Abstract
The soluble epoxide hydrolase (sEH) is a potential pharmacological target for treating hypertension, vascular inflammation, pain, cancer and other diseases. However, there is not a simple, inexpensive and reliable method to estimate levels of active sEH in tissues. Toward developing such an assay, a polyclonal-variable domain of heavy chain antibody (VHH) sandwich immunoassay was developed. Ten VHHs, which are highly selective for native human sEH, were isolated from a phage displayed library. The ten VHHs have no significant cross-reactivity with human microsomal epoxide hydrolase, rat and mouse sEH, and denatured human sEH. There is a high correlation between protein levels of the sEH determined by the ELISA and the catalytic activity of the enzyme in S9 fractions of human tissues (liver, kidney and lung). The VHH based ELISA appears to be a new reliable method for monitoring the sEH, and may be useful as a diagnostic tool for diseases influenced by sEH. This study also demonstrates the broad utility of VHH in biochemical and pharmacological research.
Keywords: Soluble epoxide hydrolase, sEH, ELISA, magnetic beads, VHH
1. Introduction
The soluble epoxide hydrolase (sEH, EC 3.3.2.10) is a member of the α/β-hydrolase fold family of enzymes and catalyzes the hydrolysis of an epoxide to its corresponding diol through the catalytic addition of a water molecule[1]. While highly expressed in the liver and kidney, sEH is also expressed in other tissues including vascular endothelium, leukocytes, red blood cells, smooth muscle cells, adipocytes, and the proximal tubule [2-4]. The sEH protein is a homo-dimer with a monomeric unit of 62.5 kDa [5]. Many recent reports have shown that treatment with potent inhibitors of sEH reduces hypertension and inflammatory responses in in vitro and in vivo experimental models, suggesting that human sEH is a promising pharmacological target for treatment of cardiovascular and other diseases [6-12]. Thus, measuring sEH levels in tissues may represent an important diagnostic tool.
A variety of activity assays are available for sEH that use liquid chromatography (LC), gas chromatography (GC), liquid chromatography mass spectrometric (LC-MS), radiometric, UV/VIS spectrophotometric or fluorometric detection [5,13,14]. Although these methods are sensitive and accurate, they have limitations. The UV/VIS spectrophotometric and fluorometric assays were designed as high throughput assays using pure enzyme; they have high background in crude homogenate. The radiometric assays require handling of radioactivity, and the best substrate is not commercially available. The LC, GC and LC-MS methods require a significant amount of sample preparation, expensive equipment, and are time consuming. So a rapid method for monitoring levels of the native sEH is still needed particularly in laboratories lacking specialized equipment.
Antibody-based immunoassays are the most commonly used type of diagnostic assay and still are one of the fastest growing technologies for the analysis of biomolecules[15]. Despite playing a very important role in diagnostic assays, monoclonal antibodies (mAb) still have some limitations. Short comings include its large size (approximately 150 kDa), relative instability and difficulty in manufacturing making them very expensive. The variable domain of heavy chain antibodies (VHHs) are small proteins (15-20 kDa) with similar specificities and affinities as mAbs. VHHs often recognize novel epitopes that are not readily accessible to mAbs because of the larger antigen-binding site of the latter [16-18]. In addition, VHHs display a high stability, high level of expression and can be easily tailored in diverse constructs, making them excellent tools in medical and biotechnological applications [19-24]. Thus, the objective of the work is to develop a VHH based ELISA for native human sEH.
In this paper, we isolated ten VHHs selective for native human sEH, and developed a polyclonal antibody-VHH sandwich enzyme-linked immunosorbent assay (ELISA). In addition, we used magnetic beads in the panning process of VHH isolation as a simple and efficient way to preserve the native structure and antigenicity of this relatively unstable protein.
2. Materials and methods
Materials
Bovine serum albumin (BSA), thyroglobulin, polyethylene glycol 8000 (PEG 8000), Tween 20, isopropyl-β-D-thiogalactopyranoside (IPT), 3, 3’, 5, 5’-tetramethylbenzidine (TMB), and (+)-biotin N-hydroxysuccinimide ester (NHS-biotin) were obtained from Sigma-Aldrich (St. Louis, MO). Helper phage M13KO7 was purchased from New England Biolabs (Ipswich, MA). Mouse anti-M13 phage mAb-horseradish peroxidase (HRP) was purchased from GE Healthcare (Piscataway, NJ). Anti-HA tag antibody-HRP was purchased from Abcam (Cambridge, MA). Chemically competent TOP10F’ cells were obtained from Invitrogen (Carlsbad, CA). The plasmid purification kit, gel purification kit, PCR purification kit, and 6xHis tag purification resins were obtained from Qiagen (Valencia, CA). Electrocompetent ER2738 E. coli cells were purchased from Lucigen Corporation (Middleton, WI). B-PER lysis solution, streptavidin magnetic beads and Zeba Spin desalting columns (MWCO 7k) were purchased from Thermo Pierce Scientific (Rockford, IL). SfiI was purchased from New England Biolabs (Ipswich, MA).
The human sEH [25], human microsomal epoxide hydrolase (mEH), mouse sEH, rat sEH, rabbit polyclonal anti-human sEH antibody and sEH null mice liver cytosol were prepared in this laboratory. The purified human sEH was boiled for 5 min to obtain the denatured human sEH.
Library construction
Blood was drawn from a three year old llama that had been immunized with 6 injections of human sEH (500 μg/dose) in incomplete Freund’s adjuvant. A VHH phage display library was built as previously described [26]. Briefly, about 108 lymphocytes were obtained from 150 mL of blood using Histopaque 1077 (Sigma) gradients. The RNA was extracted and retrotranscribed using the primer oligo (dT). The cDNA was then used as template for PCR amplification of the VH and VHH genes using the forward primers VH1, VH3, VH4 and the reverse primer JH (Table S-1 in Supporting Information) [27]. The amplified DNA was digested with SfiI and cloned into pComb3× a kind gift from Dr. Carlos Barbas (The Scripps Research Institute, La Jolla, USA). The ligated vector was electroporated into highly competent ER2738 E. coli cells. The cells were cultured and the phagemid-borne phage library displaying the VH/VHH repertoire was rescued by superinfection with helper phage M13KO7 (Pharmacia Biotech, Uppsala, Sweden).
