Abstract
The purpose of this study was to develop and assess a new quantitative four-view mammographic image feature based fusion model to predict the near-term breast cancer risk of the individual women after a negative screening mammography examination of interest. The dataset included fully-anonymized mammograms acquired on 870 women with two sequential full-field digital mammography (FFDM) examinations. For each woman, the first “prior” examination in the series was interpreted as negative (not recalled) during the original image reading. In the second “current” examination, 430 women were diagnosed with pathology verified cancers and 440 remained negative (“cancer-free”). For each of four bilateral craniocaudal and mediolateral oblique view images of left and right breasts, we computed and analyzed eight groups of global mammographic texture and tissue density image features. A risk prediction model based on three artificial neural networks was developed to fuse image features computed from two bilateral views of four images. The risk model performance was tested using a ten-fold cross-validation method and a number of performance evaluation indices including the area under the receiver operating characteristic curve (AUC) and odds ratio (OR). The highest AUC = 0.725±0.026 was obtained when the model was trained by gray-level run length statistics (RLS) texture features computed on dense breast regions, which was significantly higher than the AUC values achieved using the model trained by only two bilateral one-view images (p < 0.02). The adjustable OR values monotonically increased from 1.0 to 11.8 as model-generated risk score increases. The regression analysis of OR values also showed a significant increase trend in slope (p < 0.01). As a result, this preliminary study demonstrated that a new four-view mammographic image feature based risk model could provide useful and supplementary image information to help predict the near-term breast cancer risk.
Keywords: Breast cancer, Computer-aided detection (CAD), Near-term breast cancer risk stratification, Mammographic density feature analysis, Full-field digital mammography (FFDM)
I. INTRODUCTION
Due to the controversy and low efficacy of current uniform population-based mammography screening, 4, 22 developing a new personalized breast cancer screening paradigm to detect more early cancers and also substantially reduce false-positive recalls has been attracting extensive research interest in the last several years. 8, 15, 32 The goal of developing and implementing a personalized cancer screening paradigm is to identify a small fraction of women with significantly higher-than-average near-term risk of developing breast cancer. As a result, instead of a uniform mammography screening protocol, each woman should have an adaptive or adjustable screening interval and/or screening methods. Specifically, only a small fraction of “high-risk” women should be screened more frequently (namely, annually or semi-annually), whereas the vast majority of women with average or lower near-term risk should be screened at longer intervals (e.g., every two to five years) until their near-term cancer risk significantly increases in new assessment.
The success of establishing an optimal personalized breast cancer screening paradigm depends on the development of a reliable risk prediction model. Although many epidemiology study-based breast cancer risk models, such as Gail, Claus, and Tyrer-Cuzick model 2, 6, have been developed to assess a long-term or lifetime relative risk of a woman developing cancer compared to an average general population risk, establishing a personalized screening mammography paradigm requires developing new risk models that have higher discriminatory power in predicting the risk of individual women developing cancer in the near term. Most of the new risk models developed for investigating the feasibility of personalized screening are primarily based on breast or mammographic density. 7 Despite the fact that breast density is the second highest risk factor behind age, 2 its discriminatory power at the individual level remains quite low and controversial. 16, 30 Subjectively rating mammographic density based on the BIRADS standard is often not accurate or reliable due to the large intra- and inter-reader variability. 3 In addition, many useful mammographic density features, such as tissue spiculation patterns and bilateral asymmetry cannot be quantitatively evaluated and compared using subjective (visual) assessment.
To overcome these issues and develop a quantitative mammographic image feature based near-term breast cancer risk model, in our recent work, we had identified a new breast cancer risk factor that relates to the bilateral mammographic density asymmetry between the left and right breasts and investigated its association with the risk of the individual women having mammography-detectable breast cancer in the next sequential screening mammography after a negative examination of interest. 45 By combining the selected image features and three popular risk factors (namely, age, family history and subjectively-rated breast density using BIRADS) used in existing epidemiology based risk models, we also trained and tested a support vector machine (SVM) classifier to predict the near-term risk/likelihood of breast cancer development, which yielded the area under a receiver operating characteristic (ROC) curve, AUC = 0.725±0.018 and maximum odds ratio of 12.34. 36
Based on the promising results of our previous studies, in this study we focused on investigating two new issues to further improve the performance and robustness of applying this new quantitative image feature biomarker or analysis method to predict near-term breast cancer risk. First, since a large number of image texture features can be computed from digital mammograms, how to identify effective features and minimize redundancy is difficult in CAD and other quantitative image analysis fields. In this study, we applied a number of popular algorithms used in medical image processing to compute mammographic image texture features including Gabor features, 26 Weber Local Descriptor (WLD) features, 11 gray-level run length texture features, 37 gray-level co-occurrence matrix (GLCM) based features, 20 and generic statistical or directional based features including a fractal dimension texture based feature 1 from the different breast regions. We then performed a comprehensive data analysis to identify a group of image features that have the optimal (or “best”) association to the near-term breast cancer risk prediction. Second, most of the previous studies of predicting breast cancer risk used only mammographic tissue or density features computed from a region of interest (ROI) that are extracted from one mammogram 23, 42 and our previous studies only used two bilateral craniocaudal (CC) view images. 36, 45 In this study, we developed and tested a new scheme to combine the mammographic density features computed from four mammograms including bilateral CC and mediolateral oblique (MLO) view images of left and right breasts. Specifically, we introduced a new “scoring fusion” approach to extract and fuse information derived from all four images. Although a four-view image based computer-aided detection (CAD) scheme of mammograms has been investigated in our previous study to reduce false positive recalls in screening mammography, 35 integrating image features computed from four-view images, to the best of our knowledge, has never been tested and applied for the breast cancer risk prediction task.
Therefore, the purpose of this study is to identify a group of highly discriminative and non-redundant image features from all four-view screening mammography images and develop a new CAD-based near-term breast cancer risk prediction model by fusing the effective image features computed from both CC and MLO view images using a new “scoring fusion” classification scheme. The details of our scheme, experimental procedures and results are reported in the following sections.
II. MATERIALS
As an ongoing research effort, we have been continuing to retrospectively collect fully-anonymized full-field digital mammography (FFDM) images from an available clinical database of University of Pittsburgh Medical Center to build a large and diverse database in the last several years. All images were acquired using Hologic Selenia FFDM systems (Hologic Inc., Bedford, MA, USA). The detail description of our institutional review board (IRB) approved image data collection protocol has been reported and different portions of this database have been used in a number of our previous studies. 35, 44, 45 In this study, from this existing database we assembled a dataset that consists of FFDM images acquired from 870 women who underwent at least two sequential FFDM screening examinations. Each examination had four FFDM images representing the CC and MLO view of the left and right breasts. From the results of the latest FFDM examination on record (or namely, the “current” images), 430 were considered “positive” cases including 366 screening detected cancer cases, 41 interval cancer cases and 23 high-risk pre-cancer cases with surgically-removed lesions. Among the remaining 440 “negative” cases, 220 were diagnosed with benign lesions or recalled but later proved to be benign or negative in the additional image workup or biopsy, and 220 were screening negative (not recalled). All benign and negative cases remained cancer-free followed by at least two next sequential FFDM screening examinations. The average ages and standard deviations were 59.5±11.5, 51.9±8.2 and 46.1±5.6 years old for the positive, benign and negative groups of women, respectively.
