as covariates to adjust for population stratification. For the other cohorts, only the components that were nominally significant (p < 0.05) with SHR were included as covariates to adjust for population stratification. The SHR Z scores were then obtained by taking the resulting residuals divided by the estimated standard deviation of the residuals. The final phenotype for each cohort/group was then obtained by rank-based inversed normalization. This was performed with custom R scripts.
Determining Overall Height Phenotype
The height residuals were obtained by performing linear regression of height against age and gender and BMI as covariates separately for each of the cohorts. Additionally, the data were analyzed separately for African Americans and European Americans. For the African-American cohorts, we also adjusted for overall European ancestry by including it as a covariate. Among the top ten principal components, the components that were nominally significant (p < 0.05) were also included as covariates to adjust for population stratification. The height Z scores were then obtained by taking the resulting residuals divided by the estimated standard deviation of the residuals. The final phenotype for each cohort group was then obtained by rank-based inversed normalization. This was performed with custom R scripts.
GCTA-REML Analysis
We used the GCTA software27 to calculate the genetic relationship matrix separately for each chromosome by combining the 3,545 African-American individuals from the ARIC, CARDIA, and CHS cohorts. We also performed the same calculations by combining the 11,943 European-American individuals from the ARIC, CARDIA, and CHS cohorts. We used genome-wide autosomal SNPs that were well imputed (INFO > 0.8) in all the African-American and European-American cohorts resulting in a total of 10,250,422 and 6,655,203 SNPs for each ancestry group, respectively. We then merged the genetic relationship matrix calculated for each chromosome to obtain a global genetic relationship matrix for both the African-American and European-American groups. We then performed GCTA-GREML analysis to estimate the variance explained by the SNPs for both ancestry groups with SHR as well as overall height.
Genome-wide Association Study
The associations were performed with the SHR phenotype as the outcome variable (see above). Only unrelated individuals were used, i.e., no pair of individuals has PI_HAT > 0.05. The association for the imputed variants with SHR was performed by a linear regression with either PLINK23 (v.1.07) or SNPtest28 (v.2.5). SNPs that were imputed poorly were discarded (INFO < 0.6). The association results for each cohort were meta-analyzed together via METAL29 with genomic control correction. Variants on the X chromosome were analyzed separately between males and females and meta-analyzed together via METAL.29 For sex-specific analysis, males and females were separately analyzed via METAL.29
Enhancer Enrichment Test
The SNPs (Table S2) were mapped from hg18 to hg19 coordinates via the UCSC liftOver tool.20 For each SNP set, 2,000 sets of matched SNPs were obtained via SNPsnap with its default settings. These matched SNPs were matched for frequency, LD, and gene proximity. Proxy SNPs for each SNP set were also identified with PLINK23 based on LD (r2 > 0.5) between SNPs (≤500 kb) in the European (CEU, GBR, TSI) populations in the 1000 Genomes Phase 1 data.26 Next, the set of permissive enhancers defined by the FANTOM5 project30 were used to screen for enhancer regions that overlap at least one SNP or its proxies in each SNP set. The enhancer enrichment p value was then calculated by taking the proportion of matched SNP sets that contain equal or higher number of enhancers compared to the observed SNP sets.
Comparison with Known Height-Associated Loci
We obtained the 697 SNPs shown to be robustly associated with overall height2 (p < 5 × 10−8) and analyzed the summary statistics of SHR from a GWAS of 21,590 individuals of European ancestry. We filtered out SNPs that were either not present in the GWAS or failed imputation (INFO < 0.6) in any of the individual cohorts, which resulted in 670 SNPs. We performed conditional and joint GWAS31 for the 670 SNPs from the GWAS summary statistics of SHR from linkage-disequilibrium (LD) estimates calculated from 174 CEU individuals from HapMap v.2.22 We then tabulated the effect sizes and p values for the conditional and joint GWAS for both height and SHR with respect to the height-increasing allele (Table S1). The QQ plots and 95% confidence intervals were generated with custom R scripts. For the symmetrical analysis, we obtained 725 SNPs that were significantly associated (p < 5 × 10−5) with SHR from a GWAS of 21,590 individuals of European ancestry. We pruned away SNPs that were in strong LD (r2 > 0.1) by using 174 CEU individuals from HapMap v.2, which resulted in 70 SNPs.
Pathway/Gene-set Enrichment Analysis via DEPICT
A comprehensive pathway/gene-set enrichment and tissue/cell type analysis was performed by applying a new method, Data-Driven Expression Prioritized Integration for Complex Traits (DEPICT),32 which has been recently applied to several studies.2,33–36 DEPICT performs gene set enrichment analysis by testing whether genes in GWAS-associated loci are enriched for reconstituted versions of known molecular pathways (jointly referred to as reconstituted gene sets). The reconstitution is accomplished by identifying genes that are co-regulated with other genes in a given gene set based on a panel of 77,840 gene expression microarrays.37 Genes that are found to be transcriptionally co-regulated with genes from the original gene set are added to the gene set and thus the reconstitution. Several types of gene sets were reconstituted: 5,984 protein molecular pathways derived from 169,810 high-confidence experimentally derived protein-protein interactions,38 2,473 phenotypic gene sets derived from 211,882 gene-phenotype pairs from the Mouse Genetics Initiative,39 737 Reactome database pathways,40 184 Kyoto Encyclopedia of Genes and Genomes (KEGG) database pathways,41 and 5,083 Gene Ontology database terms.42 In total, 14,461 gene sets were assessed for enrichment in genes in associated regions. DEPICT also facilitated tissue and cell type enrichment analysis by testing whether the genes in associated regions were highly expressed in any of 209 MeSH annotations for 37,427 microarrays on the Affymetrix U133 Plus 2.0 Array platform.