Preparation of magnetic beads for VHH selection
A 50μL aliquot of NHS-biotin (32 μM) in phosphate buffer (PB buffer, 0.01 mol/L phosphate, pH 7.4) was added to 50μL human sEH (32 μM) in PB buffer and stirred at 4 °C for 4 h. After the reaction, the solution was purified using Zeba Spin desalting columns (molecular weight cutoff [MWCO], 7,000; Pierce) to remove excess NHS-biotin. The biotinylated human sEH (biotin-HsEH, 16 μM) was stored at 4 °C.
Streptavidin-coated magnetic beads (100 μL) were added to 1 mL PB buffer (containing 2% BSA; PBB), then mixed and centrifuged at 12,000 rpm for 30 seconds. The pellet was washed twice with 1 mL PBB buffer. The beads were resuspended in 100 μL of PBB buffer. The streptavidin magnetic beads (50 μL) were then mixed with the biotin-HsEH (50 μL) and incubated for 10-15 min at 4 °C. The mixture was centrifuged at 12,000 rpm for 30 seconds at 4 °C. The pellet was washed three times with 500 μL of PBB. The beads were resuspended in 50 μL PBB and stored at 4 °C.
Selection of phage anti-human sEH VHH clones
For each sample, one well of a microtiter plate (Nunc MaxiSorp) was coated with 300 μL of 2% BSA in PB buffer overnight at 4 °C. To avoid the selection of VHHs with affinity to streptavidin magnetic particles, the phage were pre-incubated with streptavidin magnetic beads in microtiter plate wells coated with BSA to remove the phages that bind to streptavidin magnetic particles and/or BSA. The phage VHH library (100 μL, 1013 c.f.u/mL) and prewashed beads (10 μL) were added into the well, and the plate was incubated for 2 h at 4 °C with gentle shaking. The plate was placed on the magnetic particle separator and allowed to stand for 15-30 seconds. The supernatant was transferred to a new 1.5 mL microfuge tube. For Method A, biotin-HsEH (10 μL, 16 μM) was added to the tube containing phage displaying VHH and incubated for 1 h at 4 °C. The prewashed streptavidin magnetic beads (10 μL) were added to the tube and incubated for 15 min at 4°C. For Method B, biotin-HsEH preincubated with streptavidin coated magnetic-beads (10 μL, 16 μM) were added to the tube containing phage displaying VHH and incubated for 1 h at 4 °C (see Electronic Supplementary Material (ESM) Fig. S1). The tube was then placed on the magnetic particle separator for 15-30 seconds. The supernatant was removed. The pellet was washed with 300 μL of PBB five times.
The bound phages were eluted by addition of 100 μL of glycine-HCl buffer (pH 2.2) with shaking for 10 min at room temperature. The tube was placed on the magnetic particle separator for 15-30 seconds. The supernatant was transferred to a new tube containing 2.5 μL of Tris base buffer (1 M). ER2738 E. coli were infected with the eluted phages and titered on LB-ampicilin (amp) agar plates (recorded as “output” titers), and 100 μL of eluted phages were used for reamplification. The reamplified phages were titered again on LB-amp agar plates (recorded as “input” titers), and 100 μL of the amplified phages was employed again in the next round of panning. For the second, third, and fourth rounds, the same procedure was used, except the concentrations of the human sEH were gradually decreased. After the final round of panning, individual clones were screened to identify positive clones by performing a phage ELISA, BSA and skim milk were used as negative controls.
Expression and purification of anti-human sEH VHHs
From the agar plate containing the fourth elution output titer, 16 individual clones from Method A and Method B were randomly selected and grown individually in cultures overnight. Duplicates of 2 mL cultures were prepared for each clone: one for plasmid extraction and another for VHH–pIII protein extraction. Both cultures for each clone were spun down at 3000 g for 10 min. For the plasmid extracted cultures, the Qiagen Mini Prep kit was employed and the sequences were submitted to the UC Davis DNA Sequencing Facility. For the protein extraction, the bacterial protein extraction reagent kit (BPER) was employed and the obtained protein was further characterized by ELISA.
The cloned plasmids pComb3×, containing the anti-human sEH VHHs, were transformed by heat shock into Top 10F’ cells. The expression and purification of VHHs was the same as previously described [28,29].
Development of sandwich ELISA method for human sEH
The method used was previously reported [28]. Briefly, each well of a microtiter plate was coated with 100 μL of rabbit polyclonal anti-human sEH antibody [30] (4.5 μg/mL) in PB saline (PBS, 0.01 mol/L phosphate, 0.137 mol/L NaCl, 3 mmol/L KCl, pH 7.4) at 4 °C overnight, and then blocked with 3% skim milk. The plate was washed three times with PBS–Twee (PBST, 0.05% Tween) and human sEH diluted serially (50 μL/well, in PBS) was added. That was followed by the addition of VHH (50 μL, 2 μg/mL) incubated at room temperature for 1 h. The plate was washed again 100 μL of anti-HA tag antibody-HRP (diluted at 1:5000 with PBS) was added and incubated another 1 h at room temperature followed by another wash step. One hundred microliters of TMB solution (400 μL of 0.6% TMB and 100 μL of 1% H2O2 in 25 mL citrate buffer, pH 5.5) was added and incubated for 15 min at room temperature. Finally, the reaction was stopped by addition of 50 μL of 2 M H2SO4, and the plate was read on an ELISA plate reader at 450 nm (Molecular Devices, Sunnyvale, CA). The description of the color formation and chemistry of the reaction is explained in ESM Fig. S2.
