Abstract
Next generation sequencing (NGS) of human bladder cancer has revealed many gene alterations compared with normal tissue, with most being predicted to be “loss of function”. However, given the high number of alterations, evaluating the functional impact of each is impractical. Here we develop and use a high-throughput, in vivo strategy to determine which alterations are loss of function in tumor growth suppressors. Genes reported as altered by NGS in bladder cancer patients were bioinformatically processed by MutationTaster and MutationAssessor, with 283 predicted as loss of function. A shRNA lentiviral library targeting these genes was transduced into T24 cells, a non-tumorigenic human bladder cancer cell line, followed by injection into mice. Tumors that arose were sequenced and the dominant shRNA constructs were found to target IQGAP1, SAMD9L, PCIF1, MED1 and KATNAL1 genes. In vitro validation experiments revealed that shRNA molecules directed at IQGAP1 showed the most profound increase in anchorage-independent growth of T24 cells. The clinical relevance of IQGAP1 as a tumor growth suppressor is supported by the finding that its expression is lower in bladder cancer compared with benign patient urothelium in multiple independent datasets. Lower IQGAP1 protein expression associated with higher tumor grade and decreased patient survival. Finally, depletion of IQGAP1 leads to increased TGFBR2 with TGFβ signaling, explaining in part how reduced IQGAP1 promotes tumor growth. These findings suggest IQGAP1 is a bladder tumor growth suppressor that works via modulating TGFβ signaling and is a potentially clinically useful biomarker.
Implications
This study used gene mutation information from patient-derived bladder tumor specimens to inform the development of a screen used to identify novel tumor growth suppressors. This included identification of the protein IQGAP1 as a potent bladder cancer growth suppressor.
Keywords: IQ motif containing GTPase activating protein 1 (IQGAP1), bladder neoplasms, Transforming Growth Factor Beta Receptor II, tumor growth suppressor
INTRODUCTION
Bladder cancer is the most common malignancy affecting the urinary system, with an expected 74 thousand new cases and 16 thousand deaths in 2015 in the United States (1). Numerous factors, including genetic polymorphisms and both genetic and epigenetic alterations are involved in tumorigenesis, growth, and metastasis in this disease (2, 3). Recent studies reporting whole-genome or whole-exome sequencing data of bladder tumors (4–7) have enabled comprehensive cataloguing of genomic alterations. Interestingly, most mutations are computationally predicted to result in proteins with reduced expression or function.
Given that such alterations number in the hundreds, evaluating their functional impact individually is impractical. Still, evaluating their function is absolutely necessary if we are to translate sequencing data into novel avenues for therapeutic intervention. Given this need, we developed and implemented a high throughput strategy that tests the hypothesis that some of these alterations are indeed loss of function in tumor growth suppressors. From the literature we gathered gene alterations discovered by next generation sequencing and identified those alterations which led to predicted loss of protein function and whose impact on in vivo tumor growth had not been previously reported in bladder cancer. We then employed RNA interference (RNAi) to reduce expression of our genes of interest and in that way, simulate loss of protein function. The effects of reduced gene expression was assessed in vivo. Therefore, plasmid-encoded short-hairpin RNAs (shRNAs) was used to study in vivo xenograft phenotypes (8), where long term target depletion is required.
A shRNA library targeting 283 candidate genes was transduced into a non-tumorigenic human bladder cell line (T24) which was then injected subcutaneously into mice. Tumors that arose were sequenced to identify the shRNA responsible for promoting tumor growth. Any gene targeted by such a shRNA represents a novel tumor growth suppressor altered in human bladder cancer. Such an approach provides a novel framework for the efficient functional evaluation of hundreds of genomic alterations found in cancer and allows us to define those that are drivers rather than passengers in regulating tumor growth.
MATERIALS AND METHODS
Cell Lines and Cell Culture
Human bladder cancer cell lines T24, 253J, UMUC3, J82, MGHU3 and HT1197 were authenticated by the University of Colorado PPSR core using an Applied Biosystems Profiler Plus Kit which analyzed 9 loci (Life Technologies 4303326). Cell ampules were resuscitated less than 2 months prior to being used in experiments in this study. Cells were cultured at 37 degrees Celsius, 5% CO2 in the following media. T24: Dulbecco’s Modified Eagle’s Medium/F12 (DMEM/F12) + 5% Fetal Bovine Serum (FBS), 253J and HT1197: Modified Eagle’s Medium (MEM) + 10% FBS + 0.1mM Nonessential Amino Acids (NEAA) + 1 mM Sodium Pyruvate, UMUC3 and MGHU3: MEM + 10% FBS + 1 mM Sodium Pyruvate, J82: MEM + 10% FBS + 0.1 mM NEAA. All reagents for media components were obtained from Invitrogen.
Pooled shRNA Library Screen and Lentiviral Transductions
T24 cells were transduced with our Lentiviral shRNA Library comprised of MISSION LentiPlex Human shRNA constructs from Sigma Life Science and The RNAi Consortium (TRC) along with 10 μg/mL polybrene for 18 hours. Forty-eight hours after transduction cells were selected with 2.5 μg/mL puromycin until stability was accomplished (approx. 4 days). Cells transduced with library were implanted subcutaneously into 6 week old NCrnu/nu mice (NCI-Frederick, Frederick, MD) at 2 × 106 cells/site. Mice that developed tumors were euthanized at the appropriate limits, tumors harvested, and DNA extracted using the DNAeasy Blood and Tissue kit (Qiagen). Polymerase chain reaction (PCR) amplification from the flanking regions of the shRNA sequence was performed using the primers LAP1, TACAAAATACGTGACGTAGAAA and LAP2, TTTGTTTTTGTAATTCTTTA. PCR reactions were subjected to agarose electrophoresis and the 350 bp bands were excised and purified using QIAquick Gel Extraction Kit (Qiagen). Products were then Sanger sequenced to identify the shRNA-encoding region.
In Vitro Cell Assays
Anchorage-independent growth in soft agar was assessed by plating 6 × 103 cells per well in 0.5% agar in 12-well plates. Colonies were stained with Nitro-BT (Sigma) for 24 hours before imaging and counting using ImageJ. Proliferation was assessed by plating 1–2 × 103 cells per well in 96-well plates. After a specific number of days relative cell abundance was measure with the CyQuant Assay (Life Technologies).
