Abstract
Enterotoxigenic Escherichia coli (ETEC) are a major cause of diarrhea worldwide, and infection of children in under-developed countries often leads to high mortality rates. Isolated ETEC expresses a plethora of colonization factors (fimbriae/pili), of which CFA/I and CFA/II, which are assembled via the alternate chaperone pathway (ACP), are among the most common. Fimbriae are filamentous structures whose shafts are primarily composed of helically arranged single pilin-protein subunits, with a unique biomechanical ability to unwind and rewind. A sustained ETEC infection, under adverse conditions of dynamic shear forces, is primarily attributed to this biomechanical feature of ETEC fimbriae. Recent understanding about the role of fimbriae as virulence factors points to an evolutionary adaptation of their structural and biomechanical features. In this work, we investigated the biophysical properties of CS2 fimbriae from the CFA/II group. Homology modeling of its major structural subunit, CotA, reveals structural clues related to the niche in which they are expressed. Using optical-tweezers force spectroscopy, we found that CS2 fimbriae unwind at a constant force of 10 pN and have a corner velocity (i.e., the velocity at which the force required for unwinding rises exponentially with increased speed) of 1300 nm/s. The biophysical properties of CS2 fimbriae assessed in this work classify them into a low-force unwinding group of fimbriae together with the CFA/I and CS20 fimbriae expressed by ETEC strains. The three fimbriae are expressed by ETEC, colonize in similar gut environments, and exhibit similar biophysical features, but differ in their biogenesis. Our observation suggests that the environment has a strong impact on the biophysical characteristics of fimbriae expressed by ETEC.
Introduction
Enterotoxigenic Escherichia coli (ETEC) diarrheal infection is considered a prevailing health problem in developing countries, since it is one of the major causes of death among infants and children. Additionally, ETEC infection is the leading cause of traveler’s diarrhea with >60% of visitors to these countries experiencing diarrhea, which in some cases can even trigger an irritable bowel syndrome (1,2). Pathogenesis of ETEC infections relies on bacterial attachment, via specialized fimbriae organelles, to the host intestine leading to subsequent release of either heat-labile (LT) or heat-stable (ST) enterotoxins (3–6).
Various strains of human ETEC express numerous serologically distinct fimbriae that are assembled either via the alternate chaperone (ACP) or classical chaperone usher pathway (CUP) (11). Colonization factor antigens (CFA)/I and CFA/II fimbriae belong to the ACP family or class 5 group, of which CFA/I fimbriae is an archetype and the most extensively studied member (7–9). The CFA/II group, which comprises three different coli surface antigens—CS1, CS2, and CS3—shares a common assembly pathway but differs from that of CFA/I fimbriae in hemagglutination properties (3,10,12,13). The CFA/II group fimbriae have been known for their role in causing diarrhea for almost 50 years, although little is known about their structural and biomechanical features (14–16). For example, micrographs of the three fimbriae indicate that CS1 and CS2 are wider and more rigid than CS3, and that CS1 is a helical filament (17–20). However, whether or not CS2 and CS3 are helix-like structures is unknown.
Sustained adhesion of both bacteria and eukaryotic cells can be facilitated by a reduction in force on their tethers. Membrane tethers are formed by some cell types to maintain cellular adhesion by reducing the load on receptor-ligand complexes. For example, a neutrophil that is attached to an inflamed endothelium cell is exposed to a drag force. The cell extends long membrane tethers, partially reducing the load experienced by the P-selection and PSGL-1 bond, thereby increasing the lifetime of the complex (21). Similarly, unwinding is believed to be an important biomechanical property of fimbriae, facilitating sustained adhesion of bacteria to their target cell (22,23). Fimbria unwinding lowers the force on the adhesin and receptor bond, thereby reducing the probability of bacteria detachment. Previous data have shown that CFA/I pilins assemble into helix-like fimbriae that are easy to unwind in comparison to, e.g., the uropathogenic E. coli (UPEC)-expressed P and Type 1 fimbriae (24). Further similarities between fimbrial unwinding and tether formation by other cell types are explained in more detail by Thomas (25).
Interestingly, fimbriae expressed in a specific microenvironment exhibit similar properties; for example, the unwinding force of CS20 is more similar to that of CFA/I than to that of P fimbriae, even though CS20 antigens share higher amino acid identity with the pilins of P fimbriae (26). Thus, the structural and biomechanical characteristics of fimbriae appear to play a determinant role in E. coli colonization and pathogenesis in a specific organ, i.e., E. coli strains sharing a common niche express fimbriae with similar biophysical properties (26). ETEC-expressing CS2 fimbriae are known to localize to the small intestine, an environment similar to that of ETEC-expressing CFA/I fimbriae of the ACP family and CS20 fimbriae of the CUP. We therefore hypothesized that CS2 fimbriae would exhibit structural and biophysical similarities to those of CFA/I and CS20.
In this work, we examined this hypothesis by investigating the biophysical strength of the CS2 fimbriae of the CFA/II group using sub-piconewton resolution force-spectroscopy instrumentation. Since a crystal structure of the major pilin subunit is lacking, we elucidated the structure of the major pilin, CotA, using homology modeling. Our data show that the CS2 fimbriae unwind at a constant force of 10 pN, demonstrating that the macromolecular structure of CS2 is helical, with weak layer-to-layer interactions. Although a three-dimensional structure of the CS2 shaft is not available, we have used a homology model of CotA and force spectroscopy results to reveal interesting biophysical features of CS2 fimbriae. Our data demonstrate similarities between these CFA/II group fimbriae and those from the CFA/I group. The results in this work, together with previous data from the literature, place ACP and CUP ETEC-expressed adhesion organelles into a low-force unwinding group.
