Abstract
Epigenetic dysfunction has been implicated in a growing list of disorders that include cancer, neurodevelopmental disorders, and neurodegeneration. Williams syndrome (WS) and 7q11.23 duplication syndrome (Dup7) are rare neurodevelopmental disorders with broad phenotypic spectra caused by deletion and duplication, respectively, of a 1.5-Mb region that includes several genes with a role in epigenetic regulation. We have identified striking differences in DNA methylation across the genome between blood cells from children with WS or Dup7 and blood cells from typically developing (TD) children. Notably, regions that were differentially methylated in both WS and Dup7 displayed a significant and symmetrical gene-dose-dependent effect, such that WS typically showed increased and Dup7 showed decreased DNA methylation. Differentially methylated genes were significantly enriched with genes in pathways involved in neurodevelopment, autism spectrum disorder (ASD) candidate genes, and imprinted genes. Using alignment with ENCODE data, we also found the differentially methylated regions to be enriched with CCCTC-binding factor (CTCF) binding sites. These findings suggest that gene(s) within 7q11.23 alter DNA methylation at specific sites across the genome and result in dose-dependent DNA-methylation profiles in WS and Dup7. Given the extent of DNA-methylation changes and the potential impact on CTCF binding and chromatin regulation, epigenetic mechanisms most likely contribute to the complex neurological phenotypes of WS and Dup7. Our findings highlight the importance of DNA methylation in the pathogenesis of WS and Dup7 and provide molecular mechanisms that are potentially shared by WS, Dup7, and ASD.
Introduction
Disorders that arise via deletion or duplication of the same set of genes provide unique insight into the effects of changes in gene dosage during development. Two such disorders are Williams syndrome1 (WS [MIM: 194050]) and 7q11.23 duplication syndrome2 (Dup7 [MIM: 609757]), which are rare neurodevelopmental disorders caused by deletion and duplication, respectively, of 25 genes in chromosomal region 7q11.23. Copy-number variation (CNV) at this locus causes symmetrical alterations in gene dosage and expression and results in distinct but overlapping phenotypic spectra.
WS is associated with a recognizable facies, characteristic cardiovascular lesions, and an array of medical problems.3 Individuals with WS usually have global cognitive impairment but show relative strength in concrete vocabulary and verbal short-term memory; they also display moderate to severe deficits in visuospatial construction.4 They often exhibit social disinhibition and lack of usual social boundaries, but non-social anxiety is common.4,5 Dup7 has not been as extensively characterized, but the emerging picture is of a syndrome that is distinct from WS and invariably associated with speech problems and language delay or disorder. Anxiety, especially social phobia, is common.6
Neuropsychiatric phenotypes are common to both WS and Dup7. Approximately 75% of individuals with Dup7 and 60% of those with WS meet DSM-IV criteria for at least one anxiety disorder.4,5 In addition, around 35% of children with Dup7 and 65% of children with WS have attention deficit hyperactivity disorder, and around 25% are diagnosed with oppositional defiant disorder or other disruptive behavior disorders.4–6 These complex neurodevelopmental disorders also include both core and associated features of autism spectrum disorder (ASD) and so provide a unique window into genes and pathways that contribute to symptoms of ASD. Features common to ASD, such as speech disorder and repetitive behaviors, have been reported in many individuals with Dup7,7,8 and 7q11.23 duplication has been independently identified as both an ASD-associated CNV9,10 and a risk factor for schizophrenia.11 We, and others, have reported that some socio-communicative difficulties in children with WS significantly overlap those in children with ASD,12–14 and some children with WS have been classified as having ASD.12–17
Research efforts have investigated the molecular bases for the complex neurological phenotypes of WS and Dup7, but the potential impact of 7q11.23 rearrangements on the epigenome has not been studied. Epigenetic modifications can be tissue specific and extremely dynamic, and they play key roles in the development of the mammalian nervous system.18 Of the 25 genes commonly deleted or duplicated in WS or Dup7, respectively, six genes and their encoded proteins have been associated with epigenetic mechanisms or complexes. Williams-Beuren syndrome chromosome region 22 (WBSCR22) contains an S-adenosyl-L-methionine binding domain typical of methyltransferases and has been associated with chromatin remodeling;19 NOP2/Sun domain family 5 (NSUN5) has been shown to act as an RNA methyltransferase;20 complete loss of bromodomain adjacent to zinc finger domain 1B (BAZ1B), a subunit of the WICH and B-WICH chromatin-remodeling complexes, has been shown to affect heterochromatin formation;21 and B cell CLL/lymphoma 7B (BCL7B) has been identified as a mammalian SWI/SNF ATP-dependent subunit of the chromatin-remodeling complex.22 The general transcription factors GTF2I (also known as TFII-I) and GTF2IRD1 have both been shown to functionally interact with the histone deacetylase HDAC3.23 Moreover, TFII-I in particular has been shown to interact with numerous regulatory complexes, including HDAC1, HDAC2, the lysine-specific demethylase 1 (LSD1) complex, and components of the polycomb complex.24,25
Given the number of 7q11.23 genes that are associated with epigenetic mechanisms and the importance of epigenetic mechanisms during neurodevelopment, we hypothesized that rearrangements of this locus would alter the epigenome of children with WS and children with Dup7. Disruptions of epigenetic mechanisms, including chromatin remodeling and DNA methylation, have been implicated in numerous neurodevelopmental and neuropsychiatric disorders, including ASD26 and schizophrenia.27 We compared the impact of gene dosage at 7q11.23 on genome-wide DNA methylation between whole-blood samples from children with WS or Dup7 and samples from age-matched typically developing (TD) control children. We identified striking, genome-wide dose-dependent changes in DNA methylation across the WS and Dup7 groups, suggesting that aberrant DNA methylation might be an important contributor to the pathophysiology of these disorders.
Subjects and Methods
Research Participants
All procedures were approved by the research ethics boards of the University of Toronto and The Hospital for Sick Children, and written informed consent was obtained from parents of all participants. 7q11.23 deletion or duplication size was determined by CNV microarray analysis and confirmed by real-time PCR analysis; only children who had classic deletions or duplications were included. Children were between 2.4 and 10.7 years of age at the time of the blood draw. Descriptive statistics for the participants’ age at blood draw are presented in Table S1.
DNA-Methylation Array Profiling
DNA was extracted from whole-blood samples with the QIAamp DNA Blood Mini Kit (QIAGEN), the ArchivePure DNA Blood Kit (5 PRIME), or the AllPrep DNA/RNA Mini Kit (QIAGEN). DNA was then subject to sodium bisulfite treatment. In brief, 1 μg of genomic DNA was converted with sodium bisulfite by the EpiTect Bisulfite Kit (QIAGEN) according to the manufacturer’s protocol. Genome-wide DNA-methylation profiling was performed with the Infinium HumanMethylation450 BeadChip Kit and arrays (Illumina) by The Centre for Applied Genomics (The Hospital for Sick Children, Toronto). Sodium-bisulfite-treated DNA was hybridized onto the BeadChip with Illumina’s Infinium HD Methylation protocol and scanned with Illumina’s iScan SQ Scanner.
