Abstract
The 16p11.2 600 kb copy-number variants (CNVs) are associated with mirror phenotypes on BMI, head circumference, and brain volume and represent frequent genetic lesions in autism spectrum disorders (ASDs) and schizophrenia. Here we interrogated the transcriptome of individuals carrying reciprocal 16p11.2 CNVs. Transcript perturbations correlated with clinical endophenotypes and were enriched for genes associated with ASDs, abnormalities of head size, and ciliopathies. Ciliary gene expression was also perturbed in orthologous mouse models, raising the possibility that ciliary dysfunction contributes to 16p11.2 pathologies. In support of this hypothesis, we found structural ciliary defects in the CA1 hippocampal region of 16p11.2 duplication mice. Moreover, by using an established zebrafish model, we show genetic interaction between KCTD13, a key driver of the mirrored neuroanatomical phenotypes of the 16p11.2 CNV, and ciliopathy-associated genes. Overexpression of BBS7 rescues head size and neuroanatomical defects of kctd13 morphants, whereas suppression or overexpression of CEP290 rescues phenotypes induced by KCTD13 under- or overexpression, respectively. Our data suggest that dysregulation of ciliopathy genes contributes to the clinical phenotypes of these CNVs.
Introduction
Copy number variants (CNVs) with rare population prevalence are major contributors to neurodevelopmental disorders. For example, the recurrent 16p11.2 BP4-BP5 (breakpoint 4-5) 600 kb deletion (MIM: 611913; Figure 1A) is one of the most frequent known genetic lesions in individuals with ASD.1–3 This CNV also predisposes to penetrant forms of obesity4–6 and macrocephaly.6,7 A series of mirror phenotypes is observed in carriers of the reciprocal duplication (MIM: 614671), who are at high risk of being underweight and microcephalic and developing schizophrenia (SCZ)6–8 as well as Rolandic epilepsy.9 Similarly, deletion and duplications show a mirroring impact on brain structures implicated in reward, language, and social cognition.10 Systematic phenotyping of deletion carriers showed a 30-point decrease in IQ, language impairment, and psychiatric co-morbidities, as well as an increased growth velocity of head circumference (HC) during infancy11 that recapitulates the pattern reported in idiopathic ASD.12
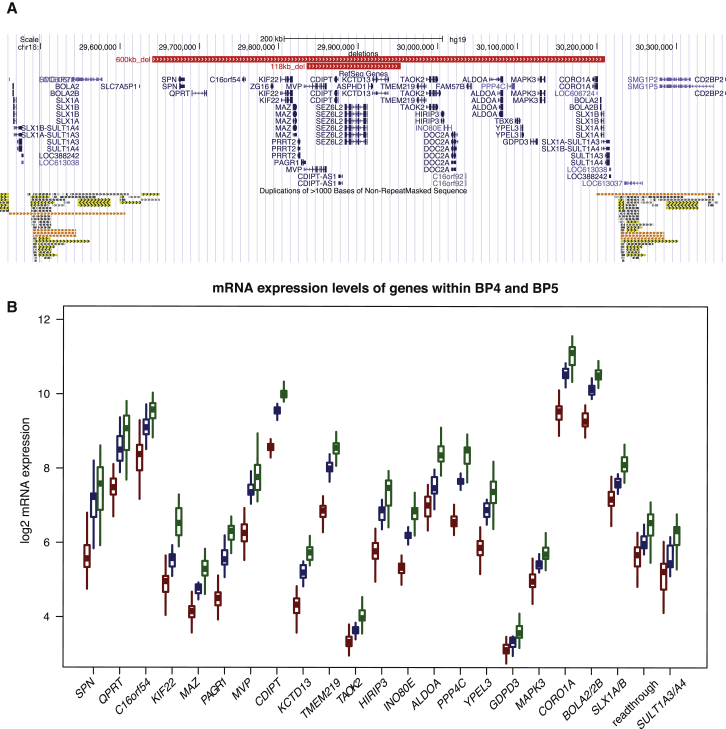
Figure 1.
Expression Level Changes in 16p11.2 Deletion and Duplication LCLs
(A) Comparison of the GRCh37/hg19 genomic boundaries of the recurrent 600 kb BP4-BP5 16p11.2 deletion with that of the 118 kb atypical deletion described in Crepel et al.15 The genes mapping within the interval are shown in blue. The low-copy repeats (LCRs) at the rearrangement breakpoints are indicated at the bottom.
(B) Boxplot distribution of relative expression levels measured by microarrays in deletion (n = 50) and duplication (n = 31) carriers (red and green, respectively) and control LCLs (n = 17, blue) of the 19 “unique” and 3 multiple-copy genes (plus the read-through transcription between SLX1A/1B [MIM: 615822/615823] and SULT1A3/A4 [MIM: 600641/615819]) expressed in LCLs and mapping within the 16p11.2 BP4-BP5 interval and its flanking LCRs, respectively. Note that the expression level of these transcripts, de facto positive controls, is positively correlated with gene dosage.
Among the 32 genes perturbed directly by the 16p11.2 600 kb CNVs, suppression of kctd13 induces macrocephaly in zebrafish embryos, whereas the reciprocal overexpression of human KCTD13 (MIM: 608947) can induce a microcephalic phenotype with concomitant reduction of neurogenesis. Microcephaly was further amplified upon co-injection of either MAPK3 (MIM: 601795) or MVP (MIM: 605088) RNAs, suggesting epistatic interactions for this phenotype in 16p11.2 CNV carriers.13 KCTD13 was associated recently with schizophrenia14 and maps within a smaller 118 kb deletion that segregates with ASD in a three-generation pedigree15 (Figure 1A). Despite these clues and other advances,16–19 the etiopathology of these lesions remains poorly understood. CNVs can impact transcriptomes by modifying the levels and timing of expression of genes localized both within the CNV20,21 and on its flanks,20,22–24 an effect that can extend over the entire length of the affected chromosome.25 Therefore, we undertook a transcriptome analysis of 16p11.2 deletion and duplication carriers and asked whether reciprocal deletion or duplication of the 16p11.2 region correlates with cellular processes or signaling pathways that might inform on involved pathomechanisms. We identified a significant enrichment of expression level changes in pathways relevant to neurodevelopment and primary ciliary functions. These results were confirmed in brains from 16p11.2 CNV mouse models, in which we detected both transcriptional dysregulation and concomitant structural ciliary defects. These results were complemented by epistasis and rescue experiments in zebrafish embryos, confirming the link between 16p11.2 dosage and cilia.
Material and Methods
Samples
The institutional review board of the University of Lausanne, Switzerland, approved this study. Participants were enrolled in the study after signing an informed consent form and being clinically assessed by their respective physicians. For the data collected through questionnaires, information was gathered retrospectively and anonymously by physicians who had ordered comparative genomic hybridization (CGH) analyses performed for clinical purposes only. Consequently, research-based informed consent was not required by the institutional review board of the University of Lausanne, which granted an exemption for this part of the data collection. Details on ascertainment and data collection were previously published.6,11 Overall cognitive functioning was assessed as published.11 Z scores were computed for all data using reference populations of European descent matched for gender, age, and geography as previously described.6
We established lymphoblastoid cell lines (LCLs) from deletion and duplication carriers as well as controls (Table S1) by transforming peripheral blood mononuclear cells with EBV. In brief, heparinized whole-blood samples of enrolled individuals were collected and processed with Ficoll-Paque Premium (GE Healthcare Bio-Sciences AB) centrifugation according to the manufacturer’s protocol, after which the mononuclear cell fractions were isolated. Mononuclear cells were incubated with an Epstein-Barr virus suspension to enable transformation. Upon transformation, the cells were cultured in RPMI 1640 medium (GIBCO), supplemented with 10% Fetal Bovine Serum (PAA, GE Healthcare) and 1% penicillin/streptomycin (GIBCO), at 37°C and 5% CO2 until a stable and exponentially growing cell line was established.