For each case, we retrospectively retrieved FFDM images acquired from the most recent (or first “prior”) FFDM screening examinations which is typically 12-18 months prior to the “current” FFDM examination. All “prior” FFDM examinations were interpreted and rated by radiologists as “negative” or “definitely benign” (i.e., screening BIRADS 1 or 2), respectively. Thus, all these “prior” examinations were not recalled. These “prior” screening benign or negative images were used as the baseline images to be analyzed in this study. Although all these baseline images were negative, they were divided into different categories based on their status changes in the next sequential FFDM examinations (or “current” images). For example, the cases were divided into subgroups of “positive” (cancer detected in “current” images) or “negative” (i.e., remained cancer-free) cases.
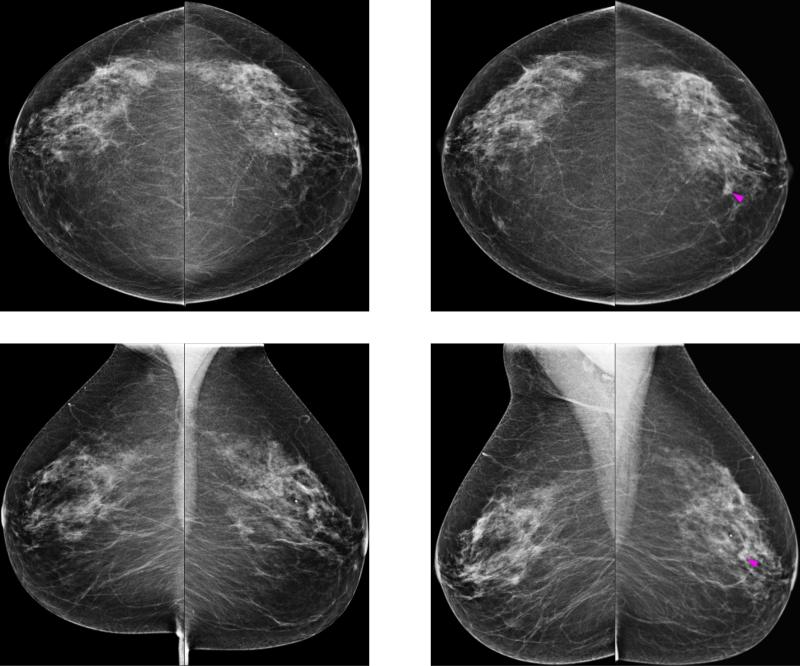
Figure 1 shows two sets of bilateral CC and MLO view images acquired from the “prior” (left) and “current” (right) FFDM screening examinations of a woman. The “prior” images were interpreted as negative and a cancer (pointed by an arrow) was detected on the “current” examination by a radiologist, which was later confirmed in biopsy and pathology examinations. From Fig. 1, we can make two observations: first, we observe that the images from the CC and MLO views demonstrated different mammographic density and texture based properties within the lesion region and the breast fibroglandular tissue background. Thus, it is likely that different and supplementary information can be extracted from both CC and MLO views, which can be integrated in a risk prediction model or classifier with an appropriate fusion method. Second, bilateral asymmetry can be observed between the two (left and right) breasts in the “prior” images and especially in the regions surrounding the developing lesion, which indicates that useful information can be extracted by computing texture based features on these images.
FIG. 1.
Example of a case in the subgroup of positive cases. It shows two sets of bilateral craniocaudal (CC) and mediolateral oblique (MLO) view mammograms acquired from the “prior” (left) and “current” (right) FFDM screening examinations of a woman. The “prior” images were interpreted as negative, and a suspicious lesion (indicated by the magenta arrow) was detected on the “current” examination by a radiologist, which was later confirmed to be malignant in biopsy and pathology examinations.
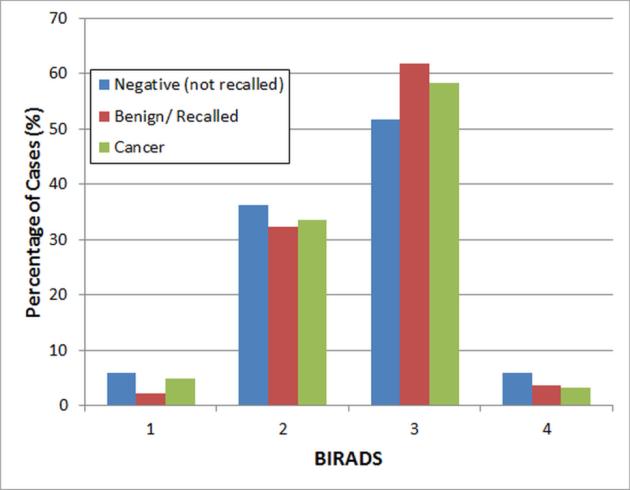
Figure 2 displays the distribution of mammographic density of the cases in our dataset, which were rated by the radiologists based on mammographic density BIRADS categories. Data analysis results based on ANOVA test showed no statistically significant differences of the mammographic density distribution (BIRADS) between the three case subgroups (p > 0.28) in our dataset.
FIG. 2.
Distributions of subjectively-rated mammographic density (BIRADS) in the three subgroups of positive, benign and negative cases.
III. METHODS
III.A. Automated breast segmentation and mammographic image feature computation
For each case in our study, two pairs of bilateral CC-view and MLO-view FFDM images of the left and right breasts were analyzed. To increase the accuracy of our scheme, we first applied an automatic segmentation scheme on each image to extract the breast regions as described in our previous publications. 35, 44 In brief, a gray level histogram of the image is plotted and an iterative searching method is used to detect the smoothest curvature between the breast tissue and background or air region. The pixels in the background were discarded and the skin region was removed by a morphological erosion operation. To detect the breast region in the MLO images, an additional step was required to detect the chest wall/pectoral muscle, following which all pixels within the pectoral muscle were discarded as they did not add beneficial information to the detection task.
After the breast segmentation step, we applied a variety of computerized schemes to explore and compute different types of image texture features. We then analyzed and compared their correlations and contributions to predict near-term cancer development risk. In the literature, various epidemiology based studies have studied the association between many mammographic texture pattern features and breast cancer risk. 17, 18, 25, 28 Various studies have also analyzed different methods of estimating percentage mammographic density and its correlation with breast cancer risk. 5, 9, 21, 24 In this study, we examined individually and combined various texture and mammographic density based features that have been proposed in the literature, as well as some new features that to our knowledge have never been examined before for breast cancer risk prediction. Specifically, we examined and compared 8 different groups of mammographic image features in this study.
The first feature group consists of the popular gray-level run length statistics (RLS) texture features computed on the whole breast region. 37, 43 The RLS features consisted of: low gray level run emphasis, high gray level run emphasis, short run high gray level emphasis, gray level non-uniformity, and run percentage. To compute the RLS features, we first reduced the gray level range of the images from 4096 to 256 gray levels resulting in an 8 bit depth gray level run length matrix. We computed four run length matrices computed along 0, 45, 90, and 135 degrees. We then computed the final feature values as the average and maximum values of the four run length features along these directions.
