Abstract
Aim
Extreme discordant phenotype and genome-wide association (GWA) approaches were combined to explore the role of genetic variants on warfarin dose requirement in Brazilians.
Methods
Patients receiving low (≤20 mg/week; n = 180) or high stable warfarin doses (≥42.5 mg/week; n = 187) were genotyped with Affymetrix Axiom® Biobank arrays. Imputation was carried out using data from the combined 1000 Genomes project.
Results
Genome-wide signals (p ≤5 × 10−8) were identified in the well-known VKORC1 (lead SNP, rs749671; OR: 20.4; p = 1.08 × 10−33) and CYP2C9 (lead SNP, rs9332238, OR: 6.8 and p = 4.4 × 10−13) regions. The rs9332238 polymorphism is in virtually perfect LD with CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910). No other genome-wide significant regions were identified in the study.
Conclusion
We confirmed the important role of VKORC1 and CYP2C9 polymorphisms in warfarin dose.
Keywords: 1000 Genomes Project, Brazilians, CYP2C9, extreme discordant phenotypes, genome-wide association study, VKORC1, warfarin
Background
Warfarin, the most commonly used anticoagulant in the world, has been the target of several pharmacogenetic/genomic studies aimed at identifying the factors that account for the large interindividual variability in dose requirement. A number of warfarin dosing algorithms, comprising genetic and nongenetic covariates emerged from these studies, but their predictive power accounts for no more than 60% of the variance in warfarin dose requirements in ambulatory patients [1–10]. Pharmacogenetic polymorphisms in VKORC1 and CYP2C9 affecting, respectively, warfarin’s pharmacodynamics and pharmacokinetics are the most informative covariates in most algorithms. Genome-wide association studies (GWAS) of warfarin dosing requirements in White/Caucasian [11,12], Japanese [13] and African–American patients [14] indicated that common SNPs with large effects on warfarin dose are unlikely to be discovered outside of the CYP2C9 and VKORC1 genes. However, the possibility that polymorphisms in other pharmacogenes may contribute in a smaller scale to the interindividual variance in warfarin dosing, may not be excluded. In this respect, conditional analyses and GWA or meta-analytic efforts including larger sample sizes may uncover novel common variants with smaller effect sizes. Indeed, a polymorphism in CYP4F2 (rs2108622) reached genome-wide significant association with warfarin dose after conditioning for VKORC1 and CYP2C9 polymorphisms [12,13]. Another study in African–Americans applied a stepwise conditional analysis and identified an association between a polymorphism in the CYP2C cluster on chromosome 10 (rs12777823) and warfarin dose [14]. In the present study, we applied the extreme discordant phenotype (EDP) methodology [15] to select Brazilian patients under stable warfarin therapy for a genome-wide association study. The EDP approach contrasts the most sensitive and the most resistant phenotype groups, which in the case of a quantitative trait such as the individual warfarin dose requirement correspond to the lower and upper ends of the dose distribution histogram. The superior power of sampling extreme phenotype individuals compared with random cohorts for GWAS of drug phenotypes has been recently reviewed [16].
Materials & methods
Study cohort
The present analyses are based on combined data from two cohorts of adult outpatients, enrolled in previously published retrospective studies designed to develop warfarin dosing algorithms for Brazilians [5,17]. The patients were attending anticoagulation clinics at two tertiary care institutions of the Brazilian Public Health System, namely the Instituto Nacional de Cardiologia Laranjeiras, a reference cardiology hospital located in Rio de Janeiro (n = 390) and the University Hospital of Universidade Federal do Rio Grande do Sul, in Porto Alegre (n = 488). Details of the original study design have been published [5,17]. The study protocols were approved by the respective institutional review boards and each patient provided a written informed consent. Patients were categorized according to the Brazilian census, which relies on self-perception of ‘race/color’, as White (branco), Brown (pardo) and Black (preto). The term Color and the color categories are capitalized to highlight their special meaning in the context of the Brazilian census. All patients were under stable warfarin dosing, defined as the prescribed weekly dose associated with the first three consecutive INR readings within the target range. The INR target range was 2–3 for all patients, except those with heart valve prosthesis, whose INR target was 2.5–3.5 [5].
