Coronin 1A is involved in cell shape dynamics and Rac1 GTPase signaling. Loss-of-function mutations in the Coro1A gene promote severe immunodeficiency. An immunodeficiency-linked Coro1A point mutant is described that becomes dysfunctional due to changes in actin-binding, actin-remodeling, and signaling activities.
Abstract
Coronin 1A (Coro1A) is involved in cytoskeletal and signaling events, including the regulation of Rac1 GTPase– and myosin II–dependent pathways. Mutations that generate truncated or unstable Coro1A proteins cause immunodeficiencies in both humans and rodents. However, in the case of the peripheral T-cell–deficient (Ptcd) mouse strain, the immunodeficiency is caused by a Glu-26-Lys mutation that targets a surface-exposed residue unlikely to affect the intramolecular architecture and stability of the protein. Here we report that this mutation induces pleiotropic effects in Coro1A protein, including the exacerbation of Coro1A-dependent actin-binding and -bundling activities; the formation of large meshworks of Coro1AE26K-decorated filaments endowed with unusual organizational, functional, and staining properties; and the elimination of Coro1A functions associated with both Rac1 and myosin II signaling. By contrast, it does not affect the ability of Coro1A to stimulate the nuclear factor of activated T-cells (NF-AT). Coro1AE26K is not a dominant-negative mutant, indicating that its pathological effects are derived from the inability to rescue the complete loss of the wild-type counterpart in cells. These results indicate that Coro1AE26K behaves as either a recessive gain-of-function or loss-of-function mutant protein, depending on signaling context and presence of the wild-type counterpart in cells.
INTRODUCTION
Coronin 1A (Coro1A; also known as tryptophan aspartate–containing coat protein, p57-coronin, and coronin-1) displays a complex structure that includes an N-terminal β-propeller domain composed of seven WD40 repeats (residues 1–352) that can bind F-actin, an adjacent C-terminal extension harboring both conserved (residues 352–392) and unique (residues 393–429) sequences relative to other family members, and a distal coiled-coil region (residues 429–461) that can interact with the actin-related proteins 2 and 3 (Arp2/3) complex (Appleton et al., 2006; Uetrecht and Bear, 2006). This protein plays key roles in the regulation of lamellipodia dynamics due to its dual ability of stimulating and inhibiting F-actin bundling and the Arp2/3 complex, respectively (Uetrecht and Bear, 2006; Galkin et al., 2008; Shiow et al., 2008). More recently, it has been shown that Coro1A is also involved in the regulation of Rac1 signaling at two different levels. On one hand, it favors the stimulation of Rac1 via F-actin–dependent interactions with p21-activated kinase (Pak), ArhGEF7 (also known as β-Pix and Cool1), and Rho GDP dissociation inhibitor (Rho GDI)–Rac1 complexes. These interactions favor the Pak-mediated phosphorylation and inactivation of Rho GDI, the release of Rac1 from phosphorylated Rho GDIs, and, subsequently, the stimulation of Rac1 at the plasma membrane by either ArhGEF7 or other upstream GDP/GTP exchange factors (Castro-Castro et al., 2011; Bustelo et al., 2012). On the other hand, Coro1A is involved in an F-actin– and myosin II–dependent step that favors the intracellular dynamics and cytoskeletal output of already activated Rac1-AhrGEF7-Pak2 complexes. Owing to the latter function, the short hairpin RNA–mediated knockdown of CORO1A transcripts leads to the sequestration of those complexes in actomyosin ring structures and the formation of large, lamella-like cell protrusions instead of the membrane ruffles typically seen in active Rac1-expressing cells (Ojeda et al., 2014). Additional functions of this protein include vesicle trafficking, neurotrophin signaling and neurite outgrowth in neurons, signaling functions in endothelial cells, macrophage lipoprotein uptake, mast cell degranulation, and lytic immune synapse formation in natural killer cells. It is also involved in the infective cycle of a number of pathogens, such as Plasmodium falciparum and Mycobacterium species inside host cells (Jayachandran et al., 2007, 2008; Foger et al., 2011; Holtta-Vuori et al., 2012; Seto et al., 2012; Liu et al., 2014; Mace and Orange, 2014; Suo et al., 2014; Terzi et al., 2014; Kim et al., 2015).
In addition to the foregoing functions, Coro1A has been the focus of recent attention due to the discovery that a number of severe human and mouse T-cell immunodeficiencies are caused by mutations in its gene (Haraldsson et al., 2008; Shiow et al., 2008, 2009; Moshous et al., 2013; Stray-Pedersen et al., 2014; Punwani et al., 2015). These observations have been confirmed using standard Coro1a-knockout mice (Foger et al., 2006; Mueller et al., 2008, 2011; Mugnier et al., 2008), although there is still disagreement on whether the immunodeficiencies observed are caused by actin-dependent or -independent mechanisms (Mueller et al., 2011). Most of the mutations found in this gene correspond to loss-of-function alleles that encode either truncated (p.P83X, p.Q262X, p.P83RfsX10, p.Q360Rfs44) or structurally unstable (p.D278V, p.V134M) mutant proteins (Haraldsson et al., 2008; Shiow et al., 2008, 2009; Moshous et al., 2013; Stray-Pedersen et al., 2014). However, the analysis of the Ptcd mouse strain revealed the presence of a mutation (E26K) in the Coro1A β-propeller domain (Shiow et al., 2008) that, according to structural criteria (Appleton et al., 2006), is unlikely to cause any obvious intrinsic structural defect in the protein. The exact localization of this mutation in the structure of the protein is shown and discussed later in Figure 5A. However, this mutant protein shows an abnormal cytosolic distribution in cells, suggesting potential defects in its interaction with the cytoskeleton and/or cytoskeletal regulators (Shiow et al., 2008). Consistent with this idea, biochemical studies have shown that Coro1AE26K exhibits increased in vitro inhibitory activity toward the Arp2/3 complex (Shiow et al., 2008). However, whether such activity is responsible for the in vivo defects of this mutant is unclear because, for example, the Coro1AE26K and the Arp2/3 complex do not seem to colocalize in cells (Shiow et al., 2008). Such activity is also at odds with data indicating that the Coro1A β-propeller domain (the region where Glu-26 is located) and the Coro1A Arp2/3-binding site are not in physical proximity, at least when coronins are forming complexes with F-actin and the Arp2/3 complex (Cai et al., 2005; Appleton et al., 2006; Galkin et al., 2008). These observations led us to study the biological features of this mutant protein. Here we report that the E26K mutation induces a gain-of-function effect that promotes exacerbated actin-binding and -bundling activities by Coro1A, as well as alterations in downstream signaling. Such property is dominant over the negative effects induced by loss-of-function mutations that disrupt association of Coro1A with actin. In vivo, Coro1AE26K promotes the formation of an extensive meshwork of filaments that is quite unusual according to staining, structural, and regulatory criteria. Coro1AE26K does not have dominant-negative effects on the normal biological activity of its wild-type counterpart. However, it cannot functionally compensate for the loss of the endogenous protein, which explains the different phenotypes exhibited by heterozygous and homozygous Ptcd mice.
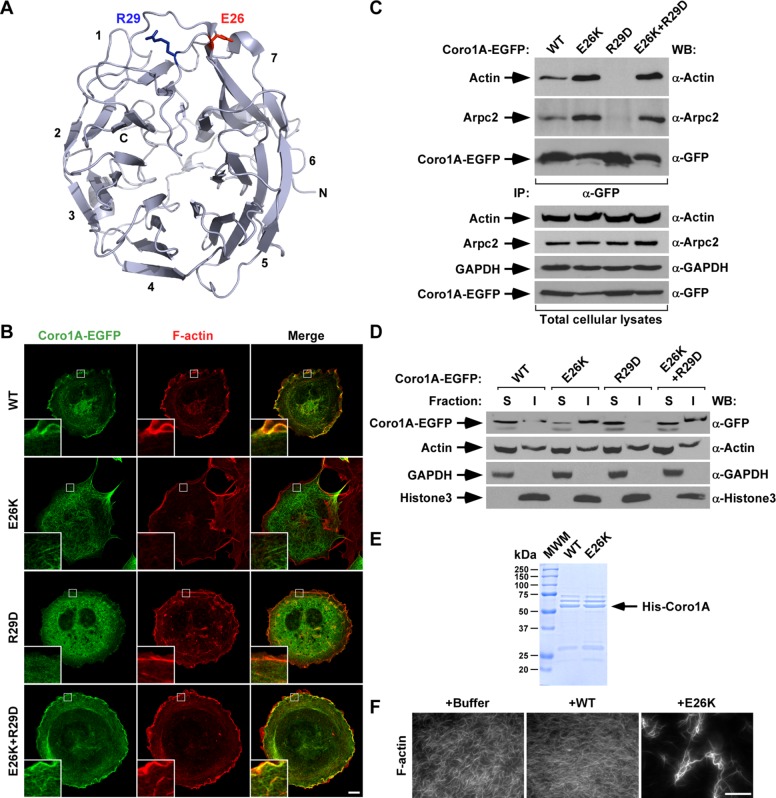
FIGURE 5:
The E26K mutation restores the association of both actin and Arpc2 to an actin binding–deficient Coro1A mutant. (A) Localization of the E26 and R29 residues (depicted as red and blue sticks, respectively) in the Coro1A β-propeller domain. Blades are numbered according to the standard nomenclature used for this type of domain. C, C-terminal end; N, N-terminal end. Illustration was generated using PyMol and the Coro1A crystal structure (Protein Data Bank: 2AQ5). (B) Representative confocal images of rhodamine-phalloidin–stained COS1 cells ectopically expressing indicated Coro1A-EGFPs (left). Coro1A proteins and F-actin are in green and red, respectively. Areas of colocalization are shown in yellow. Insets, enlarged images of the indicated cell areas (white open squares). Scale bar, 10 μm. (C) Anti-EGFP immunoprecipitates obtained from COS1 cells expressing the indicated Coro1A-EGFPs (top) were analyzed by Western blot to detect the amount of coimmunoprecipitated endogenous actin (top) and Arpc2 (second from top) in each experimental condition. As control, filters were immunoblotted with antibodies to EGFP (third from top) to visualize the amount of immunoprecipitated Coro1A-EGFP obtained in each sample. Amount of actin (fourth from top), Arpc2 (fifth from top), and Coro1A-EGFPs (bottom) present in lysates before the immunoprecipitation step was determined by immunoblot using aliquots of the same cell lysates used for the immunoprecipitation experiment. Antibodies used in each immunoblot analyses are indicated on the right. (D) Distribution of indicated Coro1A-EGFPs (top) and control endogenous proteins (remaining images) in Triton X-100–soluble (S) and –insoluble (I) fractions obtained from transiently transfected COS1 cells. Monitored proteins and antibodies used in immunoblots are shown on the left and right, respectively. Similar results were obtained in two independent experiments. (E) Coomassie-stained gel showing aliquots of His-tagged Coro1 proteins (arrow) purified from E. coli that were used in experiments presented in F. (F) Representative images of in vitro polymerized and phalloidin-stained F-actin upon incubation under the indicated experimental conditions for 15 min. Scale bar, 30 μm.