Statistical Analysis
Image files were pre-processed with the Illumina GenomeStudio (v.2011.1) Methylation Module (v.1.9.0) with background correction and Illumina-based normalization. Quality control was performed in GenomeStudio and included assessing the efficiency of sodium bisulfite conversion, extension, hybridization, and labeling. With the use of the Illumina Methylation Analyzer (IMA)28,29 and DMRcate30 packages in the R statistical environment, normalized data were further filtered for excluding poorly performing probes, probes that mapped to sex chromosomes, cross-reactive probes, and probes that overlapped SNPs with a minor allele frequency (MAF) of ≥0.5 and fell at least 2 bp from the end of the probe. Principal-component analysis (PCA) and volcano plots were generated in the R statistical environment. Logit-transformed beta values (M-values) were calculated in the IMA and used for statistical modeling.
To account for potential differences in blood cell populations, we approximated the cellular composition of each sample by using an adjusted Houseman algorithm available in the minfi package31 in the R statistical environment. We incorporated the composition of granulocytes, CD8+ T cells, CD4+ T cells, B cells, natural killer cells, and monocytes as covariates in a linear regression model by using the limma package in R,32 and we applied Benjamini-Hochberg correction to correct for multiple testing.33 This analysis revealed significant differences in the estimate of cellular composition between the three groups (Figure S1; Table S2), although there is currently no clinical evidence to suggest that either children with WS or children with Dup7 show differences in blood cell composition. It is therefore likely that at least some of the detected differences are caused by 7q11.23 dose-dependent changes in DNA methylation at the CG sites used for the analysis.
We used Kruskal-Wallis with Dunn’s post hoc test when comparing DMP methylation levels between the three cohorts as a measure of the significance of genotype. Each probe had been previously identified as significantly differentially methylated (DM) in relation to probes from TD control children.
Many probes were identified as significantly DM and clustered together, suggesting the presence of a DM region (DMR). To test for this, we performed DMR analysis by using the R package DMRcate.30 We incorporated the same limma model we used to identify DM positions (DMPs) into this analysis and used estimated blood cell counts as covariates in the linear model. We used the default DMRcate settings: a lamda of 1,000, scaling factor of 2, Benjamini-Hochberg correction for multiple testing, and p value cutoff of 0.05. Significant DMRs were retained if they contained two or more probes and the maximum change in methylation was >10%. A list of significant DMRs can be found in Table S3.
Pyroseqencing Validation
Sodium bisulfite pyrosequencing analysis was performed on selected DMRs according to a method described previously.34 Primers were designed with PyroMark Assay Design Software (QIAGEN) to target at least one DM CpG, and sodium-bisulfite-converted DNA remaining from samples previously hybridized to the array were used. Amplicons were analyzed with a PyroMark Q24 pyrosequencer (QIAGEN), and methylation levels (%) were quantified with PyroMark Q24 Software (QIAGEN). A list of the primer sequences from the pyrosequencing assay is shown in Table S4.
Expression Analysis
RNA was extracted from whole blood collected in Tempus Blood Collection Tubes (Life Technologies) with the QiaAmp RNA Blood Mini Kit (QIAGEN) according to the manufacturer’s protocol. 500 ng of RNA was converted to cDNA with Superscript III Reverse Transcriptase (Life Technologies) and the addition of a final RNase H (Life Technologies) treatment. In preparation for real-time PCR, cDNA samples were diluted at 1:100 in nuclease-free water. Real-time PCR was performed with Power SYBR Green PCR Master Mix (Life Technologies) and assessed with the ViiA 7 detection system (Life Technologies). Standard curves were generated for each primer set, and detected values were normalized to the housekeeping genes HPRT1 and TBP. Primer sequences from the real-time PCR assay are shown in Table S5.
GO Enrichment Analysis
For accounting for differences in the number of probes per gene, GO enrichment analysis was performed with Genomic Regions Enrichment of Annotations35 with the single-nearest-gene association setting. All array probes that were retained after filtering were used as a background dataset.
ENCODE Analysis
ENCODE datasets36 were retrieved and filtered to exclude peaks containing hotspots (1,628 ENCODE peaks included). Intersections of DM CpGs from each group (WS CpGs hyper- or hypomethylated and Dup7 CpGs hyper- or hypomethylated in relation to control CpGs) and ENCODE peaks were calculated with 1,000 permutations. Enrichment p values were calculated with the R function pnorm. Enrichment Z scores were calculated for the 50 most significant enrichments.
Enrichment Analysis of the Transcription Factor Motif
Probes were ranked on the basis of decreasing differential methylation (delta-beta cutoff = 10%, p < 0.05), and the sequence flanking 100 bp around the CG probe was obtained (201 bp). For identifying significantly enriched transcription factor motifs, the ranked sequences were tested with Analysis of Motif Enrichment (AME)37 in the MEME suite; the rank-sum test was used for enrichment, and sequences were tested against JASPAR CORE Vertebrata. Enrichment analysis of known motifs was performed with AME with the same statistical parameters. Sequences containing specific motifs were identified with Find Individual Motif Occurrences (FIMO)38 under the default settings.
Results
Genome-wide, Symmetrical DNA-Methylation Differences in Children with WS and Children with Dup7
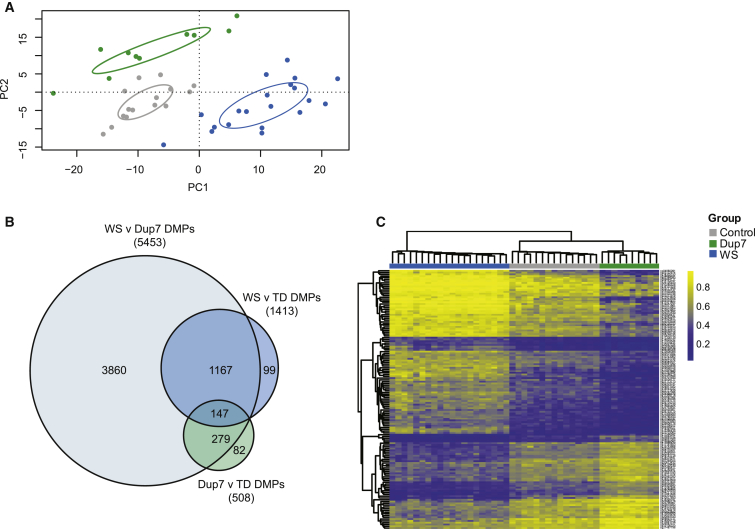
In this study, we assessed genome-wide DNA methylation in whole blood from 20 children with WS, 10 children with Dup7, and 15 TD children by using the Infinium HumanMethylation450 BeadChip array (Illumina). After removal of cross-reactive and SNP-containing probes and adjustment for blood cell composition, PCA revealed distinct differences in DNA methylation between the three groups, such that the WS cohort showed a more significant difference in DNA-methylation profiles than the Dup7 cohort (Figure 1A). Comparing each case cohort to TD control individuals, we identified 1,413 CG probes that were significantly DM between WS and TD and 508 CG probes that were DM between Dup7 and TD by using a methylation-difference delta-beta cutoff of 10% and a Benjamini-Hochberg-adjusted p ≤ 0.05 (Figure 1B). Using the same cutoff, comparison between the WS and Dup7 cohorts identified 5,453 DMPs (Figure 1B), the vast majority of which showed dose-dependent methylation changes.
Figure 1.
Differential Genome-wide DNA Methylation between TD Children and WS- and Dup7-Affected Children
WS and Dup7 cohorts can be distinguished from the TD control group and from each other on the basis of genome-wide DNA methylation.
(A) PCA revealed three distinct groups, corresponding to children with WS (blue), TD control children (gray), and children with Dup7 (green).