Materials and Methods
Bacterial strains and growth condition
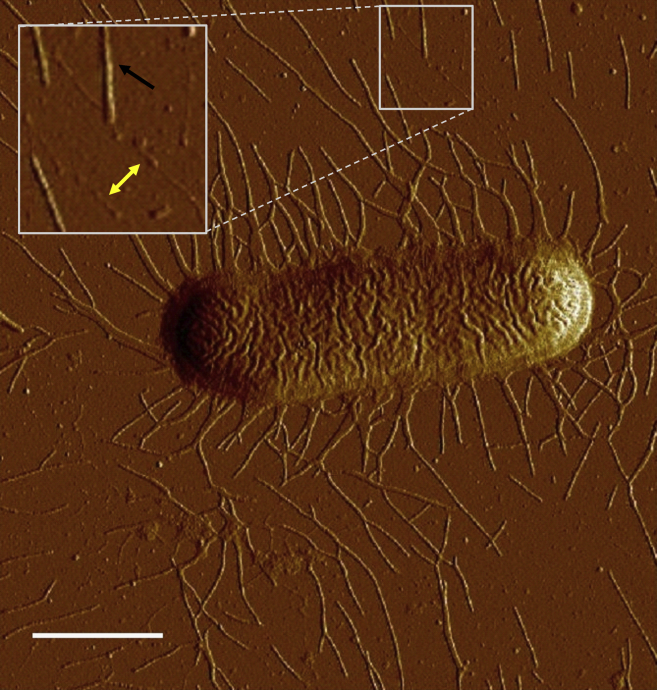
The C91F strain of ETEC-expressing CS2 fimbriae was used in this study. For expression of CS2 fimbriae, C91F strain was grown on CFA plates at 37°C overnight, restreaked on CFA plates, and again grown overnight at 37°C before analysis. Expression of CS2 fimbriae was confirmed by atomic force microscopy (AFM) (see Fig. 1).
Figure 1.
AFM image of a C91F ETEC bacterium expressing CS2 fimbriae. The AFM micrograph shows a single C91F bacterium expressing CS2 fimbriae. The black arrow points to a fimbria in its helical state, whereas the double yellow arrow points to unwound fimbriae. Scale bar, 1.0 μm. To see this figure in color, go online.
Sample preparation and force spectroscopy measurement
Force spectroscopy experiments were performed using optical tweezers with subpiconewton sensitivity. The setup of the instrumentation and assay is described in detail elsewhere (27,28). Briefly, the OT setup was constructed around an inverted microscope (Olympus IX71, Olympus, Center Valley, PA) with a high-NA oil immersion objective (UplanFl 100×, NA 1.35, Olympus). An Nd:YVO4 laser (Millennia IR, Spectra Physics, Santa Clara, CA) that operates at 1064 nm in CW mode and runs with an output power of 1.0 W was used for trapping. The position of a trapped bead, and thereby the force, was monitored by projecting the beam of a low-power fiber-coupled HeNe laser (operating at 632.8 nm) onto a position-sensitive detector (L20 SU9, Sitek Electro Optics, Partille, Sweden).
Bacteria expressing CS2 fimbriae were resuspended in phosphate-buffered saline (PBS; 1×, pH 7.4) and diluted in PBS to a suitable concentration (maximum of 10 bacteria in the field of view) for experiments. Before making the flow chamber, a 1:500 suspension of 9.5 μm carboxylate-modified latex beads (product no.2-10000, Interfacial Dynamics, Portland, OR) in Milli-Q water was prepared. These larger-sized beads were used to mount bacteria away from the coverslips to avoid any interactions with the surface. Ten microliters of the bead-water suspension was dropped onto 24 × 60 mm coverslips (no.1, Knittel Glass, Braunschweig, Germany) and put into the oven for 60 min at 60°C to immobilize the beads to the surface (28). To facilitate adhesion of the bacteria to the beads, a solution of 20 μL of 0.01% poly-L-lysine (catalog no. P4832, Sigma-Aldrich, St. Louis, MO) was added to the coverslips, which, after 45 min incubation at 60°C, were stored in a dust-free box until use. A ring of vacuum grease (Dow Corning, Midland, MI) was added around the area containing the poly-L-lysine-coated beads on one of the coverslips. Gently, a 3 μL suspension of bacteria and a 3 μL suspension of probe beads (surfactant-free 2.5 μm white amidine polystyrene latex beads, product no. 3-2600, Invitrogen, Carlsbad, CA) was dropped onto the area, which was then sealed by placing a 20 × 20 mm coverslip (no.1, Knittel Glass) on top.
An experiment was performed by trapping a single bacterium, at low laser power to make sure that the bacterium was not harmed, and firmly mounting it onto a poly-L-lysine-coated 10 μm latex bead, as described in Axner et al. (29). Subsequently, a 2 μm amidine bead was trapped and the stiffness of the trap was calibrated using the power spectrum method (30). The stability of the setup, as well as the optimal calibration time, was measured using the Allan variance method for optical tweezers (31). The trapped bead was thereafter attached to a fimbria by moving the bead in proximity to the mounted bacterium, but making sure that the bead was sufficiently far away from the bacterium to prevent a multitude of fimbriae attaching. With a fimbria attached to the bead (sometimes two to five fimbriae) the piezo-stage (PI-P5613CD, Physik Instruments, Auburn, MA) was translated using an in-house designed LabView program.