Migration assays were performed using insert chambers for 24-well plates having a PET membrane with 8 μm pores (Falcon). Cells (6 × 104) were suspended in 0.5 ml of fresh serum-free medium, plated into the chambers and placed into wells containing 0.75 ml/well of medium plus 5–10% FBS and incubated 18 hours. Cells not migrating to the underside of the membrane were removed by scrubbing the interior side of membrane with cotton swabs and the undisturbed migrating cells on the underside were fixed and stained using a crystal violet-20% methanol solution. After drying, the membranes were photographed and migrating cells quantitated using ImageJ. Invasion assays were performed using insert chambers for 24-well plates containing a Matrigel Basement Membrane-like matrix over an 8 μm pore PET membrane (BD Biosciences). Following membrane rehydration 1.5 × 104 cells, suspended in 0.5 ml of fresh serum-free medium, were plated into the hydrated chambers and placed into wells containing 0.75 ml/well of medium plus 5–10% FBS and incubated at 37°C for 22 hours. The membranes were treated as in the migration assays and cells quantitated.
Sphere assays were performed using ultra low attachment round bottom 96-well plates (Corning #7007). Cells (1 × 104) were suspended in 200 μL/well and plated. After 7 days incubation at 37°C the wells were photographed and sphere area determined in 2D using ImageJ.
qPCR
Total RNA was isolated from cells with a RNeasy Plus Mini kit (Qiagen). Single-stranded cDNAs were synthesized from total RNA using iScript™ cDNA Synthesis Kit (Bio-Rad). qPCR reactions were then composed of cDNA, specific primers and iQ SYBR Green Supermix (Bio-Rad) and run on a iQ5 Real-Time PCR Detection System (Bio-Rad). Gene expressions values were normalized to beta-actin controls.
Antibodies
Antibodies were obtained from the following sources: PCIF1 (Sigma - AB1407847); IQGAP1 (Life Technologies 33-8900); SAMD9L (Sigma - HPA019461); GAPDH (Cell signaling - 2118); pERK1/2 (Cell Signaling - 4377); ERK1/2 (Cell Signalling 9107) α-actinin (Cell Signalling - 3134); TGFBR2 (Abcam - ab186838).
Western Blot Analysis and Immunohistochemistry
Cell lysates were subjected to SDS-PAGE followed by transfer to polyvinylidene fluoride (PVDF) membranes (9). Membranes were blocked with 5% non-fat milk and incubated with primary antibodies overnight at 4°C. Membranes were then washed before adding the appropriate secondary antibody. Following 1 hour incubation and washing the blots were imaged using chemiluminescence reagent (Pierce) and a ChemiDoc Imaging Station (Bio-Rad). Densitometry was performed with ImageJ.
IHC was optimized with shRNA IQGAP1 depleted and pelleted/FFPE human bladder cells and performed and scored (0-absent; 1+-10–25%; 2+->25–50%; 3+->50–75%; 4+->75% stained cells). For statistical analysis scores were collapsed into 2 categories (0–2 and 3–4) on all tissue samples by a board certified pathologist as described (10).
In Vivo Tumor Study
T24 cells (2 × 106) or 253J (1 × 106) were implanted subcutaneously, 4 sites/mouse into five 6 week old NCrnu/nu mice (NCI-Frederick, Frederick, MD) (tumors analyzed = 20) and measured for allograft tumor development starting 1-week post-implantation and thereafter every 3–4 days via external measurement in two dimensions with a digital caliper to calculate tumor volumes using the equation (L × W2)/2. All animals used in this study were treated according to University of Colorado Denver and Institutional Animal Care and Use Committee (IACUC) guidelines. Animal protocols were reviewed and accepted in IACUC protocol number B-93410(12)1F.
Statistical Analysis
Data was analyzed using two-tailed Student t test with unequal variances. Error bars denote standard deviation. Values provided are the mean ± SEM and the differences were considered significant if P < 0.05. Kaplan-Meier curves for patients with low and high IQGAP1 expression groups were generated and the corresponding log-rank p-values calculated as previously (8).
RESULTS
In vivo functional RNAi screen identifies novel putative tumor growth suppressors
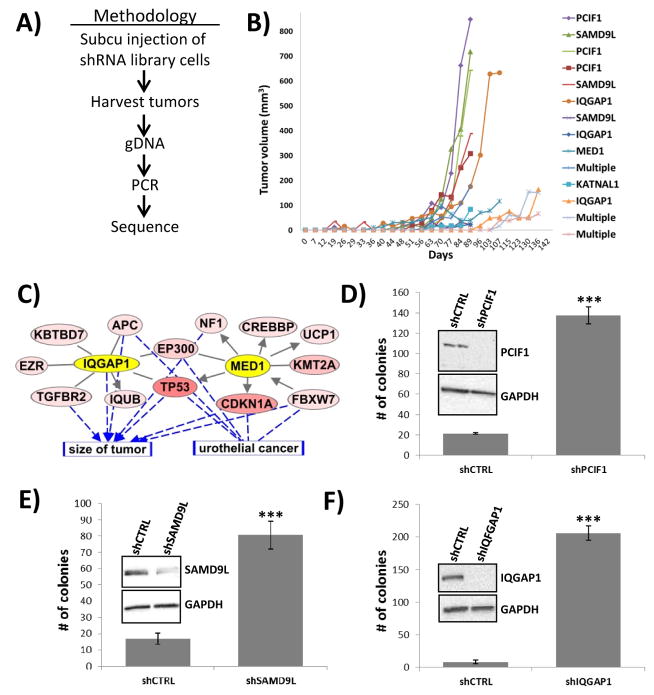
We compiled a list of genes which were reported as altered by next generation sequencing of tumors from bladder cancer patients (4–7). This list was refined to only include those genes whose protein product was predicted to be loss of function as determined by MutationTaster (11) or MutationAssessor (12) tools. To determine if loss of function of these gene products leads to promotion of tumor growth we first constructed an shRNA library containing 3 unique shRNA molecules targeting each of the 283 genes in our list. The use of shRNA molecules allows for the simulation of loss of protein function within a high throughput analysis. The shRNA molecules were introduced into T24 cells as a MISSION LentiPlex Pooled shRNA library. T24 human bladder cancer cells were chosen in this study because, while they grow well in vitro, they are non-tumorigenic at moderate inocula when implanted subcutaneously in immunocompromised mice (13). At 19 weeks after subcutaneous injection (Fig. 1A) we noted 14 of 20 sites had developed palpable tumors. These tumors were excised, genomic DNA extracted and PCR amplification performed to amplify the shRNA coding region of the genomically integrated shRNA lentiviral vector. Subsequent sequencing of the PCR reaction from each tumor allowed for identification of specific shRNAs molecules that were responsible for driving the growth of the tumor.
