Abstract
The constitutive androstane receptor (CAR) is initially defined as a xenobiotic nuclear receptor that protects the liver from injury. Detoxification of damaging chemicals is achieved by CAR-mediated induction of drug-metabolizing enzymes and transporters. More recent research has implicated CAR in energy metabolism, suggesting a therapeutic potential for CAR in metabolic diseases, such as type 2 diabetes and obesity. A better understanding of the mechanisms by which CAR regulates energy metabolism will allow us to take advantage of its effectiveness while avoiding its side effects. This review summarizes the current progress on the regulation of CAR nuclear translocation, upstream modulators of CAR activity, and the crosstalk between CAR and other transcriptional factors, with the aim of elucidating how CAR regulates glucose and lipid metabolism.
Keywords: nuclear receptor, constitutive androstane receptor, xenobiotic metabolism, metabolic diseases, energy metabolism
Introduction
CAR (NR3I1) belongs to the nuclear receptor superfamily that exhibits enriched expression in the small intestine, liver, and gallbladder1. CAR was initially defined as a xenobiotic nuclear receptor that mediates the hepatic detoxification of foreign chemicals and endogenous bile acids2. Together with other xenobiotic nuclear receptors, such as the pregnane X receptor (PXR), CAR senses xenobiotic stimuli and elicits their clearance by transactivating cytochrome P450 (CYP) enzymes and phase 2 conjugating enzymes, as well as drug transporters (eg, CYP2Bs, MRPs, and UGTs)3,4. On the promoters of these target genes, CAR directly binds to the proximal xenobiotic responsive element (XRE) and/or the distal phenobarbital responsive enhancer module (PBREM), where it heterodimerizes with the retinoid X receptor (RXR)5,6. Up-regulation of these target genes leads to the elimination of toxic chemicals and drug metabolites and prevents hepatotoxicity. Meanwhile, CAR-induced drug-metabolizing enzymes and transporters are also responsible for the removal of hydrophobic bile acids from the liver. Detoxification of bile acids to less damaging metabolites and the export of bile acids protect hepatocytes from cholestasis7,8,9,10.
Microarray analysis has suggested that CAR not only induces drug-metabolizing genes but also influences the expression of genes involved in glucose/lipid metabolism as well as those responsible for hepatocyte proliferation11,12,13. Follow-up pharmacological and biochemical studies have expanded our understanding of the pathophysiological function of CAR beyond xenobiotic metabolism. Activation of CAR improves glucose and lipid metabolism in metabolic diseases, which highlights it as a promising therapeutic target for metabolic syndromes14. However, obstacles exist for the clinical use of CAR activators due to the concerns of drug-drug interaction, hepatomegaly and liver carcinogenesis15. A better understanding of the mechanism by which CAR modulates energy metabolism will help in overcoming these obstacles. This review is dedicated to the molecular basis of the regulation of CAR activity and CAR's crosstalk with other proteins. We will also discuss how we may be able to better harness the metabolic benefits of CAR by better understanding its mechanistic properties.
Nuclear translocation of CAR
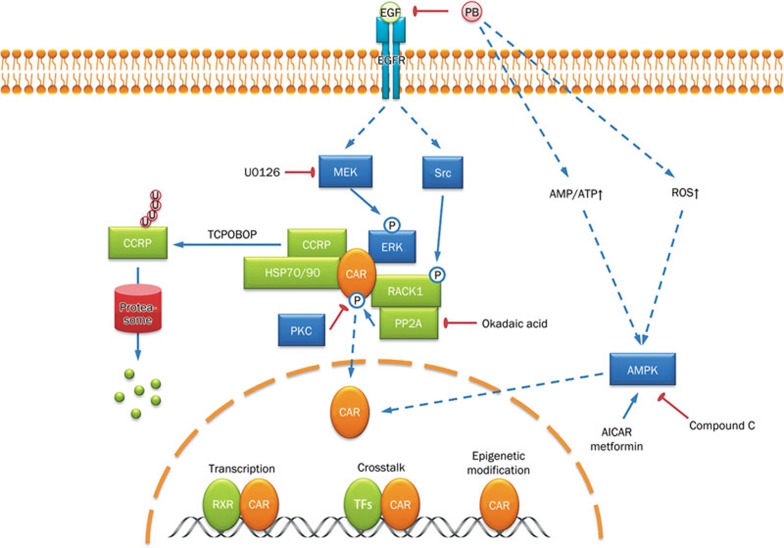
The response of CAR to xenobiotic exposure is very fast. For example, Cyp2b10 gene expression peaks within one hour upon acute treatment with 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene,3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP) in mice12. Such a rapid response is achieved primarily by massive nuclear translocation of CAR from the cytoplasm16. Therefore, substantial efforts have been devoted to delineate the mechanism of the cytoplasmic retention and nuclear translocation of CAR (Figure 1). In the past decade, Masa Negishi's group and others have identified several key proteins that participate in the phenobarbital-induced nuclear translocation of CAR. Phosphorylation and de-phosphorylation play an important role in regulating the nuclear translocation of CAR. Human CAR residue threonine 38 (corresponding to mouse CAR residue threonine 48) is a conserved protein kinase C (PKC) target site. A mutation study showed that T38A, which is unable to be phosphorylated, mimics the effect of phenobarbital treatment. In contrast, T38D, which resembles phosphorylated CAR, is largely retained in the cytoplasm17. These results suggested that phosphorylation at threonine 38 is a repressive signal for the nuclear translocation of CAR. However, further studies are needed to elucidate the upstream pathways of PKC. Hepatic growth factors negatively regulate the phenobarbital-mediated induction of CYP2B through activation of signaling cascades, including mitogen-activated protein kinase kinase (MEK) and extracellular signal-regulated kinase (ERK). Moreover, active ERK sequesters phosphorylated CAR in the cytoplasm18. Alternatively, okadaic acid, a protein phosphatase 2A (PP2A) inhibitor, diminishes the phenobarbital-mediated induction of CYP2B expression in primary rat hepatocytes, suggesting that PP2A is the phosphatase for CAR and is involved in the activation of CAR19. In a more recent study, the PP2A core enzyme was found to be recruited to phosphorylated CAR by the receptor for activated C kinase 1 (RACK1) and to de-phosphorylate CAR20. RACK1 seems to preferentially bind to phosphorylated threonine 38. However, de-phosphorylation of CAR is not limited to threonine 38, considering that the de-phosphorylation of serine 202 is also required for CAR nuclear translocation21. RACK1 is negatively regulated by Src kinase, which functions downstream of the epidermal growth factor receptor (EGFR) pathway. Phosphorylation of RACK1 by Src prevents its interaction with CAR. Phenobarbital, demonstrated as a potential antagonist of EGFR, competes with EGF binding to EGFR, therefore abrogating the blockade of RACK1 by activating the EGFR cascade20. Taken together, the two signaling branches downstream of EGFR, MEK-ERK, and Src-RACK1, integrate at cytoplasmic CAR to inhibit its nuclear translocation. Note that one of the protein phosphatase 1 catalytic subunits, PPP1R16A, interacts with CAR at the cell membrane and prevents nuclear translocation of CAR, which is dependent on phosphatase activity22. Moreover, the mitogen-activated protein kinase (MAPK) p38 also regulates human CAR activity on the CYP2B6 gene23. This additional evidence adds further complexity to the phosphorylation-dependent nuclear translocation of CAR.
