Abstract
Numerous biological functions of a cell, including polarization, differentiation, division, and migration, rely on its ability to endure mechanical forces generated by the cytoskeleton on the nucleus. Coupling of the cytoskeleton and nucleoskeleton is ultimately mediated by LINC complexes that are formed via a strong interaction between SUN- and KASH-domain-containing proteins in the nuclear envelope. These complexes are mechanosensitive and essential for the transmission of forces between the cytoskeleton and nucleoskeleton, and the progression of cellular mechanotransduction. Herein, using molecular dynamics, we examine the effect of tension on the human SUN2-KASH2 complex and show that it is remarkably stable under physiologically relevant tensile forces and large strains. However, a covalent disulfide bond between two highly conserved cysteine residues of SUN2 and KASH2 is crucial for the stability of this interaction and the transmission of forces through the complex.
Introduction
Mechanical linker of nucleoskeleton and cytoskeleton (LINC) complexes play a central role in cellular mechanotransduction (1) by providing a physical linkage between the interior of the nucleus and the cytoplasm. Tethering of the extracellular matrix (ECM), the cytoskeleton, and the nucleoskeleton mediated by these complexes allows for a direct transmission of forces to the nucleus. Transmission of forces through LINC complexes is essential for several biological functions of the cell, including polarization, differentiation, division, and migration, which are dependent on nuclear deformation and positioning (2–4). Furthermore, LINC complexes are directly connected to focal adhesions through filamentous actin bundles, resulting in ultrafast mechanotransduction (4–6). The distribution of nuclear pore complexes on the surface of the nucleus (7,8) has also been attributed to elements of the LINC complex. Additionally, components of LINC complexes have been implicated in several human diseases, including laminopathies and muscular disorders such as Emery-Dreifuss muscular dystrophy and dilated cardiomyopathy (9–15), and hearing loss (16).
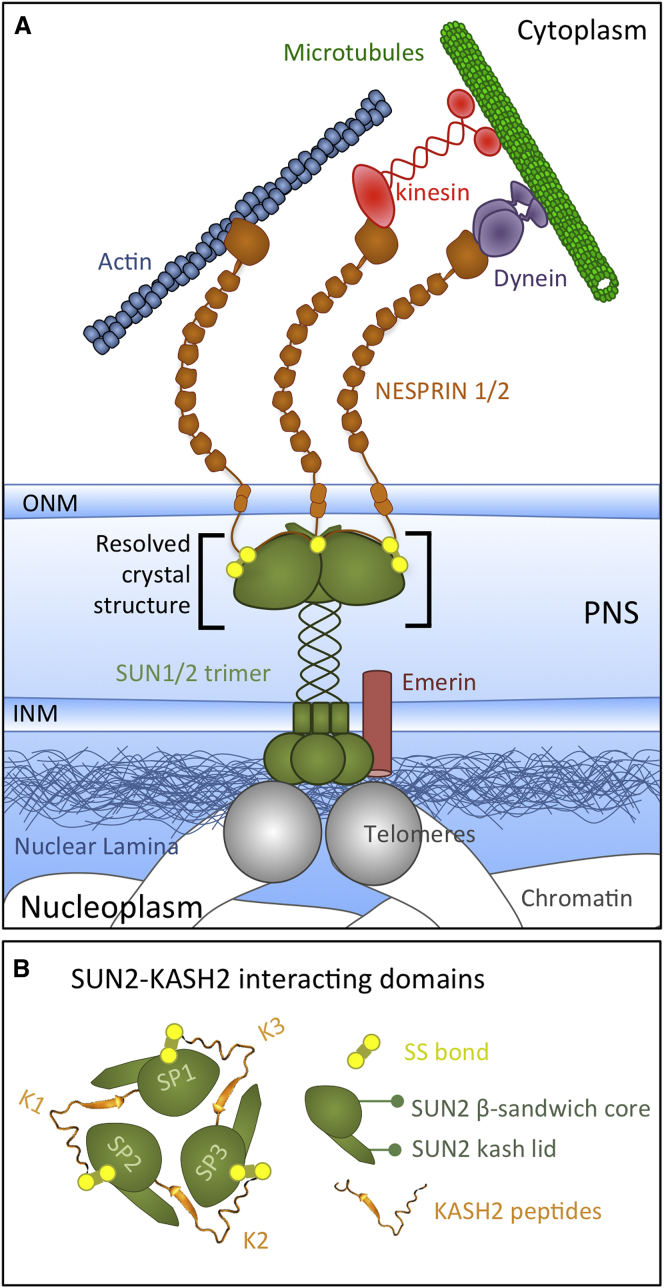
LINC complexes are formed by the interaction of SUN (Sad-1 and Unc) and KASH (Klarsicht, ANC-1, Syne Homology)-domain-containing proteins in the perinuclear space (PNS) (17) (Fig. 1). In mammalian cells, widely expressed SUN-domain-containing proteins include SUN1 and SUN2, which interact with various members of KASH-domain proteins including NESPRIN1–4 (Nuclear Envelope Spectrin repeat proteins 1–4). These complexes are structural and load-bearing elements in the cell and experience both extracellular and intracellular mechanical forces through their linkage to various elements of the cytoskeleton by NESPRIN proteins (Fig. 1 A). Furthermore, they couple the cytoskeleton to the nucleoskeleton through their interaction with nuclear lamina in the nucleoplasm (Fig. 1 A). The increased gap between the inner nuclear membrane (INM) and outer nuclear membrane (ONM) in SUN1 and SUN2 mutated cells (18) suggests that SUN proteins are subject to tensile forces maintaining the gap between the INM and ONM. Moreover, recent studies showed the initiation of mechanotransduction events (e.g., emerin phosphorylation) through the direct application of forces on SUN-KASH complexes in isolated nuclei (19) (Fig. 1).