Fig. 1. RNA interference screen reveals candidate tumor growth suppressors in bladder cancer.
(A) Flow diagram of methodology for using shRNA library to identify tumor growth suppressors in vivo. (B) From the 14 tumors that arose from shRNA library transduced T24 cells, 11 of them were driven by the presence of a dominant shRNA molecule that was targeting either PCIF1, SAMD9L, IQGAP1, MED1, or KATNAL1. (C) Analyzing the top 5 hits from the screen using Ingenuity Pathway Analysis revealed strong biological connections between the genes IQGAP1 and MED1 and the cellular processes of tumor growth and urothelial cancer. (D–F) Treatment of T24 cells with shRNA molecules targeting either PCIF1, SAMD9L or IQGAP1 leads to dramatic increases in anchorage independent growth. *** p<0.001.
While the PCR reactions from 3 of the tumors yielded poorly defined sequence in the shRNA region, suggesting a heterogeneous pool of cells and shRNA molecules, the reactions from 11 of the tumors were unambiguous (Fig. 1B). These shRNA molecules were shown to target putative tumor suppressor genes, as their activity was responsible for promoting tumor outgrowth. The single shRNA constructs identified were targeting PDX1 C-terminal inhibiting factor 1 (PCIF1, 3 tumors), sterile alpha motif domain containing 9-like (SAMD9L, 3 tumors), IQ motif containing GTPase activating protein 1 (IQGAP1, 3 tumors), mediator complex subunit 1 (MED1, 1 tumor) and katanin p60 subunit A-like 1 (KATNAL1, 1 tumor).
To assess the relationship between these 5 genes and bladder cancer, we performed a bioinformatics analysis of all somatic mutations found in the data set from The Cancer Genome Atlas (5). This analysis revealed, most notably, that IQGAP1 and MED1 are biologically connected to many important genes mutated in bladder tumors, with both connected to TP53 and EP300 (Fig. 1C). In addition, many of the mutated genes in this network have previously been shown to be associated with tumor size and are found to promote human urothelial cancer growth (Fig. 1C). Thus, our novel approach appears to have identified 5 bladder cancer tumor growth suppressors.
IQGAP1 depletion increases anchorage independent tumor cell growth
To begin validation of these results we assessed the ability of each shRNA, found in more than one tumor, to promote anchorage independent growth of T24 cells in soft agar assays. Knockdown of PCIF1, which in the screen had yielded the largest tumors at the highest rate (Fig. 1B), led to a 6-fold increase in colony formation in soft agar assays relative to control cells (Fig. 1D). Similarly, knockdown of SAMD9L led to a 5-fold increase in soft colony growth (Fig. 1E). Most dramatic was the ability of IQGAP1 knockdown to promote soft agar colony growth (16-fold increase) (Fig. 1F).
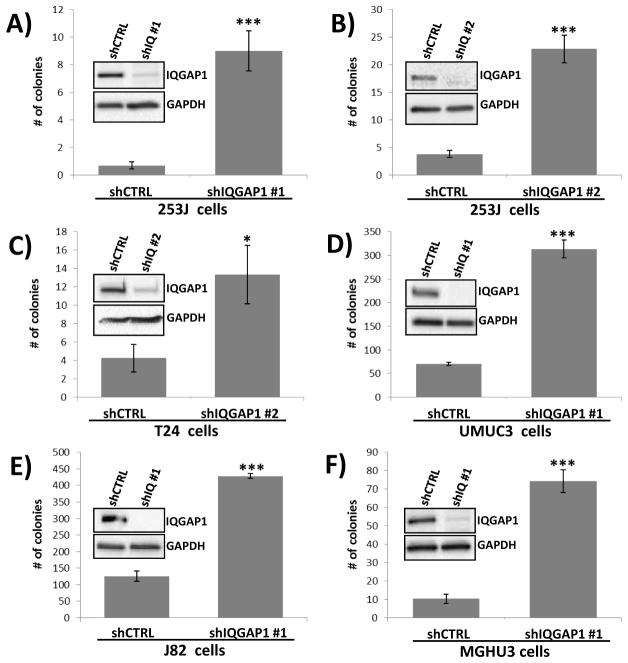
Since IQGAP1 expression correlates with tumor size in urothelial cancer and is biologically connected to many other genes altered in this disease (Fig. 1C), and its loss in T24 cells exhibits the highest increase in anchorage independent growth (Fig. 1F), we decided to focus additional studies only on IQGAP1. As a means of confirming that the changes in phenotype associated with the IQGAP1 shRNA construct used above were not the result of an off-target effect we tested the shRNA construct #1 in another bladder cancer cell line. Knockdown of IQGAP1 in 253J bladder cancer cells also resulted in increased anchorage independent growth (Fig. 2A). The use of a second IQGAP1 shRNA construct also led to dramatic increases in anchorage independent growth in both 253J (Fig. 2B) and T24 (Fig. 2C) cells. In fact, knockdown of IQGAP1 was observed to increase colony formation in an additional 3 bladder cancer cell lines we tested (Fig. 2D–F). Thus, using either of two constructs to deplete cells of IQGAP1 in several bladder cancer cell lines we conclude that loss of IQGAP1 strongly leads to increased ability of cells to grow in an anchorage independent manner.
Fig. 2. Depletion of IQGAP1 with multiple shRNA molecules in multiple bladder cancer cell lines consistently leads to increases in anchorage independent growth.