Extreme discordant phenotype
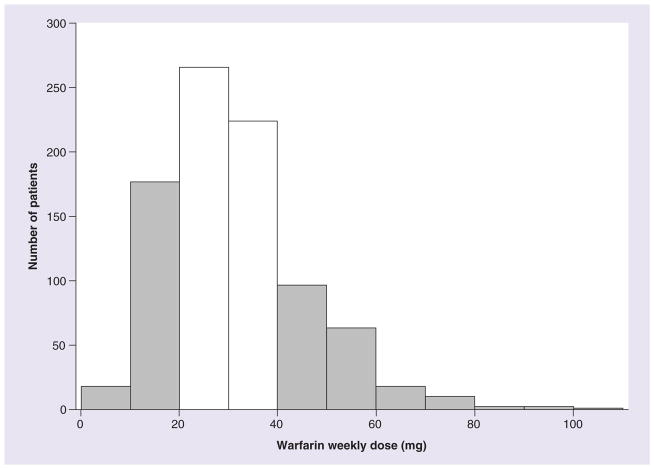
The target pharmacogenetic phenotype was the stable warfarin weekly dose, which ranged from 5 to 110 mg in the overall cohort. A frequency distribution histogram of the individual warfarin dose was constructed (Figure 1) and patients at opposite ends of the distribution were selected for genotyping with Affymetrix Axiom Biobank microarray. The cut-off points for the low-dose and high-dose groups were set at ≤20 mg/week (n = 180) and ≥42.5 mg/week (n = 187), respectively. Demographic and clinical characteristics of the patients in the two groups are shown in Table 1.
Figure 1. Histogram showing the frequency distribution of weekly warfarin dose in the Brazilian sample.
Patients at opposite ends of the distribution (gray bars) were selected for genotyping with the Affymetrix Axiom® Biobank
Table 1.
Demographics and clinical characteristics of the study cohort.
| Variable | Low dose (n = 180) (Mean [SD]) | High dose (n = 187) (Mean [SD]) |
|---|---|---|
| Warfarin weekly dose (mg) | 16.2 (3.1) | 55.2 (2.7) |
| INR | 2.5 (0.6) | 2.7 (0.6) |
| Age (years) | 65.8 (13.9) | 54.2 (13.7) |
| Weight (kg) | 67.5 (15.1) | 76.9 (16.0) |
|
| ||
| n (%) | n (%) | |
|
| ||
| Gender | ||
| – Male | 87 (48.3) | 106 (56.7) |
| – Female | 93 (51.7) | 81 (43.3) |
| BMI | 25.7 (3.9) | 27.3 (4.8) |
|
| ||
| Cohort | ||
| Rio de Janeiro | 119 (66.1) | 126 (67.4) |
| Porto Alegre | 61 (33.9) | 61 (32.6) |
|
| ||
| Race/color | ||
| White | 153 (85.0) | 139 (74.3) |
| Brown | 18 (10.0) | 27 (14.4) |
| Black | 9 (5.0) | 10 (120.7) |
| Yellow | 0 | 1 (0.5) |
|
| ||
| Admixture proportions (%) | ||
| European | 81.7 (17.8) | 72.4 (23.2) |
| African | 11.7 (15.9) | 18.2 (21.4) |
| Native American | 6.6 (7.1) | 9.4 (8.5) |
|
| ||
| Indication for warfarin | ||
| Atrial fibrillation | 97 (53.9) | 55 (29.4) |
| Heart valve prosthesis | 28 (15.6) | 88 (47.1) |
| Atrial fibrillation and heart valve prosthesis | 18 (10.0) | 12 (6.4) |
| Thromboembolic disease | 17 (9.4) | 18 (9.6) |
|
| ||
| Comorbidities† | ||
| Systemic hypertension | 119 (66.1) | 102 (54.4) |
| Rheumatic fever | 35 (19.4) | 27 (14.4) |
| Dislipidemia | 30 (16.7) | 35 (18.7) |
| Diabetes mellitus | 20 (11.1) | 27 (14.4) |
|
| ||
| Comedication† | ||
| ACE inhibitors | 96 (53.3) | 91 (48.7) |
| Aspirin | 39 (21.7) | 41 (21.9) |
| Amiodarone | 31 (17.2) | 10 (5.3) |
| Beta blockers | 79 (43.9) | 59 (31.6) |
| Ca-entry blockers | 25 (13.9) | 13 (7.0) |
| Digoxin | 56 (31.1) | 38 (20.3) |
| Isosorbide | 20 (11.1) | 10 (5.3) |
| Loop diuretics | 103 (57.2) | 77 (41.2) |
| Losartan | 22 (12.2) | 13 (7.0) |
| Metformin | 10 (5.6) | 20 (10.7) |
| Omeprazol | 35 (19.4) | 23 (12.3) |
| Simvastatin | 51 (28.3) | 40 (21.4) |
| Spironolactone | 20 (11.1) | 17 (9.1) |
Frequency >5% in the combined groups.
Genotyping
The sample was genotyped with the Affymetrix Axiom® Biobank array (Affymetrix, CA, USA) following standard protocols. Genotype calling was done with the Affymetrix PowerTools (APT) software package, using the AxiomGT1/brlmm-p algorithm and the manufacturer recommended calling pipeline. The Biobank array has more than 650,000 variants, including 265,000 exome coding snps and indels, 70,000 novel loss-of-function SNPs and indels, 23,000 eQTLs, 2000 markers of pharmacogenomic relevance and 246,000 genome-wide association markers designed to ensure good genome-wide coverage in major populations, which make it possible to carry out imputation in order to increase the number of markers in the statistical analyses.