Figure 1.
Regulation of CAR nuclear translocation and activity. CAR cytoplasmic retention is regulated by upstream phosphorylation cascades and cytoplasmic chaperones. Phenobarbital antagonizes EGFR and de-suppresses CAR, and TCPOBOP triggers ubiquitination and proteasomal degradation of CCRP. Nuclear CAR is also regulated by AMPK and ROS and elicits its effect through transcriptional activity, crosstalk with other transcriptional factors, and epigenetic modifications.
In addition to phosphorylation and de-phosphorylation, several other proteins are also involved in the cytoplasmic retention of CAR. Heat shock protein 90 (HSP90), a chaperone protein that serves as a cellular stress sensor, is associated with the glucocorticoid receptor (GR) and aryl hydrocarbon receptor (AhR). Together with immunophilin or immunophilin-like protein, HSP90 governs the nuclear translocation of GR and AhR in a ligand-dependent manner. Similarly, CAR is also associated with HSP9024. Cytoplasmic CAR retention protein (CCRP) forms a ternary complex with HSP90 and CAR and retains CAR in the cytoplasm25. Intriguingly, several facts strongly imply that cytoplasmic retention by the CCRP-HSP90 complex is an adaptive mechanism that leads to the accumulation of CAR in the cytoplasm to quickly respond to a second exposure of stress. First, the HSP90-CAR complex recruits PP2A in the presence of phenobarbital, indicating that the ternary complex is necessary for CAR de-phosphorylation, which is a critical event for CAR nuclear translocation24. Second, the CAR-HSP90-CCRP ternary complex is associated with the cellular skeleton, and inhibition of HSP90 or disruption of the microtubule network disturbs the nuclear translocation of CAR, further demonstrating that the complex is essential for the nuclear translocation of CAR26. Third, the accumulation of CAR in the cytoplasm is not derived from nuclear exclusion because CCRP does not affect the nuclear content of CAR25. A recent study has explained how the components of the ternary complex are coordinated27. Upon cellular stresses (eg, thermal stress), CAR is enriched in the CAR-HSP70/90-CCRP ternary complex. Upon treatment of TCPOBOP, CCRP undergoes ubiquitination and proteasomal degradation, which facilitates the nuclear translocation of CAR.
Although the molecular mechanism for the nucleo-cytoplasmic translocation of CAR has been established based on the studies discussed above, questions still remain for the regulation of CAR nuclear translocation. AMP-activated protein kinase (AMPK) has been long known to mediate phenobarbital-induced CAR activation28,29,30. However, the results from the mechanistic study of the AMPK-dependent activation of CAR leave much to be desired. Phenobarbital seems to increase the AMP/ATP ratio and promote liver kinase B1 (LKB1)-mediated AMPK activation31. Another pathway involving the microRNA miR-122 is also associated with AMPK activation32. Because of the lack of evidence on AMPK downstream targets, it remains unclear how AMPK activation influences CAR nuclear translocation. Pharmacological activation of AMPK seems to down-regulate HSP70 and EGFR and activate PP2A, suggesting that AMPK may trigger CAR nuclear translocation through the regulation of suppressive proteins33,34,35. In addition, how some of the coactivators drive ligand-independent nuclear translocation of CAR in vivo is still unclear. Steroid receptor coactivator 2 (SRC2/GRIP1) induces the phenobarbital-independent nuclear translocation of CAR36. Peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP/MED1), one of the components of the mediator complex, is also required for the nuclear translocation of CAR in vivo37. Whether those coactivators trigger CAR nuclear translocation by disruption of the retention complex or independently through other mechanisms remains to be understood.
Transcriptional regulation of CAR
Whereas immediate early responses leading to the nuclear translocation and activation of CAR quickly defend the intracellular environment against external xenobiotic insults, long-term protection through CAR can be augmented by transcriptional up-regulation of the CAR gene itself. Many chemicals from drugs and herbal medicines as well as environmental exposure have been shown to increase the expression of CAR38. Under those circumstances, AhR is likely an upstream transcriptional factor that positively regulates CAR39. Glucocorticoids (eg, dexamethasone) efficiently induce CAR expression in human primary hepatocytes at a nanomolar dose40. A distal glucocorticoid response element (GRE) was found on the CAR gene promoter, suggesting that CAR is a direct target gene of GR. Thyroid hormones, through activation of the thyroid receptor (TR), are able to induce CAR expression at physiological concentrations41. Alternatively, activated CAR affects serum thyroid hormone concentrations and influences thyroid-follicular cell proliferation42. All-trans retinoic acid, a metabolite of vitamin A, was also found to up-regulate CAR mRNA through the retinoic acid receptor (RAR)43. Endogenous hormones and endobiotic metabolites appear to be very important for the maintenance of basal CAR expression.