The second group of features consisted of the RLS features computed on the dense breast region, whereby we had defined segmentation method of the dense breast region in our previous publications. 10, 36 The same features and computation methodology were used to compute the features for the dense breast region as for the whole breast region.
In the third feature group, we examined Gabor filter features 26 computed on the whole breast region. Gabor features are highly popular for various tasks including content-based mammogram retrieval 41 and face recognition. 19 In particular, the spatial response profiles of Gabor filters are similar to that of the mammalian vision receptive field. 26 An important advantage of Gabor filters is that they can withstand photometric disturbances, e.g. image noise, low resolution, and monochrome as they have optimal Heisenberg resolution in space and spatial frequency 13 and are thus useful for mammographic texture analysis. Using the method proposed in Refs. 20, 33, 41, we computed the following features for this group: contrast, difference variance, difference entropy, sum of squares: variance, and dissimilarity. The fourth feature group consisted of the Gabor filter features computed on the dense breast region.
In the fifth feature group, we examined a group of new features based on the Weber Local Descriptor (WLD) descriptors proposed by Chen et al. 11 In Ref. 11, the WLD features outperformed other widely-used features (including Gabor and SIFT) in experimental results on popular texture databases (e.g., Brodatz and KTH-TIPS2-a), and produced good performances compared to best-known results on some human face detection experiments (e.g., MIT+CMU frontal face test set and CMU profile test set). The WLD descriptor is a simple, yet powerful and robust local descriptor. It consists of two separate components – differential excitation and orientation – and is inspired by Weber's Law, which is a law developed according to the perception of human beings. It states that the change of a stimulus (such as pixel intensity, lighting) that will be just noticeable is a constant ratio of the original stimulus. When the change is smaller than the constant ratio of the original stimulus, a human eye (vision system) can recognize it as background noise rather than a valid signal. Thus, for a given pixel, we computed the differential excitation component of the WLD descriptor as a ratio between two terms: 11 (1) the relative intensity differences of a current pixel against its 3×3 neighbors; (2) the intensity of the current pixel. With the differential excitation component, the local salient patterns in the image can be extracted. 11 We also computed the gradient orientation at the location of each pixel. We computed the contrast, difference variance, difference entropy, sum of squares: variance, and dissimilarity WLD features using the method proposed in Refs. 20, 33, 41.
The sixth feature group consisted of gray level co-occurrence matrix (GLCM) features computed on the whole breast region (namely, homogeneity defined in Ref. 33, homogeneity defined in Matlab®, normalized inverse difference, normalized inverse difference moment, energy, and maximum probability). 12, 20, 33 We computed these features in four directions: 0, 45, 90, and 135 degrees and at distance d = 1. In Refs. 27, 38, it was reported that the GLCM based features computed at d > 1 were strongly correlated. Similar to Refs. 27, 38, we reduced the gray level range of the images from 4096 to 256 gray levels to calculate the GLCM matrix, and computed the average and maximum values of the features along the four directions. The seventh feature group consisted of the GLCM features computed on the dense breast regions.
In the eighth feature group, we computed some generic statistical gray value features. We included a percentage density (PD) measure computed as the number of pixels within the breast region that corresponded with the maximum intensity value within the breast divided by the area (number of pixels) of the segmented breast region. 17, 40 We also examined for the first time a new fractal texture feature (average fractal dimension) that has been proved useful for differentiating between advanced (aggressive) and early-stage (non-aggressive) lung tumors in contrast-enhanced computed tomography images 1 in our risk prediction scheme. In addition, we computed the mean gradient direction 17 and the mean y-axis directional gradient by utilizing the Sobel operator. Finally, we included the regional descriptor of the area of number of pixels of the segmented breast region.
Each image feature was computed separately from the two view images of the left and right breasts. Thus, each view (e.g., CC or MLO) of a case has two corresponding sets of features. In our previous studies, 36, 40 we applied a subtraction method to compute the absolute difference of two matched corresponding feature values computed from the CC view images of the left and right breasts. In this study, we examined a different approach, namely we retained the maximum corresponding features (feature with the higher value from either the left or right breast) computed from the CC view images of both breasts and also the maximum features computed from the MLO view images of both breasts. Namely, we observed in our preliminary experiments that abnormalities that start to develop in one breast can be detected more sensitively by extracting the maximum features of both breasts. The maximum features can detect the structural and textural changes within the breast regions due to abnormalities that are starting to develop, but have not matured or developed fully yet. In contrast, a subtraction operation between the bilateral features of two breasts might not be sensitive enough to detect small or subtle abnormal changes that occur only within a very small region or locality of the abnormal breast, e.g. in the vicinity of the lesion(s). In summary, total number of 10, 10, 10, 10, 10, 12, 12, and 5 image features were explored and computed from each bilateral pair of CC or MLO view images for feature groups 1 to 8, respectively.
III.B. Optimization of a scoring fusion ANN on the CC and MLO view features
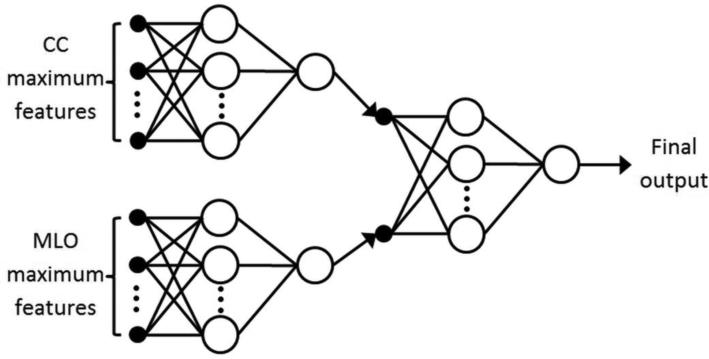
To combine image features extracted from all four images of CC and MLO views for the task of predicting near-term breast cancer risk, we propose a two-stage “scoring fusion” classification scheme as shown in Fig. 3: At the first stage, two artificial neural networks (ANNs) are trained separately and individually on the maximum features extracted from the CC and MLO views, respectively. At the second stage of the scheme, a subsequent ANN is optimized to adaptively assign/adjust appropriate weights to the outputs of the CC and MLO based ANNs from the previous stage. All three ANNs were strictly feedforward ANNs with no recurrent loops.
FIG. 3.
A two-stage “scoring fusion” ANN classification scheme is depicted (modified from Ref. 35), whereby the final classification score is derived by optimally fusing the information from two ANNs trained on maximum features extracted from the craniocaudal (CC) and mediolateral oblique (MLO) views, respectively.
We trained the ANN classifiers by the gradient descent with momentum and adaptive learning backpropagation algorithm. 31 In order to derive the optimal number of hidden nodes to utilize in the hidden layer of the ANNs, we analyzed the networks’ performances for different numbers of hidden nodes in the hidden layer: At the first stage (i.e., the CC and MLO based ANNs), the number of hidden nodes was varied between 2 to 10. For the next/subsequent “scoring fusion” ANN, this range was 1 to 5. Namely, we employed a higher range for the CC and MLO view image based ANNs as they had more input features compared with only two features for the scoring fusion ANN. We performed the training and testing of our scheme in a ten-fold cross-validation framework as explained in Sec. III.C. To identify the optimal topology of the ANN in particular the number of hidden neurons, for each cycle of the cross-validation scheme, we trained 150 ANNs initialized with random weights on the training subset and selected the ANNs that produced the highest area under a receiver operating characteristic (ROC) curve (AUC) results on the training subset. Other parameters related to the ANN training are as follows: Number of training iterations (500), training momentum (0.9), and learning rate (0.01). Namely, we utilized a high ratio of the training momentum to the learning rate and a limited number of iterations in order to reduce “overfitting” and maintain classifier robustness, and to reduce the overall computation time. We also used the hyperbolic tangent activation function at the ANN hidden nodes and the linear activation function at the output node. We repeated this training process 9 times using the image features computed from each of 8 image feature groups and the combination of all features from 8 groups to identify the optimal number of hidden neurons for each ANN trained using image features computed from bilateral CC or MLO view images.