RESULTS
Coro1AE26K promotes formation of filaments with unusual staining properties
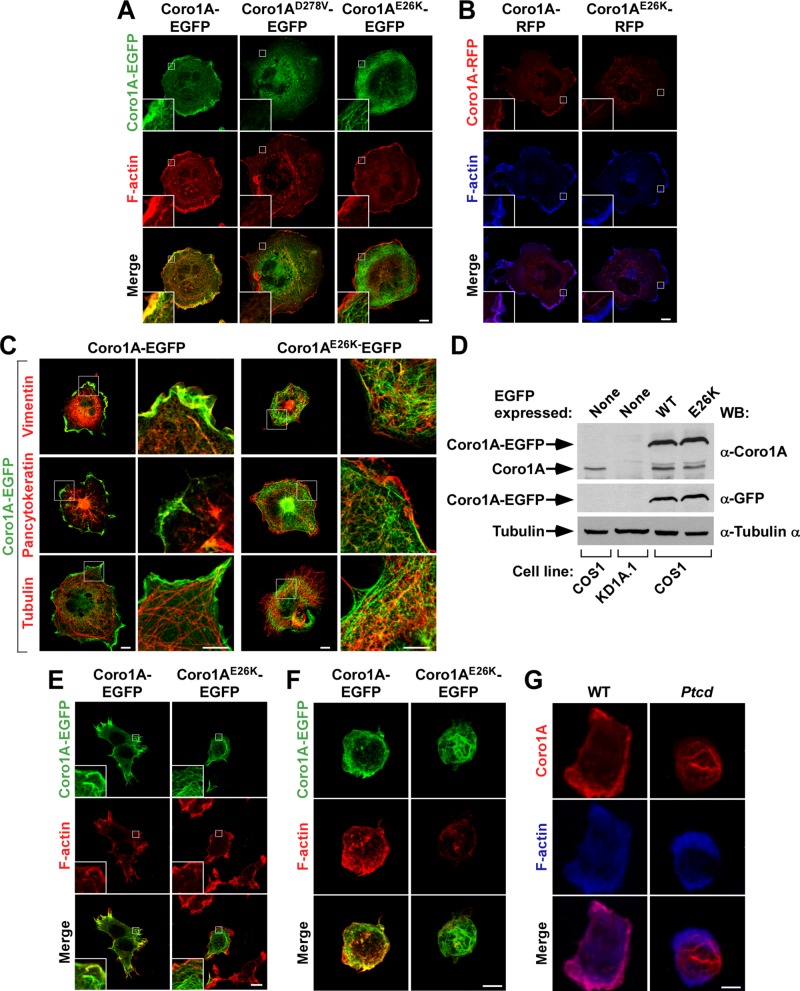
To evaluate the effect of the E26K mutation in Coro1A function, we first transfected COS1 cells with vectors encoding either enhanced green fluorescent protein (GFP)– or red fluorescent protein (RFP)–tagged versions of Coro1AE26K and, upon staining with fluorescence-labeled versions of phalloidin to decorate the cytoskeleton, analyzed them by confocal microscopy. For comparative purposes, we studied in parallel COS1 cells ectopically expressing wild-type Coro1A and Coro1AD278V, a protein harboring a missense mutation in a residue that, due to its location in the Coro1A structure (Appleton et al., 2006), probably disrupts the functionality of the β-propeller domain. We chose those cells because they are Coro1A dependent (Castro-Castro et al., 2011; Bustelo et al., 2012; Ojeda et al., 2014) and, in addition, their large cytoplasms facilitate the visualization of cytoskeleton-related processes. Using this strategy, we observed that wild-type Coro1A-EGFP (Figure 1A, left) and Coro1A-RFP (Figure 1B, left) localize preferentially in membrane ruffle–rich areas and, in addition, throughout the cytoplasm of transfected cells. In some cases, these proteins are found in reticular structures throughout the cytoplasm (Figure 1A). Such localization is disrupted in the case of Coro1AD278V-EGFP mutant (Figure 1A, middle), which shows a predominant cytoplasmic distribution, probably as a consequence of lack of actin binding. This distribution is similar to that previously described for another actin-deficient mutant version of this protein (Coro1AR29D; Castro-Castro et al., 2011). By contrast, we unexpectedly found that both Coro1AE26K-EGFP (Figure 1A, right) and Coro1AE26K-RFP (Figure 1B, right) show a quite idiosyncratic distribution, since they both localize in meshworks of very thick filaments uniformly distributed throughout the cytoplasm. The shape of these structures, which are not detected in control cells, is slightly different depending on the C-terminal tag, since the Coro1AE26K-RFP decorated filaments are usually shorter and more fusiform (Figure 1B, right) than those found in Coro1AE26K-EGFP–expressing cells (Figure 1A, right). These filaments are resistant to conventional staining with F-actin–binding molecules (phalloidin; Figure 1, A and B) and antibodies to either myosin IIB (Supplemental Figure S1) or a number of intermediate filament (vimentin, cytokeratins) and microtubule (α-tubulin) markers (Figure 1C). The amount of ectopic expression of the wild-type and E26K mutant versions of Coro1A was similar in these cells (Figure 1D, top and middle; compare third and fourth lanes), indicating that the differential subcellular distribution exhibited by each of those two chimeric proteins could not be just attributed to either different amounts of ectopic expression or differential overexpression relative to endogenous Coro1A (Figure 1D, top; compare first with third and fourth lanes). As expected, the endogenous protein was missing in CORO1A-knockdown COS1 cells (KD1A.1; Figure 1D, top; compare first and second lanes; Castro-Castro et al., 2011; Ojeda et al., 2014). Coro1AE26K-EGFP also forms phalloidin-negative filaments when ectopically expressed in both HEK 293T (Figure 1E) and Jurkat cells (Figure 1F), indicating that this effect is not cell-type specific. As in the case of COS1 cells, these bundles are located away from areas of active cytoskeletal reorganization (Figure 1, E and F). This feature is most notable in Jurkat cells, because the Coro1AE26K-decorated filaments are specifically located in the cortex of the cell body that protrudes away from the substratum (Figure 1F). To rule out artifacts derived from either the protein overexpression or C-terminal tags used in the foregoing experiments, we next investigated the distribution of endogenous Coro1A in thymocytes obtained from Ptcd mice. To this end, we attached these cells to coverslips coated with antibodies to mouse CD3, fixed them, and stained them with both phalloidin and antibodies to Coro1A. Using confocal immunofluorescence microscopy analysis, we found that the endogenous Coro1AE26K also displays a cortical distribution in phalloidin-negative filaments similar to that previously observed in Coro1AE26K-EGFP–expressing Jurkat cells (Figure 1G, right). By contrast, the endogenous Coro1A present in wild-type cells shows the expected distribution in phalloidin-positive membrane ruffles that emanate from the thymocyte/substrate contact zone (Figure 1G, left). These results indicate that the E26K mutation promotes a switch in the normal function of Coro1A, leading to the formation of thick, Coro1AE26K-decorated filaments that are located away from active areas of cytoskeletal reorganization.
FIGURE 1:
Ectopic and endogenous Coro1AE26K decorate a phalloidin-negative filament meshwork. (A–C) Representative confocal images of COS1 cells expressing the indicated EGFP-tagged (A and C, green signals) and RFP-tagged (B, red signals) Coro1A versions (top) and stained with rhodamine-labeled phalloidin (A, red signals), Alexa Fluor 635–labeled phalloidin (B, blue signals), or antibodies to the indicated proteins (C, red signals). Potential colocalization areas between Coro1A proteins and F-actin had to be seen in either yellow (A, bottom) or purple (B, bottom). Potential colocalization areas between Coro1A proteins and vimentin, pancytokeratins, or α-tubulin had to be seen in yellow (C). Scale bars, 10 μm. Insets, enlarged images of the indicated cell areas (white open squares). (D) Total cellular lysates obtained from parental COS1 cells, CORO1A knockdown (KD1A.1) COS1 cells, and parental COS1 cells (bottom) transiently expressing EGFPs fused to either wild type (WT, top) or the Coro1AE26K (E26K, top) mutant were analyzed by Western blot (WB) analysis using the indicated antibodies (right) to detect the amount of endogenous (top) and ectopic Coro1A-EGFPs (top and middle) present in them. The amount of α-tubulin in each cell lysate was used as internal loading control (bottom). (E) Representative confocal images of HEK 293T cells expressing indicated EGFP-tagged Coro1A versions (top, green) and stained with rhodamine-labeled phalloidin (red). Potential colocalization areas between Coro1A proteins and F-actin had to be seen in yellow. Scale bars, 10 μm. Insets, enlarged images of the indicated cell areas (white open squares). (F) Confocal Z-stack three-dimensional (3D) reconstructions of Jurkat cells expressing the indicated ectopic Coro1A proteins (green signals) and stained with rhodamine-labeled phalloidin (red signals). Potential areas of colocalization between Coro1A and F-actin had to be seen in yellow (bottom). Scale bar, 5 μm. (G) Confocal Z-stack 3D reconstructions of thymocytes isolated from mice of indicated genotypes (top) and stained with antibodies to Coro1A (first and third rows, red signals) and Alexa Fluor 635–labeled phalloidin (middle and bottom, blue signals). Potential areas of colocalization between Coro1A and F-actin had to be seen in purple (bottom). Scale bar, 2.5 μm.
Coro1AE26K shows increased interactions with actin in immunoprecipitation experiments
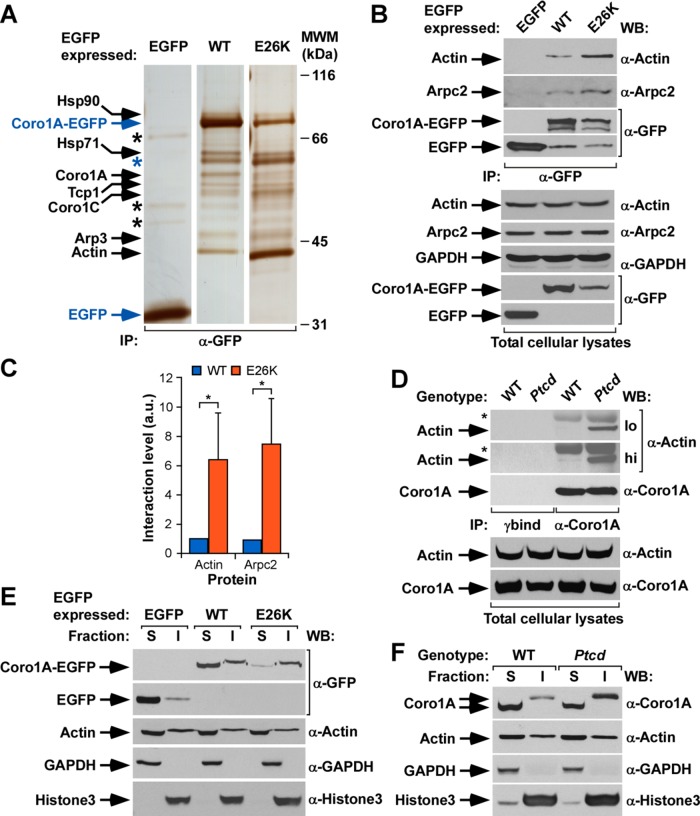
Given the lack of staining with usual markers for cytoskeletal, intermediate filaments and microtubules, we next carried out proteomics experiments to get clues about the possible nature of the filaments generated by Coro1AE26K. To this end, we compared by electrophoresis and mass spectrometry the protein complexes formed by EGFP-tagged wild-type Coro1A and Coro1AE26K in COS1 cells. As negative control, we performed similar experiments using the nonchimeric EGFP. These analyses revealed that the two Coro1A-EGFPs, but not the EGFP alone, can associate with the heat shock proteins of 90 (Hsp90, Mowse score 130; see Materials and Methods) and 71 (Hsp71, Mowse score 219) kDa, the T-complex protein 1 chaperonin (Tpc1, Mowse score 107), Coro1C (Mowse score 156), actin isoforms (β and γ; scores 258 and 276, respectively), and the Arp3 subunit of the Arp2/3 complex (Mowse score 205; Figure 2A). We also detected a band corresponding to bovine serum albumin (BSA) that was nonspecifically brought down in the immunoprecipitation of both Coro1A-EGFPs but not with the nonchimeric EGFP bait (Figure 2A, blue asterisk, Mowse score 173). Whereas most of those proteins coimmunoprecipitated at similar amounts with both the wild-type and E26K versions of Coro1A, we found that the actin isoforms and, to a much lesser extent, the Arp3 subunit and Coro1C associated at much higher amounts with the mutant Coro1A version (Figure 2A, compare lanes 2 and 3). This observation suggests that the E26K mutation probably increases either the affinity or the intracellular formation of Coro1A–actin complexes. The increased in vivo association of Coro1AE26K-EGFP with endogenous actin and Arp2/3 complex proteins (Arpc2, also known as p34Arc) was confirmed using coimmunoprecipitation experiments in transiently transfected COS1 cells (Figure 2, B, top, and C). This is even more conspicuous when these coimmunoprecipitation analyses are performed using endogenous proteins present in cell lysates derived from wild-type and homozygous Ptcd thymocytes (Figure 2D, top). Consistent with these results, we found that Coro1AE26K-EGFP and endogenous Coro1AE26K are preferentially localized in detergent- insoluble fractions of COS1 cells (Figure 2E) and Ptcd thymocytes (Figure 2F), respectively. These fractions are typically enriched in cytoskeletal and nuclear components and exclude proteins loosely attached to these cell structures. By contrast, wild-type Coro1A is distributed in both the cytosolic and insoluble fractions under both experimental conditions (Figure 2, E and F). Despite the differential distribution shown by Coro1AE26K in these experiments, we did not observe any enrichment of actin in the insoluble pellets obtained from Coro1AE26K-expressing COS1 cells (Figure 2E, third from top) and Ptcd thymocytes (Figure 2F, second from top). This suggests that the mutant protein recruits only a relatively small fraction of the total F-actin present in cells. The detection of other proteins in the expected cytosolic (ectopic EGFP, endogenous glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) and insoluble (endogenous histone H3) fractions confirmed the proper purification of those fractions in both COS1 cells (Figure 2E, bottom) and Ptcd thymocytes (Figure 2F, bottom).
