A Molecular Tumor Board (MTB) was established to interpret individual patients’ tumor genetic profiles and provide treatment recommendations. For the majority of cases evaluated in its first year, the MTB was able to provide treatment recommendations based on targetable genetic alterations. Increasing awareness of molecular profiling and targeted therapies by both clinicians and patients will improve acceptance and adherence to treatments that could significantly improve outcomes.
Keywords: Molecular tumor board, Targeted therapy, Genetic profiling, Next-generation sequencing, Precision medicine
Abstract
Background.
Although genetic profiling of tumors is a potentially powerful tool to predict drug sensitivity and resistance, its routine use has been limited because clinicians are often unfamiliar with interpretation and incorporation of the information into practice. We established a Molecular Tumor Board (MTB) to interpret individual patients’ tumor genetic profiles and provide treatment recommendations.
Patients and Methods.
DNA from tumor specimens was sequenced in a Clinical Laboratory Improvement Amendments-certified laboratory to identify coding mutations in a 50-gene panel (n = 34) or a 255-gene panel (n = 1). Cases were evaluated by a multidisciplinary MTB that included pathologists, oncologists, hematologists, basic scientists, and genetic counselors.
Results.
During the first year, 35 cases were evaluated by the MTB, with 32 presented for recommendations on targeted therapies, and 3 referred for potential germline mutations. In 56.3% of cases, MTB recommended treatment with a targeted agent based on evaluation of tumor genetic profile and treatment history. Four patients (12.5%) were subsequently treated with a MTB-recommended targeted therapy; 3 of the 4 patients remain on therapy, 2 of whom experienced clinical benefit lasting >10 months.
Conclusion.
For the majority of cases evaluated, the MTB was able to provide treatment recommendations based on targetable genetic alterations. The most common reasons that MTB-recommended therapy was not administered stemmed from patient preferences and genetic profiling at either very early or very late stages of disease; lack of drug access was rarely encountered. Increasing awareness of molecular profiling and targeted therapies by both clinicians and patients will improve acceptance and adherence to treatments that could significantly improve outcomes.
Implications for Practice:
Case evaluation by a multidisciplinary Molecular Tumor Board (MTB) is critical to benefit from individualized genetic data and maximize clinical impact. MTB recommendations shaped treatment options for the majority of cases evaluated. In the few patients treated with MTB-recommended therapy, disease outcomes were positive and support genetically informed treatment.
Introduction
Although cancer cells can harbor aberrations in many genes, it is becoming clear that there are a limited number of “driver genes” that, when mutated or deregulated, promote cancer phenotypes. Alterations in such driver genes can predict cancer sensitivity or resistance to targeted therapies, such as EGFR or KRAS mutations and epidermal growth factor receptor (EGFR) kinase inhibitors or monoclonal antibodies [1]. Clinical trials testing novel targeted agents (e.g., inhibitors of phosphatidylinositol 3-kinase [PI3K] or CDK4/6) are using somatic variants as inclusion/exclusion criteria based on evidence that such variants are associated with tumor response. With the expansion of “genetically informed” or “basket” trials, it is becoming increasingly important to incorporate tumor genetic profiling into routine clinical care to inform treatment decisions.
Mounting clinical evidence indicates that genetically informed (“matched”) anticancer therapy provides improved clinical benefit compared with noninformed (“nonmatched”) therapy. In one analysis, 379 patients with advanced cancer harboring an aberration in ≥1 gene (of 7–12 tested) were enrolled into clinical studies with matching (n = 175) or nonmatching (n = 116) agents [2]. The matched group showed a significantly increased objective response rate (ORR; 27% vs. 5%), time-to-treatment failure (5.2 vs. 2.2 months), and overall survival (OS; 13.4 vs. 9 months) compared with the nonmatched group. Analysis of a similar cohort showed improvements in ORR (12% vs. 5%), progression-free survival (3.9 vs. 2.2 months), and OS (11.4 vs. 8.6 months) in a matched group (n = 143) versus a nonmatched group (n = 236) [3]. The adaptive phase II BATTLE trial also indicated that matched therapy provides superior clinical results compared with nonmatched or traditional chemotherapies [4].
Tumor genetic profiling can provide vast amounts of data for each patient. Identified genetic variants must be curated and evaluated to determine whether they may be clinically important and therapeutically actionable (targetable). This information must be distilled and communicated to the referring oncologist. We established a Molecular Tumor Board (MTB) to analyze and interpret patient cases with genetic alterations and to guide treatment decisions. Herein, we provide our framework for the MTB, our format for case evaluation, a summary of 1 year of cases (n = 35), and our experience with anticipated obstacles such as data interpretation and drug access.