(B) A Venn diagram shows the total number of DMPs between WS and TD groups, between Dup7 and TD groups, and between WS and Dup7 groups. Analysis of differential methylation in each cohort resulted in 1,413 DMPs between WS and TD groups, 508 DMPs between Dup7 and TD groups, and 5,453 DMPs between WS and Dup7 groups (adjusted p ≤ 0.05, delta-beta ≥ 10%). Comparison of the WS and Dup7 datasets to the TD control dataset resulted in 147 sites sharing significant differential methylation.
(C) A heatmap shows methylation changes for all 147 overlapping DMPs, all of which showed dose-dependent changes in DNA methylation.
DMPs were found genome-wide, and there was no specific chromosomal enrichment (Figure S2). Comparison of the DMPs within each cohort revealed 147 probes that were significantly DM between TD control individuals and both WS and Dup7 groups (Figure 1B), and hierarchical clustering of these probes revealed a distinct gene-dose-dependent response, such that the WS-affected children showed more gain of methylation and the Dup7-affected children showed more loss of methylation than did control children (Figure 1C). Only one of these probes, cg19313311, fell within the 7q11.23 CNV. Most of these probes that were DM in children with WS showed symmetrical methylation patterns in Dup7. Importantly, the methylation levels across these 147 probes were sufficient for correctly clustering each individual within his or her correct genotypic cohort (Figure 1C). These 147 DMPs corresponded to 56 unique genes, 3 pseudogenes, and 1 long non-coding RNA (Table S6). Many probes were not significantly DM by >10% between TD control individuals and each cohort, but they were significantly DM by >10% between the WS and Dup7 groups.
Dose-Dependent Changes in DNA Methylation across Genes Important for Brain Development and Function
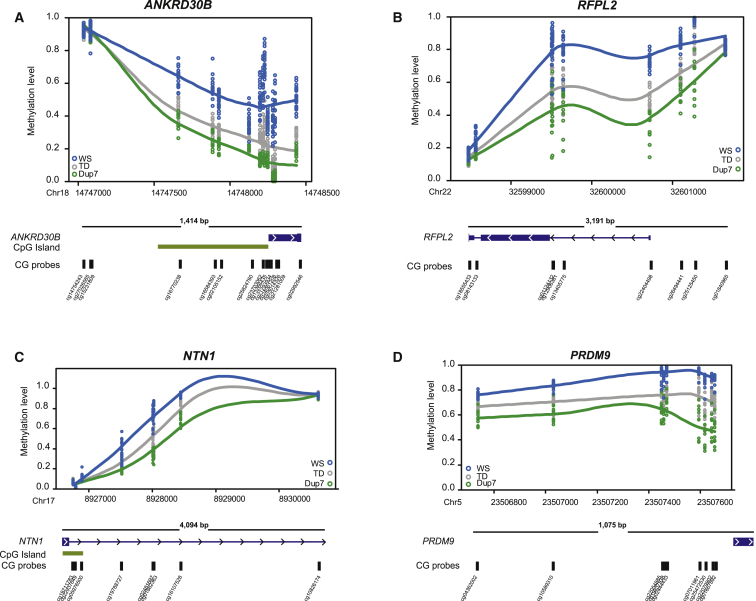
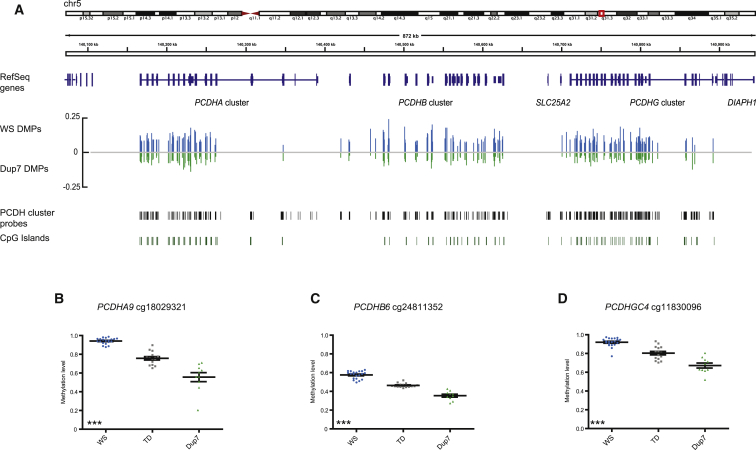
The majority of genes associated with DMPs were not expressed in our starting tissue—whole blood—but were expressed in other tissues, particularly in the developing or adult brain (DMPs are listed in Table S7). One of the most DM genes was ankyrin repeat domain 30B (ANKRD30B), which showed striking dose-dependent DNA-methylation changes spanning 11 different probes across the promoter (Figure 2A). Found only in primates, ANKRD30B is expressed in the brain, breast, and testis, but its function is unknown.39 Consistent dose-dependent differential methylation was also observed across the promoter region of ret finger protein-like 2 (RFPL2 [MIM: 605969]; Figure 2B), a member of the primate-specific chromosome 22 RFPL cluster, which shows high expression during the onset of neurogenesis and within the developing neocortex and has been linked to primate cortical evolution.40 We saw 282 DMPs spanning the entire protocadherin (PCDH) cluster in chromosomal region 5q31.3 (Figure 3A). The PCDHs are homophillic cell-adhesion proteins present predominantly in the developing brain and confer single-cell neuron identity.41 We also saw changes in DNA methylation outside promoter regions, such as at probe cg05374271, found in an active transcription start site of the regulator of G protein signaling RGS2 (MIM: 600861), which has been previously associated with anxiety,42 panic disorder,43 and schizophrenia44 (Figure S3A). HDAC4 (MIM: 605314), an ASD candidate gene,45 also showed altered DNA methylation (Figure S3C). Differential methylation across ANKRD30B, RFPL2, and RGS2 was confirmed by pyrosequencing (Figure S4), and real-time PCR analysis of RGS2 and HDAC4, two of only a few DM genes expressed in blood, demonstrated that changes in methylation were associated with reduced gene expression in the WS group (Figures S3B and S3D). Additional dosage-sensitive genes of interest include the neuronal migration gene NTN1 (MIM: 60161446; Figure 2C) and PRDM9 (MIM: 609760; Figure 2D), which encodes a histone methyltransferase that activates recombination hotspots during meiosis47 and is rapidly evolving in primates.48
Figure 2.
Dose-Dependent DMRs in WS- and Dup7-Affected Children
Scatter plots fitted with Loess curves depict dose-dependent DNA-methylation changes between the WS (blue), Dup7 (green), and TD (gray) groups. Methylation levels for each sample are shown on the basis of the position of each CG probe within the gene. A schematic of the gene and the corresponding CG probes are shown below each graph.
(A) A dose-dependent DMR was observed across 14 CpG sites in ANKRD30B (mean DMR p = 1.13e−28).
(B) Two DMRs spanning nine CpG sites within the promoter region of RFPL2 (mean DMR p = 6.64e−9 at chr22: 32,598,479–32,599,648 and p = 3.06e−35 at chr22: 32,600,722–32,601,654).
(C) One DMR spanning eight CpG sites within the promoter region of NTN1 (mean DMR p = 2.94e−3).
(D) One DMR spanning nine CpG sites within the promoter region of PRDM9 (mean DMR p = 3.10e−76).
Figure 3.
