Abstract
The inter-subject alignment of functional MRI (fMRI) data is important for improving the statistical power of fMRI group analyses. In contrast to existing anatomically-based methods, we propose a novel multi-subject algorithm that derives a functional correspondence by aligning spatial patterns of functional connectivity across a set of subjects. We test our method on fMRI data collected during a movie viewing experiment. By cross-validating the results of our algorithm, we show that the correspondence successfully generalizes to a secondary movie dataset not used to derive the alignment.
1 Introduction
Functional MRI (fMRI) studies of human neuroanatomical organization commonly analyze fMRI data across a population of subjects. The effective use of this data requires deriving a spatial correspondence across the set of subjects, i.e., the data must be aligned, or registered, into a common coordinate space. Current inter-subject registration techniques derive this correspondence by aligning anatomically-defined features, e.g. major sulci and gyri, across subjects, either in the volume or on extracted cortical surfaces. Talairach normalization [1], for example, derives a piecewise affine transformation by matching a set of major anatomical landmarks in the brain volume. More advanced techniques match a denser set of anatomical features, such as cortical curvature [2], and derive nonlinear transformations between a reference space and each subject’s cortical surface.
It is known, however, that an accurate inter-subject functional correspondence cannot be derived using only anatomical features, since the size, shape and anatomical location of functional loci vary across subjects [3], [4]. Because of this deficiency in current alignment methods, it is common practice to spatially smooth each subject’s functional data prior to a population based analysis. However, this incurs the penalty of blurring the functional data within and across distinct cortical regions. Thus, the functional alignment of multi-subject fMRI data remains an important problem.
We propose to register functional loci directly by using anatomical and functional data to learn an inter-subject cortical correspondence. This approach was first explored in [5], where subject cortices were registered by maximizing the inter-subject correlation of the functional response elicited by a common stimulus (a movie viewing). In essence, the correspondence was selected to maximize the correlation of the fMRI time series between subjects. This relies on the functional response being time-locked with the experimental stimulus. Large regions of visual and auditory cortex stimulated by a movie viewing do indeed show consistent inter-subject synchrony [6]. However, other areas in the intrinsic [7] or default [8] system fail to exhibit significant correlations across repeated stimulus trials. The technique of [5] is hence not expected to improve alignment in these intrinsic regions.
In contrast to [5], we propose to achieve inter-subject alignment by aligning intra-subject patterns of cortical functional connectivity. By functional connectivity, we mean within-subject similarity of the temporal response of remote regions of cortex [9]. This can be estimated from fMRI data, for example, by correlating the functional time series between pairs of cortical nodes within a subject. This yields a dense set of functional features for each subject from which we learn an inter-subject correspondence. Unlike other functional connectivity work (see e.g. [10]), we define connectivity between pairs of cortical nodes rather than with respect to anatomical regions of interest. Our approach is inspired by studies showing that the patterns of functional connectivity in the intrinsic network are consistent across subjects [7], [11]. This suggests that our method has the potential to learn an inter-subject functional correspondence within both extrinsic and intrinsic cortical networks.
In summary, we formulate a multi-subject cortical alignment algorithm that minimizes the difference between functional connectivity vectors of corresponding cortical nodes across subjects. We do so by learning a dense-deformation field on the cortex of each subject, suitably regularized to preserve cortical topology [2]. Our key contributions are: a) the novel alignment objective, b) a principled algorithm for accomplishing the alignment, and c) experimental verification on fMRI data.
The paper is organized as follows. In §2 we formulate the multi-subject alignment problem, followed by a detailed exposition of the algorithm in §3 and§4. Finally, we exhibit results of the algorithm applied to multi-subject fMRI data in §5 and draw conclusions in§6.
2 Formulation of the Multi-Subject Alignment Problem
For each subject we are given volumetric anatomical MRI data and fMRI data. The anatomical data is used to extract a two-dimensional surface model of cortex. This greatly facilitates cortical based analysis and subsequent visualization [12], [13], [14]. Cortex is segmented, then each cortical hemisphere is inflated to obtain a smooth surface, which is projected to the sphere, S2, represented by a discrete spherical mesh Ms = {pk ∈ S2; 1 ≤ k ≤ Nv/2}. The two cortical hemispheres are hence modeled by the disjoint union S = S2 ⊎ S2, represented by the corresponding disjoint union of mesh points M = Ms ⊎ Ms. Anatomical cortical features, such as cortical curvature, are functions Da : S → RNa sampled on M. Thus, our analysis is restricted to cortex only.