III.C. Classification methodology and experimental setup
Next, to test the performance of this two-layer ANN based “scoring fusion” classifier we applied a ten-fold cross-validation method in which the sum of 430 positive cases and 440 negative and benign cases were randomly divided into 10 exclusive subgroups/partitions. In each validation (namely, training and testing) cycle, nine partitions were used to train the classifier and the trained classifier was subsequently applied to the remaining partition. For each case, the classifier generated a corresponding risk score of predicting cancer development in the near term. In this study, a higher risk score indicates a higher probability of the woman having breast cancer detectable in the next sequential FFDM screening examination, or vice versa. This process was repeatedly executed 10 times with the 10 different combinations of partitions. Thus, each of the cases in our dataset was tested once with a corresponding “scoring fusion” classifier-generated risk score. To evaluate the contribution of each of 8 feature groups discussed in Sec. III.A, we repeated the ten-fold cross-validation experiments for each individual feature group as well as combining all features and computed Spearman's rank correlation coefficient along with the corresponding p-values to assess the degree of association between each feature group.
To evaluate the accuracy and potential clinical utility of our breast cancer risk prediction model, we assessed two major performance components, namely the discrimination and calibration components of our new image feature based risk model. 34 Specifically, we used an area under ROC curve (AUC) and an adjusted odds ratio (OR) to assess the discrimination and calibration performance of our model, respectively. First, we computed AUC values along with the corresponding 95% confidence intervals (CIs) using a ROC curve fitting program that uses the expanded binormal model and maximum likelihood estimation method (ROCKIT http://metz-roc.uchicago.edu/MetzROC/software, University of Chicago, 1998). We computed and compared AUC values of the ANN based risk prediction using different image feature sets, as well as using the ANN trained with a different single view (either CC or MLO) and the final “scoring fusion” based ANN classification scheme. From the comparison, we identified one optimal feature set and the prediction model that yielded highest risk prediction performance.
Second, using the optimal risk prediction model identified in the previous step based on AUC value comparison, we computed and analyzed the ORs of the prediction results. In this process, we sorted the model-generated risk prediction scores in ascending order and selected five threshold values to segment all the cases into five subgroups/bins with an equal number of cases within each subgroup. We then used a publically-available statistical computing software package (R version 2.1.1, http://www.r-project.org) to compute and analyze the adjusted ORs in all subgroups including an OR increasing risk trend. The adjusted ORs and the corresponding 95% CIs at subgroups 2 to 5 were computed using the cases in subgroup 1 as a baseline/reference.
Since an operational threshold on the scheme or model generated classification or prediction score is required to apply a CAD scheme or a risk model in the clinical practice, in this study we applied a threshold of 0.6 on the risk model-generated risk prediction scores, which is in the middle range of the ANN-generated scores as we used a linear activation function in the ANN. Using this operational threshold, we generated a confusion matrix and then compute the overall prediction accuracy, as well as the positive and negative predictive values of applying our risk model to our testing dataset. In addition, to test the potential performance dependency of our new risk prediction model on different clinical or demographic information, we stratified the cases within our dataset into different subgroups based on different criteria (i.e., density BIRADS categories). We then computed and analyzed the AUC values, prediction sensitivity and specificity levels of our new risk prediction model in different subgroups.
IV. RESULTS
Table 1 displays the AUC results obtained for each of 8 feature groups as discussed in Sec. III.A. From Table 1, the highest AUC result was obtained using the run length features computed on the dense breast region (AUC = 0.725±0.026), while the lowest-performing features were yielded using WLD and the generic statistical features. From Table 1, we also observed that using a “scoring fusion” based ANN approach to combine two classification scores generated using CC and MLO view image features always yielded higher AUC results than the individual CC and MLO based ANNs except for the RLS features computed on the whole breast region and the WLD features. For RLS computed on the whole breast region and WLD, the AUC result computed using the MLO view ANN was much lower than that of the CC view ANN; this likely caused the AUC result of the “scoring fusion” classifier to decrease. Also, the AUC result of combining all 79 features (AUC = 0.695±0.075) was lower than the AUC result of using RLS features computed on the dense breast region. This indicates that many of the features are significantly correlated or redundant, which was confirmed by calculating Spearman's rank correlation coefficient between the different feature groups (as shown in Tables 2 and 3). From Tables 2 and 3, we observed that significant correlations were obtained between all feature groups except between the RLS features computed on the dense breast region and the generic statistical features, between WLD and GLCM features computed on the dense breast region, and between GLCM computed on the dense breast region and the generic statistical features.
TABLE 1.
Average AUC results and corresponding standard deviation intervals computed over the ten-fold cross-validation experiments for 9 sets of ANNs optimized on the CC and MLO views and the proposed scoring fusion method using the image features from 8 feature groups and all 79 features, respectively.
| Feature group | Number of features | AUC |
||
|---|---|---|---|---|
| CC | MLO | Scoring fusion | ||
| RLS whole | 10 | 0.702±0.072 | 0.639±0.078 | 0.696±0.076 |
| RLS dense | 10 | 0.701±0.039 | 0.671±0.043 | 0.725±0.026 |
| Gabor whole | 10 | 0.643±0.050 | 0.618±0.076 | 0.676±0.047 |
| Gabor dense | 10 | 0.666±0.052 | 0.611±0.038 | 0.682±0.041 |
| WLD | 10 | 0.655±0.041 | 0.549±0.057 | 0.637±0.054 |
| GLCM whole | 12 | 0.677±0.036 | 0.612±0.037 | 0.686±0.049 |
| GLCM dense | 12 | 0.634±0.052 | 0.617±0.071 | 0.658±0.058 |
| Generic statistical | 5 | 0.622±0.042 | 0.585±0.065 | 0.636±0.049 |
| All | 79 | 0.681±0.063 | 0.654±0.080 | 0.695±0.075 |
TABLE 2.
Spearman's rank correlation coefficient between different feature groups. The diagonal coefficient values in the table are equivalent and were thus omitted (–).
| Feature group | Spearman's rank correlation coefficient |
||||||
|---|---|---|---|---|---|---|---|
| RLS dense | Gabor whole | Gabor dense | WLD | GLCM whole | GLCM dense | Generic statistical | |
| RLS whole | 0.19 | 0.15 | 0.18 | 0.09 | 0.19 | 0.18 | 0.07 |
| RLS dense | – | 0.22 | 0.18 | 0.11 | 0.27 | 0.12 | 0.05 |
| Gabor whole | – | – | 0.12 | 0.08 | 0.15 | 0.15 | 0.08 |
| Gabor dense | – | – | – | 0.07 | 0.21 | 0.13 | 0.10 |
| WLD | – | – | – | – | 0.09 | 0.06 | 0.36 |
| GLCM whole | – | – | – | – | – | 0.17 | 0.07 |
| GLCM dense | – | – | – | – | – | – | 0.04 |
TABLE 3.