Cross-Reactivity of VHH
The selectivity of the VHH ELISA was evaluated by determining the cross-reactivity with human mEH, rat sEH, mouse sEH, and denatured human sEH. A 100 μL solution of rabbit polyclonal anti-human sEH antibody in PBS (4.5 μg/mL) was coated on a 96-well microtiter plate at 4 °C overnight. After washing and blocking, 50 μL of human mEH (5 μg/mL), rat sEH (3 μg/mL), mouse sEH (6 μg/mL), denatured human sEH (3 μg/mL) or native human sEH (5 μg/mL) in PBS were added, followed by the addition of 50 μL of VHH (2μg/mL) in PBS. The anti-HA tag-HRP was added, and the development of the color was the same as described above. All analyses were repeated 3 times.
Matrix effects
The sEH null mice liver cytosol [31] was used to evaluate the matrix effect in a tissue sample. The cytosol was spiked with 100, 200, 300 and 500 ng/mL native and denatured human sEH, respectively.
Our laboratory studies the impact of inhibition of sEH by chemical modulators. We wanted to determine if the VHH could detect the interaction of sEH inhibitor with the native protein. To the native human sEH (100, 200, 300 and 500 ng/mL in PBS buffer) was added sEH inhibitor 1728 (1 μM) [32,33]. VHH B3 was used to analyze the samples. All the samples were directly analyzed without dilution.
Human sample analysis
VHH B3 was selected to analyze the human samples. The sandwich ELISA was performed as described above. S9 fractions of pooled (4–50 persons) human tissue samples (Xenotech LLC, Lenexa, KS) were diluted in PBS buffer to the appropriate concentration.
Effect of VHHs on human sEH
Human sEH has two distinct enzyme activities epoxide hydrolase (Cterm-EH) and phosphatase (Nterm-phos). The effect of VHHs on epoxide hydrolase activity and phosphatase activity in vitro were evaluated by using the method described previously [1,34]. For the epoxide hydrolase activity (cyano-(2-methoxynaphthalen-6-yl)-methyl trans-(3-phenyl-oxyran-2-yl)-methyl carbonate; CMNPC; 5 μM) was used as the substrate, the concentration of human sEH was 1 nM. For the phosphatase activity, AttoPhos (25 μM) was used as the substrate, the concentration of human sEH was 2 nM, and each clone was evaluated for its ability to inhibit the epoxide hydrolase or phosphate catalytic activity, respectively.
RESULTS AND DISCUSSION
Selection of anti-human sEH phage-displayed VHHs
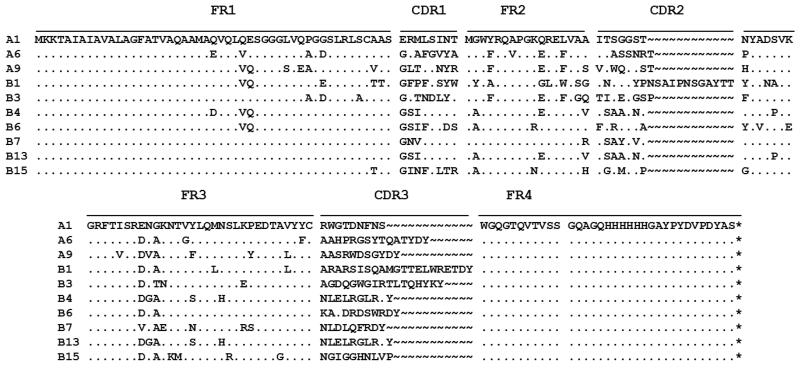
Bio-panning for proteins or small molecules from libraries of phage-displayed VHHs library is commonly done in 96-well microtiter plates [28,29,35,36]. The targeted antigen is generally immobilized on the plate by physical adsorption to select potent phage-displayed VHHs. It works very well for stable proteins and small analytes. However, 90% of monoclonal and 75% of polyclonal antibodies are denatured by physical adsorption [37,38]. To avoid the denaturation of the human sEH on a plastic plate by physical adsorption, the library was screened using biotinylated human sEH and streptavidin-coated magnetic beads. This method minimized the denaturation of the human sEH, and sufficiently exposed the epitopes of human sEH on magnetic beads in solution. The molar ratio of human sEH and NHS-biotin was 1:1. Compared to untreated enzyme, the human sEH did not lose any activity after biotinylation. Two methods were used in parallel for the selection of VHHs. In Method A, the free biotinylated human sEH was first mixed with the phage-displayed VHHs, then the streptavidin-coated magnetic beads were added to capture the sEH-VHH complexes. In Method B, the biotinylated human sEH was first mixed with streptavidin-coated magnetic beads, the mixture was then added to the phage-displayed VHHs. Overall, both methods performed similarly (Table 1). In total, three unique sequences (A1, A6 and A9) were obtained from Method A, and seven unique sequences (B1, B3, B4, B6, B7, B13 and B15) were obtained from Method B (Fig. 1). It is interesting that both methods yielded different sets of sequences. In theory, both approaches should yield similar results. Because of the large size of sEH (130 kDa), one would expect many possible epitopes and thus many VHHs with varying affinities for sEH in the library. However, neither method was able to select out all of them, suggesting that utilizing different screening approaches can be important.