Several studies have also shown that CAR expression is highly inducible during the feeding-fasting switch. Fasting-dependent induction of CAR is mediated primarily by hepatocyte nuclear factor 4α (HNF4α) and PPARα44,45. Whereas fasted WT mice exhibit a higher CAR mRNA level compared with fed mice, this fasting response is almost completely attenuated in either HNF4α or PPARα knockout mice. HNF4α and PPARα responsive elements were found on the promoter of CAR, indicating that HNF4α and PPARα directly bind to the promoter of CAR and induce CAR expression. Moreover, PPARγ coactivator 1α (PGC1α) coactivates HNF4α and PPARα on the CAR promoter and coactivates CAR on the Cyp2b10 promoter and possibly others, which may contribute to the amplification of CAR downstream genes45. In addition, the expression of CAR and some of its downstream target genes fluctuates in accordance with the diurnal rhythm, indicating that the circadian rhythm also has an impact on the expression of CAR46,47. RAR-related orphan receptors (RORs) and Rev-erbs are master regulators for the circadian rhythm48. Whether the circadian expression of CAR is mediated by RORs and/or Rev-erbs would be interesting to determine. Last but not least, miR-137 negatively regulates CAR mRNA, suggesting that microRNA-mediated gene silencing is another mechanism governing CAR expression49.
Modulators of CAR activity
As a typical nuclear receptor, CAR is composed of the N-terminal AF1 ligand-independent domain, DNA-binding domain (DBD), and ligand-binding domain (LBD). Crystal structure analysis revealed that the LBD amino acids on CAR form a pocket that is responsible for the high binding affinity for the Leu-X-X-Leu-Leu (LXXLL) motif, a conserved motif found in many nuclear receptor coactivators50. A number of coactivators have been found to physically interact with CAR51. The three members of p160 coactivators, SRC1, SRC2, and SRC3, as well as PGC1α have been reported to enhance the activity of CAR based on reporter gene assays52,53,54,55,56,57. The function of individual coactivators seems to be redundant, at least for the induction of mouse Cyp2b1058. However, an in vivo deficiency of SRC3 impaired TCPOBOP-induced hepatic hyperplasia and drug-metabolizing enzymes, indicating that the coactivation of CAR by different coactivators can be involved in different genomic contexts59. Loss-of-function studies have identified more coactivators that are essential for the full activity of CAR. For example, a dominant negative fragment of activating signal cointegrator (ASC2/NcoA6) abolished TCPOBOP-induced gene expression and prevented acetaminophen-induced liver toxicity60. In PBP knockout mice, the nuclear translocation of CAR is blocked in the presence of phenobarbital37. Loss of the protein growth arrest and DNA-damage-inducible β (GADD45β) in hepatocytes attenuated Cyp2b10 induction by TCPOBOP61. The LXXLL-binding pocket can also be targeted by some corepressors. Like many other nuclear receptors, CAR is also bound and repressed by the following common corepressors: nuclear receptor corepressor 1 (NCoR1), NCoR2/SMRT, and DAX-1 (NR0B1)53,54,57,62. Collectively, the LXXLL-mediated direct interaction of CAR and its cofactors is very important for the activity of CAR. More importantly, because CAR shares multiple coregulators with other transcriptional factors, coregulator exchange and competition may influence many cellular processes when CAR is activated63.
CAR has also been shown to functionally crosstalk with many transcriptional factors. The outcome of crosstalk is either stimulatory or inhibitory. The transcriptional factors that potentiate the activity of CAR include HNF4α, CCAAT-enhancer-binding protein α (C/EBPα), activating transcription factor 5 (ATF5), nuclear factor (erythroid-derived 2)-like 2 (Nrf2), and hypoxia-inducible factor 1 (Hif1)64,65,66,67. Much of the crosstalk occurs between specific genes. Whether such crosstalk could be extended to the regulation of other genes needs to be further tested. CAR inducible genes are overlapped with AhR, PXR, PPARα, and Nrf2, indicating that CAR's crosstalk with these transcriptional factors will contribute to the coordinated regulation of xenobiotic detoxification, fasting responses, and anti-oxidant responses68. Overlapping gene induction could be due to different cis-acting elements on specific target genes, which may result in some additive effects. Nevertheless, with the help of additional factors, crosstalk between CAR and other transcriptional factors will be facilitated and will exhibit synergistic effects67,69,70,71. However, several CAR inhibitory factors, such as sterol regulatory element-binding protein 1 (SREBP1), liver X receptor (LXR), farnesoid X receptor (FXR), estrogen receptor (ER), and histone deacetylase 1 (HDAC1), also exist72,73,74,75,76. The inhibitory crosstalk attenuates CAR activity and down-regulates drug-metabolizing enzymes. Conversely, CAR-mediated suppression of certain transcriptional factors, such as HNF4α and LXR, leads to inhibition of glucose and lipid metabolism. In general, the mutual inhibitory effects are due primarily to competitive binding to the conserved elements on the DNA and/or the coactivators that are shared. As further discussed, the crosstalk between CAR and nuclear transcriptional factors may provide links between xenobiotic metabolism and energy metabolism.
CAR in energy metabolism
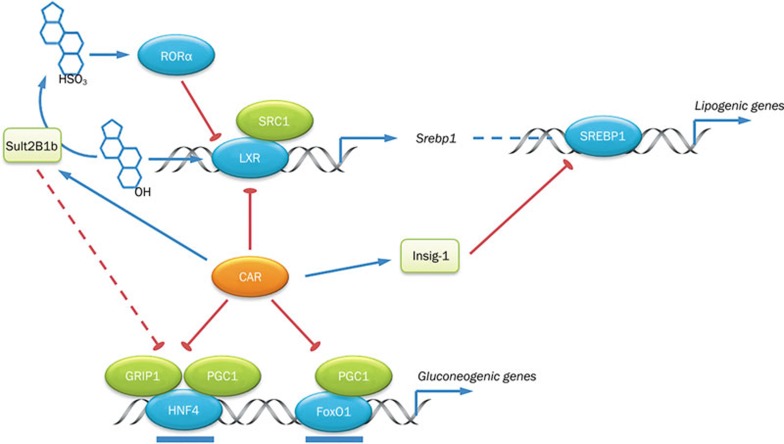
CAR has been long implicated in xenobiotic metabolism. The role of CAR in energy metabolism did not receive much attention until several studies within the past few years demonstrating that CAR activation influences glucose and lipid metabolism (Figure 2). Hyperglycemia and dyslipidemia are the hallmarks of obesity-associated type 2 diabetes. Activation of CAR by the synthetic agonist TCPOBOP increased insulin sensitivity and ameliorated liver steatosis in both high fat diet (HFD)- and leptin deficiency-induced obesity77,78. Several studies have shown that pharmacological activation and the genetic gain-of-function of CAR leads to lower hepatic triglyceride levels, whereas loss of CAR causes basal accumulation of triglycerides in the liver79. The LXR-SREBP pathway plays a central role in hepatic lipogenesis by transducing the genes involved in fatty acid biosynthesis and lipid uptake80. Crosstalk between CAR and LXR causes mutual repression through coactivator-binding competition. Consequently, the expression of LXR target genes are down-regulated, including lipogenic genes, such as Srebp1, Acc1, Fas, and Scd175. CAR also directly binds to SREBP1, which suggests potential crosstalk between CAR and SREBP1 in the context of lipogenesis, although further functional and mechanistic studies are needed to prove this association76.