Dose-Dependent DNA-Methylation Changes Spanning the PCDH Cluster
(A) Epigenetic information spanning the PCDH cluster shows a total of 282 significant DMPs between the WS group (blue) and the Dup7 group (green). The WS group had 105 significant DMPs, of which 60 were DM by >10% in comparison to the TD group, and all showed hypermethylation in relation to the TD group. The Dup7 group contained 30 significant DMPs, of which 4 had a loss of methylation of >10% in relation to the TD group.
(B) Methylation levels of probe cg18029321 (within PCDHA9) showed a 18.5% gain of methylation in the WS group (blue; adjusted p = 0.002) and a 20% loss of methylation in the Dup7 group (green; adjusted p = 0.018) in relation to the TD group (gray).
(C) Methylation levels of probe cg24811352 (within PCDHB6) showed a 11.2% gain of methylation in the WS group (blue; adjusted p = 0.009) and a 10.9% loss of methylation in the Dup7 group (green; adjusted p = 9.08e−4) in relation to the TD group (gray).
(D) Methylation levels of probe cg11830096 (within PCDHGC4) showed a 11.6% gain of methylation in the WS group (blue; adjusted p = 0.03) and a 13.2% loss of methylation in the Dup7 group (green; adjusted p = 3.15e−2) in relation to the TD group (gray). Significance of the genotype effect was measured by the Kruskal-Wallis test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Some genes were significantly DM between TD control individuals and only one affected group, but we observed a non-significant trend in the other group. For example, MKRN3 (MIM: 603856) and NDN (MIM: 602117), both of which lie within 15q11–13, the imprinted region associated with Prader-Willi (MIM: 176270) and Angelman (MIM: 105830) syndromes49 (Figure S5A), were DM in the Dup7 group (decreased methylation) and showed a trend toward increased methylation in the WS group (Figures S5B and S5C). Many other probes within 15q11–13 were symmetrically DM in the WS and Dup7 cohorts but did not meet statistical significance or were not DM by a minimum of 10% between the affected cohorts and the TD control group (Figure S5A). DMR analysis identified all of the above genes as containing significant DMRs, which were represented by the individually significant DMPs (Table S3).
Cohort-Specific Changes in DNA Methylation
A few loci showed altered DNA methylation in either the WS or the Dup7 cohort, but not in both groups. For example, the imprinted homeobox 4A (HOXA4 [MIM: 142953])50 showed consistent hypermethylation only in the WS group, but the methylation levels in the Dup7 group were indistinguishable from those in the TD control group (Figure S6A). In contrast, HOXB7 (MIM: 142962) showed hypomethylation in the Dup7 group, but no significant increase in methylation was seen in the WS group (Figure S6B).
GO Enrichment in WS and Dup7 DM Gene Sets
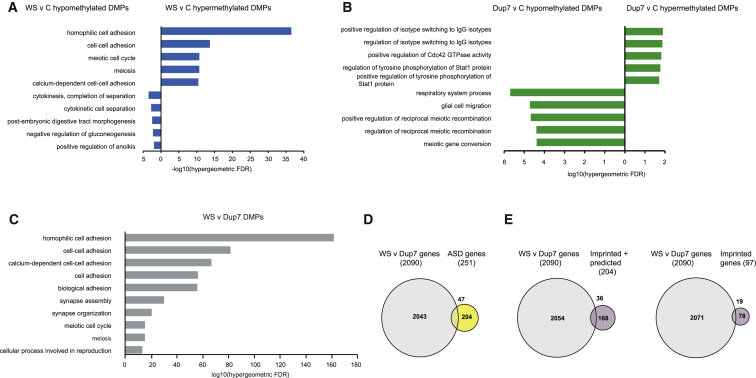
Given that many DM genes were related to neurodevelopment, we assessed whether the DM genes were enriched with specific Gene Ontology (GO) terms. Using stringent assessment parameters that correct for differences in the number of probes per gene on this array, we observed that the genes hypermethylated in the WS group showed significant enrichment of GO terms relating to cell adhesion (Figure 4A), synapse assembly (p = 9.14e−5), synapse organization (p = 3.10e−2), and synaptic transmission (p = 1.89e−3) (Table S8A), suggesting that aberrant DNA methylation might alter the expression of genes important to synapse biology, a system consistently linked with neurodevelopmental disorders, particularly ASD.45 Hypomethylated WS-associated genes were enriched with terms relating to cytokinesis (Figure 4A). In comparison, hypermethylated Dup7-associated genes were enriched with GO terms relating to immunoglobulin isotype switching, regulation of Cdc42, and regulation of STAT1 (Figure 4B), whereas hypomethylated genes within the Dup7 cohort showed enrichment of terms relating not only to meiosis and glial cell migration (Figure 4B) but also to axonal fasciation (p = 1.75e−4), neuron recognition (p = 6.95e−3), and axon extension (p = 1.18e−3) (Table S8C). The WS-versus-Dup7 dataset showed enrichment of multiple terms relevant to brain development and function (Figure 4C; Table S8E). A list of all enriched terms identified can be found in Table S8.
Figure 4.
Analysis of GO Terms and ASD and Imprinted Gene Sets Reveals Enrichment of Neuronal Functions and CTCF Binding Sites
(A) GO enrichment analysis of WS hypermethylated DMPs (right, blue) and WS hypomethylated DMPs (left, blue).
(B) GO enrichment analysis of Dup7 hypermethylated DMPs (right, green) and Dup7 hypomethylated DMPs (left, green) identified enrichment in diverse functions.
(C) GO enrichment analysis of DMPs between the WS and Dup7 groups.
(D) Comparative analysis of the symmetrically DM genes between the WS and Dup7 groups and known ASD candidate genes identified 47 shared genes (hypergeometric p = 3.367e−5).
(E) Comparative analysis of the symmetrically DM genes between the WS and Dup7 groups and imprinted and provisionally imprinted genes (n = 204) identified 36 genes (hypergeometric p = 4.867e−3). When the analysis was restricted to firmly established imprinted genes (n = 97), 19 genes were identified (hypergeometric p = 2.510e−3).
WS and Dup7 DM Gene Set Is Enriched with ASD-Associated and Imprinted Genes
Given the associations between pathway enrichments and specific DM genes and neurodevelopment and ASD, as well as the phenotypic features of WS and Dup7, we assessed the overlap between our DM gene set and known ASD candidate genes from SFARI Gene. We found significant enrichment of ASD candidate genes, such that there were 47 known ASD candidates in the symmetrically DM set (hypergeometric p = 3.37e−5; Figure 4D). These included 13 high-confidence ASD-associated genes ranked by SFARI as having the strongest possible evidence of involvement in ASD (categories 1S and 2S in Table S9). Examples included HDAC4 (three DM probes), SHANK2 (MIM: 603290; three DM probes), DYRK1A (MIM: 600855; two DM probes), SHANK3 (MIM: 606230; one DM probe), NRXN1 (MIM: 600565; one DM probe), CNTNAP2 (MIM: 604569; one DM probe), and ANKRD11 (MIM: 611192; one DM probe). The remaining candidates were ranked as having lower confidence by SFARI but still had supporting evidence of involvement in ASD. These included EIF4E (MIM: 133440; nine DM probes; Figure S7A), DPP6 (MIM: 126141; eight DM probes; Figure S7B), DLGAP2 (MIM: 605438; four DM probes), NRXN2 (MIM: 600566; two DM probes), NRXN3 (MIM: 600567; two DM probes), OXTR (MIM: 167055; two DM probes), and NLGN1 (MIM: 600568; one DM probe). A list of all enriched ASD-associated genes can be found in Table S9.