FIGURE 2:
The Coro1AE26K mutant has increased F-actin binding capabilities. (A) Silver-stained polyacrylamide electrophoresis gel showing the proteins that coimmunoprecipitate with indicated ectopic proteins (top) in COS1 cells. Proteins of interest are indicated on the left, with the bait proteins highlighted in blue. Only mass spectrometry hits with scores >56 were considered (see Materials and Methods). The Coro1A band likely corresponds to proteolytic fragments of the baits used. Black asterisks mark nonspecific bands detected in the control EGFP immunoprecipitation. The blue asterisk highlights a contaminant BSA band present in the coimmunoprecipitations made with the Coro1A baits (see text). Molecular weight markers (MWMs) are shown on the right. IP, immunoprecipitation. (B) EGFP immunoprecipitates (IPs) obtained from COS1 cells expressing the indicated ectopic proteins (top) were analyzed by Western blot to detect the amount of coimmunoprecipitated endogenous actin (top) and Arpc2 (second from top) in each experimental condition. As control, filters were immunoblotted with antibodies to EGFP (third and fourth from top) to visualize the amount of immunoprecipitated EGFP obtained in each sample. Amount of actin (fifth from top) and Arpc2 (sixth from top), GAPDH (seventh from top), and ectopic EGFPs (bottom two ) present in cell supernatants before the immunoprecipitation step was determined by Western blot analysis of aliquots of the total cell lysates used in the experiments. (C) Quantitation of the coimmunoprecipitation of indicated Coro1A proteins with endogenous actin and Arpc2 obtained in experiments shown in B. Values normalized according to the total amount of Coro1A-EGFP immunoprecipitated in each case. *p ≤ 0.05 (n = 3 independent experiments). a.u., arbitrary units. (D) Coro1A immunoprecipitates obtained from thymocytes isolated from WT or Ptcd mice were analyzed by Western blot to detect the amount of coimmunoprecipitated endogenous actin (top). As control, filters were immunoblotted with antibodies to Coro1A (third from top) to determine the amount of Coro1A immunoprecipitated in each sample. Amount of actin (fourth from top) and Coro1A (bottom) present in lysates before the immunoprecipitation step was determined by immunoblot analysis of aliquots of total cell lysates used in the immunoprecipitation step. hi, high exposure; lo, low exposure; *immunoglobulin heavy chain of the antibody used to immunoprecipiate Coro1A. γbind, immunoprecipitation done with beads in the absence of the antibody to Coro1A. (E) Distribution of indicated ectopic EGFPs (top) in Triton X-100–soluble (S) and –insoluble (I) fractions of transiently transfected COS1 cells. The distribution of Coro1A-EGFP and nonchimeric EGFP is shown in the first and second from the top, respectively. As control, we detected the distribution of endogenous proteins including actin (third from top), GAPDH (fourth from top), and histone H3 (bottom). (F) Distribution of endogenous Coro1A (WT) and Coro1AE26K (Ptcd) proteins present in thymocytes in the indicated Triton X-100–soluble (S) and –insoluble (I) fractions (top). Other markers used in this experiment are described in E. Similar results were obtained in two additional experiments. In B and D–F, antibodies used in the immunoblot analyses are indicated on the right of the appropriate Western blot strip.
Coro1AE26K filaments show intervening F-actin–rich segments
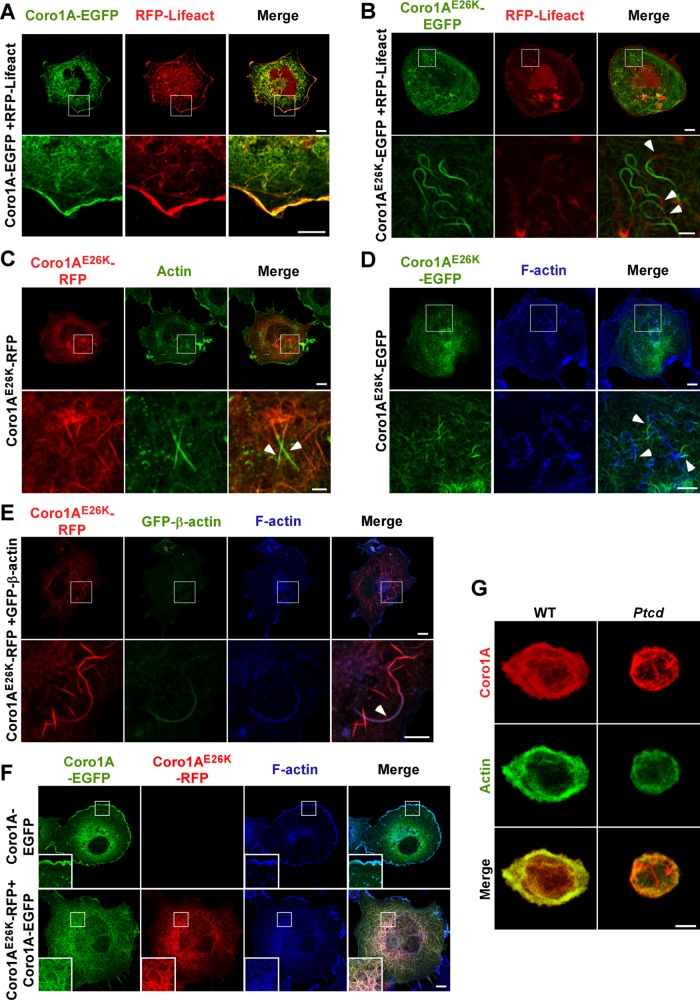
The foregoing results led us to consider whether the filaments induced by Coro1AE26K could represent F-actin filaments that, for unknown reasons, could not be stained by phalloidin. To assess this possibility, we reanalyzed the structure of filaments present in Coro1AE26K-expressing COS1 cells using other techniques that could facilitate the visualization of F-actin enriched structures. As a first experimental avenue, we used RFP-LifeAct, a fusion protein that contains a C-terminal 17–amino acid sequence derived from the Saccharomyces cerevisiae Abp140 actin-binding protein (Riedl et al., 2008). When expressed in cells, this bioreporter allows the visualization of F-actin filaments in both living and fixed cells using standard epifluorescence detection techniques (Riedl et al., 2008). Consistent with this, we observed that Coro1A-EGFP and RFP-LifeAct colocalize in juxtamembrane areas of the transfected COS1 cells (Figure 3A). As a second experimental alternative, we stained Coro1AE26K-EGFP–expressing cells using antibodies to an epitope common to all actin isoforms. We observed using those two avenues that the Coro1AE26K-EGFP–decorated filaments are still mostly negative for both RFP-LifeAct–derived (Figure 3B) and actin-derived (Figure 3C) fluorescence signals. However, a more detailed examination of cells indicated that these fibers are interrupted in some cases by small Coro1AE26K-EGFP–negative segments that do show RFP-LifeAct epifluorescence (Figure 3B, arrows) and actin immunoreactivity (Figure 3C, arrows). Similar results are observed when Coro1AE26K-expressing COS1 cells are stained with Alexa Fluor 635–labeled phalloidin (Figure 3D, arrows) or cotransfected with a GFP-β-actin–encoding plasmid (Figure 3E, arrows). The latter technique was the only one that could find clear areas of colocalization of Coro1AE26K and β-actin in COS1 cells (Figure 3E). By contrast, we observed, using coexpression experiments, that Coro1AE26K does not block the incorporation of wild-type Coro1A in those filaments (Figure 3F). We did not find any substantial colocalization of Coro1AE26K with both endogenously (Supplemental Figure S2A, second column from left) and ectopically (Supplemental Figure S2B) expressed γ-actin, an actin isoform that forms highly stable filaments that are resistant, for example, to latrunculin-induced depolymerization (Baranwal et al., 2012). As control, endogenous γ-actin does show colocalization with ectopically EGFPs fused to wild-type Coro1A (Supplemental Figure S2A, first column from left) and the constitutively active Rac1Q61L and Vav1 oncoprotein (Supplemental Figure S2A, third and fourth columns from left, respectively) in these experiments. Similarly to COS1 cells, the Coro1AE26K-decorated filaments present in Ptcd thymocytes are also poorly stained with antibodies to actin (Figure 3G). Collectively these results indicate that the E26K mutation favors the formation of both Coro1A–F-actin complexes and Coro1A-decorated filaments in cells. These filaments may represent F-actin structures that cannot be properly recognized by common F-actin binding reagents or, alternatively, structures composed of independent Coro1AE26K- and F-actin–positive segments (see Discussion).
FIGURE 3:
Coro1AE26K-nucleated filaments contain F-actin. (A, B) Confocal images of COS1 cells coexpressing RFP-LifeAct (A and B, red signals) in the presence of either Coro1A-EGFP (A, green signals) or Coro1AE26K-EGFP (B, green signals). In A and B, enlarged images of indicated cell areas (white open squares) are shown at the bottom. Potential areas of colocalization between RFP-LifeAct and Coro1A proteins had to be seen in yellow. Some of the filament areas found positive for RFP-LifeAct epifluorescence are indicated by arrowheads (B). Scale bars, 10 μm (top), 5 μm (bottom); same scales in C–F. (C) Confocal images of Coro1AE26K-RFP–expressing COS1 cells stained with antibodies to actin. Ectopic Coro1A and endogenous actin proteins are seen in red and green, respectively. Areas of colocalization had to be seen in yellow in the right column. Enlargements of indicated cell areas (white open squares) are shown in the appropriate bottom images. Some of the filament areas found positive for actin immunoreactivity are indicated by arrows. (D) Coro1AE26K-EGFP–expressing cells were stained with Alexa Fluor 635–phalloidin and analyzed by confocal microscopy. Enlargements of the indicated cell areas (top, white open squares) are shown in the appropriate bottom images. Coro1AE26K-EGFP and F-actin signals are seen in green and blue, respectively. Areas of colocalization had to be seen in white. Arrowheads show segments of filaments that are phalloidin-positive (bottom). (E) COS1 cells coexpressing Coro1AE26K-RFP and GFP–β-actin were stained with Alexa Fluor 635–phalloidin and analyzed by confocal microscopy. Enlargements of the indicated cell areas (top, white open squares) are shown in the appropriate bottom images. Epifluorescence signals for Coro1A and β-actin are shown in red and green color, respectively. F-actin signals are seen in blue. Areas of colocalization for those two proteins are seen in purple. Arrowhead shows a filament segment that displays significant colocalization between these two proteins (bottom). (F) COS1 cells expressing the indicated combination of wild-type and Coro1AE26K proteins (top) were stained with Alexa Fluor 635–labeled phalloidin and analyzed by confocal microscopy. RFP and EGFP epifluorescence signals are seen in red and green, respectively. F-actin–rich areas are in blue. Areas of colocalization between RFP- and EGFP-tagged proteins are in yellow. Colocalization areas for RFP, EGFP, and F-actin had to be seen in white. Insets, enlarged images of the indicated cell areas (white open squares). (G) Confocal Z-stack 3D reconstructions of thymocytes isolated from mice of indicated genotypes (top) and stained with antibodies to Coro1A (top and bottom; red signals) and to actin (middle and bottom; green signals). Potential areas of colocalization between Coro1A and actin had to be seen in yellow (bottom). Scale bar, 2.5 μm.
Coro1AE26K-induces highly stable and functionally inert filaments
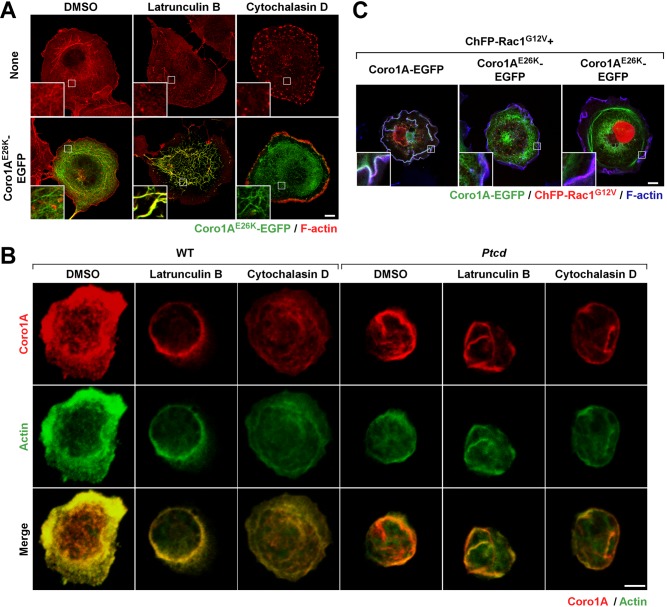
Further experiments indicated that the filaments formed by both ectopically and endogenously expressed Coro1AE26K proteins are more stable than the conventional F-actin structures found in control cells, as determined by their resistance to conventional treatments with cytoskeleton-disrupting agents such as latrunculin B and cytochalasin D (Figure 4, A and B). Of interest, the latrunculin B treatment of COS1 cells (Figure 4A, compare bottom, left and middle) and Ptcd thymocytes (Figure 4B, compare fourth and fifth columns from left) favors the staining of some of the partially collapsed Coro1AE26K-EGFP–positive filaments with phalloidin, further indicating that these structures contain F-actin at least in some intervening segments. Some staining with phalloidin is also observed when Ptcd thymocytes, but not COS1 cells ectopically expressing Coro1AE26K-EGFP (Figure 4A, compare bottom, left and right ), are treated with cytochalasin D (Figure 4B, compare fourth and sixth columns from left).