RNA Preparation
For RNA isolation, exponentially growing LCLs were harvested and resuspended in TRIzol reagent (Ambion), after which the manufacturer’s protocol for RNA isolation was performed. Upon RNA isolation, RNA clean-up was performed with the RNeasy Mini Kit (QIAGEN), including the DNase treatment step, according to the manufacturer’s protocol. RNA quality and concentration were determined with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and a 2100 Bioanalyzer platform (Agilent).
Microarray Data Analysis
For each sample, 100 ng of total RNA was processed according to the Affymetrix GeneChip 3′ IVT Express Kit manual (P/N 702646 Revision 6) and hybridized to the Affymetrix GeneChips Human Genome U133+ PM 24 array plates (GEO platform GPL13158). The Affymetrix GeneTitan Instrument was used for automated array processing. Raw data were generated and extracted from the scanned images for analysis with the Affymetrix GeneChip Command Console (AGCC) Software and Expression Console Software packages. Data were annotated according to the NetAffx Analysis Center. All microarrays considered in this analysis were performed in concomitance and, based on clustering analyses and principal component analysis, we can exclude that the array plate is affecting our results. The Robust Multi-array Average (RMA) approach was employed for the creation and normalization of the summarized probe set signals. We applied a non-specific filter to discard probe sets with low variability and low signal: we retained 23,602 probe sets whose SD was greater than median of SD of all probe sets and maximum signal greater than the median of maximum signal of all probe sets. To reduce a potential bias toward genes with multiple probe sets, for the modular analysis, we kept only one probe set with the highest variance per gene, for a total of 15,112 probe sets, 11,830 with Entrez Id and 3,282 without Entrez Id.
Differential Expression Analysis
Genes (represented by 23,602 probe sets) were tested for differential expression via the moderated t-statistic as implemented in Limma.26 We evaluated a dosage model in which CNV was treated as a numerical variable to reflect a dosage effect, relatedness was taken into account by providing an estimate of the within-family correlation via the duplicateCorrelation function in Limma, and the model was adjusted for age and gender (Model 1: gene ∼cnv+age+gender with the cnv covariate coded as −1, 0, 1 and specified within-family correlation). We exploited topGO, DAVID GO, g:Profiler, and Thomson Reuters MetaCore and EnrichR to assess whether the DE genes were related to specific pathways, enriched in specific protein domains and OMIM terms.27–29 Similarly, we used the SFARI Gene list, the union of the genes cataloged in Girard et al.30 and Xu et al.,31,32 and the GWAS hits for BMI33 to assess enrichment for ASD, SCZ, and BMI genes, respectively.
Weighted Gene Co-expression Network Analysis
We used the residual matrix of gene expression corrected for relatedness or for relatedness, age, and gender to build co-expression networks with the WGCNA package in R.34 A Pearson correlation matrix between the residuals of all gene pairs was calculated and then an adjacency matrix was defined by the soft-thresholding approach aij = cor|x,xj|b with b = 6, which represent an approximate scale-free topology network with the scale-free fitting index R2 greater than 0.85. As a preprocessing step in the construction of the network, we selected the 50% of the genes with the highest connectivity. The blockwiseModules function was used for network construction and module detection. Each module was summarized by its first principal component (designated as eigengene) and denoted by a color. Each module eigengene was correlated with the age, gender, CNV, and Z scores of the anthropometric measures, Pearson correlation and the corresponding Benjamini-Hochberg adjusted p value were reported for each module.
Transcriptome Profiling of Mice Models
We sequenced RNA and profiled the transcriptome from the cortical tissue of mice harboring deletion and duplication of the 7qF3 region of synteny conservation with 16p11.2 as described.35 The mouse models were created at the Cold Spring Harbor Laboratory by A. Mills and colleagues, as previously described in Horev et al.,36 and provided by the Jackson Laboratories (stock numbers 013128 and 013129). Sixteen mice were selected from two litters of an engineered line with deletion or duplication of the mouse 16p11.2 equivalent region plus four additional genes found in the human immediately outside of the segmental duplication, centromeric to the CNV (Cd2bp2, Tbc1d10b, Mylpf, Sept1). Within each litter, four mice with CNVs were compared to four wild-type littermates (eight total wild-type mice). The design was fully sex balanced for each genotype. Dissection of mouse cortex was performed simultaneously for all mice at 8 weeks of age. RNA-seq libraries were prepared and sequenced as detailed in Blumenthal et al.35 Raw sequence data were quality checked with fastQC and aligned with TopHat2 using default parameters against the RefSeq transcriptome and mm10 genome (UCSC Genome Browser, accessed September 2012). All aligned reads were analyzed with RNA-SeqQC37 to ensure uniformity of alignment rate, duplication rate, evenness of coverage, GC content, and rRNA content. Reads aligned to the ERCC synthetic RNAs were used to check the linearity of dose response within and between samples, to determine the abundance limits of gene expression that could be accurately quantified in our experiment (defined as the average abundance of the lowest detectable ERCC transcript from all samples), and to assess the power to detect differential expression at varying fold-changes. Raw gene level counts were tabulated with the BEDTools suite38 against the mouse RefSeq transcriptome (accessed February 2013). Genes with read counts below our detection threshold in any individual sample were excluded from analysis in all samples, and raw counts were normalized with the total number of counts generated per sample prior to statistical analysis. Gene level read counts generated by RNA-seq are over-dispersed relative to the Poisson; therefore, we chose the negative binomial distribution to model these data.39,40 The mouse cortex data were analyzed with negative binomial generalized linear models (GLMs). The dispersion parameter for each gene was estimated by an iterative weighted least-squares approach for the GLM analysis. All analyses were conducted in R with the MASS package. Differential expression analysis was performed by fitting a factorial ANOVA with genotype and sex as the factors. We used FDR corrected p values (Benjamini-Hochberg FDR) for thresholds of statistical significance, though nominally significant results were retained for exploratory analyses. We used all genes with expression levels above our detection thresholds as the background list for enrichment analyses (14,435 mouse genes with human orthologs). The mRNA levels of Bbs4, Bbs7, and Cep290 correlated with number of copies of the CNV, and expression of Bbs12 and Ccdc28b were modified in the cortex of deletion mice (all p < 0.10).
Primary Cilium Immunostaining
Mice were backcrossed for three generations into a C57BL/6 background; brain tissue was obtained from mice perfused with 4% PFA. After an overnight infiltration in 30% sucrose, 50 μm floating frozen sections were cut with a cryostat (Leica). All staining was performed in a 24-well plate with an antibody against ACIII (Santa Cruz) and DAPI (Roche). Sections were placed on a glass slide and coverslipped with mounting medium (Millipore). Image acquisition was performed on a Zeiss LSM 710 confocal microscope, and images with a Z plane of ∼20 μm were processed with Imaris software. Cilia trace images were obtained from a maximum image projection of each z stack. Measurements were scored blind to the genotype and obtained from three separate sections from three different animals per genotype. The standard error of the mean was plotted.
Morpholino, Immunostaining, and Embryo Manipulations
Zebrafish (Danio rerio) embryos were raised and maintained as described.41 Splice-blocking MOs against kctd13 (5′-TCTAAGGGTACACGCCTGACCTGTA-3′) and cep290 (5′-TTACGGCTCGGCATACCTTTGATGC-3′) were designed and obtained from Gene Tools as described.13 We injected 10 ng and 6 ng of kctd13 MO and cep290 MO, respectively, into wild-type zebrafish embryos at the 1- to 2-cell stage. For rescue experiments, the human wild-type mRNAs were cloned into the pCS2 vector and transcribed in vitro with the SP6 Message Machine kit (Ambion); 100 pg of the human wild-type mRNAs were coinjected with the MOs. Suppression of endogenous message was shown by PCR amplification of cDNA reverse transcribed from extracted total mRNA as previously described.13 Whole-mount immunostaining with HuC/D was performed for investigating neuronal development at a cellular level, a surrogate for the evaluation of normal neurogenesis.42 Anti-HuC/D (A21271, Invitrogen) is a marker for post-mitotic neurons, i.e., cells positive for HuC and HuD (also known as ELAVL3 and ELAVL4). Embryos were fixed in 4% PFA overnight and stored in 100% methanol at −20°C. After rehydration in PBS, PFA-fixed embryos were washed in immunofluorescence (IF) buffer (0.1% Tween-20 and 1% BSA in PBS 1×) for 10 min at room temperature. The embryos were incubated in the blocking buffer (10% FBS and 1% BSA in PBS 1×) for 1 hr at room temperature. After two washes in IF buffer for 10 min each, embryos were incubated in the first antibody solution, 1:500 anti-HuC/D, in blocking solution overnight at 4°C. After two washes in IF buffer for 10 min each, embryos were incubated in the secondary antibody solution, 1:500 Alexa Fluor goat anti-mouse IgG (A21207, A11001, Invitrogen), in blocking solution for 1 hr at room temperature. Stained embryos were scored visually at 2 dpf and classified as normal (bilateral), ectopic, unilateral, or absent/reduced on the basis of the relative expression of the HuC/D marker in the forebrain with that of age-matched controls from the same clutch. Measurements of the distance between the convex tip of the eye cups as a read-out for head size was performed at 4.5 dpf as described.13 All the experiments were repeated three times and chi-square or Fisher exact tests were performed to determine the significance of the rescue compared to overexpressants and morphants. We blindly scored ∼100 injected embryos for each measure.