We also saw enrichment of imprinted genes in our DM gene set when we used either a list that included predicted imprinted genes, such as HOXA4 (hypergeometric p = 4.867e−3), or a set that included only validated imprinted genes (hypergeometric p = 2.51e−3) (Figure 4E; Table S10).
ENCODE Enrichment Analysis of DM Probes
In addition to the DMRs that were associated with specific genes, 28%–46% of DMPs mapped within non-genic loci, particularly those in the WS hypermethylated and Dup7 hypomethylated datasets (Figure S8). Enrichment analysis using ENCODE datasets36 identified significant dose-dependent enrichment of H3K9me3 and of regions binding CCCTC-binding factor (CTCF) across multiple different cell lines (Figure S9), suggesting an enrichment of long-range regulatory elements.51
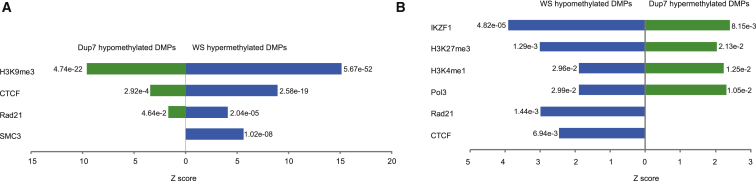
When focusing on a cell line that more closely resembles that of our whole-blood DNA source (human B lymphocyte cell line GM12878), we again identified highly significant dose-dependent enrichment of H3K9me3, CTCF, and members of the cohesion complex within hypermethylated probes in WS and hypomethylated probes in Dup7 (Figure 5A). Hypermethylated probes within the Dup7 cohort and hypomethylated probes within the WS cohort revealed enrichment of different regulatory elements, including IKZF1, an important transcriptional regulator of hematopoiesis,52 and H3K27me3, a repressive chromatin mark important in cellular differentiation and lineage specification53 (Figure 5B). We again saw enrichment of CTCF but only within the WS hypomethylated probes (Figure 5B). When focusing on a human embryonic stem cell line (H1-hESC), we observed even greater enrichment of H3K9me3 and CTCF and of additional transcription factors and histone marks (Figure S10). We again detected enrichment of H3K27me3 within the WS hypomethylated probe set and also detected dose-dependent enrichment of the methyltransferase EZH2, which is responsible for the methylation of H3K2754 (Figure S10). We observed consistent results when we tested the DMPs in the WS and Dup7 datasets (data not shown).
Figure 5.
Enrichment Analysis Using ENCODE Data Identifies Significant Dose-Dependent Enrichment of CTCF Binding Sites
(A) DMP enrichment analysis using ENCODE data from the human B lymphocyte cell line GM12878 revealed a dose-dependent enrichment of the H3K9me3 histone mark and the CTCF and Rad21 binding sites (WS hypermethylated, blue, right; Dup7 hypomethylated, green, left). WS hypermethylated probes also showed enrichment of binding sites for SMC3, a cohesion-complex member that is often found in association with CTCF and Rad21.
(B) Dose-dependent enrichment of the transcriptional regulator IKZF1 binding site, the H3K27me3 and H3K4me1 histone marks, and the Pol3 binding site was identified in WS hypomethylated probes (left, blue) and Dup7 hypermethylated probes (right, green). CTCF and Rad21 binding sites were also enriched, but only in the WS hypomethylated probes.
In concordance with our findings from the ENCODE datasets, we also saw enrichment of CTCF binding consensus sequences within the WS hypermethylated dataset when we used AME (Table S11). Using this tool, we also found enrichment of other transcription factor binding sites, such as those for the early growth response transcription factors EGR1 and EGR2, which have been associated with neuronal plasticity55 and myelination,56 and those for FOXP1 and FOXP2, both of which have been associated with developmental speech disorders and/or ID57 (Table S12). Because three transcription factors are contained within the common 7q11.23 deletion and duplication, we investigated whether their putative consensus sequences were enriched in our datasets. We observed significant enrichment of motifs associated with TFII-I58 and GTF2IRD159 binding in WS hypermethylated probes and in Dup7 hyper- and hypomethylated probes, and we also observed enrichment of the MLXIPL binding ChORE motif,60 but only in the WS hypermethylated dataset (Table S12). GO enrichment analysis using DM genes independently identified a TFII-I consensus sequence within the symmetrically opposite gene set (WS versus Dup7; Table S11).
Discussion
We have identified striking gene-dose-dependent DNA-methylation differences in two symmetrical CNVs. The vast majority of DMPs showed symmetrical changes in DNA methylation between the WS and Dup7 groups, although both the total number of DMPs and the degree of differential methylation appeared greater in the WS cohort, suggesting that loss of 7q11.23 has a more profound effect on DNA methylation than gain of the same region. Dose-dependent transcriptional dysregulation was recently identified in WS and Dup7 human induced pluripotent stem cells (iPSCs),61 but we found many more dose-dependent changes in DNA methylation, and in fact our stringent adjustment of the data for cell composition most likely excluded even more significant DMPs.
Aberrant DNA methylation has been implicated in numerous neurological disorders, particularly ASD26 and schizophrenia,27 and in WS- and Dup7-affected children, we identified aberrant DNA methylation spanning many genes and loci implicated in brain development or psychiatric disorders. Genes that were symmetrically DM were enriched with biological processes relevant to the development and function of the nervous system and were enriched with ASD candidate genes. At this point, we do not know whether these changes will be conserved in other cell types, specifically neurons, but if changes in DNA methylation occur very early in development, they could be present in all tissues. Analyses of genome-wide DNA methylation in iPSC-derived neurons would go some way toward addressing this question.
The altered DNA methylation identified across regions of tightly regulated transcriptional control, such as the PCDH cluster and the 15q11–13 region, is particularly intriguing. PCDH genes have a unique mode of regulation where allele-specific alternative splicing is controlled by DNA methylation across transcript-specific promoters.62 It has been hypothesized that this mode of regulation facilitates the unique and combinatorial patterns of PCDHs on the neuronal cell surface and thus leads to patterns of cell identity in neurons of the developing brain.41 Previous studies showed that DNA methylation across this locus, as well as CTCF and cohesion binding,63,64 regulates the monoallelic and combinatorial expression of specific PCDH isoforms,65 and genetic variation across the locus has previously been associated with ASD,66 ID,67 schizophrenia, and bipolar disorder.68
CNVs within 15q11–13, the imprinted region associated with Prader-Willi and Angelman syndromes, have also been associated with autism69 and schizophrenia,70 and epigenetic dysregulation of the normally biallelically expressed GABAA receptors within this locus has been identified in ASD.71 In our DM gene set, the enrichment of imprinted genes, including MKRN3 and NDN at 15q11–13, suggests that CNVs at 7q11.23 affect either the erasure or the re-establishment of the genomic imprint after fertilization.72
Given the degree of differential methylation we observed in children with WS and children with Dup7, it is likely that one or more genes within 7q11.23 is actively involved in the targeting or regulation of DNA methylation at specific genomic regions. TFII-I has recently been shown to associate with DNA methyltransferase 3-like (DNMT3L),73 suggesting that it might be involved in targeting DNMT3L to specific genomic locations, thereby increasing the catalytic activity of the active methyltransferases DNMT3A and DNMT3B at those sites.74 A separate study also identified an interaction between TFII-I and DNMT3B in iPSCs.61 Interestingly, both the PCDH cluster and DPP6 were shown to undergo DNMT3B-mediated methylation,75,76 and so it is possible that gene dosage of GTF2I might be involved in the DM profiles we detected at these loci. TFII-I has also been shown to bind to the promoter region of EZH2, encoding a member of the polycomb repressive complex 2 (PRC2), in embryonic stem cells,77 which is the same cell type in which we observed enrichment of EZH2 binding sites and regions containing H3K27me3-marked chromatin, a characteristic of transcriptional silencing by PRC2, within DM probes.