Table 1. Summary of the panning conditions, input and output titer.
| Panning | Concentration of sEH (μM) |
Method A | Method B | ||
|---|---|---|---|---|---|
|
| |||||
| Input (pfu/ml) |
Output (pfu/ml) |
Input (pfu/ml) |
Output (pfu/ml) |
||
| 1st | 16 | 5×1013 | 9 ×109 | 5×1013 | 9 ×109 |
| 2nd | 16 | 3.6×1013 | 1 ×1010 | 7.5 ×1013 | 1.4×109 |
| 3rd | 8 | 1.6×1013 | 1 ×1010 | 1.4 ×1013 | 1 ×1010 |
| 4th | 4 | 4.7 ×1013 | 1 ×1010 | 2.8 ×1013 | 1 ×1010 |
Phage input and output number: Ten μL of the phage input or output was diluted in sterile LB medium from each panning cycle and mixed with 90 μL of E. coli ER2738 cells with OD600 = 0.5-1. The mixture was incubated for 15 min at 37 °C without shaking and then plated on a LB plate with carbenicillin (100 μg/mL) . The number of colonies was counted the next day.
Fig. 1.
Alignment of amino acid sequences of selected VHH phage clones. Sixteen clones from Method A are divided into three groups: A1, A6 and A9. Sixteen clones from Method B are divided into seven groups: B1, B3, B4, B6, B7, B13 and B15. The deduced amino acid sequences of the ten clones are given in the single-letter code. (~) Absence of amino acid residues; (.) residues identical to those of clones in A1.
Development of the sandwich ELISA method to quantify human sEH
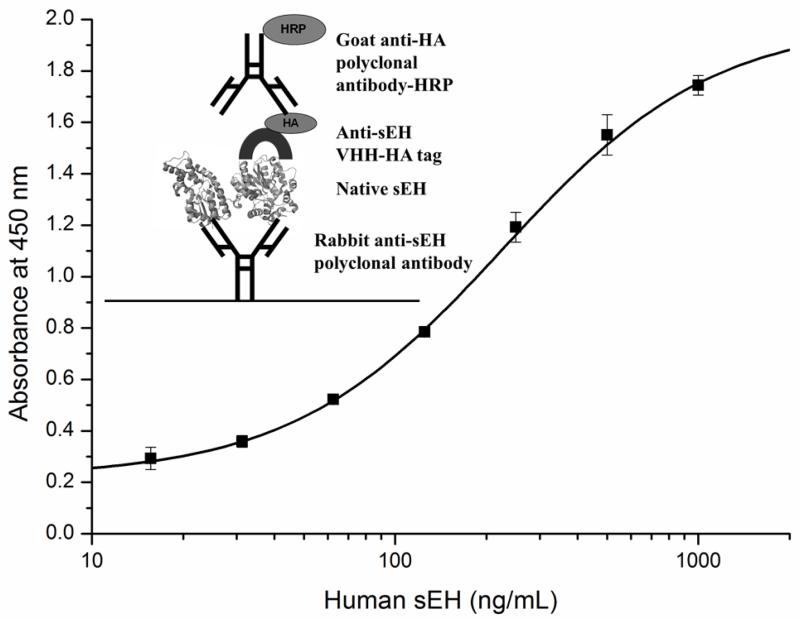
A sandwich ELISA requires two epitope-specific antibodies for the analyte, the capture antibody and the detection antibody. Since the polyclonal antibody detects both the native and the denatured sEH of multiple species (human, mouse, rat and horse), it was used as the capture antibody to detect the native human sEH. The VHH was used as the detection antibody. The format of the sandwich ELISA is shown in Fig. 2. Native human sEH was used in a standard curve with concentrations from 15.6 to 1,000 ng/mL. Among the 10 VHHs, B3 VHH was observed to detect the lowest amount of human sEH compared to the other clones (Table 2). Hence, it was used in further studies to analyze the human sEH in various samples. To our knowledge, there are no other antibodies for ELISA of native sEH. The existing polyclonal antibody is mostly used for western blot analysis, and was recently reported to have a limit of detection around 10 ng of purified human sEH loaded into a gel [39]. With a theoretical loading volume of 50 μL, and a detection limit of 15.6 ng/mL, our developed sandwich ELISA should be able to detect down to 0.8 ng of human sEH, suggesting a greater sensitivity than available polyclonal antibodies against the human sEH.
Fig. 2.
The sandwich ELISA format and calibration curve for human sEH with rabbit polyclonal anti-human sEH antibody (4.5 μg/mL) as capture antibody, VHH B3 (2 μg/mL) as detector antibody and anti-HA tag-HRP (1:5000) as secondary antibody. Error bars are standard deviations of the mean of three well replicates.
Table 2. Performance of 10 VHHs in the sandwich ELISA assay.
| VHH clones | SC50* (ng/mL) |
Linear range (ng/mL) |
VHH clones | SC50* (ng/mL) |
Linear range (ng/mL) |
|---|---|---|---|---|---|
| A1 | 246 | 86-700 | B4 | 373 | 123-1121 |
| A6 | 515 | 151-1750 | B6 | 1481 | 292-7514 |
| A9 | 268 | 104-688 | B7 | 356 | 113-1127 |
| B1 | 341 | 108-1084 | B13 | 463 | 141-1515 |
| B3 | 225 | 74-684 | B15 | 179 | 100-321 |
Sandwich ELISA conditions: Rabbit polyclonal anti-human sEH antibody (4.5 μg/mL) as capture antibody, VHHs (2 μg/mL) as detector antibody and anti-HA tag-HRP (1:5000) as secondary antibody. Error bars are standard deviations of the mean of three well replicates.
The 50% saturation of the signal (SC50).