Simons Variation in Individuals Project
The 307 Simons VIP exomes were sequenced on Illumina HiSeq 2000 systems and SNPs and indels were called in a single large batch by GATK v.2.6. Variants were called from VCF files, annotated with SNPEFF, and processed to include only high-quality alternate variants passing GATK filters. We further restricted the number of variants by including only those that passed a series of thresholds to reduce false positives: read depth ≥ 10, genotype quality ≥ 30, phred scale likelihood > 30, and allele balance (defined as the ratio of alternate reads to total reads) < 0.1 for homozygous reference individuals, > 0.9 for homozygous alternate individuals, and between 0.3 and 0.7 for heterozygous individuals. Finally, all variants with more than 1% missing genotypes were removed from the analysis.
Results
We profiled the transcriptome of lymphoblastoid cell lines (LCLs) of 50 deletion and 31 duplication carriers with BP4-BP5 boundaries, as well as of 17 control individuals (Table S1) and tested for genes associated with the 16p11.2 CNV by using a dosage effect model and moderated t-statistics. We identified 1,527 significantly differentially expressed (DE) probe sets (6.5% out of 23,602 analyzed probe sets) representing 1,188 unambiguously annotated genes (FDR ≤ 1%; Table S2). This set includes all 22 genes mapping within the CNV with detectable expression in LCLs (Figures 1A and 1B; Table S3). Consistent with published data from LCLs6,43 and adipose tissue,5 the expression levels of these transcripts were positively correlated with gene dosage (Figure 1B). Thus, in contrast to some other genomic disorders,22,43 none of the 16p11.2 CNV genes appear subject to dosage compensation. In agreement with previous studies reporting that CNVs also affect the expression of nearby flanking genes,20,22–25,43 we identified neighboring normal-copy genes with significantly modified expression (Figure S1). Within DE genes, we observed a significant overrepresentation of genes implicated in processes involving RNA metabolism and expression (Table S4). The top-10 list of MetaCore enrichments contains also five pathways involved in developmental signaling: glucocorticoid receptor (FDR 3.99 × 10−5), TGF-β receptor (FDR 9.15 × 10−5), VEGF (FDR 9.15 × 10−5), BMP (FDR 9.91 × 10−5), and FGFR signaling (FDR 1.29 × 10−4) (Tables S5 and S6). We also searched for overrepresentation of Human Phenotype Ontology and found enrichment for “abnormality of skull size” (HP:0000240; Benjamini-Hochberg FDR 1.06 × 10−2), “abnormality of hindbrain morphology” (HP:0011282; FDR 5.00 × 10−2), “abnormality of the metencephalon” (HP:0011283; FDR 5.00 × 10−2), “abnormality of the cerebellum” (HP:0001317; FDR 5.00 × 10−2), and “truncal obesity” (HP:0001956; FDR 3.12 × 10−2) (Table S7). The search for enrichment of OMIM terms identified Bardet-Biedl syndrome (BBS [MIM: 209900]) as the highest hit (Fisher’s exact test: nominal p = 1.2 × 10−4, Bonferroni adjusted p = 4.32 × 10−3 [EnrichR OMIM gene set] and nominal p = 2.52 × 10−6, Bonferroni adjusted p = 4.69 × 10−4 [EnrichR expanded OMIM gene-set]). For accuracy and because some tools were not recently updated (e.g., DAVID), we manually compiled a curated list of genes associated with the overlapping BBS, Joubert (JBTS [MIM: 213300]), and Meckel-Gruber (MIM: 249000) syndromes and identified a significant enrichment of known BBS genes (6 genes out of 16 tested, OR [95% CI] = 5.18 [1.54–15.76], p = 4.0 × 10−3, Bonferroni adjusted p = 0.012), JBTS genes (5/16, OR [95% CI] = 3.92 [1.07–11.26], p = 2.0 × 10−2, Bonferroni adjusted p = 0.06), and BBS and JBTS genes together (10/30, OR [95% CI] = 4.33 [1.80–9.71], p = 6.2 × 10−4, Bonferroni adjusted p = 0.0019) (Tables S8 and S9). These syndromes are associated with abnormal structures or functions of the primary cilium, an essential component required to sense the environment in signal transduction.44,45 The 1,188 DE genes are also significantly enriched for genes included in SFARI Gene, an annotated list of genes enriched in autism genes (47 DE genes among 313 SFARI genes with detectable expression; Fisher test, OR [95% CI] = 1.63 [1.16–2.25], p = 3.8 × 10−3; Bonferroni adjusted p = 0.011). We found no enrichment for genes associated with SCZ or GWAS hits for BMI (body mass index) (Table S10; Material and Methods). Taken together our data suggest (1) that the profiling of LCL, a non-neural tissue, can identify pathways relevant for ASD and head size and (2) that other genes in our DE list could be relevant to brain pathophysiologies.
We complemented our approach with unsupervised assessments via Weighted Gene Co-expression Network Analysis (WGCNA)34,46 and identified five modules for which expression correlated with the number of CNV copies (Figure S2; Tables S11 and S12). The “light green” module (51 genes, Pearson 0.95, BH FDR adjusted p = 28.0 × 10−49) was anticorrelated with weight (p = 6.0 × 10−4), BMI (p = 3.0 × 10−7), and HC (p = 6.0 × 10−5; all FDR adjusted) and included all 22 imbalanced 16p11.2 genes expressed in LCLs, confirming the association of 16p11.2 genes with mirror effects on body weight and HC6,7 and providing additional confidence for the specificity of our data. The second most correlated module (264 genes, Pearson −0.65, FDR adjusted p = 5 × 10−12) regrouped genes involved in RNA biosynthesis/regulation (GO enrichment p = 4.3 × 10−6), gene expression/transcription (p = 1.7 × 10−5), and cilium morphogenesis (p = 1.3 × 10−2). This “black” module correlated positively with BMI (0.41, FDR adjusted p = 4.0 × 10−4) and HC (0.3, FDR adjusted p = 3.0 × 10−2), supporting the association between changes in the expression of ciliopathy genes and these anthropometric features. Correction for gender, age, and kinship had no substantial influence on definition of these modules and their correlations (Material and Methods).