P-values for testing the hypothesis of no correlation between different feature groups against the alternative that there is a nonzero correlation. Each p-value corresponds to the values of Spearman's rho in Table 2. The diagonal p-values are equivalent and were thus omitted (–).
| Feature group |
p-value |
||||||
|---|---|---|---|---|---|---|---|
| RLS dense | Gabor whole | Gabor dense | WLD | GLCM whole | GLCM dense | Generic statistical | |
| RLS whole | <0.0001 | <0.0001 | <0.0001 | 0.008 | <0.0001 | <0.0001 | 0.045 |
| RLS dense | – | <0.0001 | <0.0001 | 0.001 | <0.0001 | 0.0003 | 0.15 |
| Gabor whole | – | – | 0.0003 | 0.02 | <0.0001 | <0.0001 | 0.01 |
| Gabor dense | – | – | – | 0.03 | <0.0001 | 0.0001 | 0.002 |
| WLD | – | – | – | – | 0.01 | 0.07 | <0.0001 |
| GLCM whole | – | – | – | – | – | <0.0001 | 0.046 |
| GLCM dense | – | – | – | – | – | – | 0.23 |
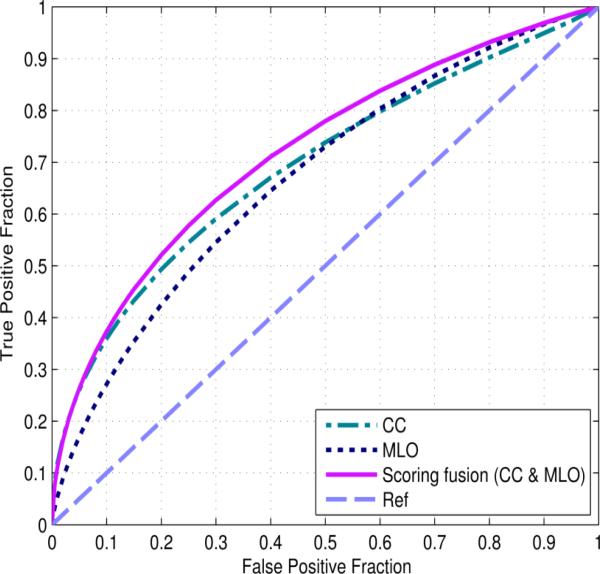
When using the group of RLS features computed on the dense breast region, the average number of hidden nodes of the bilateral CC and the MLO view image based ANNs were 8.5±1.1 and 8.9±1.1 in the 10-fold cross-validation experiment, respectively. The average mean squared error (MSE) of the ANN training results was 0.183±0.003. Figure 4 displays the three ROC curves of the CC, MLO and “scoring fusion” based ANNs using the RLS features computed on the dense breast region. The corresponding average AUC results with standard deviation intervals computed across the ten folds of the cross-validation experiments are 0.701±0.039, 0.671±0.043 and 0.725±0.026, respectively. Using DeLong's test for paired samples, 14 we analyzed significant differences at the 5% significance level between the “scoring fusion” ANN and the CC based ANN (p = 0.015), and between the “scoring fusion” ANN and the MLO based ANN (p < 0.001). The performance of the ANN trained by features computed from the MLO view was not significantly different with that of the ANN trained by features computed from the CC view (p = 0.29), which demonstrates that our new “scoring fusion” scheme can effectively combine information extracted and derived from both CC and MLO views. This result also shows that mammograms acquired from different projection views contain supplementary information and optimally combining the image features from two views has potential to increase prediction power of the new cancer risk assessment models.
FIG. 4.
Three receiver operating characteristic (ROC) curves generated by the output of three ANNs trained using the run length features computed on the dense breast region from the individual CC and MLO views as well as the proposed scoring fusion method of both views.
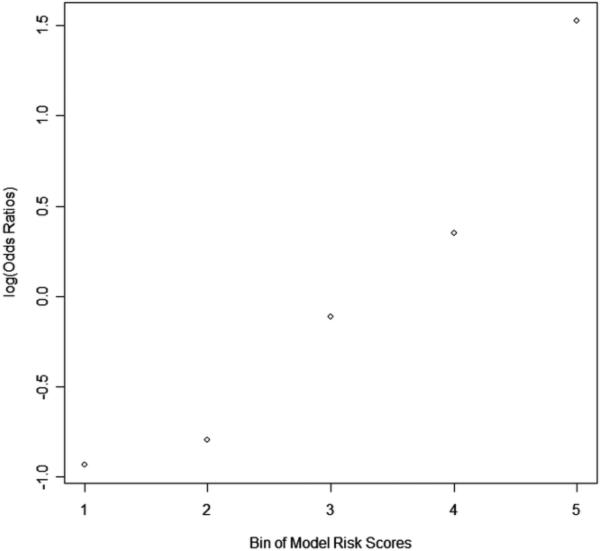
When using the prediction scores generated by the optimal ANN using the second group of features (RLS in the dense region) to compute adjusted ORs and corresponding 95% CIs for five subgroups/bins of cases, the results in Table 4 showed an increasing trend of the OR values as a function of increasing our two view “scoring fusion” model-generated risk scores to predict between the positive (cancer) and negative (cancer free) cases. Namely, the ORs increased from 1.00 in subgroup 1 to 11.77 in subgroup 5 (with a 95% CI of 7.07 to 19.59). As shown in Figure 5, the slope of the regression trend line between the risk scores generated by our risk model and the adjusted ORs is significantly different from zero (p = 0.009), which shows that as the risk prediction score generated by our “scoring fusion” model increases, the risk of women developing breast cancer in the next sequential screening examination also increases.
TABLE 4.
Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) at five subgroups (bins) with increasing values of the risk scores generated by our “scoring fusion” classifier.
| Subgroup | Number of cases (Positive – Negative) | Adjusted OR | 95% CI |
|---|---|---|---|
| 1 | 49 – 125 | 1.00 | Baseline |
| 2 | 54 – 120 | 1.15 | [0.72, 1.82] |
| 3 | 82 – 92 | 2.27 | [1.46, 3.55] |
| 4 | 102 – 72 | 3.61 | [2.31, 5.65] |
| 5 | 143 – 31 | 11.77 | [7.07, 19.59] |
FIG. 5.
Trend of the increase in odds ratios (ORs) with the increase in risk scores generated by our trained “scoring fusion” classifier. The slope of the regression trend line between the adjusted ORs and the risk scores is significantly different from the zero slope (p = 0.009).
Table 5 shows the confusion matrix obtained by applying an operational threshold of 0.6 on the risk prediction score of the “scoring fusion” ANN trained using the RLS features computed on the dense breast region. At this operational threshold, the overall prediction accuracy was 65.4% in which 569 of 870 cases were correctly classified, whereas 34.6% (301 of 870) were misclassified. The positive predictive value was 48.1% (207 of 430), whereas the negative predictive value was 82.3% (362 of 440) at this particular operational threshold and testing dataset.