Quality control prior to imputation
Prior to imputation, SNPs were removed from the initial list of autosomal markers based on the following criteria: 1/Markers classified as CallRateBelowThreshold, OffTargetVariants or Other by the program SNPolisher, 2/minor allele frequency <1%, 3/Hardy–Weinberg p-values <10–5 in the low-dose or high-dose sample and 4/missing genotyping rate per SNP > 5%. All the QC steps were carried out with the program PLINK [18].
Population stratification & admixture estimates
In order to evaluate population stratification we used the program EIGENSOFT [19] to perform a principal component (PC) analysis of the Mexico City sample, after pruning markers showing linkage disequilibrium. The samples from Brazil were plotted jointly with samples from other population groups, including Europeans (CEU), Africans (YRI), East Asians (CHB), Native Americans (Nahua and Maya from Mexico, and Andeans from South America) and other admixed groups from the Americas (Mexican Americans from LA, Puerto Ricans and Colombians). In addition to the PC analyses, we also estimated admixture proportions based on genome-wide data using the program ADMIXTURE [20].
Imputation
Imputation was carried out with the program IMPUTE v2 [21], using the combined 1000 Genomes project samples as the reference sample (1000 Genomes Phase I integrated haplotypes, NCBI build b37, release date December 2013, no singletons). Imputations were carried out using a two-step strategy: 1/prephasing using the program SHAPEIT [22] and 2/Imputation from the reference panel into the estimated haplotypes with IMPUTE v2 [21,23].
Exclusion of individuals prior to association tests
Prior to the association tests, four individuals were excluded from the dataset: three individuals due to cryptic relatedness (pi-Hat > 0.2, based on an analysis with PLINK; [18]) and one individual showing evidence of substantial East Asian ancestry based on the PC plots. For the pairs of individuals showing cryptic relatedness, we removed the individual with the highest genotype missingness rate.
Association tests
In order to evaluate the association of genetic markers with warfarin dose (low- vs high-dose groups), we used the program SNPTEST v2 [24], including as covariates sex, age, BMI, treatment with amiodarone and the first two PCA scores. In order to control for genotype uncertainty, we used the ml method, which uses multiple Newton–Raphson iterations to estimate the parameters in the missing data likelihood for the model. In addition to this initial analysis, we also carried out a conditional analysis including in the models the genotypes of VKORC1 (rs749671) and CYP2C9 (CYP2C9*2 and CYP2C9*3).
Identification of markers to follow-up
Based on the results of the conditional analyses, we identified a list of highly suggestive regions (p < 10−5), and the lead SNPs for each region were followed up in 798 samples of European ancestry, from five previously published cohorts [6,25–28], 1611 samples of Japanese origin [13] and 191 samples of African–American ancestry, from two cohorts [29–31].
Gene-based & pathway association tests
We carried out gene-based and pathway association tests using the program KGG3 [32]. For the gene-based association tests, we used the GATES approach [33], using a window of 10Kb for the extended gene region length. This test takes into account the LD patterns between the SNPs, which in this case were defined by the patterns observed in the Puerto Rican 1000 Genomes sample, which is the closest sample to the Brazilian sample in terms of admixture proportions (e.g., European, African and Native American contributions). For the pathway association tests. we used the HYST [34] test implemented in the KGG3 program. We only report pathways that are significant using the HYST test and also a hypergeometric test enrolling genes with a p-value of 0.001. We also performed a pathway analysis using an alternative program, GSA-SNP [35], using the PAGE (parametric analysis of gene set enrichment) method [36]. We used the second best SNP to define the gene p-values, and used a padding window of 10 kb. The program was run using both Gene Ontology (GO) and KEGG gene sets.
eQTL analyses
We used the eQTL browser [37] to download eQTL datasets generated in lymphoblastoid cell lines (LCLs) [38,39] and human liver [40]. We then evaluated if any of the eQTLs in the aforementioned tissues (here defined as markers associated with gene expression levels with p-values ≤10−5) are associated with warfarin dose in our study, after Bonferroni correction to take into account multiple tests (e.g., total number of eQTLS relevant for each tissue).
Results
Characterization of admixture in the Brazilian sample
The PC analysis shows that the Brazilian sample is primarily the result of admixture between European and African populations (Supplementary Figure 1). The Brazilian individuals are located between the relatively tight clusters corresponding to European and West African individuals. However, there is also evidence of Native American ancestry in the sample, based on the presence of individuals that are located toward the cluster defined by the Native American populations. The placement of the Brazilian individuals in the plot, primarily between the European and African clusters, contrasts with the position of the Mexican Americans from LA, which are primarily located between the Native American and European clusters. The samples from Colombia and Puerto Rico also show clear evidence of admixture. We estimated the admixture proportions in the Brazilian sample using the program ADMIXTURE [20]. The average European, African and Native American ancestry in the Brazilian sample were estimated as 76.8, 15.1 and 8.1%, respectively. The estimates of individual ancestry obtained with the program ADMIXTURE are highly correlated with the scores of the first two PC-axes (Supplementary Figure 2). The scores of PC1 are highly correlated with the African contribution (R2 = 0.9921), and the scores of PC2 are highly correlated with the Native American contribution (R2 = 0.9802).