FIGURE 4:
Coro1AE26K limits F-actin filament turnover. (A) Confocal images of Coro1AE26K-EGFP–expressing COS1 cells treated with the indicated compounds (top) and stained with rhodamine-labeled phalloidin. Insets, enlarged image of the indicated cell areas (white open squares). Color code for confocal signals is indicated at the bottom. Areas of colocalization are seen in yellow (bottom). Scale bar, 10 μm. (B) Confocal Z-stack 3D reconstructions of thymocytes isolated from mice of indicated genotypes (top) and treated with indicated compounds (top) and stained with antibodies to Coro1A (top and bottom row) and to actin (second and third from top). Color code for confocal signals is indicated at the bottom. Areas of colocalization between Coro1A and actin are seen in yellow (bottom row). Scale bar, 2.5 μm. (C) Effect of ChFP-Rac1G12V in the cytoskeleton of Coro1A-EGFP– (left) and Coro1AE26K-EGFP–expressing (second and third from left) COS1 cells. To visualize F-actin, transfected cells were stained with Alexa Fluor 635–phalloidin. Insets, enlarged image of the indicated cell areas (white open squares). Color code for confocal signals is indicated at the bottom. Areas of triple colocalization are seen in white. Scale bar, 10 μm.
Confirming the high stability of these filaments, we found that they cannot be disassembled upon the ectopic coexpression of a constitutively active, Cherry fluorescent protein (ChFP)–tagged version of Rac1 (G12V mutant) in COS1 cells (Figure 4C). As a consequence, the Coro1AE26K-EGFP remains filament bound under those conditions and, unlike the case of wild-type Coro1A-EGFP (Figure 4C, left, and Supplemental Videos S1 and S2), does not show any detectable translocation to the large membrane ruffles induced by Rac1G12V (Figure 4C, middle and right, and Supplemental Videos S3 and S4). Coro1AE26K-EGFP decreases the size, but does not eliminate, the Rac1G12V-induced membrane ruffling generated in those cells (Figure 4C and Supplemental Videos S3 and S4), suggesting that Coro1AE26K does not act as a dominant-negative mutant even under overexpression conditions. This stability is also found in the case of endogenous Coro1AE26K, since the F-actin filaments positive for this protein are excluded from the ruffles and cell/substratum contact zone of Ptcd thymocytes (Figures 1F and 4B). Finally, we observed that the Coro1AE26K-nucleated filaments also remain intact upon the addition of a myosin II inhibitor (blebbistatin) to COS1 cells (Supplemental Figure S3). Collectively these results indicate that Coro1AE26K represents a nondominant, gain-of-function mutant protein that leads to the formation of a highly stable and functionally inert meshwork of filaments in cells.
The E26K mutation has a dominant effect on Coro1A actin- binding properties
The Glu-26 residue is located on a surface-exposed Coro1A region that also contains Arg-29, a residue critical for the actin-binding properties of this protein (Castro-Castro et al., 2011; Ojeda et al., 2014; Figure 5A). Given the strong positive effect of the E26K mutation on the actin-binding properties of Coro1A, we decided to investigate whether it could rescue the loss-of-function effect induced by the R29D missense mutation. Unlike the case of Coro1AE26K (Figure 5B, second row from top), we observed that Coro1AR29D cannot associate to the F-actin cytoskeleton when expressed in COS1 cells (Figure 5B, compare first and third rows from top), does not bind to actin and Arpc2 when tested in coimmunoprecipitation experiments (Figure 5C, compare lane 3 with lanes 1 and 2), and, as a consequence, is found preferentially localized in detergent-soluble cell fractions (Figure 5D, compare lanes 5 and 6 with lanes 1 and 2 and lanes 3 and 4). This is consistent with previous observations made with both Coro1AR29D and Coro1BR30D mutant proteins (Cai et al., 2007; Castro-Castro et al., 2011; Ojeda et al., 2014). By contrast, proteins with the compound E26K and R29D mutations do not show the foregoing defects and behave similarly to the single-Coro1AE26K mutant protein when transfected in COS1 cells (Figure 5, B–D). However, the activity of the Coro1AE26K+R29D seems to be lower than that shown by Coro1AE26K, as judged by the detection of a small fraction of Coro1AE26K+R29D in active areas of membrane ruffling (Figure 5B, compare second and fourth from top).
To confirm that the effect of the E26K mutation is in-cis, we next compared the activity of histidine (His)-tagged Coro1A and Coro1AE26K proteins purified from Escherichia coli (Figure 5E) on the organization of in vitro–polymerized actin. We observed that whereas the addition of the wild-type protein does not induce any overt effect on F-actin (Figure 5F), Coro1AE26K under the same conditions promotes the aggregation of polymerized actin filaments into thicker and larger bundles (Figure 5F). Taken together, these results suggest that the E26K mutation has a direct, intrinsic effect on the actin- binding and -bundling activity of Coro1A.
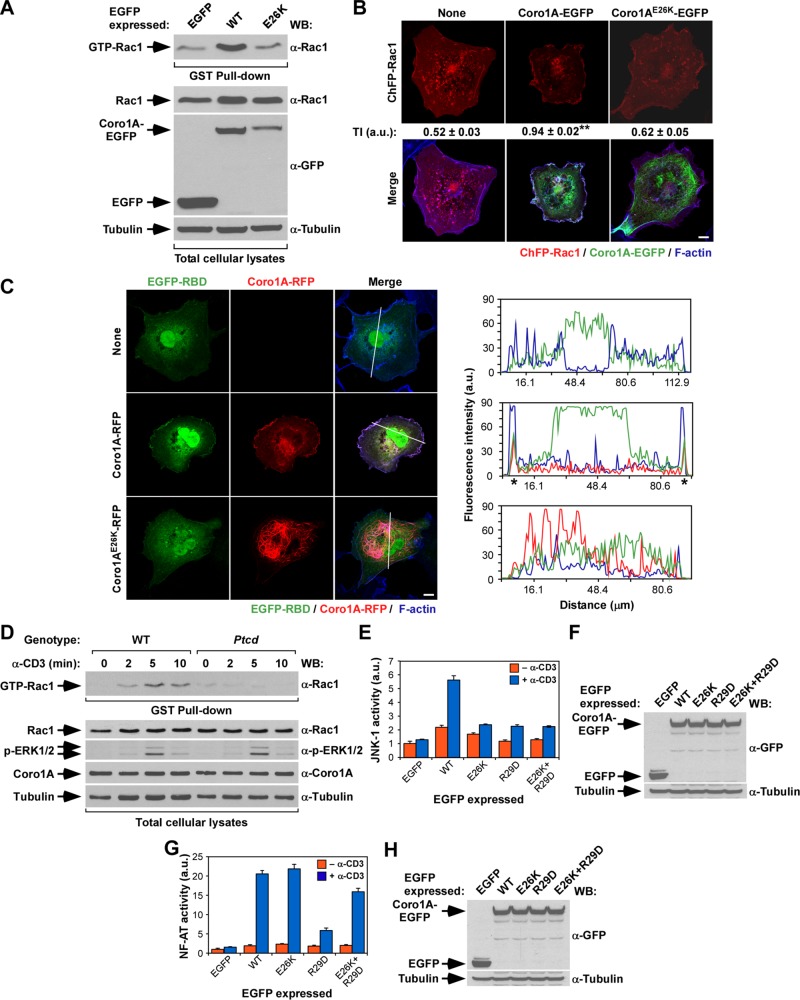
The E26K mutation specifically eliminates the Coro1A-dependent activation of Rac1
We recently showed that Coro1A participates in a signaling loop that favors the activation of Rac1 in some cell types (Castro-Castro et al., 2011; Bustelo et al., 2012; Ojeda et al., 2014). To assess the effect of the E26K mutation in this process, we first used pull-down experiments with the glutathione S-transferase (GST) fused to the Rac1-binding domain (RBD) of Pak to measure the activation of endogenous Rac1 induced upon the ectopic expression of either Coro1AE26K-EGFP or its wild-type counterpart. Unlike the case of wild-type Coro1A (Figure 6A, compare first and second lanes from left; Castro-Castro et al., 2011), we found that Coro1AE26K does not induce any significant activation of Rac1 when ectopically expressed in COS1 cells (Figure 6A, compare lanes 2 and 3). Consistent with this, we found that Coro1A, but not Coro1AE26K, induces the translocation of both the ChFP-labeled Rac1 (Figure 6B) and the EGFP-labeled Pak RBD (Figure 6C) from the cytosol to the plasma membrane when ectopically expressed in COS1 cells (Castro-Castro et al., 2011). This defect is conserved in Ptcd thymocytes, because we found, using GST-RBD pull-down experiments, that the T-cell receptor–mediated activation of Rac1 is severely compromised in these cells (Figure 6D, top). It is also specific, because T-cell receptor–stimulated Ptcd thymocytes show Erk phosphorylation kinetics similar to that exhibited by wild-type cells (Figure 6D, third from top). In relation with the foregoing responses, we showed previously, using luciferase-based reporter plasmids, that Coro1A, but not the actin binding–deficient Coro1AR29D mutant protein, can promote the stimulation of both c-Jun N-terminal kinase (JNK) and nuclear factor of activated T-cells (NF-AT) when ectopically expressed in those cells (Castro-Castro et al., 2011). JNK is a well-known downstream element of the Rac1 route, whereas the stimulation of NF-AT requires the orchestration of more complex signaling outputs, including T-cell receptor–, phospholipase C-γ–, and calcineurin phosphatase–dependent signals (Woodrow et al., 1993a, b; Lopez-Lago et al., 2000; Kaminuma et al., 2001). Coro1A elicits these two responses using an actin- and ArhGEF7-dependent mechanism (Castro-Castro et al., 2011). Unlike the case of wild-type Coro1A (Castro-Castro et al., 2011), we observed that the ectopic expression of Coro1AE26K-EGFP does not lead to any detectable levels of JNK activation in either nonstimulated or CD3-stimulated Jurkat cells (Figure 6, E and F). Consistent with this, and unlike the case of the cytoskeleton-related readouts performed in COS1 cells (Figure 5), this mutation could not restore the activation of this downstream signaling pathway when combined with the R29D loss-of-function mutation (Figure 6, E and F). By contrast, we observed that Coro1AE26K does promote high levels of NF-AT activity in Jurkat cells (Figure 6, G and H). Furthermore, the inclusion of the E26K mutation in Coro1AR29D overcomes the deleterious effects that the latter mutation induces in that biological response (Figure 6, G and H). Thus these results indicate that the E26K mutation induces different effects in actin binding (elevation), the Rac1-JNK pathway (inactivation), and the NF-AT route (normal signaling).