(A) Depletion of IQGAP1 with shRNA #1 in 253J bladder cancer cells led to a dramatic increase in anchorage independent growth. (B) Depletion of IQGAP1 with shRNA #2 elicited the same phenotypes as shRNA #1. (C) As in Figure 1 with IQGAP1 shRNA #1, the presence of IQGPA1 shRNA #2 in T24 cells leads to IQGAP1 depletion and an increase in soft agar colonies. (D–F) Treatment of UMUC3, J82 and MGHU3 bladder cancer cell lines with IQGAP1 shRNA #1 leads to IQGAP1 depletion and dramatic increases anchorage independent growth. * p<0.05, *** p<0.001.
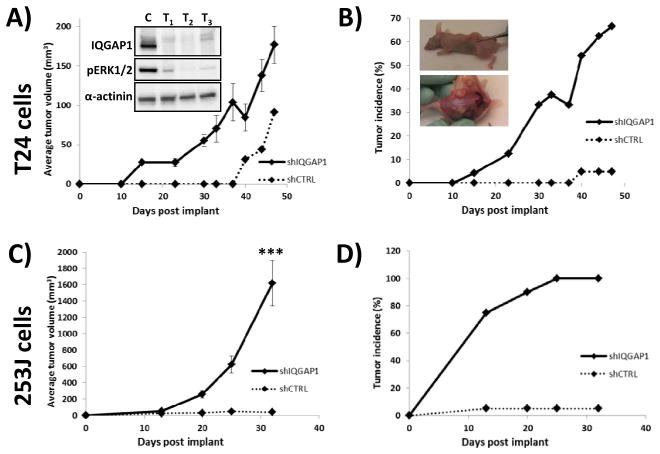
IQGAP1 depletion enhances 253J and T24 tumor growth in vivo
We next examined the effect of IQGAP1 depletion on subcutaneous mouse xenograft growth kinetics using 253J and T24 cells, both of which have poor subcutaneous tumorigenicity and growth in immunocompromised mice (13, 14). In T24 implanted mice the shIQGAP1 cohort reached IACUC protocol limits on tumor size at day 47 post-implant. Therefore, at that time all mice in the experiment were euthanized. At this time point the tumor volume for the shIQGAP1 cohort ranged from 42 to 358 mm3 for an average volume of 177 mm3 while the shCTRL cohort had 1 tumor at 91 mm3 (Fig. 3A). While the average tumor volume appears to be only a 2-fold increase it is important to note that T24 cells depleted of IQGAP1 had a 67% tumor take (16 tumors from 24 injections) while control cells only had a 5% incidence (1 tumor from 21 injections) (Fig. 3B). Thus, loss of IQGAP1 expression was important for promoting tumor growth but even more important for promoting tumorigenicity. Western blotting of tumor lysates confirmed IQGAP1 expression remained reduced in vivo (Inset, Fig. 3A). Lysates were obtained from tumors (T1–T3) of similar size, irrespective of time points, to ensure similarity of the tumor microenvironment. The tumor lysates also revealed a reduction in phosphorylated ERK1/2. This observation is consistent with previous reports (15–17) that IQGAP1 binds ERK2 and modulates its activity due to the fact that IQGAP1 has been shown to physically interact with MEK and ERK proteins (16, 18). The decrease in tumor volume from day 30 to 40 was the result of newly measurable tumors on day 40 being added to the analysis. We also analyzed 253J cells and discovered that 253J cells depleted of IQGAP1 had an even more dramatic increase in tumorigenicity and growth compared with IQGAP1 depletion in T24 (Fig. 3C). This necessitated termination of group analysis at 32 days. At this time point the average shIQGAP1 tumor volume was 1375 mm3 versus 33 mm3 for shCTRL (Fig. 3C). The shCTRL cohort had only a 5% tumor incidence compared to 100% in the shIQGAP1 cohort (Fig. 3D). These results provide additional validation of our screen and additional support for our hypothesis that IQGAP1 functions as a tumor suppressor in bladder tissue.
Fig. 3. IQGAP1 knockdown leads to dramatic increases in tumor outgrowth in vivo.
(A) In validation of the screening results, T24 cells expressing IQGAP1 shRNA exhibit significant increases in tumor outgrowth after subcutaneous injection in mice. Analysis of all the tumors following animal sacrifice at day 47 post-implant confirmed continued knockdown of IQGAP1 (inset). Consistent with the literature, depletion of IQGAP1 also lead to a decrease in activated Erk (inset). As the shCTRL cells only produced 1 tumor, statistics cannot be performed. (B) Not only does loss of IQGAP1 in T24 cells increase tumor growth rate and volume but also increases the % of tumor incidence to a rate of nearly 70% by day 47, relative to only 10% for control cells. The inset necropsy picture illustrates representative tumors in mice at time of harvest. (C–D) The ability of IQGAP1 depletion to promote tumor outgrowth and incidence is even more dramatic in 253J cells. In 253J cells the experiment was halted at 32 weeks at which point there was a 100% tumor take with shIQGAP1 cells. *** p<0.001.
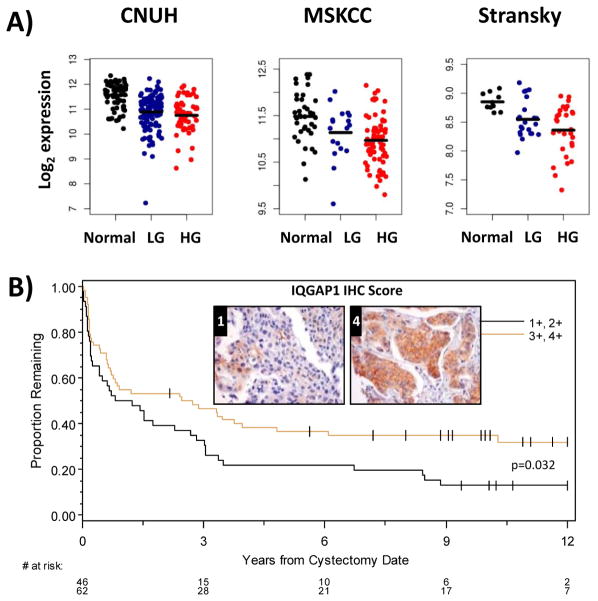
Reduced IQGAP1 level in human bladder cancer is associated with worse prognosis
To investigate if human bladder cancer has lower levels of IQGAP1 relative to normal tissue we analyzed IQGAP1 mRNA expression in several human bladder cancer patient microarray data sets (CNUH (19), MSKCC (20) and Stransky (21)). This analysis revealed that IQGAP1 mRNA expression was in fact lower in tumor versus normal tissues in all three data sets (Fig. 4A). To analyze IQGAP1 protein levels in bladder cancer tumors we optimized and then performed immunohistochemistry analysis on human bladder tumor tissue microarrays from patients that underwent radical cystectomy. Patient clinical management and follow up has been previously described (22). Lower IQGAP1 protein levels were associated with decreased survival rates among patients who had undergone cystectomy (Fig. 4B). These data from bladder cancer patients are consistent with both our in vitro and in vivo results and strongly suggest that IQGAP1 plays a clinically relevant role as a tumor growth suppressor in bladder cancer and illustrates its potential value as a prognostic indicator.