GWAS of warfarin maintenance dose
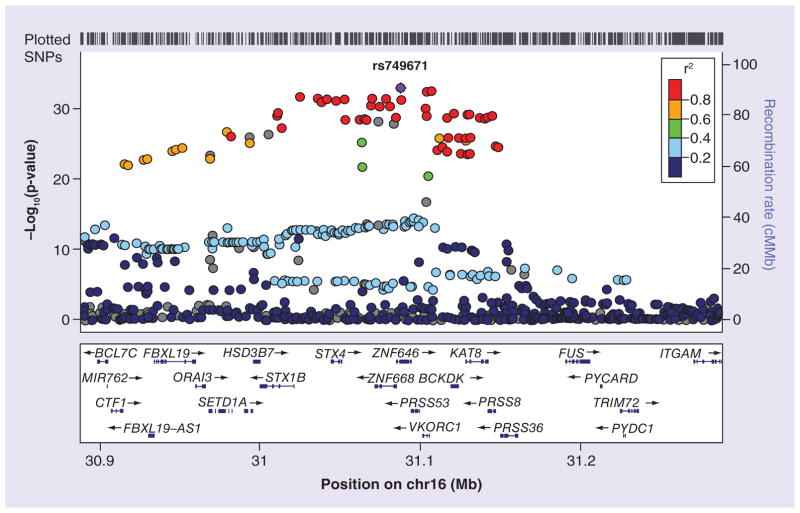
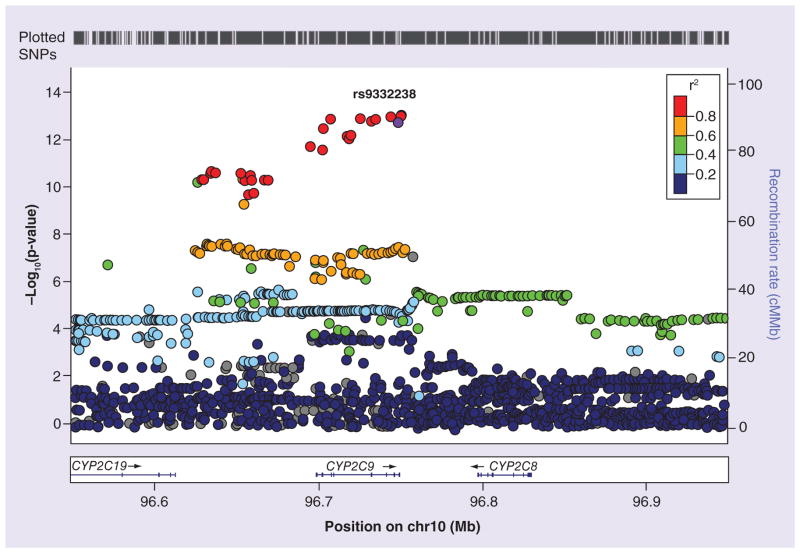
We evaluated the association of directly genotyped and imputed genetic markers with warfarin dose (low vs high dose) including as covariates sex, age, BMI, treatment with amiodarone and the first two PC scores. The Manhattan and QQ plots corresponding to this analysis are shown in Supplementary Figures 3 & 4, respectively. There was little evidence of inflation of p-values (lambda = 1.03). We observed genome-wide significant signals (p ≤ 5 × 10−8) in the well-known VKORC1 and CYP2C9 regions. Figures 2 and 3 show the regional plots for the VKORC1 and CYP2C9 regions, respectively. In the VKORC1 region, the marker showing the strongest effect was rs749671. For this SNP, the G allele was strongly associated with high warfarin dose (G allele, OR: 20.4 [14.3–29.0]; p = 1.08 × 10−33). In the CYP2C9 region, the strongest effect was observed for rs9332238. For this marker, the G allele was associated with high warfarin dose (G allele, OR: 6.8 [5.0–9.1]; p = 4.4 × 10−13), although the nearby marker rs4918798 had the lowest p-value (p = 2.3 × 10−13). A detailed analysis of the linkage disequilibrium (LD) patterns in the CYP2C9 region indicated that rs9332238 (and to a lesser extent rs4918798) is in almost perfect LD with two known functional SNPs, CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910) (Table 2). With the exception of a single haplotype, the minor allele of rs9332238 (allele A) is always associated with CYP2C9*2 (rs1799853 T allele) or *3 (rs1057910 C allele). The ORs [CI] and p-values observed in the meta-analysis for CYP2C9*2 were 0.21 [0.16–0.30]; p = 2.1 × 10−7 and for CYP2C9*3 0.16 [0.10–0.26]; p = 1.8 × 10−5, respectively. Aside from VKORC1 and CYP2C9, no other regions surpassed genome-wide significance in our analysis. Supplementary Table 1 shows additional information about the main signals identified in the VKORC1 and CYP2C9 regions.