AFM
Bacteria expressing CS2 were resuspended in 50 μL of filtered Milli-Q water. Of that suspension, 10 μL was then placed onto a freshly cleaved ruby red mica sheet (Goodfellow Cambridge, Cambridge, United Kingdom). The cells were incubated for 5 min at room temperature before being placed into a desiccator for ∼2 h. Images were then collected with a Nanoscope V Multimode8 AFM setup (Bruker, Billerica, MA) using Bruker ScanAsyst mode with Bruker ScanAsyst-air probe oscillated at a resonant frequency of 50–90 kHz (32).
The Sticky-chain model for helix-like biopolymers
The unwinding velocity, , of helix-like biopolymer under tensile force, F, can be described using the opening and closing rates of the layer-to-layer interactions, and , and the opening length, , of a subunit,
| (1) |
where A and B represent the open and closed states, respectively (33,34). Using the nomenclature defined by Zakrisson et al. (22), the unwinding velocity can be rewritten as
| (2) |
where is the thermal bond opening rate, , where k is the Boltzmann constant, is the temperature, is the energy difference between the ground and transition state, and is the distance between the transition state and the open state. From this expression, it is possible to denote the corner velocity, , as the highest extension velocity that can be used without the need to include the dynamic behavior of the polymer, i.e., for low extension velocities, the opening and closing rates are in balance, whereas for high extension velocities, the closing rate can be neglected. This gives the expression
| (3) |
By combining Eqs. 2 and 3, the unwinding velocity can finally be expressed using the corner velocity,
| (4) |
Results
Atomic force imaging of CS2 fimbriae
We used AFM to image CS2 fimbriae at high magnification. A representative micrograph of a single cell grown under normal conditions is shown in Fig. 1. The average length of fimbriae was measured from AFM micrographs, such as that shown in Fig. S1, using ImageJ software. CS2 fimbriae were arranged peritrichously on the bacterial cell surface and had a length of 0.88 ± 0.34 μm (n = 270 samples). These fimbriae were found in two morphologies, with most fimbriae attached to the cell being wider structures (black arrow) and only a few being narrower structures (yellow arrow). This suggested that the wider structures had intact quaternary structure, indicating that layer-to-layer interactions of CS2 fimbriae are strong enough to be maintained during sample preparation. However, the few narrower structures identified in the micrographs suggested that CS2 fimbriae can be unwound. Thus, these findings called for an investigation of the properties of the major pilin subunit.
Homology modeling of the major structural subunit, CotA, using CfaB as a template
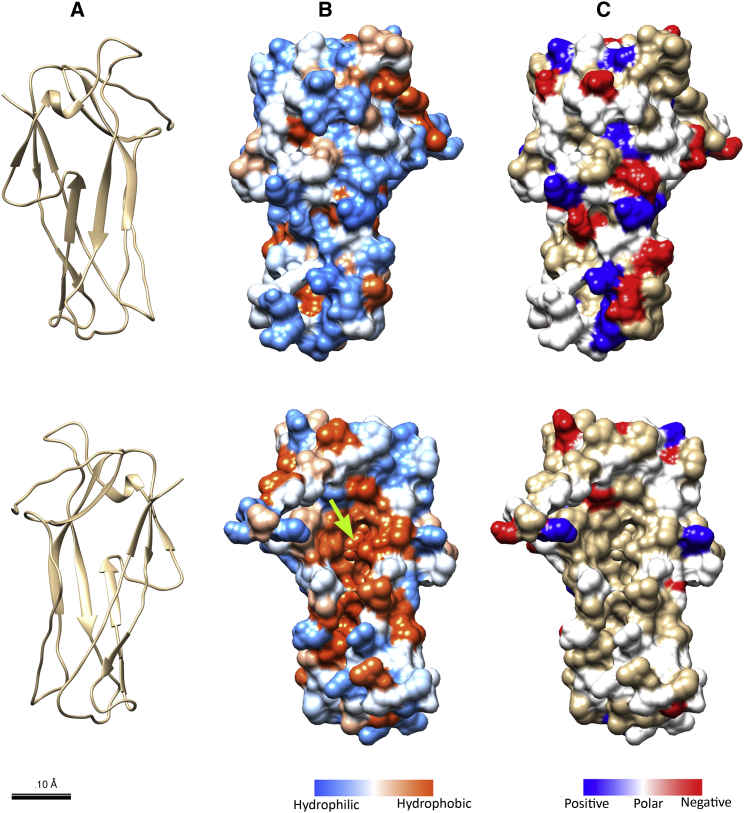
CS2 fimbriae share a close genetic and pathological relationship with CFA/I fimbriae of the class 5 family. Each CS2 fimbria consists of continuously repeating major pilin subunits (CotA) and a tip-localized minor subunit (CotD). The mature CotA protein is composed of 147 amino acids and shows significant homology to CfaB, the major pilin subunit of CFA/I fimbriae (PDB ID 3F84) (23). The sequence alignment presented in Fig. S2 was performed using the Clustal Omega algorithm (35); these two homologous pilin subunits share 51% identity and 80% similarity. Utilizing this high similarity, the CotA structure was homology-modeled using MODELER software (36). Fig. 2 shows a model of the outer (upper) and inner (lower) representation of the CotA subunit as a ribbon structure and as surface views of the hydrophobicity and charge distribution, respectively. The homology model of CotA was superimposed on the CfaB template and the root mean-square difference between the backbone C-α atoms was assessed to 0.37 Å. Moreover, the surface properties of CotA and CfaB, the hydrophobicity and the electrostatic potential, were thereafter examined using University of California San Francisco Chimera software (37). To investigate the surface potential of the model and the template, we colored the negative and positive residues red and blue, respectively, as seen in the surface views of the charge distributions of two faces for both CfaB and CotA, presented side by side in Fig. S3. In the upper row, the inner surface of the hydrophobic groove (tan) is visible running approximately vertically, in the center of the subunit. According to the charge distribution, CotA is slightly more negatively charged than CfaB.