FIGURE 6:
The E26K mutation impairs the Coro1A-mediated activation of Rac1. (A) Top, activation levels of endogenous Rac1 upon expression in COS1 cells of indicated EGFPs. Bottom, aliquots of total cell lysates used in the foregoing experiment were immunoblotted to monitor the amount of total Rac1 (second from top) and EGFPs (third from top) present in the lysates used in the pull-down experiments. α-Tubulin was used as loading control (bottom; n = 3). Note that the E26K mutant shows lower abundance here due to its preferential localization in the nonsoluble fraction (see Figure 2). (B) Amount of cytosolic–plasma membrane translocation of ChFP-Rac1 in the presence of the indicated Coro1A-EGFPs (top). On transfection of appropriate plasmids, cells were fixed, stained with Alexa Fluor 635–labeled phalloidin, and subjected to confocal microscopy to visualize ChFP-Rac1 (top and bottom, red signals), Coro1A-EGFP proteins (bottom, green signals), and F-actin (bottom, blue signals). Areas of colocalization between Rac1, Coro1A, and F-actin are shown in white (bottom). Scale bar, 10 μm. The Rac1 translocation index (TI) is indicated below the first row of panels (see Materials and Methods for determination of this index). **p ≤ 0.01 compared with control cells (n = 3). (C) Left, levels of translocation of EGFP fused to the Rac1-binding domain of Pak1 (EGFP-RBD) from the cytosol to the plasma membrane in the presence of indicated Coro1A-RFP proteins (left). On transfection, cells were fixed, stained with fluorochrome-labeled phalloidin, and subjected to confocal analysis to visualize the EGFP-RBD bioreporter (left, green signals), Coro1A-RFP (middle, red signals) and F-actin (right, blue signals). Areas of colocalization of the three proteins are shown in white (right). The white lanes present on cells (right) highlight the cell sections selected in each case to measure the distribution of fluorescence intensities for EGFP-RBD (green lines), Coro1A-RFP (red lines), and F-actin (blue lines) in the graphs on the right. (D) Top, GST-RBD pull down showing the amount of GTP-Rac1 present in wild-type and Ptcd thymocytes under the indicated stimulation conditions (top). Bottom, aliquots of the same cell lysates were analyzed by Western blot to detect the abundance and/or phosphorylation levels of proteins indicated on the left (bottom four). The antibodies used in the immunoblot analysis are indicated on the right. (E–H) Activation of JNK (E) and NF-AT (G) induced by the indicated ectopically expressed proteins in nonstimulated (–α-CD3) and CD3-stimulated (+α-CD3) Jurkat cells. Data represent mean and SD of a representative experiment performed in triplicate. The abundance of ectopic proteins (F and H, top) and α-tubulin (loading control; F and H, bottom) in the JNK (F) and NF-AT (H) experiments is shown. Same results were obtained in a second independent experiment, also performed in triplicate.
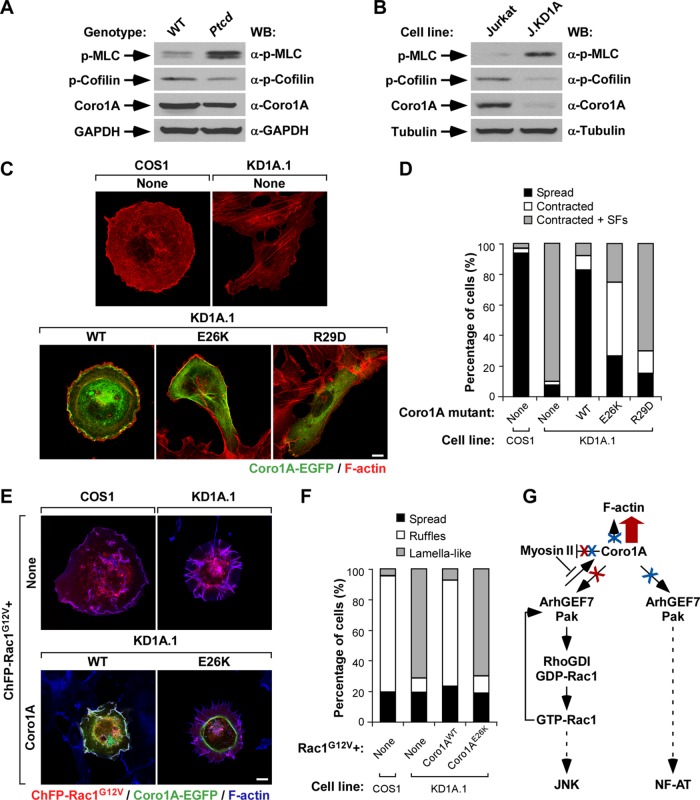
The E26K mutation impairs the Coro1A-dependent myosin II inactivation step
In addition to favoring Rac1 stimulation, we recently showed that Coro1A is important for proper myosin II inactivation. Owing to this, the depletion of Coro1A leads to elevated amounts of myosin light chain phosphorylation, increased actomyosin contractility, and the formation of highly elongated cells that containing large amounts of phospho-myosin light chain–positive stress fibers (Ojeda et al., 2014). These defects can be eliminated using the myosin II inhibitor blebbistatin (Ojeda et al., 2014). This regulatory step is also particularly important for the signaling of active Rac1/ArhGEF7/Pak complexes because, in the absence of Coro1A, these complexes become trapped in actomyosin-rich structures and promote the formation of lamella-like cell protrusions instead of membrane ruffles and lamellipodia (Ojeda et al., 2014). The E26K mutation also impairs the Coro1A-mediated myosin II regulatory step, because Ptcd thymocytes show higher amounts of myosin light chain phosphorylation than do controls (Figure 7A). This effect is comparable to that seen in Jurkat cells upon the short hairpin RNA–mediated depletion of the endogenous Coro1A protein (Figure 7B; Ojeda et al., 2014). In both cases, this effect is not linked to parallel elevations in the amount of phosphorylated (inactive) cofilin (Figure 7, A and B; Ojeda et al., 2014). Consistent with these results, we observed that the ectopic expression of Coro1AE26K-EGFP could not eliminate the actomyosin-driven elongated shape and stress fiber formation typically exhibited by CORO1A-deficient COS1 cells (Figure 7, C and D). Similar effects are seen when these cells are transfected with the inactive Coro1AR29D mutant (Figure 7, C and D; Ojeda et al., 2014). However, as expected (Ojeda et al., 2014), these cells do acquire normal shapes and cytoskeletal organization when transfected with wild-type Coro1A (Figure 7, C and D). Also consistent with the lack of proper myosin II inactivation, we found that the ectopic expression of Coro1AE26K-EGFP does not eliminate the defective cytoskeletal responses exhibited by CORO1A-knockdown cells upon the expression of active versions of Rac1 (Figure 7, E and F). As a result, these cells exhibit the characteristic formation of lamellar structures composed of radial extensions of F-actin filaments decorated by both phalloidin and active Rac1 (Figure 7, E and F). In agreement with the high stability previously observed for the Coro1AE26K-induced filaments (Figure 4), we observed that both Coro1AE26K and the Coro1AE26K-decorated filaments are not present in any of those structures (Figure 7E, bottom). On the contrary, they accumulate under those conditions in a thin and multifilament ring-like perinuclear structure (Figure 7E, bottom). These Coro1AE26K-positive filaments remain resistant to phalloidin staining despite being located in overlapping positions to the actomyosin ring (Figure 7E, bottom right). Taken together, these results indicate that Coro1AE26K behaves as a noncompetitive gain-of-function or loss-of-function mutant protein, depending on the expression status of Coro1A.
FIGURE 7:
The E26K mutation affects the Coro1A-mediated regulation of myosin II and downstream Rac1-Pak signaling. (A, B) Total cellular extracts from thymocytes from mice of indicated genotypes (A, top) and the indicated Jurkat T-cell lines (B, top) were analyzed by Western blot to detect the amount of phospho-myosin light chain (MLC; top), phospho-cofilin (second from top), and Coro1A (third from top). Loading controls used were GAPDH (A, bottom) and α-tubulin (B, bottom). Antibodies used in the immunoblot analyses are indicated on the right. Jurkat, wild-type Jurkat cells; J.KD1A, CORO1A-knockdown Jurkat cells; p-, phospho. (C, D) Representative images (C) and quantification (D; n = 3; 150 cells/transfection) of the effect of indicated Coro1A-EGFPs in the cytoskeletal and morphological phenotype of CORO1A-knockdown COS1 cells (referred to as KD1A.1 cells). In C, color code for confocal signals is indicated below the images. Colocalization of Coro1A-EGFP and F-actin is shown in yellow. Scale bar, 10 μm. (E, F) Representative images (E) and quantification (F; n = 3; 150 cells/condition) of the effect of indicated Coro1A-EGFPs in the cytoskeletal defects shown by ChFP-Rac1G12V–expressing CORO1A-knockdown COS1 cells. In E, the code for confocal signals is indicated below the images. Areas of colocalization of active Rac1, Coro1A-EGFPs proteins, and F-actin are shown in white. Scale bar, 10 μm. (G) Summary of results obtained in this work. The depicted pathway is based on observations made in this work and previous publications (Castro-Castro et al., 2011; Ojeda et al., 2014). In all cases, activation and inactivation steps are indicated by arrows and blunted lanes, respectively. Broken lanes indicate signaling cascades. Enhanced and reduced activities exhibited by the Coro1AE26K mutant protein are indicated by thick arrows and crosses (both in red), respectively. Effects induced by the actin-binding deficient Coro1AR29D mutation are shown in blue. RhoGDI, Rho GDP dissociation inhibitor. See Discussion for further details.
DISCUSSION
We showed here that Coro1AE26K, a mutant version of Coro1A found in immunodeficient Ptcd mice, causes quite variegated effects (gain of function, inhibition, neutral) in the normal functions of this protein. The gain-of-function effect leads to the formation of a highly stable and functionally inert meshwork of Coro1AE26K-decorated filaments in cells that remain segregated away from active sites of actin remodeling. This effect is associated with exacerbated in vivo actin-binding and in vitro bundling activities of the mutant protein (Figure 7G, thick red arrow). We detected these filaments in both transfected COS1 cells and thymocytes obtained from Ptcd mice, indicating that this phenotype cannot be solely the consequence of the overexpression of the ectopically expressed mutant protein. Consistent with this, we showed that the wild-type and E26K versions of Coro1A, which are expressed at comparable amounts in cells, do not induce the same cellular and signaling phenotypes when overexpressed in cells. It is also unlikely that they represent nonspecific aggregation events of the mutant protein, because 1) other null Coro1A mutant proteins remain cytosolic and do not form this type of networks; 2) the formation of nonspecific aggregates does not seem consistent with the preservation of some of the biological activities of the wild-type protein by Coro1AE26K (NF-AT stimulation) and, of greater importance, with the functional rescue observed when this mutation is combined with Coro1A mutants that cannot bind actin; 3) it is unclear why a nonspecific aggregate would specifically trap actin and not other potential binding partners; and 4) there is always a correlation between the cytoskeletal phenotype observed and the increased association of Coro1A mutants with actin in vivo.
Of interest, these filaments can only be sparsely stained with phalloidin, LifeAct, and other actin-binding reagents. Taking into consideration that a single Coro1A molecule can bind to two actin protomers in one filament and an additional one located in the opposite F-actin strand (Galkin et al., 2008), we favor the possibility that these structures are abnormal Coro1AE26K-F-actin structures that cannot be stained with standard markers due to a Coro1AE26K-mediated steric effect. A similar lack of staining with phalloidin and LifeAct is found, for example, in cytoskeletal structures induced by cofilin mutants (McGough et al., 1997) and in actin rods (Nishida et al., 1987; Munsie et al., 2009), a type of cofilin-rich, short F-actin filament structure formed in some stressed cells and pathological conditions (Bamburg et al., 2010). However, rods can be effectively stained with antibodies to actin (Nishida et al., 1987; Munsie et al., 2009). Alternatively, it can be argued that the structures detected in the present study can be composed of independent segments of Coro1AE26K-positive and F-actin–positive filaments. Although we cannot formally exclude this possibility, this model seems at odds with the high amounts of actin found in Coro1AE26K immunoprecipitates, the direct correlation of actin coimmunoprecipitation levels with the formation of those structures by each of the Coro1A mutant proteins used in our study, and the localization of the E26K mutation in a Coro1A region potentially involved in active actin binding (see later discussion). Furthermore, the F-actin–rich areas present in those filaments do seem to contain smaller amounts of the Coro1AE26K mutant proteins. The observation that Coro1AE26K can stimulate the NF-AT route also seems to stand against the formation of filaments lacking actin, since we previously showed that this biological response requires the physical interaction of Coro1A with F-actin (Castro-Castro et al., 2011).