Patients and Methods
Human Subjects
The data reported herein have been classified as “not human subjects research” by the Dartmouth College Institutional Review Board.
Tumor Genetic Profiling
All testing was performed in the Clinical Laboratory Improvement Amendments-certified Dartmouth-Hitchcock Medical Center and Norris Cotton Cancer Center Pathology Shared Resource Laboratory using validated methods [5]. After a pathologist’s review, macrodissection was performed to ensure ≥10% tumor cellularity. DNA was extracted from eight 4-μm sections of formalin-fixed paraffin-embedded tumor tissue using the Gentra PureGene Blood Kit Plus (Qiagen, Hilden, Germany, http://www.qiagen.com) and quantified using PicoGreen (Promega, Madison, WI, http://www.promega.com). Barcoded libraries were prepared from 10 ng of DNA using the Ion AmpliSeq Library Kit 2.0 with Ion AmpliSeq Cancer Hotspot Panel v2 as per manufacturer’s protocol (Life Technologies, Rockville, MD, http://www.lifetech.com). This method generates 207 polymerase chain reaction amplicons covering 2,855 COSMIC-cited mutations in 50 cancer-related genes (supplemental online Table 1). Libraries were sequenced using the Ion Torrent Personal Genome Machine System and Ion 318 Chips (Life Technologies). Sequencing reads were aligned to hg19, and variants were called using Torrent Suite Variant Caller Plugin v4.0. Variant annotation was performed using Golden Helix SNP and Variation Suite software v.8.2.1. At this point, filters were applied to remove benign polymorphisms and noncoding and synonymous variants. Our thresholds for calling a variant were ≥5% allelic frequency and ≥500-fold coverage; in our facility, tumor DNA is always sequenced to >1,000-fold average coverage. A report detailing variants detected in the tumor and resultant amino acid changes was included in each patient’s medical record. One tumor specimen was sent to Foundation Medicine (Cambridge, MA, http://www.foundationmedicine.com) for genetic profiling using the Foundation One platform, which probed for mutations and copy number alterations in 236 cancer-related genes and 47 introns of 19 genes involved in rearrangements.
Results
MTB Format
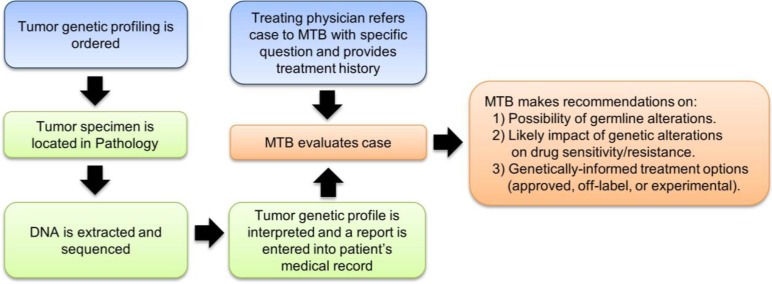
The MTB meets monthly to discuss clinical, laboratory, and scientific information pertinent to patient management. The MTB is comprised of molecular and anatomic pathologists, medical oncologists, hematologists, genetics counselors, and basic science researchers with expertise in cancer genetics, oncogenic signaling pathways, and molecular therapeutics. Each month, three to five patient cases are presented: the clinical history, surgical pathology, and genetic findings are discussed, and treatment and referral recommendation(s) are proposed (Fig. 1). The MTB is also a forum to educate and disseminate information on relevant clinical trials and clinical laboratory updates and to discuss ethical considerations, evaluation of novel analysis software, and challenges in interpretation and application of genetic data.
Figure 1.
Overview of workflow for tumor genetic profiling and MTB case evaluation.
Abbreviation: MTB, Molecular Tumor Board.