The fMRI volumeric data is first aligned with the anatomical scan, then mapped onto S. This assigns each mesh node pk ∈ M a “volumetric cortical voxel” vk ∈ ℝ3, with associated functional time series fk ∈ ℝNt. The functional time series data is then a function Df: S → RNt sampled on M.
As indicated in the introduction, we do not directly register the fMRI time series but instead register the functional connectivity derived from the time series. Let σ(f1, f2) denote a similarity measure on pairs of time series f1, f2 ∈ RNt. A useful example is empirical correlation: σ(f1, f2) = corr(f1, f2); another possibility is an estimate of the mutual information between the pairwise entries of f1, f2. Define the functional connectivity of the fMRI data under σ as the map C(pi, pj) = σ(Df(pi), Df(pj)), i.e., the similarity of the functional times series at the pairs of cortical nodes. Functional connections both within and across cortical hemispheres are considered. Functional connectivity can be conceptualized as the adjacency matrix of an edge-weighted graph on all cortical nodes. The edge between nodes pi, pj is weighted by the pairwise similarity measure σ(fi, fj) codifying the functional similarity of pi and pj. In the case of correlation, C is the correlation matrix of the time series data. For typical values of Nv (≈ 72, 000), the functional connectivity data structure is huge. Hence we need efficient mechanisms for working with C.
We are given the data discussed above for Ns subjects. Subject k’s training data is specified by samples of the functions Da,k : Sk → RNa, Df,k : Sj → RNt, and the derived functional connectivity Ck, all sampled on the mesh Mk, k = 1, …, Ns. Our objective is to learn a relation consisting of Ns-tuples of corresponding points across the set of cortices. To do so, we could select a node from M1 for subject 1 and learn the corresponding points on the cortices of the remaining Ns − 1 subjects through smooth and invertible mappings gk : S1 → Sk, k = 2, …, Ns. However, this arbitrarily and undesirably gives special status to one subject. Instead, we introduce a reference model Sref = S2 ⊎ S2 with mesh Mref. For each node p ∈ Mref on Sref, we seek to learn the Ns-tuple of corresponding points (g1(p), g2(p), …, gNs(p)), parameterized by gk : Sref → Sk, k = 1, …, Ns.
In general terms, we can now summarize our task as follows: use the functional connectivity data Ck, in conjunction with the anatomical data Da,k, k = 1, …, Ns, to estimate warping functions {gk : k = 1, …, Ns}, subject to specified regularity conditions, that bring some specified balance of anatomy and functional connectivity into alignment across subjects. That said, for the remainder of the paper we restrict attention to aligning only functional connectivity across subjects. There is no doubt that anatomy must be an integral part of a full solution; but that aspect is not new, and is already well understood. Restricting attention to the alignment of functional connectivity will allow us to concentrate on the most novel and important aspects of our approach.
To proceed, assume a reference connectivity Cref, such that for each subject k = 1, …, Ns,
| (1) |
where Ck(gk(pi), gk(pj)) = σ(Df,k(gk(pi)), Df,k(gk(pj))), and εk is zero-mean random noise. Since gk(p) may not be a mesh point, computation of Df,k(gk(p)) requires interpolation of the time series using mesh nodes in a neighborhood of gk(p). This will be important as we proceed.