Figure 2.
Inhibitory effect of CAR on glucose and lipid metabolism. CAR regulates gluconeogenic and lipogenic genes through competitive binding to cis-acting elements, coactivator quenching, and/or induction of suppressive genes (Sult2B1b and Insig-1).
CAR can also influence lipid biosynthesis through downstream target genes. Insulin-induced gene-1 (Insig-1) is an endoplasmic reticulum-bound cholesterol sensor that suppresses the proteolytic activation of SREBPs when sterols are abundant81. CAR binds to the DR4 element on the Insig-1 promoter and directly induces the expression of Insig-1, therefore preventing SREBP1 from entering the nucleus to induce lipogenic genes79. Unlike Insig-1 that directly targets the activation of SREBP1, sulfotransferase 2B1b (Sult2B1b), another CAR responsive gene, inhibits Srebp1 expression and hepatic lipogenesis through enzymatic deactivation of LXR ligands. Sult2B1b belongs to the subfamily of cytosolic sulfotransferases, mediating the sulfonation of oxysterols such as 22-hydroxycholesterol, 24S-hydroxycholesterol, 25-hydroxycholesterol, 27-hydroxycholesterol, and 24, 25-epoxycholesterol82. Those endogenous oxysterols are potent ligands for LXR activation. Sulfonation of oxysterols reduces their capacity to activate LXR and down-regulates the LXR-SREBP pathway83. Consistently, the TCPOBOP-induced reduction of lipogenic genes is abolished in Sult2B1b knockout mice78. Liver-specific overexpression of Sult2B1b, either by adenoviral delivery or transgenic strategy, ameliorates dyslipidemia in a diabetic mouse model84,85. In addition to deactivating LXR ligands, the products of sulfonation, such as 25-hydroxycholesterol-3-sulfate, have been reported to decrease lipid accumulation and inflammation86,87,88,89. Cholesterol sulfates, which are converted from cholesterol by Sult2B1b, have also been reported to be potent agonists for RORα90. Inhibitory crosstalk between RORα and LXR could be another reason for the down-regulation of lipid accumulation when Sult2B1b is overexpressed91.
Activation of CAR not only tightly regulates lipid metabolism but also impacts hepatic glucose metabolism. Diabetic mice displayed better glucose tolerance when treated with TCPOBOP78. Improved glucose tolerance is due primarily to the suppression of hepatic gluconeogenesis rather than glucose disposal in white adipose tissue and skeletal muscle, as shown by a hyperinsulinemic-euglycemic clamp study in ob/ob mice. In agreement with lower hepatic glucose production, the two rate-limiting gluconeogenic enzymes Pepck and G6pase were down-regulated by CAR activation. Several models were proposed to explain the inhibition of gluconeogenesis by CAR. In the first model, CAR competes with forkhead box protein O1 (FoxO1) and HNF4α on the insulin responsive sequence (IRS) and DR1 element, respectively, on the promoters of Pepck and G6pase92. In the second model, CAR competitively binds to SRC2/GRIP1 and PGC1α, which are two coactivators of HNF4α, thus diminishing the expression of gluconeogenic genes93. HNF4α-mediated gluconeogenic transactivation is also controlled by nuclear HNF4α exclusion. Acetylation of HNF4α is an essential event for HNF4α translocation into the nucleus. Indeed, cholesterol sulfate and Sult2B1b were recently reported to inhibit gluconeogenesis through the de-acetylation of HNF4α85. Therefore, up-regulation of Sult2B1b by CAR may have also played a role in the suppression of gluconeogenesis.
Insulin resistance is associated with elevated plasma lipoproteins, very-low-density lipoprotein (VLDL) production, and plasma low-density lipoprotein (LDL)94,95. Subsequent atherosclerotic lesions are a serious cause for the cardiovascular complications that arise in patients with type 2 diabetes. CAR activation has been shown to decrease VLDL secretion and plasma cholesterol concentration in apolipoprotein A-I (apoA-I) transgenic mice and Ldlr−/− mice, partially through the down-regulation of apoA-I and up-regulation of Vldlr, respectively96,97. Excessive cholesterol levels in the liver are also eliminated in the form of bile salts into the feces98. In this process, the genes responsible for the conversion of cholesterol to bile salts and bile acid hydration, conjugation, and export are up-regulated in response to TCPOBOP in the liver. In the intestine, bile acid excretion is further facilitated by the inhibition of bile acid reabsorption machinery, most likely through the inhibition of LXR. In addition, VLDL secretion and lipid homeostasis are also maintained by HNF4α99. The inhibitory effect of CAR on HNF4α may explain its benefits on cholesterol metabolism. Overall, CAR activation attenuates the development of atherosclerotic lesions and has a therapeutic potential for prevention of diabetes-associated cardiovascular diseases.
Despite the fact that CAR activation lowers plasma cholesterol levels, the genes involved in de novo cholesterol biosynthesis are up-regulated by CAR activation100. Considering that hepatic cholesterols are prone to be metabolized into bile acids, the up-regulation of cholesterol biosynthesis could be a compensatory mechanism to maintain intracellular cholesterol homeostasis101. Interestingly, although de novo cholesterol and triglyceride biosynthesis share the same transcriptional factors such as SREBP, cholesterol biosynthesis seems to be selectively elevated. One explanation is that lipogenic genes are also directly regulated by other nuclear transcription factors, such as LXR and carbohydrate-responsive element-binding protein (ChREBP), but whether crosstalk between CAR and those transcription factors are necessary for the inhibitory effect on triglyceride accumulation needs to be further investigated102,103. In addition, preferential de novo cholesterol biosynthesis can be achieved by selective activation of SREBP2104. Whether CAR interplays with SREBP2 to activate cholesterol biosynthesis remains to be determined.