Our results are further supported by our recent observation that genes with modified chromosomal contacts to BP4-BP5 600 kb 16p11.2 interval genes in CNV carriers cell lines are also enriched (OR = 1.5, p = 0.030) for ciliary genes listed in van Dam et al.47 and for genes associated with specific ciliopathies (A.R., unpublished data). These findings and some commonalities between the phenotypes of BBS and 16p11.2 CNV carriers (see below) prompted us to further explore the possibility of a potential contributory role for ciliary dysfunction to aspects of the 16p11.2 pathology. Cognizant of the potential pitfalls of drawing conclusions from LCLs, we profiled the transcriptome of cortical tissues of mouse models engineered to carry one or three copies of the 16p11.2 orthologous region36 (n = 16, i.e., 4 deletion, 4 duplication, and 8 wild-type mice, fully balanced for gender) by RNA sequencing.35 Using generalized linear models to contrast wild-type expression against the abnormal copy states, we found marked overlap with human LCL findings. Of the 311 SFARI genes (excluding KCTD13 and MVP that map within the 16p11.2 CNV region), 299 were quantified in mouse cortex, in particular all 45 human LCL DE genes. Of those 45 genes, we observed differential expression (p < 0.05) of 13 genes in mouse cortex deletion data alone (where both studies are best powered; Fisher’s exact test for enrichment against all genes differentially expressed in the mouse cortex, p = 0.011) and of 14 genes in the deletion and duplication combined data (p = 0.092). These genes include CEP290 (MIM: 610142), YTHDC2, TOP1 (MIM: 126420), UBE3A (MIM: 601623), RB1CC1 (MIM: 606837), GNAS (MIM: 139320), TBL1XR1 (MIM: 608628), DCUN1D1 (MIM: 605905), ARID1B (MIM: 614556), BCL2 (MIM: 151430), ROBO1 (MIM: 602430), EPC2 (MIM: 611000), CYFIP1 (MIM: 606322), and SNX19. At a differential expression threshold of p < 0.10, we observed 17 of 45 LCL DE genes in the deletion data alone (p = 0.019), and 20/45 in the deletion and duplication combined data (p = 0.099). These findings include the previously described genes along with BBS4 (MIM: 600374), RORA (MIM: 600825), SNRPN (MIM: 182279), CD44 (MIM: 107269), AFF4 (MIM: 604417), and EIF4E (MIM: 133440). In particular, we observed a significant enrichment of perturbed expression of genes involved in both JBTS (p = 2.6 × 10−4 and 2.9 × 10−3) and BBS (p = 1.4 × 10−3 and 1.4 × 10−2, in deletion and duplication models, respectively). Cep290 expression was increased in deletion (p = 8.0 × 10−3) and decreased in duplication (p = 8.0 × 10−2) animals. WGCNA of murine cortex RNA-seq data further uncovered that the 149 genes-strong module that includes the 16p11.2 interval orthologous genes was enriched for genes involved in flagellar function (p = 4.3 × 10−2). These results from different tissues and species further support the notion that 16p11.2 rearrangements alter ciliary gene expression.
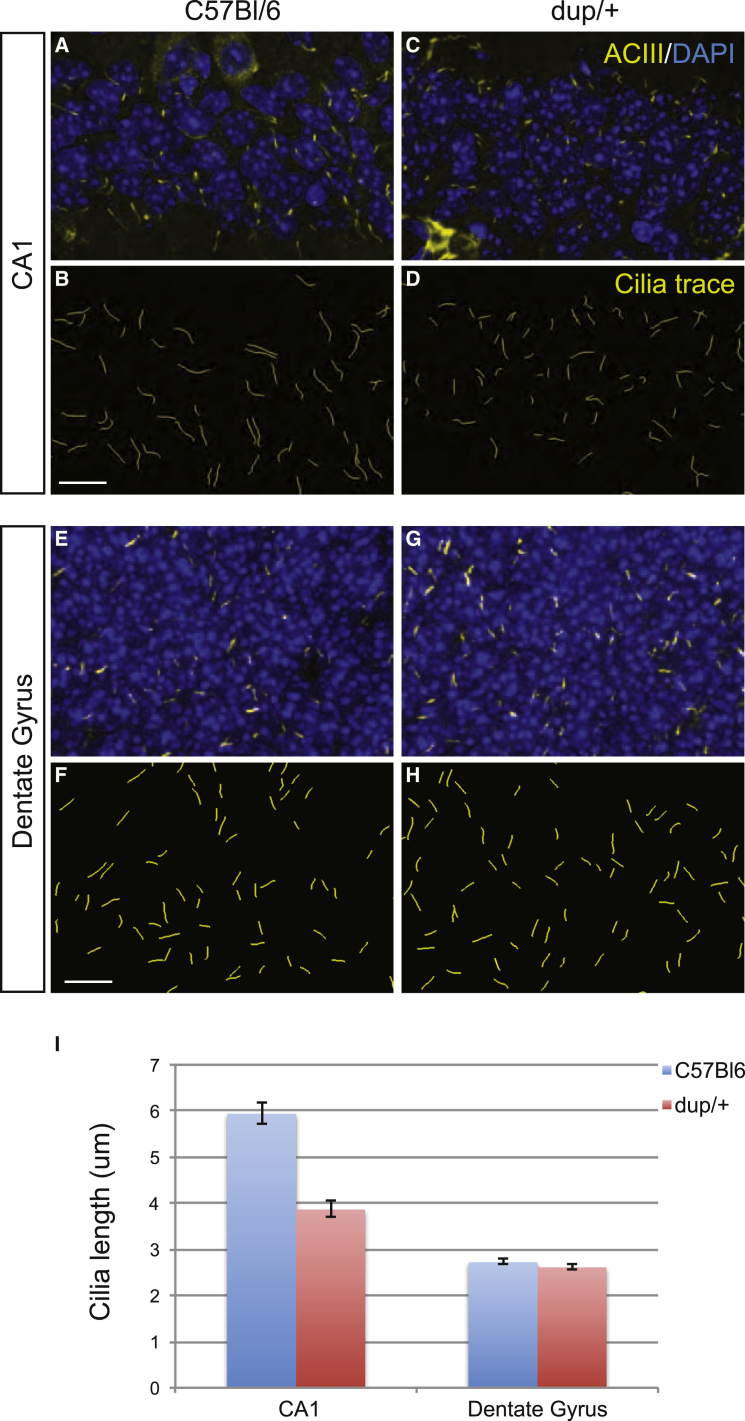
Although our expression analyses are correlative, they are compatible with a potential contributory role for ciliary dysfunction to aspects of the 16p11.2 pathology. If this is accurate, we posited that (1) given that dysfunction of several ciliopathy proteins is associated with structural ciliary defects, we should be able to observe these and (2) if dysregulation of ciliopathy proteins is a phenotypic driver, then anti-regulation of these genes should rescue aspects of the 16p11.2 pathology. Mice knocked-out for genes that contribute loss-of-function mutations to ciliopathy-affected individuals show significant defects in hippocampal primary cilia length.48–52 Because this brain structure is relevant to ASD and SCZ53 and shows 16p11.2 dosage-dependent volume changes,54 we asked whether dosage-dependent ciliary phenotypes could be observed in CA1, CA3, and the dentate gyrus of mouse models engineered to carry either one or three copies of the 16p11.2 orthologous region.36 Although there was no significant difference in animals carrying the deletion, we found a CA1-specific significant shortening of neuronal cilia in the duplication model (cilia median length decrease from 5.9 to 3.9 μm [p < 1.0 × 10−3]; Figure 2). This suggests that the observed transcriptional changes affect ciliary length in discrete hippocampal regions.
Figure 2.
Ciliary Length Analyses in Hippocampal Tissue from C57BL/6 and 16p11.2 dup/+ Mice
(A–H) Staining of cilia in the CA1 (A, C) and dentate gyrus (E, G) regions of the hippocampus via ACIII (yellow) and counterstained with DAPI (blue) in C57BL/6 controls (A, E) and 16p11.2 duplication models (C, G). CA1 (B, D) and dentate gyrus (F, H) ciliary length analyses were performed by plotting pixel information from ACIII staining in C57BL/6 controls (A, E) and 16p11.2 duplication models (C, G).
(I) Total ciliary length comparisons were measured and plotted for both the CA1 and dentate gyrus (SEM was plotted; ∗∗∗p < 0.001).