Results
GCTA-GREML Analysis to Determine Heritability of SHR
We performed GCTA-GREML analysis12 to estimate the variance explained by the autosomal SNPs from 3,545 African Americans and 11,943 European Americans by using unrelated individuals from the ARIC,13 CHS,15 CARDIA,16 and FHS17 cohorts. The results show that SHR is highly heritable; the phenotypic variance explained by the autosomal SNPs is estimated to be 0.38 (95% CI: 0.22–0.55; p = 4.96 × 10−6) and 0.26 (95% CI: 0.21–0.31; p = 1.92 × 10−24) for African Americans and European Americans, respectively (Table 1). We also tested overall height: the phenotypic variance is estimated to be 0.77 (95% CI: 0.62–0.92; p = 1.66 × 10−20) and 0.45 (95% CI: 0.4–0.5; p = 1.74 × 10−72) for African Americans and European Americans, respectively (Table 1). The variance explained for European Americans on overall height is similar to what was previously estimated.2,43
Table 1.
Estimation of Phenotypic Variance of SHR Explained by Genetic Relationships from Autosomal SNPs
| Phenotype | No. of SNPs | h2(SE) | LRT | p Value |
|---|---|---|---|---|
| African Americans (n = 3,545) | ||||
| SHR | 10,250,422 | 0.387 (0.0845) | 19.53 | 4.96 × 10−6 |
| Height | 10,250,422 | 0.768 (0.0780) | 84.79 | 1.66 × 10−20 |
| European Americans (n = 11,943) | ||||
| SHR | 6,655,203 | 0.26 (0.0267) | 102.73 | 1.92 × 10−24 |
| Height | 6,655,203 | 0.449 (0.0265) | 322.84 | 1.74 × 10−72 |
| European-American Males (n = 5,477) | ||||
| SHR | 6,655,203 | 0.366 (0.0552) | 46.81 | 3.92 × 10−12 |
| Height | 6,655,203 | 0.485 (0.0550) | 83.54 | 3.11 × 10−20 |
| European-American Females (n = 6,466) | ||||
| SHR | 6,655,203 | 0.188 (0.0454) | 19.15 | 6.05 × 10−6 |
| Height | 6,655,203 | 0.454 (0.0465) | 100.85 | 4.96 × 10−24 |
The results from GCTA-REML analysis performed on both African- and European-American individuals for SHR as well as overall height using autosomal variants successfully imputed (INFO > 0.8) in all cohorts.
We further investigated gender-specific heritability of SHR by performing GCTA-GREML analysis separately on European-American males (n = 5,477) and European-American females (n = 6,466). The results show that SHR is significantly more heritable in males than in females; the phenotypic variance explained for males is estimated to be 0.37 (95% CI: 0.26–0.47; p = 3.92 × 10−12) whereas for females, it is estimated to be 0.188 (95% CI: 0.10–0.28; p = 6.05 × 10−6) (Table 1). Overall height showed no significant gender-specific heritability.
Comparing SHR between African and European Americans
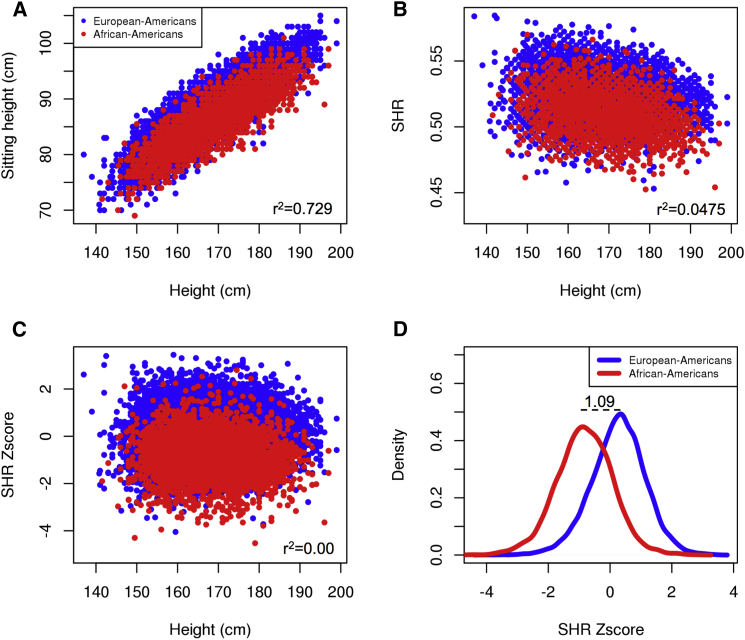
We tested whether SHR differs across populations as previously reported.11 We used the ARIC,13 CARDIA,16 and CHS15 cohorts because they include both African Americans and European Americans, resulting in a total sample size of 11,230 European Americans and 3,545 African Americans. We observed that African Americans have lower sitting heights than European Americans (Figures 1A–1C): SHR is 1.09 SD lower in African Americans than European Americans (Figure 1D), consistent with earlier studies.
Figure 1.
Comparing the Distribution of SHR between European Americans and African Americans
Plots of sitting height and height of individuals, both European Americans and African Americans, from combining the ARIC, CARDIA, and CHS cohorts. Red points indicate individuals of African ancestry; blue points indicate individuals of European ancestry.
(A) Values of sitting height and height in centimeters.
(B) Values of SHR and height in centimeters.
(C) Values of SHR Z scores (adjusted for height, sex, BMI, age, and cohort) and height in centimeters.
(D) The histograms of the SHR Z scores of African Americans (red line) and European Americans (blue line). The dotted horizontal line indicates the mean difference (in standard deviation units) between the two distributions.
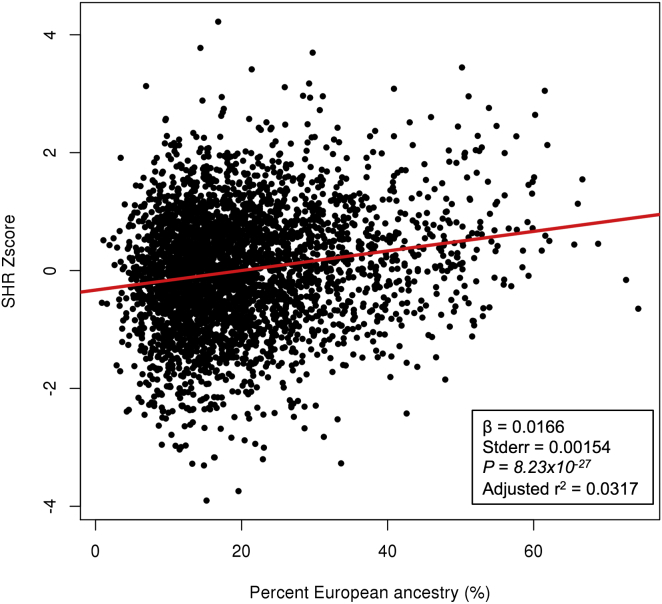
To address whether genetic factors could explain the basis for the SHR difference between populations, we tested whether the degree of European admixture in African Americans is correlated with SHR because African Americans have variable amount of European admixture, with a mean of about 20%.44,45 If genetic factors contribute to the difference between populations, the degree of overall European admixture within African Americans should be positively correlated with SHR. We observed a very strong correlation between overall European admixture and SHR in the African-American individuals (Figure 2); on average, a 1% increase in European admixture is correlated with a 0.0166 SD increase in SHR (p = 8.23 × 10−27). This result can explain most if not all of the SHR difference between African Americans and European Americans, suggesting that the basis for this difference is mainly genetic.