TABLE 5.
A confusion matrix obtained when applying a threshold of 0.6 on the “scoring fusion” classifier generated risk/probability scores.
| Actual ↓ |
Negative cases | Positive cases |
|---|---|---|
| Negative cases | 362 | 78 |
| Positive (cancer) cases | 223 | 207 |
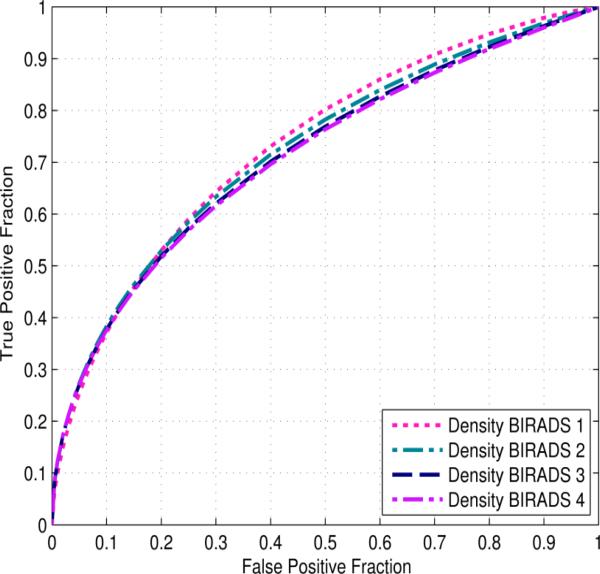
Figure 6 displays the ROC curves of applying our risk prediction scheme to four subgroups of cases divided by mammographic density BIRADS categories. The AUC values are 0.734±0.079, 0.725±0.029, 0.715±0.023, and 0.712±0.090 for four subgroups of BIRADS 1 to 4, respectively. The sensitivity levels of our risk prediction scheme at specificity levels of 80%, 85%, 90%, and 95% on the cases stratified by density BI-RADS ratings are displayed in Table 6. The results show no significant performance dependency of our risk prediction model on the cases in different density BIRADS categories. This was confirmed by DeLong's test for unpaired samples (p > 0.68 for all comparisons).
FIG. 6.
Four ROC curves of our CAD scheme applied to four case subgroups rated in four different mammographic density BIRADS categories.
TABLE 6.
Sensitivity levels of our CAD scheme at four specificity levels by stratifying the testing cases according to the four mammographic density BIRADS categories.
| Specificity | 95% | 90% | 85% | 80% |
|---|---|---|---|---|
| Density BI-RADS 1 | 25.3% | 37.1% | 45.8% | 53.0% |
| Density BI-RADS 2 | 27.4% | 38.4% | 46.4% | 52.9% |
| Density BI-RADS 3 | 27.1% | 37.7% | 45.5% | 51.9% |
| Density BI-RADS 4 | 27.1% | 37.6% | 45.3% | 51.5% |
V. DISCUSSION
This study is part of our continuing effort to develop and optimize new quantitative image analysis methods to help more accurately predict near-term breast cancer risk. Comparing to the previous studies in this field, this study has several unique characteristics and distinct study results or observations. First, the vast majority of studies related to the image-based breast cancer risk prediction in the literature 17, 18, 25, 28 only examined and computed mammographic image features from a ROI or one image per patient. Since mammograms are two-dimensional projection images with severe overlapping fibro-glandular tissue, the image features can be distorted and quite different in CC and MLO view images. Hence, using only one ROI or one image has limitations. In this study, we developed a first new image-based near-term breast cancer risk prediction model that combines image features computed from four mammograms together using two unique steps. (1) We compared and selected the maximum feature value computed from two bilateral (CC or MLO) view images of the left and right breasts. (2) We built a scoring fusion ANN to combine the prediction scores generated from the image features of CC and MLO views separately. Our study results demonstrated that our “scoring fusion” ANN based classifier can derive and combine useful and supplementary information from both CC and MLO view images to significantly increase risk prediction performance comparing to the results obtained using the risk prediction scores generated using the ANN trained by the image features computed separately from CC or MLO view images (p ≤ 0.02).
Second, comparing to majority of previous studies using image features to predict breast cancer risk, our experiments and data analysis were conducted based on a much larger and diverse dataset involving 870 FFDM cases (430 malignant, 220 benign/recalled and 220 negative), which consists of 3480 digital mammograms altogether. In addition, we optimized the parameters in ANN training (i.e., the ratio of training momentum and learning rate, as well as training iterations) to reduce over-fitting of ANNs. Our studies shown that using 500 training iterations was sufficient for this task. Increasing training iterations to 1,000 yielded lower performance (e.g., AUC = 0.719±0.030) when using the second feature group as shown in Table 1. Hence, due to the large dataset and optimal training parameters, the results of our study might be more reliable and robust. The prediction results (e.g., AUC value) of this study are also consistent with our previous studies using a small dataset.45
Third, our new image feature based risk model aims to predict near-term cancer risk (i.e., the risk of a woman having mammography-detectable cancer in the next sequential FFDM screening examination in this study). As a result, our model does not directly compete with the existing epidemiology based breast cancer risk prediction models 2 including the Gail Model, the Tyrer-Cuzick Model and other newly-reported risk models based on mammographic density image 45 and histologic features, 29 which focus on predicting long-term or lifetime risk of a woman as compared to the general population. The prediction results of our new risk model with AUC = 0.725±0.026 and an adjusted OR of 11.77 with a 95% CI of 7.07 to 19.59 in subgroup 5 were significantly higher than using the subjectively rated mammographic density (BIRADS) (as shown in Figure 2) and other existing risk factors. 2 The primary reason of our model being able to predict near-term risk lies in that our model detects and analyzes the variation and/or increases of bilateral mammographic image feature asymmetry, which is a useful and early sign of leading to develop cancer. The existing lifetime risk models only provide a fixed estimation score, which is not sensitive to the near-term cancer risk variation.
Fourth, in our previous studies, 36, 45we computed the bilateral mammographic image feature differences based on absolute feature value subtractions between the left and right breasts. In this study, we tested a new approach to extract image features with higher discriminatory power from the two bilateral images, which is based on the observation in our preliminary experiments. We selected the maximum value of a feature that is computed separately from the two bilateral images of the left and right breasts to represent the final feature value. This approach has an advantage to maintain both the overall mammographic density information (since mammographic density is a well-known breast cancer risk factor) and (2) the bilateral difference information (whereby we have demonstrated that it is also an important risk factor to predict near-term breast cancer risk).
Fifth, although a large number of different types of image features have been computed and reported in the literature to assess mammographic density and/or tissue structures, whether the different types of features are redundant or how to select optimal and robust image features has not been well investigated. In this study, we examined 8 groups of popular textural and mammographic density based features and compared the effectiveness and correlations of these groups of features by training a two-layer ANN-based risk model on the individual feature groups as well as a combination of all feature groups. From the comparison results, we observed that (1) the run length features computed on the dense breast regions are most effective for breast cancer risk prediction; (2) many of the popular features reported in literature, such as GLCM and PD, and new features examined in this study, such as WLD and an average fractal dimension based feature yielded very comparable prediction performance due to higher correlation among these features. Thus, the performance of the ANN trained using all features combined was lower than the ANN trained only on the run length features computed on the dense breast region. In addition, we also tested a fast and accurate sequential forward floating selection (SFFS) based feature selection method 39 to select optimal features from 8 feature groups. The ANN classifier generated an AUC = 0.716±0.056, which was higher than using all 79 features, but lower than using the second group of RLS features (as shown in Table 1).