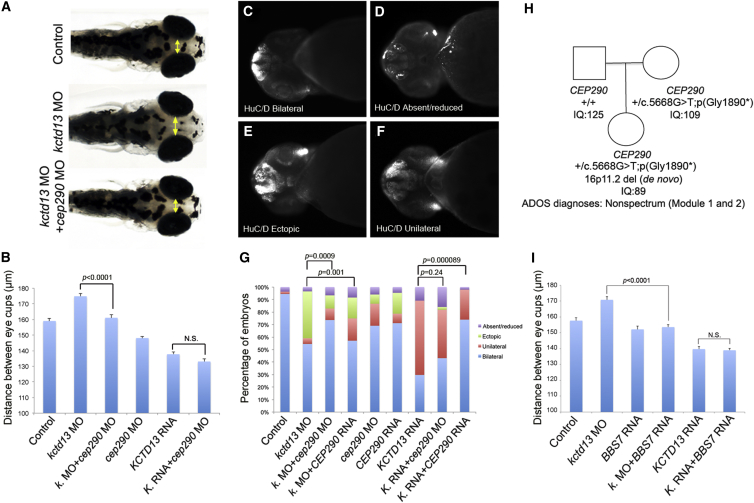
To test directly whether the transcriptional dysregulation of cilia genes might be a driver of the pathology, we turned to zebrafish. We have shown previously that reciprocal under- and overexpression of KCTD13 induces macro- and microcephaly with concomitant defects in neurogenesis and bilateral symmetry of post-mitotic neurons.13 This kctd13 morpholino-induced (MO) macrocephaly was rescued significantly by BBS7 (MIM: 607590) mRNA (p < 1.0 × 10−4), whereas overexpression of BBS7 alone induced no appreciable phenotypes (Figure 3I). Next, we tested CEP290, whose expression profile is opposite to BBS7. We combined overexpression of human KCTD13 with either cep290 MO or human CEP290 RNA and asked whether suppression or overexpression of CEP290 might rescue head size and expression neurogenesis. Embryos co-injected with kctd13 MO and cep290 MO showed a normal head size, indistinguishable from sham-injected embryos (p < 1.0 × 10−4; Figures 3A and 3B). To probe the cellular basis of this correction further, we stained injected embryos with HuC/D antibody (a marker of early postmitotic neurons) and binned them into four classes: bilateral, unilateral, ectopic, and reduced/absent expression (Figures 3C–3F). At 2 days post-fertilization (dpf), a time at which kctd13 overexpressants are anatomically indistinguishable from sham-injected embryos,13 we observed a loss of bilateral expression of HuC/D expression in the forebrain (Figure 3G). Masked qualitative scoring of embryos at 2 dpf injected with KCTD13 RNA and CEP290 RNA showed a significant rescue of bilateral HuC/D expression in the forebrain (Fisher exact test p = 9.0 × 10−5), whereas co-injection with cep290 MO showed no difference compared to embryos injected with KCTD13 RNA alone (p = 0.24; Figure 3G). Reciprocally, we scored embryos injected with kctd13 MO with either human CEP290 RNA or MO (Figure 3G). Suppression of kctd13 led to the ectopic expression of HuC/D concomitant with an increased number of newborn neurons and macrocephaly as described.13 Injection of kctd13 MO and cep290 MO yielded a significant rescue of bilateral expression of HuC/D, which was no longer expressed ectopically but was instead restricted to the forebrain, similar to HuC/D expression in controls (chi-square test p = 9.0 × 10−4; Figure 3G). This restricted expression of HuC/D in the forebrain was consistent with the normal head size of these double morphants observed at 4.5 dpf (Figures 3A and 3B). In contrast, injection of CEP290 RNA failed to rescue the ectopic expression of HuC/D seen in kctd13 morphants. Taken together, these data suggest that overexpression or suppression of CEP290 alleviates the neuroanatomical defects observed in the context of an over- or underexpression of the 16p11.2 CNV driver KCTD13, respectively.
Figure 3.
Ciliopathy Gene Suppression or Overexpression Rescues the Neuroanatomical Defects Associated with KCTD13 Imbalance
(A) From top to bottom, dorsal views of representative 4.5 dpf controls, embryos injected with kctd13 MO, and co-injected with kctd13 and cep290 MOs. The yellow double-head arrows mark the distance between the convex tip of the eyecups, a proxy for head size.
(B) Bar graph represents the distance between the eyecups for controls and injected embryos. SEM was plotted. Student’s t test was performed and the corresponding p value is denoted on the bar graph.
(C–F) Representative photographs show ventral views of zebrafish embryos at 2 dpf stained with HuC/D. Embryos were binned into four classes: normal bilateral expression (C), absence or reduced expression (D), ectopic expression (E), and unilateral expression (F).
(G) Percentage of embryos with normal bilateral HuC/D protein levels (blue) or unilateral HuC/D (red), ectopic (green), and absent/reduced protein levels (purple) in the anterior forebrain in embryo batches injected with kctd13, cep290 MOs, and/or KCTD13, CEP290 mRNAs. HuC/D levels in the anterior forebrain of the embryo injected with the kctd13 MO are considerably increased compared to those of the control embryo. This defect was rescued significantly by co-injection of cep290 MO. Likewise, HuC/D levels in the anterior forebrain of the embryo injected with the KCTD13 mRNA are considerably lower than those of the control embryo. This defect was rescued significantly by co-injection of full-length human CEP290 mRNA. Although significant differences were observed in unilateral and ectopic classes for embryos injected with CEP290 RNA and kctd13 MO compared to kctd13 morphants, the total percentage of abnormal embryos remained the same. Chi-square test was performed and the corresponding p values are denoted on the bar graph.
(H) Two-generation pedigree shows mother-daughter transmission of a CEP290 nonsense variant and a de novo 16p11.2 deletion identified in the daughter. The genotypes, IQ, and ADOS diagnoses are indicated (see also Table S13).
(I) Bar graph represents the distance between the eyecups for controls and embryos injected with kctd13 morpholino (MO), BBS7 RNA, and co-injected with kctd13 MO and BBS7 RNA. SEM was plotted. Student’s t test was performed and the corresponding p value is denoted on the bar graph.
Our studies predict that loss of CEP290 might have a protective effect to the 16p11.2 pathology in humans. To assess this, we sequenced the exomes of 307 individuals of the Simons VIP collection carrying 16p11.2 deletion or duplication.11 We identified a 61-month-old female of northern European descent with a de novo 16p11.2 deletion and a maternally inherited nonsense mutation c.5668G>T in CEP290 (GenBank: NM_025114.3), encoding p.(Gly1890∗) (Figure 3H). This allele, which is absent from >8,000 control exomes, is the second most common CEP290 mutation reported in literature. It was found in homozygosity in one55 and in compound heterozygosity with other deleterious CEP290 alleles in five JBTS-affected families.56,57 This individual had normal HC and weight (both 50th centile), tested negative for ASD (ADOS modules 1 and 2), had no reported seizures, and had mild reduction of IQ compared to her parents (full-scale IQ [FSIQ] 89; non-verbal IQ 86; Table S13). These results should be compared to the deletion phenotypes previously described.11 In brief, 16p11.2 BP4-BP5 deletions increase on average the HC and the BMI by +0.6 and +0.8 SD at 5 years of age, respectively (i.e., approximately one-third and 20% of deletion carriers have HC and BMI equal or below 50th percentile, respectively). Furthermore, the FSIQ of de novo 16p11.2 BP4-BP5 deletion carriers equals 83. Although the young carrier identified here was screened originally by array CGH because of concerns relating to expressive language development, she has a verbal IQ of 93, i.e., 1 SD above the mean of 16p11.2 deletion carriers (n = 42, mean verbal IQ = 74, SD = 17.511). We are cautious at interpreting data from a single young CNV carrier. Both follow up of this individual and identification of additional cases are required to investigate the modulation of 16p11.2-related phenotypes by mutations affecting ciliary function; nonetheless, these observations are compatible with the in vivo prediction that reduction of CEP290 might have an ameliorating effect on key phenotypes associated with the 16p11.2 deletion.