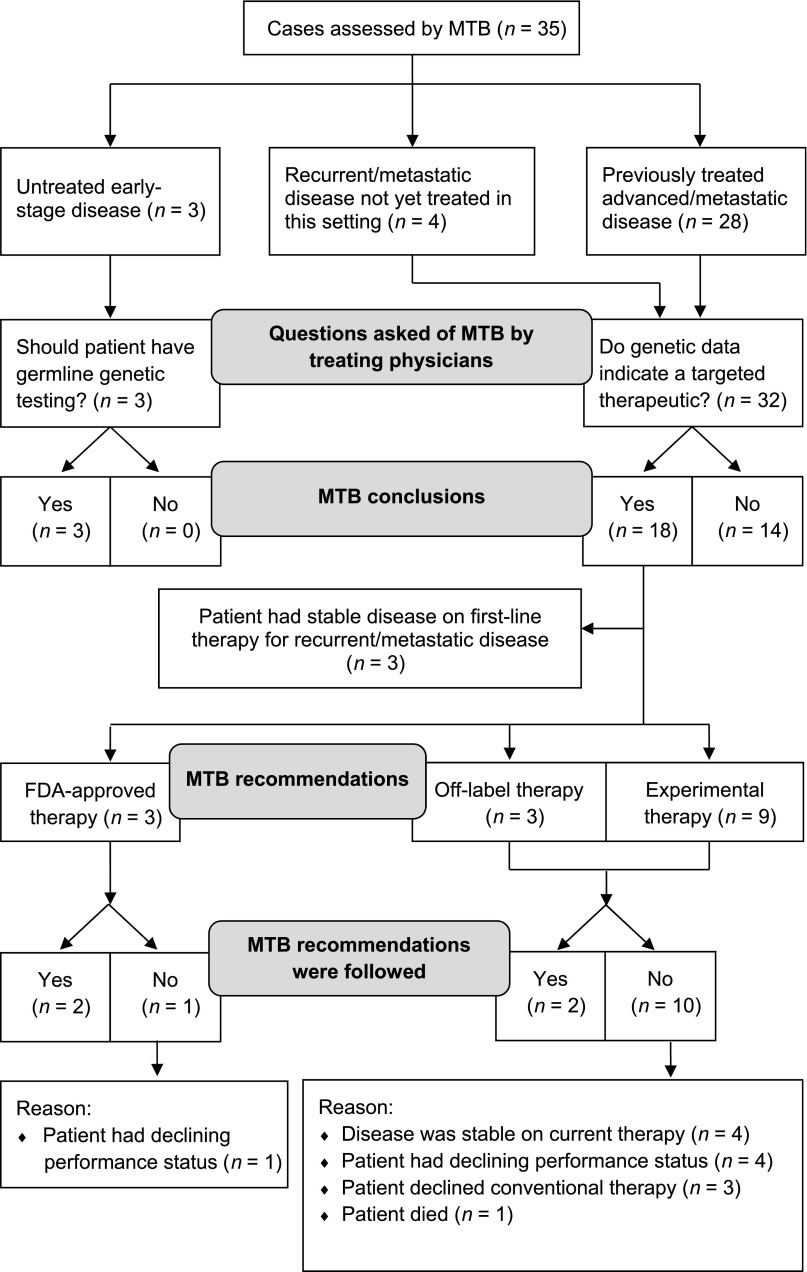
At the request of a treating physician, a case that has undergone tumor DNA sequencing is referred to the MTB with a specific question to be addressed. In our first 35 cases, the majority (91.4%) of referrals requested recommendations on targeted therapies, and the MTB recommended treatment with a targeted agent in 56.3% (18 of 32) of these cases (Fig. 2). Three cases were referred to the MTB to assess the risk of germline variants.
Figure 2.
CONSORT flow diagram of patient cases evaluated by MTB.
Abbreviation: MTB, Molecular Tumor Board.
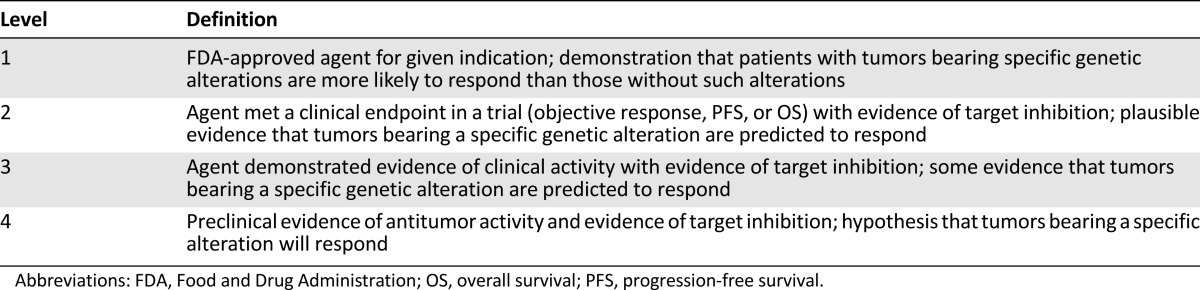
Each case was assigned to a member of the MTB to review publically available tools and databases (e.g., cBio Cancer Genomics Portal [6]) to determine the frequency of a given mutation in large patient populations. We then use databases (e.g., PubMed, COSMIC [7], Google, MutationAssessor [8], UniProt [9], ClinVar [10], and dbSNP [11]) to determine (a) whether a given mutation was previously observed and evaluated, (b) potential for germline mutation, (c) relevant pathway(s) that may be affected by the mutation, (d) available drugs (approved, off-label, or experimental) targeting the affected pathway(s), and (e) the level(s) of evidence (i.e., preclinical in vitro [cell-free vs. cell culture], preclinical in vivo, clinical case report, clinical trial, and phase [Table 1]) supporting a mutation-induced change in protein function and/or drug sensitivity (at the protein, pathway, cellular, or tumoral level). Supplemental online Table 2 summarizes the genetic aberrations identified and the MTB recommendations that were conveyed to the treating physicians.