The reason for the observed increased interaction of ectopic and endogenous Coro1AE26K proteins with actin isoforms in cells is unknown. However, it is worth noting that the Glu-26 residue is located in one the blades of the Coro1A β-propeller domain that, according to evolutionary conservation criteria and overall basic electrostatic charge, may harbor one of the Coro1A actin-binding sites (Appleton et al., 2006). This blade also contains another key residue (Arg-29, Appleton et al., 2006), which, when mutated into aspartate, eliminates the interaction of Coro1A with actin (Castro-Castro et al., 2011). Electron microscopy and in silico modeling experiments suggest that this region may contact two actin protomers present in opposite strands of the F-actin filament (Galkin et al., 2008). It can be speculated, therefore, that the Glu-26 residue could have a fine-tuning role in the regulation of the affinity of the Coro1A/F-actin complexes in order to make possible proper cycles of complex formation and disassembly during cytoskeletal processes. If this were the case, the E26K mutation would break such a mechanism by favoring the formation of long-lasting Coro1A/actin complexes and highly stable F-actin filaments. This is consistent with previous observations indicating that actin-binding proteins use basic residues to facilitate electrostatic interactions with acidic-rich regions of actin (Tang and Janmey, 1996; Amann et al., 1998). Alternatively, it could be argued that this mutation could facilitate the interaction with actin via the elimination of inhibitory interactions with other amino acid sequences in Coro1A. This scenario, for example, has been seen in the case of the K255E mutation found in α-actinin-4, a cross-linker actin protein involved in focal segmental glomerulosclerosis (Kaplan et al., 2000; Weins et al., 2007). We believe that this possibility is unlikely because, if that were the case, the inclusion of the E26K mutation could not result in the rescue of actin binding of actin-deficient mutant versions. In any case, the elucidation of the atomic structure of such complexes will be required to fully understand the gain-of-function effect of this mutation. Of importance, Glu-26 is highly evolutionarily conserved (either as such or as an aspartate residue), thus indicating that it will probably play similar roles in most coronin family members. Previous results showed that Coro1AE26K has a more potent Arp2/3 inhibitory activity than the wild-type counterpart when tested in vitro (Shiow et al., 2008). We surmise that this effect must be an indirect consequence of the increased association of the mutant protein with actin in vivo, because crystal and electron microscopy structural data indicate that Glu-26 and the rest of the first blade of the β-propeller domain are not in physical proximity to the C-terminal coiled-coil region that regulates Arp2/3 binding (Cai et al., 2005; Appleton et al., 2006; Galkin et al., 2008). Such interpretation is consistent with previous data indicating that the binding of some coronins to F-actin facilitates better interactions with the Arp2/3 complex in cells (Cai et al., 2005).
Unlike the foregoing defects, we found that the E26K mutation leads to the inactivation of the Coro1A-dependent pathways that contribute to the stimulation of both Rac1 and the downstream Rac1-JNK pathway, the regulation of cytoskeletal changes induced by active Rac1-ArhGEF7-Pak2 complexes, and the inactivation of myosin IIB (Figure 7G, steps with red crosses). However, it does not impair the Coro1A-mediated stimulation of the transcriptional factor NF-AT in Jurkat cells, further indicating that the effects of this mutation are signaling-branch specific and cannot be attributed to the formation of some kind of fully inert intracellular aggregates (Figure 7G). The preservation of the latter function is to some extent puzzling because we previously showed that the Coro1A-mediated stimulation of the Rac1-JNK and NF-AT pathways are both F-actin and ArhGEF7 dependent (Castro-Castro et al., 2011). This indicates that the NF-AT route is probably regulated by Coro1A using an ArhGEF7-dependent but Rac1-independent mechanism (Figure 7G). This interpretation is consistent with previous reports indicating that ArhGEF7 can stimulate NF-AT in T-cells independently of Rac1 using a Pak- and phospholipase C-γ–dependent pathway (Kuhne et al., 2000; Phee et al., 2005). Alternatively, it could be speculated that an ArhGEF7-Cdc42-Pak–dependent pathway not affected by the E26K mutation could modulate this route (Manser et al., 1998; Rosenberger and Kutsche, 2006). This possibility seems unlikely, given that Coro1A does not promote the membrane translocation, activation, or effector functions of Cdc42 (Castro-Castro et al., 2011; Ojeda et al., 2014). Regardless of the cause, these data are interesting because they suggest that the previously described F-actin–dependent activation of the Rac1-JNK axis by Coro1A specifically depends on cell rheological status, active cytoskeletal dynamics, or the subcellular localization where such a stimulation step takes place. By contrast, the F-actin–dependent stimulation of the NF-AT pathway by both Coro1A and Coro1AE26K seems to exclusively require the assembly of any type of cytoskeleton regardless of its mechanical, turnover, and subcellular localization properties (Figure 7G).
Despite all of those cytoskeletal and signaling defects, we found that the expression of the Coro1AE26K is not deleterious, provided that the expression of the endogenous Coro1A protein is preserved in cells. Thus we observed that Coro1AE26K-expressing cells do not show the typical morphological changes exhibited by CORO1A-knockdown cells under both basal and constitutive Rac1 signaling conditions. Furthermore, we could detect only mild defects in the dynamics of membrane-ruffling formation when constitutively active versions of Rac1 were coexpressed with Coro1AE26K in COS1 cells. However, Coro1AE26K does not compensate for the loss of endogenous wild-type protein, and, as a consequence, it cannot rescue any of the cell shape and cytoskeletal defects exhibited by CORO1A-knockdown cells under both basal and constitutively active Rac1 expression conditions. Taken together, these results suggest that the pathological effects induced by Coro1AE26K in the immunodeficient Ptcd mouse strain are caused by a nonconventional gain-of-function effect that, due to the trapping of the mutant protein in nonproductive, signaling inert filaments, does not act in a dominant manner over the wild-type protein. These results probably explain the lack of overt defects that had been observed in heterozygous Ptcd mice (Shiow et al., 2008). Whether the Coro1AE26K mutant protein can act in a dominant manner in cells in which the actin concentration could represent a more limiting factor remains to be determined. To date, this mutation has been only found in immunodeficient mice. Because it is not a lethal one, it would be interesting to see whether this or analogous mutations arise in the near future in humans and, if so, whether they exert similar or distinctive phenotypes to those shown by unstable or loss-of-function mutants. It would be also interesting to carry out a comprehensive mutational approach in Coro1A to identify further amino acid residues and subdomains involved in this cytoskeletal and signaling phenotype.
Taken together, these results provide a new mechanistic scenario to explain the contribution of the Coro1A E26K mutation to the pathology of Ptcd mice and, in addition, reveal its implication in the nucleation of a quite unconventional filament network in cells. This mutant version might represent and interesting tool to further dissect the structural and signaling roles of Coro1A in cells.
MATERIALS AND METHODS
Generation of mammalian and bacterial expression vectors
Mammalian expression vectors encoding wild-type human Coro1A-EGFP (pCor-GFP[WT]), Coro1A-RFP (pCor-RFP[WT]), and Coro1AR29D-EGFP (pACC53) were previously described (Castro-Castro et al., 2011). Those encoding Coro1AE26K-EGFP (pVOS15) and Coro1AD278V-EGFP (pVOS38) were generated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) to incorporate either the E26K or the D278V single point mutations into the human CORO1A cDNA present in pCor-GFP[WT]. Sequences of mutagenesis primers were 5′-CCG ACC AGT GCT ATA AAG ATG TGC GCG TC-3z (forward, E26K mutation underlined), 5′-GAC GCG CAC ATC TTT ATA GCA CTG GTC GG-3′ (reverse, E26K mutation underlined), 5′-GCC CTT CTT TGA CCC TGT CAC CAA CAT CGT CTA CC-3′ (forward, D278V mutation underlined), and 5′-GGT AGA CGA TGT TGG TGA CAG GGT CAA AGA AGG GC-3′ (reverse, D278V mutation underlined). The expression vector encoding the RFP-tagged E26K Coro1A mutant (pVOS30) was obtained by site-directed mutagenesis using pCor-RFP[WT] as template and the oligonucleotides described. To generate the plasmid encoding Coro1AE26K+R29D-EGFP (pVOS32), we mutagenized the pVOS15 using oligonucleotides 5′-CCA GTG CTA TAA AGA TGT GGA CGT CTC ACA GAC CAC C-3′ (forward, mutations underlined) and 5′-GGT GGT CTG TGA GAC GTC CAC ATC TTT ATA GCA CTG G-3′ (reverse, mutations underlined). The pRFP-Ruby-N1-LifeAct (Ojeda et al., 2014) plus vectors encoding ChFP-tagged Rac1 (pmCherry-Rac1; Castro-Castro et al., 2011) and ChFP-tagged Rac1G12V (pmCherry-Rac1G12V; Ojeda et al., 2014) were provided by M. A. del Pozo (Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain). The expression vectors encoding EGFP-Rac1Q61L (pNM42), EGFP-Vav1Onc (N-terminal deletion lacking the N-terminal CH and Ac regions, pNM108), and the Pak1 RBD domain fused to EGFP (pJRC45) were previously described (Lopez-Lago et al., 2000; Ruiz et al., 2007; Castro-Castro et al., 2011). The GFP-β-actin (pGreen-FP-vector-actin; 558721)– and mCherry-γ-actin (mCherry-Gamma-Actin2-C-18; 55049)–encoding expression vectors were from BD PharMingen (San Jose, CA) and Addgene (Cambridge, MA), respectively. The pFA2-cJun (JNK activation assays) luciferase reporter plasmid was from Agilent Technologies. The pNF-AT-Luc (used in NF-AT activation assays) was provided by G. Crabtree (Stanford University Medical School, Stanford, CA). The control pRL-SV40 plasmid encoding the Renilla luciferase under the control of a constitutive promoter was from Promega (Durham, NC).
To generate the bacterial expression vectors encoding His-tagged Coro1A (pVOS33) and Coro1AE26K (pVOS34), XhoI/BamHI-digested fragments obtained from pCor-GFP[WT] and pVOS15 were ligated independently into different aliquots of the XhoI-BglII-linearized pTrcHisA plasmid (Invitrogen, Carlsbad, CA). All plasmids were sequence verified in the Genomics and Proteomics Unit of our Center. The vector (pGST-Pak-BD) encoding the GST-Rac1–binding domain of Pak1 was provided by R. A. Cerione (Department of Molecular Medicine, College of Veterinary Medicine, Cornell University, Ithaca, NY).
Immunological reagents
Rhodamine- and Alexa Fluor 635–labeled phalloidin were obtained from Molecular Probes/Invitrogen. Rabbit polyclonal antibodies to Coro1A, myosin IIB, and Arpc2 were purchased from Encor Biotechnology (Gainesville, FL), Covance (Princeton, NJ), and Merck Millipore (Darmstadt, Germany), respectively. Mouse monoclonal antibodies to vimentin and rabbit polyclonal antibodies to histone H3 were from Abcam (Cambridge, UK). Mouse monoclonal antibodies to pancytokeratin, rabbit polyclonal antibodies to (GADPH, and goat antibodies to all actin isoforms were obtained from Santa Cruz Biotechnology (Dallas, TX). Mouse monoclonal antibodies to γ-actin, α-tubulin, and GFP were from Serotech (Burlington, ON, Canada), Calbiochem/Merck Millipore, and Covance, respectively. The mouse and hamster antibodies to human and mouse CD3 were from Merck Millipore and BD PharMingen, respectively. Rabbit polyclonal antibodies to phospho-myosin light chain, phospho-cofilin, and phospho-Erk were from Cell Signaling (Boston, MA). When required, appropriate Cy2- and Cy3-labeled secondary antibodies were used (Jackson ImmunoResearch, West Grove, PA).
Cell lines and tissue culture
COS1 and HEK 293T cells were maintained at 37°C and a 5% CO2 atmosphere in DMEM supplemented with 10% fetal bovine serum (FBS) plus 1% l-glutamine plus 100 U/ml of both penicillin and streptomycin (Sigma-Aldrich, St. Louis, MO). When indicated, COS1 cells were incubated with 2 μM cytochalasin D (15 min; Sigma-Aldrich), 10 μM latrunculin B (20 min; Enzo Life Sciences, Farmingdale, NY), or 50 μM blebbistatin (2 h; Sigma-Aldrich). Jurkat cells were maintained as described in RPMI-1640 supplemented with 10% FBS and 100 U/ml penicillin and streptomycin. When necessary, Jurkat cells were stimulated for the indicated periods of time with antibodies to human CD3 (10 μg/ml; Castro-Castro et al., 2011; Barreira et al., 2014). The stable CORO1A-knockdown COS1 (KD1A.1 cell clone) and Jurkat (J.KD1A cell pool) cells were generated, characterized, and cultured as described elsewhere (Castro-Castro et al., 2011; Ojeda et al., 2014).
Transient transfections
COS1 and HEK 293T cells were grown onto six-well plates and transfected using Lipofectamine 2000 (Invitrogen). To that end, 1 μg of the appropriate plasmid DNA and 3 μl of Lipofectamine 2000 were diluted separately in 100 μl of serum-free DMEM, and the two solutions were mixed, incubated for 20 min at room temperature, and then added onto cells. Transfected cells were trypsinized 24 h later, seeded onto 0.001% polylysine (Sigma-Aldrich)-coated coverslips, and fixed 24 h later. To transfect Jurkat cells, 20 μg of plasmid DNA was added to 2 × 106 cells diluted in 2 ml of R buffer (Invitrogen) and the mix subjected to two electroporation cycles at 1.7 mV (20 ms each) using the Neon transfection system (Invitrogen). Electroporated cells were transferred to standard culture media for 36 h and subsequently plated onto 0.001% polylysine-coated coverslips, allowed to settle for 20 min, fixed, and stained.