Discussion
The 16p11.2 rearrangements provide an opportunity to investigate molecular mechanisms underlying the co-morbidity triad of neurodevelopmental disorders, energy imbalance, and HC alterations. Our study of 98 human cell lines identified gene expression perturbations of developmental pathways including ciliopathy loci and ASD genes. We are well aware of the limitations of the study of LCLs, which might not be the best target tissue for many genes, for instance genes whose expression specificity resides in other cell lineages. These experiments are nevertheless worth pursuing, because pattern of expressions in peripheral tissue might be used as a first-tier biomarker, for example. Importantly, dysregulated pathways could be validated in an appropriate animal model. Accordingly, we validated our transcriptome results in the cerebral cortex of 16p11.2 mouse models. The mouse model harboring duplication of the orthologous 7qF3 region concomitantly showed structural ciliary defects specifically in the CA1 area of the hippocampus similar to what was reported in the Bbs4−/− animals.52 Whereas the CA2 region is essential for social memory,58 CA1 is associated with spatial memory,59 an endophenotype of schizophrenia60,61 associated with the 16p11.2 duplication.8 The association between 16p11.2 600 kb BP4-BP5 rearrangements and transcript perturbations of ciliopathy and autism spectrum disorder genes is further supported by the results of our study of the chromatin conformation changes induced by these CNVs. We uncovered that long-range chromosomal contacts of genes located within the 600 kb interval are similarly enriched for genes associated with autism (both SFARI-ASD and de novo ASD genes) and the primary cilium, as well as for genes that are differentially expressed in 16p11.2 CNV carrier cell lines, suggesting that chromosomal contacts are necessary for co-regulation (A.R., unpublished data). The primary cilium has been the focus of intensive studies and emerging data show that apart from specific ciliopathy syndromes, this important developmental organelle plays a widespread role in human genetic disease. For example, prior evidence does exist that ciliary dysfunction might contribute to multiple neuropsychiatric disorders beyond the simple correlation of clinical overlap. Suppression of candidate genes for intellectual disability, SCZ, and ASD in cells resulted in disturbed ciliogenesis in 56% of tested genes.62 Consistent with this observation, several well-established ASD genes are directly involved in ciliary biology (e.g., AHI1 [MIM: 608894], DISC1 [MIM: 605210], CTNNB1 [MIM: 116806]) or their knockdowns result in cilia loss (e.g., KATNAL2 [MIM: 614697], NRXN1 [MIM: 600565], FOXP1 [MIM: 605515], CHD7 [MIM: 608892]).47 Our findings suggest a new link between ciliopathy and ASDs. Several bona fide ciliopathy loci have been associated with discrete 16p11.2 CNV phenotypes.63–69 Our genetic interaction experiment supports this notion further, because changes in expression of either BBS7 or CEP290 modified the neurodevelopmental defects induced by KCTD13 dosage imbalance. Although the mechanisms of causality or rescue remain unknown, recent studies have shown that a subset of ciliopathy proteins regulate paracrine signals such as Wnt and Shh through the regulation of proteasomal function proximal to the primary cilium.70–72 KCTD13 is an ubiquitin ligase that targets RhoA,73 offering a possible mechanistic link. Other genes whose products participate in proteasomal function have been implicated in ASDs, including CUL3 (MIM: 603136)74,75 and UBE3A.76,77 Of note, mice null for Cep290, whose expression is anti-correlated to the BBS genes in the context of the 16p11.2 transcriptome, have been shown to rescue phenotypes induced by ablation of Bbs6.78 We hypothesize that antagonistic regulation of either broad proteasomal activity or specific proteasome-dependent signaling pathways might underlie the observed reciprocal rescue.
Our studies also have potential implications for the clinical evaluation and management of 16p11.2 CNV carriers. First, the dysregulation of ciliopathy genes in this CNV predicts, beside the reported obesity, hyperphagia, posterior fossa abnormalities, cognitive impairment, and social communication deficits,11,79,80 plus additional co-morbidities that include anomalies of urogenital tract, renal dysfunction, and retinal defects. Several studies already described renal defects and high incidence of genital malformations involving Müllerian ducts in 16p11.2 CNV carriers;11,81–84 these phenotypes warrant further attention. Second, cataloging and interpreting deleterious variations at ciliopathy loci and other DE genes identified in this study might inform on the observed variability in clinical expressivity of this CNV,6,11 which in turn might assist in carrier management. The discovery of an individual carrying a de novo 16p11.2 deletion and a heterozygous null CEP290 allele supports this idea. Finally, our data raise the possibility that genes that are overexpressed in either deletion or duplication carriers offer a route to therapeutic development either through direct or indirect suppression of their function. This approach is reminiscent of spinal muscular atrophy, where expression studies in LCLs were used as a screen to identify modifiers.85,86
Consortia
16p11.2 European Consortium members are Eugenia Migliavacca, Katrin Männik, Louise Harewood, Maria Nicla Loviglio, Robert Witwicki, Gérard Didelot, Ilse van der Werf, Ali A. Alfaiz, Marianna Zazhytska, Giuliana Giannuzzi, Jacqueline Chrast, Aurélien Macé, Sven Bergmann, Zoltan Kutalik, Loyse Hippolyte, Anne M. Maillard, Vanessa Siffredi, Flore Zufferey, Danielle Martinet, Frédérique Bena, Anita Rauch, Sonia Bouquillon, Joris Andrieux, Bruno Delobel, Odile Boute, Bénédicte Duban-Bedu, Cédric Le Caignec, Bertrand Isidor, Jean Chiesa, Boris Keren, Brigitte Gilbert-Dussardier, Renaud Touraine, Dominique Campion, Caroline Rooryck Thambo, Michèle Mathieu-Dramard, Ghislaine Plessis, Frank Kooy, Hilde Peeters, Katrin Ounap, Anneke T. Vulto-van Silfhout, Bert B. de Vries, Ellen van Binsbergen, Mieke M. van Haelst, Ann Nordgren, Mafalda Mucciolo, Alessandra Renieri, Evica Rajcan-Separovic, John A. Philipps III, Richard J. Ellis, Jacques S. Beckmann, Sébastien Jacquemont, and Alexandre Reymond. The affiliations and email addresses of the members of the 16p11.2 European Consortium are listed in Table S14.
Acknowledgments
We thank the members of the Lausanne Genomic Technologies Facility, Patrick Meylan, and Alexandre Thiery for technical help. We are grateful to all the families participating to the Simons Variation in Individuals Project (Simons VIP), as well as to the Simons VIP working group. K.M. was awarded a scholarship from the Swiss Scientific Exchange NMS Program, S.J. is recipient of a Bursary Professor fellowship of the Swiss National Science Foundation (SNSF), and C.G., E.C.O., and M.E.T. are grantees of NARSAD Young Investigator Grants from the Brain and Behavior Research Foundation. A.A.A. was awarded a scholarship from the Saudi Arabian National Guard Health Affairs. This work is supported by the Simons Foundation (grants SFARI274424 to A.R., SFARI239983 to N.K., SFARI238504 to M.E.T., and SFARI240021 to J.F.G.), the Swiss National Science Foundation (grant 31003A_160203 to A.R.), a specific SNSF Sinergia grant (CRIS FN CRSII33-133044 to A.R.), the Leenaards Foundation Prize (S.J. and A.R.), by NIH P50MH094260 (N.K.) and MH095867 (M.E.T.), and the Nancy Lurie Marks Family Foundation (J.F.G. and M.E.T.). N.K. is a Distinguished Brumley Professor. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: April 30, 2015
Footnotes
Supplemental Data include 2 figures and 14 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.04.002.
Contributor Information
Nicholas Katsanis, Email: katsanis@cellbio.duke.edu.
Alexandre Reymond, Email: alexandre.reymond@unil.ch.
16p11.2 European Consortium:
Eugenia Migliavacca, Katrin Männik, Louise Harewood, Maria Nicla Loviglio, Robert Witwicki, Gérard Didelot, Ilse van der Werf, Ali A. Alfaiz, Marianna Zazhytska, Giuliana Giannuzzi, Jacqueline Chrast, Aurélien Macé, Sven Bergmann, Zoltan Kutalik, Loyse Hippolyte, Anne M. Maillard, Vanessa Siffredi, Flore Zufferey, Danielle Martinet, Frédérique Bena, Anita Rauch, Sonia Bouquillon, Joris Andrieux, Bruno Delobel, Odile Boute, Bénédicte Duban-Bedu, Cédric Le Caignec, Bertrand Isidor, Jean Chiesa, Boris Keren, Brigitte Gilbert-Dussardier, Renaud Touraine, Dominique Campion, Caroline Rooryck Thambo, Michèle Mathieu-Dramard, Ghislaine Plessis, Frank Kooy, Hilde Peeters, Katrin Ounap, Anneke T. Vulto-van Silfhout, Bert B. de Vries, Ellen van Binsbergen, Mieke M. van Haelst, Ann Nordgren, Mafalda Mucciolo, Alessandra Renieri, Evica Rajcan-Separovic, John A. Philipps, III, Richard J. Ellis, Jacques S. Beckmann, Sébastien Jacquemont, and Alexandre Reymond
Accession Numbers
Our data were deposited in the NCBI Gene Expression Omnibus under accession number GEO: GSE57802 and in the NIH-supported National Database for Autism Research (NDAR) under study #344.