Figure 2.
Association of Global European Admixture with SHR in African-American Individuals
The association of global European admixture (x axis) with SHR (y axis) for African-American individuals in the ARIC, CARDIA, and CHS cohorts (n = 3,545). The correlation (red line) obtained from linear regression is shown with the corresponding effect size (β), standard deviation, p value, and r2 (inset).
GWASs of SHR in African Americans and European Americans
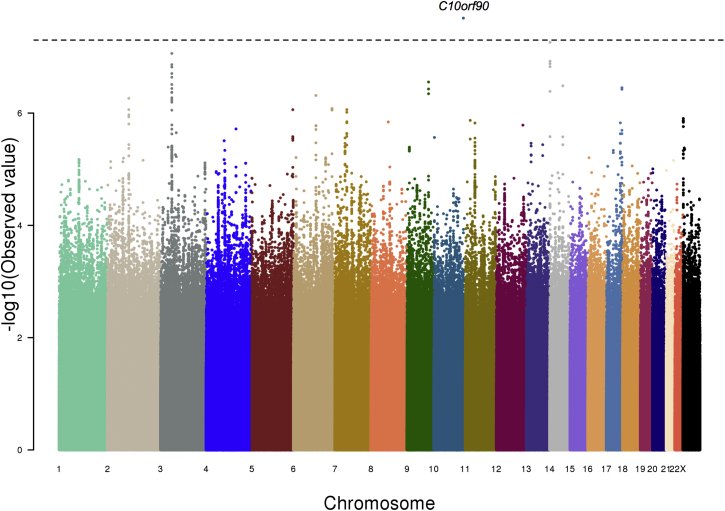
To test for specific genetic loci associated with SHR, we performed genome-wide association of SHR on imputed genotypes of the 3,545 African-American individuals from ARIC,13 CARDIA,16 and CHS15 cohorts. A single locus, rs10736877, reached genome-wide significance (p = 2.03 × 10−8) (Table 2; Figures 3 and S1). rs10736877 is a common variant that is directly genotyped (allele frequency = 51%, effect size = −0.13 SD) and is not in strong LD with its neighbors and thus none of its neighbors show significant associations with SHR. rs10736877 is upstream of C10orf90, the function of which is still unknown. Thus, more samples are required to confirm whether rs10736877 is truly associated with SHR in African Americans.
Table 2.
Genome-wide Significant SNPs
| Variant | Location (chr-pos)a | Alleles | Freq |
SHR |
Geneb | |
|---|---|---|---|---|---|---|
| Beta (SE) | p | |||||
| African Americans | ||||||
| rs10736877 | chr10: 128,373,707 | A/G | 0.51 | −0.1311 (0.0234) | 2.03 × 10−8 | C10orf90 |
| European Ancestry | ||||||
| rs6931421 | chr6: 80,880,138 | T/G | 0.68 | 0.0586 (0.0103) | 1.45 × 10−8 | BCKDHB |
| rs1722141 | chr7: 46,016,662 | A/G | 0.22 | −0.0667 (0.0121) | 3.31 × 10−8 | IGFBP3 |
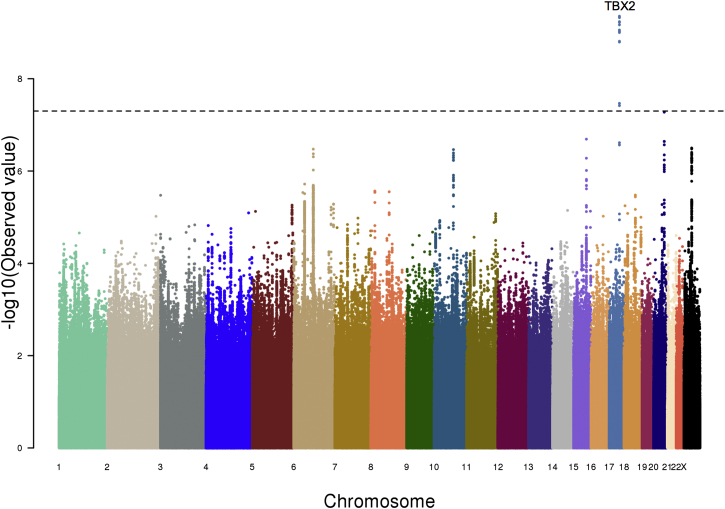
| rs882367 | chr17: 59,494,574 | C/T | 0.33 | −0.0778 (0.0104) | 8.42 × 10−14 | TBX2 |
| rs140449984 | chr18: 8,321,896 | T/TAGA… | 0.0747 | −0.1043 (0.0188) | 2.91 × 10−8 | PTPRM |
| rs228836 | chr20: 50,065,648 | G/A | 0.41 | 0.0608 (0.0099) | 8.48 × 10−10 | NFATC2 |
| Male-Specific | ||||||
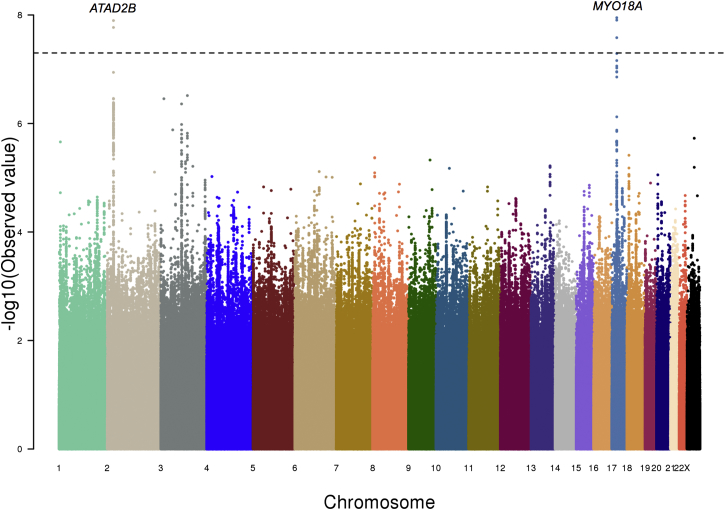
| rs35832626 | chr2: 24,054,647 | T/C | 0.41 | 0.0902 (0.0159) | 1.27 × 10−8 | ATAD2B |
| rs34638952 | chr17: 27,537,410 | A/T | 0.64 | −0.0906 (0.0159) | 1.12 × 10−8 | MYO18A |
The table shows the variants that reached genome-wide significance (p < 5 × 10−8) when tested for their association with SHR.