Given (1), we estimate g by maximizing a regularized log likelihood:
| (2) |
where Reg(gk) constrains each warping function gk to be smooth and invertible. Here, we will focus on the log likelihood term and delay the discussion of regularization to §3. Optimization of (2) is complicated by the fact that Cref is a latent variable, so it must be estimated along with g. We use Expectation-Maximization to iteratively alternate between computing an expectation of Cref (E-step), and a maximum likelihood estimate of g given both the observed and estimated unobserved data (M-step) [15]. In the E-step, the expectation of Cref, C̄ref, conditioned on the current estimate of g, ĝ, is computed by averaging the connectivity across subjects:
| (3) |
In the M-step, the estimate ḡ is refined to maximize the likelihood of the full data:
| (4a) |
| (4b) |
where we have assumed that the noise in (1) is i.i.d. Gaussian. Because (4b) decouples, we can optimize over each subject’s warp separately, i.e., these optimizations can be done in parallel:
| (5) |
However, an interesting alternative is to perform these sequentially with an E-step after each that updates the reference estimate C̄ref. This also allows some other interesting adaptations. We note:
| (6a) |
where
| (7) |
is the leave-one-out template for subject k, which is indepedendent of gk. Thus, we replace (5) by:
| (8) |
From (5) and (8) we observe that the multi-subject alignment problem reduces to a sequence of pairwise registrations, each of which registers one subject to an average of connectivity matrices. If we use (5), each round of pairwise registrations can be done in parallel and the results used to update the average template. The difficulty is the computational update of C̄ref. Alternatively, using (8) we do the pairwise registrations sequentially and compute a new leave-one-out template after each registration. This is the approach we pursue. An algorithm for solving the pairwise registration is derived in the next section and we examine the computation of leave-one-out templates in §4.
3 Pairwise Cortical Alignment
We now develop an algorithm for aligning one subject, with connectivity CF, to a reference, with connectivity CR, with CF, CR ∈ RNv×Nv. For concreteness, from this point forward we let σ(f1, f2) = corr(f1, f2) and assume that the time series have zero mean and unit norm.
A function g: MR → SF maps a reference mesh point pi ∈ MR to g(pi) ∈ SF. By interpolating the floating subject’s times series at the points g(pi) ∈ SF we obtain the associated warped functional connectivity: . We seek ĝ that best matches C̃F to CR in the sense:
| (9) |
Here ||·||f is the matrix Frobenius norm and the regularization term Reg(g) serves as a prior over the space of allowable mappings. In the following steps, we examine how to efficiently solve (9).
Step 1: Parameterizing the dependence of C̃F on the warp
We first develop the dependence of the matrix C̃F on the warping function g. This requires specifying how the time series at the warped points g(pi) ∈ SF is interpolated using the time series data { , i = 1, …, Nv} at the mesh points { , i = 1, …, Nv}. Here, we employ linear interpolation with a spherical kernel Φ: , p ∈ SF. The kernel should be matched to the following specific objectives: (a) The kernel should be monomodal. Since the gradient of the registration objective depends on the derivative of the interpolation kernel, this will reduce the likelihood of the algorithm converging to a local minimum; (b) The support of the kernel should be finite. This will limit interpolation complexity. However, as the size of the support decreases, so will the capture range of the algorithm. At the initial stages of the algorithm, the kernel should have a broad extent, due to higher initial uncertainty, and become increasingly more localized as the algorithm converges. Thus, (c) The support of the kernel should be easily adjustable.
With these considerations in mind, we select Φ(p, pi) to be a spherical radial basis function Φi : S2 → ℝ centered at pi ∈ S2 and taking the form: Φi(p) = ϕ(d(p, pi)), p ∈ S2, where ϕ: [0, π] → ℝ and d(p, pi) is the spherical geodesic distance between p and pi [16]. Then Φi(p) is monomodal with a maximum at pi, it depends only on the distance between p and pi and is radially symmetric. In detail, we employ the particular spherical radial basis function:
| (10) |
where r is a fixed parameter, and (a)+ = a1{a ≥ 0}. Φi(p) has two continuous derivatives and its support is {p ∈ S2: d(p, pi) < 2sin−1(r/2)}. Note that the support can be easily adjusted through the parameter r. So the kernel has all of our desired properties.
We can now make the dependence of C̃F on g more explicit. Let . Then where A = [Φi(g(pj))] is the Nv × Nv matrix of interpolation coefficients dependent on g and the interpolation kernel. Next, noting that , we use A to write the post-warp correlation matrix as:
| (11) |
where D = diag(d1, d2, …, dNv) serves to normalize the updated data to unit norm: dj = ||fF(g(pj))||−1. Finally, we use à = AD to write:
| (12) |
Here, (12) encodes the dependence of the registration objective on g through the matrix Ã. It is also important to note that since the interpolation kernel is locally supported, Ã is a sparse matrix.