Table 1.
Levels of evidence supporting targeted therapies recommended by Molecular Tumor Board
Disease Settings Evaluated by Molecular Tumor Board
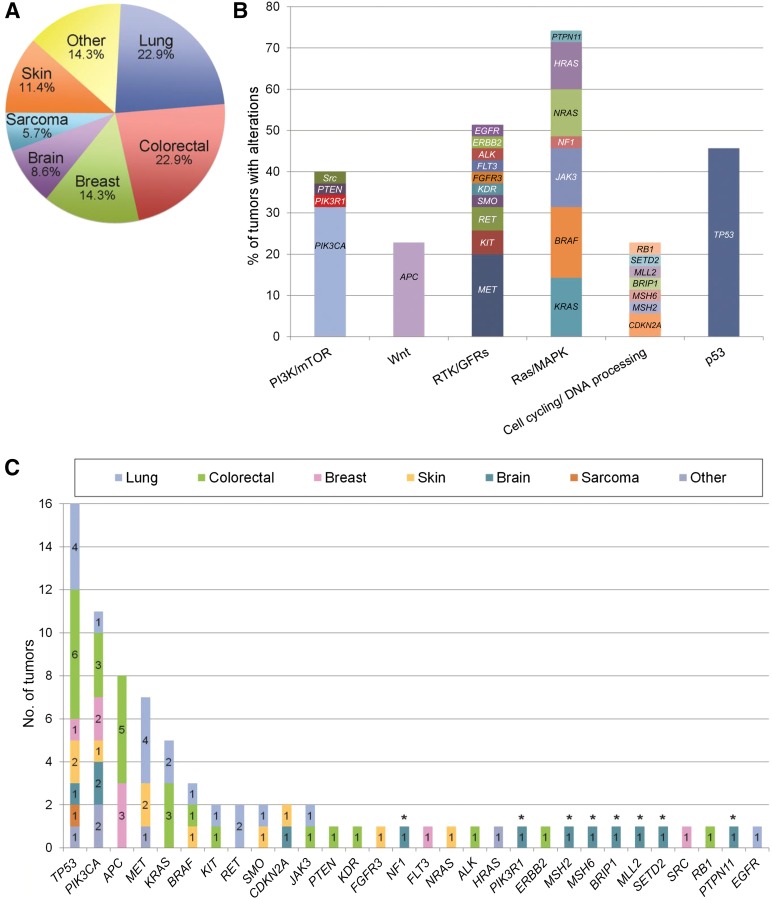
Among the 35 cases evaluated, 3 were patients with early-stage disease who had not yet started treatment, 4 were patients with recurrent or metastatic disease that had not yet been treated in the advanced/metastatic setting, and 28 were patients previously treated for advanced/metastatic disease (median of 2 lines of prior therapy; range of 1–7). In 74.3% of cases (26 of 35), tumor specimens from biopsies performed within 1 year prior to DNA sequencing were used. In the other 9 cases, archived tumor specimens from biopsies performed 2–7 years prior to sequencing were used (median of 3 years). The most common types of cancer evaluated by MTB included lung, colorectal, and breast carcinomas (Fig. 3A). Across 30 genes, 71 different aberrations were found (Fig. 3B), and 63 aberrations were observed only once. Therapeutically actionable alterations were detected in 71.4% of cases (25 of 35). Grouping alterations into functionally relevant pathways (outlined in [12]), we found that potentially actionable alterations frequently occur in the Ras/mitogen-activated protein kinase, PI3K/mammalian target of rapamycin (mTOR), and receptor tyrosine kinase/growth factor receptor pathways (Fig. 3C).
Figure 3.
Tumor types and mutated genes evaluated by the Molecular Tumor Board. (A): Distribution of tumor types among 35 cases. (B): Incidence of aberrations by gene. Colors indicate tumor type in which aberration was identified. (C): Genes were grouped into pathways as in [12], and frequencies of alterations were calculated. ∗, Genes were analyzed by Foundation Medicine for only one patient.
Abbreviations: GFR, growth factor receptor; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; RTK, receptor tyrosine kinase.