Figure 1.
(A) Schematic representation of the SUN1/2-KASH1/2 complex, which acts as a bridge between the cytoskeleton and nucleoskeleton. SUN domains of SUN1/2 interact with three KASH peptides of NESPRIN1/2 in the PNS. The nucleoplasmic domains of SUN1/2 interact with nuclear lamina and telomeres. In the cytoplasm, NESPRIN1/2 proteins interact with various cytoskeletal components, including actin filaments, through their actin-binding domains, and microtubules through motor proteins, namely, kinesin and dynein. Cytoskeletal forces are transmitted through SUN-KASH complexes and result in mechanotransduction events such as Emerin phosphorylation. The crystal structure of a segment of the SUN2-KASH2 complex has been resolved. (B) Schematic representation of SUN2-KASH2 interactions. Each KASH peptide (orange) interacts with the KASH lid of one SUN2 protomer and the β-sandwich core of the neighboring SUN2 protomer (green). To see this figure in color, go online.
The crystal structure of human SUN2 in complex with the KASH2 peptide of NESPRIN2 revealed a trimeric SUN2 structure, where three SUN2 protomers interact with three independent ∼22 aa KASH2 peptides and form an overall hexameric structure (20,21) (Figs. 1 B and 2, A and B). Each SUN2 protomer consists of a SUN domain (SUN2540–717) and a minimal helical coiled-coil region (SUN2525–540), which are necessary for SUN2 trimerization and KASH binding (20) (Figs. 1 B and 2 A). The SUN domain consists of a β-sandwich core and an ∼20 aa β-hairpin extending from this domain, forming the KASH lid (SUN2567–587) (Fig. 2 C) (20). Each KASH2 peptide interacts with the KASH lid of one SUN2 protomer (KASH6884–6872) and the β-sandwich core of the neighboring SUN2 protomers (KASH6872–6862) (Fig. 1 B). A unique feature of this complex is the perfect alignment of two highly conserved cysteine residues, CYS563 of SUN2 and CYS6862 of KASH2 (22), resulting in the formation of a disulfide bond (SS bond) between the two proteins and highly stabilizing the SUN2-KASH2 interaction (Figs. 1 B and 2 C). Although this bond has been shown to be dispensable for SUN-KASH binding in vitro (20), the high conservation of both cysteine residues suggests their physiological importance. Herein, we show that under physiologically relevant forces, the stability of the SUN2-KASH2 interaction depends on the intermolecular SS bond between SUN2 and KASH2. Furthermore, the transmission of forces through these complexes is disrupted upon disruption of this bond.
Figure 2.
Crystal structure of the wild-type human SUN2-KASH2 hexamer (PDB ID: 4DXS). (A) Side view of a SUN2 trimer (green) containing a SUN domain and a helical coiled-coil region in complex with three KASH2 peptides (orange). (B) Top view of a SUN2-KASH2 hexamer. (C) The SUN2-KASH2 interactions, including several nonbonded interactions, terminated with an interaction between CYS563 of SUN2 and CYS6862 of KASH2, which form a disulfide bond (SS bond) (yellow). Each KASH peptide (orange) interacts with the KASH lid of one SUN2 protomer and the β-sandwich core of the neighboring SUN2 protomer (green). SMD simulations were carried out by applying 25 pN tensile forces on CYS6862 of each KASH2 peptide in the SUN2-KASH2 hexamer (the site of force application is represented as a pink triangle). To see this figure in color, go online.