Sixth, unlike the general screening mammography and the current lesion based CAD schemes of mammograms in which the detection or classification performance decreases as the mammographic density level increases (from BIRADS 1 to 4), our data analysis results showed that our risk prediction model was not mammographic density dependent. The risk prediction performance maintains a relatively stable level across all four density BIRADS categories (Table 6). This is another advantage of our new risk model using the bilateral mammographic image feature asymmetry analysis and two-view information fusion approach.
Last, when using an operational threshold of 0.6 on the ANN-generated risk prediction scores, we observed that the negative predictive value (82.3%) was substantially higher than the positive predictive value (48.1%). This indicates that under this threshold, the prediction result is more accurate in identifying the women with low risk of having mammography-detectable cancers in the next sequential FFDM screening. As a result, these women can be screened at a longer interval (such as, every two years rather than every one year). Due to the lower cancer detection yield in annual breast cancer screening (i.e., < 5 cancers detected in every 1000 screened women), achieving high negative predictive value is more important or has higher clinical impact in improving efficacy of breast cancer screening, which can exclude a large number of low-risk women from unnecessary frequent screening and thus reduce the false-positive recalls. This also shows that the different evaluation criterion should be used to evaluate the performance of the quantitative image feature analysis based cancer risk models and the conventional lesion-based CAD schemes of mammograms.
Despite the promising results and new observations, this is a laboratory based retrospective data analysis study with a number of limitations. First, although we used a relatively large and diverse image dataset, it may have case selection bias. The ratio between positive and negative cases also does not represent the actual cancer prevalence ratio in general screening practice. Hence, the performance and robustness of our new risk model needs to be further tested in future prospective or cohort studies. Second, this is a preliminary technology development study. Its clinical utility has not been tested. For example, by adjusting the operational threshold, we can adjust the positive and negative predictive values of our risk prediction model. What is an optimal and clinically-acceptable operational threshold has not been determined. Third, our model was only applied to predict the risk of having mammography-detectable cancer in the next sequential FFDM screening examinations following a negative screening of interest. Whether the similar model can be developed and applied to predict risk in a relatively longer time period (i.e., 2 to 5 years) has not been tested. Fourth, we have a limited tracking time of two additional follow-up FFDM examinations for the benign and negative cases maintaining cancer-free status. Therefore, in this preliminary study, we built a new near-term breast cancer risk prediction model based on mammographic image features, which needs to be further examined before it can be clinically acceptable to help establish an optimal, personalized breast cancer screening paradigm.
ACKNOWLEDGMENTS
This study is supported in part by Grant R01 CA160205 from the National Cancer Institute, National Institutes of Health. The authors also acknowledge the support received from the Peggy and Charles Stephenson Cancer Center, University of Oklahoma.
Footnotes
CONFLICT OF INTEREST
There are no conflicts of interest in relation to this work.
REFERENCES
- 1.Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans. Biomed. Eng. 2008;55:1822–1830. doi: 10.1109/TBME.2008.919735. [DOI] [PubMed] [Google Scholar]
- 2.Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J. Natl. Cancer Inst. 2010;102:680–691. doi: 10.1093/jnci/djq088. [DOI] [PubMed] [Google Scholar]
- 3.Berg WA, Campassi C, Langenberg P, Sexton MJ. Breast Imaging Reporting and Data System: inter- and intraobserver variability in feature analysis and final assessment. AJR Am. J. Roentgenol. 2000;174:1769–1777. doi: 10.2214/ajr.174.6.1741769. [DOI] [PubMed] [Google Scholar]
- 4.Berlin L, Hall FM. More mammography muddle: emotions, politics, science, costs, and polarization. Radiology. 2010;255:311–316. doi: 10.1148/radiol.10100056. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand KA, Tamimi RM, Scott CG, Jensen MR, Pankratz VS, Visscher D, Norman A, Couch F, Shepherd J, Fan B, Chen YY, Ma L, Beck AH, Cummings SR, Kerlikowske K, Vachon CM. Mammographic density and risk of breast cancer by age and tumor characteristics. Breast Cancer Res. 2013;15:R104. doi: 10.1186/bcr3570. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boughey JC, Hartmann LC, Anderson SS, Degnim AC, Vierkant RA, Reynolds CA, Frost MH, Pankratz VS. Evaluation of the Tyrer-Cuzick (International Breast Cancer Intervention Study) model for breast cancer risk prediction in women with atypical hyperplasia. J. Clin. Oncol. 2010;28:3591–3596. doi: 10.1200/JCO.2010.28.0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd NF, Martin LJ, Yaffe MJ, Minkin S. Mammographic density and breast cancer risk: current understanding and future prospects. Breast Cancer Res. 2011;13:223. doi: 10.1186/bcr2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brawley OW. Risk-based mammography screening: an effort to maximize the benefits and minimize the harms. Ann. Intern. Med. 2012;156:662–663. doi: 10.7326/0003-4819-156-9-201205010-00012. [DOI] [PubMed] [Google Scholar]
- 9.Byng JW, Boyd NF, Fishell E, Jong RA, Yaffe MJ. The quantitative analysis of mammographic densities. Phys. Med. Biol. 1994;39:1629–1638. doi: 10.1088/0031-9155/39/10/008. [DOI] [PubMed] [Google Scholar]
- 10.Chang Y-H, Wang X-H, Hardesty LA, Chang TS, Poller WR, Good WF, Gur D. Computerized assessment of tissue composition on digitized mammograms. Acad. Radiol. 2002;9:899–905. doi: 10.1016/s1076-6332(03)80459-2. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Shan S, He C, Zhao G, Pietikainen M, Chen X, Gao W. WLD: A Robust Local Image Descriptor. IEEE Trans. Pattern Anal. Mach. Intell. 2010;32:1705–1720. doi: 10.1109/TPAMI.2009.155. [DOI] [PubMed] [Google Scholar]
- 12.Clausi DA. An analysis of co-occurrence texture statistics as a function of grey level quantization. Can. J. Remote Sens. 2002;28:45–62. [Google Scholar]
- 13.Daugman JG. Complete discrete 2-D Gabor transforms by neural networks for image analysis and compression. IEEE Trans. Acoust., Speech, Signal Process. 1988;36:1169–1179. [Google Scholar]
- 14.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;11:837–845. [PubMed] [Google Scholar]
- 15.Gail MH. Personalized estimates of breast cancer risk in clinical practice and public health. Stat. Med. 2011;30:1090–1104. doi: 10.1002/sim.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H Gail M., Mai PL. Comparing breast cancer risk assessment models. J. Natl. Cancer Inst. 2010;102:665–668. doi: 10.1093/jnci/djq141. [DOI] [PubMed] [Google Scholar]