Figure 2.
Regional plot of the VKORC1 region.
Figure 3.
Regional plot of the CYP2C9 region.
Table 2.
Linkage disequilibrium patterns observed between CYP2C9*2 (rs1799853), CYP2C9*3 (rs1057910) and rs9332238.
| Haplotype (rs1799853, rs1057910, rs9332338) | Expected | Estimated |
|---|---|---|
| C A G | 0.6463 | 0.7980 |
| C A A | 0.1621 | 0.0014 |
| C C G | 0.0463 | 0.0014 |
| C C A | 0.0116 | 0.0655 |
| T A G | 0.0997 | 0.0000 |
| T A A | 0.0250 | 0.1337 |
| T C G | 0.0071 | 0.0000 |
| T C A | 0.0018 | 0.0000 |
The total number of haplotypes in the sample is 718. Note that, with the exception of a single haplotype, the minor allele of rs9332238 (A allele) is always associated with either CYP2C9*2 (rs1799853 T allele) or CYP2C9*3 (rs1057910 C allele).
GWAS conditioning on VKORC1 & CYP2C9 & follow-up of suggestive signals
We carried out conditional analysis including as covariates the VKORC1 rs749671 SNP and CYP2C9*2 and CYP2C9*3, in addition to the covariates used in the initial analysis. The Manhattan and QQ plots are depicted in Supplementary Figures 5 and 6. No genome-wide significant regions were identified in this analysis, but there were several regions with suggestive signals (p < 10−5). We analyzed in more detail these regions and, based on the regional plots and the imputation scores, we identified a number of markers to follow-up in three available datasets corresponding to GWA studies in European, African–American and Japanese samples. The European and African–American GWA studies used stable warfarin dose as a quantitative trait, and did not follow the EDP approach that we used in our study. In order to make these two studies directly comparable to our Brazilian study, the European and African–American samples were classified in two categories, following the same approach used in our study: a low-dose category including individuals with stable warfarin weekly dose less than 20 mg and a high-dose category including individuals with weekly dose greater than 42.5 mg/week. A logistic regression was then used to evaluate the association of the lead SNPs with warfarin dose, conditioning on the same markers used as covariates in our study. The Japanese GWA study used a similar EDP approach to that employed in our study, classifying individuals into low and high warfarin categories. However, Japanese typically require lower warfarin dosage than individuals from other populations, and the weekly warfarin thresholds used in the Japanese study were lower than the thresholds defined in our study: 7 mg/week for the low dose category and 28 mg/week for the high-dose category. Supplementary Table 2 shows information about the suggestive regions identified in our conditional analysis. The Table provides information about the lead SNP in each region, and the p-values and ORs observed in the Brazilian sample, as well as the European, African–American and Japanese samples. We could not replicate any of the suggestive signals observed in our conditional analysis. Two of the markers (rs10916661 and rs6897106) were nominally significant in the African–American sample, but the direction of effect is discordant in both studies.
Gene-based & pathway-based analyses
We carried out gene-based and pathway-based analyses using the programs KGG3 and GSA-SNP (for more details, see Materials & methods section). As expected, the KGG3 program identified genome-wide significant results for genes in the VKORC1 and CYP2C9 regions. In the analysis taking into account LD patterns (based on the Puerto Rican 1000 Genomes sample), we identified three additional genome-wide significant genes: LINC00511 and NDEL1, located on different regions of chromosome 17, and POU4F3, located on chromosome 5. Similar results were obtained when assuming independence between markers. These three genes show clusters of markers showing highly suggestive signals in our SNP analysis. The gene-based results are depicted in Supplementary Table 3. The pathway analysis using the program KGG3, based on 1320 gene sets extracted from several pathway databases, primarily highlighted gene sets related to the Cytochrome P450 pathway. The complete list of pathways that were both significant using the HYST and hypergeometric tests are depicted in Supplementary Table 4. The lists of pathways identified by the program GSA-SNP, based on KEGG and Gene Ontology (GO) gene sets are provided in Supplementary Tables 5 and 6.
eQTL analyses
We evaluated if eQTLs reported for LCL and human liver are associated with warfarin dose in our study, after Bonferroni correction for multiple testing. For LCL, we used two different datasets: one based on RNA-seq [31], and the other based on gene expression arrays [33]. For human liver, we used the array-based dataset generated by Schadt [34]. For LCL, the only eQTL reaching significance was rs36063822 (p = 4.4 × 10−7), which is associated with the expression of the SRCAP gene on chromosome 16. For liver, two eQTLs reached Bonferroni corrected significance. The first eQTL is rs4889606, which is one of the top signals of our GWA study (p = 1.0 × 10−29). This SNP is located on the STX1B gene on chromosome 16 and is a cis-eQTL strongly associated with the expression of the nearby VKORC1 gene (1.6 × 10−23). The second eQTL is rs2289442 (3.2 × 10−7), which is associated with the expression of the RNF40 gene on chromosome 16.