Figure 2.
Homology modeling of the major structural subunit CotA, showing the model with its outer (upper) and inner surfaces (lower). The model is represented as a ribbon structure (A), a surface hydrophobicity map (B), and a surface-charge map (C). The hydrophobic groove is clearly visible (green arrow) in the central part of (B) (lower). Scale bar (for all), 10 Å. To see this figure in color, go online.
Residues in both the model and the template were also selected and shown according to their hydrophobicity and hydrophilicity, as shown in Fig. S4. Since the structural model of a pilus filament has not yet been determined, we are not able to make an optimal fit of a subunit’s position and exactly determine the inner and outer surfaces. However, by assuming that CotA and CfaB subunits have similar orientations, it is possible to estimate the inner and outer surfaces. Using this assumption, the model indicates that the majority of the area of the outer surfaces is hydrophilic, whereas the inner surfaces are hydrophobic, which is in accordance with other helix-like adhesion fimbriae. Finally, the deep hydrophobic groove of the subunit is clearly visible in the center of the two structures.
With information from both the CS2 macromolecular structure, assessed from static AFM images, and the major pilin subunit, using homology modeling, the next step was to gain information regarding the dynamic properties via force spectroscopy measurements.
CS2 fimbrial response to tensile force
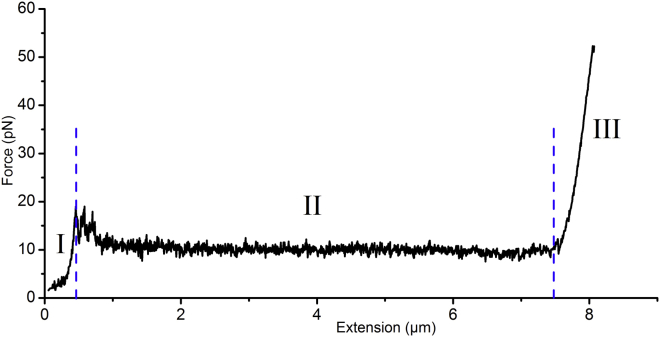
Mechanical response of the CS2 fimbriae to force was measured using optical-tweezers force spectroscopy under steady-state conditions (38). According to our homology modeling data, CotA showed a negative charge distribution on the outer surface, suggesting the possibility of a strong nonspecific interaction with positively charged amidine microspheres. Our test run confirmed a strong enough interaction between the two, and therefore, we used amidine microspheres for our force spectroscopy measurements. A representative force curve of a CS2 fimbria with three clearly distinct regions is presented in Fig. 3. CS2 first responds to tensile stress by a linearly increasing force, i.e., stretching layers gives a response similar to that of a Hookean spring, and we denote this as region I. After reaching a threshold in the structural resistance of the layers, a transient change to a constant force response at ∼10 pN can be seen; this is denoted as region II, and it originates from unwinding of the individual shaft subunits. We marked this region by two dashed blue lines in the representative data shown in Fig. 3. The average unwinding force was calculated from 90 independent measurements and assessed to 10 ± 1.5 pN. After complete unwinding, the force increases linearly with fimbria extension to ∼7.8 μm, shown as region III. As seen in the rewinding curve in Fig. 4 (blue curve), a drop in force at ∼6.9–6.8 μm is required to nucleate recoiling of the helical filament. The need for nucleation to begin helical rewinding supports a model in which the helical region is fully unwound before entering region III, and this region can be attributed to stretching of subunits already aligned in an open coil.
Figure 3.
Force spectroscopy measurement of a CS2 fimbria. The black curve represents the unwinding force response of a CS2 fimbria at a velocity of 0.1 μm/s. The unwinding force response shows three distinct regions: region I, a linear increase of the force; region II, a constant-force plateau (region between the blue dashed lines); and region III. Initially, the curve shows force peaks that originate from weakly attached fimbriae that detached with extension. To see this figure in color, go online.
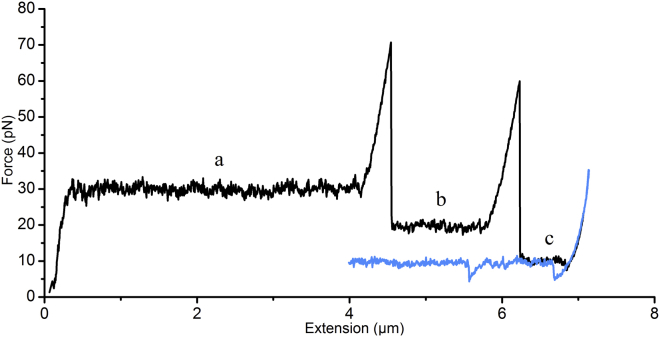
Figure 4.
Force spectroscopy measurement of multiple CS2 fimbriae. The unwinding (black) and rewinding (blue) responses of three simultaneously attached fimbriae, where (a) all three fimbriae are simultaneously extended until the shortest fimbria detaches at ∼4.3 μm, resulting in a force drop to 20 pN; (b) the two remaining fimbriae are further extended until the shorter detaches at ∼6 μm, with a corresponding drop in force to 10 pN; and (c) a single fimbria remains. The blue curve shows the corresponding rewinding response of the single fimbria. To see this figure in color, go online.