Web Resources
The URLs for data presented herein are as follows:
Bioconductor, http://www.bioconductor.org
FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc
OMIM, http://www.omim.org/
SFARI, https://sfari.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 2.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K., Saeed S., Hamilton-Shield J., Clayton-Smith J., O’Rahilly S. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinawi M., Liu P., Kang S.H., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy S.E., Makarov V., Kirov G., Addington A.M., McClellan J., Yoon S., Perkins D.O., Dickel D.E., Kusenda M., Krastoshevsky O., Wellcome Trust Case Control Consortium Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinthaler E.M., Lal D., Lebon S., Hildebrand M.S., Dahl H.H., Regan B.M., Feucht M., Steinböck H., Neophytou B., Ronen G.M., 16p11.2 European Consortium. EPICURE Consortium. EuroEPINOMICS Consortium 16p11.2 600 kb duplications confer risk for typical and atypical Rolandic epilepsy. Hum. Mol. Genet. 2014;23:6069–6080. doi: 10.1093/hmg/ddu306. [DOI] [PubMed] [Google Scholar]
- 10.Maillard A.M., Ruef A., Pizzagalli F., Migliavacca E., Hippolyte L., Adaszewski S., Dukart J., Ferrari C., Conus P., Mannik K. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol. Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courchesne E., Mouton P.R., Calhoun M.E., Semendeferi K., Ahrens-Barbeau C., Hallet M.J., Barnes C.C., Pierce K. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 13.Golzio C., Willer J., Talkowski M.E., Oh E.C., Taniguchi Y., Jacquemont S., Reymond A., Sun M., Sawa A., Gusella J.F. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crepel A., Steyaert J., De la Marche W., De Wolf V., Fryns J.P., Noens I., Devriendt K., Peeters H. Narrowing the critical deletion region for autism spectrum disorders on 16p11.2. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2011;156:243–245. doi: 10.1002/ajmg.b.31163. [DOI] [PubMed] [Google Scholar]
- 16.Wood H. Genetics: expanding the spectrum of neurological disorders associated with PRRT2 mutations. Nat. Rev. Neurol. 2012;8:657. doi: 10.1038/nrneurol.2012.240. [DOI] [PubMed] [Google Scholar]
- 17.Sparrow D.B., McInerney-Leo A., Gucev Z.S., Gardiner B., Marshall M., Leo P.J., Chapman D.L., Tasic V., Shishko A., Brown M.A. Autosomal dominant spondylocostal dysostosis is caused by mutation in TBX6. Hum. Mol. Genet. 2013;22:1625–1631. doi: 10.1093/hmg/ddt012. [DOI] [PubMed] [Google Scholar]
- 18.de Anda F.C., Rosario A.L., Durak O., Tran T., Gräff J., Meletis K., Rei D., Soda T., Madabhushi R., Ginty D.D. Autism spectrum disorder susceptibility gene TAOK2 affects basal dendrite formation in the neocortex. Nat. Neurosci. 2012;15:1022–1031. doi: 10.1038/nn.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaker-Lee A., Gupta S., McCammon J.M., De Rienzo G., Sive H. Zebrafish homologs of genes within 16p11.2, a genomic region associated with brain disorders, are active during brain development, and include two deletion dosage sensor genes. Dis. Model. Mech. 2012;5:834–851. doi: 10.1242/dmm.009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrichsen C.N., Vinckenbosch N., Zöllner S., Chaignat E., Pradervand S., Schütz F., Ruedi M., Kaessmann H., Reymond A. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 21.Chaignat E., Yahya-Graison E.A., Henrichsen C.N., Chrast J., Schütz F., Pradervand S., Reymond A. Copy number variation modifies expression time courses. Genome Res. 2011;21:106–113. doi: 10.1101/gr.112748.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merla G., Howald C., Henrichsen C.N., Lyle R., Wyss C., Zabot M.T., Antonarakis S.E., Reymond A. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am. J. Hum. Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reymond A., Henrichsen C.N., Harewood L., Merla G. Side effects of genome structural changes. Curr. Opin. Genet. Dev. 2007;17:381–386. doi: 10.1016/j.gde.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricard G., Molina J., Chrast J., Gu W., Gheldof N., Pradervand S., Schütz F., Young J.I., Lupski J.R., Reymond A., Walz K. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 27.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Reimand J., Arak T., Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girard S.L., Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., Dionne-Laporte A., Spiegelman D., Henrion E., Diallo O. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 31.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu B., Roos J.L., Dexheimer P., Boone B., Plummer B., Levy S., Gogos J.A., Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vimaleswaran K.S., Tachmazidou I., Zhao J.H., Hirschhorn J.N., Dudbridge F., Loos R.J. Candidate genes for obesity-susceptibility show enriched association within a large genome-wide association study for BMI. Hum. Mol. Genet. 2012;21:4537–4542. doi: 10.1093/hmg/dds283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:e17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 35.Blumenthal I., Ragavendran A., Erdin S., Klei L., Sugathan A., Guide J.R., Manavalan P., Zhou J.Q., Wheeler V.C., Levin J.Z. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am. J. Hum. Genet. 2014;94:870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horev G., Ellegood J., Lerch J.P., Son Y.E., Muthuswamy L., Vogel H., Krieger A.M., Buja A., Henkelman R.M., Wigler M., Mills A.A. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc. Natl. Acad. Sci. USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund S.P., Nettleton D., McCarthy D.J., Smyth G.K. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat. Appl. Genet. Mol. Biol. 2012;11:11. doi: 10.1515/1544-6115.1826. [DOI] [PubMed] [Google Scholar]
- 40.Soneson C., Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics. 2013;14:91. doi: 10.1186/1471-2105-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westerfield M. Univ. of Oregon Press; Eugene: 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 42.Beunders G., Voorhoeve E., Golzio C., Pardo L.M., Rosenfeld J.A., Talkowski M.E., Simonic I., Lionel A.C., Vergult S., Pyatt R.E. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 2013;92:210–220. doi: 10.1016/j.ajhg.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo R., Sanders S.J., Tian Y., Voineagu I., Huang N., Chu S.H., Klei L., Cai C., Ou J., Lowe J.K. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am. J. Hum. Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerdes J.M., Davis E.E., Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh E.C., Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henrichsen C.N., Csárdi G., Zabot M.T., Fusco C., Bergmann S., Merla G., Reymond A. Using transcription modules to identify expression clusters perturbed in Williams-Beuren syndrome. PLoS Comput. Biol. 2011;7:e1001054. doi: 10.1371/journal.pcbi.1001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dam T.J., Wheway G., Slaats G.G., Huynen M.A., Giles R.H., SYSCILIA Study Group The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2:7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breunig J.J., Sarkisian M.R., Arellano J.I., Morozov Y.M., Ayoub A.E., Sojitra S., Wang B., Flavell R.A., Rakic P., Town T. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2008;105:13127–13132. doi: 10.1073/pnas.0804558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Y.G., Spassky N., Romaguera-Ros M., Garcia-Verdugo J.M., Aguilar A., Schneider-Maunoury S., Alvarez-Buylla A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat. Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 50.Klinger M., Wang W., Kuhns S., Bärenz F., Dräger-Meurer S., Pereira G., Gruss O.J. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol. Biol. Cell. 2014;25:495–507. doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkins C.E., Aviles G.D., East M.P., Kahn R.A., Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agassandian K., Patel M., Agassandian M., Steren K.E., Rahmouni K., Sheffield V.C., Card J.P. Ciliopathy is differentially distributed in the brain of a Bardet-Biedl syndrome mouse model. PLoS ONE. 2014;9:e93484. doi: 10.1371/journal.pone.0093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung C., Yu K., Fung G., Leung M., Wong C., Li Q., Sham P., Chua S., McAlonan G. Autistic disorders and schizophrenia: related or remote? An anatomical likelihood estimation. PLoS ONE. 2010;5:e12233. doi: 10.1371/journal.pone.0012233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qureshi A.Y., Mueller S., Snyder A.Z., Mukherjee P., Berman J.I., Roberts T.P., Nagarajan S.S., Spiro J.E., Chung W.K., Sherr E.H., Buckner R.L., Simons VIP Consortium Opposing brain differences in 16p11.2 deletion and duplication carriers. J. Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., International Joubert Syndrome Related Disorders Study Group Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 56.Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 57.Brancati F., Barrano G., Silhavy J.L., Marsh S.E., Travaglini L., Bielas S.L., Amorini M., Zablocka D., Kayserili H., Al-Gazali L., International JSRD Study Group CEP290 mutations are frequently identified in the oculo-renal form of Joubert syndrome-related disorders. Am. J. Hum. Genet. 2007;81:104–113. doi: 10.1086/519026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hitti F.L., Siegelbaum S.A. The hippocampal CA2 region is essential for social memory. Nature. 2014;508:88–92. doi: 10.1038/nature13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsien J.Z., Huerta P.T., Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 60.Glahn D.C., Therman S., Manninen M., Huttunen M., Kaprio J., Lönnqvist J., Cannon T.D. Spatial working memory as an endophenotype for schizophrenia. Biol. Psychiatry. 2003;53:624–626. doi: 10.1016/s0006-3223(02)01641-4. [DOI] [PubMed] [Google Scholar]
- 61.Piskulic D., Olver J.S., Norman T.R., Maruff P. Behavioural studies of spatial working memory dysfunction in schizophrenia: a quantitative literature review. Psychiatry Res. 2007;150:111–121. doi: 10.1016/j.psychres.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 62.Marley A., von Zastrow M. A simple cell-based assay reveals that diverse neuropsychiatric risk genes converge on primary cilia. PLoS ONE. 2012;7:e46647. doi: 10.1371/journal.pone.0046647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Acs P., Bauer P.O., Mayer B., Bera T., Macallister R., Mezey E., Pastan I. A novel form of ciliopathy underlies hyperphagia and obesity in Ankrd26 knockout mice. Brain Struct. Funct. 2014 doi: 10.1007/s00429-014-0741-9. Published online March 16, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cukier H.N., Dueker N.D., Slifer S.H., Lee J.M., Whitehead P.L., Lalanne E., Leyva N., Konidari I., Gentry R.C., Hulme W.F. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol. Autism. 2014;5:1. doi: 10.1186/2040-2392-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 66.Janssen S., Ramaswami G., Davis E.E., Hurd T., Airik R., Kasanuki J.M., Van Der Kraak L., Allen S.J., Beales P.L., Katsanis N. Mutation analysis in Bardet-Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Hum. Genet. 2011;129:79–90. doi: 10.1007/s00439-010-0902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otto E.A., Hurd T.W., Airik R., Chaki M., Zhou W., Stoetzel C., Patil S.B., Levy S., Ghosh A.K., Murga-Zamalloa C.A. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shalata A., Ramirez M.C., Desnick R.J., Priedigkeit N., Buettner C., Lindtner C., Mahroum M., Abdul-Ghani M., Dong F., Arar N. Morbid obesity resulting from inactivation of the ciliary protein CEP19 in humans and mice. Am. J. Hum. Genet. 2013;93:1061–1071. doi: 10.1016/j.ajhg.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stratigopoulos G., Martin Carli J.F., O’Day D.R., Wang L., Leduc C.A., Lanzano P., Chung W.K., Rosenbaum M., Egli D., Doherty D.A., Leibel R.L. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 2014;19:767–779. doi: 10.1016/j.cmet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 71.Ezratty E.J., Stokes N., Chai S., Shah A.S., Williams S.E., Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y.P., Tsai I.C., Morleo M., Oh E.C., Leitch C.C., Massa F., Lee B.H., Parker D.S., Finley D., Zaghloul N.A. Ciliopathy proteins regulate paracrine signaling by modulating proteasomal degradation of mediators. J. Clin. Invest. 2014;124:2059–2070. doi: 10.1172/JCI71898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y., Yang Z., Meng M., Zhao Y., Dong N., Yan H., Liu L., Ding M., Peng H.B., Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell. 2009;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 74.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nurmi E.L., Bradford Y., Chen Y., Hall J., Arnone B., Gardiner M.B., Hutcheson H.B., Gilbert J.R., Pericak-Vance M.A., Copeland-Yates S.A. Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics. 2001;77:105–113. doi: 10.1006/geno.2001.6617. [DOI] [PubMed] [Google Scholar]
- 77.Smith S.E., Zhou Y.D., Zhang G., Jin Z., Stoppel D.C., Anderson M.P. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 2011;3:03ra97. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rachel R.A., May-Simera H.L., Veleri S., Gotoh N., Choi B.Y., Murga-Zamalloa C., McIntyre J.C., Marek J., Lopez I., Hackett A.N. Combining Cep290 and Mkks ciliopathy alleles in mice rescues sensory defects and restores ciliogenesis. J. Clin. Invest. 2012;122:1233–1245. doi: 10.1172/JCI60981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baker K., Northam G.B., Chong W.K., Banks T., Beales P., Baldeweg T. Neocortical and hippocampal volume loss in a human ciliopathy: A quantitative MRI study in Bardet-Biedl syndrome. Am. J. Med. Genet. A. 2011;155A:1–8. doi: 10.1002/ajmg.a.33773. [DOI] [PubMed] [Google Scholar]
- 80.Brinckman D.D., Keppler-Noreuil K.M., Blumhorst C., Biesecker L.G., Sapp J.C., Johnston J.J., Wiggs E.A. Cognitive, sensory, and psychosocial characteristics in patients with Bardet-Biedl syndrome. Am. J. Med. Genet. A. 2013;161A:2964–2971. doi: 10.1002/ajmg.a.36245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nik-Zainal S., Strick R., Storer M., Huang N., Rad R., Willatt L., Fitzgerald T., Martin V., Sandford R., Carter N.P. High incidence of recurrent copy number variants in patients with isolated and syndromic Müllerian aplasia. J. Med. Genet. 2011;48:197–204. doi: 10.1136/jmg.2010.082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sampson M.G., Coughlin C.R., 2nd, Kaplan P., Conlin L.K., Meyers K.E., Zackai E.H., Spinner N.B., Copelovitch L. Evidence for a recurrent microdeletion at chromosome 16p11.2 associated with congenital anomalies of the kidney and urinary tract (CAKUT) and Hirschsprung disease. Am. J. Med. Genet. A. 2010;152A:2618–2622. doi: 10.1002/ajmg.a.33628. [DOI] [PubMed] [Google Scholar]
- 83.Sanna-Cherchi S., Kiryluk K., Burgess K.E., Bodria M., Sampson M.G., Hadley D., Nees S.N., Verbitsky M., Perry B.J., Sterken R. Copy-number disorders are a common cause of congenital kidney malformations. Am. J. Hum. Genet. 2012;91:987–997. doi: 10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sandbacka M., Laivuori H., Freitas É., Halttunen M., Jokimaa V., Morin-Papunen L., Rosenberg C., Aittomäki K. TBX6, LHX1 and copy number variations in the complex genetics of Müllerian aplasia. Orphanet J. Rare Dis. 2013;8:125. doi: 10.1186/1750-1172-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oprea G.E., Kröber S., McWhorter M.L., Rossoll W., Müller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ackermann B., Kröber S., Torres-Benito L., Borgmann A., Peters M., Hosseini Barkooie S.M., Tejero R., Jakubik M., Schreml J., Milbradt J. Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum. Mol. Genet. 2013;22:1328–1347. doi: 10.1093/hmg/dds540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.