Conclusions and perspectives
CAR is regulated not only by environmental cues, such as xenobiotic exposure and circadian rhythm, but also by endogenous metabolic signaling. High basal activity, rapid drug response, and versatile gene induction make CAR an intriguing therapeutic target. Recent research has revealed that CAR, the formerly known xenobiotic nuclear receptor, exhibits previously unknown functions in energy metabolism. Most of the metabolic benefits of CAR are observed in metabolic disease models, such as diet-induced obesity, suggesting that CAR inhibits certain factors that are highly inducible in metabolic disorders. Indeed, through the crosstalk with transcriptional factors involved in gluconeogenesis and lipogenesis, activation of CAR ameliorates hyperglycemia and dyslipidemia associated with metabolic diseases and increases systemic insulin sensitivity. The modes of action for such inhibitory effects may include the following: (1) competitive binding to cis-acting elements; (2) coactivator quenching; and (3) induction of suppressive genes, leading to direct inhibition of certain transcriptional factors, deactivation of pro-diabetic agonists or production of anti-diabetic substances, and changes in metabolite profiles. In addition, the metabolic benefits can also be transmitted to offspring due to permanent epigenetic switches105,106. In summary, CAR is a potential therapeutic target for the prevention and treatment of metabolic diseases. However, the potential to stimulate carcinogenesis presents an obstacle for clinical use. Moving forward, mechanistic studies that dissociate the beneficial effects with the unwanted side effects will be interesting. Future work for the discovery of activators that are selective for the metabolic benefits will also be important. Accumulating evidence shows that CAR is necessary for normal function in tissues in which the expression of CAR is relatively low. The human CAR agonist CITCO induces the expression of drug transporters at the blood-brain barrier, indicating that the expression of CAR in endothelial cells is important for drug resistance107,108,109. The association of CAR single-nucleotide polymorphisms with bone mineral density was also reported, and loss of CAR increased bone mass, suggesting that CAR is essential for osteoblast/osteoclast homeostasis110,111.
Acknowledgments
Our original work described in this article was supported in part by National Institutes of Health grants DK083952, HD073070, DK099232, and ES023438 (to Wen XIE). This work was also supported by the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions, China. Wen XIE is the Joseph Koslow Endowed Chair in Pharmaceutical Sciences at the University of Pittsburgh School of Pharmacy.
References
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006; 126: 789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 2000; 407: 920–3. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 2002; 62: 638–46. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Simon CM, Pierce AM, Safe S, Blumberg B, et al. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev 2000; 14: 3014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. Characterization of a phenobarbital-responsive enhancer module in mouse P450 Cyp2b10 gene. J Biol Chem 1997; 272: 14943–9. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol 1998; 18: 5652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, et al. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol 2004; 65: 292–300. [DOI] [PubMed] [Google Scholar]
- Uppal H, Toma D, Saini SP, Ren S, Jones TJ, Xie W. Combined loss of orphan receptors PXR and CAR heightens sensitivity to toxic bile acids in mice. Hepatology 2005; 41: 168–76. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem 2004; 279: 49517–22. [DOI] [PubMed] [Google Scholar]
- Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, et al. Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc Natl Acad Sci U S A 2005; 102: 2063–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, et al. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol 2002; 61: 1–6. [DOI] [PubMed] [Google Scholar]
- Locker J, Tian J, Carver R, Concas D, Cossu C, Ledda-Columbano GM, et al. A common set of immediate-early response genes in liver regeneration and hyperplasia. Hepatology 2003; 38: 314–25. [DOI] [PubMed] [Google Scholar]
- Columbano A, Ledda-Columbano GM, Pibiri M, Cossu C, Menegazzi M, Moore DD, et al. Gadd45β is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology 2005; 42: 1118–26. [DOI] [PubMed] [Google Scholar]
- Moore DD. Nuclear receptors reverse McGarry's vicious cycle to insulin resistance. Cell metabolism 2012; 15: 615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, et al. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol 2005; 19: 1646–53. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol 1999; 19: 6318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osabe M, Negishi M. Active ERK1/2 protein interacts with the phosphorylated nuclear constitutive active/androstane receptor (CAR; NR1I3), repressing dephosphorylation and sequestering CAR in the cytoplasm. J Biol Chem 2011; 286: 35763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Moore R, Negishi M. Extracellular signal-regulated kinase is an endogenous signal retaining the nuclear constitutive active/androstane receptor (CAR) in the cytoplasm of mouse primary hepatocytes. Mol Pharmacol 2007; 71: 1217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu JS, Omiecinski CJ. An okadaic acid-sensitive pathway involved in the phenobarbital-mediated induction of CYP2B gene expression in primary rat hepatocyte cultures. J Pharmacol Exp Ther 1997; 282: 1122–9. [PubMed] [Google Scholar]
- Mutoh S, Sobhany M, Moore R, Perera L, Pedersen L, Sueyoshi T, et al. Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci Signal 2013; 6: ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinpour F, Moore R, Negishi M, Sueyoshi T. Serine 202 regulates the nuclear translocation of constitutive active/androstane receptor. Mol Pharmacol 2006; 69: 1095–102. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Moore R, Sugatani J, Matsumura Y, Negishi M. PPP1R16A, the membrane subunit of protein phosphatase 1β, signals nuclear translocation of the nuclear receptor constitutive active/androstane receptor. Mol Pharmacol 2008; 73: 1113–21. [DOI] [PubMed] [Google Scholar]