We observed that the force response of multiple fimbriae was additive, as seen for other helix-like fimbriae (39). Fig. 4 shows the extension of three fimbriae of different lengths. Similar to that of a single fimbria, the force response first increases linearly with extension, but with an increased stiffness, since three fimbriae are attached to the bead and the force is equally shared between these fimbriae. The transition to the constant-force plateaus throughout the measurement took place for even multiples of a single fimbria. That is, first all three fimbriae unwound, giving rise to a plateau force of 30 pN; at an extension of ∼4.5 μm, the shortest fimbria was fully unwound and entered region III before it detached from the bead. The two remaining fimbriae continued to unwind at ∼20 pN for ∼1 μm until the second fimbria reached region III and detached. Eventually, the longest fimbria unwound an additional ∼0.5 μm until it entered region III. At this point, we reversed the motion of the stage and allowed the fimbria to rewind (Fig. 4, blue curve). The rewinding force curve continued along the unwinding curve except for a 5 pN force drop that occurred between 6.8 and 6.6 μm and originated from the lack of the nucleation kernel that is required for regenerating the helical form of the fimbrial shaft after complete unwinding. An additional force measurement that showed multiple fimbrial binding in the first sequence with four consecutive unwinding and rewinding sequences is presented in Fig. S5.
Dynamic force responses of CS2
To investigate the dynamic response of CS2 fimbriae, we applied dynamic force spectroscopy (DFS) using optical tweezers (28). Before a DFS measurement, a fimbria was slowly unwound to measure the total length of the unwinding region (region II) and to remove any possibility of multifimbrial binding to the bead. With the known length of region II, the fimbria was allowed to partially rewind to make sure that a 2-μm-long region could be unwound without reaching region III. The 2-μm-long region was thereafter unwound at five different velocities, i.e., 0.1, 0.4, 1.6, 6.4, and 25.6 μm/s, and the corresponding position and force response were sampled at 5 kHz. Between each pull, the fimbria was allowed to rewind under steady-state conditions (0.1 μm/s) and a short pause of 2 s was introduced before the next pull. An example of a DFS measurement of a CS2 fimbria is shown in Fig. S6, with raw data represented by the gray curves and the corresponding mean values of the plateau force for each measured velocity represented by the dashed black lines. The data were cropped in the x-scale at 1.0 μm for better visualization.
In Fig. 5, the mean unwinding force versus extension velocity for all analyzed CS2 fimbriae (n = 30) is summarized. In the graph, two distinct regions of structural response to extension velocity can be identified. For low velocities, <∼1.0 μm/s, the force response is independent of extension velocity and the unwinding force is ∼10 pN. For extension velocities >∼1.0 μm/s, the unwinding force increases logarithmically with increasing velocity. This particular transition point is denoted the corner velocity, (28). Thus, for velocities below the corner velocity, fimbria extension occurs independent of speed and the experiment is performed under steady-state conditions, whereas for velocities above the corner velocity ( < ) fimbriae enter a dynamic response region (39).
Figure 5.
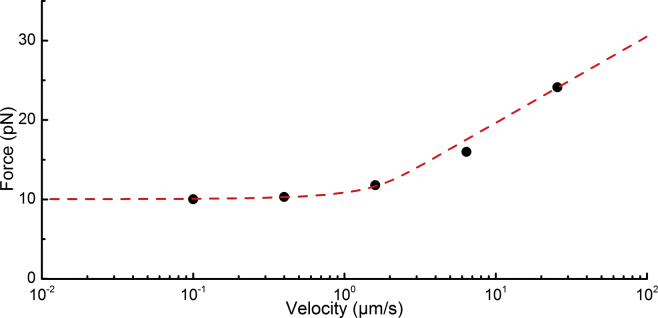
Dynamic force spectroscopy measurement of CS2 fimbria. The quaternary structure of an individual CS2 fimbria was unwound at velocities of 0.1, 0.4, 1.6, 6.4, and 25.6 μm/s for a distance of ∼2 μm and the corresponding force responses were sampled at 5 kHz. Each data point (black dots) shows the average unwinding force of fimbriae at five distinct velocities. For low velocities, i.e., velocities below the corner velocity, the unwinding force is independent of speed and amounts to 10 pN. The red dashed line shows a fit of the helix-like polymer model to the data, yielding the corner velocity, nm/s, and the bond length, nm, respectively. To see this figure in color, go online.
To find the bond length, the distance from the ground state to the transition barrier, , and the corner velocity, , of CS2 fimbriae, and to see whether the sticky-chain model (33), which describes the behavior of a helix-like polymer under force, could be fitted to the force-versus-extension velocity data, we numerically fitted the full set of rate equations, i.e., Eq. 4. The fit is shown by the red dashed line in Fig. 5, with set to 293 K. The corresponding model parameters were set according to the best fit of the model to our experimental data: nm/s, nm, and nm.
Discussion
Adhesion of pathogenic bacteria to host cells is the initial step of colonization. Adhesion fimbriae are thus key virulence factors, and a detailed understanding of their structural and functional role in bacterial adhesion is essential for elucidating the mechanism of infection. Many fimbriae have the ability to extend and contract when the bacteria are exposed to external forces, and it has been shown that unwinding of fimbriae help the bacteria to withstand shear forces by reducing the load on the adhesin protein at the fimbrial tip (22,40,41). CFA/I and CS20 fimbriae expressed by ETEC that colonize the small intestine, are helical and can be unwound to several times their native length. Whether or not CS2 fimbriae exhibit structural and biophysical similarities to CFA/I and CS20 fimbriae, i.e., whether CS2 should be classified as helix-like fimbriae that can be unwound and thereby be significantly elongated, was to our knowledge not known before this study.