Cross-reactivity
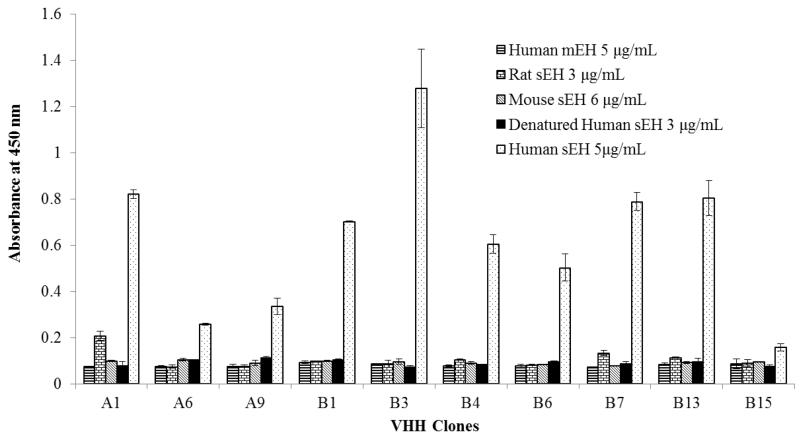
The resulting sandwich ELISA was tested against human mEH, rat sEH, mouse sEH, and denatured human sEH (Fig. 3). None of the VHHs showed cross-reactivity with human mEH, mouse sEH and denatured human sEH, even at high concentrations. The VHH A1 and B7 displayed a small amount of cross-reactivity with rat sEH at 3 μg/mL. The cross-reactivity result indicated the specificity of the VHHs in the sandwich ELISA for native human sEH.
Fig. 3.
Cross-reactivity of VHH clones with human mEH, rat sEH, mouse sEH, and denatured human sEH. Rabbit polyclonal anti-human sEH antibody (4.5 μg/mL) as capture antibody, VHH A1, A6, A9, B1, B3, B4, B6, B7, B13 and B15 (2 μg/mL) as detector antibody and anti-HA tag-HRP (1:5000) as secondary antibody. Error bars are standard deviations of the mean of three well replicates.
Matrix effect
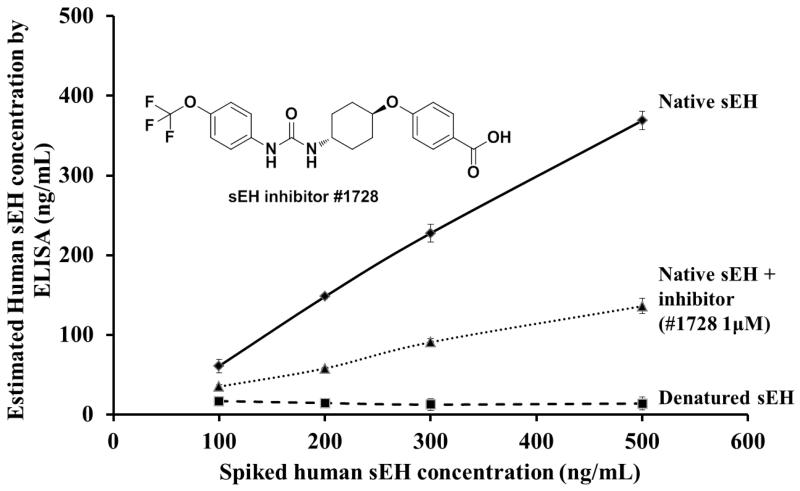
Matrix effects, inevitable in sample analysis, may affect the performance of the assay or cause some false positive results. To study matrix effects, sEH null mice liver cytosol was spiked with native and denatured human sEH. The cytosol samples were directly analyzed without dilution. The developed sandwich ELISA accurately detected native sEH and did not detect the denatured sEH (Fig. 4), similarly to the cross-reactivity study (Fig. 3). The results suggest that the developed sandwich ELISA is a robust and convenient method to detect native human sEH in tissue extracts with no or little matrix effect.
Fig. 4.
Estimated concentration of human sEH in spiked samples. The native and denatured human sEH (100, 200, 300 and 500 ng/mL) were spiked into sEH null mice liver cytosol. The native human sEH (100, 200, 300 and 500 ng/mL) with 1 μM 1728 inhibitor was spiked into PBS. Each sample was detected three times.
The effect of sEH inhibitor on the assay was also evaluated. When the native human sEH in PBS buffer was added to sEH inhibitor (1728, 1 μM), the sEH concentration detected by the sandwich ELISA was lower than without inhibitor (Fig. 4). This suggested that the inhibitor influenced the interaction between antibody and native human sEH. The development method can distinguish native from native inhibited sEH enzyme. Since the VHHs do not recognize denatured and inhibited enzyme, it suggests that they are recognizing the 3-dimentional folding of the enzyme. In this study, we used the native human sEH as the immunogen to generate and screen the library. None of the VHH generated recognized the denatured enzyme, only the VHHs against the native human sEH were identified. However, if one uses denatured sEH as immunogen or perhaps as the selection protein along with the native form, other VHHs that recognize the primary structure of the human sEH might be found.
Analysis of tissue samples
To study the utility of the sandwich ELISA, the concentration of sEH was determined with the sandwich ELISA in human tissue S9 samples (liver, kidney, and lung) (Table 3), and compared with the enzyme activity and Western blot method [39]. There are no significant variations between the enzyme activity and sandwich ELISA method. The Western blot method showed a higher concentration of human sEH than the other methods, because it could detect both denatured and native sEH in the samples. The good correlations between enzyme activity and the sandwich ELISA indicated the utility of the developed method.
Table 3. The concentration of sEH in the S9 fraction of pooled (4–50 persons) human tissues.
| Human tissues |
sEH (nM) |
||
|---|---|---|---|
| estimated from activity# Mean ± SD |
estimated from Western-blot# Mean ± SD |
estimated from ELISA Mean ± SD |
|
| Liver | 420 ± 89 | 500± 150 | 386 ± 48 |
| Kidney | 44 ± 3 | 80 ± 15 | 44.4 ± 1.6 |
| Lung (N.S.*) | 3.0 ± 0.3 | 22 ± 6 | 6.1 ± 0.3 |
| Lung (S.*) | 2.8 ± 0.3 | 21 ± 4 | 5.9 ± 0.1 |
Concentrations estimated from the specific activity using [3H] t -DPPO as substrate ([S]final = 50 μM), by Western blot and sandwich ELISA using recombinant purified human sEH as a standard. Results are average ± SD (n = 3).