Discussion
We carried out a GWA study of warfarin maintenance dose in a Brazilian sample. Given our limitations regarding the number of samples that could be analyzed in the study, we applied the EDP approach in order to maximize the statistical power to identify loci with significant effects on warfarin dose [16]. The cutoffs for inclusion in the low- and high-dose warfarin dose were defined as at ≤20 mg/week and ≥42.5 mg/week, respectively. We identified genome-wide significant associations in the VKORC1 and CYP2C9 regions. In the VKORC1 region, there were multiple markers with p-values <10−30 (Figure 2). The top signal was rs749671, with a p-value of 1.08 × 10−33 and an odds ratio estimate of 20.4 for the G allele. The SNP rs9923231, which has been associated with warfarin dose in previous studies [12,13], showed a similar p-value (p = 3.38 × 10−33). These two markers are in very strong LD in the Brazilian samples (r2 = 0.976) as well as in the European (r2 = 1.0) and East Asian samples (r2 = 0.901). Therefore, we seem to be capturing the same signal that has been observed in these populations. Given that our study is based on very dense imputed data generated on 1000 Genomes reference files, we carried out annotations of the top variants (p-value < 10−28) identified in the VKORC1 region using SNP-Nexus [41] and SNVrap [42]. These results are summarized in Supplementary Table 7. A number of variants are worth mentioning. In our analyses with SNP-Nexus, our top signal, rs749671 was identified as a conserved site based on PHAST and GERP++. Another of our top signals, rs749670 (p = 5 × 10−32), is a nonsynonymous polymorphism and the program Polyphen indicates, based on the analysis of one of the transcripts, that it is possibly damaging. This variant was also estimated to be a conserved site based on PHAST and GERP++. The SNP rs14235 (p = 4.25 × 10−31) was identified as a conserved transcription binding site, and labeled as evolutionary conserved by PHAST and GERP++. The annotation portal SNVrap provides functional prediction scores that are based on multiple annotation sources. Our top variant, rs749671, as well as rs749670 and rs2303223, were classified as deleterious by SNVrap. Finally, our eQTL analysis identified another SNP, rs4889606 (p = 1.02 × 10−29), which is strongly associated with the expression of the VKORC1 gene in human liver.
In the CYP2C9 region, the marker showing the highest OR was rs9332238 (G allele, OR: 6.8 and p = 4.4 × 10−13), although there were a few other markers with slightly lower p-values (Figure 3). Interestingly, the two major functional variants reported in this region, CYP2C9*2 and CYP2C9*3, showed substantially lower p-values (p = 2.1 × 10−7 and p = 1.8 × 10−5, respectively) than rs9332238. A detailed analysis of LD between the three markers indicates that rs9332238 (as well as other polymorphisms, such as rs4918798, our top marker in terms of p-value) may be capturing the effects of both CYP2C9*2 and CYP2C9*3. The minor allele of rs9332238 is always associated with CYP2C9*2 or CYP2C9*3 (but never with both, see Table 2). A similar situation was reported by Takeuchi et al. [12], who described in a European sample that their top signal in the CYP2C9 region, rs4917639, was almost perfectly associated with the ‘composite’ CYP2C9*2 and *3 allele. In our sample, rs9332238 (minor allele frequency 19.6%) shows a stronger association with the ‘composite’ of CYP2C9*2 (frequency 13.1%) and *3 (frequency 6.6%) than rs4917639 (frequency 22.2%) or rs4918798 (frequency 21.9%).