Materials and Methods
Modeled system
The crystal structure of the SUN2-KASH2 complex was obtained from the Protein Data Bank (PDB ID: 4DXS (20)) and visualized using VMD software. The structure was trimerized using MakeMultimer.py online software. The system was solvated in water, neutralized with counterions, and subsequently ionized with an ion concentration of 150 mM of KCl and 50 μM of CaCl2. The concentration of Ca2+ in the nuclear envelope is estimated to be similar to that of the endoplasmic reticulum lumen, which is known to be in the micromolar range (23–25).
Steered molecular-dynamics simulations
We conducted steered molecular-dynamics (SMD) simulations using NAMD scalable MD (26) with the CHARMM27 force field. To simulate tensile cytoskeletal forces applied on KASH-domain-containing proteins, we applied a constant 25 pN force directly to the end residue of each KASH2 peptide (CYS6862) in the direction parallel to the central symmetry axis of the SUN2 trimer (perpendicular to the INM and ONM; see Fig. 3). The 25 pN direct tensile forces on groups of SUN-KASH complexes resulted in stiffening of the nuclei (19). To observe changes using SMD simulations on nanosecond timescales, we applied the same 25 pN force to a single SUN2-KASH2 complex. Furthermore, the Cα carbons of GLY522 on the helical coiled coil of the SUN2 trimer were fixed in all simulations (see Fig. 3). This was rationalized by the fact that the SUN2 protein interacts with KASH-domain-containing proteins at its C-terminus, it is tethered to INM through its transmembrane domain, and it interacts with nuclear lamins and chromatin in the nucleus at its N-terminus (18,27,28). We simulated the covalent disulfide bond between CYS6862 and CYS563 using a patch command in NAMD. To study the role of this bond in force transmission and endurance, we mutated CYS563 of SUN2 to alanine (this structure is referred to as C563A in the text). Additionally, to isolate and study the role of the disulfide bond without any potential local changes in the complex due to the alanine substitution, we conducted SMD on a model in which the patch between the two cysteines was not created (i.e., CYS563 and CYS6862 were included, but not covalently bound with an SS bond). We conducted a total of three SMD simulation runs on the wild-type (WT) structure (referred to in the text as WT simulations). Each simulation contained three KASH peptides interacting with three SUN protomers, resulting in a total of nine interacting pairs. Similarly, four simulation runs were performed for both the mutated structure (C563A) and the structure with a disrupted SS bond (denoted as SS−), resulting in 12 SUN-KASH interacting pairs in each case. An independent equilibration run was performed for each simulation run. The system was minimized at 5000 steps and equilibrated for ∼2 ns with a time step of 2 fs. The temperature and pressure of the system were held constant at 1 atm and 310 K using Langevin’s piston and Hoover’s method during minimization and equilibration (26). A time step of 2 fs was used and the cutoff distance for nonbonded interactions was 1.2 nm. Periodic boundary conditions were applied in all three directions. SUN2 mutations were modeled using VMD.
Figure 3.
Extension of the SUN2-KASH2 complex (green, WT; blue, C563A) to 100% of their initial lengths under 25 pN forces. (A) CYS6862 of each KASH2 peptide (orange) was pulled with a constant force of 25 pN in both cases using SMD, resulting in an ∼100% increase in the length of both structures. Distinct conformational changes were observed in the WT and C563A structures. In the WT complex (right, green), forces on the KASH2 peptide are translated directly to the helical coiled-coil region of SUN2, resulting in conformational changes in this region, whereas the SUN domain remains intact. Conversely, the C563A mutated structure experiences minimal conformational changes in its coiled-coil region. Pink spheres and triangles represent constraints at the C-terminal ends and the sites of force application, respectively. (B) RMSF of all SUN2 protomer residues (SUN2525–717) averaged over all simulation runs (with three protomers in each simulation run) for 100% elongation of the complex. The RMSF is highest in the helical CC (SUN2525–540) region of the WT SUN2, but peaks in the KASH lid (SUN2567–587) region of the C563A structure. (C) Comparison of the dynamic cross correlations of all SUN2 protomer residues averaged over all simulation runs (with three protomers in each simulation run), showing that (i) the negative correlation between the helical CC (SUN2525–540) and KASH lid (SUN2567–587) is reduced with the C563A mutation, (ii) the dynamic cross correlation of the KASH lid with other SUN2 residues is reduced and changes from a positive correlation to a negative correlation as a result of the C563A mutation, and (iii) the negative dynamic cross correlation of the helical CC region with other SUN2 residues in the C563A structure is significantly reduced. To see this figure in color, go online.