hg19 annotation.
Candidate gene within the locus.
Figure 3.
GWAS of SHR of African Americans
The GWAS results of SHR of African-American individuals (n = 3,545) shown as a Manhattan plot. The x axis marks the chromosomes and their relative positions of each variant tested and the y axis indicates the –log10 p value. The candidate gene within the locus that reached genome-wide significance is also shown. The horizontal dashed line represents the threshold of genome-wide significance (p < 5 × 10−8).
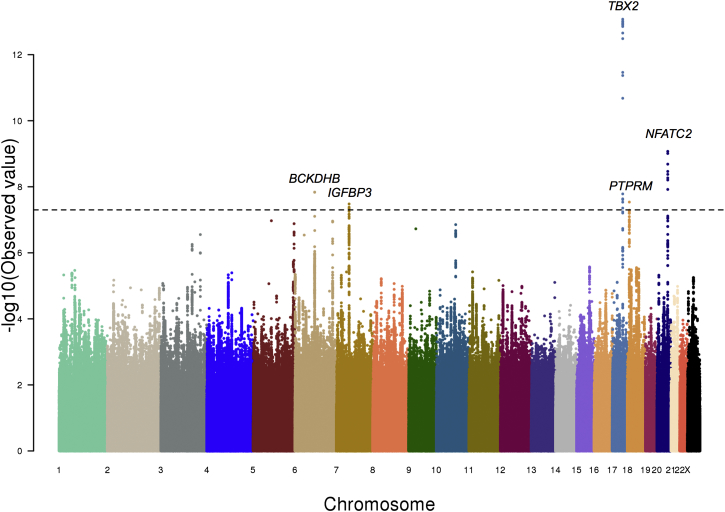
We also performed genome-wide association of sitting height ratio (SHR) in a total of 21,590 individuals of European ancestry (ARIC,13 CARDIA,16 CHS,15 FHS,17 B58C,18 and ALSPAC19). We identified five independent loci reaching genome-wide significance (p < 5 × 10−8) (Table 2; Figures 4 and S2). Of the five lead SNPs, two (rs6931421 and rs882367) are in strong LD with variants known to be significantly associated with overall height.1,2 Interestingly, rs1722141 is in the IGFBP3 (MIM 146732) locus, which has different SNPs significantly associated with overall height (Table 2); rs1722141 is not itself associated with height and is not in LD with the height-associated SNPs. The remaining two loci (rs140449984 and rs228836) have not been associated with overall height.
Figure 4.
GWAS of SHR of Individuals with Ancestry from Europe
The GWAS results of SHR of individuals with ancestry from Europe (n = 21,590) shown as a Manhattan plot. The x axis marks the chromosomes and their relative positions of each variant tested and the y axis indicates the –log10 p value. Candidate genes within loci that reached genome-wide significance are also shown. The horizontal dashed line represents the threshold of genome-wide significance (p < 5 × 10−8).
Because SHR is significantly different between men and women and their overall heritability were shown to be significantly different, we reanalyzed the European ancestry genome-wide association data for SHR separately for females (n = 12,965) and males (n = 8,625). Although we did not find any additional significant loci for the female-specific analysis (Figures 5 and S3), two new loci (rs34638952, rs35832626) reached genome-wide significance (p < 5 × 10−8) in the male-specific analysis (Table 2; Figures 6 and S4). One of these, rs35832626, has also been shown to be significantly associated with overall height, but not in any gender-specific manner2 (Table 2).
Figure 5.
Female-Specific GWAS of SHR of Individuals with Ancestry from Europe
The GWAS results of SHR using only the female individuals with ancestry from Europe (n = 12,965) shown as a Manhattan plot. The x axis marks the chromosomes and their relative positions of each variant tested and the y axis indicates the –log10 p value. The candidate gene within the locus that reached genome-wide significance is also shown. The horizontal dashed line represents the threshold of genome-wide significance (p < 5 × 10−8).
Figure 6.
Male-Specific GWAS of SHR of Individuals with Ancestry from Europe
The GWAS results of SHR using only the male individuals with ancestry from Europe (n = 8,625) shown as a Manhattan plot. The x axis marks the chromosomes and their relative positions of each variant tested and the y axis indicates the –log10 p value. Candidate genes within loci that reached genome-wide significance are also shown. The horizontal dashed line represents the threshold of genome-wide significance (p < 5 × 10−8).