Fig. 4. IQGAP1 expression suppresses tumorigenicity and mortality in bladder cancer patients.
(A) Dot plots of IQGAP expression in bladder cancer patient cohorts as reported by CNUH (N = 255), MSKCC (N = 129), and Stransky (N = 62). Analyzing expression across normal and tumor samples demonstrates that normal tissues have higher IQGAP1 expression (FC, fold change). Black lines correspond to mean expression for each group. Wilcoxon Rank-Sum test were used to generate p values and were < 0.001 in all three cohort analyses. (B) Kaplan-Meier examination of IQGAP1 protein expression using tissue microarray analysis (TMA) on samples obtained from bladder cancer patients undergoing cystectomy indicates that patients grouped in the 2 lower staining scores have decreased survival post-cystectomy (log-rank p-value shown) Representative stained tissue are presented for IHC scores 0, 1 and 4.
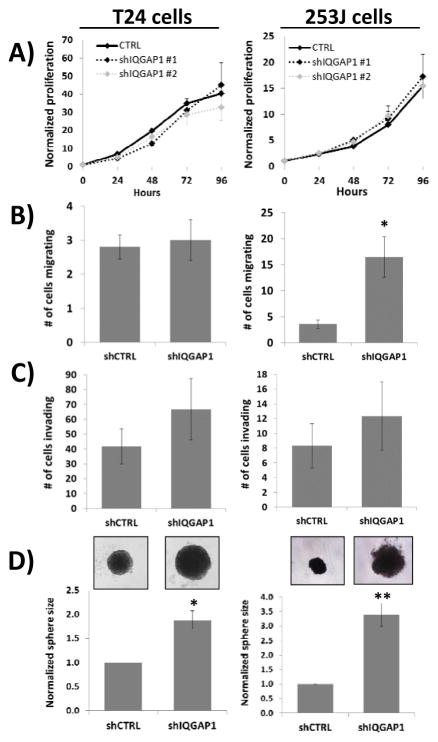
IQGAP1 marginally affects in vitro anchorage dependent growth, migration and invasion
All of the analyses above reporting on the biological impact of IQGAP1 depletion were in the context of either anchorage independent growth or in vivo. In sharp contrast, IQGAP1 knockdown showed little effect on cells when they were analyzed in monolayer. Cells which stably express shIQGAP1 to knockdown IQGAP1 expression (Figure 1F and 2A) exhibited the same rate of proliferation relative to shCTRL when grown in monolayer (Fig. 5A). Second, there was no difference in the migration ability of shIQGAP1 versus shCTRL T24 cells (Fig. 5B), although IQGAP1 depletion did impact 253J migration. Third, there was no significant difference between control cells and cells depleted of IQGAP1 in invasion assays, in either T24 or 253J cell lines (Fig. 5C). Thus, in 3 different monolayer in vitro assays we saw little to no difference in control cells compared with cells depleted of IQGAP1 in 2 bladder cancer cell lines. Consistent with this finding we discovered that pERK1/2 levels were also unaltered in monolayer (Supplemental Fig. 1), while pERK1/2 levels were dramatically decreased in xenograft tumors (Fig. 4A). Finally, as another means of assessing in vitro cell growth we evaluated the impact of IQGAP1 depletion in 3 dimensional sphere growth assays performed with non-adherent plates (23). Both T24 and 253J cells readily formed tumor spheres (Fig 5D). Furthermore, depletion of IQGAP1 in either cell line led to a significant increase in sphere volume, indicative of increased growth (Fig. 5D). Together, these experiments show that IQGAP1 depletion has by far its greatest impact when cells are grown in three dimensions, whether in vitro or in vivo.
Fig. 5. Loss of IQGAP1 expression has the most profound effect on cell growth when analyzed in 3-dimensional assays.
(A) Proliferation assays revealed that loss of IQGAP1 from either T24 or 253J cells has little impact on cell growth in monolayer. (B) Migration assays reveal that loss of IQGAP1 in T24 cells has no impact while there is a statistically significant increase in migration with loss of IQGAP1 in 253J cells. (C) Invasion assays reveal loss of IQGAP1 has no statistically significant effect in either T24 or 253J cells. (D) When grown as tumor spheres both T24 and 253J cells lacking IQGAP1 display a significant increase in cell growth as measured by sphere area. Representative spheres are shown in insets for each cell line. * p<0.05, ** p<0.01.