TFII-I was recently identified as a novel binding partner of CTCF, and this interaction was required for the targeting of CTCF to specific genomic regions in cancer cell lines.78 We observed highly significant enrichment of DMPs over CTCF binding sites in many different cell types, suggesting that these regions are susceptible to aberrant DNA methylation in WS and Dup7 in either a direct or indirect manner. CTCF has been implicated in chromosome looping, monoallelic gene expression, and the regulation of developmentally restricted or lineage-specific genes, making it a key regulator of the genome.51 Notably, CTCF binding is sensitive to DNA-methylation levels, suggesting that the aberrant DNA methylation identified in WS and Dup7 could affect CTCF-based regulation. Given the associations with TFII-I and various epigenetic regulators, this protein is an attractive candidate for mediating at least some of the DNA-methylation changes we observed in children with WS and children with Dup7, although other genes at 7q11.23 could be involved. Further studies are required to fully elucidate which gene or combination of genes within 7q11.23 is responsible for the dose-dependent DNA-methylation changes observed in WS and Dup7.
Although our research focused on neuropsychiatric phenotypes, some DM genes might contribute to other symptoms seen in WS or Dup7. Hypertension is common in WS, but not all the risk can be attributed to deletion of ELN or copy number of the modifying NCF1.3 RGS2, which was DM and showed decreased expression in the WS group, has been linked to vascular remodeling and hypertension, given that RGS2 levels correlate inversely with blood pressure.79 It is therefore possible that altered methylation and subsequent expression of RGS2 contribute to hypertension in WS. The promoter of HOX4A, which showed increased methylation only in the WS group, has been shown to become hypermethylated in hematopoietic cancers, particularly those arising from the B cell lineage.80 Several cases of B cell lymphoma have recently been reported in WS,81 and changes in the regulation of HOXA4 could be a contributing factor to their pathogenesis or prognosis. Early puberty is also seen in WS,3 and mutations in MKRN3, which is in the region associated with Prader-Willi and Angelman syndromes, have been associated with precocious puberty.82
In conclusion, we report evidence of genome-wide, symmetrical, and dose-dependent changes in DNA methylation in reciprocal deletion (WS) and duplication (Dup7) syndromes. The DM genes were enriched with those involved in neurological development, function, and disease, even though the analysis was performed on whole-blood DNA. Our data suggest that altered DNA methylation might affect neurodevelopment in WS and Dup7 and contribute to their distinct phenotypic expression, particularly features of ASD. Identification of the specific 7q11.23 genes in which mutations cause altered DNA methylation will improve our understanding of the relationship and dynamics between chromatin remodeling and DNA methylation not only in relation to the 7q11.23 region but also throughout the genome. Further analyses in individuals with WS and individuals with Dup7 could reveal identifiable epigenomic signatures that will give insight into the pathobiology of these disorders (and perhaps similar neurodevelopmental disorders) and could lead to improvements in prognosis and treatments.
Acknowledgments
We thank Elaine Tam for technical assistance with collecting blood samples, Youliang Lou for performing the pyrosequencing assays, and The Centre for Applied Genomics for providing DNA samples from typically developing children. E.S. was supported by an Autism Speaks Weatherstone Predoctoral Fellowship (9093). This work was funded by grants from the Canadian Institutes for Health Research (MOP77720) to L.R.O., from the Simons Foundation Autism Research Initiative (238896) to C.B.M and L.R.O., and from the National Institute of Neurological Disorders and Stroke (R01 NS35102) and the National Institute of Child Health and Development (R37 HD29957) to C.B.M.
Published: July 9, 2015
Footnotes
Supplemental Data include 10 figures and 12 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.05.019.
Accession Numbers
The Gene Expression Omnibus (GEO) accession number for the raw and normalized Illumina 450K methylation data reported in this paper is GEO: GSE66552.
Web Resources
The URLs for data presented herein are as follows:
SFARI Gene, https://gene.sfari.org/autdb/
Geneimprint, http://www.geneimprint.com/
JASPAR CORE, http://jaspar.genereg.net
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Ewart A.K., Morris C.A., Atkinson D., Jin W., Sternes K., Spallone P., Stock A.D., Leppert M., Keating M.T. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat. Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 2.Somerville M.J., Mervis C.B., Young E.J., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Pérez-Jurado L.A. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris C.A. Williams syndrome. In: Pagon R.A., Adam M.P., Ardinger H.H., Bird T.D., Dolan C.R., Fong C.T., Smith R.J.H., Stephens K., editors. GeneReviews(R) University of Washington; 2013. [Google Scholar]
- 4.Mervis C.B., Velleman S.L. Children with Williams syndrome: Language, Cognitive, and Behavioral Characteristics and Their Implications for Intervention. Perspect. Lang. Learn. Educ. 2011;18:98–107. doi: 10.1044/lle18.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leyfer O., Woodruff-Borden J., Mervis C.B. Anxiety disorders in children with williams syndrome, their mothers, and their siblings: implications for the etiology of anxiety disorders. J. Neurodev. Disord. 2009;1:4–14. doi: 10.1007/s11689-009-9003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mervis C.B., Klein-Tasman B.P., Huffman M.J., Velleman S.L., Pitts C.H., Henderson D.R., Woodruff-Borden J., Morris C.A., Osborne L.R. Children with 7q11.23 duplication syndrome: Psychological characteristics. Am. J. Med. Genet. A. 2015;167:1436–1450. doi: 10.1002/ajmg.a.37071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg J.S., Brunetti-Pierri N., Peters S.U., Kang S.H., Fong C.T., Salamone J., Freedenberg D., Hannig V.L., Prock L.A., Miller D.T. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet. Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- 8.Van der Aa N., Rooms L., Vandeweyer G., van den Ende J., Reyniers E., Fichera M., Romano C., Delle Chiaie B., Mortier G., Menten B. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur. J. Med. Genet. 2009;52:94–100. doi: 10.1016/j.ejmg.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Devlin B., Scherer S.W. Genetic architecture in autism spectrum disorder. Curr. Opin. Genet. Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulle J.G., Pulver A.E., McGrath J.A., Wolyniec P.S., Dodd A.F., Cutler D.J., Sebat J., Malhotra D., Nestadt G., Conrad D.F., Molecular Genetics of Schizophrenia Consortium Reciprocal duplication of the Williams-Beuren syndrome deletion on chromosome 7q11.23 is associated with schizophrenia. Biol. Psychiatry. 2014;75:371–377. doi: 10.1016/j.biopsych.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asada K., Itakura S. Social phenotypes of autism spectrum disorders and williams syndrome: similarities and differences. Front. Psychol. 2012;3:247. doi: 10.3389/fpsyg.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein-Tasman B.P., Mervis C.B., Lord C., Phillips K.D. Socio-communicative deficits in young children with Williams syndrome: performance on the Autism Diagnostic Observation Schedule. Child Neuropsychol. 2007;13:444–467. doi: 10.1080/09297040601033680. [DOI] [PubMed] [Google Scholar]