Comparison with Known Height-Associated Loci
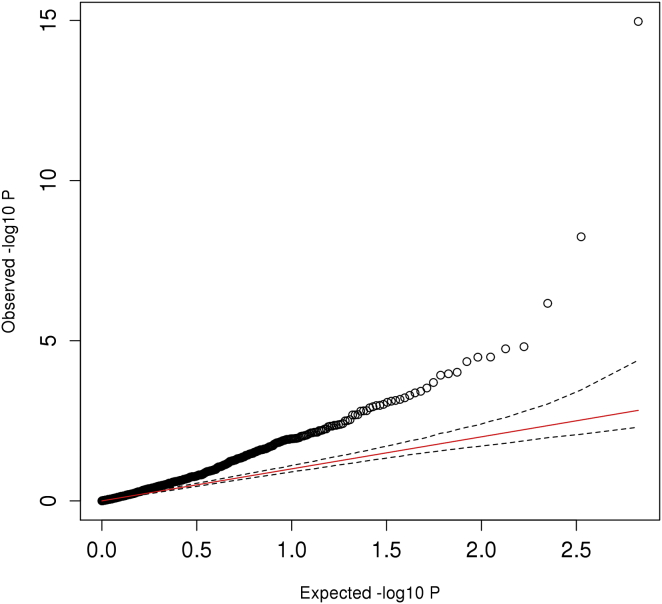
We examined the association statistics of SHR for the SNPs that were reported to be significantly associated with overall height. Among the 697 height-associated SNPs, only 670 SNPs were robustly imputed (INFO > 0.6) in all of our cohorts. We performed conditional and joint GWASs31 for the 670 SNPs from the GWAS summary statistics of SHR. We tabulated the effect sizes and p values for both height and SHR with respect to the height-increasing allele (Table S1). We observed a significant enrichment of height-associated SNPs that were also nominally significant with SHR (p < 0.05). A total of 130 SNPs reached nominal significance whereas only 33.5 were expected under the null hypothesis (p = 1.16 × 10−60) (Figure 7; Table S2). This result indicates that many of these 130 SNPs have a disproportionate effect on the long bones or spine/head lengths to cause a change not only in SHR, but also in overall height. Among these 130 SNPs, 71 of them had the height-increasing allele predicted to decrease SHR (SNPlongbones), which indicate that they act disproportionately on the long bones, whereas 59 had the height-increasing allele predicted to increase SHR (SNPheadspine), which indicate that they act disproportionately to affect head or spine length.
Figure 7.
QQ Plot of the 670 Height-Associated SNPs with SHR
The plot shows the observed –log10 p values of SHR (y axis) for the 670 height-associated SNPs and compare them with the expected –log10 p values (x axis) under the null hypothesis. The red line is the trend line under the null hypothesis and the dashed lines indicate the 95% confidence interval under the null hypothesis.
We further performed a symmetric analysis and investigated the SNPs that were associated with SHR (p < 5 × 10−5) for their association with overall height. After pruning for LD, we obtained 70 such SNPs. Of these 70 SNPs, 32 were nominally significant (p < 0.05) for overall height whereas only 3.5 were expected under the null hypothesis (p = 3.07 × 10−23). Of these 32 SHR-increasing alleles, 14 were associated with increased height and 18 were associated with decreased height (Table S3).
Enhancer Enrichment Analysis
We analyzed the genome-wide significant SNPs (p < 5 × 10−8) to determine whether there are any overlaps with known enhancer regions. We used the set of permissive and robust enhancers as defined by the FANTOM5 project.30 We obtained the genomic region surrounding the lead SNPs by taking the other SNPs significant for SHR (p < 5 × 10−6) spanning the region (Table S4). The SNP rs10736877 found to be genome-wide significant from African Americans had no other SNPs in LD spanning the region and was dropped from this analysis. Among the seven remaining genome-wide significant loci, six overlapped with at least one known permissive or robust enhancer region (Table S5).
Next, we investigated the 130 height-associated SNPs that were nominally significant (p < 0.05) for SHR. Using a randomized permutation procedure, we tested whether the genomic regions surrounding these 130 SNPs have significant enrichment with the set of permissive enhancers as defined by the FANTOM5 project.30 We also tested the remaining 540 height-associated SNPs that were not nominally significant for SHR (p > 0.05) in the same way. We observed that although both groups of SNPs had significant enrichment for enhancer regions (p < 0.05), the 130 SNPs are more significantly associated (p = 0.0005) with enhancer regions than the 540 SNPs (p = 0.003) (Table S6). We further divided the 130 SNPs into the 71 SNPlongbones and 59 SNPheadspine SNPs and tested them separately for enrichment of enhancer regions. Again, although we observed that both the SNPlongbones and SNPheadspine SNPs were significantly enriched for enhancer regions (p < 0.05), the SNPlongbones showed more significant enrichment (p = 0.0045) than the SNPheadspine SNPs (p = 0.0465) (Table S6). These results suggest that there is more overlap with annotated regulatory regions for height-associated SNPs that are also associated with SHR, especially those that were predicted to act disproportionately on the long bones (SNPlongbones).
Pathway and Tissue Enrichment Analysis by DEPICT
We used a recently available method, Data-Driven Expression Prioritized Integration for Complex Traits (DEPICT),32 to analyze the various groups of SNPs. We first used the list of SNPs that were significant from the earlier GWAS of 21,950 individuals of European ancestry. Because there were only five genome-wide significant SHR loci, which is too few loci to utilize DEPICT, we used a more liberal significance threshold (p < 5 × 10−6), resulting in 21 distinct genomic loci (Table S7). Interestingly, although there was no significant tissue enrichment (Table S9), a single pathway/gene set, labeled as “short tibia” (MGI MP: 0002764) was significantly enriched (p = 5.46 × 10−8; false discovery rate [FDR] < 0.05) (Table S8). With just these 21 loci, DEPICT also successfully prioritized (FDR < 0.05) a candidate causal gene in three of these loci (NSD1 [MIM 606681], DMP1 [MIM 600980], and IBSP [MIM 147563]) (Table S10).