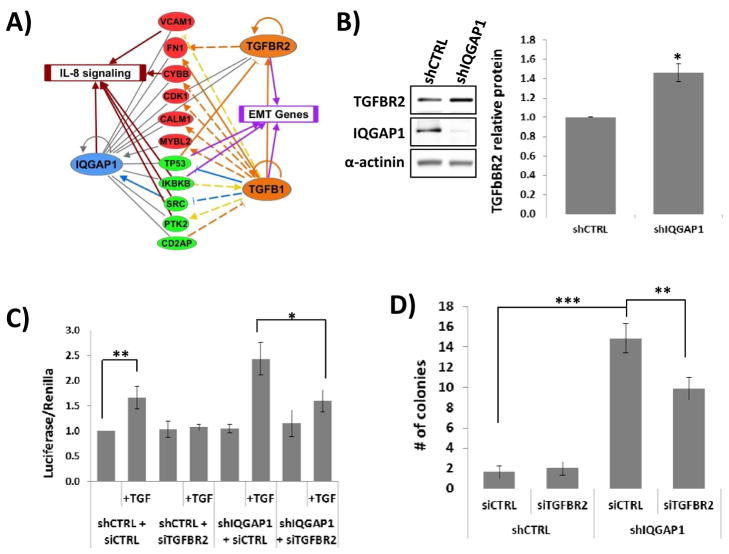
Inhibition of TGFBR2 reverses growth phenotypes associated with IQGAP1 depletion
IQGAP1 is a major scaffold protein involved in a number of cellular processes and associates with over 90 direct or indirect binding partners (24, 25). As an initial approach to understanding the mechanisms by which loss of IQGAP1 leads to dramatic increases in tumorigenicity and tumor growth we undertook a bioinformatics assessment to identify genes/proteins that are associated with both IQGAP1 and advanced bladder tumors. Of 909 genes identified as differentially expressed (FDR<0.05) in a majority of 7 microarray studies of muscle invasive bladder tumors (19–21, 26–28), we identified 22 genes/proteins with a biological connection to IQGAP1. We then used QIAGEN’s Ingenuity Pathway Analysis to look for other genes/proteins that were connected to these 22 genes/proteins. We discovered that half of them were also connected to TGF-β signaling, with direct and indirect associations with the growth factor TGF-β1 and its receptor TGFBR2 (Fig. 6A). These genes/proteins were also associated with the biological process of cell viability (except CALM1), while all but two of them (CALM1 and CYBB) are connected to cell morphology using the IPA Knowledge Base. The probability of selecting these genes associated with cell viability and cell morphology by random chance, given all possible human genes, is 1.2 × 10−13 and 2.8 × 10−13 respectively (Fisher’s exact test). We also note that six of these genes (CYBB, IKBKB, IQGAP1, PTK2, SRC, and VCAM1) are associated with canonical IL-8 signaling (p = 1.1 × 10−9, Fisher’s exact test), a pathway that is associated with the epithelial–mesenchymal transition (EMT) in many types of tumors (29). In addition, TP53, IKBKB, TGFB1 and TGFBR2 are strongly associated with EMT (p = 9.0 × 10−6, Fisher’s exact test). A literature search revealed that IQGAP1 has been shown to bind TGFBR2, following TGF-β stimulation, which promotes SMURF1 association with TGFBR2 and subsequent ubiquitination/degradation of the receptor (30). Additionally, dysregulated TGF-β signaling has been shown to be pro-oncogenic (31). Therefore, we hypothesized that IQGAP1 loss results in increased TGFBR2 protein levels and subsequently increases TGF-β signaling to promote cell growth. We found that depletion of IQGAP1 did in fact lead to increased TGFBR2 protein levels in bladder cancer cells (Fig. 6B). Using a reporter of TGF-β signaling (3TP-Lux) we also found that shIQGAP1 cells have higher levels of TGF-β-induced activity (Fig. 6C). As hypothesized, treatment of cells with TGFBR2 siRNA (Supplemental Fig. 2A) led to an abolishment of the increased TGF-β signaling (Fig. 6C). To determine if these observations had biological relevance we performed a rescue experiment in soft agar assays using cells transfected with TGFBR2 siRNA. Knockdown of TGFBR2 reversed the increase in soft agar colonies induced by IQGAP1 depletion in 253J cells (Fig. 6D) and T24 cells (Supplemental Fig. 2B). This reduction in colony formation demonstrates that the ability of IQGAP1 to function as a tumor suppressor in bladder cancer is attributable in part to its ability to reduce TGF-β signaling.
Fig. 6. Loss of IQGAP1 expression enhances TGF-β signaling.
(A) The use of Ingenuity Pathway Analysis identified genes and pathways which had a biological connection to IQGAP1. The most dramatic association was found between IQGAP1 and TGFBR2, as well as its ligand TGF-β. (B) Loss of IQGAP1 from 253J cells leads to a statistically significant increase in TGFBR2 protein levels. The bar graph averages the results from 3 independent experiments. (C) The use of a TGF-β reporter assay revealed that loss of IQGAP1 from 253J cells leads to enhanced TGF-β signaling. Furthermore, this enhanced signaling was attenuated with the concurrent knockdown of TGFBR2. (D) Knockdown of TGFBR2 expression in 253J cells results in abrogation of the previously noted increase in soft agar colony formation that results from loss of IQGAP1. * p<0.05, ** p<0.01, *** p<0.001.
DISCUSSION
The work presented here makes two important contributions to the field of cancer biology. First, it provides a proof of principle that mutational analysis of human tumors can inform the design of an shRNA library, which in turn can be used for the discovery of tumor suppressor genes. More broadly, this approach allows high throughput evaluation of genomic changes in cancer to determine which are functional, thus contributing to the classification of these into driver and passenger alterations. Second, the work led to the identification of 5 novel tumor suppressor gene candidates in bladder cancer, 3 of which we validated in this study. Since the novel candidate tumor growth suppressor IQGAP1 had a bioinformatics association with another candidate, as well as an association with tumor growth and urothelial cancer, we further characterized the role of this candidate in bladder tissue. We found that depletion of IQGAP1 in 5 different bladder cancer cell lines, using 2 different shRNA molecules, led to dramatic increases in growth in 3-dimensional based in vitro assays. Interestingly, it was only in the 3-dimensional/anchorage independent assays that we were able to phenocopy the increased growth observed in vivo with depletion of IQGAP1. Why the loss of IQGAP1 has little to no impact on cell growth/movement in a monolayer environment remains unclear. IQGAP1 normally functions as a scaffold protein near the plasma membrane. Perhaps with cell-cell contact being dramatically reduced in a monolayer setting relative to a 3-dimensional setting the cell has less of a need for IQGAP1, and thus, its loss has little to no impact on cell growth/movement. Future experiments designed to tease out the mechanism behind the ability of IQGAP1 to suppress tumor growth will have to take this important consideration into account.