- 17.Gierach GL, Li H, Loud JT, Greene MH, Chow CK, Lan L, Prindiville SA, Eng- Wong J, Soballe PW, Giambartolomei C, Mai PL, Galbo CE, Nichols K, Calzone KA, Olopade OI, Gail MH, L M. Giger. Relationships between computer-extracted mammographic texture pattern features and BRCA1/2 mutation status: a cross-sectional study. Breast Cancer Res. 2014;16:424. doi: 10.1186/s13058-014-0424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häberle L, Wagner F, Fasching P, Jud S, Heusinger K, Loehberg C, Hein A, Bayer C, Hack C, Lux M, Binder K, Elter M, Münzenmayer C, Schulz-Wendtland R, Meier-Meitinger M, Adamietz B, Uder M, Beckmann M, Wittenberg T. Characterizing mammographic images by using generic texture features. Breast Cancer Res. 2012;14:1–12. doi: 10.1186/bcr3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghighat M, Zonouz S, Abdel-Mottaleb M. Identification Using Encrypted Biometrics. In: Wilson R, Hancock E, Bors A, Smith W, editors. Computer Analysis of Images and Patterns. Springer Berlin; Heidelberg: 2013. pp. 440–448. [Google Scholar]
- 20.Haralick RM, Shanmugam K, Dinstein I. Texture features for image classification. IEEE Trans. Syst. Man, Cybern. 1973;3:610–621. [Google Scholar]
- 21.Heine JJ, Scott CG, Sellers TA, Brandt KR, Serie DJ, Wu FF, Morton MJ, Schueler BA, Couch FJ, Olson JE, Pankratz VS, Vachon CM. A novel automated mammographic density measure and breast cancer risk. J. Natl. Cancer Inst. 2012;104:1028–1037. doi: 10.1093/jnci/djs254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen KJ. Is the tide turning against breast screening? Breast Cancer Res. 2012;14:107–107. doi: 10.1186/bcr3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Giger ML, Olopade OI, Chinander MR. Power Spectral Analysis of Mammographic Parenchymal Patterns for Breast Cancer Risk Assessment. J. Digit. Imaging. 2008;21:145–152. doi: 10.1007/s10278-007-9093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Szekely L, Eriksson L, Heddson B, Sundbom A, Czene K, Hall P, Humphreys K. High-throughput mammographic-density measurement: a tool for risk prediction of breast cancer. Breast Cancer Res. 2012;14:R114. doi: 10.1186/bcr3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manduca A, Carston MJ, Heine JJ, Scott CG, Pankratz VS, Brandt KR, Sellers TA, Vachon CM, Cerhan JR. Texture Features from Mammographic Images and Risk of Breast Cancer. Cancer Epidemiol. Biomarkers Prev. 2009;18:837–845. doi: 10.1158/1055-9965.EPI-08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcelja S. Mathematical description of the responses of simple cortical cells. J. Opt. Soc. Am. 1980;70:1297–1300. doi: 10.1364/josa.70.001297. [DOI] [PubMed] [Google Scholar]
- 27.Mudigonda NR, Rangayyan RM, Desautels JE. Gradient and texture analysis for the classification of mammographic masses. IEEE Trans. Med. Imaging. 2000;19:1032–1043. doi: 10.1109/42.887618. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen M, Karemore G, Loog M, Raundahl J, Karssemeijer N, Otten JD, Karsdal MA, Vachon CM, Christiansen C. A novel and automatic mammographic texture resemblance marker is an independent risk factor for breast cancer. Cancer Epidemiol. 2011;35:381–387. doi: 10.1016/j.canep.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Pankratz VS, Degnim AC, Frank RD, Frost MH, Visscher DW, Vierkant RA, Hieken TJ, Ghosh K, Tarabishy Y, Vachon CM, Radisky DC, Hartmann LC. Model for Individualized Prediction of Breast Cancer Risk After a Benign Breast Biopsy. J. Clin. Oncol. 2015 doi: 10.1200/JCO.2014.55.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passaperuma K, Warner E, Hill KA, Gunasekara A, Yaffe MJ. Is mammographic breast density a breast cancer risk factor in women with BRCA mutations? J. Clin. Oncol. 2010;28:3779–3783. doi: 10.1200/JCO.2009.27.5933. [DOI] [PubMed] [Google Scholar]
- 31.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323:533–536. [Google Scholar]
- 32.Schousboe JT, Kerlikowske K, Loh A, Cummings SR. Personalizing mammography by breast density and other risk factors for breast cancer: analysis of health benefits and cost-effectiveness. Ann. Intern. Med. 2011;155:10–20. doi: 10.7326/0003-4819-155-1-201107050-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soh LK, Tsatsoulis C. Texture analysis of SAR sea ice imagery using gray level co- occurrence matrices. IEEE Trans. Geosci. Remote Sens. 1999;37:780–795. [Google Scholar]
- 34.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan M, Pu J, Zheng B. Reduction of false-positive recalls using a computerized mammographic image feature analysis scheme. Phys. Med. Biol. 2014;59:4357–4373. doi: 10.1088/0031-9155/59/15/4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan M, Zheng B, Ramalingam P, Gur D. Prediction of near-term breast cancer risk based on bilateral mammographic feature asymmetry. Acad. Radiol. 2013;20:1542–1550. doi: 10.1016/j.acra.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X. Texture information in run-length matrices. IEEE Trans. Image Proc. 1998;7:1602–1609. doi: 10.1109/83.725367. [DOI] [PubMed] [Google Scholar]
- 38.Varela C, Timp S, Karssemeijer N. Use of border information in the classification of mammographic masses. Phys. Med. Biol. 2006;51:425–441. doi: 10.1088/0031-9155/51/2/016. [DOI] [PubMed] [Google Scholar]
- 39.Ververidis D, Kotropoulos C. Fast and accurate sequential floating forward feature selection with the Bayes classifier applied to speech emotion recognition. Signal Process. 2008;88:2956–2970. [Google Scholar]
- 40.Wang X, Lederman D, Tan J, Wang XH, Zheng B. Computerized prediction of risk for developing breast cancer based on bilateral mammographic breast tissue asymmetry. Med. Eng. Phys. 2011;33:934–942. doi: 10.1016/j.medengphy.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei C-H, Li Y, Li C-T. IEEE International Conference on Multimedia and Expo. IEEE; Beijing: 2007. Effective Extraction of Gabor Features for Adaptive Mammogram Retrieval. pp. 1503–1506. [Google Scholar]
- 42.Wei J, Chan HP, Wu YT, et al. Association of computerized mammographic parenchymal pattern measure with breast cancer risk: a pilot case-control study. Radiology. 2011;260:42–49. doi: 10.1148/radiol.11101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei X. Gray Level Run Length Matrix Toolbox v1.0. Beijing Aeronautical Technology Research Center; 2007. [16 February 2015]. http://www.mathworks.com/matlabcentral/fileexchange/17482-gray-level-run-length-matrix-toolbox. [Google Scholar]
- 44.Zheng B, Sumkin JH, Zuley ML, Lederman D, Wang X, Gur D. Computer-aided detection of breast masses depicted on full-field digital mammograms: a performance assessment. Br. J. Radiol. 2012;85:e153–e161. doi: 10.1259/bjr/51461617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng B, Sumkin JH, Zuley ML, Wang X, Klym AH, Gur D. Bilateral mammographic density asymmetry and breast cancer risk: a preliminary assessment. Eur. J. Radiol. 2012;81:3222–3228. doi: 10.1016/j.ejrad.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]