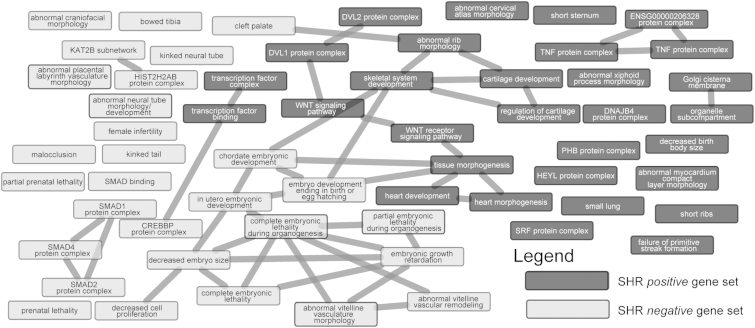
We next analyzed the set of 130 height-associated SNPs that were nominally associated with SHR (Table S2). We also separately analyzed a set of 130 height-associated SNPs that have the least significant SHR p values (Table S11). We used both sets of SNPs as inputs and analyzed them with DEPICT. For the 130 SNPs that were nominally associated with SHR, DEPICT reported a substantial number (>500) of pathway/gene sets that were significantly enriched (FDR < 0.05) (Table S12). Many of these gene sets had belonged to pathways involved in bone development, e.g., “abnormal rib morphology,” “chondrocyte differentiation,” “abnormal skeleton morphology,” etc. The “short tibia” gene set found previously was also significant. There were also a number of cell/tissue types (ten types) that showed significant enrichment (FDR < 0.05) (Table S13). These cell/tissue types included the tissues that are known to be broadly involved in skeletal growth, like cartilage, chondrocytes, and spine. In contrast, the 130 SNPs least associated with SHR had much fewer significant pathways or gene sets that are directly associated with skeletal or bone development and appear to be more strongly associated with pathways involved in early development, e.g., “embryonic growth retardation,” “decreased embryo size,” “decreased cell proliferation,” etc. (Table S14). These 130 SNPs also showed significant enrichment in some cell/tissue types (three types) (FDR < 0.05), but none of those directly involved in skeletal growth, i.e., cartilage, chondrocytes, and spine, showed significant enrichment (Table S15). By comparing the top 30 pathways or gene sets discovered from either set of SNPs, we demonstrate that the height-associated SNPs nominally associated with SHR are tied more with pathways involved in skeletal, bone, or cartilage development whereas the height-associated SNPs least associated with SHR are tied more to pathways involved in embryonic development (Figure 8).
Figure 8.
Pathway Analysis by DEPICT of Height-Associated SNPs
Comparing the top 30 gene sets obtained by DEPICT for the 130 known height-associated SNPs also nominally associated with SHR (SHR-positive gene set) as well as the 130 known height-associated SNPs least associated with SHR (SHR-negative gene set). A line connecting two gene sets indicates that the gene sets have a Pearson correlation coefficient > 0.6.
Finally, we analyzed the 71 SNPlongbones and 59 SNPheadspine SNPs separately with DEPICT. The SNPlongbones SNPs yielded a number of pathways/gene sets (>30) that were significant (FDR < 0.05) (Table S16). Although the number of significant pathways/gene sets were much smaller than those obtained from using all the 130 SNPs, the results were similar, i.e., the significant gene sets generally belonged to pathways involved in bone development like “abnormal rib morphology,” “skeletal system development,” and “short ribs.” There were also three cell/tissue types that were enriched, i.e., “cartilage,” “mesenchymal stem cells,” and “osteoblasts” (FDR < 0.05) (Table S17), implicating the tissues involved in cartilage and bone development. In contrast, there were no significant results (FDR < 0.05) from analyzing the SHRheadspine SNPs (Tables S18 and S19). This suggests that the genes disproportionately affecting the long bones are more tied to known skeletal growth pathways than are genes disproportionately affecting the head/spine, although this could be due to having fewer SNPheadspine loci than SNPlongbones loci. Nonetheless, these results demonstrate that the height-associated SNPs can be stratified into different biological groups by examining their respective effects on SHR.
Discussion
Unlike overall height, body proportion or sitting height ratio (SHR) has not been well studied and less is known about its underlying genetics. Here, we have provided evidence that SHR is heritable and we have identified loci that are significantly associated with SHR. We also showed that the lower SHR in African-ancestry populations is consistent with an effect of genetic factors. However, the GWAS performed on African-American samples did not identify loci that either explains much of the heritability of SHR nor much of the difference between European- and African-ancestry populations. These results suggest that the difference in SHR is polygenic, i.e., due to many loci.
The results from the GWAS of individuals of European ancestry yielded more genome-wide significant results, probably because of the larger number of samples. The most significant locus (rs882367) has been shown to be significantly associated with overall height.1 Although the most plausible candidate gene in this locus (TBX2 [MIM 600747]) has been demonstrated to affect heart development,46,47 it has also been suggested to cause limb abnormalities and skeletal malformations.48,49 Another significant locus (rs1722141), although not significantly associated with overall height, contains multiple SNPs, not correlated to rs1722141, that are strongly associated with height. The likely gene underlying this locus (IGFBP3 [MIM 146732]) is known to bind to and modulate IGF by increasing IGF half-life and inhibiting or potentiating IGF binding to the IGF receptor,50 which has been directly tied to skeletal growth.51 The SNPs that are associated with SHR, but not overall height, have also been shown to be significantly correlated with circulating levels of IGFBP3,52 suggesting that circulating IGFBP3 levels might influence body proportion.
Some of the SNPs that showed suggestive significance (p < 5 × 10−6) are in loci with genes underlying Mendelian causes of growth disorders and/or skeletal dysplasias that are known to affect body proportion. One example, rs2011077 (p = 2.52 × 10−7), is near NSD1, a known cause of Sotos syndrome (MIM 117550), which is characterized by extreme overgrowth in childhood, large head size, and advanced bone age. Individuals affected with Sotos syndrome have a decreased upper to lower body segment ratio as compared to age- and gender-matched controls.53
We also demonstrated that SHR is a useful tool to understand the underlying biology of height-associated loci. Although height and SHR are orthogonal traits, we showed that there are significant overlaps between the loci affecting SHR and the loci affecting overall height. Height-increasing alleles that cause a reduction in SHR have to have disproportionate effect such that the increase in long bone length is relatively larger than the increase in the spine and/or head length. In contrast, height-increasing alleles that cause an increase in SHR have a relatively larger effect on the spine and/or head. From the pathway analysis by DEPICT, we have shown that the loci affecting height disproportionately through the long bones (SNPlongbones) are involved in fundamental processes of bone and cartilage development whereas loci in the other sets are less clearly connected to these processes.
Recent work has raised the possibility that adjusting one trait for a highly correlated covariate can lead to false-positive “mirror” associations at variants that are strongly associated with the covariate.54 This phenomenon will cause such variants to be associated with the trait but have a direction of effect that is opposite of the covariate. This phenomenon does not explain our result because our SHR associations are divided between SNPs that are associated with increased and decreased height. Furthermore, SHR is only weakly correlated with height and not all height-associated SNPs at genome-wide significance are associated with SHR.