Fimbriae are stabilized by layer-to-layer interactions, implying that the unwinding force of a fimbria is related to the number of layer-to-layer interactions (24). Since neither the macromolecular structure of the CS2 fimbriae nor the crystal structure of the major pilin subunit, CotA, has been solved, we first looked at AFM micrographs to elucidate the macromolecular structure and to identify any possible morphologies. The expressed CS2 fimbriae were found to be ∼1 μm in length and were, primarily, in a wider state, thus suggesting that they are in a wound state. However, the presence of a few observed narrow-width segments suggested that CS2 could make a morphological change from a wide to a narrow structure by breaking layer-to-layer interactions formed by subunits in adjacent layers.
To investigate the properties of individual subunits, we aligned the major protein, CotA, using the CFA/I major protein, CfaB, as a template. Comparison of amino acid composition of the two structures showed 51% identity and 80% similarity. Our subsequent homology model revealed a slightly higher negative surface potential of CotA compared with that of CfaB (23). The negative surface was thereafter confirmed by force spectroscopy measurements with positively charged amidine beads.
Since it has been shown earlier that for some fimbriae, niche identity, rather than amino acid composition of the major subunit, conveys the physical properties of adhesion fimbriae, e.g., the unwinding force of CS20 (15 pN) is more similar to that of CFA/I (7.5 pN) than to that of P fimbriae (28 pN) even though CS20 fimbriae share a higher amino acid identity with P fimbriae (22.8% identity) than with CFA/I fimbriae (19.7% identity) (26), we investigated this correlation using the force spectroscopy technique. Force spectroscopy measurements on single fimbriae at steady state revealed that CS2 is highly flexible and unwinds at a constant force of 10 pN. First, unwinding and rewinding at a constant force suggests that CS2 has a helix-like macromolecular structure, similar to both CFA/I and CS20 (33). In addition, the CS2 force response, with a pattern of linear increase, constant-force plateau, linear increase, is in line with what has been observed for other helix-like fimbriae, such as P, Type 1, and Type 3 fimbriae (42,43). This specific force response was also modeled, in a recent work, using a rigid-body model assembled into a helical structure exposed to tensile force (44). Second, the unwinding force level, which is slightly higher than that for CFA/I and slightly lower than that for CS20, places CS2 interestingly between these other two ETEC-expressed fimbriae colonizing the small intestine.
To analyze whether the dynamic properties of CS2 were similar to those of CFA/I, we carried out DFS measurements. The unwinding velocity was increased in steps to measure the unwinding force required for a given velocity. A physical model describing the force response of a helix-like polymer was fitted to the mean unwinding force for each velocity to assess the corner velocity as well as the bond length. The corner velocity provides information regarding the dynamics of the fimbriae, i.e., fimbriae with a high corner velocity can be unwound at high velocities without responding to an increase in resistance. This implies that a bacterium will go with the flow up to the corner velocity of a fimbria, with the unwinding force as the only resisting force. A high corner velocity thereby provides a higher force-buffering capability than a low corner velocity, which suggests that fast fluctuating forces are more easily damped out. The model fitted the DFS data well, as can be seen in Fig. 5. The corner velocity of CS2 (1300 ± 200 nm/s) is similar to that of CFA/I (1400 ± 200 nm/s). Also, the bond length of CS2, derived from DFS measurements, yielded a value of ΔxAT = 0.86 ± 0.1 nm, which is similar to the bond length of CFA/I, ΔxAT = 1.1 ± 0.1 nm. From the data presented above, we can thus conclude that the similarity of pilin subunits, unwinding forces, corner velocities, and bond lengths strongly indicates that CFA/I and CS2 that are expressed in the same niche are structurally and biophysically similar fimbriae.
This study, together with data from the literature, suggests that helix-like fimbriae expressed by, e.g., pathogenic E. coli and Klebsiella pneumonia, where the latter is associated with respiratory tract infections, could be categorized into three fimbrial mechanical groups: the low- (∼15 pN), medium- (∼30 pN), and high-force (∼60 pN) unwinding fimbriae. ETEC fimbriae such as CS2, CFA/I, and CS20 require <15 pN unwinding force, despite differences in their assembly mechanism (see Table S1 and our previous work (24,26)); UPEC and meningitis-associated strains of E. coli express fimbriae requiring 21–30 pN of unwinding force (33,39,42); however, Type 3 fimbriae expressed by K. pneumonia in the respiratory tract require 66 pN of unwinding force (43). One should also note that the T4 fimbriae expressed by Streptococcus pneumonia, which also colonize the respiratory tract, are significantly stiffer than UPEC- and ETEC-expressed fimbriae (45). The analogy between the niche and the biomechanical features of fimbriae has been suggested in earlier studies, and the results in this study support that hypothesis by placing CS2 in the low-force unwinding group of fimbriae, where other ETEC-expressed fimbriae are also found.
Author Contributions
N.M. carried out the force spectroscopy experiments, DFS measurement, and length analysis of fimbriae; B.S. prepared the strains; M.A. and E.B. carried out the homology modeling, M.A. and J.Z. performed the modeling, and J.Z. developed the software used in the force spectroscopy and DFS measurements. All authors contributed to the planning of the study and interpretation of the results. N.M., B.S., E.B., and M.A. drafted the main manuscript text and all authors reviewed the final version of the manuscript.
Acknowledgments
We are grateful to Dr. Stephen J. Savarino for providing the reagents in this work and to Monica Persson for assistance with AFM micrographs.