Root mean-square fluctuation and dynamic cross-correlation calculations
The Bio3D R package was used for root mean-square fluctuation (RMSF) and dynamic cross-correlation analyses (29). In both cases, the calculations were averaged over the three SUN2 protomers that formed the SUN2 trimer in each simulation run, after 100% elongation of the complexes. In total, nine data sets were averaged in the WT structure, and 12 sets were averaged in the C563A and SS− simulations.
Interaction energy calculations
The interaction energies were calculated between each SUN protomer and KASH peptide independently. Each KASH peptide interacted distinctly with only two neighboring SUN protomers and these interaction energies were reported separately. Only the SUN domain of each SUN2 protomer (residues 545–618) was used in the energy calculations, as the helical coiled coil did not interact with the KASH peptide.
Sequence alignment
Sequences of human and mouse SUN and NESPRIN proteins were aligned using UniProt (30).
Results
The disulfide bond between CYS563 of SUN2 and CYS6862 of KASH2 is required for the transmission of forces through the SUN-KASH complex
Conformational changes induced by 25 pN tensile forces on the end residue (CYS6862) of each KASH peptide in the WT and C563A complex are shown in Fig. 3 A. Distinct deformation behaviors were observed in the two complexes. The single C563A mutation constrained the deformation of the SUN-KASH complex to the SUN domain, and forces on the KASH peptide were no longer able to transmit to the helical coiled-coil region, as is evident from the elongated structures in Fig. 3 A. The deformation mechanisms of the WT and C563A complexes are compared in Fig. 3, where the complex is extended up to 100% of its initial length in both cases. The average RMSFs of all SUN2 residues (SUN2525–718) are shown in Fig. 3 B. In the WT structure, the helical coiled-coil region of SUN2 comprising residues SUN2525-540 experiences the highest fluctuations. On the contrary, in the C563A structure, the fluctuations peak in the KASH lid region (SUN2567–587), and minimal fluctuations are observed in the helical coiled-coil region.
To determine how the C563A mutation affects the overall dynamics of SUN2 structure, we evaluated residue-to-residue dynamic cross-correlations for the three SUN2 protomers that formed each SUN2 trimer, and averaged them over all simulation runs (Fig. 3 C). A comparison of the dynamic cross-correlation plots for the WT and C563A structures clearly shows that the overall correlations of SUN2 residues are significantly reduced for C563A. Furthermore, the correlation between the KASH lids and the SUN2 helical coiled-coil region are notably reduced in the C563A structure (Fig. 3 C, i). Note that the main SUN-KASH interactions occur in this region, namely, the KASH lid of SUN2. Another significant difference in the plots is the change in the cross-correlation values of residues in the KASH lid region from a positive correlation with all other SUN2 residues in WT SUN2 to a negative correlation in the C563A structure (Fig. 3 C, ii). Negative cross correlations correspond to movement of the residues in opposite directions (29). Finally, the negative cross correlations between the helical coiled-coil region and all other SUN2 residues were remarkably lower in the C563A structure, suggesting a decoupling between the coiled-coil region and the SUN domain.
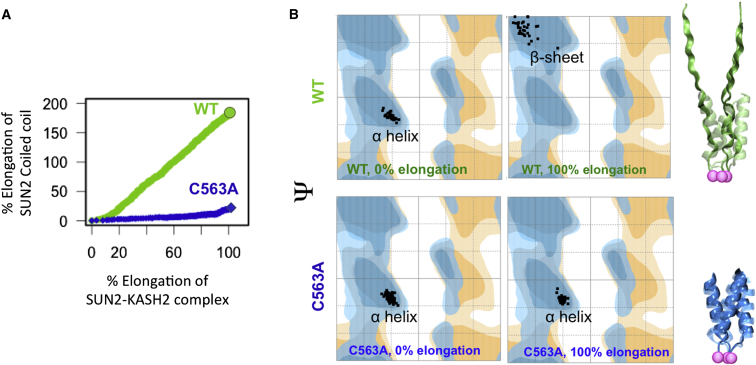
To further expand on this decoupling, we compared the conformational changes in the helical coiled-coil region between the WT and C563A structures. In the WT SUN2-KASH2 complex, the percent elongation of the helical coiled-coil region was linearly correlated with the percent elongation of the entire SUN2-KASH2 complex (Fig. 4), where ∼100% elongation resulted in ∼176% elongation of the coiled-coil region. On the other hand, the SUN domain remained entirely intact throughout the SMD simulation. However, with the C563A mutation, the transmission of forces to the helical coiled-coil region was interrupted and the coiled-coil region remained unstretched upon 100% extension of the complex (Fig. 4 B). Furthermore, deformation was confined to the SUN domain, where the most prominent changes were seen in the KASH lids (Fig. 3, A and B). This is further exemplified by the changes in the backbone rotational angles of residues in the coiled-coil region of WT SUN2 (SUN2525–540), as shown in the Ramachandran plots in Fig. 4 B (31). The ψ-ϕ angles of residues in the helical coiled-coil region significantly changed after 100% elongation of the molecule (Fig. 4 B). In contrast, this conformational change was not observed in the C563A structure.