Isolation and culturing of thymocytes from Ptcd mice
All animal work was done following the protocols approved by the bioethics committees of both the University of Salamanca and the Consejo Superior de Investigaciones Científicas. Cataract-Shionogi (CTS/Shi) mice carrying the Ptcd locus (Ikegami et al., 1988; Shiow et al., 2008) in the B6 genetic background were provided by J. Serody (University of North Carolina at Chapel Hill, Chapel Hill, NC). Genotyping of these mice was done using a quantitative reverse transcription-PCR approach with three simultaneous primers to distinguish the natural wild-type and Ptcd alleles of the Coro1A gene according their specific cycle threshold (Ct) amplification values. Oligonucleotides used were 5′-CAA GGC TGA CCA GTG CTA AG-3′ (forward for wild-type sequence), 5′-CAA GGC TGA CCA GTG CTA AA-3′ (forward for Ptcd allele sequence), and 5′-GTT GAC AGC GCA GAA GCC A-3′ (reverse, common for both alleles). The two forward oligonucleotides will yield different Ct depending on their specific match with either wild-type or mutant DNA sequences.
To isolate thymocytes from those mice, single-cell suspensions were obtained by mechanical homogenization of thymi in 3 ml of phosphate-buffered saline (PBS) solution supplemented with 2% BSA and 0.5 mM EDTA, washed once by low-speed centrifugation and resuspension in the buffer, subjected to 0.17 M NH4Cl lysis to eliminate erythrocytes, and washed twice as described. For immunofluorescence experiments, final cell pellets were resuspended in PBS solution, seeded onto mouse anti–CD3-coated coverslips (2 μg/ml; BD Biosciences), allowed to settle for 10 min, fixed, and stained as described. When indicated, cells were treated with latrunculin B and cytochalasin D as described. For immunoblot and biochemical determinations, thymocytes were kept in suspension and, when indicated, stimulated with hamster antibodies to mouse CD3 (10 μg/ml) for the indicated periods of time.
Immunofluorescence experiments
Samples were fixed, permeabilized, stained, and mounted as previously described for most of the confocal microscopy experiments presented in this work (Castro-Castro et al., 2011). However, in the case of γ-actin localization experiments, we followed a specific protocol (Dugina et al., 2009). Briefly, cells were fixed in 1% paraformaldehyde in prewarmed DMEM for 30 min, treated with MetOH at −20ºC for 5 min, permeabilized in TBS-T (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Tween-20) for 10 min, washed three times with TBS-T, blocked in a 2% BSA in TBS-T for 30 min, and then stained with antibodies to γ-actin for 3 h and the secondary antibody (diluted in blocking solution) for 1 h. In all cases, stained preparations were mounted onto microscope slides using Mowiol (Calbiochem) and analyzed using a Leica SP5 confocal microscope and a 63× objective (Leica). Microscope images were exported and processed for presentation using Canvas 9.0.4 software (ACD Systems, Seattle, WA). When required, profiles of fluorescence intensity along cells were made using ImageJ software (National Institutes of Health, Bethesda, MD). The Rac1 translocation index obtained in each experimental condition was calculated as previously described (Castro-Castro et al., 2011). To this end, ChFP-positive cells were selected from confocal images obtained under the indicated experimental conditions and scored 0 for no detectable Rac1 in the plasma membrane, 1 for weak Rac1 plasma membrane staining, and 2 for high Rac1 localization in the plasma membrane. The Rac1 translocation index to the plasma membrane was calculated as (y + 2z)/(x + y + z), where x, y, and z are the numbers of cells scored as 0, 1, or 2, respectively. When histograms are included, they refer to the percentage of a given phenotype in the total population of fluorescent protein–expressing cells. In these cases, we define as “spread,” “contracted,” and “contracted + stress fibers” cells displaying typical morphology of COS1 cells, cells with smaller areas than control ones, and cells with reduced cell size plus the presence of prominent stress fibers, respectively. When using the term “ruffles,” we refer to cells displaying the large membrane ruffles typically elicited by active versions of Rac1. Only cells displaying ruffles in more than half of the cell perimeter were included in this category. The term “lamella-like” is used to describe cells displaying unpolarized cytoplasmic extensions that were supported by radial, phalloidin- and Rac1-decorated F-actin filaments. Quantification was done using 150 cells in three independent experiments.
Proteomics analyses
On GFP-trap immunoprecipitation and electrophoretic fractionation of protein complexes, desired silver-stained gel slices were manually excised, destained with a 1:1 solution of 15 mM K3[Fe(CN)6] (Merck) and 50 mM Na2S2O3 (Merck) for 15 min, rinsed with water, and dehydrated with 100% CH3CN (Fisher). Plugs were then treated stepwise with 10 mM dithiothreitol (Sigma-Aldrich) in 25 mM (NH4)HCO3 (Sigma-Aldrich) at 56ºC for 45 min and 55 mM iodoacetamide (Sigma-Aldrich) in 50 mM (NH4)HCO3 at room temperature for 30 min and rinsed with 100% CH3CN and 20 mM (NH4)HCO3. Modified porcine trypsin (Promega) at a final concentration of 3 ng/μl in 20 mM (NH4)HCO3 was added to gel pieces and incubated at 37°C for 16 h. Plugs were then acidified by adding 1 μl of 5% HCOOH (Merck), incubated for 15 min at 37ºC, and finally dried in a speed vacuum system. For matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) peptide mass fingerprinting, a 0.5-μl aliquot of matrix solution (5 g/l 2,5-dihydroxybenzoic acid in a 33% aqueous solution of CH3CN containing 0.1% CF3CO2H [Merck]) plus 1 μl of the aforementioned peptide extraction solution was manually loaded onto a 400-μm-diameter AnchorChip target plate probe (Bruker Daltonics, Billerica, MA) and allowed to dry at room temperature. Samples were analyzed on an Ultraflex MALDI-TOF mass spectrometer (Bruker Daltonics) using an acquisition mass range of 600–4000 Da. To this end, the equipment was first externally calibrated using protonated mass signals from a standard peptide calibration mixture containing seven peptides (Peptide Calibration Standard #206196; Bruker Daltonics) covering the m/z range 1000–3200. Every spectrum was internally calibrated using selected signals arising from trypsin autoproteolysis (842.510- and 2211.105-Da peptides) to reach a typical mass measurement accuracy of ±30 ppm. Each raw spectrum was opened in FlexAnalysis 3.0 (Bruker Daltonics) software and processed and analyzed using the following parameters: signal-to-noise threshold of 1, Savitzky–Golay algorithm for smoothing, tangential algorithm for baseline subtraction, and centroid algorithm for monoisotopic peak assignment. In all cases, resolution was >9000. All known contaminants (trypsin autoproteolysis and known keratin peaks) were excluded during the process.
Peaks were sent to the Mascot Server (version 2.1) using Bio Tools 3.0 (Bruker Daltonics) software and searched against SwissProt database (SwissProt 2011_09; 532,146 sequences; 188,719,038 residues) for protein identification. Searches were carried out using the following parameters: Homo sapiens species (20,249 sequences), up to one missed tryptic cleavage, mass accuracy of 100 ppm, MH+ monoisotopic masses, carbamidomethyl cysteine as fixed modification, and methionine oxidation as variable modification. Mowse scores with a value >56 for SwissProt database were considered as significant (p < 0.05). All proteomics studies were carried out by technical personnel of the Genomics and Proteomics Unit of our Center.
Immunoprecipitation, pull-down, and blotting experiments
Cell lysis, electrophoresis, and Western blot techniques were done exactly as previously described (Castro-Castro et al., 2011). Immunoprecipitations of EGFP-tagged proteins were carried out using the GFP-trap reagent (ChromoTek, Planneg-Martinsried, Germany). To analyze Triton X-100–soluble and –insoluble fractions, cells were lysed as described, and pellets obtained were solubilized in 80 mM Tris-HCl (pH 6.8) containing 2% SDS, 10% glycerol, 0.02% bromophenol, and 5% mercaptoethanol. The determination of Rac1-GTP levels in transfected COS1 cells and CD3-stimulated thymocytes from control and Ptcd mice was done using pull-down experiments with the GST-Rac1 binding domain of Pak1 fusion protein as previously indicated (Castro-Castro et al., 2011). In all cases, proteins and total cellular extracts were analyzed by SDS–PAGE and immunoblot following standard procedures. In all cases, protein signals in Western blot analyses were revealed using chemiluminescence with horseradish peroxidase coupled to secondary antibodies (GE Healthcare Life Biosciences, Buckinghamshire, UK).
Video recordings
Subconfluent cells plated onto 35-mm culture plates were transfected with ChFP-Rac1G12V and either pCor-GFP (WT) or pVOS15 (encoding Coro1AE26K-EGFP) plasmids and, 24 h later, analyzed using a 20× objective (Olympus) in a DeltaVision microscope (Image Solutions). Images were taken using the Resolve3D softWoRx-Aquire software (version 5.5.0). Capture settings were 0.8-s exposure time with 1-s intervals. Videos are played at 10 frames/s.
Protein purification from E. coli
Exponentially growing cultures E. coli DH5α cells containing either the pVOS33 or pVOS34 plasmid were treated with 1 mM isopropyl-β-d-1-thiglucopyranoside (Roche Applied Science, Basel, Switzerland) for 2 h at 30°C. Cells were then harvested by centrifugation at 5500 rpm for 15 min at 4°C and lysed by sonication in lysis buffer (50 mM phosphate buffer, pH 8.0, 300 mM NaCl, and 10 mM imidazole). After centrifugation at 10,000 rpm for 10 min at 4°C, supernatants were collected and incubated in batch with nickel-nitriloacetic acid agarose beads (Qiagen, Limburg, The Netherlands) at 4°C under continuous rotation. After 2 h, beads were collected by centrifugation and washed three times in lysis buffer with 20 mM imidazole, and proteins were eluted in lysis buffer containing 250 mM imidazole. The GST-Rac1 binding domain of Pak1 protein was induced as described and purified using glutathione-coated beads (GE Healthcare). Beads with the purified protein were stored frozen until experimental use. In all cases, protein concentration was determined by running aliquots of both the purified His-tagged protein and GST fusion protein–coated beads in SDS–PAGE in the presence of known concentrations of BSA as standard and stored at –20°C until further use.
Studies with in vitro–polymerized actin
F-actin was obtained by adding 5 μl of a 10-fold-concentrated actin polymerization buffer (100 mM Tris-HCl, pH 7.5, containing 1 M KCl, 20 mM MgCl2, and 10 mM ATP) to 45 μl of a purified rabbit muscle actin solution (2 μg/μl; A2522; Sigma-Aldrich) followed by a 1-h-long incubation at room temperature. The solution obtained was incubated with rhodamine-labeled phalloidin (70 nM) for 20 min at room temperature and subsequently supplemented with 1 μM indicated His-tagged Coro1A proteins purified from E. coli. After 15 min at room temperature, aliquots of the incubation mix were placed between a microscope slide and a coverslip, sealed, and analyzed by immunofluorescence microscopy using a 63× objective (Leica) in a Leica DM 6000B microscope.
JNK and NF-AT luciferase reporter assays
These techniques have been described elsewhere (Barreira et al., 2014).
Image processing
Images were assembled and processed for final figure presentation using Canvas 9.0.4 (ACD Systems).
Statistical analyses
Data from at least three independent experiments were analyzed using the Student's t test. p ≤ 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
We thank M. Blázquez and personnel of both the Centro de Investigación del Cáncer Microscopy and the Genomics and Proteomics units for technical assistance. We also thank M. Dosil for comments on the manuscript. This work has been supported by grants to X.R.B. from the Castilla-León Autonomous Government (CSI101U13), the Spanish Ministry of Economy and Competitiveness (SAF2012-31371, RD12/0036/0002), the Solórzano Foundation, and the Ramón Areces Foundation. Spanish government–sponsored funding is partially supported by the European Regional Development Fund.