This work was supported by the National Institutes of Health (grants GM05722 and RR025434 to E.B.), the Swedish Research Council (grant 621-2013-5379 to M.A.), and the Carl Trygger Foundation (to M.A.).
Editor: Jennifer Curtis.
Footnotes
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00507-X.
Supporting Material
References
- 1.Steffen R. Epidemiology of traveler’s diarrhea. Clin. Infect. Dis. 2005;41(Suppl 8):S536–S540. doi: 10.1086/432948. [DOI] [PubMed] [Google Scholar]
- 2.DuPont H.L. Systematic review: the epidemiology and clinical features of travellers’ diarrhoea. Aliment. Pharmacol. Ther. 2009;30:187–196. doi: 10.1111/j.1365-2036.2009.04028.x. [DOI] [PubMed] [Google Scholar]
- 3.Smyth C.J. Two mannose-resistant haemagglutinins on enterotoxigenic Escherichia coli of serotype O6:K15:H16 or H-isolated from travellers’ and infantile diarrhoea. J. Gen. Microbiol. 1982;128:2081–2096. doi: 10.1099/00221287-128-9-2081. [DOI] [PubMed] [Google Scholar]
- 4.Rodas C., Mamani R., Iniguez V. Enterotoxins, colonization factors, serotypes and antimicrobial resistance of enterotoxigenic Escherichia coli (ETEC) strains isolated from hospitalized children with diarrhea in Bolivia. Braz. J. Infect. Dis. 2011;15:132–137. doi: 10.1016/s1413-8670(11)70158-1. [DOI] [PubMed] [Google Scholar]
- 5.Valvatne H., Sommerfelt H., Grewal H.M. Identification and characterization of CS20, a new putative colonization factor of enterotoxigenic Escherichia coli. Infect. Immun. 1996;64:2635–2642. doi: 10.1128/iai.64.7.2635-2642.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyofo B.A., Subekti D.S., Lesmana M. Toxins and colonization factor antigens of enterotoxigenic Escherichia coli among residents of Jakarta, Indonesia. Am. J. Trop. Med. Hyg. 2001;65:120–124. doi: 10.4269/ajtmh.2001.65.120. [DOI] [PubMed] [Google Scholar]
- 7.Gaastra W., Sommerfelt H., Grewal H.M. Antigenic variation within the subunit protein of members of the colonization factor antigen I group of fimbrial proteins in human enterotoxigenic Escherichia coli. Int. J. Med. Microbiol. 2002;292:43–50. doi: 10.1078/1438-4221-00189. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay S., Tchesnokova V., Savarino S.J. Adaptive evolution of class 5 fimbrial genes in enterotoxigenic Escherichia coli and its functional consequences. J. Biol. Chem. 2012;287:6150–6158. doi: 10.1074/jbc.M111.303735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anantha R.P.R.P., McVeigh A.L.A.L., Savarino S.J. Evolutionary and functional relationships of colonization factor antigen I and other class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect. Immun. 2004;72:7190–7201. doi: 10.1128/IAI.72.12.7190-7201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cravioto A., Scotland S.M., Rowe B. Hemagglutination activity and colonization factor antigens I and II in enterotoxigenic and non-enterotoxigenic strains of Escherichia coli isolated from humans. Infect. Immun. 1982;36:189–197. doi: 10.1128/iai.36.1.189-197.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuccio S.-P., Bäumler A.J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knutton S., Lloyd D.R., McNeish A.S. Adhesion of enterotoxigenic Escherichia coli to human small intestinal enterocytes. Infect. Immun. 1985;48:824–831. doi: 10.1128/iai.48.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D.J., Jr., Evans D.G.J., Graham D.Y. Mannose-resistant hemagglutination of human erythrocytes by enterotoxigenic Escherichia coli with colonization factor antigen II. J. Clin. Microbiol. 1988;26:1626–1629. doi: 10.1128/jcm.26.9.1626-1629.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans D.G., Silver R.P., Gorbach S.L. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect. Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans D.G., Evans D.J., Jr., Tjoa W. Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect. Immun. 1977;18:330–337. doi: 10.1128/iai.18.2.330-337.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans D.G., Evans D.J., Jr., DuPont H.L. Detection and characterization of colonization factor of enterotoxigenic Escherichia coli isolated from adults with diarrhea. Infect. Immun. 1978;19:727–736. doi: 10.1128/iai.19.2.727-736.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda T., Cakir N., Miwatani T. Purification and characterization of the CS2 pili of colonization factor antigen II produced by human enterotoxigenic Escherichia coli. Microbiol. Immunol. 1989;33:265–275. doi: 10.1111/j.1348-0421.1989.tb01976.x. [DOI] [PubMed] [Google Scholar]
- 18.Tobias J., Svennerholm A.-M., Lebens M. Construction and expression of immunogenic hybrid enterotoxigenic Escherichia coli CFA/I and CS2 colonization fimbriae for use in vaccines. Appl. Microbiol. Biotechnol. 2010;87:1355–1365. doi: 10.1007/s00253-010-2577-4. [DOI] [PubMed] [Google Scholar]
- 19.Sjöberg P.O., Lindahl M., Wadström T. Purification and characterization of CS2, a sialic acid-specific haemagglutinin of enterotoxigenic Escherichia coli. Biochem. J. 1988;255:105–111. doi: 10.1042/bj2550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galkin V.E., Kolappan S., Craig L. The structure of the CS1 pilus of enterotoxigenic Escherichia coli reveals structural polymorphism. J. Bacteriol. 2013;195:1360–1370. doi: 10.1128/JB.01989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King M.R., Heinrich V., Hammer D.A. Nano-to-micro scale dynamics of P-selectin detachment from leukocyte interfaces. III. Numerical simulation of tethering under flow. Biophys. J. 2005;88:1676–1683. doi: 10.1529/biophysj.104.051805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakrisson J., Wiklund K., Andersson M. Helix-like biopolymers can act as dampers of force for bacteria in flows. Eur. Biophys. J. 2012;41:551–560. doi: 10.1007/s00249-012-0814-8. [DOI] [PubMed] [Google Scholar]