In our initial analysis, aside from VKORC1 and CYP2C9, no other regions reached genome-wide significance. In previous GWA studies of warfarin dose, the application of conditional analysis including VKORC1 and CYP2C9 in the statistical models uncovered novel variants in CYP4F2 and the CYP2C cluster on chromosome 10 [12–14]. We carried out statistical analysis conditioning on VKORC1 rs749671, CYP2C9*2 and CYP2C9*3. We did not observe any genome-wide significant signals in this analysis (Supplementary Figures 5 & 6). Based on the regional plots and imputation scores, we selected a number of suggestive SNPs (p < 10−5) for follow-up in European and African–American samples, which were analyzed using the same EDP approach used in our study. We could not replicate any of the suggestive signals (Supplementary Table 2). The ideal approach for replication would have been to analyze the markers in another Brazilian sample, but no such sample was available to us. The results of the replication do not have a straightforward interpretation, given that the patterns of LD may differ between these populations and the Brazilian admixed groups. The lack of replication could be due to a number of factors, including: 1) the signals identified are false positives, 2) lack of statistical power, particularly considering that the effect sizes observed in our study may be inflated (winner’s curse), and 3) discordant patterns of LD in samples of different ancestry [43]. In our gene-based association tests using the GATES approach32, in addition to VKORC1 and CYP2C9, three other genes, LINC00511, NDEL1 and POU4F3, reached genome-wide significance. To our knowledge, these genes have not been associated with warfarin dose in any previous studies. We evaluated the association of the lead SNPs in these genes in the European and African–American samples, but none of the SNPs was nominally significant in these samples (data not shown). As expected, our pathway analyses using KEGG annotations identified genesets involved in the metabolism of xenobiotics, driven by genome-wide signals observed on chromosome 10, where CYP2C9 and other genes of the cytochrome P450 CYP2C cluster are located. The pathway analyses based on GO annotations also highlighted genesets involved in drug metabolism process, including VKORC1 and CYP2C9.
In conclusion, here we present the first GWA study of warfarin dose in the Brazilian population. In agreement with previous studies in other populations, the strongest effects on warfarin dose were observed for VKORC1 and CYP2C9. No other genome-wide significant regions were identified in the study, and we could not replicate a number of suggestive signals in European and African–American samples. This may be related to two important limitations of this study: the sample size of the discovery sample was relatively small, in spite of the fact that we used an EDP approach to maximize our power to detect significant effects, and no Brazilian samples were available for replication, therefore complicating the interpretation of the replication results. Further GWA studies or meta-analytic efforts with larger samples sizes will be necessary to identify other potential regions with smaller effects on warfarin dose in this population.
Future perspective
The complex structure of the Brazilian population, highly heterogeneous and admixed, implies that extrapolation of pharmacogenomic data from candidate gene or GWA studies in well-defined ethnic groups (e.g., European and North American Caucasians) may not be appropriate to the majority of Brazilians [44]. This prompted the present study, which extended to Brazilians the GWAS evidence that common SNPs with large effects on warfarin dose appear to be restricted to the CYP2C9 and VKORC1 regions [11]. One of the major strengths of this study was the availability of imputed data based on the dense 1000 Genomes reference dataset, which made it possible to evaluate in detail the effect of common variants in these regions. Based on our annotations of the top signals, several SNPs within the VKORC1 region could have putative functional effects, and it would be important to explore through functional assays or studies in animal models the specific role of these variants in warfarin dose. In CYP2C9, we identified a polymorphism, rs749671, that is virtually in perfect association with the CYP2C9*2 and *3 alleles. It would be interesting to examine the impact of including rs749671 – rather than CYP2C9*2 and *3 – as a covariate in warfarin dosing algorithms for Brazilians. Finally, further GWA studies or meta-analytic efforts with independent cohorts of Brazilian patients will be necessary to validate the suggestive signals with smaller effects on warfarin dose in this population.
Supplementary Material
Executive summary.
Background
Warfarin is the most commonly used anticoagulant in the world, and several genome-wide association (GWA) studies in diverse populations have explored the role of genetic variants on warfarin dose.
Here we report the first GWA study in Brazilian patients, from two cohorts who had been previously enrolled in warfarin pharmacogenomic trials using candidate gene approaches.
The extreme discordant phenotype (EDP) methodology was applied to select patients receiving low (≤20 mg/week, n = 180) or high stable warfarin doses (≥42.5 mg/week, n = 187). Sampling extreme phenotype individuals confers higher power for GWAS of drug phenotypes, compared with random cohorts.
Genome-wide signals are identified in the VKORC1 & CYP2C9 regions
In the VKORC1 region, the lead SNP was rs749671 (OR: 20.4; p = 1.08 × 10−33), whereas in the CYP2C9 region, the strongest effect was observed for rs9332238 (OR: 6.8; p = 4.4 × 10−13).
The rs9332238 SNP is in virtually perfect LD with two known functional SNPs, namely CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910).
No other genome-wide significant regions were identified in the initial GWA analysis.
GWAS analyses conditioning for VKORC1 rs749671, CYP2C9*2 & CYP2C9*3 disclosed suggestive signals
Conditional analyses identified a list of highly suggestive regions (p < 10−5).
Because no samples from Brazilian patients were available to us for validation, the lead SNPs for each region were followed up in European, East Asian and African–American samples, which were analyzed using the EDP approach.
We could not replicate any of the suggestive signals in these samples.
Conclusion
In agreement with previous studies in other populations, the strongest effects on warfarin dose were observed for VKORC1 and CYP2C9. No other genome-wide significant regions were identified in the study.
Suggestive signals observed in conditional analyses could not be replicated in European, East Asian and African–American samples. This may be related to two important limitations of this study: the sample size of the discovery sample was relatively small, in spite of the fact that we used an EDP approach to maximize our power to detect significant effects, and no Brazilian samples were available for replication, which hinders the interpretation of the replication results.