Impact of Molecular Tumor Board on Treatment Decisions
The recommendations of the MTB are summarized in Figure 2. Further germline testing was recommended in three of three cases, all of whom had early-stage disease. Of the four patients with recurrent or metastatic disease not previously treated, all received standard-of-care therapy (three had potentially targetable genetic alterations). In 28 cases of previously treated advanced/metastatic disease, the MTB recommended treatment with standard-of-care therapy or non-genetically-informed clinical trials in 13 cases (with recommended future treatment with a targeted agent in 2 of these cases), consideration of treatment with a genetically informed Food and Drug Administration-approved therapy in 3 cases, treatment with a specific targeted agent(s) off-label (off-study) in 3 cases, and treatment with a specific targeted agent(s) in a clinical trial in 9 cases.
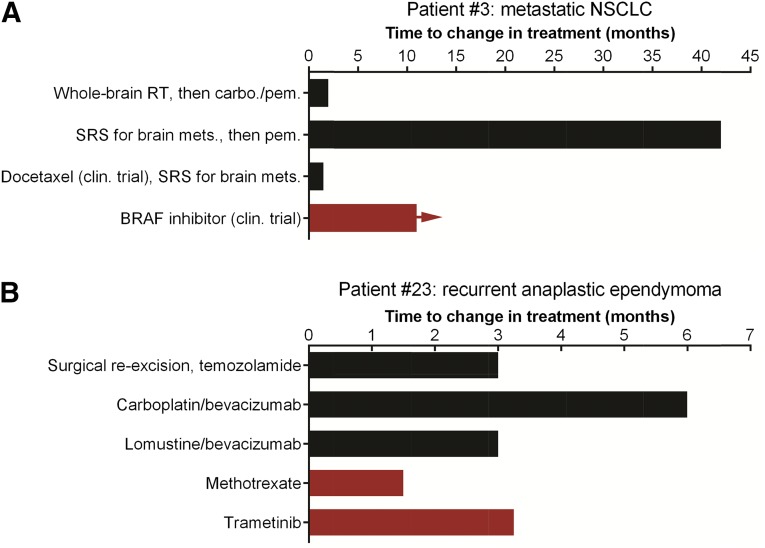
Among the 12 patients with previously treated metastatic disease for whom MTB recommended off-label or experimental treatment with a specific targeted agent(s), 2 patients were subsequently treated with a MTB-recommended therapy (patients 3 and 23). Herein, we provide brief treatment histories of these two cases in which MTB recommendations altered treatment decisions to begin to gauge the effect of a MTB on individual patients’ disease courses (Fig. 4). Although the number of cases is small, the outcomes are notably improved compared with expectations on standard treatments for patients with similar stages of disease.
Figure 4.
Treatment history of the two patients whose management was altered by MTB recommendations. Treatments used are indicated along the y-axis. Times to change in treatment are indicated along the x-axis. Red columns indicate MTB-recommended therapy; horizontal arrow indicates continued benefit from therapy at the time of this writing. (A): Patient 3 with metastatic NSCLC. (B): Patient 23 with recurrent anaplastic ependymoma.
Abbreviations: carbo., carboplatin; clin., clinical; mets., metastasis; NSCLC, non-small cell lung cancer; pem., pemetrexed; RT, radiation therapy; SRS, stereotactic radiosurgery.
Patient 3 is a 51-year-old male diagnosed with stage IV lung adenocarcinoma with symptomatic brain metastases (Fig. 4A). Molecular profiling of a lung tumor revealed mutations in BRAF (p.V600E; 8.5% allelic frequency) and MET (p.T992I; 49% allelic frequency) (supplemental online Table 2). Case reports and interim results of a phase II trial indicate that BRAF p.V600E-mutant lung cancers frequently respond to BRAF inhibition [13–15]. The MTB recommended treatment with a BRAF inhibitor. This patient was then enrolled in a clinical trial testing a BRAF inhibitor. He has remained on therapy for >10 months and is tolerating the medication well with continued radiographic response and clinical benefit.