N.S. nonsmoker; S. smoker.
Data from reference [39].
Effect of VHHs on human sEH
The sEH protein in mammals is a 125-kD dimer composed of two identical 62-kDa monomers arranged in an antiparallel fashion [40-42]. The N- and C-terminal regions of sEH are separated by a proline-rich linker [43]. The C-terminal contains the EH activity, and the smaller N-terminal is a phosphatase (EC 3.1.3.76) [44]. In addition, it was shown that only the sEH dimer is active [45]. Inhibition of sEH activity by either pharmacological tools or gene deletion has been shown to be protective in multiple models of cardiovascular diseases, thus identifying sEH as a valuable therapeutic target [46,47].
The effect of VHHs on the EH activity and phosphatase activity have been studied using the CMNPC and AttoPhos as substrates, respectively (Table 4). Clone A1 decreased the EH activity. Clone B3 increased the EH activity. Clone A6 decreased the phosphatase activity. Clone B13 and B15 increased the phosphatase activity. The other VHHs had little effect on either the EH or phosphatase activity. None of the clones appeared to have a significant effect on catalytic activity large enough to be of biochemical utility, but they show the possibility of allosteric regulation of both catalytic activities of the sEH. It may be useful to study pathological changes of human sEH in various human diseases.
Table 4. Effect VHH on human sEH enzymatic activities.
| VHH clones | Concentration of VHH (μM) |
Remaining activity |
|
|---|---|---|---|
| EH activity (%) |
Phosphatase activity (%) |
||
| A1 | 0.71 | 66.4 ± 1.1 | 103 ± 4 |
| A6 | 0.44 | 87.6 ± 2.7 | 66.9 ± 3.0 |
| A9 | 0.75 | 94.7 ± 1.6 | 94.4 ± 7.3 |
| B1 | 1.02 | 93.6 ± 2.8 | 107 ± 1 |
| B3 | 1.11 | 124 ± 6 | 108 ± 3 |
| B4 | 0.49 | 95.4 ± 2.2 | 103 ± 5 |
| B6 | 0.51 | 96.6 ± 2.8 | 96.0 ± 5.1 |
| B7 | 2.35 | 83.2 ± 2.3 | 111 ± 8 |
| B13 | 1.76 | 85.4 ± 3.9 | 146 ±7 |
| B15 | 1.89 | 81.5 ± 3.6 | 133 ± 10 |
EH activity: Human sEH= 1nM, CMNPC as substrate=5 μM
Phosphatase activity: Human sEH= 2 nM, AttoPhos as substrate [S]=25 μM
Results are the average and standard deviation of triplicate determinations.
Conclusions
In the present study, ten VHHs highly selective for the native human sEH were obtained using magnetic beads for selection. The use of magnetic beads offers an efficient method for panning the phage-displayed VHHs to obtain antibodies for the native protein. A polyclonal-VHH based sandwich ELISA has been developed for native human sEH. There is a high correlation between protein levels of the sEH determined by the ELISA and the catalytic activity of the enzyme in S9 fractions of human tissues (liver, kidney and lung). The VHH based ELISA appears to be a new reliable method for monitoring the sEH, and may be used as a diagnostic tool for diseases influenced by sEH. In addition, the VHHs are a potential tool to study the structure and function of human sEH.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) Superfund Research Program grant P42 ES04699, NIEHS R01 ES02710, FCE 6812 ANII (Agencia Nacional de Investigación e Innovación, Uruguay) and TW05718 Fogarty Center NHI. MR is recipients of scholarships from ANII and CSIC, Uruguay.
References
- 1.Jones PD, Wolf NM, Morisseau C, Whetstone P, Hock B, Hammock BD. Fluorescent substrates for soluble epoxide hydrolase and application to inhibition studies. Analytical biochemistry. 2005;343(1):66–75. doi: 10.1016/j.ab.2005.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman JW, Morisseau C, Hammock BD. Epoxide hydrolases: their roles and interactions with lipid metabolism. Progress in lipid research. 2005;44(1):1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Morisseau C, Hammock BD. Gerry Brooks and epoxide hydrolases: four decades to a pharmaceutical. Pest management science. 2008;64(6):594–609. doi: 10.1002/ps.1583. [DOI] [PubMed] [Google Scholar]
- 4.Taeye BM, Morisseau C, Coyle J, Covington JW, Luria A, Yang J, Murphy SB, Friedman DB, Hammock BB, Vaughan DE. Expression and regulation of soluble epoxide hydrolase in adipose tissue. Obesity. 2010;18(3):489–498. doi: 10.1038/oby.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez GA, Morisseau C, Hammock BD, Christianson DW. Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis. Biochemistry. 2004;43(16):4716–4723. doi: 10.1021/bi036189j. [DOI] [PubMed] [Google Scholar]
- 6.Imig JD, Zhao X, Capdevila JH, Morisseau C, Hammock BD. Soluble epoxide hydrolase inhibition lowers arterial blood pressure in angiotensin II hypertension. Hypertension. 2002;39(2):690–694. doi: 10.1161/hy0202.103788. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X, Yamamoto T, Newman JW, Kim I-H, Watanabe T, Hammock BD, Stewart J, Pollock JS, Pollock DM, Imig JD. Soluble epoxide hydrolase inhibition protects the kidney from hypertension-induced damage. Journal of the American Society of Nephrology. 2004;15(5):1244–1253. [PubMed] [Google Scholar]
- 8.Jung O, Brandes RP, Kim I-H, Schweda F, Schmidt R, Hammock BD, Busse R, Fleming I. Soluble epoxide hydrolase is a main effector of angiotensin II–induced hypertension. Hypertension. 2005;45(4):759–765. doi: 10.1161/01.HYP.0000153792.29478.1d. [DOI] [PubMed] [Google Scholar]