Figure 4.
The SS bond between CYS563 of SUN2 and CYS6862 of KASH2 is required for transmission of forces through the SUN2-KASH2 complex. (A) Elongation of the minimal helical coiled-coil region of SUN2 in response to tensile forces on the KASH2 peptide in the WT (green) and mutated (C563A, blue) structures as a function of the % elongation in the entire SUN2-KASH2 complex. With the C563A mutation, no forces are transmitted to the helical coiled-coil region of SUN2, resulting in no elongation in this region, whereas the coiled-coil region of SUN2 is elongated to >150% of its initial length in the presence of the SS bond. (B) Ramachandran plot of residues in the helical coiled-coil region before (left) and after (right) elongation of the SUN2-KASH2 complex. Changes in the Ψ versus Φ angles for these residues indicate conformational changes in the helical coiled-coil region of WT SUN2 upon the application of tensile forces to the KASH2 peptide. No such changes are seen in the mutated structure (C563A, bottom). To see this figure in color, go online.
We next explored whether the changes seen in the C563A mutated structure were mainly due to the inhibition of SS bond formation between C563 and C6862. We conducted similar SMD simulations on the WT structure, but with the SS bond between C563A and C6862 inhibited (SS− structure). We found that the simulations on the SS− structures yielded results similar to those obtained with the C563A mutation (Figs. S1 and S2 in the Supporting Material), suggesting that the main contribution of this mutation is disulfide inhibition. Force transmission to the helical coiled-coil region was inhibited and changes were mainly limited to the KASH lids in the absence of the SS bond, as is evident from the deformed crystal structures (Fig. S1 B) as well as the RMSF values in the SS− versus WT structures (Fig. S1 B).
The stability of the SUN2-KASH2 interaction is highly maintained under tensile forces
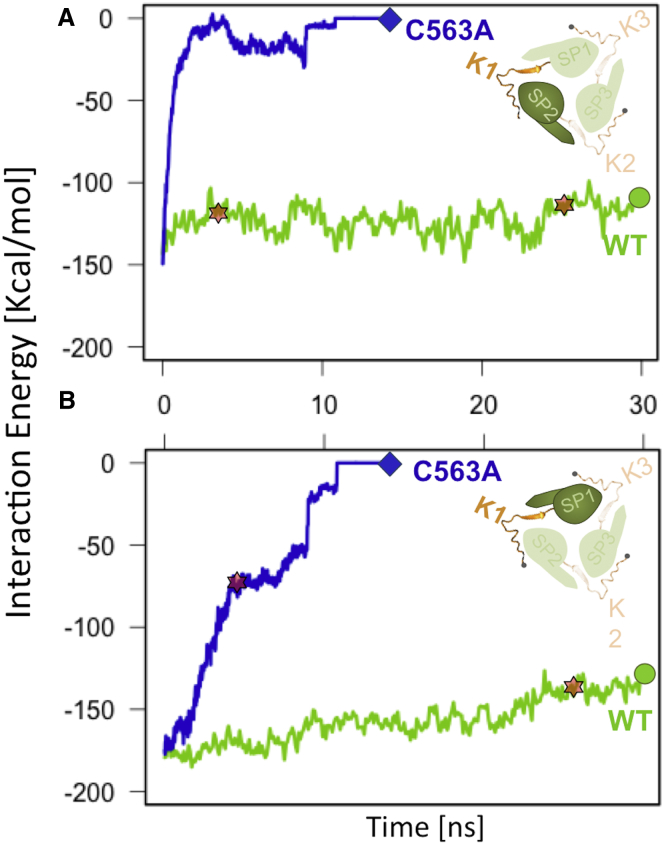
Next, we examined the effect of tensile forces on other nonbonded interactions of SUN2 and KASH2 in the WT, C563A, and SS− structures. As noted above, each KASH2 peptide interacts with the KASH lid of one SUN2 protomer and the β-sandwich core of the neighboring protomer (Figs. 1 B and 2 C). The average total nonbonded (electrostatic and van der Waals) interaction energies between a KASH peptide and each of the two WT SUN protomers with which it interacts were evaluated during the application of 25 pN tensile forces to the terminal residue of each KASH peptide. The results showed minimal fluctuations in the total nonbonded energies, indicating that the stability of the interactions was preserved even with an elongation of the complex to 100% of its original length (Fig. 5, A and B). The KASH peptide maintained a strong ∼−145 kcal/mol average nonbonded interaction with the SUN2 KASH lid, as shown in Fig. 5 B. The KASH peptide also interacted with the SUN β-sandwich core at −122 kcal/mol (Fig. 5 A). The WT SUN-KASH complex elongated to 100% of its original length after 28.6 ns of SMD simulations; the elongated structure is shown in Fig. 2 A. The head domain of SUN2 remained entirely intact throughout this force-induced elongation of the complex to 100% of its original length, maintaining a stable SUN2-KASH2 interaction.