Step 2: Efficient Representation/Computation of the Registration Objective
We now consider the Nv × Nv matrices CF and CR. At a spatial resolution of 2 mm, the spherical model of human cortex can yield Nv ≈ 72, 000 total mesh points. In this situation, direct computation with CF and CR is prohibitive. Hence we need an efficient way to represent and compute the objective (12).
For fMRI data it is reasonable to assume that Nt ≪ Nv. Hence, since the data has been centered, the rank of and of is at most Nt − 1. For simplicity, we make the reasonable assumption that rank(TF) = rank(TR) = d. Then CF and CR can be efficiently represented by compact d-dimensional SVDs and . Moveover, these can be computed directly from SVDs of the data matrices: and . In detail:
VF = VTF, VR = VTR, , and .
The above representation avoids computing CF and CR, but we must also show that it enables efficient evaluation of (12). To this end, introduce the following linear transformation:
| (13) |
where , are orthogonal with the Nv − d columns of and forming orthonormal bases for range(VF)⊥ and range(VR)⊥, respectively. Write B as:
| (14) |
with B1 ∈ ℝd×d, B2 ∈ ℝd×Nv, B3 ∈ ℝ(Nv−d)×d and B4 ∈ ℝ(Nv−d)×(Nv−d). Substituting (13) and (14) into (12) and simplifying yields:
| (15) |
with
| (16) |
The d × d matrix B1 is readily computed since VF, VR are of manageable size. Computation of the d×Nv matrix B2 depends on . This has ON columns spanning the Nv − d dimensional subspace null(CR). Since there is residual freedom in the choice of and B2 is large, its selection merits closer examination. Now (16) can be viewed as a projection of the rows of onto the columns of VR and . The columns of lie in null(CR) and . Hence a QR-factorization yields d ON vectors in null(CR). Choosing these as the first d columns of , yields B2 = [R, 0], i.e., B2 is very sparse.
In summary, we have derived the following efficient means of evaluating the objective. By onetime preprocessing of the time series data we obtain ΣF, ΣR and VF, VR. Then given a warp g, we compute: the interpolation matrix Ã, , and finally B2 via QR factorization of . Then we evaluate (15).
Step 3: The Transformation Space and Regularization
We now examine the specification of g in greater detail. We allow each mesh point to move freely (locally) in two directions. The use of such nonlinear warp models for inter-subject cortical alignment has been validated over, for example, rigid-body transformations [17]. To specify g, we first need to set up a coordinate system on the sphere. Let U = {(φ, θ); 0 < φ < π, 0 < θ < 2π}. Then the sphere can be parameterized by x: U → ℝ3 with x(φ, θ) = (sin φ cos θ, sin φ sin θ, cos φ). Here, φ is a zenith angle measured against one of the principal axes, and θ is an azimuthal angle measured in one of the projective planes (i.e., xy-plane, xz-plane, or yz-plane). Note that x omits a semicircle of S2; so at least two such parameterizations are required to cover the entire sphere [18].
Consider pi ∈ S2 parameterized by x(φ, θ) such that pi = x(φi, θi). Then the warp field at pi is:
| (17) |
for displacements Δφi and Δθi. The warp g is thus parameterized by: {φ̃i, θ̃i, i = 1, …, Nv}.
The warp g must be regularized to avoid undesired topological distortions (e.g. folding and excessive expansion) and to avoid over-fitting the data. This is achieved by adding a regularization term to the objective that penalizes such distortions. There are several ways this can be done. Here we follow [14] and regularize g by penalizing both metric and areal distortion. The metric distortion term penalizes warps that disrupt local distances between neighboring mesh nodes. This has the effect of limiting the expansion/contraction of cortex. The areal distortion term seeks to preserve a consistent orientation of the surface. Given a triangularization of the spherical mesh, each triangle is given an oriented normal vector that initially points radially outward from the sphere. Constraining the oriented area of all triangles to be positive prevents folds in the surface [14].
Step 4: Optimization of the objective
We optimize (3) over g by gradient descent. Denote the objective by S(g), let ãij = aijdj be the (i, j)-th entry of à = AD and . From the parameterization of the warp (17), we see that ãij = Φi(x(φ̃j, θ̃j))||TF a(x(φ̃j, θ̃j))||−1 depends only on the warp parameters of the jth mesh node, φ̃j and θ̃j. Then, by the chain rule, the partial derivative of S(g) with respect to φ̃j is given by:
| (18) |
A similar expression is obtained for the partial derivative with respect to θ̃j. Since the interpolation kernel is supported locally, the summation in (18) is taken over a small number of terms. A full expression for ∂S/∂φ̃j is given in the supplemental, and that of ∂Reg(g)/∂φ̃j in [14].