- Saito K, Moore R, Negishi M. p38 Mitogen-activated protein kinase regulates nuclear receptor CAR that activates the CYP2B6 gene. Drug Metab Disposition 2013; 41: 1170–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. Identification of the nuclear receptor CAR: HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett 2003; 548: 17–20. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol 2003; 64: 1069–75. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Miyama Y, Ando M, Inouye Y. Dependence on the microtubule network and 90-kDa heat shock protein of phenobarbital-induced nuclear translocation of the rat constitutive androstane receptor. Mol Pharmacol 2010; 77: 311–6. [DOI] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. Coordinated regulation of nuclear receptor CAR by CCRP/DNAJC7, HSP70 and the ubiquitin-proteasome system. PLoS One 2014; 9: e96092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencurel F, Stenhouse A, Hawley SA, Friedberg T, Hardie DG, Sutherland C, et al. AMP-activated protein kinase mediates phenobarbital induction of CYP2B gene expression in hepatocytes and a newly derived human hepatoma cell line. J Biol Chem 2005; 280: 4367–73. [DOI] [PubMed] [Google Scholar]
- Rencurel F, Foretz M, Kaufmann MR, Stroka D, Looser R, Leclerc I, et al. Stimulation of AMP-activated protein kinase is essential for the induction of drug metabolizing enzymes by phenobarbital in human and mouse liver. Mol Pharmacol 2006; 70: 1925–34. [DOI] [PubMed] [Google Scholar]
- Shindo S, Numazawa S, Yoshida T. A physiological role of AMP-activated protein kinase in phenobarbital-mediated constitutive androstane receptor activation and CYP2B induction. Biochem J 2007; 401: 735–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blättler SM, Rencurel F, Kaufmann MR, Meyer UA. In the regulation of cytochrome P450 genes, phenobarbital targets LKB1 for necessary activation of AMP-activated protein kinase. Proc Natl Acad Sci U S A 2007; 104: 1045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizu R, Shindo S, Yoshida T, Numazawa S. MicroRNA-122 down-regulation is involved in phenobarbital-mediated activation of the constitutive androstane receptor. PLoS One 2012; 7: e41291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CM, Su YH, Lin HY, Lin JN, Liu LC, Ho CT, et al. Demethoxycurcumin modulates prostate cancer cell proliferation via AMPK-induced down-regulation of HSP70 and EGFR. J Agric Food Chem 2012; 60: 8427–34. [DOI] [PubMed] [Google Scholar]
- Jung JH, Lee JO, Kim JH, Lee SK, You GY, Park SH, et al. Quercetin suppresses HeLa cell viability via AMPK-induced HSP70 and EGFR down-regulation. J Cell Physiol 2010; 223: 408–14. [DOI] [PubMed] [Google Scholar]
- Kim KY, Baek A, Hwang JE, Choi YA, Jeong J, Lee MS, et al. Adiponectin-activated AMPK stimulates dephosphorylation of AKT through protein phosphatase 2A activation. Cancer Res 2009; 69: 4018–26. [DOI] [PubMed] [Google Scholar]
- Min G, Kemper JK, Kemper B. Glucocorticoid receptor-interacting protein 1 mediates ligand-independent nuclear translocation and activation of constitutive androstane receptor in vivo. J Biol Chem 2002; 277: 26356–63. [DOI] [PubMed] [Google Scholar]
- Guo D, Sarkar J, Ahmed MR, Viswakarma N, Jia Y, Yu S, et al. Peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP) but not PPAR-interacting protein (PRIP) is required for nuclear translocation of constitutive androstane receptor in mouse liver. Biochem Biophys Res Commun 2006; 347: 485–95. [DOI] [PubMed] [Google Scholar]
- Chang TK, Waxman DJ. Synthetic drugs and natural products as modulators of constitutive androstane receptor (CAR) and pregnane X receptor (PXR). Drug Metab Rev 2006; 38: 51–73. [DOI] [PubMed] [Google Scholar]
- Patel RD, Hollingshead BD, Omiecinski CJ, Perdew GH. Aryl-hydrocarbon receptor activation regulates constitutive androstane receptor levels in murine and human liver. Hepatology 2007; 46: 209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: consequences on cytochrome P450 gene regulation. Mol Pharmacol 2000; 58: 1441–50. [DOI] [PubMed] [Google Scholar]
- Ooe H, Kon J, Oshima H, Mitaka T. Thyroid hormone is necessary for expression of constitutive androstane receptor in rat hepatocytes. Drug Metab Disposition 2009; 37: 1963–69. [DOI] [PubMed] [Google Scholar]
- Qatanani M, Zhang J, Moore DD. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology 2005; 146: 995–1002. [DOI] [PubMed] [Google Scholar]
- Saito K, Kobayashi K, Mizuno Y, Furihata T, Chiba K. Constitutive androstane/active receptor is a target of retinoic acid receptor in humans. Biochem Pharmacol 2010; 80: 129–35. [DOI] [PubMed] [Google Scholar]
- Wieneke N, Hirsch-Ernst KI, Kuna M, Kersten S, Püschel GP. PPARα-dependent induction of the energy homeostasis-regulating nuclear receptor NR1i3 (CAR) in rat hepatocytes: potential role in starvation adaptation. FEBS Lett 2007; 581: 5617–26. [DOI] [PubMed] [Google Scholar]
- Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor α, and the coactivator peroxisome proliferator-activated receptor γ coactivator-1α. J Biol Chem 2006; 281: 26540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Yeager RL, Klaassen CD. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab Dispos 2009; 37: 106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006; 126: 801–10. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell 2008; 134: 728–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takwi AA, Wang YM, Wu J, Michaelis M, Cinatl J, Chen T. miR-137 regulates the constitutive androstane receptor and modulates doxorubicin sensitivity in parental and doxorubicin-resistant neuroblastoma cells. Oncogene 2014; 33: 3717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, et al. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell 2004; 16: 893–905. [DOI] [PubMed] [Google Scholar]
- Plevin MJ, Mills MM, Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci 2005; 30: 66–9. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)] benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol 2000; 20: 2951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, et al. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol 2002; 22: 5270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyrkkärinne J, Windshügel B, Mäkinen J, Ylisirniö M, Peräkylä M, Poso A, et al. Amino acids important for ligand specificity of the human constitutive androstane receptor. J Biol Chem 2005; 280: 5960–71. [DOI] [PubMed] [Google Scholar]