The use of RNAi in high throughput in vivo screening is a very powerful and economically practical method to rapidly identify novel genes of interest. There are a number of shRNA library systems (reviewed in (32, 33)) and we used the TRC shRNA library system (34–36). Furthermore, we employed a focused library as opposed to whole genome or module libraries since our genes of interest were determined by bladder cancer patient mutations followed by a bioinformatic prediction of which mutations lead to loss of function proteins. Another unique aspect of our screen was that we were only interested in identifying shRNA molecules which were present in harvested tumors, as the targets of these shRNAs represented candidate tumor growth suppressor genes. This is in contrast to determining which shRNAs were lost, as these represent proto-oncogenes, or determining the levels of all shRNA molecules in the tumors as a means of assessing the effects of gene expression on tumor formation. Yet another unique aspect of our screen was the identification of the dominant shRNA molecules in each tumor in a low throughput, more cost effective fashion as detailed in the methods section. There are any number of routes a study can take from a standard high throughput, RNAi in vivo screen and here we have detailed our unique in vivo approach which successfully yielded a number of novel tumor growth suppressors in bladder cancer.
This study was also able to determine that the ability of IQGAP1 to suppress tumorigenicity in bladder cancer cell lines is due in part to its ability to regulate TGF-β signaling. Unfortunately, the complete mechanism by which IQGAP1 acts to suppress tumorigenicity in bladder cancer is likely more complex than just having a relationship with TGF-β signaling. IQGAP1 has a large number of binding partners and, when they are dysregulated, these partners themselves have been shown to contribute to cancer (25). In addition to the screen, and subsequent in vitro and in vivo validation, we found a statistically significant correlation between IQGAP1 expression and bladder cancer patient outcomes. Such findings demonstrate a clinically relevant aspect of IQGAP1 expression and label the protein as a potential biomarker for advanced disease. Furthermore, the fact that IQGAP1 expression was found to be inversely proportional to grade suggests that IQGAP1 has an ability to inhibit tumor progression (24, 25).
Identification of a patient mutation in IQGAP1 led to the incorporation of the gene in our RNAi library. An analysis of The Cancer Genome Atlas revealed 3 additional patient missense mutations (5). This prevalence suggests that mutational status is less of a factor than decreases in gene expression. This is exactly what we observed in our study. IQGAP1 mRNA expression was lower in tumor versus normal in all 3 patient datasets we analyzed. Additionally, an immunohistochemistry analysis of patients following radical cystectomy revealed that lower IQGAP1 protein levels correlated with decreased survival. Thus, our data show that decreased expression of IQGAP1, and not necessarily mutational status, correlates strongly with negative prognosis. As a large scaffolding protein this would not be surprising as its reduced expression would impact over 90 other proteins while an intact protein with missense mutations would be expected to impact fewer, assuming the protein stability and conformation was unchanged.
It is certainly worth noting that, in contrast with our findings in bladder cancer cells and patients, IQGAP1 is actually up-regulated in other cancers and has been found to play a significant role in cellular functions often exaggerated in cancer (24). For example, IQGAP1 protein contributes to increased proliferation and tumorigenicity of breast epithelial cells (37). Interestingly, a similar dual role has been demonstrated between bladder and breast cancer for the protein RhoGDI2. In bladder cancer, reduced expression of RhoGDI2 is associated with decreased patient survival (38), while in breast cancer, higher RhoGDI2 expression is associated with decreased patient survival (39). IQGAP1 protein also shows increased expression in aggressive vs. less-aggressive glioma neoplasms, and has higher expression in tumor vs. normal tissue of glioma, lung and colorectal cancers (24). It has also been shown to be overexpressed in hepatocellular carcinoma and promote proliferation through Akt activation (40). With these studies labeling IQGAP1 as an oncogene we were surprised to discover IQGAP1 has an opposite function in bladder cancer. However, supportive of our discoveries is the finding that IQGAP1 has the ability to act as a suppressor of TGFBR2-mediated myofibroblastic activation and metastatic growth in liver (30). Additionally, while IQGAP1 knockout mice have no increase in tumor incidence or progression, they do exhibit increased late-onset gastric hyperplasia (41). This, together with the inverse association of IQGAP1 levels with tumor progression, indicates that this protein may be more important in this aspect of malignancy than in tumor formation and may also have organ-specific roles in cancer progression.
Supplementary Material
Acknowledgments
Financial Support: Supported by National Institutes of Health grant CA143971 to DT.
Footnotes
Conflict of Interest: The authors declare no conflict of interest
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Dancik GM, Theodorescu D. Pharmacogenomics in bladder cancer. Urologic oncology. 2014;32(1):16–22. doi: 10.1016/j.urolonc.2013.09.007. Epub 2013/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaig TW, Theodorescu D. Bladder cancer in 2011: the dawn of personalized medicine. Nature reviews Urology. 2012;9(2):65–6. doi: 10.1038/nrurol.2011.220. Epub 2011/12/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nature genetics. 2013;45(12):1464–9. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nature genetics. 2013;45(12):1459–63. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon DA, Kim JS, Bondaruk J, Shariat SF, Wang ZF, Elkahloun AG, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nature genetics. 2013;45(12):1428–30. doi: 10.1038/ng.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guin S, Pollard C, Ru Y, Ritterson Lew C, Duex JE, Dancik G, et al. Role in tumor growth of a glycogen debranching enzyme lost in glycogen storage disease. Journal of the National Cancer Institute. 2014;106(5) doi: 10.1093/jnci/dju062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahmood T, Yang PC. Western blot: technique, theory, and trouble shooting. North American journal of medical sciences. 2012;4(9):429–34. doi: 10.4103/1947-2714.100998. Epub 2012/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Said N, Frierson HF, Sanchez-Carbayo M, Brekken RA, Theodorescu D. Loss of SPARC in bladder cancer enhances carcinogenesis and progression. The Journal of clinical investigation. 2013;123(2):751–66. doi: 10.1172/JCI64782. Epub 2013/01/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11(4):361–2. doi: 10.1038/nmeth.2890. Epub 2014/04/01. [DOI] [PubMed] [Google Scholar]