To help avoid local minima we take a multi-resolution optimization approach [19]. The registration is run on a sequence of spatial resolutions r1 > r2 > ··· > rM, with rM given by the original resolution of the data. The result at resolution rm is used to initialize the alignment at resolution rm+1. The alignment for rm is performed by matching the kernel parameter r in (10) to rm. Note that the reference dataset is also spatially smoothed at each rm by the transformation in (11), with A = [a(p1) a(p2) ··· a(pNv)]. The pairwise algorithm is summarized as Algorithm 1 in Figure 1.
Figure 1.
The registration algorithms.
4 Multi-Subject Alignment: Computing Leave-one-out Templates
We now return to the multi-subject alignment problem, which is summarized as Algorithm 2 in Figure 1. It only remains to discuss efficient computation of the leave-one-out-template (7). Since C̄k is an average of Ns − 1 positive semi-definite matrices each of rank d, the rank d̄ of C̄k is bounded as follows d ≤ d̄ ≤ (Ns − 1)d. Assume that C̃n, the connectivity matrix of subject n after warp gn (see (11)), has an efficient d ≪ Nv dimensional SVD representation .
To compute the SVD for C̄k, we exploit the sequential nature of the multi-subject alignment algorithm by refining the SVD of the leave-one-out template for subject k−1, , computed in the previous iteration. This is achieved by expressing C̄k in terms of C̄k−1:
| (19) |
and computing matrix decompositions for the singular vectors of C̃k−1 and C̃k in terms of V̄k−1:
| (20a) |
| (20b) |
where , for j = k − 1, k, projects the columns of Ṽj onto the columns of V̄k−1. The second term of (20a), Qk−1Rk−1, is the QR-decomposition of the residual components of Ṽk−1 after projection onto range(V̄k−1). Since C̄k−1 is an average of positive semi-definite matrices that includes C̃k, we are sure that range(Ṽk) ⊆ range(V̄k−1), (supplementary material).
Using the matrix decompositions (20a) and (20b), C̄k in (19) above can be expressed as:
| (21) |
where G is the symmetric (d̄ + d) × (d̄ + d) matrix:
| (22) |
We now compute the SVD of . Then, using (21), we obtain the SVD for C̄k as:
| (23) |
For a moderate number of subjects, (d̄ + d) ≤ Nsd ≪ Nv, this approach is more efficient than a brute-force . Additionally, it works directly on the singular values Σ̃k and vectors Ṽk of each warped connectivity matrix C̃k, alleviating the need to store large Nv × Nv matrices.
5 Experimental Results
We tested the algorithm using fMRI data collected from 10 subjects viewing a movie split into 2 sessions separated by a short break. The data was preprocessed following [5]. For each subject, a structural scan was acquired before each session, from which the cortical surface model was derived (§2) and then anatomically aligned to a template using FreeSurfer (Fischl, http://surfer.nmr.mgh.harvard.edu). Similar to [5], we find that anatomical alignment based on cortical curvature serves as a superior starting point for functional alignment over Talairach alignment.
First, functional connectivity was found for each subject and session: Ck,i, k = 1, …, Ns, i = 1, 2. These were then aligned within subjects, Ck,1 ↔ Ck,2, and across subjects, Ck,1 ↔ Cj,2, using Algorithm 1. Since the data starts in anatomical correspondence, we expect small warp displacements within subject and larger ones across subjects. The mean intra-subject warp displacement was 0.72 mm (σ = 0.48), with 77% of the mesh nodes warped less than 1 mm and fewer than 1.5% warped by more than the data spatial resolution (2 mm). In contrast, the mean inter-subject warp displacement was 1.46 mm (σ = 0.92 mm), with 22% of nodes warped more than 2 mm. See Figures 2(a)–(b).
Figure 2.