Patient 23 is a 45-year-old male with recurrent anaplastic ependymoma (Fig. 4B). He has a family history of Muir-Torre syndrome, a cancer-predisposing condition. DNA sequencing of the initial baseline tumor specimen revealed mutations in NF1, PIK3R1, PTPN11, CDKN2A, TP53, MSH2, MSH6, BRIP1, MLL2, and SETD2 and several other mutations of unknown significance (supplemental online Table 2). An obstacle in recommending investigational therapies for this patient is the exclusion of primary brain tumors in most phase I trials. Although germline DNA was not analyzed, we speculate that the DNA mismatch repair defects in MSH2 (deletion of exons 1–3) and/or MSH6 (p.F1088fs*5) were associated with his history of Muir-Torre syndrome [16]. Preclinical evidence indicates that MSH2 deficiency induces synthetic lethality with the anti-folate methotrexate [17]. A phase II clinical trial is ongoing to test the efficacy of methotrexate in patients with MSH2-deficient colorectal cancer (clinicaltrials.gov NCT00952016). The NF1 p.N1465fs* and PTPN11 (Shp-2) p.V428M mutations are projected to activate Ras signaling and confer sensitivity to mTOR and MEK inhibitors [18–20]. The loss-of-function truncating mutation in PIK3R1 (p.R301*), which encodes the p85α regulatory subunit of PI3K, may create a neomorph that increases c-Jun N-terminal kinase and extracellular signal-regulated kinase signaling [21]. The MTB recommended off-label treatment with the MEK inhibitor trametinib and consideration of treatment with methotrexate as it crosses the blood-brain/tumor barrier [22, 23]. While seeking insurance carrier approval of coverage for trametinib, the patient was treated with methotrexate starting 5 days after MTB case discussion and continuing for 6 weeks, which provided stable disease and improvement of symptoms (left arm and leg function). At the time of disease progression on methotrexate, off-label trametinib therapy was initiated, and the disease was stabilized for 10 weeks before progression. Further therapy was not administered because of declining performance status, and the patient was referred for home hospice care.
Discussion
Establishment of a MTB has provided a streamlined resource for clinicians to help interpret tumor genetic profiles, infer drug sensitivity and resistance, recommend anticancer therapies, and assess whether germline genetic testing is warranted. Our experience has shown that (a) a MTB requires expertise from medical oncologists, molecular pathologists, cancer geneticists, genetic counselors, and biologists with expertise in cancer molecular biology and therapeutics; (b) a MTB has become a critical resource relied upon by treating physicians to incorporate genetic data in therapeutic decisions; (c) turnaround times for genetic analysis and MTB recommendations do not delay treatment; and (d) attempts to obtain coverage for off-label use of targeted therapeutics are frequently successful but may result in delays in treatment (2–4 weeks).
Key issues we have identified in using tumor genetic profiling to inform disease management include: (a) prioritization of genetic aberrations by potential impact and contraindications of simultaneously detected aberrations; (b) determination of the optimum set of genes to evaluate; (c) lack of detection of copy number alterations or gene rearrangements with our current sequencing platform; (d) interpretation of variants of unknown significance; and (e) decreased ability to distinguish somatic from germline alterations with tumor-only sequencing. We also found that other circumstances often influence treatment decisions, including eligibility for clinical trials (e.g., brain tumors are frequently an exclusion criterion in phase I studies) and patient preferences for treatment (e.g., interest in or ability to travel for clinical trials). Such issues contributed to the low rate (4 of 15 cases, 26.7%) of acceptance of MTB-recommended targeted therapy in patients with previously treated advanced disease (Fig. 2). Similar to our experience, a recent report from the University of California San Diego MTB indicated that genetic profiling altered treatment decisions in 35.3% of cases (12 of 34); other cases were not informed by molecular diagnostics because patients had stable disease on current therapy (n = 13); the drug was unavailable because of cost, clinical trial ineligibility, or distance to travel (n = 7); lack of actionable alterations (n = 1); or near-term death (n = 1) [24]. A team from Vanderbilt University also reported that 17.5% of patients (18 of 103) with tumor genetic profiling received matched therapy; 35% of patients received standard (nonmatched) therapy, 12% had no evidence of disease following surgery or therapy, 10% enrolled in nonmatched trials, and 13% deteriorated clinically [25].
With the rapid expansion of the number of anticancer drug targets and availability of “genetically informed” clinical trials, there will be an increasing need to consider genetic alterations in terms of pathways rather than just the targets themselves (as in Fig. 3C). Although the 50-gene panel used herein for mutation detection may soon not provide sufficient genetic detail for therapeutic decision making, the overabundance of genetic information such as will occur with whole-exome sequencing will be overwhelming without a properly designed analysis and interpretation infrastructure. The issue of incorporating copy number alteration detection into our sequencing pipeline will soon be resolved by technological advances. For germline testing, a cost-effective approach is to refer MTB cases if inherited alterations are suspected based on disease type, patient characteristics, symptoms, family history, and/or tumor genetic profile. As we move forward with ever-increasing amounts of individualized genetic data, we will continue to assign genes and mutations to tiers for prioritization; genes encoding drug targets (e.g., MET) and common downstream signaling nodes (e.g., PI3K, MEK) and mutations shown to confer drug sensitivity/resistance will be prioritized as most informative. Ultimately, bioinformatics pipelines must be implemented to comprehensively analyze individual tumor genetic profiles to infer pathway activation and likelihood of drug sensitivity/resistance.