- 14.Klein-Tasman B.P., Phillips K.D., Lord C., Mervis C.B., Gallo F.J. Overlap with the autism spectrum in young children with Williams syndrome. J. Dev. Behav. Pediatr. 2009;30:289–299. doi: 10.1097/DBP.0b013e3181ad1f9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tordjman S., Anderson G.M., Botbol M., Toutain A., Sarda P., Carlier M., Saugier-Veber P., Baumann C., Cohen D., Lagneaux C. Autistic disorder in patients with Williams-Beuren syndrome: a reconsideration of the Williams-Beuren syndrome phenotype. PLoS ONE. 2012;7:e30778. doi: 10.1371/journal.pone.0030778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tordjman S., Anderson G.M., Cohen D., Kermarrec S., Carlier M., Touitou Y., Saugier-Veber P., Lagneaux C., Chevreuil C., Verloes A. Presence of autism, hyperserotonemia, and severe expressive language impairment in Williams-Beuren syndrome. Mol. Autism. 2013;4:29. doi: 10.1186/2040-2392-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lincoln A.J., Searcy Y.M., Jones W., Lord C. Social interaction behaviors discriminate young children with autism and Williams syndrome. J. Am. Acad. Child Adolesc. Psychiatry. 2007;46:323–331. doi: 10.1097/chi.0b013e31802b9522. [DOI] [PubMed] [Google Scholar]
- 18.Gapp K., Woldemichael B.T., Bohacek J., Mansuy I.M. Epigenetic regulation in neurodevelopment and neurodegenerative diseases. Neuroscience. 2014;264:99–111. doi: 10.1016/j.neuroscience.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 19.Jangani M., Poolman T.M., Matthews L., Yang N., Farrow S.N., Berry A., Hanley N., Williamson A.J., Whetton A.D., Donn R., Ray D.W. The methyltransferase WBSCR22/Merm1 enhances glucocorticoid receptor function and is regulated in lung inflammation and cancer. J. Biol. Chem. 2014;289:8931–8946. doi: 10.1074/jbc.M113.540906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schosserer M., Minois N., Angerer T.B., Amring M., Dellago H., Harreither E., Calle-Perez A., Pircher A., Gerstl M.P., Pfeifenberger S. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015;6:6158. doi: 10.1038/ncomms7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Culver-Cochran A.E., Chadwick B.P. Loss of WSTF results in spontaneous fluctuations of heterochromatin formation and resolution, combined with substantial changes to gene expression. BMC Genomics. 2013;14:740. doi: 10.1186/1471-2164-14-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Middeljans E., Wan X., Jansen P.W., Sharma V., Stunnenberg H.G., Logie C. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS ONE. 2012;7:e33834. doi: 10.1371/journal.pone.0033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tussié-Luna M.I., Bayarsaihan D., Seto E., Ruddle F.H., Roy A.L. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc. Natl. Acad. Sci. USA. 2002;99:12807–12812. doi: 10.1073/pnas.192464499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crusselle-Davis V.J., Zhou Z., Anantharaman A., Moghimi B., Dodev T., Huang S., Bungert J. Recruitment of coregulator complexes to the beta-globin gene locus by TFII-I and upstream stimulatory factor. FEBS J. 2007;274:6065–6073. doi: 10.1111/j.1742-4658.2007.06128.x. [DOI] [PubMed] [Google Scholar]
- 25.Hakimi M.A., Dong Y., Lane W.S., Speicher D.W., Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 26.Ladd-Acosta C., Hansen K.D., Briem E., Fallin M.D., Kaufmann W.E., Feinberg A.P. Common DNA methylation alterations in multiple brain regions in autism. Mol. Psychiatry. 2014;19:862–871. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Numata S., Ye T., Herman M., Lipska B.K. DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front. Genet. 2014;5:280. doi: 10.3389/fgene.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pidsley R., Y Wong C.C., Volta M., Lunnon K., Mill J., Schalkwyk L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., Yan L., Hu Q., Sucheston L.E., Higgins M.J., Ambrosone C.B., Johnson C.S., Smiraglia D.J., Liu S. IMA: an R package for high-throughput analysis of Illumina’s 450K Infinium methylation data. Bioinformatics. 2012;28:729–730. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters, T., and Buckley, M. (2014). DMRcate: Illumina 450K methylation array spatial analysis methods. R package version 1.2.0, http://www.bioconductor.org/packages/release/bioc/html/DMRcate.html.
- 31.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 34.Tost J., Gut I.G. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 35.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLeay R.C., Bailey T.L. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jäger D., Stockert E., Güre A.O., Scanlan M.J., Karbach J., Jäger E., Knuth A., Old L.J., Chen Y.T. Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res. 2001;61:2055–2061. [PubMed] [Google Scholar]
- 40.Bonnefont J., Nikolaev S.I., Perrier A.L., Guo S., Cartier L., Sorce S., Laforge T., Aubry L., Khaitovich P., Peschanski M. Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. Am. J. Hum. Genet. 2008;83:208–218. doi: 10.1016/j.ajhg.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thu C.A., Chen W.V., Rubinstein R., Chevee M., Wolcott H.N., Felsovalyi K.O., Tapia J.C., Shapiro L., Honig B., Maniatis T. Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smoller J.W., Paulus M.P., Fagerness J.A., Purcell S., Yamaki L.H., Hirshfeld-Becker D., Biederman J., Rosenbaum J.F., Gelernter J., Stein M.B. Influence of RGS2 on anxiety-related temperament, personality, and brain function. Arch. Gen. Psychiatry. 2008;65:298–308. doi: 10.1001/archgenpsychiatry.2007.48. [DOI] [PubMed] [Google Scholar]
- 43.Otowa T., Shimada T., Kawamura Y., Sugaya N., Yoshida E., Inoue K., Yasuda S., Liu X., Minato T., Tochigi M. Association of RGS2 variants with panic disorder in a Japanese population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2011;156B:430–434. doi: 10.1002/ajmg.b.31178. [DOI] [PubMed] [Google Scholar]
- 44.Campbell D.B., Lange L.A., Skelly T., Lieberman J., Levitt P., Sullivan P.F. Association of RGS2 and RGS5 variants with schizophrenia symptom severity. Schizophr. Res. 2008;101:67–75. doi: 10.1016/j.schres.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto D., Delaby E., Merico D., Barbosa M., Merikangas A., Klei L., Thiruvahindrapuram B., Xu X., Ziman R., Wang Z. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarting G.A., Raitcheva D., Bless E.P., Ackerman S.L., Tobet S. Netrin 1-mediated chemoattraction regulates the migratory pathway of LHRH neurons. Eur. J. Neurosci. 2004;19:11–20. doi: 10.1111/j.1460-9568.2004.03094.x. [DOI] [PubMed] [Google Scholar]