Figure 5.
The disulfide bond between CYS563 of SUN2 and CYS6862 of KASH2 is required for the stability of the SUN2-KASH2 complex under tensile forces. Average nonbonded interaction energies (electrostatic and van der Waals) are shown for KASH peptides with the two neighboring SUN protomers it interacts with (inset) in the mutated (C563A, blue) and WT (green) structures. (A and B) KASH peptides interact with the KASH lid (A) and β-sandwich core (B) of two neighboring SUN protomers. The point at which the SUN2-KASH2 complex experiences a 100% elongation is also shown for the WT and C563A structures (stars). The absence of the SS bond results in a 100% elongation of the complex in less than ∼3.2 ns of pulling, and a complete dissociated of the KASH2 peptide with the KASH lid (A) and β-sandwich core (B) in the first 10 ns. To see this figure in color, go online.
The disulfide bond between CYS563 of SUN2 and CYS6862 of KASH2 is required for the stability of the SUN2-KASH2 interaction under tensile forces
The C563A mutation and the disruption of the SS bond between two interacting cysteine residues of SUN2 and KASH2 in MD simulations both resulted in a full dissociation of the KASH2 peptide from the SUN2 trimer (Figs. 5 and S3). The average total nonbonded interaction energies between the KASH peptides and the β-sandwich core of the SUN trimers were reduced abruptly after the application of 25 pN tensile forces (Figs. 5 and S3). The KASH peptides initially interacted strongly with the β-sandwich core at ∼−177 kCal/mol (Fig. 5 B); however, this interaction showed high instability under forces in the C563A (Fig. 5 A) and SS− structures (Fig. S3 A). A full dissociation of KASH with the β-sandwich core was observed after ∼3 ns, with a 0 kcal/mol interaction energy after this detachment (Fig. 5 A). The KASH peptide remained in contact with the KASH lid a few nanoseconds longer (Fig. 4 B), with an initial ∼−171 kCal/mol interaction, a linear decrease in interaction energy, and a full dissociation after ∼10 ns of SMD simulations.
Discussion
LINC complexes are anchored to several elements of the cytoskeleton through their KASH-domain-containing proteins. For example, NESPRIN1 and NESPRIN2 contain actin-binding domains that directly bind to the actin cytoskeleton (32) and have also been observed to interact and colocalize with dynein and kinesin complexes (33), connecting the nucleoplasm to actin filaments as well as microtubules (Fig. 1 A). Through focal adhesion proteins, the actin cytoskeleton is also linked to the ECM (17,34). SUN1 and SUN2 proteins interact with type A and B lamins at their nucleoplasmic N-terminal domain (13,18,27), and several members of the NESPRIN protein family at their C-terminus in the PNS. In this way, the nucleoskeleton is coupled with the ECM and extracellular forces can be translated to the nucleus directly through the SUN-KASH complexes. Furthermore, all intracellular actomyosin generates contractile forces, and forces generated by other motor proteins (i.e., dynein and kinesin) are also translated to the nucleus through these complexes, underscoring their potential role in mechanotransduction through the nuclear envelope.
Our results show that despite a strong ∼−801 Kcal/mol average nonbonded interaction between SUN2 and three KASH2 peptides, the covalent disulfide bond between CYS563 of SUN2 and CYS6862 of KASH2 is necessary for the stability of this interaction under tensile forces. The SS bond is not required for the SUN2-KASH2 interaction (20); however, our results suggest that after the initial anchorage of KASH peptides onto SUN proteins through nonbonded interactions (Fig. 6 A), the SS bond forms (Fig. 6 B) to allow the initiation of force transmission in the subsequent linkage of NESPRIN proteins to the cytoskeleton (Fig. 6 C). Furthermore, upon potential clustering of LINC complexes, forces of magnitudes up to hundreds of piconewtons can conceivably be endured by these complexes and utilized for nuclear positioning and chromosome organization (Fig. 6 C).