- Ueda A, Matsui K, Yamamoto Y, Pedersen LC, Sueyoshi T, Negishi M. Thr176 regulates the activity of the mouse nuclear receptor CAR and is conserved in the NR1I subfamily members PXR and VDR. Biochem J 2005; 388: 623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Sakai N, Kanaya E, Jingami H. Activation of orphan nuclear constitutive androstane receptor requires subnuclear targeting by peroxisome proliferator-activated receptor γ coactivator-1α. A possible link between xenobiotic response and nutritional state. J Biol Chem 2003; 278: 11344–50. [DOI] [PubMed] [Google Scholar]
- Jyrkkärinne J, Mäkinen J, Gynther J, Savolainen H, Poso A, Honkakoski P. Molecular determinants of steroid inhibition for the mouse constitutive androstane receptor. J Med Chem 2003; 46: 4687–95. [DOI] [PubMed] [Google Scholar]
- Xia J, Liao L, Sarkar J, Matsumoto K, Reddy JK, Xu J, et al. Redundant enhancement of mouse constitutive androstane receptor transactivation by p160 coactivator family members. Arch Biochem Biophys 2007; 468: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Chen Q, Xu Y, Zhou Q, Zhu J, Zhang H, et al. SRC-3 is required for CAR-regulated hepatocyte proliferation and drug metabolism. J Hepatol 2012; 56: 210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Lee S, Yeom SY, Kim GH, Lee JW, Kim SW. Characterization of activating signal cointegrator-2 as a novel transcriptional coactivator of the xenobiotic nuclear receptor constitutive androstane receptor. Mol Endocrinol 2005; 19: 1711–9. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Negishi M. The antiapoptotic factor growth arrest and DNA-damage-inducible 45 β regulates the nuclear receptor constitutive active/androstane receptor-mediated transcription. Drug Metab Dispos 2008; 36: 1189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenzana EM, Chen T, Kannuswamy M, Sell BE, Strom SC, Li Y, et al. The orphan nuclear receptor DAX-1 functions as a potent corepressor of the constitutive androstane receptor (NR1I3). Mol Pharmacol 2012; 82: 918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 2000; 14: 121–41. [PubMed] [Google Scholar]
- Pascual M, Gómez-Lechón MJ, Castell JV, Jover R. ATF5 is a highly abundant liver-enriched transcription factor that cooperates with constitutive androstane receptor in the transactivation of CYP2B6: implications in hepatic stress responses. Drug Metab Dispos 2008; 36: 1063–72. [DOI] [PubMed] [Google Scholar]
- Ashino T, Ohkubo-Morita H, Yamamoto M, Yoshida T, Numazawa S. Possible involvement of nuclear factor erythroid 2-related factor 2 in the gene expression of Cyp2b10 and Cyp2a5. Redox Biology 2014; 2: 284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizu R, Shindo S, Yoshida T, Numazawa S. Cross-talk between constitutive androstane receptor and hypoxia-inducible factor in the regulation of gene expression. Toxicol Lett 2013; 219: 143–50. [DOI] [PubMed] [Google Scholar]
- Benet M, Lahoz A, Guzmán C, Castell JV, Jover R. CCAAT/enhancer-binding protein α (C/EBPα) and hepatocyte nuclear factor 4α (HNF4α) synergistically cooperate with constitutive androstane receptor to transactivate the human cytochrome P450 2B6 (CYP2B6) gene: application to the development of a metabolically competent human hepatic cell model. J Biol Chem 2010; 285: 28457–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksunes LM, Klaassen CD. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 2012; 40: 1366–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhu QS, Zhong S, Levy D. Transcription of the human microsomal epoxide hydrolase gene (EPHX1) is regulated by an HNF-4α/CAR/RXR/PSF complex. Biochim Biophys Acta 2013; 1829: 1000–9. [DOI] [PubMed] [Google Scholar]
- Inoue K, Negishi M. Early growth response 1 loops the CYP2B6 promoter for synergistic activation by the distal and proximal nuclear receptors CAR and HNF4α. FEBS Lett 2009; 583: 2126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surapureddi S, Rana R, Reddy JK, Goldstein JA. Nuclear receptor coactivator 6 mediates the synergistic activation of human cytochrome P-450 2C9 by the constitutive androstane receptor and hepatic nuclear factor-4α. Mol Pharmacol 2008; 74: 913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renga B, Migliorati M, Mencarelli A, Cipriani S, D'Amore C, Distrutti E, et al. Farnesoid X receptor suppresses constitutive androstane receptor activity at the multidrug resistance protein-4 promoter. Biochim Biophys Acta 2011; 1809: 157–65. [DOI] [PubMed] [Google Scholar]
- Min G, Kim H, Bae Y, Petz L, Kemper JK. Inhibitory cross-talk between estrogen receptor (ER) and constitutively activated androstane receptor (CAR) CAR inhibits ER-mediated signaling pathway by squelching p160 coactivators. J Biol Chem 2002; 277: 34626–33. [DOI] [PubMed] [Google Scholar]
- Takizawa D, Kakizaki S, Horiguchi N, Tojima H, Yamazaki Y, Ichikawa T, et al. Histone deacetylase inhibitors induce cytochrome P450 2B by activating nuclear receptor constitutive androstane receptor. Drug Metab Dispos 2010; 38: 1493–8. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Wada T, Zhang B, Khadem S, Ren S, Kuruba R, et al. A functional cross-talk between liver X receptor-α and constitutive androstane receptor links lipogenesis and xenobiotic responses. Mol Pharmacol 2010; 78: 666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Looser R, Kaufmann M, Meyer UA. Sterol regulatory element binding protein 1 interacts with pregnane X receptor and constitutive androstane receptor and represses their target genes. Pharmacogenet Genomics 2008; 18: 325–37. [DOI] [PubMed] [Google Scholar]
- Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem 2009; 284: 25984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A 2009; 106: 18831–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Looser R, Kaufmann M, Blättler SM, Rencurel F, Huang W, et al. Regulatory cross-talk between drug metabolism and lipid homeostasis: constitutive androstane receptor and pregnane X receptor increase Insig-1 expression. Mol Pharmacol 2008; 73: 1282–9. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 2002; 109: 1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002; 110: 489–500. [DOI] [PubMed] [Google Scholar]
- Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J 1997; 11: 206–16. [DOI] [PubMed] [Google Scholar]
- Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab 2007; 5: 73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Xu L, Kakiyama G, Runge-Morris MA, Hylemon PB, Yin L, et al. Sulfation of 25-hydroxycholesterol by SULT2B1b decreases cellular lipids via the LXR/SREBP-1c signaling pathway in human aortic endothelial cells. Atherosclerosis 2011; 214: 350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Cheng Q, Xu L, Yan J, Jiang M, He J, et al. Cholesterol sulfate and cholesterol sulfotransferase inhibit gluconeogenesis by targeting hepatocyte nuclear factor 4α. Mol Cell Biol 2014; 34: 485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Xu L, Rodriguez-Agudo D, Li X, Heuman DM, Hylemon PB, et al. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1 signaling pathway. Am J Physiol Endocrinol Metab 2008; 295: E1369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen S, Ma Y, Kim JK, Rodriguez-Agudo D, Heuman DM, et al. 25-Hydroxycholesterol-3-sulfate attenuates inflammatory response via PPARγ signaling in human THP-1 macrophages. Am J Physiol Endocrinol Metab 2012; 302: E788–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kim JK, Bai Q, Zhang X, Kakiyama G, Min HK, et al. 5-Cholesten-3β,25-diol 3-sulfate decreases lipid accumulation in diet-induced nonalcoholic fatty liver disease mouse model. Mol Pharmacol 2013; 83: 648–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Bai Q, Rodriguez-Agudo D, Hylemon PB, Heuman DM, Pandak WM, et al. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids 2010; 45: 821–32. [DOI] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, et al. X-ray structure of the hRORα LBD at 1.63 Å: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure 2002; 10: 1697–707. [DOI] [PubMed] [Google Scholar]
- Xiao L, Xie X, Zhai Y. Functional crosstalk of CAR-LXR and ROR-LXR in drug metabolism and lipid metabolism. Adv Drug Del Rev 2010; 62: 1316–21. [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 2004; 24: 7931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Fang S, Bae Y, Kemper JK. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1α. J Biol Chem 2006; 281: 14537–46. [DOI] [PubMed] [Google Scholar]
- Haidari M, Leung N, Mahbub F, Uffelman KD, Kohen-Avramoglu R, Lewis GF, et al. Fasting and postprandial overproduction of intestinally derived lipoproteins in an animal model of insulin resistance. Evidence that chronic fructose feeding in the hamster is accompanied by enhanced intestinal de novo lipogenesis and apob48-containing lipoprotein overproduction. J Biol Chem 2002; 277: 31646–55. [DOI] [PubMed] [Google Scholar]
- Federico LM, Naples M, Taylor D, Adeli K. Intestinal insulin resistance and aberrant production of apolipoprotein b48 lipoproteins in an animal model of insulin resistance and metabolic dyslipidemia: evidence for activation of protein tyrosine phosphatase-1B, extracellular signal-related kinase, and sterol regulatory element-binding protein-1c in the fructose-fed hamster intestine. Diabetes 2006; 55: 1316–26. [DOI] [PubMed] [Google Scholar]
- Masson D, Qatanani M, Sberna AL, Xiao R, de Barros JPP, Grober J, et al. Activation of the constitutive androstane receptor decreases HDL in wild-type and human apoA-I transgenic mice. J Lipid Res 2008; 49: 1682–91. [DOI] [PubMed] [Google Scholar]
- Sberna AL, Assem M, Xiao R, Ayers S, Gautier T, Guiu B, et al. Constitutive androstane receptor activation decreases plasma apolipoprotein B-containing lipoproteins and atherosclerosis in low-density lipoprotein receptor-deficient mice. Atertio Thromb Vasc Biol 2011; 31: 2232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sberna AL, Assem M, Gautier T, Grober J, Guiu B, Jeannin A, et al. Constitutive androstane receptor activation stimulates faecal bile acid excretion and reverse cholesterol transport in mice. J Hepatol 2011; 55: 154–61. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001; 21: 1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojima H, Kakizaki S, Yamazaki Y, Takizawa D, Horiguchi N, Sato K, et al. Ligand dependent hepatic gene expression profiles of nuclear receptors CAR and PXR. Toxicol Lett 2012; 212: 288–97. [DOI] [PubMed] [Google Scholar]
- Režen T, Tamasi V, Lövgren-Sandblom A, Björkhem I, Meyer UA, Rozman D. Effect of CAR activation on selected metabolic pathways in normal and hyperlipidemic mouse livers. BMC Genomics 2009; 10: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, et al. Role of LXRs in control of lipogenesis. Genes Dev 2000; 14: 2831–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 2006; 4: 107–10. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest 1998; 101: 2331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama H, Hiramatsu Y. Treatment with constitutive androstane receptor ligand during pregnancy prevents insulin resistance in offspring from high-fat diet-induced obese pregnant mice. Am J Physiol Endocrinol Metab 2012; 303: E293–300. [DOI] [PubMed] [Google Scholar]
- Chen WD, Fu X, Dong B, Wang YD, Shiah S, Moore DD, et al. Neonatal activation of the nuclear receptor CAR results in epigenetic memory and permanent change of drug metabolism in mouse liver. Hepatology 2012; 56: 1499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slosky LM, Thompson BJ, Sanchez-Covarrubias L, Zhang Y, Laracuente ML, Vanderah TW, et al. Acetaminophen modulates P-glycoprotein functional expression at the blood-brain barrier by a constitutive androstane receptor-dependent mechanism. Mol Pharmacol 2013; 84: 774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmen J, Tozakidis IE, Bele P, Galla HJ. Constitutive androstane receptor upregulates Abcb1 and Abcg2 at the blood-brain barrier after CITCO activation. Brain Res 2013; 1501: 68–80. [DOI] [PubMed] [Google Scholar]
- Wang X, Sykes DB, Miller DS. Constitutive androstane receptor-mediated up-regulation of ATP-driven xenobiotic efflux transporters at the blood-brain barrier. Mol Pharmacol 2010; 78: 376–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano T, Usui T, Shiraki M, Ouchi Y, Inoue S. Association of a single nucleotide polymorphism in the constitutive androstane receptor gene with bone mineral density. Geriatr Gerontol Int 2009; 9: 235–41. [DOI] [PubMed] [Google Scholar]
- Cho HY, Jung JY, Park H, Yang JY, Jung S, An JH, et al. In vivo deletion of CAR resulted in high bone mass phenotypes in male mice. J Cell Physiol 2014; 229: 561–71. [DOI] [PubMed] [Google Scholar]