- 9.Schmelzer KR, Kubala L, Newman JW, Kim I-H, Eiserich JP, Hammock BD. Soluble epoxide hydrolase is a therapeutic target for acute inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(28):9772–9777. doi: 10.1073/pnas.0503279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inceoglu B, Wagner K, Schebb NH, Morisseau C, Jinks SL, Ulu A, Hegedus C, Rose T, Brosnan R, Hammock BD. Analgesia mediated by soluble epoxide hydrolase inhibitors is dependent on cAMP. Proceedings of the National Academy of Sciences. 2011;108(12):5093–5097. doi: 10.1073/pnas.1101073108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D, Li N, He Y, Timofeyev V, Lu L, Tsai H-J, Kim I-H, Tuteja D, Mateo RKP, Singapuri A. Prevention and reversal of cardiac hypertrophy by soluble epoxide hydrolase inhibitors. Proceedings of the National Academy of Sciences. 2006;103(49):18733–18738. doi: 10.1073/pnas.0609158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodani SD, Hammock BD. The 2014 Bernard B. Brodie Award Lecture Epoxide Hydrolases: Drug Metabolism to Therapeutics for Chronic Pain. Drug Metabolism and Disposition. 2015 doi: 10.1124/dmd.115.063339. dmd. 115.063339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doderer K, Schmid RD. Fluorometric assay for determining epoxide hydrolase activity. Biotechnology letters. 2004;26(10):835–839. doi: 10.1023/b:bile.0000025887.36874.33. [DOI] [PubMed] [Google Scholar]
- 14.Bhatnagar T, Manoj KM, Baratti JC. A spectrophotometric method to assay epoxide hydrolase activity. Journal of biochemical and biophysical methods. 2001;50(1):1–13. doi: 10.1016/s0165-022x(01)00162-2. [DOI] [PubMed] [Google Scholar]
- 15.Borrebaeck CA. Antibodies in diagnostics–from immunoassays to protein chips. Immunology today. 2000;21(8):379–382. doi: 10.1016/s0167-5699(00)01683-2. [DOI] [PubMed] [Google Scholar]
- 16.Conrath KE, Lauwereys M, Wyns L, Muyldermans S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. Journal of Biological Chemistry. 2001;276(10):7346–7350. doi: 10.1074/jbc.M007734200. [DOI] [PubMed] [Google Scholar]
- 17.Stijlemans B, Conrath K, Cortez-Retamozo V, Van Xong H, Wyns L, Senter P, Revets H, De Baetselier P, Muyldermans S, Magez S. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies African trypanosomes as paradigm. Journal of Biological Chemistry. 2004;279(2):1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 18.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(12):4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frenken LG, van der Linden RH, Hermans PW, Bos JW, Ruuls RC, de Geus B, Verrips CT. Isolation of antigen specific llama V HH antibody fragments and their high level secretion by Saccharomyces cerevisiae. Journal of biotechnology. 2000;78(1):11–21. doi: 10.1016/s0168-1656(99)00228-x. [DOI] [PubMed] [Google Scholar]
- 20.Arbabi-Ghahroudi M, Tanha J, MacKenzie R. Prokaryotic expression of antibodies. Cancer and Metastasis Reviews. 2005;24(4):501–519. doi: 10.1007/s10555-005-6193-1. [DOI] [PubMed] [Google Scholar]
- 21.Ghahroudi MA, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS letters. 1997;414(3):521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 22.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, Muyldermans S, Wyns L, Matagne A. Single - domain antibody fragments with high conformational stability. Protein Science. 2002;11(3):500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyldermans S. Nanobodies: natural single-domain antibodies. Annual review of biochemistry. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 24.Saerens D, Ghassabeh GH, Muyldermans S. Single-domain antibodies as building blocks for novel therapeutics. Current opinion in pharmacology. 2008;8(5):600–608. doi: 10.1016/j.coph.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Przybyla-Zawislak BD, Srivastava PK, Vázquez-Matías J, Mohrenweiser HW, Maxwell JE, Hammock BD, Bradbury JA, Enayetallah AE, Zeldin DC, Grant DF. Polymorphisms in human soluble epoxide hydrolase. Molecular pharmacology. 2003;64(2):482–490. doi: 10.1124/mol.64.2.482. [DOI] [PubMed] [Google Scholar]
- 26.Tabares-da Rosa S, Rossotti M, Carleiza C, Carrión F, Pritsch O, Ahn KC, Last JA, Hammock BD, González-Sapienza G. Competitive selection from single domain antibody libraries allows isolation of high-affinity antihapten antibodies that are not favored in the llama immune response. Analytical chemistry. 2011;83(18):7213–7220. doi: 10.1021/ac201824z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarebski LM, Urrutia M, Goldbaum FA. Llama single domain antibodies as a tool for molecular mimicry. Journal of molecular biology. 2005;349(4):814–824. doi: 10.1016/j.jmb.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Bever CR, Majkova Z, Dechant JE, Yang J, Gee SJ, Xu T, Hammock BD. Heterologous antigen selection of camelid heavy chain single domain antibodies against tetrabromobisphenol A. Analytical chemistry. 2014;86(16):8296–8302. doi: 10.1021/ac5017437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bever CR, Majkova Z, Radhakrishnan R, Suni I, McCoy M, Wang Y, Dechant J, Gee S, Hammock BD. Development and Utilization of Camelid VHH Antibodies from Alpaca for 2, 2′, 4, 4′–Tetrabrominated Diphenyl Ether Detection. Analytical chemistry. 2014;86(15):7875–7882. doi: 10.1021/ac501807j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Z, Davis BB, Morisseau C, Hammock BD, Olson JL, Kroetz DL, Weiss RH. Vascular localization of soluble epoxide hydrolase in the human kidney. American Journal of Physiology-Renal Physiology. 2004;286(4):F720–F726. doi: 10.1152/ajprenal.00165.2003. [DOI] [PubMed] [Google Scholar]