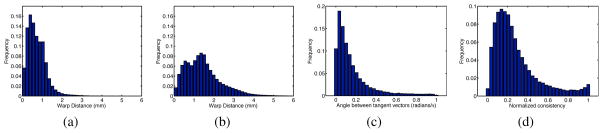
Consistency Histograms. (a) Intra-subject warp distances; (b) Inter-subject warp distances; (c) Angle between warp vectors across sessions; (d) Across-session normalized warp consistency measure WNC.
In a separate analysis, each subject was aligned to its leave-one-out template on each session using Algorithm 1, yielding a set of warps gk,i(pj), k = 1, …, Ns, i = 1, 2, j = 1, …, Nv. To evaluate the consistency of the correspondence derived from different sessions, we compared the warps gk,1 to gk,2 for each subject k. Here, we only consider nodes that are warped by at least the data resolution. This analysis provides a measure of the sensitivity to noise present in the fMRI data. At node pj, we compute the angle 0 ≤ θ ≤ π between the warp tangent vectors of gk,1(pj) and gk,2(pj). This measures the consistency of the direction of the warp across sessions: smaller values of θ suggest a greater warp coherence across sessions. Figure 2(c) shows a histogram of θ averaged across the cortical nodes of all 10 subjects. The tight distribution centered near θ = 0 suggests significant consistency in the warp direction across sessions. In particular, 93% of the density for θ lies inside π/2, 81% inside π/4, and 58% inside π/8. As a secondary comparison, we compute a normalized consistency measure WNC(pj) = d(gk,1(pj), gk,2(pj))/(d(gk,1(pj), pj) + d(gk,2(pj), pj)), where d(·, ·) is spherical geodesic distance. The measure takes variability in both warp angle and magnitude into account; it is bounded between 0 and 1, and WNC(pj) = 0 only if gk,1(pj) = gk,2(pj). A histogram for WNC is given in 2(d); WNC exhibits a peak at 0.15, with a mean of 0.28 (σ = 0.22).
Finally, Algorithm 2 was applied to the first session fMRI data to learn a set of warps g = (g1, …, gNs) for 10 subjects. The alignment required approximately 10 hours on a Intel 3.8GHz Nehalem quad-core processor with 12GB RAM. To evaluate the alignment, we apply the warps to the held out second session fMRI data, where subjects viewed a different segment of the movie. This warping yields data { } for each subject k, with interpolation performed in the original volume to avoid artificial smoothing. The cross-validated inter-subject correlation ISC(pi) is the mean correlation of each subject’s functional time series with the mean time series of the other subjects:
| (24) |
We also compute the mean inter-subject correlation, .
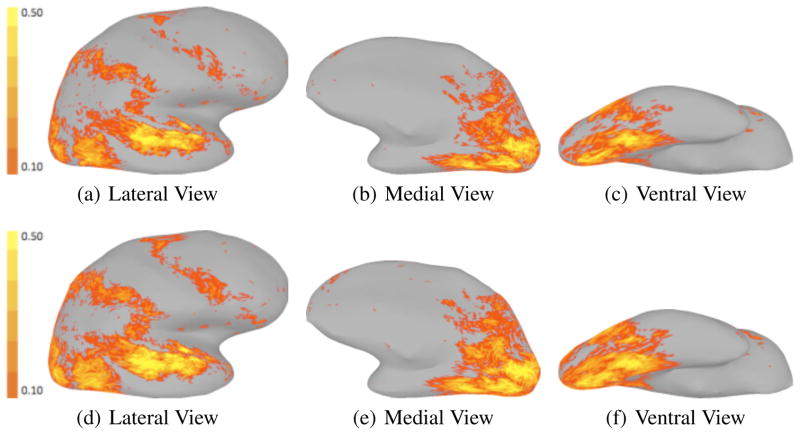
We compare the cross-validated ISC map with the ISC map of the second session movie viewing computed under anatomical correspondence. Mean ISC improved by 18%, from 0.072 to 0.085. In addition, the number of significant inter-subject correlations (ISC(pi) > 0.1, P < 0.01) increased by 22.9%, from 19, 362 to 23, 789. Figure 3 shows the ISC maps computed under anatomical alignment and functional alignment on the inflated right cortical hemisphere. As expected, the areas of improvement in inter-subject correlation are consistent with the extrinsic regions of cortex [6].
Figure 3.
Map of ISC on right cortical hemisphere, alignment: anatomical (top), functional (bottom).