- 47.Parvanov E.D., Petkov P.M., Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz J.J., Roach D.J., Thomas J.H., Shendure J. Primate evolution of the recombination regulator PRDM9. Nat. Commun. 2014;5:4370. doi: 10.1038/ncomms5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabinovitz S., Kaufman Y., Ludwig G., Razin A., Shemer R. Mechanisms of activation of the paternally expressed genes by the Prader-Willi imprinting center in the Prader-Willi/Angelman syndromes domains. Proc. Natl. Acad. Sci. USA. 2012;109:7403–7408. doi: 10.1073/pnas.1116661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hannula-Jouppi K., Muurinen M., Lipsanen-Nyman M., Reinius L.E., Ezer S., Greco D., Kere J. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. Epigenetics. 2014;9:351–365. doi: 10.4161/epi.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 53.Coskun V., Tsoa R., Sun Y.E. Epigenetic regulation of stem cells differentiating along the neural lineage. Curr. Opin. Neurobiol. 2012;22:762–767. doi: 10.1016/j.conb.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Veyrac A., Besnard A., Caboche J., Davis S., Laroche S. The transcription factor Zif268/Egr1, brain plasticity, and memory. Prog. Mol. Biol. Transl. Sci. 2014;122:89–129. doi: 10.1016/B978-0-12-420170-5.00004-0. [DOI] [PubMed] [Google Scholar]
- 56.O’Donovan K.J., Tourtellotte W.G., Millbrandt J., Baraban J.M. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 57.Bacon C., Rappold G.A. The distinct and overlapping phenotypic spectra of FOXP1 and FOXP2 in cognitive disorders. Hum. Genet. 2012;131:1687–1698. doi: 10.1007/s00439-012-1193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segura-Puimedon M., Sahún I., Velot E., Dubus P., Borralleras C., Rodrigues A.J., Valero M.C., Valverde O., Sousa N., Herault Y. Heterozygous deletion of the Williams-Beuren syndrome critical interval in mice recapitulates most features of the human disorder. Hum. Mol. Genet. 2014;23:6481–6494. doi: 10.1093/hmg/ddu368. [DOI] [PubMed] [Google Scholar]
- 59.Lazebnik M.B., Tussie-Luna M.I., Roy A.L. Determination and functional analysis of the consensus binding site for TFII-I family member BEN, implicated in Williams-Beuren syndrome. J. Biol. Chem. 2008;283:11078–11082. doi: 10.1074/jbc.C800049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cairo S., Merla G., Urbinati F., Ballabio A., Reymond A. WBSCR14, a gene mapping to the Williams--Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum. Mol. Genet. 2001;10:617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- 61.Adamo A., Atashpaz S., Germain P.L., Zanella M., D’Agostino G., Albertin V., Chenoweth J., Micale L., Fusco C., Unger C. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 62.Kaneko R., Kato H., Kawamura Y., Esumi S., Hirayama T., Hirabayashi T., Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 2006;281:30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 63.Golan-Mashiach M., Grunspan M., Emmanuel R., Gibbs-Bar L., Dikstein R., Shapiro E. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 2012;40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monahan K., Rudnick N.D., Kehayova P.D., Pauli F., Newberry K.M., Myers R.M., Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc. Natl. Acad. Sci. USA. 2012;109:9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dallosso A.R., Hancock A.L., Szemes M., Moorwood K., Chilukamarri L., Tsai H.H., Sarkar A., Barasch J., Vuononvirta R., Jones C. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5:e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anitha A., Thanseem I., Nakamura K., Yamada K., Iwayama Y., Toyota T., Iwata Y., Suzuki K., Sugiyama T., Tsujii M. Protocadherin α (PCDHA) as a novel susceptibility gene for autism. J. Psychiatry Neurosci. 2013;38:192–198. doi: 10.1503/jpn.120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimojima K., Isidor B., Le Caignec C., Kondo A., Sakata S., Ohno K., Yamamoto T. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am. J. Med. Genet. A. 2011;155A:732–736. doi: 10.1002/ajmg.a.33891. [DOI] [PubMed] [Google Scholar]
- 68.Pedrosa E., Stefanescu R., Margolis B., Petruolo O., Lo Y., Nolan K., Novak T., Stopkova P., Lachman H.M. Analysis of protocadherin alpha gene enhancer polymorphism in bipolar disorder and schizophrenia. Schizophr. Res. 2008;102:210–219. doi: 10.1016/j.schres.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cook E.H., Jr., Lindgren V., Leventhal B.L., Courchesne R., Lincoln A., Shulman C., Lord C., Courchesne E. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am. J. Hum. Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- 70.Krefft M., Frydecka D., Adamowski T., Misiak B. From Prader-Willi syndrome to psychosis: translating parent-of-origin effects into schizophrenia research. Epigenomics. 2014;6:677–688. doi: 10.2217/epi.14.52. [DOI] [PubMed] [Google Scholar]
- 71.Hogart A., Nagarajan R.P., Patzel K.A., Yasui D.H., Lasalle J.M. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum. Mol. Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adalsteinsson B.T., Ferguson-Smith A.C. Epigenetic control of the genome-lessons from genomic imprinting. Genes (Basel) 2014;5:635–655. doi: 10.3390/genes5030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pacaud R., Sery Q., Oliver L., Vallette F.M., Tost J., Cartron P.F. DNMT3L interacts with transcription factors to target DNMT3L/DNMT3B to specific DNA sequences: role of the DNMT3L/DNMT3B/p65-NFκB complex in the (de-)methylation of TRAF1. Biochimie. 2014;104:36–49. doi: 10.1016/j.biochi.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 74.Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 75.Sheikh M.A., Malik Y.S., Yu H., Lai M., Wang X., Zhu X. Epigenetic regulation of Dpp6 expression by Dnmt3b and its novel role in the inhibition of RA induced neuronal differentiation of P19 cells. PLoS ONE. 2013;8:e55826. doi: 10.1371/journal.pone.0055826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toyoda S., Kawaguchi M., Kobayashi T., Tarusawa E., Toyama T., Okano M., Oda M., Nakauchi H., Yoshimura Y., Sanbo M. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82:94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Makeyev A.V., Enkhmandakh B., Hong S.H., Joshi P., Shin D.G., Bayarsaihan D. Diversity and complexity in chromatin recognition by TFII-I transcription factors in pluripotent embryonic stem cells and embryonic tissues. PLoS ONE. 2012;7:e44443. doi: 10.1371/journal.pone.0044443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peña-Hernández R., Marques M., Hilmi K., Zhao T., Saad A., Alaoui-Jamali M.A., del Rincon S.V., Ashworth T., Roy A.L., Emerson B.M., Witcher M. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc. Natl. Acad. Sci. USA. 2015;112:E677–E686. doi: 10.1073/pnas.1416674112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsang S., Woo A.Y., Zhu W., Xiao R.P. Deregulation of RGS2 in cardiovascular diseases. Front. Biosci. (Schol. Ed.) 2010;2:547–557. doi: 10.2741/s84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strathdee G., Sim A., Parker A., Oscier D., Brown R. Promoter hypermethylation silences expression of the HoxA4 gene and correlates with IgVh mutational status in CLL. Leukemia. 2006;20:1326–1329. doi: 10.1038/sj.leu.2404254. [DOI] [PubMed] [Google Scholar]
- 81.Guenat D., Quentin S., Rizzari C., Lundin C., Coliva T., Edery P., Fryssira H., Bermont L., Ferrand C., Soulier J. Constitutional and somatic deletions of the Williams-Beuren syndrome critical region in non-Hodgkin lymphoma. J. Hematol. Oncol. 2014;7:82. doi: 10.1186/s13045-014-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abreu A.P., Dauber A., Macedo D.B., Noel S.D., Brito V.N., Gill J.C., Cukier P., Thompson I.R., Navarro V.M., Gagliardi P.C. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N. Engl. J. Med. 2013;368:2467–2475. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.