Figure 6.
Transmission of forces through LINC complexes. (A) A SUN trimer interacts with three KASH peptides from NESPRIN proteins, forming several nonbonded interactions. (B) The SUN-KASH complex is subject to cytoskeletal forces through the direct interactions of NESPRIN proteins with the actin cytoskeleton, and with microtubules through kinesin and dynein. Interprotein SS bonds further stabilize the SUN-KASH interaction under mechanical forces and allow the transmission of cytoskeletal forces to the helical coiled-coil region of SUN proteins. (C) Potential clustering of SUN-KASH complexes into higher-order complexes will allow for the transmission of forces on the order of hundreds of piconewtons, as required for nuclear positioning or the organization of chromatin. Besides intracellular motor protein-dependent forces, extracellular forces can be transmitted directly to the nucleus through integrins and focal adhesion proteins that also interact with the actin cytoskeleton, such as talin and vinculin. To see this figure in color, go online.
This stable covalent bond between SUN2 and KASH2 suitably positions the KASH peptide to allow for all other interacting residues to remain aligned during force application to the KASH peptide and continue their nonbonded interactions. As a result, the SUN domain and KASH peptide complex show minimal fluctuations and conformational changes, and forces are directly translated to the coiled-coil region, resulting in conformational changes in this region (Fig. 3). The full coiled-coil region of a full-length SUN1/2 protein is predicted to be ∼40 nm in length (22) and serves as a possible structural component in translating forces between the SUN domain and the nucleoplasm domain of SUN. Coiled coils are also found in several other structural and motor proteins, and are known to be load-bearing structural motifs involved in force transmission between various protein domains (35). Furthermore, coiled coils have highly elastic properties (36); for example, coiled-coil regions in Myosin II have been shown to be truly elastic structures and to refold against forces up to 30 pN on short timescales (37). These properties make SUN1 and SUN2 proteins very suitable elastic load-bearing components under the constant application and release of cytoskeletal forces on the nuclear envelope.
Among human and mouse SUN proteins, SUN1 and SUN2 contain the cysteine residue that can form disulfide bonds with four members of the NESPRIN family (NESPRIN1–4) (Fig. 7). However, testis-specific SUN3–5 proteins, which are small spermiogenesis-specific proteins (38–40) and are predicted to have shorter coiled-coil regions than SUN1/2, lack this cysteine residue. Studies have shown that shorter coiled coils unfold under lower forces (41), which may suggest that LINC complexes with shorter coiled coils are not suitable for bearing and transmitting large forces, thus explaining the absence of the disulfide bond in these complexes. Our results suggest that the nature and strength of the SUN-KASH interaction may determine two distinct roles for LINC complexes: load bearing and transmission versus mere anchorage of the nucleus.
Figure 7.
Sequence of human and mouse SUN- and KASH-domain-containing proteins. SUN1 and SUN2 contain a cysteine residue that can form an SS bond with the cysteine of KASH1–4, and assist in load bearing and force transmission to the nucleus. Testis-specific SUN3–5 lack the cysteine, suggesting a potentially distinct functional role for these SUN proteins. To see this figure in color, go online.
Author Contributions
Z.J., H.S., and M.R.K.M. conceived and designed the experiments, analyzed the data, and wrote the manuscript. Z.J. and H.S. performed the experiments. M.R.K.M. contributed reagents, materials, and analysis tools.
Acknowledgments
We thank Mehrdad Mehrbod for technical assistance and fruitful discussions.
This work was supported by grants from the National Science Foundation (CAREER award CBET-0955291 to M.R.K.M.) and the Natural Sciences and Engineering Research Council of Canada (to Z.J.). This research used resources of the National Energy Research Scientific Computing Center, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Editor: Andrew McCulloch.
Footnotes
Three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00668-2.