Of the nine cases that used archived tumor specimens for DNA sequencing, seven patients had received treatment with at least one line of systemic therapy between the time of tumor biopsy and DNA sequencing; thus, treatment that occurred during the intervening time period may have altered tumor genetic profiles. However, the question remains: are primary or recurrent/metastatic tumors most appropriate for testing? Reports of treatment-associated genetic evolution within tumors are beginning to emerge, and evidence indicates that paired primary versus recurrent gliomas have drastically different genetic profiles [26]. In contrast, primary hepatocellular carcinomas have mutational profiles similar to paired asynchronous (>2-year interval) lung metastases [27]. Similar observations were made in paired primary colorectal carcinomas versus asynchronous metastases [28]. Because the status of commonly mutated genes (e.g., KRAS, BRAF) is concordant between most primary versus asynchronous metastatic tumors, it has been suggested that genetic profiling of archived diagnostic tumor specimens is acceptable. Although this notion may be suitable for select tumor types and therapies, expanded use of targeted therapies, which are being shown to induce novel mutations, may necessitate rebiopsy to obtain current tumor tissue for molecular analysis. A well-known example of treatment-induced mutation is EGFR inhibitor-induced acquisition of the EGFR p.T790M mutation that confers secondary drug resistance [1]. Examples of acquired secondary resistance mutations have also been associated with treatment with inhibitors of mTORC1 and BRAF [29, 30]. Thus, rebiopsy of a tumor for genetic profiling to inform treatment decisions, particularly following disease progression on a targeted agent, is often warranted.
Conclusion
This report of our MTB experience is intended to be a resource for other medical centers as they establish similar panels of specialists to maximize use of tumor genetic profiling. Although future integration of other molecular profiles, including (phospho)proteomics and transcriptomics, into clinical decision making will make data interpretation more complex, such information is also expected to highlight therapeutically relevant alterations to simplify treatment decisions. A well-designed MTB will evolve along with the technology to ensure that patients receive the best possible treatment without unnecessary costs or risks.
See http://www.TheOncologist.com for supplemental material available online.
This article is available for continuing medical education credit at CME.TheOncologist.com.
Supplementary Material
Acknowledgments
We thank the patients and staff of the Norris Cotton Cancer Center and the Dartmouth-Hitchcock Medical Center, the Molecular Pathology Laboratory, and the Norris Cotton Cancer Center Pathology Translational Research Shared Resource. This work was supported by the Norris Cotton Cancer Center.
Footnotes
For Further Reading: Maria Schwaederle, Barbara A. Parker, Richard B. Schwab et al. Molecular Tumor Board: The University of California San Diego Moores Cancer Center Experience. The Oncologist 2014;19:631–636.
Implications for Practice: This study relates the authors' experience with the initiation of molecular tumor board meetings, which are a new vehicle for managing patients with complex malignancies on whom molecular diagnostics have been performed. This experience could be of significant importance to oncologists who are increasingly faced with advanced molecular diagnostic data, yet have minimal training in genomics. This article should help clinicians to handle practical issues related to setting up and efficiently utilizing molecular tumor board meetings. The article also aims at helping oncologists and health care systems understand and address practical, logistical, and scientific issues, such as the challenges associated with interpretation of molecular testing for patients with advanced cancer.