Further GWA studies or meta-analytic efforts with larger sample sizes will be necessary to identify other potential regions with smaller effects on warfarin dose in this population.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
The research was supported in part by grants from Financiadora de Estudos e Projetos (Finep 01.08.01230.00), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (Faperj) to GS-K. TK was supported by NIH NIGMS R24 GM61374. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 3.Wen MS, Lee M, Chen JJ, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clin Pharmacol Ther. 2008;84:83–89. doi: 10.1038/sj.clpt.6100453. [DOI] [PubMed] [Google Scholar]
- 4.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Perini JA, Struchiner CJ, Silva-Assunção E, et al. Pharmacogenetics of warfarin: development of a dosing algorithm for brazilian patients. Clin Pharmacol Ther. 2008;84:722–728. doi: 10.1038/clpt.2008.166. First study of warfarin pharmacogenetics in Brazilian patients. The present study enrolled some of these patients. [DOI] [PubMed] [Google Scholar]
- 6.International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenzini P, Wadelius M, Kimmel S, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther. 2010;87:572–578. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limdi NA, Wadelius M, Cavallari L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369:2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 11•.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. The first warfarin GWA study, in which it was suggested that common SNPs with large effects on warfarin dose were unlikely to be discovered outside of the CYP2C9 and VKORC1 genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Takeuchi F, McGinnis R, Bourgeois, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. Conditional analysis revealed a genome-wide association study (GWA) signal in CYP4F2 (rs2108622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha PC, Mushiroda T, Takahashi A, et al. Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet. 2010;19:4735–4744. doi: 10.1093/hmg/ddq389. [DOI] [PubMed] [Google Scholar]
- 14.Perera MA, Cavallari LH, Limdi NA, et al. Genetic variants associated with warfarin dose in African–American individuals: a genome-wide association study. Lancet. 2013;382:790–796. doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Nebert DW. Extreme discordant phenotype methodology: an intuitive approach to clinical pharmacogenetics. Eur J Pharmacol. 2000;410:107–120. doi: 10.1016/s0014-2999(00)00809-8. Description of the extreme discordant phenotype methodology. [DOI] [PubMed] [Google Scholar]
- 16.Gurwitz D, McLeod HL. Genome-wide studies in pharmacogenomics: harnessing the power of extreme phenotypes. Pharmacogenomics. 2013;14:337–339. doi: 10.2217/pgs.13.35. [DOI] [PubMed] [Google Scholar]
- 17•.Botton MR, Bandinelli E, Rohde LE, Amon LC, Hutz MH. Influence of genetic, biological and pharmacological factors on warfarin dose in a Southern Brazilian population of European ancestry. Br J Clin Pharmacol. 2011;72:442–450. doi: 10.1111/j.1365-2125.2011.03942.x. Patients from this cohort were enrolled in the present GWA study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchini J, Howie B, Myers S, McVean G, Donnelly A new multipoint method for genome-wide association studies via imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 22.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 23.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen AL, Al-Zubiedi S, Zhang JE, et al. Genetic and environmental factors determining clinical outcomes and cost of warfarin therapy, a prospective study. Pharmacogenet Genomics. 2009;19:800–812. doi: 10.1097/FPC.0b013e3283317ab5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I, Burmester JK. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics. 2004;14:539–547. doi: 10.1097/01.fpc.0000114760.08559.dc. [DOI] [PubMed] [Google Scholar]
- 28.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 29.Limdi NA, McGwin G, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African–American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limdi NA, Beasley TM, Baird MF, et al. Kidney function influences warfarin responsiveness and hemorrhagic complications. J Am Soc Nephrol. 2009;20:912–921. doi: 10.1681/ASN.2008070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallari LH, Langaee TY, Momary KM, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 32.KGG: A systematic biological knowledge-based mining system for genome-wide association studies. http://statgenpro.psychiatry.hku.hk/limx/kgg/
- 33.Li MX, Gui HS, Kwan JS, Sham PC. GATES, a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li MX, Kwan JS, Sham PC. HYST, A hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91:478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam D, Kim J, Kim SY, Kim S. GSA-SNP, a general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010;28:W749–W754. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S-Y, Volsky DJ. PAGE, parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NCBI eQTL browser. http://www.ncbi.nlm.nih.gov/projects/gap/eqtl/index.cgi#.
- 38.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SNP nexus. http://www.snp-nexus.org/
- 42.SNVrap: Quick annotation web portal. http://jjwanglab.org/snvrap.
- 43.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Suarez-Kurtz G. Pharmacogenetics in the Brazilian population. Front Pharmacol. 2010;1:118. doi: 10.3389/fphar.2010.00118. An analysis of the pharmacogenomic implications of the Brazilian population structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.