Abbreviations used:
- ArhGEF7
Arh GDP/GTP exchange factor 7
- Arp2/3
actin-related proteins 2 and 3
- Arpc2
actin-related protein 2/3 complex subunit 2
- a.u.
arbitrary units
- BSA
bovine serum albumin
- ChFP
cherry fluorescent protein
- Coro1A
coronin 1A; Coro1B, coronin 1B; Coro1C, coronin 1C
- EGFP
enhanced green fluorescent protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GDI
GDP dissociation inhibitor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- Hsp71
heat shock protein of 71 kDa
- Hsp90
heat shock protein of 90 kDa
- I
insoluble
- IP
immunoprecipitation
- JNK
c-Jun N-terminal kinase
- MLC
myosin light chain
- MWM
molecular weight marker
- NF-AT
nuclear factor of activated T-cells
- Pak
p21-activated kinase
- Ptcd
peripheral T-cell deficient
- RBD
Rac1-binding domain of Pak
- RFP
red fluorescent protein
- S
soluble
- Tpc1
T-complex protein 1
- WB
Western blot
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0052) on June 24, 2015.
V.O. carried out most of the experimental work reported in this article, analyzed data, and generated figures. J.R.-V. carried out the experiments shown in Figures 1, C and D, and 3A and Supplemental Figure S1. M.B. performed luciferase reporter assays in Jurkat cells (Figure 6, E–H). X.R.B. directed the work, analyzed data, and wrote the manuscript
The authors declare that they have no competing interests.
REFERENCES
- Amann KJ, Renley BA, Ervasti JM. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J Biol Chem. 1998;273:28419–28423. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- Appleton BA, Wu P, Wiesmann C. The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14:87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bernstein BW, Davis RC, Flynn KC, Goldsbury C, Jensen JR, Maloney MT, Marsden IT, Minamide LS, Pak CW, et al. ADF/Cofilin-actin rods in neurodegenerative diseases. Curr Alzheimer Res. 2010;7:241–250. doi: 10.2174/156720510791050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranwal S, Naydenov NG, Harris G, Dugina V, Morgan KG, Chaponnier C, Ivanov AI. Nonredundant roles of cytoplasmic beta- and gamma-actin isoforms in regulation of epithelial apical junctions. Mol Biol Cell. 2012;23:3542–3553. doi: 10.1091/mbc.E12-02-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreira M, Fabbiano S, Couceiro JR, Torreira E, Martinez-Torrecuadrada JL, Montoya G, Llorca O, Bustelo XR. The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins. Sci Sig. 2014;7:ra35. doi: 10.1126/scisignal.2004993. [DOI] [PubMed] [Google Scholar]
- Bustelo XR, Ojeda V, Barreira M, Sauzeau V, Castro-Castro A. Rac-ing to the plasma membrane. The long and complex work commute of Rac1 during cell signaling. Small GTPases. 2012;3:1–7. doi: 10.4161/sgtp.19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280:31913–31923. doi: 10.1074/jbc.M504146200. [DOI] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. J Cell Sci. 2007;120:1779–1790. doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, Couceiro JR, Pimentel-Muinos FX, Del Pozo MA, Bustelo XR. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011;30:3913–3927. doi: 10.1038/emboj.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugina V, Zwaenepoel I, Gabbiani G, Clement S, Chaponnier C. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J Cell Sci. 2009;122:2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- Foger N, Jenckel A, Orinska Z, Lee KH, Chan AC, Bulfone-Paus S. Differential regulation of mast cell degranulation versus cytokine secretion by the actin regulatory proteins Coronin1a and Coronin1b. J Exp Med. 2011;208:1777–1787. doi: 10.1084/jem.20101757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foger N, Rangell L, Danilenko DM, Chan AC. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science. 2006;313:839–842. doi: 10.1126/science.1130563. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Brieher W, Kueh HY, Mitchison TJ, Egelman EH. Coronin-1A stabilizes F-actin by bridging adjacent actin protomers and stapling opposite strands of the actin filament. J Mol Biol. 2008;376:607–613. doi: 10.1016/j.jmb.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsson MK, Louis-Dit-Sully CA, Lawson BR, Sternik G, Santiago-Raber ML, Gascoigne NR, Theofilopoulos AN, Kono DH. The lupus-related Lmb3 locus contains a disease-suppressing Coronin-1A gene mutation. Immunity. 2008;28:40–51. doi: 10.1016/j.immuni.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtta-Vuori M, Vainio S, Kauppi M, Van Eck M, Jokitalo E, Ikonen E. Endosomal actin remodeling by coronin-1A controls lipoprotein uptake and degradation in macrophages. Circ Res. 2012;110:450–455. doi: 10.1161/CIRCRESAHA.111.256842. [DOI] [PubMed] [Google Scholar]
- Ikegami H, Makino S, Harada M, Eisenbarth GS, Hattori M. The cataract Shionogi mouse, a sister strain of the non-obese diabetic mouse: similar class II but different class I gene products. Diabetologia. 1988;31:254–258. doi: 10.1007/BF00290594. [DOI] [PubMed] [Google Scholar]
- Jayachandran R, Gatfield J, Massner J, Albrecht I, Zanolari B, Pieters J. RNA interference in J774 macrophages reveals a role for coronin 1 in mycobacterial trafficking but not in actin-dependent processes. Mol Biol Cell. 2008;19:1241–1251. doi: 10.1091/mbc.E07-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran R, Sundaramurthy V, Combaluzier B, Mueller P, Korf H, Huygen K, Miyazaki T, Albrecht I, Massner J, Pieters J. Survival of mycobacteria in macrophages is mediated by coronin 1-dependent activation of calcineurin. Cell. 2007;130:37–50. doi: 10.1016/j.cell.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Kaminuma O, Deckert M, Elly C, Liu YC, Altman A. Vav-Rac1-mediated activation of the c-Jun N-terminal kinase/c-Jun/AP-1 pathway plays a major role in stimulation of the distal NFAT site in the interleukin-2 gene promoter. Mol Cell Biol. 2001;21:3126–3136. doi: 10.1128/MCB.21.9.3126-3136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, Pollak MR. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- Kim GY, Kim H, Lim HJ, Park HY. Coronin 1A depletion protects endothelial cells from TNFalpha-induced apoptosis by modulating p38beta expression and activation. Cell Sig. 2015;27:1688–1693. doi: 10.1016/j.cellsig.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Kuhne MR, Ku G, Weiss A. A guanine nucleotide exchange factor-independent function of Vav1 in transcriptional activation. J Biol Chem. 2000;275:2185–2190. doi: 10.1074/jbc.275.3.2185. [DOI] [PubMed] [Google Scholar]
- Liu C, Wang Y, Zhang H, Cheng S, Charreyre C, Audonnet JC, Chen P, He Q. Porcine coronin 1A contributes to nuclear factor-kappa B (NF-kappaB) inactivation during Haemophilus parasuis infection. PLoS One. 2014;9:e103904. doi: 10.1371/journal.pone.0103904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lago M, Lee H, Cruz C, Movilla N, Bustelo XR. Tyrosine phosphorylation mediates both activation and downmodulation of the biological activity of Vav. Mol Cell Biol. 2000;20:1678–1691. doi: 10.1128/mcb.20.5.1678-1691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace EM, Orange JS. Lytic immune synapse function requires filamentous actin deconstruction by Coronin 1A. Proc Natl Acad Sci USA. 2014;111:6708–6713. doi: 10.1073/pnas.1314975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshous D, Martin E, Carpentier W, Lim A, Callebaut I, Canioni D, Hauck F, Majewski J, Schwartzentruber J, Nitschke P, et al. Whole-exome sequencing identifies Coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated B-cell lymphoproliferation. J Allergy Clin Immunol. 2013;131:1594–1603. doi: 10.1182/blood-2012-07-440339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P, Liu X, Pieters J. Migration and homeostasis of naive T cells depends on coronin 1-mediated prosurvival signals and not on coronin 1-dependent filamentous actin modulation. J Immunol. 2011;186:4039–4050. doi: 10.4049/jimmunol.1003352. [DOI] [PubMed] [Google Scholar]
- Mueller P, Massner J, Jayachandran R, Combaluzier B, Albrecht I, Gatfield J, Blum C, Ceredig R, Rodewald HR, Rolink AG, Pieters J. Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat Immunol. 2008;9:424–431. doi: 10.1038/ni1570. [DOI] [PubMed] [Google Scholar]
- Mugnier B, Nal B, Verthuy C, Boyer C, Lam D, Chasson L, Nieoullon V, Chazal G, Guo XJ, He HT, et al. Coronin-1A links cytoskeleton dynamics to TCR alpha beta-induced cell signaling. PLoS One. 2008;3:e3467. doi: 10.1371/journal.pone.0003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie LN, Caron N, Desmond CR, Truant R. Lifeact cannot visualize some forms of stress-induced twisted F-actin. Nat Methods. 2009;6:317. doi: 10.1038/nmeth0509-317. [DOI] [PubMed] [Google Scholar]
- Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, Sakai H. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci USA. 1987;84:5262–5266. doi: 10.1073/pnas.84.15.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda V, Castro-Castro A, Bustelo XR. Coronin1 proteins dictate rac1 intracellular dynamics and cytoskeletal output. Mol Cell Biol. 2014;34:3388–3406. doi: 10.1128/MCB.00347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phee H, Abraham RT, Weiss A. Dynamic recruitment of PAK1 to the immunological synapse is mediated by PIX independently of SLP-76 and Vav1. Nat Immunol. 2005;6:608–617. doi: 10.1038/ni1199. [DOI] [PubMed] [Google Scholar]
- Punwani D, Pelz B, Yu J, Arva NC, Schafernak K, Kondratowicz K, Makhija M, Puck JM. Coronin-1A: immune deficiency in humans and mice. J Clin Immunol. 2015;35:100–107. doi: 10.1007/s10875-015-0130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G, Kutsche K. AlphaPIX and betaPIX and their role in focal adhesion formation. Eur J Cell Biol. 2006;85:265–274. doi: 10.1016/j.ejcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Santos E, Bustelo XR. RasGRF2, a guanosine nucleotide exchange factor for Ras GTPases, participates in T-cell signaling responses. Mol Cell Biol. 2007;27:8127–8142. doi: 10.1128/MCB.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Tsujimura K, Koide Y. Coronin-1a inhibits autophagosome formation around Mycobacterium tuberculosis-containing phagosomes and assists mycobacterial survival in macrophages. Cell Microbiol. 2012;14:710–727. doi: 10.1111/j.1462-5822.2012.01754.x. [DOI] [PubMed] [Google Scholar]
- Shiow LR, Paris K, Akana MC, Cyster JG, Sorensen RU, Puck JM. Severe combined immunodeficiency (SCID) and attention deficit hyperactivity disorder (ADHD) associated with a Coronin-1A mutation and a chromosome 16p11.2 deletion. Clin Immunol. 2009;131:24–30. doi: 10.1016/j.clim.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow LR, Roadcap DW, Paris K, Watson SR, Grigorova IL, Lebet T, An J, Xu Y, Jenne CN, Foger N, et al. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat Immunol. 2008;9:1307–1315. doi: 10.1038/ni.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray-Pedersen A, Jouanguy E, Crequer A, Bertuch AA, Brown BS, Jhangiani SN, Muzny DM, Gambin T, Sorte H, Sasa G, et al. Compound heterozygous CORO1A mutations in siblings with a mucocutaneous-immunodeficiency syndrome of epidermodysplasia verruciformis-HPV, molluscum contagiosum and granulomatous tuberculoid leprosy. J Clin Immunol. 2014;34:871–890. doi: 10.1007/s10875-014-0074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo D, Park J, Harrington AW, Zweifel LS, Mihalas S, Deppmann CD. Coronin-1 is a neurotrophin endosomal effector that is required for developmental competition for survival. Nat Neurosci. 2014;17:36–45. doi: 10.1038/nn.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JX, Janmey PA. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J Biol Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- Terzi YK, Kocaefe YC, Ayter S. Coronin 1A inhibits neurite outgrowth in PC12 cells. Neurosci Lett. 2014;582:38–42. doi: 10.1016/j.neulet.2014.08.044. [DOI] [PubMed] [Google Scholar]
- Uetrecht AC, Bear JE. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Weins A, Schlondorff JS, Nakamura F, Denker BM, Hartwig JH, Stossel TP, Pollak MR. Disease-associated mutant alpha-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci USA. 2007;104:16080–16085. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow M, Clipstone NA, Cantrell D. p21ras and calcineurin synergize to regulate the nuclear factor of activated T cells. J Exp Med. 1993a;178:1517–1522. doi: 10.1084/jem.178.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow MA, Rayter S, Downward J, Cantrell DA. p21ras function is important for T cell antigen receptor and protein kinase C regulation of nuclear factor of activated T cells. J Immunol. 1993b;150:3853–3861. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.