Author Contributions
Conception/Design: Laura J. Tafe, Todd W. Miller, Mary D. Chamberlin
Collection and/or assembly of data: Laura J. Tafe, Joel A. Lefferts, Jason R. Pettus, Jonathan D. Marotti, Kasia J. Bloch, Konstantin H. Dragnev, Camilo E. Fadul, Gary N. Schwartz, Clinton R. Morgan, Britt M. Holderness, Jason D. Peterson, Gregory J. Tsongalis, Todd W. Miller, Mary D. Chamberlin
Data analysis and interpretation: Laura J. Tafe, Ivan P. Gorlov, Francine B. de Abreu, Joel A. Lefferts, Xiaoying Liu, Jason R. Pettus, Jonathan D. Marotti, Kasia J. Bloch, Vincent A. Memoli, Arief A. Suriawinata, Konstantin H. Dragnev, Camilo E. Fadul, Gary N. Schwartz, Clinton R. Morgan, Britt M. Holderness, Jason D. Peterson, Gregory J. Tsongalis, Todd W. Miller, Mary D. Chamberlin
Manuscript writing: Laura J. Tafe, Ivan P. Gorlov, Francine B. de Abreu, Joel A. Lefferts, Xiaoying Liu, Jason R. Pettus, Jonathan D. Marotti, Kasia J. Bloch, Vincent A. Memoli, Arief A. Suriawinata, Konstantin H. Dragnev, Camilo E. Fadul, Gary N. Schwartz, Clinton R. Morgan, Britt M. Holderness, Jason D. Peterson, Gregory J. Tsongalis, Todd W. Miller, Mary D. Chamberlin
Final approval of manuscript: Laura J. Tafe, Ivan P. Gorlov, Francine B. de Abreu, Joel A. Lefferts, Xiaoying Liu, Jason R. Pettus, Jonathan D. Marotti, Kasia J. Bloch, Vincent A. Memoli, Arief A. Suriawinata, Konstantin H. Dragnev, Camilo E. Fadul, Gary N. Schwartz, Clinton R. Morgan, Britt M. Holderness, Jason D. Peterson, Gregory J. Tsongalis, Todd W. Miller, Mary D. Chamberlin
Disclosures
Joel A. Lefferts: Roche Molecular Diagnostics (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Duffy MJ, O’Donovan N, Crown J. Use of molecular markers for predicting therapy response in cancer patients. Cancer Treat Rev. 2011;37:151–159. doi: 10.1016/j.ctrv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: The MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsimberidou AM, Wen S, Hong DS, et al. Personalized medicine for patients with advanced cancer in the phase I program at M.D. Anderson: Validation and landmark analyses. Clin Cancer Res. 2014;20:4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: Personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsongalis GJ, Peterson JD, de Abreu FB, et al. Routine use of the Ion Torrent AmpliSeq™ Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2014;52:707–714. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 6.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): A resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UniProt Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balko JM, Giltnane JM, Wang K, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid S, Siano M, Joerger M, et al. Response to dabrafenib after progression on vemurafenib in a patient with advanced BRAF V600E-mutant bronchial adenocarcinoma. Lung Cancer. 2015;87:85–87. doi: 10.1016/j.lungcan.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Robinson SD, O’Shaughnessy JA, Cowey CL, et al. BRAF V600E-mutated lung adenocarcinoma with metastases to the brain responding to treatment with vemurafenib. Lung Cancer. 2014;85:326–330. doi: 10.1016/j.lungcan.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Planchard D, Mazieres J, Riely GJ, et al. Interim results of phase ii study brf 113928 of dabrafenib in braf v600e mutation-positive non-small cell lung cancer (nsclc) patients. J Clin Oncol. 2013;31:8009a. [Google Scholar]
- 16.South CD, Hampel H, Comeras I, et al. The frequency of Muir-Torre syndrome among Lynch syndrome families. J Natl Cancer Inst. 2008;100:277–281. doi: 10.1093/jnci/djm291. [DOI] [PubMed] [Google Scholar]
- 17.Martin SA, McCarthy A, Barber LJ, et al. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO Mol Med. 2009;1:323–337. doi: 10.1002/emmm.200900040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irving J, Matheson E, Minto L, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124:3420–3430. doi: 10.1182/blood-2014-04-531871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee S, Crouse NR, Emnett RJ, et al. Neurofibromatosis-1 regulates mTOR-mediated astrocyte growth and glioma formation in a TSC/Rheb-independent manner. Proc Natl Acad Sci USA. 2011;108:15996–16001. doi: 10.1073/pnas.1019012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao JC, Shah MH, Ito T, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung LW, Yu S, Zhang D, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26:479–494. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassman AB, Abrey LE, Shah GD, et al. Systemic high-dose intravenous methotrexate for central nervous system metastases. J Neurooncol. 2006;78:255–260. doi: 10.1007/s11060-005-9044-6. [DOI] [PubMed] [Google Scholar]
- 23.Tetef ML, Margolin KA, Doroshow JH, et al. Pharmacokinetics and toxicity of high-dose intravenous methotrexate in the treatment of leptomeningeal carcinomatosis. Cancer Chemother Pharmacol. 2000;46:19–26. doi: 10.1007/s002800000118. [DOI] [PubMed] [Google Scholar]
- 24.Schwaederle M, Parker BA, Schwab RB, et al. Molecular tumor board: The University of California-San Diego Moores Cancer Center experience. The Oncologist. 2014;19:631–636. doi: 10.1634/theoncologist.2013-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DB, Dahlman KH, Knol J, et al. Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. The Oncologist. 2014;19:616–622. doi: 10.1634/theoncologist.2014-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang L, Lee J, Park CK, et al. Whole-genome sequencing of matched primary and metastatic hepatocellular carcinomas. BMC Med Genomics. 2014;7:2. doi: 10.1186/1755-8794-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagle N, Grabiner BC, Van Allen EM, et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N Engl J Med. 2014;371:1426–1433. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.