- 23.Li Y.-F., Poole S., Bullitt E. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA. 2009;106:10793–10798. doi: 10.1073/pnas.0812843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson M., Björnham O., Bullitt E. A structural basis for sustained bacterial adhesion: biomechanical properties of CFA/I pili. J. Mol. Biol. 2012;415:918–928. doi: 10.1016/j.jmb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas W. Catch bonds in adhesion. Annu. Rev. Biomed. Eng. 2008;10:39–57. doi: 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
- 26.Mortezaei N., Epler C.R., Bullitt E. Structure and function of enterotoxigenic Escherichia coli fimbriae from differing assembly pathways. Mol. Microbiol. 2015;95:116–126. doi: 10.1111/mmi.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fällman E., Schedin S., Axner O. Optical tweezers based force measurement system for quantitating binding interactions: system design and application for the study of bacterial adhesion. Biosens. Bioelectron. 2004;19:1429–1437. doi: 10.1016/j.bios.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 28.Andersson M., Fällman E., Axner O. Dynamic force spectroscopy of E. coli P pili. Biophys. J. 2006;91:2717–2725. doi: 10.1529/biophysj.106.087429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Axner O., Andersson M., Björnham O., Castelain M., Klinth J.E. Assessing bacterial adhesion on an individual adhesin and single pili level using optical tweezers. In: Linke D., Goldman A., editors. Bacterial Adhesion. Springer; Berlin: 2011. pp. 301–313. [DOI] [PubMed] [Google Scholar]
- 30.Berg-Sørensen K., Flyvbjerg H. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 2004;75:594–612. [Google Scholar]
- 31.Andersson M., Czerwinski F., Oddershede L.B. Optimizing active and passive calibration of optical tweezers. J. Opt. 2011;13:044020. [Google Scholar]
- 32.Balsalobre C., Morschhäuser J., Uhlin B.E. Transcriptional analysis of the sfa determinant revealing mmRNA processing events in the biogenesis of S fimbriae in pathogenic Escherichia coli. J. Bacteriol. 2003;185:620–629. doi: 10.1128/JB.185.2.620-629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson M., Fällman E., Axner O. A sticky chain model of the elongation and unfolding of Escherichia coli P pili under stress. Biophys. J. 2006;90:1521–1534. doi: 10.1529/biophysj.105.074674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersson M., Axner O., Fällman E. Physical properties of biopolymers assessed by optical tweezers: analysis of folding and refolding of bacterial pili. ChemPhysChem. 2008;9:221–235. doi: 10.1002/cphc.200700389. [DOI] [PubMed] [Google Scholar]
- 35.Sievers F., Wilm A., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iovino M., Falconi M., Desideri A. Molecular dynamics simulation of the antimicrobial salivary peptide histatin-5 in water and in trifluoroethanol: a microscopic description of the water destructuring effect. J. Pept. Res. 2001;58:45–55. doi: 10.1034/j.1399-3011.2001.00885.x. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Axner O., Björnham O., Andersson M. Unraveling the secrets of bacterial adhesion organelles using single-molecule force spectroscopy. In: Gräslund A., Rigler R., Widengren J., editors. Single Molecule Spectroscopy in Chemistry, Physics and Biology: Nobel Symposium. Springer; Berlin: 2010. pp. 337–362. [Google Scholar]
- 39.Castelain M., Ehlers S., Axner O. Fast uncoiling kinetics of F1C pili expressed by uropathogenic Escherichia coli are revealed on a single pilus level using force-measuring optical tweezers. Eur. Biophys. J. 2011;40:305–316. doi: 10.1007/s00249-010-0648-1. [DOI] [PubMed] [Google Scholar]
- 40.Miller E., Garcia T., Oberhauser A.F. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zakrisson J., Wiklund K., Andersson M. The shaft of the type 1 fimbriae regulates an external force to match the FimH catch bond. Biophys. J. 2013;104:2137–2148. doi: 10.1016/j.bpj.2013.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson M., Uhlin B.E., Fällman E. The biomechanical properties of E. coli pili for urinary tract attachment reflect the host environment. Biophys. J. 2007;93:3008–3014. doi: 10.1529/biophysj.107.110643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F.-J., Chan C.-H., Hsu L. Structural and mechanical properties of Klebsiella pneumoniae type 3 fimbriae. J. Bacteriol. 2011;193:1718–1725. doi: 10.1128/JB.01395-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zakrisson J., Wiklund K., Andersson M. Rigid multibody simulation of a helix-like structure: the dynamics of bacterial adhesion pili. Eur. Biophys. J. 2015 doi: 10.1007/s00249-015-1021-1. Published online April 8, 2015. [DOI] [PubMed] [Google Scholar]
- 45.Castelain M., Koutris E., Axner O. Characterization of the biomechanical properties of T4 pili expressed by Streptococcus pneumoniae—a comparison between helix-like and open coil-like pili. ChemPhysChem. 2009;10:1533–1540. doi: 10.1002/cphc.200900195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.