6 Conclusion
We have proposed a novel cortical registration algorithm that produces a functional correspondence across a set of subjects. The algorithm uses the fMRI data directly to align the spatial patterns of functional response elicited by a movie viewing. Despite the high-dimensionality of the data under consideration, the algorithm is efficient in both space and time complexity.
By comparing the inter-subject alignments derived from different fMRI experimental sessions, we show that the correspondence is consistent and robust to noise and variability in the fMRI temporal response. We also cross-validate the correspondence on independent test data that was not used to derive the alignment. On the test data, the algorithm produces a consistent increase in inter-subject correlation of fMRI time series, suggesting that functional alignment of extrinsic regions of cortex that are directly driven by the movie viewing experiment, such as visual and auditory areas, is improved considerably. Further testing is warranted to evaluate improvement in intrinsic areas of cortex whose response is not temporally synchronized with the experimental stimulus.
Supplementary Material
References
- 1.Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain. Thieme Publishing Group; 1988. [Google Scholar]
- 2.Fischl B, Tootell RBH, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson JDG, Myers R, Frackowiak RSF, Hajnal JV, Woods RP, Mazziotta JC, Shipp S, Zeki S. Area v5 of the human brain: evidence from a combined study using positron emission tomography and magnetic resonance imaging. Cerebral Cortex. 1993;3:79–94. doi: 10.1093/cercor/3.2.79. [DOI] [PubMed] [Google Scholar]
- 4.Rademacher J, Caviness VS, Steinmetz H, Galaburda AM. Topographical variation of the human primary cortices: implications for neuroimaging, brain mapping and neurobiology. Cerebral Cortex. 1995;3:313–329. doi: 10.1093/cercor/3.4.313. [DOI] [PubMed] [Google Scholar]
- 5.Sabuncu MR, Singer BD, Conroy B, Bryan RE, Ramadge PJ, Haxby JV. Function-based inter-subject alignment of human cortical anatomy. [Access published on May 6, 2009];Cerebral Cortex Advance. doi: 10.1093/cercor/bhp085. [DOI] [Google Scholar]
- 6.Hasson U, Nir Y, Fuhrmann G, Malach R. Intersubject synchronization of cortical activity during natural vision. Science. 2004;303:1634–1640. doi: 10.1126/science.1089506. [DOI] [PubMed] [Google Scholar]
- 7.Golland Y, Bentin S, Gelbard H, Benjamini Y, Heller R, Nir Y, Hasson U, Malach R. Extrinsic and intrinsic systems in the posterior cortex of the human brain revealed during natural sensory stimulation. Cerebral Cortex. 2007;17:766–777. doi: 10.1093/cercor/bhk030. [DOI] [PubMed] [Google Scholar]
- 8.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. PNAS. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friston KJ. Functional and effective connectivity in neuroimaging. Human Brain Mapping. 1994;2:56–78. [Google Scholar]
- 10.Greicius Michael D, Krasnow Ben, Reiss Allan L, Menon Vinod. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. PNAS. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent JL, Snyder AZ, Fox MD, Shannon BJ, Andrews JR, Raichle ME, Buckner RL. Coherent spontaneous activity identifies a hippocampal-parietal memory network. J Neurophysiol. 2006;96:3517–3531. doi: 10.1152/jn.00048.2006. [DOI] [PubMed] [Google Scholar]
- 12.Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. J Am Med Inform Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. i. segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 14.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. ii. inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 15.McLachlan GJ, Krishnan T. The EM Algorithm and Extensions. Wiley; 1997. [Google Scholar]
- 16.Fasshauer GE, Schumaker LL. Scattered data fitting on the sphere. Proceedings of the international conference on mathematical methods for curves and surfaces II; 1998. pp. 117–166. [Google Scholar]
- 17.Ardekani BA, Bachman AH, Strother SC, Fujibayashi Y, Yonekura Y. Impact of inter-subject image registration on group analysis of fmri data. International Congress Series. 2004;1265:49–59. [Google Scholar]
- 18.Do Carmo M. Differential Geometry of Curves and Surfaces. Prentice Hall; 1976. [Google Scholar]
- 19.Bajcsy R, Kovacic S. Multiresolution elastic matching. Computer Vision, Graphics, and Image Processing. 1989;46:1–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.