Supporting Material
References
- 1.Kaminski A., Fedorchak G.R., Lammerding J. The cellular mastermind(?)—mechanotransduction and the nucleus. Prog. Mol. Biol. Transl. Sci. 2014;126:157–203. doi: 10.1016/B978-0-12-394624-9.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lombardi M.L., Jaalouk D.E., Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundersen G.G., Worman H.J. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khatau S.B., Hale C.M., Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambliss A.B., Khatau S.B., Wirtz D. The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci. Rep. 2013;3:1087. doi: 10.1038/srep01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D.-H., Khatau S.B., Wirtz D. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci. Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q., Pante N., Roux K.J. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamali T., Jamali Y., Mofrad M.R.K. Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int. Rev. Cell Mol. Biol. 2011;287:233–286. doi: 10.1016/B978-0-12-386043-9.00006-2. [DOI] [PubMed] [Google Scholar]
- 9.Méjat A., Misteli T. LINC complexes in health and disease. Nucleus. 2010;1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folker E.S., Baylies M.K. Nuclear positioning in muscle development and disease. Front. Physiol. 2013;4:363. doi: 10.3389/fphys.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meinke P., Nguyen T.D., Wehnert M.S. The LINC complex and human disease. Biochem. Soc. Trans. 2011;39:1693–1697. doi: 10.1042/BST20110658. [DOI] [PubMed] [Google Scholar]
- 12.Meinke P., Mattioli E., Shackleton S. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014;10:e1004605. doi: 10.1371/journal.pgen.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque F., Mazzeo D., Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isermann P., Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puckelwartz M.J., Kessler E.J., McNally E.M. Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell. Cardiol. 2010;48:600–608. doi: 10.1016/j.yjmcc.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn H.F., Brownstein Z., Avraham K.B. The LINC complex is essential for hearing. J. Clin. Invest. 2013;123:740–750. doi: 10.1172/JCI66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahed Z., Shams H., Mofrad M.R.K. Mechanotransduction pathways linking the extracellular matrix to the nucleus. Int. Rev. Cell Mol. Biol. 2014;310:171–220. doi: 10.1016/B978-0-12-800180-6.00005-0. [DOI] [PubMed] [Google Scholar]
- 18.Crisp M., Liu Q., Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilluy C., Osborne L.D., Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sosa B.A., Rothballer A., Schwartz T.U. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Shi Z., Zhou Z. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 2012;22:1440–1452. doi: 10.1038/cr.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sosa B.A., Kutay U., Schwartz T.U. Structural insights into LINC complexes. Curr. Opin. Struct. Biol. 2013;23:285–291. doi: 10.1016/j.sbi.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen O.H., Gerasimenko O.V., Tepkin A.V. The calcium store in the nuclear envelope. Cell Calcium. 1998;23:87–90. doi: 10.1016/s0143-4160(98)90106-3. [DOI] [PubMed] [Google Scholar]
- 24.Grygorczyk C., Grygorczyk R. A Ca2+- and voltage-dependent cation channel in the nuclear envelope of red beet. Biochim. Biophys. Acta. 1998;1375:117–130. doi: 10.1016/s0005-2736(98)00142-4. [DOI] [PubMed] [Google Scholar]
- 25.Matzke A.J.M., Weiger T.M., Matzke M. Ion channels at the nucleus: electrophysiology meets the genome. Mol. Plant. 2010;3:642–652. doi: 10.1093/mp/ssq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haque F., Lloyd D.J., Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart-Hutchinson P.J., Hale C.M., Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant B.J., Rodrigues A.P.C., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 30.Magrane M., Consortium UniProt. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovell S.C., Davis I.W., Richardson D.C. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q., Ragnauth C., Roberts R.G. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- 33.Zhang X., Lei K., Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mofrad M.R.K., Kamm R.D. Cambridge University Press; Cambridge/New York: 2014. Cellular mechanotransduction diverse perspectives from molecules to tissues. [Google Scholar]
- 35.Kreuzer S.M., Elber R. Coiled-coil response to mechanical force: global stability and local cracking. Biophys. J. 2013;105:951–961. doi: 10.1016/j.bpj.2013.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolgemuth C.W., Sun S.X. Elasticity of α-helical coiled coils. Phys. Rev. Lett. 2006;97:248101. doi: 10.1103/PhysRevLett.97.248101. [DOI] [PubMed] [Google Scholar]
- 37.Schwaiger I., Sattler C., Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat. Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 38.Frohnert C., Schweizer S., Hoyer-Fender S. SPAG4L/SPAG4L-2 are testis-specific SUN domain proteins restricted to the apical nuclear envelope of round spermatids facing the acrosome. Mol. Hum. Reprod. 2011;17:207–218. doi: 10.1093/molehr/gaq099. [DOI] [PubMed] [Google Scholar]
- 39.Shao X., Tarnasky H.A., van der Hoorn F.A. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 1999;211:109–123. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- 40.Göb E., Schmitt J., Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS One. 2010;5:e12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadeghi S., Emberly E. Length-dependent force characteristics of coiled coils. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:061909. doi: 10.1103/PhysRevE.80.061909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.