- 12.Robinson PN, Kohler S, Oellrich A, Wang K, Mungall CJ, Lewis SE, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome research. 2014;24(2):340–8. doi: 10.1101/gr.160325.113. Epub 2013/10/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gildea JJ, Golden WL, Harding MA, Theodorescu D. Genetic and phenotypic changes associated with the acquisition of tumorigenicity in human bladder cancer. Genes, chromosomes & cancer. 2000;27(3):252–63. doi: 10.1002/(sici)1098-2264(200003)27:3<252::aid-gcc5>3.0.co;2-9. Epub 2000/02/19. [DOI] [PubMed] [Google Scholar]
- 14.Dinney CP, Fishbeck R, Singh RK, Eve B, Pathak S, Brown N, et al. Isolation and characterization of metastatic variants from human transitional cell carcinoma passaged by orthotopic implantation in athymic nude mice. The Journal of urology. 1995;154(4):1532–8. Epub 1995/10/01. [PubMed] [Google Scholar]
- 15.Cheung KL, Lee JH, Shu L, Kim JH, Sacks DB, Kong AN. The Ras GTPase-activating-like protein IQGAP1 mediates Nrf2 protein activation via the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)-ERK pathway. The Journal of biological chemistry. 2013;288(31):22378–86. doi: 10.1074/jbc.M112.444182. Epub 2013/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. The Journal of biological chemistry. 2004;279(17):17329–37. doi: 10.1074/jbc.M308405200. Epub 2004/02/19. [DOI] [PubMed] [Google Scholar]
- 17.Stuart DD, Sellers WR. Targeting RAF-MEK-ERK kinase-scaffold interactions in cancer. Nature medicine. 2013;19(5):538–40. doi: 10.1038/nm.3195. Epub 2013/05/09. [DOI] [PubMed] [Google Scholar]
- 18.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Molecular and cellular biology. 2005;25(18):7940–52. doi: 10.1128/MCB.25.18.7940-7952.2005. Epub 2005/09/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Molecular cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. Epub 2010/01/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24(5):778–89. doi: 10.1200/JCO.2005.03.2375. Epub 2006/01/25. [DOI] [PubMed] [Google Scholar]
- 21.Lindgren D, Frigyesi A, Gudjonsson S, Sjodahl G, Hallden C, Chebil G, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer research. 2010;70(9):3463–72. doi: 10.1158/0008-5472.CAN-09-4213. Epub 2010/04/22. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Moissoglu K, Wang H, Wang X, Frierson HF, Schwartz MA, et al. Src phosphorylation of RhoGDI2 regulates its metastasis suppressor function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5807–12. doi: 10.1073/pnas.0810094106. Epub 2009/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & development. 2003;17(10):1253–70. doi: 10.1101/gad.1061803. Epub 2003/05/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS letters. 2009;583(12):1817–24. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cellular signalling. 2012;24(4):826–34. doi: 10.1016/j.cellsig.2011.12.005. Epub 2011/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, et al. Bladder cancer outcome and subtype classification by gene expression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(11):4044–55. doi: 10.1158/1078-0432.CCR-04-2409. Epub 2005/06/03. [DOI] [PubMed] [Google Scholar]
- 27.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer research. 2004;64(11):4040–8. doi: 10.1158/0008-5472.CAN-03-3620. Epub 2004/06/03. [DOI] [PubMed] [Google Scholar]
- 28.Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, et al. Regional copy number-independent deregulation of transcription in cancer. Nature genetics. 2006;38(12):1386–96. doi: 10.1038/ng1923. Epub 2006/11/14. [DOI] [PubMed] [Google Scholar]
- 29.Palena C, Hamilton DH, Fernando RI. Influence of IL-8 on the epithelial-mesenchymal transition and the tumor microenvironment. Future Oncol. 2012;8(6):713–22. doi: 10.2217/fon.12.59. Epub 2012/07/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Billadeau DD, Abdelhakim H, Leof E, Kaibuchi K, Bernabeu C, et al. IQGAP1 suppresses TbetaRII-mediated myofibroblastic activation and metastatic growth in liver. The Journal of clinical investigation. 2013;123(3):1138–56. doi: 10.1172/JCI63836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massague J. A very private TGF-beta receptor embrace. Molecular cell. 2008;29(2):149–50. doi: 10.1016/j.molcel.2008.01.006. Epub 2008/02/05. [DOI] [PubMed] [Google Scholar]
- 32.Gargiulo G, Serresi M, Cesaroni M, Hulsman D, van Lohuizen M. In vivo shRNA screens in solid tumors. Nature protocols. 2014;9(12):2880–902. doi: 10.1038/nprot.2014.185. Epub 2014/11/21. [DOI] [PubMed] [Google Scholar]
- 33.Hu G, Luo J. A primer on using pooled shRNA libraries for functional genomic screens. Acta biochimica et biophysica Sinica. 2012;44(2):103–12. doi: 10.1093/abbs/gmr116. Epub 2012/01/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–98. doi: 10.1016/j.cell.2006.01.040. Epub 2006/03/28. [DOI] [PubMed] [Google Scholar]
- 35.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nature methods. 2006;3(9):715–9. doi: 10.1038/nmeth924. Epub 2006/08/25. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, et al. A public genome-scale lentiviral expression library of human ORFs. Nature methods. 2011;8(8):659–61. doi: 10.1038/nmeth.1638. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jadeski L, Mataraza JM, Jeong HW, Li Z, Sacks DB. IQGAP1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. The Journal of biological chemistry. 2008;283(2):1008–17. doi: 10.1074/jbc.M708466200. Epub 2007/11/06. [DOI] [PubMed] [Google Scholar]
- 38.Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF., Jr Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(11):3800–6. doi: 10.1158/1078-0432.CCR-03-0653. Epub 2004/06/03. [DOI] [PubMed] [Google Scholar]
- 39.Moon HG, Jeong SH, Ju YT, Jeong CY, Lee JS, Lee YJ, et al. Up-regulation of RhoGDI2 in human breast cancer and its prognostic implications. Cancer research and treatment: official journal of Korean Cancer Association. 2010;42(3):151–6. doi: 10.4143/crt.2010.42.3.151. Epub 2010/10/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. IQGAP1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation by Akt activation. Experimental & molecular medicine. 2010;42(7):477–83. doi: 10.3858/emm.2010.42.7.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li QW, Chakladar A, Bronson RT, Bernards A. Gastric Hyperplasia in Mice Lacking the Putative Cdc42 Efector IQGAP1. Molecular and cellular biology. 2000;20(2):697–701. doi: 10.1128/mcb.20.2.697-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.