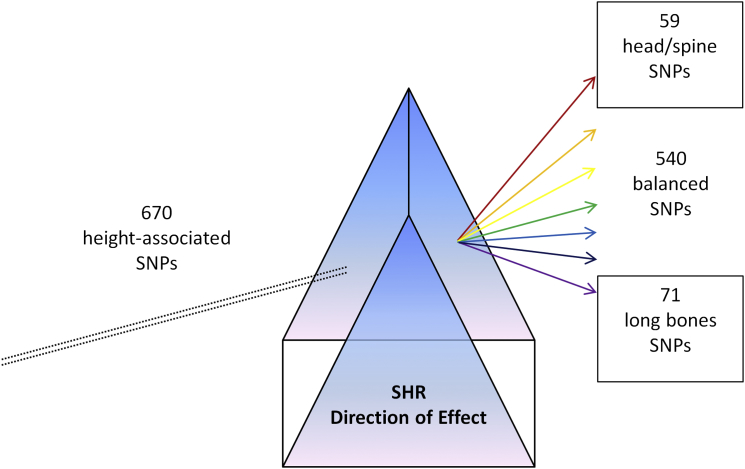
Overall, this study is a large-scale whole-genome experiment to address the underlying genetic basis for differences in body proportion by SHR. We uncovered several loci that are significantly associated with SHR and showed that many loci associated with height also alter SHR. These results suggest that SHR is also polygenic and that further studies of larger sample sizes are required to explain the full genetic spectrum of SHR. Finally, we demonstrated that SHR can be used as a prism (Figure 9) to partition the height-associated loci into those that affect the head/spine, the long bones, or have no effect on SHR and showed that different groups of loci are enriched for different pathways and tissue types. This suggest that in general, the use of orthogonal but related phenotypes can be used as a tool to dissect the variants into different functional classes so as to better understand the biological mechanism underlying the trait or disease.
Figure 9.
SHR as a Prism to Dissect the Height-Associated SNPs into Functional Classes
This diagram illustrates the use of an orthogonal phenotype like SHR to partition the known variants associated with overall height into various functional classes (head/spine, long bones, and balanced).
Acknowledgments
This publication is the work of the authors and Y.C. and J.N.H. will serve as guarantors for the contents of this paper. This research was specifically funded by NIH grant R01DK075787 and March of Dimes grant 6-FY12-393 to J.N.H. R.M.S. was supported by an NIH-NHLBI K99 award (#1K99HL122515-01A1) and advanced postdoctoral fellowship award from the Juvenile Diabetes Research Foundation (JDRF # 3-APF-2014-111-A-N). Acknowledgments for each of the cohorts used in this study can be found in the Supplemental Data.
Published: April 9, 2015
Footnotes
Supplemental Data include GIANT consortium information, cohort-specific acknowledgments, 4 figures, and 19 tables, and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.02.018.
Contributor Information
Yingleong Chan, Email: rigel@broadinstitute.org.
Joel N. Hirschhorn, Email: joelh@broadinstitute.org.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
ALSPAC data dictionary, http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/
GENE PRISM, http://www.broadinstitute.org/∼rigel/prism/
OMIM, http://www.omim.org/
SNPsnap, http://www.broadinstitute.org/mpg/snpsnap/index.html
Supplemental Data
References
- 1.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredriks A.M., van Buuren S., van Heel W.J.M., Dijkman-Neerincx R.H., Verloove-Vanhorick S.P., Wit J.M. Nationwide age references for sitting height, leg length, and sitting height/height ratio, and their diagnostic value for disproportionate growth disorders. Arch. Dis. Child. 2005;90:807–812. doi: 10.1136/adc.2004.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Arriba Muñoz A., Domínguez Cajal M., Rueda Caballero C., Labarta Aizpún J.I., Mayayo Dehesa E., Ferrández Longás Á. Sitting height/standing height ratio in a Spanish population from birth to adulthood. Arch. Argent. Pediatr. 2013;111:309–314. doi: 10.5546/aap.2013.eng.309. [DOI] [PubMed] [Google Scholar]
- 5.Krakow D., Rimoin D.L. The skeletal dysplasias. Genet. Med. 2010;12:327–341. doi: 10.1097/GIM.0b013e3181daae9b. [DOI] [PubMed] [Google Scholar]
- 6.Stokes D.C., Pyeritz R.E., Wise R.A., Fairclough D., Murphy E.A. Spirometry and chest wall dimensions in achondroplasia. Chest. 1988;93:364–369. doi: 10.1378/chest.93.2.364. [DOI] [PubMed] [Google Scholar]
- 7.Hertel N.T., Müller J. Anthropometry in skeletal dysplasia. J. Pediatr. Endocrinol. Metab. 1994;7:155–161. doi: 10.1515/jpem.1994.7.2.155. [DOI] [PubMed] [Google Scholar]
- 8.Gunnell D., Whitley E., Upton M.N., McConnachie A., Smith G.D., Watt G.C.M. Associations of height, leg length, and lung function with cardiovascular risk factors in the Midspan Family Study. J. Epidemiol. Community Health. 2003;57:141–146. doi: 10.1136/jech.57.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilling K., Lawlor D.A., Davey Smith G., Chambless L., Szklo M. The relation between components of adult height and intimal-medial thickness in middle age: the Atherosclerosis Risk in Communities Study. Am. J. Epidemiol. 2006;164:136–142. doi: 10.1093/aje/kwj184. [DOI] [PubMed] [Google Scholar]
- 10.Lawlor D.A., Ebrahim S., Davey Smith G. The association between components of adult height and type II diabetes and insulin resistance: British Women’s Heart and Health Study. Diabetologia. 2002;45:1097–1106. doi: 10.1007/s00125-002-0887-5. [DOI] [PubMed] [Google Scholar]
- 11.Bogin B., Varela-Silva M.I. Leg length, body proportion, and health: a review with a note on beauty. Int. J. Environ. Res. Public Health. 2010;7:1047–1075. doi: 10.3390/ijerph7031047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Lee S.H., Goddard M.E., Visscher P.M. Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Methods Mol. Biol. 2013;1019:215–236. doi: 10.1007/978-1-62703-447-0_9. [DOI] [PubMed] [Google Scholar]
- 13.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 14.Korn J.M., Kuruvilla F.G., McCarroll S.A., Wysoker A., Nemesh J., Cawley S., Hubbell E., Veitch J., Collins P.J., Darvishi K. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fried L.P., Kronmal R.A., Newman A.B., Bild D.E., Mittelmark M.B., Polak J.F., Robbins J.A., Gardin J.M. Risk factors for 5-year mortality in older adults: the Cardiovascular Health Study. J. Am. Med. Assoc. 1998;279:585–592. doi: 10.1001/jama.279.8.585. [DOI] [PubMed] [Google Scholar]