- 31.Luria A, Morisseau C, Tsai H-J, Yang J, Inceoglu B, De Taeye B, Watkins SM, Wiest MM, German JB, Hammock BD. Alteration in plasma testosterone levels in male mice lacking soluble epoxide hydrolase. American Journal of Physiology-Endocrinology and Metabolism. 2009;297(2):E375–E383. doi: 10.1152/ajpendo.00131.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammock BD, Wagner K, Inceoglu B. The soluble epoxide hydrolase as a pharmaceutical target for pain management. Pain management. 2011;1(5):383–386. doi: 10.2217/pmt.11.47. [DOI] [PubMed] [Google Scholar]
- 33.Hwang SH, Wecksler AT, Zhang G, Morisseau C, Nguyen LV, Fu SH, Hammock BD. Synthesis and biological evaluation of sorafenib-and regorafenib-like sEH inhibitors. Bioorganic & medicinal chemistry letters. 2013;23(13):3732–3737. doi: 10.1016/j.bmcl.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morisseau C, Sahdeo S, Cortopassi G, Hammock BD. Development of an HTS assay for EPHX2 phosphatase activity and screening of nontargeted libraries. Analytical biochemistry. 2013;434(1):105–111. doi: 10.1016/j.ab.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Xu Y, Xiong Y-h, Tu Z, Li Y-p, He Z-y, Qiu Y-l, Fu J-h, Gee SJ, Hammock BD. VHH Phage-Based Competitive Real-Time Immuno-Polymerase Chain Reaction for Ultrasensitive Detection of Ochratoxin A in Cereal. Analytical chemistry. 2014;86(15):7471–7477. doi: 10.1021/ac501202d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P, Li G, Yan J, Hu Y, Zhang C, Liu X, Wan Y. Bactrian camel nanobody-based immunoassay for specific and sensitive detection of Cry1Fa toxin. Toxicon. 2014;92:186–192. doi: 10.1016/j.toxicon.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 37.Butler J, Ni L, Nessler R, Joshi K, Suter M, Rosenberg B, Chang J, Brown W, Cantarero L. The physical and functional behavior of capture antibodies adsorbed on polystyrene. Journal of immunological methods. 1992;150(1):77–90. doi: 10.1016/0022-1759(92)90066-3. [DOI] [PubMed] [Google Scholar]
- 38.Butler J, Ni L, Brown W, Joshi K, Chang J, Rosenberg B, Voss E. The immunochemistry of sandwich ELISAs—VI. Greater than 90% of monoclonal and 75% of polyclonal anti-fluorescyl capture antibodies (CAbs) are denatured by passive adsorption. Molecular immunology. 1993;30(13):1165–1175. doi: 10.1016/0161-5890(93)90135-x. [DOI] [PubMed] [Google Scholar]
- 39.Morisseau C, Wecksler AT, Deng C, Dong H, Yang J, Lee KSS, Kodani SD, Hammock BD. Effect of soluble epoxide hydrolase polymorphism on substrate and inhibitor selectivity and dimer formation. Journal of lipid research. 2014;55(6):1131–1138. doi: 10.1194/jlr.M049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 41.Pinot F, Skrabs M, Compagnon V, Salaün J, Benveniste I, Schreiber L, Durst F. omega-Hydroxylation of epoxy-and hydroxy-fatty acids by CYP94A1: possible involvement in plant defence. Biochemical Society Transactions. 2000;28(6):867–870. [PubMed] [Google Scholar]
- 42.Kiyohara C, Otsu A, Shirakawa T, Fukuda S, Hopkin JM. Genetic polymorphisms and lung cancer susceptibility: a review. Lung cancer. 2002;37(3):241–256. doi: 10.1016/s0169-5002(02)00107-1. [DOI] [PubMed] [Google Scholar]
- 43.Baxter S, Choong D, Campbell I. Microsomal epoxide hydrolase polymorphism and susceptibility to ovarian cancer. Cancer letters. 2002;177(1):75–81. doi: 10.1016/s0304-3835(01)00782-0. [DOI] [PubMed] [Google Scholar]
- 44.To-Figueras J, Gené M, Gómez-Catalán J, Piqué E, Borrego N, Caballero M, Cruellas F, Raya A, Dicenta M, Corbella J. Microsomal epoxide hydrolase and glutathione S-transferase polymorphisms in relation to laryngeal carcinoma risk. Cancer letters. 2002;187(1):95–101. doi: 10.1016/s0304-3835(02)00406-8. [DOI] [PubMed] [Google Scholar]
- 45.Nelson JW, Subrahmanyan RM, Summers SA, Xiao X, Alkayed NJ. Soluble epoxide hydrolase dimerization is required for hydrolase activity. Journal of Biological Chemistry. 2013;288(11):7697–7703. doi: 10.1074/jbc.M112.429258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Koerner IP, Noppens R, Grafe M, Tsai H-J, Morisseau C, Luria A, Hammock BD, Falck JR, Alkayed NJ. Soluble epoxide hydrolase: a novel therapeutic target in stroke. Journal of Cerebral Blood Flow & Metabolism. 2007;27(12):1931–1940. doi: 10.1038/sj.jcbfm.9600494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, Otsuka T, Sugo N, Ardeshiri A, Alhadid YK, Iliff JJ, DeBarber AE, Koop DR, Alkayed NJ. Soluble epoxide hydrolase gene deletion is protective against experimental cerebral ischemia. Stroke. 2008;39(7):2073–2078. doi: 10.1161/STROKEAHA.107.508325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.