- 16.Gardin J.M., Wagenknecht L.E., Anton-Culver H., Flack J., Gidding S., Kurosaki T., Wong N.D., Manolio T.A. Relationship of cardiovascular risk factors to echocardiographic left ventricular mass in healthy young black and white adult men and women. The CARDIA study. Coronary Artery Risk Development in Young Adults. Circulation. 1995;92:380–387. doi: 10.1161/01.cir.92.3.380. [DOI] [PubMed] [Google Scholar]
- 17.Wilson P.W.F., Castelli W.P., Kannel W.B. Coronary risk prediction in adults (the Framingham Heart Study) Am. J. Cardiol. 1987;59:91G–94G. doi: 10.1016/0002-9149(87)90165-2. [DOI] [PubMed] [Google Scholar]
- 18.Strachan D.P., Rudnicka A.R., Power C., Shepherd P., Fuller E., Davis A., Gibb I., Kumari M., Rumley A., Macfarlane G.J. Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int. J. Epidemiol. 2007;36:522–531. doi: 10.1093/ije/dyl309. [DOI] [PubMed] [Google Scholar]
- 19.Fraser A., Macdonald-Wallis C., Tilling K., Boyd A., Golding J., Davey Smith G., Henderson J., Macleod J., Molloy L., Ness A. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int. J. Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaneau O., Zagury J.-F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 25.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 29.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., FANTOM Consortium An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pers T.H., Karjalainen J.M., Chan Y., Westra H.-J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., Genetic Investigation of ANthropometric Traits (GIANT) Consortium Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geller F., Feenstra B., Carstensen L., Pers T.H., van Rooij I.A.L.M., Körberg I.B., Choudhry S., Karjalainen J.M., Schnack T.H., Hollegaard M.V. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat. Genet. 2014;46:957–963. doi: 10.1038/ng.3063. [DOI] [PubMed] [Google Scholar]
- 34.Van der Valk R.J.P., Kreiner-Møller E., Kooijman M.N., Guxens M., Stergiakouli E., Sääf A., Bradfield J.P., Geller F., Hayes M.G., Cousminer D.L. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum. Mol. Genet. 2015;24:1155–1168. doi: 10.1093/hmg/ddu510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., ADIPOGen Consortium. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GEFOS Consortium. GENIE Consortium. GLGC. ICBP. International Endogene Consortium. LifeLines Cohort Study. MAGIC Investigators. MuTHER Consortium. PAGE Consortium. ReproGen Consortium New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fehrmann R.S.N., Karjalainen J.M., Krajewska M., Westra H.-J., Maloney D., Simeonov A., Pers T.H., Hirschhorn J.N., Jansen R.C., Schultes E.A. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 38.Lage K., Karlberg E.O., Størling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tümer Z., Pociot F., Tommerup N. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 39.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Mouse Genome Database Group The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croft D., O’Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.-M., Doumbo O. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bryc K., Durand E.Y., Macpherson J.M., Reich D., Mountain J.L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Habets P.E.M.H., Moorman A.F.M., Clout D.E.W., van Roon M.A., Lingbeek M., van Lohuizen M., Campione M., Christoffels V.M. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrelson Z., Kelly R.G., Goldin S.N., Gibson-Brown J.J., Bollag R.J., Silver L.M., Papaioannou V.E. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 48.Ballif B.C., Theisen A., Rosenfeld J.A., Traylor R.N., Gastier-Foster J., Thrush D.L., Astbury C., Bartholomew D., McBride K.L., Pyatt R.E. Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am. J. Hum. Genet. 2010;86:454–461. doi: 10.1016/j.ajhg.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radio F.C., Bernardini L., Loddo S., Bottillo I., Novelli A., Mingarelli R., Dallapiccola B. TBX2 gene duplication associated with complex heart defect and skeletal malformations. Am. J. Med. Genet. A. 2010;152A:2061–2066. doi: 10.1002/ajmg.a.33506. [DOI] [PubMed] [Google Scholar]
- 50.Weinzimer S.A., Gibson T.B., Collett-Solberg P.F., Khare A., Liu B., Cohen P. Transferrin is an insulin-like growth factor-binding protein-3 binding protein. J. Clin. Endocrinol. Metab. 2001;86:1806–1813. doi: 10.1210/jcem.86.4.7380. [DOI] [PubMed] [Google Scholar]
- 51.Yakar S., Rosen C.J., Beamer W.G., Ackert-Bicknell C.L., Wu Y., Liu J.-L., Ooi G.T., Setser J., Frystyk J., Boisclair Y.R., LeRoith D. Circulating levels of IGF-1 directly regulate bone growth and density. J. Clin. Invest. 2002;110:771–781. doi: 10.1172/JCI15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng I., DeLellis Henderson K., Haiman C.A., Kolonel L.N., Henderson B.E., Freedman M.L., Le Marchand L. Genetic determinants of circulating insulin-like growth factor (IGF)-I, IGF binding protein (BP)-1, and IGFBP-3 levels in a multiethnic population. J. Clin. Endocrinol. Metab. 2007;92:3660–3666. doi: 10.1210/jc.2007-0790. [DOI] [PubMed] [Google Scholar]
- 53.Agwu J.C., Shaw N.J., Kirk J., Chapman S., Ravine D., Cole T.R. Growth in Sotos syndrome. Arch. Dis. Child. 1999;80:339–342. doi: 10.1136/adc.80.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aschard H., Vilhjálmsson B.J., Joshi A.D., Price A.L., Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 2015;96:329–339. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.